Abstract
Background
FUN14 domain-containing 1 (FUNDC1), as a novel member of mitochondria-associated endoplasmic reticulum (ER) membranes associates with mitochondrial division and mitophagy. However, the expression profile and functional roles of FUNDC1 remain largely unclear in human cancer biology, including breast cancer (BC).
Methods
Immunohistochemistry and western blot analysis were used to determine the expression of FUNDC1 and BMI1 polycomb ring finger oncogene (BMI1). CCK8, cell counting and transwell assays were used to analyze cell proliferation, migration and invasion, respectively. Luciferase reporter and chromatin immunoprecipitation (ChIP) assays were used to detect the transcriptional regulation of Nuclear factor of activated T-cells, cytoplasmic 1 (NFATC1). The prognostic merit of NFATC1 expression was assessed by Kaplan-Meier assay.
Findings
Immunohistochemistry revealed strong immunostaining for FUNDC1 in cytoplasmic and nuclear membrane distribution in BC tissues as compared with normal breast epithelium. Kaplan–Meier survival analysis showed worse outcome for BC patients with high FUNDC1 expression. In vitro assay of gain- and loss-of-function of FUNDC1 suggested that FUNDC1 could stimulate BC cell proliferation, migration and invasion. Furthermore, elevated FUNDC1 level promoted Ca2+ cytosol influx from ER and extracellular, as well as NFATC1 nuclear translocation and activity. Nuclear NFATC1 bound to the BMI1 gene promoter and transcriptionally upregulated its expression. Notably, BMI1 overexpression could rescue the loss of function of FUNDC1. Co-expression of FUNDC1 and BMI1 in BC patients predicted worse prognosis than without either expression.
Interpretation
FUNDC1 might promote BC progression by activating the Ca2+–NFATC1–BMI1 axis. This pathway may be promising for developing multiple targets for BC therapy.
Keywords: FUNDC1, Breast cancer, Calcium, NFATC1, BMI1
Research in context.
Evidence before this study
The mitochondria-associated endoplasmic reticulum (ER) membranes (MAMs), is a molecular platform for enrichment of enzymes, which plays a major role on calcium (Ca2+) transport and signaling, as well as proteins with oncogenic functions. Increasing evidence indicates that aberrant expression or localization of MAMs may account for the progression and metastasis of cancers. Recently, FUN14 domain-containing 1 (FUNDC1) has been identified as a novel member of MAMs and associated with hypoxia related cardiac dysfunction and heart failure via Ca2+ transport, mitochondrial fission and autophagy. However, the expression profile and functional roles of FUNDC1 remain largely unclear in human cancer biology, including breast cancer (BC), which is the most common cancer among women globally.
Added value of this study
In this study, we demonstrated FUNDC1 promoted breast cancer proliferation, migration and invasion. Mechanistically, elevated FUNDC1 level promoted Ca2+ release from ER and extracellular into cytosol, NFATC1 nuclear translation and activity, which consequently resulting in oncogenic BMI1 (BMI1 Proto-Oncogene, Polycomb Ring Finger) transcriptional upregulation. Notably, BMI1 overexpression could sufficient rescue the loss of function of FUNDC1 in BC. Co-expression of FUNDC1 and BMI1 in BC patients predicted worse prognosis.
Implications of all the available evidence
Elevated FUNDC1 could be sever a novel diagnostics and/or prognostics marker for breast cancer. Furthermore, since FUNDC1 promote BC progression by activating the Ca2+–NFATC1–BMI1 axis, inhibition of FUNDC1 or its pathway could represent a candidate therapeutic target for BC.
Alt-text: Unlabelled Box
1. Introduction
Worldwide, breast cancer (BC) is the most common cancer among women [1]. In China, it is also the most frequently newly diagnosed cancer among about 4292,000 invasive cancers in woman, representing about 15% in 2015 [2]. Advances in early detection and individualized medical treatment have improved the survival rate of patients with BC. However, BC is still the leading cause of cancer deaths in women younger than 45 years [2]. These observations highlight the importance of early diagnosis to improve therapy, molecular diagnostics and the development of new prognostic biomarkers for BC.
Mitochondria-associated endoplasmic reticulum (ER) membranes (MAMs) are highly specialized subcellular compartments that are shaped by ER subdomains juxtaposed with mitochondria [3]. MAMs supply a molecular platform for enrichment of enzymes involved in calcium (Ca2+) transport and signaling, lipid synthesis and transport and proteins with oncogenic functions [4,5]. Increasing evidence indicates that aberrant expression or localization of MAMs may account for the onset as well as progression and metastasis of cancer including BC. For example, RNA-dependent protein kinase (PKR)-like ER kinase (PERK) has been identified as a key MAM component [6]. PERK-dependent signaling participates in tumor initiation and expansion to preserve redox homeostasis and promote tumor growth in MDA-MB-468 and T47D cell lines [7]. Silencing PERK reduces tumor growth and restores the sensitivity to chemotherapy in resistant tumor xenografts [8]. Notably, ER oxidoreductin 1-α (ERO1-α) has been extensively studied because of its high expression in various tumors [9] and its association with poor prognosis in BC [10]. Mechanistically, ERO1-α controls ER redox homeostasis and regulates ER Ca2+ flux and consequent cytoplasm Ca2+ accumulation. Cancer cells appear to be drawn to these constitutive ER-mitochondrial Ca2+ fluxes for proliferation, migration and survival while inhibiting cell death [[11], [12], [13]]. In addition, an increasing number of proto-oncogenes and tumor suppressors are found to affect Ca2+-signaling pathways by directly modulating intracellular Ca2+-transport systems with critical functions in cell survival and cell death [14].
The dynamic import–export balance of calcium regulates nuclear factor of activated T cells 1 (NFATC1) activity and nuclear trans-localization by calmodulin-dependent phosphatase [15]. Once in the nucleus, NFATC1 bind to specific sequences located in the regulatory regions of target genes such as vascular endothelial growth factor A (VEGFA), VEGF receptor (VEGFR), and cyclooxygenase-2 expression in endothelial cells and switch on their expression [[16], [17], [18]]. A recent study showed that the Ca2+–NFATC1 pathway is activated in diagnosed cases of BC and is essential to the tumorigenic and metastatic potential of mammary tumor cell lines. Thus, pharmacological inhibition of the Ca2+–NFATC1 pathway at different levels could be of therapeutic interest for BC patients [19].
Recently, FUN14 domain-containing protein 1 (FUNDC1) has been identified as a new outer mitochondrial membrane protein localized in MAMs [20,21]. FUNDC1 regulates the MAM formation responsible for Ca2+ release from the ER into mitochondria and cytosol in mouse cardiomyocytes. Moreover, overexpression of FUNDC1 was sufficient to induce mitophagy in several cancer cell lines [[22], [23], [24]]. However, the expression profile of FUNDC1 in the development of BC and its function remain unclear. Here, we aimed to explore the relation between the expression of FUNDC1 and prognosis/ prognostics for patients with BC. We also investigated the possible biological mechanisms of FUNDC1 in the sensitivity of BC cells.
2. Materials and methods
2.1. BC specimens and cell lines
102 cases of primary BC specimens were obtained randomly from who underwent BC surgery at the Cancer Hospital of Shantou University Medical College, China, between March 2012 and March 2014. The clinical characteristics of the patients were analyzed retrospectively. Histology evaluation by HE staining was confirmed by two pathologists who were blinded to the clinical data of patients. Normal epithelium sections of the breast were collected from 44 age-matched controls without BC. This sample size was calculated by a public service of creative research systems survey software. The use of clinical samples in this study was approved by the hospital research ethics committee on animal and human experimentations. Written informed consent was obtained from each participant.
BC cell lines used in this study, including SKBR3, BT-549, MDA-MB-231, MCF-7 and MDA-MB-453 cells, were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Accordance with the manufacturer's protocol, all cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Corning, Manassas, VA, USA) with 10% fetal bovine serum (FBS), 100 units/mL penicillin, and 100 μg/mL streptomycin and grown in a humidified incubator in a 5% CO2. All cell lines were authenticated and confirmed negative for mycoplasma contamination by providers.
2.2. Immunohistochemistry
BC tissue sections were deparaffinized and treated with citrate antigen repair buffer (pH 6.0) to antigen repair, with 1% hydrogen peroxide to block endogenous peroxidase activity, with 3% bovine serum albumin (BSA) for serum blocking. Tissue sections were then incubated with FUNDC1 (1:500, Abcam: ab173226) and BMI1 (1:500, R&D System: MAB33342), as well as their coordinate secondary antibody. Rabbit IgG (Santa Cruz Biotechnology) was used as negative control. Staining was displayed with DAKO DBA solution. Harris hematoxylin was used to re-stain the nucleus. The software automatically identified colors on the tissue slice and set all dark brown =3, brown yellow = 2, light yellow = 1, blue nucleus = 0, and extent of stained cells (0–5% = 0; 5–25% = 1; 26–50% = 2; 51–75% = 3 and 76–100% = 4). The final score was determined by multiplying the intensity score and the score for the extent of stained cells, generating a score that ranged from 0 to 12. The staining results were classified as negative (score 0; −), low (score 1–4; +), moderate (score 5–8; ++), and high (score 9–12; +++). The results were evaluated by two independent pathologists.
2.3. Transient cell transfections
Cells were cultured to 60% confluence and transiently transfected with (i) FUNDC1 or BMI1 cDNA ORF Clone (Sino Biological, USA) or a negative control pcDNA™3.1/CAT (Thermo Fisher Scientific, USA) by using Lipofectamine 3000 reagent (Invitrogen, USA) and (ii) siRNA-FUNDC1, -BMI1, -NFATC1, -STIM1, or control siRNA (Santa Cruz Biotechnology, USA) by using Lipofectamine RNAiMax (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer's protocol.
2.4. RNA extraction and real-time quantitative RT-PCR (qRT-PCR)
Total RNA was isolated from cultured cells by using Trizol reagent (Invitrogen) following the manufacturer's protocol. cDNA was synthesized from 1.0 μg total RNA with the PrimeScript Real-Time RT-PCR reagent kit (Takara Biotechnology [DALIAN] Co.) as previous described [25,26]. The samples were amplified by using the 7300 Realtime PCR System (Applied Biosystems). Relative mRNA expression was analyzed according to the comparative Ct method and normalized to that of β-actin. The primer sequences are listed in Table S1.
2.5. Luciferase reporter assays
BMI1 promoter reporter vectors were generated by PCR amplification and inserted into pGl3-basic plasmid as we previous reported [27,28]. BMI1 promoter reporter vectors contain the fragment of two NFATC1 motifs. Mutation and deletion of NFATC1 motifs was generated by using the Q5 Site-Directed Mutagenesis Kit (NEB) as the manufacturer's protocol. All constructs were confirmed by Sanger sequencing. The nucleotide sequences of primers are in Table S1. Cells grown in 96-well plates were transfected with BMI1 reporter vector and SV-40-Renilla-Luc in the presence of Lipofectamine 3000 Reagent (Invitrogen). At 24 h after transfection, cell extracts were prepared with passive lysis buffer. Luminescence was measured with the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Luciferase reporter activities were calculated as luciferase normalized to Renilla luciferase luminescence.
2.6. Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed as our previously described [27,29]. Briefly, treated MCF-7 cells at 90% to 100% confluence in 10-cm dishes were fixed with 1% formaldehyde and sonicated into 100- to 500-bp fragments. Soluble chromatin was precipitated with anti-NFAT1 (Abcam: ab2722) or a control non-immune IgG (Cell Signaling Technology: #2729). For real-time ChIP-PCR, the SYBR green system was used with the ABI Prism 7300 sequence detector (Applied Biosystems). Data are reported as relative fold enrichment. Primers used for ChIP qPCR of the BMI1 promoter are in Table S1.
2.7. Cell proliferation assay
For cell proliferation assay, the Cell Counting Kit-8 (CCK8, Sigma, 96,992) was used: cells (1–2 × 103/well) were plated into 96-well plates for 24 h and transfected with siRNA or pcDNA. After incubation for 24, 48 and 72 h, cells were stained with 20 μL CCK8. Absorbance was measured at 450 nm wavelength by using a microplate reader. Cell number counting was also used: 0.4 × 106 cells were seeded onto 6-well plates. After 24 h, cells were transfected with siRNA or pcDNA focused on FUNDC1 or BMI1, respectively. Then, cell number was monitored by using a Beckman Z1 Coulter Cell and Particle counter for three consecutive days.
2.8. Transwell migration and invasion assay
Cell invasion and migration of cells were evaluated by Transwell assay. For invasion, cell-culture inserts (0.8 μm, BD Biosciences) were coated with 40 mL Matrigel and dried overnight. For migration assays, inserts were not coated. Inserts were rehydrated with Opti-MEM (Invitrogen) for 2 h and 40,000 cells per insert were seeded in Opti-MEM Reduced Serum Media. Complete medium was used in the lower chamber. After 24 h of migration or invasion, cells remaining on the upper side of membrane were gently wiped off, and cells that migrated to the lower side of the membranes were fixed with 4% paraformaldehyde for 30 min and stained with 1% crystal violet. Images of stained cells in five random fields were captured by using an optical microscope (Olympus, Japan) and counted.
2.9. Western blot analysis
Total protein from treated cells was extracted with RIPA buffer (Sigma) and quantified by using the BCA Protein Assay Kit (Thermo Fisher Scientific). A 20-μg amount of protein was loaded and separated on 12% SDS-PAGE. After transferring to a polyvinylidene fluoride (PVDF) membrane (Millipore), the membrane was incubated overnight at 4 °C with antibody for FUNDC1 (1:500, Abcam:ab173226), BMI1 (1:1000, R&D System: MAB33342), NFATC1 (1:500, Abcam: ab2722), NFATC1 (phospho S54) (1:500, Abcam: ab200819), STIM1(1:500 Proteintech:11565-1-AP), ORAI1 (1:1000 Proteintech: 13130-1-AP), TRPC1 (1:500 Proteintech: 19482-1-AP) or mouse monoclonal antibody against beta ACTIN(1:2000, Abcam: ab8227) and GAPDH (1:1000; Santa Cruz Biotechnology: sc-47724. USA). After incubation with peroxidase-conjugated anti-mouse or rabbit IgG (Santa Cruz Biotechnology) at room temperature for 1 h, bound proteins were visualized by using ECL (Pierce) and detected by using BioImaging Systems (UVP Inc., Upland, CA). The relative protein levels were calculated by normalizing to beta-ACTIN or GAPDH protein as a loading reference by using ImageJ.
2.10. Immunofluorescence staining
Immunofluorescence staining was performed on cultured MCF-7 cells. After treatment, cells were fixed with 4% PFA, then permeabilized with 0.3% Triton/PBS. Cells were blocked in 1% BSA and incubated with the indicated primary antibody (NFATC1, Abcam) overnight at 4 °C. The next day, cells were washed three times in PBS, then incubated with the secondary antibody conjugated to Alexa Fluor 555 (donkey anti-rabbit, Invitrogen) for 1 h at room temperature. Cells were washed three times in PBS and stained with DAPI (Sigma) for 15 min to label nuclei. Fluorescence was observed under a Leica SP8 confocal laser scanning microscope.
2.11. Calcium measurement
For measuring cytoplasm Ca2+ levels [Ca2+]cyt. FUNDC1 overexpressed MCF-7 cells or FUNDC1 knock-down SKBR3 cells as well as their control cells were incubated with 5 μM green-5 N fluorescent probe (CaG5N, Thermo Fisher Scientific, Waltham, MA) for 30 min at room temperature and trypsinized. Then, cells were subjected to FACS analysis by using a FACScanto II flow cytometer (BD Bioscience, San Jose, CA). In total, 10,000 event cells per each independent experiment were used for further quantitative analysis by FlowJo (v10) software.
For ER Ca2+ [Ca2+]ER measurement, treated cells were incubated with 5 μM mag-fura-2 AM (Thermo Fisher Scientific, Waltham, MA) in DMEM for 45 min at room temperature, then perfused with HBSS buffer. Cells were washed with intracellular-like medium (ICM: 125 mM KCl, 19 mM NaCl, 10 mM Hepes, and 1 mM EGTA), and permeabilized by exposure to 25 μM β-escin in ICM for 2–3 min. Then, cells were switched to ICM containing 100 nM Ca2+ and 1.5 mM Magnesium ATP. Ratiometric Ca2+ measurements were performed by using the Myocyte Calcium & Contractility Recording System (IonOptix, MA). Fura-2 ratio values were converted into Ca2+ concentrations by using [Ca2+] = Kd·β·(R-Rmin)/(Rmax-R).
2.12. Kaplan-Meier assay
The prognostic merit of NFATC1 mRNA expression was appraised by using an online database of Kaplan-Meier Plotter (www.kmplot.com), with which including gene expression data and survival information for BC patients from the Gene Expression Omnibus database. To analyze the overall survival of patients with BC, patient samples were divided into two groups by median expression (high vs. low expression) and assessed by a Kaplan-Meier survival plot, estimating the hazard ratio (HR) with 95% confidence intervals (CI) and log-rank p value. The Affymetrix ID is valid: 202265_at (FUNDC1).
2.13. Correlation analysis with an online database
The correlation module computed the association between NFATC1 and BMI1 mRNA expression in tissues of BC patients from the online databases bc-GenExMiner v4.0 (Breast Cancer Gene-Expression Miner v4.0), cBioPortal (www.cbioportal.org), and GEPIA (Gene Expression Profiling Interactive Analysis, http://gepia.cancer-pku.cn/), as well as in BC cell lines by using the CCLE database (https://portals.broadinstitute.org/ccle/home).
2.14. Statistical analysis
All data are presented as mean ± SD. All in vitro experiments were performed in triplicate and repeated at least twice independently. Statistical analyses were performed using SPSS statistical software program 20.0 (IBM, Armonk, NY, USA) and GraphPad Prism version 6.0 (GraphPad Software). Student's t-test or Mann–Whitney U test was used to compare means between two groups. Two-way ANOVA was used to compare growth curves. The association of FUNDC1 expression with patient survival was analyzed by the Kaplan-Meier survival curve and log-rank test. Correlation analysis was involved the Pearson and Kendall correlation coefficients. Variance similar between the groups was statistically compared. P < 0.05 was considered statistically significant.
3. Results
3.1. Elevated expression of FUNDC1 was positively associated with worse disease progression in BC
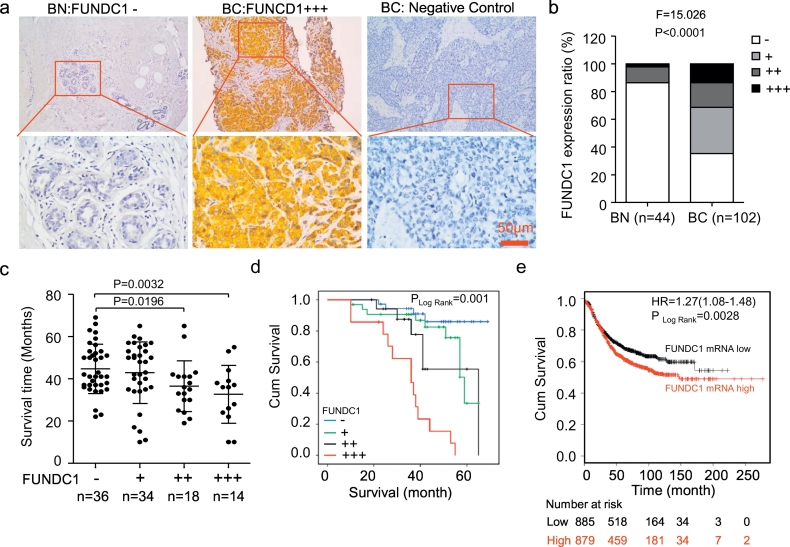
We found positive immunostaining for FUNDC1 in the cytoplasm and membrane of 66/102 (64.71%) BC tissues, with absent/weak immunostaining in the normal breast epithelium (Fig. 1a and b). FUNDC1 expression was positively correlated with pathological tumor size (r = 0.254, P = 0.013), stage (r = 0.297, P = 0.002), metastasis (r = 0.387, P < 0.0001) and death (r = 0.263, P = 0.008) of BC patients, respectively. No relationship was found with age, menopause, differentiation, relapse, lymph-node metastasis, as well as the expression of P53 and KI67 (Table 1).
Fig. 1.
Elevated FUNDC1 expression is associated with poor survival in BC. (a) Representative FUNDC1 negative and positive immunochemistry staining in tissues of normal breast (BN) and breast cancer (BC). IgG was the negative control for staining. (b) Distribution of FUNDC1 expression in BC and BN tissues. (c) Scatter diagram of overall survival time for patients with and without FUNDC1 expression. Staining results were classified as negative (score 0; −), low (score 1–4; +), moderate (score 5–8; ++), and high (score 9–12; +++) expression. (d-e) Kaplan-Meier analysis of overall survival in patients with high FUNDC1 expression predicted poor progression in 102 cases of clinical BC patients (d) and online database of Kaplan-Meier Plotter (e).
Table 1.
Correlation of FUNDC1 protein expression and clinical characteristics of patients with breast cancer (BC). Statistical analysis involved chi-square test and correlation analysis with the Kendall correlation coefficient. Bold values indicate significant correlation between FUNDC1 level and variables.
| Characteristic | No. of cases | FUNDC1 |
r | P value | |||
|---|---|---|---|---|---|---|---|
| − | + | ++ | +++ | ||||
| Age, years | <50 (n = 54) | 13 | 21 | 12 | 8 | −0.184 | 0.064 |
| ≥50 (n = 48) | 23 | 13 | 6 | 6 | |||
| Menopause | No (n = 60) | 16 | 23 | 13 | 8 | −0.137 | 0.169 |
| Yes (n = 42) | 20 | 11 | 5 | 6 | |||
| Tumor size, cm | <10 (n = 48) | 22 | 21 | 5 | 3 | 0.254 | 0.013 |
| ≥10 (n = 48) | 14 | 11 | 11 | 8 | |||
| Type | 1 (n = 85) | 33 | 27 | 14 | 11 | 0.069 | 0.493 |
| 2 (n = 9) | 1 | 6 | 2 | 0 | |||
| 3 (n = 6) | 2 | 1 | 2 | 1 | |||
| Differentiation | I (n = 31) | 17 | 10 | 3 | 1 | 0.221 | 0.030 |
| II (n = 25) | 6 | 8 | 7 | 4 | |||
| III (n = 46) | 13 | 16 | 8 | 9 | |||
| Stage | I (n = 19) | 10 | 11 | 1 | 0 | 0.297 | 0.002 |
| II (n = 38) | 14 | 13 | 6 | 5 | |||
| III + IV (n = 45) | 12 | 10 | 11 | 9 | |||
| Lymph node metastasis | No (n = 44) | 17 | 14 | 5 | 8 | 0.043 | 0.673 |
| Yes (n = 57) | 19 | 20 | 12 | 6 | |||
| Metastasis | No (n = 77) | 33 | 27 | 11 | 6 | 0.387 | <0.001 |
| Yes (n = 25) | 3 | 7 | 7 | 8 | |||
| KI67 level | <30% (n = 42) | 14 | 17 | 8 | 3 | 0.051 | 0.614 |
| ≥30% (n = 55) | 22 | 16 | 10 | 9 | |||
| P53 level | −(n = 41) | 14 | 16 | 4 | 3 | 0.175 | 0.086 |
| +(n = 26) | 11 | 15 | 1 | 3 | |||
| ++/++(n = 30) | 11 | 4 | 9 | 6 | |||
| Relapse | No (n = 81) | 30 | 28 | 14 | 9 | 0.144 | 0.151 |
| Yes (n = 20) | 4 | 5 | 4 | 7 | |||
| Death | No (n = 73) | 28 | 25 | 12 | 5 | 0.263 | 0.008 |
| Yes (n = 29) | 8 | 9 | 6 | 9 | |||
The mean survival was significantly shorter for patients with strong and medium than absent/weak immunostaining for FUNDC1 (32.7 ± 13.7 and 36.5 ± 12.0 vs 44.7 ± 11.7/42.8 ± 14.6 months) (Fig. 1c). On Kaplan-Meier survival analysis, BC patients with high FUNDC1 protein level had a worse outcome than those with low expression (P = 0.001, Fig. 1d). Further prognostic assay with the online Kaplan-Meier Plotter database also showed the increasing of FUNDC1 mRNA level was significantly correlated with worse overall survival (HR = 1.27; 95% CI: 1.08–1.48, p = 0.0028) (Fig. 1e). Therefore, FUNDC1 was frequently upregulated in human BC and implicated in the pathogenesis and progression of BC.
4. FUNDC1 functionally promoted BC proliferation and migration
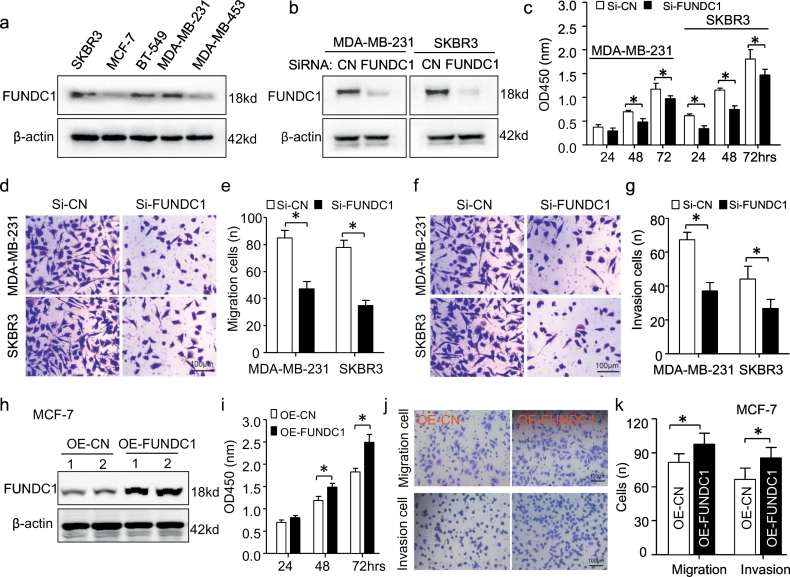
To investigate the functional roles of FUNDC1 in BC, we screened the protein expression of FUNDC1 in five BC cell lines and found high FUNDC1 expression in SKBR3, BT-549 and MDA-MB-231 cells but low expression in MCF-7 and MDA-MB-453 cells (Fig. 2a). Then, we established transient FUNDC1 knockdown models in MDA-MB-231 and SKBR3 cells with a pool of siRNA sequences. Successful knockdown of FUNDC1 was confirmed at the protein level (Fig. 2b). Knockdown of FUNDC1 significantly suppressed BC-cell proliferation (Fig. 2c) and inhibited BC cell migratory and invasion ability (Fig. 2d–g). Next, we successfully overexpressed endogenous FUNDC1 by transfecting pcDNA-FUNDC1 in MCF-7 cells (Fig. 2h). FUNDC1 overexpression could promote BC proliferation, migration and invasion (Fig. 2i–k). Thus, consistent with above clinical study, FUNDC1 functional promoted BC proliferation and metastatic growth in vitro.
Fig. 2.
FUNDC1 promotes breast cancer proliferation, migration and invasion. (a) Profile expression of FUNDC1 protein in five breast cancer cell lines by western blot assay (n = 3). (b) FUNDC1 expression was successfully knocked down by siRNA-FUNDC1 transfection for 24 h in MDA-MB-231 and SKBR3 cells (n = 3). (c–g) Cell proliferation (c), migration (d,e) and invasion (f,g) of MDA-MB-231 and SKBR3 cells with or without FUNDC1 knockdown were measured by CCK-8 assay and transwell assay, respectively (n = 6). (h) Western blot analysis of overexpression of FUNDC1 (OE-FUNDC1) by pc-DNA transfection for 24 h in MCF-7 cells (n = 3). (i–k) Cell proliferation (i), migration and invasion (k) of MCF-7 cells with or without FUNDC1 overexpression measured by CCK-8 assay and transwell assay (n = 5). Data are mean ± SD. *P < 0.05.
5. FUNDC1-induced cytosol Ca2+ influx and NFATC1 activity
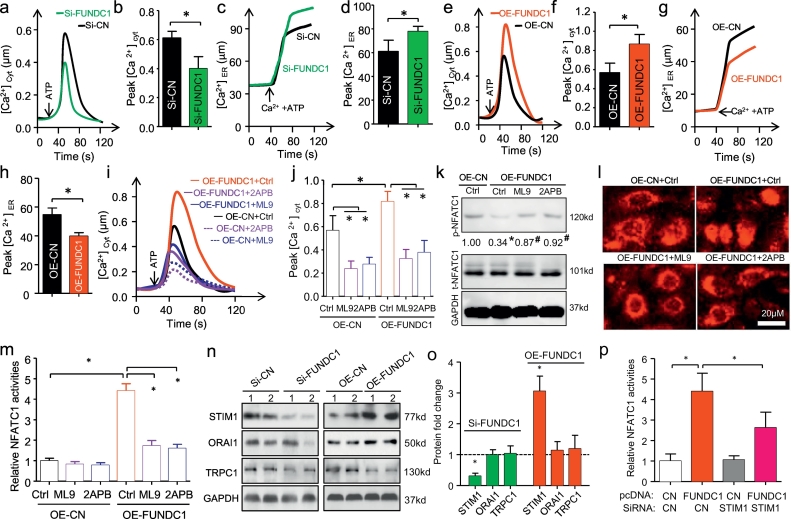
To assess the impact of FUNDC1 on Ca2+ signaling and NFATC1 activity in BC, we first evaluated Ca2+ cytoplasmic [Ca2+]cyt and ER storage [Ca2+]ER levels by FUNDC1 gain- or loss-of-function assay. FUNDC1 knockdown decreased [Ca2+]cyt level and accumulated [Ca2+]ER level in SKBR3 cells (Fig. 3a–d). Conversely, FUNDC1 overexpression in MCF-7 cells increased [Ca2+]cyt level but reduced [Ca2+]ER level (Fig. 3e–h). Therefore, FUNDC1 regulate Ca2+ release from ER to cytosol. On the other hand, store-operated calcium entry (SOCE) also is a major mechanism for increasing of cytosolic Ca2+ concentration and has been reported to play an important role in breast tumorigenesis [30]. We next explored whether the SOCE was correlated with the FUNDC1 activating Ca2+ signaling and NFATC1 activity. To this end, we found that two SOCE blockers, ML9 and 2-aminoethoxydiphenyl borate (2APB) could inhibited the cytosol Ca2+ influx at the both of the basal level and FUNDC1 over-expression modulated level (Fig. 3i,j). Subsequently, over-expression FUNDC1-induced NFATC1 de-phosphorylation and NFATC1 translocation. Interestingly, the NFATC1 activity were significantly prevented by ML9 and 2APB pretreatment (Fig. 3k–m). STIM (stromal interaction molecule), ORAI and TRPC (canonical transient receptor potential channel) protein families are the main modulators for SOCE and have been reported to be responsible for store-operated Ca2+ influx [30]. The western blot results showed that FUNDC1 knockdown selectively and significantly downregulated STIM1 expression in SKBR3 cells, and STIM1 upregulation was detected by FUNDC1 overexpression in MCF-7 cells (Fig. 3o,n). No alters of ORAI1 and TRPC1 were found by FUNDC1 regulation. Moreover, STIM1 silence by siRNA transfection could inhibit FUNDC1 over-expression induced NFATC1-luciferase activation in MCF-7 cells (Fig. 3p). Thus, the STIM1-Ca2+-NFATC1 signaling pathway was also involved in FUNDC1-induced BC progression.
Fig. 3.
FUNDC1 promotes cytosol Ca2+ influx, and NFATC1 translocation and activation. (a,b) Representative and quantification of Fura-2 measurement of [Ca2+]cyt level in response to ATP (0.1 mM) in SKBR3 cells with and without FUNDC1 knockdown (n = 4). (c,d) Representative recordings and quantification of Ca2+ ER [Ca2+]ER level during store filling initiated by the addition of 100 nmol/L Ca2+ and 1.5 mM ATP in SKBR3 cells (n = 4). (e–h) [Ca2+] measurements in cytosol (e,f) and ER (g,h) by ATP stimulation in MCF-7 cells with and without FUNDC1 overexpression (n = 4). (i,j) Effect of the SOCE inhibitors (50 μM of ML9 or 2APB) treatment on [Ca2+]cyt level in response to ATP (0.1 mM) in MCF-7 cells with and without over-expression (OE) FUNDC1 (n = 4). (k) Western blot analysis of de-phosphorylation of NFATC1 by FUNDC1 overexpression. Value is the relative ratio of phosphorylation (p)-NFATC1 and total (t)-NFATC1 (n = 3). *P < 0.05, with OE-CN Ctrl. #P < 0.05, with OE-FUDNC1 Ctrl. (l) Representative immune-staining of subcellular localization of NFATC1 with FUNDC1 overexpression in MCF-7 cells after treatment with 50 μM ML9 or 2APB (n = 5). (m) NFATC1 activation in MCF-7 determined by transfection with an NFATC1-luciferase reporter showing stimulation by FUNDC1 overexpression, with rescued by pre-treatment with store-operated Ca2+ entry blockers ML9 and 2APB (n = 6). (n,o) Western blot assay shown that the regulation of three major SOCE members by FUNDC1 in SKBR3 and MCF-7 cells (n = 3). (p) STIM1 silence by siRNA cloud inhibit FUNDC1 over-expression induced NFATC1-luciferase activation in MCF-7 cells (n = 3). Data are mean ± SD. *P < 0.05.
6. FUNDC1-induced Ca2+–NFATC1 signaling activated BMI1 expression
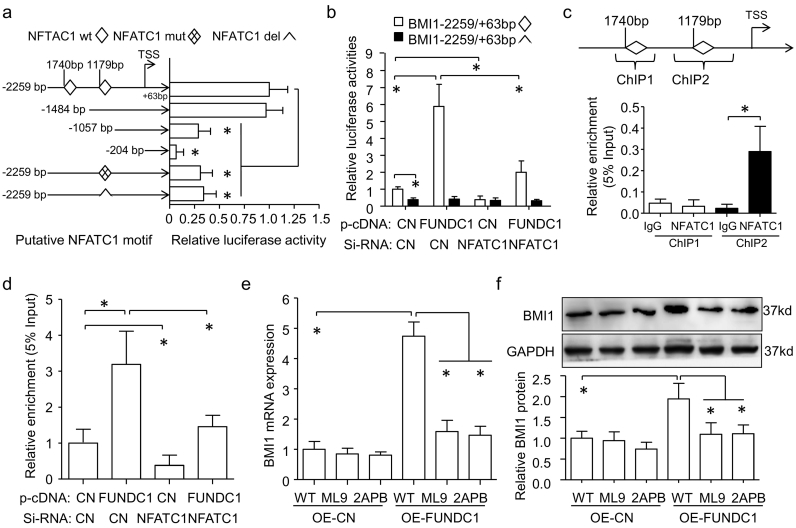
BMI1 is frequently overexpressed in BC and confers poor prognosis [31,32]. By bioinformatics assay, we found two NFATC1 motifs on the human BMI1 gene promoter, which prompted us to explore the regulation of BMI1 by FUNDC1. We then generated serial deletion constructs containing NFATC1 motifs as well as NFATC1 mutation and deletion plasmids based on BMI1-2259/+63-Luc plasmids. Dual-luciferase reporter assay revealed the NFATC1 motif at −1179 bp was required for BMI1 transcriptional activity, because plasmids with than without this motif had higher transcriptional activation. Specifically, BMI1 transcriptional activity could be slightly repressed by individual mutation of the NFATC1 motif at −1179 bp (46.71 ± 11.12%) or deletion (71.11 ± 11.36%) (Fig. 4a). Overexpression of FUNDC1 upregulated BMI1 activity with the wild-type NFATC1 motif at −1179 bp but not its deletion. Notably, this upregulation could be largely blocked by NFATC1 knockdown in MCF-7 cells (Fig. 4b). Moreover, NFATC1 could occupy the BMI1 promoter fragment only with the NFATC1 motif at −1179 bp, and greater enrichment of NFATC1 on the MBI1 promoter was found in FNUDC1-overexpressed MCF-7 cells. NFATC1 knockdown could inhibit this enrichment (Fig. 4c, d). As expected, FNUDC1 overexpression induced BMI1 activity and NFATC1 enrichment contributed to elevating BMI1 mRNA and protein expression, which was prevented by ML9 and 2APB, two SOCE blockers (Fig. 4e, f). Therefore, FUNDC1-induced Ca2+–NFATC1 signaling promoted BMI1 expression.
Fig. 4.
NFATC1 activates BMI1. (a) Serial deletion constructs containing the human BMI1 gene promoters, as well as BMI1-2259/+63bp-Luc plasmids with NFATC1 motif wildtype (wt), mutation (mut) or deletion (del) were transfected into MCF-7 cells for 24 h. Renilla plasmid was co-transfected as a transfection control. The mean luciferase activity of BMI1-2259/+63-Luc plasmids with NFATC1 motif wildtype was set as 1 (n = 6). Dual-luciferase reporter assay of NFATC1 motif at −1179 bp is required for BMI1 transcriptional activity. TSS, transcription start site. (b) MCF-7 cells were transfected with pcDNA-FUNDC1 or siRNA-NFATC1 for 24 h and then with BMI1-2259/+63-Luc plasmids with NFATC1 motif wild type or mutation for another 24 h. BMI1 promoter activities were measured by luciferase activity, normalized to that of Renilla (n = 6). (c) Quantitative chromatin immunoprecipitation (q-ChIP) assay showing that NFATC1 could bind to the fragment with NFATC1 motif at −1179 bp but not −1740 bp in MCF-7 cells; normal rabbit IgG was a control (n = 4). (d) q-ChIP analysis of enrichment of NFATC1 motif by overexpression of FUNDC1 and/or knockdown of NFATC1 (n = 4). (e,f) Increasing BMI1 mRNA and protein expression with overexpression of FUNDC1 was prevented by ML9 or 2APB pre-treatment of MCF-7 cells (n = 3). Data are mean ± SD. *P < 0.05.
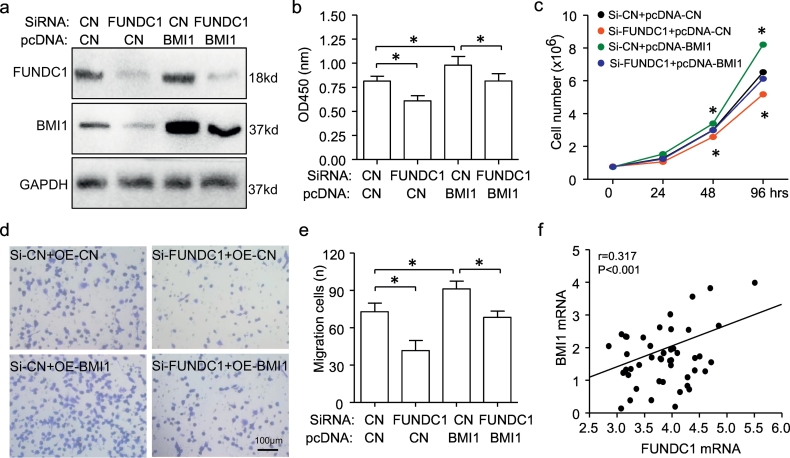
7. FUNDC1 regulated BC proliferation and migration by BMI1
We next determined the role of FUNDC1-BMI1 in BC proliferation and migration [32]. BMI1 protein expression was indeed reduced with FUNDC1 knockdown and recovered with pcDNA-BMI1 transfection in SKBR3 cells (Fig. 5a). Notably, CCK8 assay and cell counting shown that overexpression of BMI1 by pcDNA-BMI1 transfection could also rescue the decreasing of cell proliferation induced by FUNDC1 knockdown (Fig. 5b, c). Moreover, a similar effect was found for cell migration (Fig. 5d, e). RNA-seq data from the CCLE database demonstrated that FUNDC1 mRNA level was correlated with BMI1 mRNA level in 47 BC cell lines (Fig. 5f). Above observation indicated that BMI1 was necessary for FUNDC1 promoting BC proliferation and migration.
Fig. 5.
BMI1 contributes to NFATC1-induced cell proliferation and migration. (a) Western blot analysis of downregulation of BMI1 at protein level on transfection with siRNA-FUNDC1 and successful recovery by co-transfection with p-cDNA BMI1 plasmid in SKBR3 cells for 48 h. GAPDH was the internal control (n = 3). (b–e) BMI1 overexpression on transfection with p-cDNA BMI1 induced SKBR3 cell proliferation and migration and rescued FUNDC1 knockdown-reduced cell proliferation and migration, detected by CCK8 assay (b, n = 6), cell counting (c, n = 6) and transwell assay (d,e, n = 5). (f) Positive correlation of FUNDC1 and BMI1 mRNA expression among 47 breast cell lines based on their RNA sequence from the CLLE database.
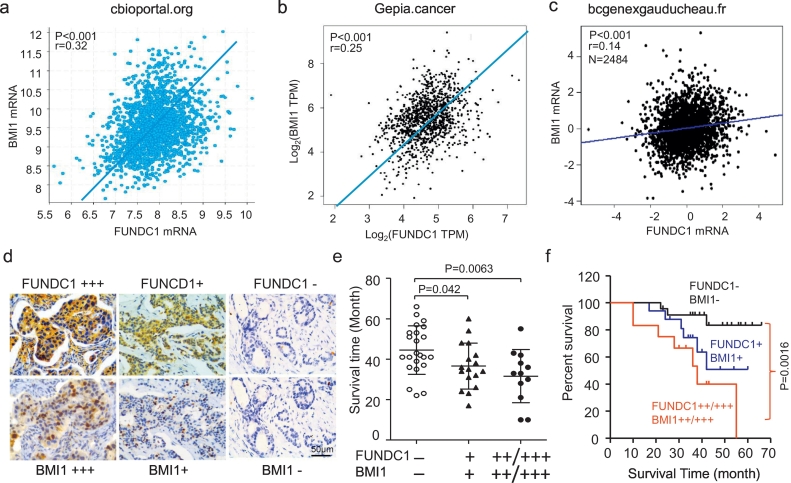
8. FUNDC1 positively related with BMI1 for BC prognosis
To study the relationship between FUNDC1 and BMI1 expression in BC patients, we queried the online databases chipportal.org, GEPIA and ba-GenExMine. In all datasets, FUNDC1 mRNA level was positively correlated with BMI1 mRNA level (Fig. 6a–c). By immunohistochemical staining of serial section, we found the comparable FUNDC1 and BMI1 protein expression (64.71%, 66/102 samples and 62.75%, 64/102 samples) in BC patients. Similarly, FUNDC1 protein level was closely and positively correlated with BMI1 protein level (r = 0.411, P < 0.001) (Table 2, Fig. 6d). Notably, BC patients with co-expression of weak and strong FUNDC1 and BMI1 staining had shorter survival time than those without either expression (36.59 ± 11.35 and 31.58 ± 13.18 vs 44.48 ± 12.01 months) (Fig. 5e). Kaplan-Meier survival analysis also showed that BC patients with high FUNDC1 and BMI1 co-staining had poor patient prognosis as compared with those without staining (P = 0.0016, Fig. 6f).
Fig. 6.
BMI1 level correlates with FUNDC1 level in progression of clinical BC. (a–c) Correlation between FUNDC1 and BMI1 mRNA expression for clinical BC patient data from the indicated online databases. (d) Representative immunohistochemical co-staining for FUNDC1 and BMI1 protein in continuous slices of BC tissue. (e) Scatter diagram showing overall survival time for patients with co-expression of FUNDC1 and BMI1 protein. (d–e) Kaplan-Meier analysis of overall survival in patients with high level of FUNDC1 and BMI1 co-expression predicting worse progression of clinical BC.
Table 2.
Correlation of FUNDC1 and BMI1 protein levels in patients with breast cancer.
| BMI1 | FUNDC1 |
Pearson's r | |||
|---|---|---|---|---|---|
| − | + | ++ | +++ | ||
| − | 26 | 5 | 5 | 0 |
r = 0.411 P < 0.001 |
| + | 1 | 10 | 4 | 1 | |
| ++ | 6 | 11 | 9 | 4 | |
| +++ | 5 | 6 | 5 | 4 | |
Staining results were classified as negative (score 0; −), low (score 1–4; +), moderate (score 5–8; ++), and high (score 9–12; +++).
9. Discussion
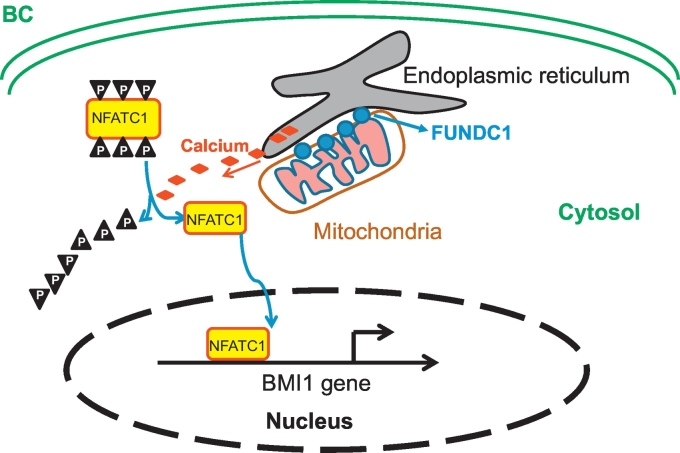
In this study, we found elevated expression of FUNDC1, the new member located in MAMs, in BC tissue as compared with normal breast tissue, and high FUNDC1 protein expression could predict worse disease progression with BC. Furthermore, FUNDC1 could promote cell proliferation, migration and invasion via the Ca2+–NFATC1–BMI1 axis (Fig. 7). We identified FUNDC1-Ca2+–NFATC1–BMI1 pathway is required and sufficient for BC progression and prognosis, which suggesting that inhibition of FUNDC1 or its pathway could represent a candidate therapeutic target for BC.
Fig. 7.
The schematic model. Elevated FUNDC1 expression in MAMs location promotes Ca2+ flux into cytoplasm, which de-phosphorylates NFATC1, transfers it to nucleus and activates BMI1 expression, further promotes the progression of BC.
MAMs, as highly specialized subcellular compartments, are biochemically distinct from pure ER and pure mitochondria. In MAMs, multiplex enzymes are related to calcium and/or lipid transport, and proteins with oncogenic/oncosuppressive functions. Recently, aberrant expression or localization of MAM-resident proteins was widely reported in rewiring normal cell signaling toward malignancy [5], such as BCL2 for hematopoietic, lung, breast and prostate cancer; GRP78 for breast and prostate cancer; HK2 for lung cancer; P53 for almost all cancer; and PTEN for lung, prostate, head, stomach, breast, and pancreas cancer [5]. Notably, the role of these oncogenes and oncosuppressors is related to modulation of Ca2+ influx. Here, we found that FUNDC1 could promote Ca2+ influx from ER and extracellular. Of note, SOCE blockers significantly inhibited FUNDC1-induced Ca2+ influx and NFATC1 signaling events. Based on above observation, we hypothesis that STIM1 mainly locates in the ER membrane and could be directly interact with FUNDC1 in MAM region. The co-expression of FUNDC1 and STIM1 in breast cancer patients indicated that the elevated FUNDC1 upregulates STIM1, which promoting more extracellular Ca2+ transfer into cytoplasm. Indeed, there is a growing body of evidence linking SOCE with a variety of breast cancer cell hallmarks, including cell survival, proliferation, migration and invasion [33,34]. Consistent with our study, transforming growth factor (TGF)-β induces cell cycle arrest in breast cancer cells by a mechanism of decreasing STIM1 expression [35]. Similarly, a recent study by Gueder and coworkers revealed that the pseudo-C-octyl glycoside 2-oxa-3-oxocastanospermine derivatives selectively decreased the expression of STIM1 at the protein level and attenuated SOCE, which results in the inhibition of MCF-7 and MDA-MB-231 cell [36]. In addition, resent study using cardiomyocytes shown that the inositol 1,4,5-trisphosphate receptors (IP3Rs) was involved into FUNDC1 regulated Ca2+ release from ER to cytosol [20]. Thus, FUNDC1 regulation of calcium flux from both of the ER and extracellular might be one of a major function of MAMs.
The implication of NFATCs in breast oncogenic processes is beginning to emerge. First, the NFATC transcription factors regulated by phosphatase calcineurin play a role in BC metastasis-promoting tumor cell invasion [37]. Second, the Ca2+–NFATC1 pathway is activated in the triple-negative ER-PR-HER2-BC subtype and is essential for the tumorigenic and metastatic potential of mammary tumor cell lines [19]. The Ca2+-NFAT pathway is also stimulated and required during angiogenesis induced by VEGF and secreted frizzle-related protein 2 in endothelial cells and may be a favorable target for inhibiting angiogenesis in solid tumors.
In our study, FUNDC1 could act as a novel stimulator for the Ca2+-NFATC1 pathway. FUNDC1 was sufficient to suppress NFATC1 phosphorylation and promote NFATC nuclear import. Importantly, nuclear NFATC1 could induce BMI1 transcription by binding to the NFATC1 motif within its proximal promoter. FUNDC1 level was correlated with BMI1 level in various cancer cell lines and clinical patients. BMI1, as an oncogene, acts a major mediator for cancer stem-cell self-renewal by regulating genes for cell cycle, stem-cell fate decisions, survival, and cellular senescence in multiple cancer models. BMI1 expression is significantly correlated with poor prognosis and survival [38] as well as aggressiveness [9] in human BC. Similarly, BMI1 overexpression sufficiently promoted cell proliferation and migration and also reversed FUNDC1 silencing-induced cell proliferation and migration in our study. Our data indicate that BMI1 as a novel and key target gene of the Ca2+–NFATC1 pathway plays a functional role in FUNDC1-induced breast carcinogenesis. Moreover, BC patients with high FNDC1 and BMI1 expression showed worse disease progression.
Ca2+ is released from the ER and then taken up into mitochondria in a “quasi-synaptic” manner. The accumulation of Ca2+ in the mitochondrial matrix has important implications for cancer processes including autophagy, metabolism, and apoptosis. Therefore, FUNDC1-stimulated Ca2+ influx has a waterfall effect on breast carcinogenesis, and the Ca2+-NFATC1-BMI1 axis might be a pathway induced by FUNDC1. The above evidence indicated that FUNDC1 is an upstream and major factor regulating BC, although we did not check the Ca2+ taken up by mitochondria and its contribution on cancer. BMI1 is sufficient to rescue the anti-carcinogenesis role of FUNDC1 silencing and is closely related to FUNDC1 level in multiple cell lines and patients. Recently, dysregulated Ca2+ homeostasis may play a role more like a “driver” than a “passenger” in carcinogenesis or tumorigenesis. Thus, targeting derailed Ca2+ signaling for cancer therapy has become an emerging research area. The known compounds or antibodies targeting Ca2+-ATPase inhibitors, voltage-gated Ca2+ channel inhibitors, TRP channel regulators, ORAI inhibitors, etc. have been studied in pre-clinical research or even clinical trials [[39], [40], [41]]. In our study, the FUNDC1–Ca2+ influx–NFATC1–BMI1 axis may suggest multiple targets for cancer treatment such as FUNDC1 knockdown, SOCE blockers, NFATC1 activity and translocation as well as BMI1 regulation. These data allow us to better understand the mechanisms of FUNDC1 involved in this process that may help improve diagnostic and therapeutic approaches to BC.
In summary, our results demonstrate that elevated FUNDC1 level could stimulate human BC proliferation and migration via Ca2+ influx as well as NFATC1 activation and translocation into the nucleus, which further binds and activates BMI1 transcription. The positive relationship between FUNDC1 and BMI1 expression further predicts worse progression of BC. Our findings provide novel mechanistic insight into how members of MAMs regulate BC, and FUNDC1 specially might be of value in developing new biomarkers for the prognosis of BC.
The following are the supplementary data related to this article
Reprehensive images of unprocessed western blot for Fig. 2h.
Complete list of primers used in this work. Sequences are from the GenBank. The bold fonts indicate mutations in the NFATC1 motif on the BMI1 promoter sequence.
Acknowledgments
Acknowledgements
We thank Laura Smales (BioMedEditing, Toronto, Canada) for critical reading and editing of the manuscript.
Funding sources
This work was supported by the grants of (1) the Medical Scientific Research Foundation of Guangdong Province, China [grant numbers: B2018222 (Jiongyu Chen)]; (2) the Key Project of Science and Technology of Shantou [grant numbers: [2018]37 (Jiongyu Chen)]; (3) the Youth Research Grant from Shantou University Medical College Cancer Hospital [grant numbers: 2018A008, (Jiongyu Chen)]; (4) the CAMS Innovation Fund for Medical Sciences (CIFMS) [Grant numbers: 2017-I2M-3-005 (Wei Cui)]; (5) the National Natural Science Foundation of China [Grant numbers: 81778272 (Wei Cui)].
Competing interests
The authors have declared that no competing interest exists.
Author contributions
C.J., Z.D., and W.L. conceived and designed the project, acquired research data, and wrote the manuscript. Z.D., C.J., W.L., Z.Y., Z.L., and P.Y. acquired research data. C.J., Z.Y., and Z.L. provided clinical tissue sections and cell lines. C.W. provided plasmids and reviewed the manuscript. Z.L. and W.L. contributed to helpful discussion and reviewed the manuscript.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Zheng R., Baade P.D. Cancer statistics in China, 2015. CA: A Cancer J Clinic. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Csordas G., Renken C., Varnai P. Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol. 2006;174(7):915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szabadkai G., Bianchi K., Varnai P. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J Cell Biol. 2006;175(6):901–911. doi: 10.1083/jcb.200608073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morciano G., Marchi S., Morganti C. Role of mitochondria-associated ER membranes in calcium regulation in cancer-specific settings. Neoplasia. 2018;20(5):510–523. doi: 10.1016/j.neo.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verfaillie T., Rubio N., Garg A.D. PERK is required at the ER-mitochondrial contact sites to convey apoptosis after ROS-based ER stress. Cell Death Differ. 2012;19(11):1880–1891. doi: 10.1038/cdd.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bobrovnikova-Marjon E., Grigoriadou C., Pytel D. PERK promotes cancer cell proliferation and tumor growth by limiting oxidative DNA damage. Oncogene. 2010;29(27):3881–3895. doi: 10.1038/onc.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salaroglio I.C., Panada E., Moiso E. PERK induces resistance to cell death elicited by endoplasmic reticulum stress and chemotherapy. Mol Cancer. 2017;16(1):91. doi: 10.1186/s12943-017-0657-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kakihana T., Nagata K., Sitia R. Peroxides and peroxidases in the endoplasmic reticulum: integrating redox homeostasis and oxidative folding. Antioxid Redox Signal. 2012;16(8):763–771. doi: 10.1089/ars.2011.4238. [DOI] [PubMed] [Google Scholar]
- 10.Kutomi G., Tamura Y., Tanaka T. Human endoplasmic reticulum oxidoreductin 1-alpha is a novel predictor for poor prognosis of breast cancer. Cancer Sci. 2013;104(8):1091–1096. doi: 10.1111/cas.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimm S. The ER-mitochondria interface: the social network of cell death. Biochim Biophys Acta. 2012;1823(2):327–334. doi: 10.1016/j.bbamcr.2011.11.018. [DOI] [PubMed] [Google Scholar]
- 12.Prevarskaya N., Skryma R., Shuba Y. Calcium in tumour metastasis: new roles for known actors. Nat Rev Cancer. 2011;11(8):609–618. doi: 10.1038/nrc3105. [DOI] [PubMed] [Google Scholar]
- 13.Roderick H.L., Cook S.J. Ca2+ signalling checkpoints in cancer: remodelling Ca2+ for cancer cell proliferation and survival. Nat Rev Cancer. 2008;8(5):361–375. doi: 10.1038/nrc2374. [DOI] [PubMed] [Google Scholar]
- 14.Akl H., Bultynck G. Altered Ca(2+) signaling in cancer cells: proto-oncogenes and tumor suppressors targeting IP3 receptors. Biochim Biophys Acta. 2013;1835(2):180–193. doi: 10.1016/j.bbcan.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Jauliac S., Lopez-Rodriguez C., Shaw L.M., Brown L.F., Rao A., Toker A. The role of NFAT transcription factors in integrin-mediated carcinoma invasion. Nat Cell Biol. 2002;4(7):540–544. doi: 10.1038/ncb816. [DOI] [PubMed] [Google Scholar]
- 16.Greenhough A., Smartt H.J., Moore A.E. The COX-2/PGE2 pathway: key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis. 2009;30(3):377–386. doi: 10.1093/carcin/bgp014. [DOI] [PubMed] [Google Scholar]
- 17.Ryeom S., Baek K.H., Rioth M.J. Targeted deletion of the calcineurin inhibitor DSCR1 suppresses tumor growth. Cancer Cell. 2008;13(5):420–431. doi: 10.1016/j.ccr.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 18.Zhao X., Wang Q., Yang S. Quercetin inhibits angiogenesis by targeting calcineurin in the xenograft model of human breast cancer. Eur J Pharmacol. 2016;781:60–68. doi: 10.1016/j.ejphar.2016.03.063. [DOI] [PubMed] [Google Scholar]
- 19.Quang C.T., Leboucher S., Passaro D. The calcineurin/NFAT pathway is activated in diagnostic breast cancer cases and is essential to survival and metastasis of mammary cancer cells. Cell Death Dis. 2015;6:e1658. doi: 10.1038/cddis.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu S., Lu Q., Wang Q. Binding of FUN14 domain containing 1 with inositol 1,4,5-trisphosphate receptor in mitochondria-associated endoplasmic reticulum membranes maintains mitochondrial dynamics and function in hearts in vivo. Circulation. 2017;136(23):2248–2266. doi: 10.1161/CIRCULATIONAHA.117.030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu W., Lin C., Wu K. FUNDC1 regulates mitochondrial dynamics at the ER-mitochondrial contact site under hypoxic conditions. EMBO J. 2016;35(13):1368–1384. doi: 10.15252/embj.201593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W., Li Y., Siraj S. FUNDC1-mediated mitophagy suppresses hepatocarcinogenesis by inhibition of inflammasome activation. Hepatology. 2019;69(2):604–621. doi: 10.1002/hep.30191. [DOI] [PubMed] [Google Scholar]
- 23.Hui L., Wu H., Wang T.W., Yang N., Guo X., Jang X.J. Hydrogen peroxide-induced mitophagy contributes to laryngeal cancer cells survival via the upregulation of FUNDC1. Clin Transl Oncol. 2018 doi: 10.1007/s12094-018-1958-5. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 24.Hirota Y., Yamashita S., Kurihara Y. Mitophagy is primarily due to alternative autophagy and requires the MAPK1 and MAPK14 signaling pathways. Autophagy. 2015;11(2):332–343. doi: 10.1080/15548627.2015.1023047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang D., Sun X., Liu J., Xie X., Cui W., Zhu Y. Homocysteine accelerates senescence of endothelial cells via DNA hypomethylation of human telomerase reverse transcriptase. Arterioscler Thromb Vasc Biol. 2015;35(1):71–78. doi: 10.1161/ATVBAHA.114.303899. [DOI] [PubMed] [Google Scholar]
- 26.Zhang D., Zhang Q., Zhou L. Comparison of prevalence, viral load, physical status and expression of human papillomavirus-16, -18 and -58 in esophageal and cervical cancer: a case-control study. BMC Cancer. 2010;10:650. doi: 10.1186/1471-2407-10-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang D., Wang Y., Lu P. REST regulates the cell cycle for cardiac development and regeneration. Nat Commun. 2017;8(1):1979. doi: 10.1038/s41467-017-02210-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Zhang D., Wu B., Wang P. Non-CpG methylation by DNMT3B facilitates REST binding and gene silencing in developing mouse hearts. Nucleic Acids Res. 2017;45(6):3102–3115. doi: 10.1093/nar/gkw1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang D., Xie X., Chen Y., Hammock B.D., Kong W., Zhu Y. Homocysteine upregulates soluble epoxide hydrolase in vascular endothelium in vitro and in vivo. Circ Res. 2012;110(6):808–817. doi: 10.1161/CIRCRESAHA.111.259325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jardin I., Lopez J.J., Salido G.M., Rosado J.A. Store-operated Ca(2+) entry in breast cancer cells: remodeling and functional role. Int J Mol Sci. 2018;19(12) doi: 10.3390/ijms19124053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y., Zhe H., Ding Z., Gao P., Zhang N., Li G. Cancer stem cell marker Bmi-1 expression is associated with basal-like phenotype and poor survival in breast cancer. World J Surg. 2012;36(5):1189–1194. doi: 10.1007/s00268-012-1514-3. [DOI] [PubMed] [Google Scholar]
- 32.Siddique H.R., Saleem M. Role of BMI1, a stem cell factor, in cancer recurrence and chemoresistance: preclinical and clinical evidences. Stem Cells. 2012;30(3):372–378. doi: 10.1002/stem.1035. [DOI] [PubMed] [Google Scholar]
- 33.Jardin I., Rosado J.A. STIM and calcium channel complexes in cancer. Biochim Biophys Acta. 2016;1863(6 Pt B):1418–1426. doi: 10.1016/j.bbamcr.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 34.Vashisht A., Trebak M., Motiani R.K. STIM and Orai proteins as novel targets for cancer therapy. A review in the theme: cell and molecular processes in cancer metastasis. Am J Physiol Cell Physiol. 2015;309(7):C457–C469. doi: 10.1152/ajpcell.00064.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng H., Wang S., Feng R. STIM1 plays an important role in TGF-beta-induced suppression of breast cancer cell proliferation. Oncotarget. 2016;7(13):16866–16878. doi: 10.18632/oncotarget.7619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gueder N., Allan G., Telliez M.S. sp(2)-Iminosugar alpha-glucosidase inhibitor 1-C-octyl-2-oxa-3-oxocastanospermine specifically affected breast cancer cell migration through Stim1, beta1-integrin, and FAK signaling pathways. J Cell Physiol. 2017;232(12):3631–3640. doi: 10.1002/jcp.25832. [DOI] [PubMed] [Google Scholar]
- 37.Yoeli-Lerner M., Yiu G.K., Rabinovitz I., Erhardt P., Jauliac S., Toker A. Akt blocks breast cancer cell motility and invasion through the transcription factor NFAT. Mol Cell. 2005;20(4):539–550. doi: 10.1016/j.molcel.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 38.Silva J., Garcia V., Garcia J.M. Circulating Bmi-1 mRNA as a possible prognostic factor for advanced breast cancer patients. Breast Cancer Res. 2007;9(4):R55. doi: 10.1186/bcr1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui C., Merritt R., Fu L., Pan Z. Targeting calcium signaling in cancer therapy. Acta Pharmaceut Sin B. 2017;7(1):3–17. doi: 10.1016/j.apsb.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brasky T.M., Krok-Schoen J.L., Liu J. Use of calcium channel blockers and breast cancer risk in the Women's Health Initiative. Cancer Epidemiol Biomark Prevent. 2017;26(8):1345–1348. doi: 10.1158/1055-9965.EPI-17-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenberg L., Rao R.S., Palmer J.R. Calcium channel blockers and the risk of cancer. JAMA. 1998;279(13):1000–1004. doi: 10.1001/jama.279.13.1000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reprehensive images of unprocessed western blot for Fig. 2h.
Complete list of primers used in this work. Sequences are from the GenBank. The bold fonts indicate mutations in the NFATC1 motif on the BMI1 promoter sequence.