Abstract
Background
The increased prevalence of cardiovascular disease (CVD) indicates a demand for novel therapeutic approaches. Proteome analysis of vascular tissues from animal models and humans with CVD could lead to the identification of novel druggable targets.
Methods
LC-MS/MS analysis of thoracic aortas from three mouse models of non-diabetic and diabetic (streptozotocin (STZ)-induced) atherosclerosis followed by bioinformatics/pathway analysis was performed. Selected findings were confirmed by proteomics analysis of human vessels from patients with CVD as well as in vitro studies (migration, proliferation, angiogenesis assays) using endothelial (HUVEC) cells.
Findings
Comparative tissue proteomics of low density lipoprotein receptor deficient (Ldlr−/−) and diabetic Ldlr−/− (Ldlr−/−STZ) with wild type (WT) animals led to the identification of 284 differentially expressed proteins in both models. Among them, 177 proteins were also differentially expressed in diabetic apolipoprotein E deficient (ApoE−/−STZ) mice, suggesting expression changes associated with atherosclerosis independent of the model used. These proteins recapitulated the hallmarks of atherosclerosis. Comparison of these findings with differentially expressed proteins in human vessels with CVD enabled shortlisting of six commonly dysregulated proteins. Among them, lysine-specific demethylase 5D (KDM5D) exhibited pronounced overexpression accompanied by a reduction in the protein levels of its substrate, the trimethylated lysine 4 of histone H3 (H3K4me3), in patients with CVD. Functional interference studies applying a KDM5 inhibitor on HUVEC reduced cell proliferation, migration and tube-forming ability in vitro.
Interpretation
This high-throughput proteomics strategy identified KDM5 histone demethylases being potentially involved in CVD, possibly by affecting H3K4 methylation.
Fund
[SysVasc, HEALTH-2013 603288], [ERA-CVD PROACT: ANR-17-ECVD-0006, 01KL1805], [FRM, DEQ20170336759].
Keywords: Cardiovascular disease, Atherosclerosis, Diabetes, Proteomics, KDM5, H3K4
Research in context.
Evidence before this study
A number of high-throughput proteomic studies using different animal models have shed light on several key mechanisms of atherosclerosis. An in depth comparative characterization of atherosclerotic animal models on different genetic backgrounds, promises to improve the validity of findings and lead to the identification of key molecules involved in cardiovascular disease.
Added value of this study
In this study, we provide a holistic, proteomics-based integrated approach using several animal models of atherosclerosis, human samples and in vitro experiments for the characterization of consistent protein changes associated with cardiovascular disease regardless of genetic background or disease aetiology. We report for the first time, a high-throughput proteomics analysis of the common proteomic alterations in three different animal models of atherosclerosis from two different genetic backgrounds (Ldlr−/− and ApoE−/−), in the presence or absence of diabetes. The specificity of these findings was further improved by comparative analysis with proteomics data of human vascular tissues, which finally led to identification of proteins and pathways common in both species. The cross-species comparison provides evidence that KDM5D is a novel candidate with increased expression in cardiovascular disease. This finding was further supported by a concomitant decrease on the expression levels of the H3K4me3, a KDM5 substrate previously associated with cardiovascular disease.
Implications of all the available evidence
Our data support the implication of KDM5 demethylases in cardiovascular disease, possibly by dysregulating the methylation status of H3K4. Furthermore, high confidence proteomic datasets are provided supporting previously identified atherosclerosis-associated changes in widely employed animal models, for further use in systems biology approaches and model selection for preclinical studies.
Alt-text: Unlabelled Box
1. Introduction
The development and progression of atherosclerotic lesions is a complex process that includes endothelial cell dysfunction [1], inflammation, fibrous cap and necrotic core formation as well as plaque destabilization and rupture [2]. Given the multifactorial phenotype of atherosclerosis, novel diagnostic and therapeutic approaches should be based on the study of multiple molecular features simultaneously [3]. High-throughput omics strategies including genomics, transcriptomics, proteomics, lipidomics and metabolomics have been applied in atherosclerosis studies [4]. Among these omics approaches, proteomics produces a stable readout directly linked to cell function and phenotype. In addition, proteins can be pharmacologically addressed, and may serve as biomarkers of disease [5]. Previous proteomics or metabolomics-based efforts to delineate molecular mechanisms of early atherosclerosis, included among others, proteome and metabolome characterization of atherosclerotic rabbit models with subsequent investigation of translatability of the findings into human disease using plasma or urine samples [6,7] as well as studies of comprehensive analysis of the proteomic architecture of human early atherosclerotic arterial tissues [8]. Nevertheless many mechanisms still remain elusive.
Well established animal models of atherosclerosis have shown to be important tools for the elucidation of the molecular mechanisms that govern atherosclerosis [9]. Among those, we and others have shown that low density lipoprotein receptor deficient (Ldlr−/−) and apolipoprotein E deficient (ApoE−/−) mice on high cholesterol diet mimic major characteristics of human dyslipidemia [9] and metabolic changes [10], supporting their frequent use as preclinical models of atherosclerotic disease. In brief, in a previous study, we employed five different models of cardiovascular disease (CVD) including the atherosclerotic Ldlr−/− and ApoE−/− animal models, the klotho-hypomorphic mice (kl/kl) and the stroke-prone spontaneously hypertensive (SHRSP) rats with or without salt feeding [10]. Comparison of the blood metabolite signature of these animals with the 26 metabolite- signature from patients with CVD (represented as carotid intima media thickness (cIMT)), identified eleven and ten metabolites in the Ldlr−/− and the ApoE−/− animals respectively having the same statistical significant regulation trend with humans [10]. Among the common blood metabolites were several phospholipids and acylcarnitines. The lower coverage that was observed when comparing the other animal models with the human metabolite signature further suggested that the Ldlr−/− and ApoE−/− models better recapitulate the human cIMT signature [10]. Even more, diabetes-accelerated atherosclerosis, characterized currently by increased prevalence and limited therapeutic options [11] can be mimicked in ApoE−/− and Ldlr−/− models by artificial induction of diabetes using streptozotocin (STZ) [12]. As these models (ApoE−/− and Ldlr−/−) are characterized by different types of lipoproteins, the atherogenic mechanisms should ideally be investigated in both genetic backgrounds [13].
With the aim to identify potential novel targets in CVD, we applied proteome analyses on vascular tissue of these frequently used mouse models of atherosclerotic disease in the presence and/or absence of diabetes. Potential targets for intervention identified with this approach were validated in human vascular tissue of patients with CVD and their impact was further assessed in vitro. Our results reveal dysregulation of histone modifiers in CVD and suggest an involvement of the KDM5 histone demethylases in vessel formation, being of potential therapeutic interest.
2. Materials and methods
2.1. Animals
Wild type (WT) (n = 5), Ldlr−/− (n = 5), Ldlr−/−STZ (n = 5) and ApoE−/−STZ (n = 3) on C57BL/6 background male mice (Charles River) were maintained in a pathogen-free environment under standard laboratory conditions. To induce diabetes, 6 week-old ApoE−/− and Ldlr−/− mice received daily intraperitoneal injections of STZ (Sigma-Aldrich) at a dose of 50 mg/kg in sodium citrate buffer (pH 4.5) for 5 consecutive days. All mice were then fed a 1.25% cholesterol enriched diet (TD.96335, ssniff Spezialdiäten) for 17 weeks with unrestricted access to water. One week before sacrifice, the mice were placed in metabolic cages for 18 h to measure diuresis and to collect urine samples that were stored at −80 °C. For blood analyses, animals underwent a 6 h fasting period prior to the blood test. Glycemia was assessed by measuring the blood glucose concentration using a glucometer. An autoanalyzer was used to measure the total cholesterol and triglyceride concentrations. At the end of the experiment, mice were euthanized after mild anesthesia with isoflurane (1.5%) followed by cervical dislocation. The thoracic aortas were removed, frozen in liquid nitrogen and stored at −80 °C until processed. All animal procedures were in accordance with the guidelines from Directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes. The project was approved by the local (Inserm/UPS US006 CREFRE) and national ethics committees under the number 02604.02.
2.2. Human vascular tissues
The clinical samples used in this study were collected at the Medical University of Graz, Austria. Human vascular tissues from peripheral vessels (iliaca communis, iliaca externa, femoralis superficialis) and coronary arteries (left anterior descending artery) were collected from multiorgan donors without CVD background (controls, n = 20) and patients with cardiovascular disease including typical cardiovascular risk factors (cases, n = 30). All patients had manifest vascular disease as key inclusion criterion (such as peripheral artery disease or coronary artery disease) and their clinical data are presented in the results section below. Controls had no evidence of vascular disease. Further details on assessing CVD are given in Lygirou et al. [14]. More than one vessel type was available for some participants. Following harvesting, the vascular tissues were snap frozen in liquid nitrogen and stored at −80 °C until preparation for proteomics analysis. The study complied with the principles outlined in the Declaration of Helsinki. Sample collection was approved by the local ethics committee (approval number: 26–355 ex 13/14), and for all individuals there was a signed written informed consent.
2.3. Sample preparation
Approximately 10–20 mg (net weight) of animal or human tissue specimens, as applicable, were homogenized in lysis buffer containing 0.1 M Tris-HCl pH 7.6 supplemented with 4% SDS and 0.1 M DTE. Homogenization was performed with the bullet blender homogenizer (Next Advance) [15]. After tissue homogenization the protein concentration of each sample was determined by the Bradford Assay. Ten micrograms of each protein sample were applied onto SDS-PAGE (5% stacking, 12% separating). Electrophoresis was stopped when samples just entered the separating gel. Gels were fixed with 30% methanol, 10% acetic acid for 30 min followed by 3 washes with water (5 min each) and stained with Coomassie Colloidal Blue overnight. Each band (one per sample) was excised from the gel and further sliced into small pieces (1–2 mm). Gel pieces were destained with 40% Acetonitrile, 50 mM NH4HCO3 and then reduced with 10 mM DTE in 100 mM NH4HCO3 for 20 min at room temperature (RT). After reduction, samples were alkylated with 54 mM Iodoacetamide in 100 mM NH4HCO3 for 20 min at RT in the dark. Samples were then washed with 100 mM NH4HCO3 for 20 min at RT, followed by another wash with 40% Acetonitrile, 50 mM NH4HCO3 for 20 min at RT and a final wash with ultrapure water under the same conditions was performed. Gel pieces were dried in a centrifugal vacuum concentrator and trypsinized overnight in the dark at RT, by adding 600 ng of trypsin per sample (trypsin stock solution: 10 ng / μl in 10 mM NH4HCO3, pH 8.5). Peptides were extracted after incubation with the following buffers: 50 mM NH4HCO3 for 15 min at RT followed by two incubations with 10% formic acid (FA), acetonitrile (1:1) for 15 min at RT. Peptides were eluted in a final volume of 600 μl and filtered with 0.22 μm pore size PVDF membrane syringe filters (Merck Millipore) before dried in a centrifugal vacuum concentrator. Dried peptides were reconstituted in mobile phase A buffer (0.1% formic acid, pH 3.5) and processed with LC-MS/MS analysis.
2.4. LC-MS/MS analysis
LC-MS/MS was performed as previously described [16]. Samples were injected into a Dionex Ultimate 3000 RSLS nano flow system configured with a Dionex 0.1 × 20 mm 5 μm C18 nano trap column. Mobile phase was 2% ACN: 0.1% FA with a flow rate of 5 μl/min. The analytical column was an Acclaim PepMap C18 nano column 75 μm × 50 cm, 2 μm 100 Å at a flow rate of 300 nl/min. The trap and nano-flow column were maintained at 35 °C. The samples were eluted with a gradient starting at 1% B for 5 min rising to 5% B at 10 min then to 25% B at 360 min and 65% B at 480 min. Mobile phase A was 0.1% FA while mobile phase B was 80% ACN:0.1% FA. The column was washed and re-equilibrated prior to each sample injection. The eluent was ionized using a Proxeon nano spray ESI source operating in positive ion mode. For mass spectrometry analysis, an Orbitrap LTQ Velos (Thermo Finnigan) was operated in MS/MS mode, scanning from 380 to 2,000 m / z. Ionization voltage was 2.6 kV and the capillary temperature was 275 °C. The resolution of ions in MS1 was 60,000 and 7,500 for higher-energy collisional dissociation (HCD) MS2. The top 20 multiply charged ions were selected from each scan for MS/MS analysis using HCD at 35% collision energy. Dynamic exclusion was enabled with a repeat count of 1, exclusion duration of 30 s.
2.5. MS data processing, quantification, illustration and functional annotation
Raw files were analyzed with Proteome Discoverer 1.4 software package (Thermo Finnigan), using the SEQUEST search engine and the UniProt human (Homo sapiens) reviewed database, downloaded on May 30, 2016, including 20,204 entries (human samples) or the UniProt mouse (Mus musculus) reviewed database, downloaded on June 26, 2015, including 16,717 entries. The search was performed using carbamidomethylation of cysteine as static and oxidation of methionine as dynamic modifications. Two missed cleavage sites, a precursor mass tolerance of 10 ppm and fragment mass tolerance of 0.05 Da were allowed. False discovery rate (FDR) validation was based on q value: target FDR (strict): 0.01, target FDR (relaxed): 0.05. Quantification was performed at the peptide level employing a clustering approach as previously described [14]. Label free quantification was performed by utilizing the precursor area values of each sample as defined by the Proteome Discoverer 1.4 software package. Samples were analyzed individually (not pooled) and assigned to distinct groups. For a small number of peptide sequences where no peptide precursor area could be retrieved (although the peptides were identified) the missing values were replaced by zero. If a peptide was not identified in a particular sample the missing values were replaced by zero. For the quantitation-statistical analysis, only the peptides which were present in at least 60% of the samples in at least one group were further considered. The precursor area values were subjected to the following normalization method within each sample prior quantification analysis: normalized peak area = (peptide peak area/sum of peptides peak area) × 106. Protein expression values were calculated as the sum of all the normalized peptide areas that were assigned for a given protein. P value ≤0.05 was considered statistically significant. Visualization of the proteomics findings in the form of heat map was performed using the Heatmapper web server (http://heatmapper.ca/) [17]. The gene names used in the heatmap, in Table 1 and Supplementary Tables (wherever indicated) were updated on December 2018 based on UniProt database. The function of the proteins was extracted from UniProt. Functional analysis was performed with the Ingenuity Pathway Analysis software (QIAGEN Inc.) using the default settings [18].
Table 1.
The top 15 (based on fold change) proteins in Ldlr−/−, Ldlr−/−STZ and ApoE−/−STZ mice that were found statistically significant upregulated compared to WT.
| Gene Symbol | Protein | Ldlr−/− vs WT |
Ldlr−/−STZ vs WT |
ApoE−/−STZ vs WT |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Rank | Log2 fold change | P value | Rank | Log2 fold change | P value | Rank | Log2 fold change | P value | ||
| Stk25 | Serine/threonine-protein kinase 25 | 1 | 9.316 | 0.011 | 2 | 9.28 | 0.011 | 1 | 9.55 | 0.032 |
| Xrn1 | 5′-3′ exoribonuclease 1 | 2 | 9.310 | 0.0097 | 1 | 9.51 | 0.0097 | 2 | 9.46 | 0.026 |
| Letm1 | Mitochondrial proton/calcium exchanger protein | 3 | 8.598 | 0.0097 | 3 | 9.04 | 0.0097 | 3 | 9.00 | 0.026 |
| Kif26b | Kinesin-like protein KIF26B | 4 | 8.097 | 0.0097 | 4 | 8.12 | 0.0097 | 5 | 7.81 | 0.026 |
| Cfl1 | Cofilin-1 | 5 | 8.028 | 0.0097 | 5 | 7.68 | 0.0097 | 4 | 7.95 | 0.026 |
| Hsd3b4 | 3 beta-hydroxysteroid dehydrogenase type 4 | 6 | 7.542 | 0.011 | 7 | 7.08 | 0.011 | 8 | 7.07 | 0.032 |
| Zfyve1 | Zinc finger FYVE domain-containing protein 1 | 7 | 7.529 | 0.0097 | 16 | 5.85 | 0.0097 | 11 | 6.96 | 0.026 |
| Tiam1 | T-lymphoma invasion and metastasis-inducing protein 1 | 8 | 7.30 | 0.044 | 46 | 3.91 | 0.018 | 12 | 6.75 | 0.026 |
| Zmat1 | Zinc finger matrin-type protein 1 | 9 | 7.28 | 0.0097 | 8 | 7.02 | 0.0097 | 7 | 7.21 | 0.026 |
| Hectd3 | E3 ubiquitin-protein ligase HECTD3 | 10 | 6.72 | 0.0097 | 6 | 7.23 | 0.0097 | 6 | 7.23 | 0.026 |
| Cenpe | Centromere-associated protein E | 11 | 6.66 | 0.0097 | 71 | 3.00 | 0.031 | 14 | 6.30 | 0.026 |
| Hpf1 | Histone PARylation factor 1 | 12 | 6.54 | 0.011 | 12 | 6.37 | 0.0012 | 24 | 5.64 | 0.036 |
| Kdm5d | Lysine-specific demethylase 5D | 13 | 6.47 | 0.0097 | 10 | 6.77 | 0.0097 | 9 | 7.07 | 0.026 |
| Ice1 | Little elongation complex subunit 1 | 14 | 6.47 | 0.011 | 11 | 6.61 | 0.034 | 13 | 6.58 | 0.032 |
| Fzd6 | Frizzled-6 | 15 | 6.31 | 0.0097 | 23 | 5.35 | 0.0097 | 19 | 6.03 | 0.026 |
| Ndufa10 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial | 20 | 5.69 | 0.0097 | 9 | 6.91 | 0.0097 | 40 | 4.64 | 0.026 |
| Iars2 | Isoleucine‐‐tRNA ligase, mitochondrial | 19 | 5.77 | 0.0097 | 13 | 6.29 | 0.0097 | 22 | 5.76 | 0.026 |
| Afg1l | AFG1-like ATPase | 18 | 5.79 | 0.0097 | 14 | 6.23 | 0.0097 | 17 | 6.09 | 0.026 |
| Ndrg2 | Protein NDRG2 | 17 | 6.15 | 0.0097 | 15 | 6.11 | 0.0097 | 15 | 6.24 | 0.026 |
| Lgals3 | Galectin-3 | 31 | 5.07 | 0.0097 | 17 | 5.82 | 0.0097 | 10 | 7.02 | 0.026 |
2.6. Cell culture and treatment with the inhibitor
Human Umbilical Vein Endothelial Cells (pooled HUVEC) were cultured in Endothelial Growth Medium-2 (EGM-2) (Lonza) and were used at passages 4–6. When required, HUVEC were starved in Endothelial Basal Medium-2 (EBM-2): EGM-2 (4∶1). For the inhibition experiments the KDM5 Histone Demethylases inhibitor, KDM5-C70, ethyl 2-(((2-((2-(dimethylamino)ethyl)(ethyl)amino)-2-oxoethyl)amino)methyl)isonicotinate (Xcessbio Biosciences) was added at different concentrations (10 μΜ, and 20 μΜ) in the growth media (complete medium or starvation medium depending on the experiment). 0.1% DMSO treated cells were used as controls since DMSO was the solvent of the inhibitor. The inhibitory activity was examined after 3 days of treatment through western blot analysis for the trimethylated lysine 4 of histone H3 (H3K4me3).
2.7. Western blot analysis
30 μg total protein from cell extracts and tissue extracts were analyzed for H3K4me3 (1/5,000 rabbit monoclonal anti-human Tri-Methyl-Histone H3 (Lys4) (C42D8), Cell Signaling Technology). Total histone H3 (1/5,000 rabbit monoclonal anti-human histone H3 (D2B12), Cell Signaling Technology) was used as a loading control. The films were scanned at a GS-800 imaging densitometer (BioRad) in transmission mode and the densitometry analysis of the results was performed using the Quantity One software (BioRad).
2.8. PCR analysis
KDM5D expression was examined in cDNA samples by performing Real-time PCR using the SYBR Green master mix (Kapa Biosystems), and specific primers for KDM5D (forward: 5′-ACAGCTACAGGCCAAACC-3′ and reverse: 5′-CAGCCCTTGGACCTTAGAAATA-3′). GAPDH was used as endogenous control.
2.9. Flow cytometry analysis
To test for potential toxicity of the KDM5-C70 inhibitor, apoptosis and necrosis were evaluated on untreated or treated for 3 days with 0.1% DMSO, 10 μM or 20 μM KDM5-C70 HUVEC using FITC Annexin V Apoptosis Detection Kit with 7-amino-actinomycin D (7-AAD) (BioLegend) according to the manufacturer's instructions. Flow cytometry was performed using the Beckman Coulter Cytomics FC 500 flow cytometer (Beckman Coulter).
2.10. MTS cell proliferation assay
The effect of the KDM5-C70 inhibitor on the proliferation rate of HUVEC was assessed using the MTS cell proliferation assay. HUVEC were initially treated for 3 days with 0.1% DMSO, 10 μM or 20 μM KDM5-C70. Subsequently, the cells were seeded in 96-well plates at a density of 1,000 cells per well and subjected to 0.1% DMSO, 10 μM or 20 μM KDM5-C70 for the following 1, 2 and 3 days. To assess any differences in the proliferation rate, the recommended amount of MTS reagent (Promega) was supplemented into each well of the 96-well plate at the indicated time points. The cells were incubated with the reagent for 3 h at 37 °C in a humidified, 5% CO2 atmosphere. The absorbance was then recorded at 490 nm with a 96-well plate reader (SPECTROstar Nano, BMG LABTECH). Three independent experiments were performed with each experiment containing five replicates.
2.11. Transwell migration assay
The impact of the KDM5-C70 inhibitor on the migratory capacity of HUVEC was assessed using the transwell assay. HUVEC were initially treated for 3 days with 0.1% DMSO, 10 μM or 20 μM KDM5-C70 and subsequently starved in EBM-2: EGM-2 (4∶1) medium for 24 h. Then, 2 × 104 cells from each category were added to the inner compartment of transwell inserts with 5 μm pores (Corning-Costar) in 100 μl starvation medium containing 0.1% DMSO, 10 μM or 20 μM KDM5-C70 respectively. As stimulus, EGM-2 medium supplemented with 18% FBS (20% EGM-2) was added at the bottom chamber of the transwell plate. Cells were allowed to migrate for 24 h. Afterwards, the non migrated cells were removed from the top of the insert with a wet cotton swab whereas the migrated cells at the bottom were fixed with 4% paraformaldehyde and stained with eosin-haematoxylin. Photographs of the stained cells were taken at 5 different fields (20×) of each insert using a Leica CTR MIC microscope. The number of the migrated cells was assessed by using the Image J software (version 1.51j8). Three independent experiments were performed with each experiment containing two replicates.
2.12. Tube formation assay
The effect of the KDM5-C70 inhibitor on the tube forming ability of HUVEC was assessed using the matrigel-based tube formation assay. HUVEC were initially treated for 3 days with 0.1% DMSO, 10 μM or 20 μM KDM5-C70 and subsequently starved for 4 h in EBM-2: EGM-2 (4:1) medium containing 0.1% DMSO, 10 μM or 20 μM KDM5-C70 respectively. Subsequently, the cells from each condition were plated at a density of 2 × 104 cells/well in 100 μl starvation medium containing 0.1% DMSO, 10 μM or 20 μM KDM5-C70, in 96-well plates coated with 50 μl of growth factor-reduced Matrigel (BD Biosciences). Plates were incubated for 16 h at 37 °C in a humidified, 5% CO2 atmosphere. Photographs from 5 different fields (5×) per well were taken using a Leica CTR MIC microscope and were analyzed by using the Angiogenesis Analyzer plug-in in Image J software (version 1.51j8). Representative photographs were also taken at 10× magnification. The angiogenesis was assessed by measuring the number of the nodes, junctions, segments as well as the total length (in pixel) which is defined as the sum of length of segments, isolated elements and branches in the analyzed area. Three independent experiments were performed with each experiment containing two replicates.
2.13. Statistical analysis
Statistical significance of the proteomic differences between WT animals and Ldlr−/− or Ldlr−/−STZ, or ApoE−/−STZ animals was determined using the 2-tailed Mann-Whitney U test on the R statistical environment (R version 3.5.1). Log2 transformation of the mean values prior to adding constant 1 to data [log2(x + 1) with x representing the raw data from the LC-MS/MS analysis] was applied in datasets containing zero values. Pearson correlation coefficient was assessed for data that passed normality after log2 transformation of the mean values. If normal distribution could not be established, Spearman correlation coefficient was assessed. Nonparametric regression analysis was performed using the Local Polynomial Regression Fitting (LOESS function in R statistical package). Statistical significance of differences between groups was determined using the 2-tailed Mann-Whitney U test. In case of multiple group analyses, the one-way and two-way ANOVA tests for randomized block experiments were performed followed by a Tukey post hoc test. Data are presented as mean ± SD (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). All statistical analyses were performed using GraphPad Prism 7 software. Principal component analysis was performed in SPSS version 25 in an unsupervised fashion. The volcano plots for proteins identified with at least two peptides (excluding proteins that were unique in one model) were created using the package EnhancedVolcano (Publication-ready volcano plots with enhanced colouring and labeling, Version: 1.1.1, Author: Kevin Blighe) on the R statistical environment (R version 3.5.1). Violin plots showing the Kernel probability density of the log2 transformed data, were created using R software and ggplot2 package.
3. Results
3.1. Proteomic profiling of animal models with atherosclerosis
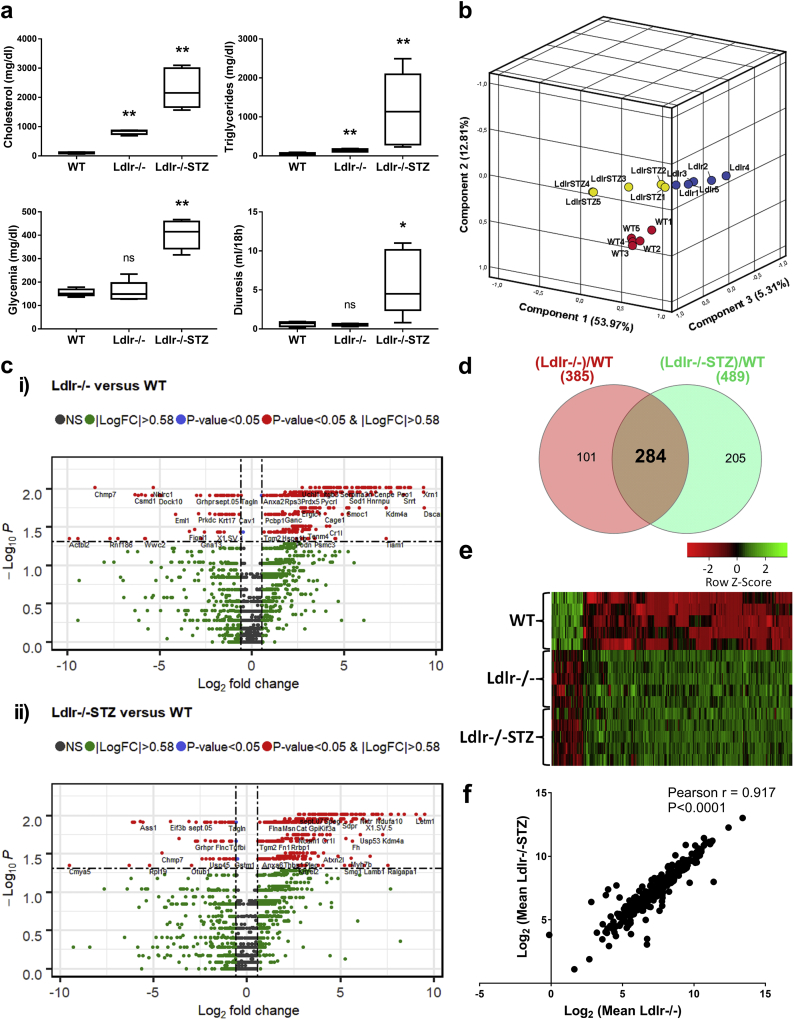
To identify proteome alterations associated with atherosclerosis, LC-MS/MS proteome analysis was performed on thoracic aortas isolated from the Ldlr−/− atherosclerotic mouse models in the absence of diabetes (Ldlr−/−, n = 5) or presence of diabetes (Ldlr−/−STZ, n = 5) in comparison to WT (n = 5) mice. In all cases, animals were evaluated for their phenotypical and biochemical characteristics to confirm presence of atherosclerosis, prior to be included in the study (Supplementary Table 1). As shown in Fig. 1a, atherosclerosis in those animals was characterized by dyslipidemia; and in the case of the diabetic animals, as expected, increased glycemia and diuresis were also observed.
Fig. 1.
Proteome analysis of the Ldlr−/− background mouse models. (a) Box-and-whiskers plots of main biochemical characteristics (cholesterol and glycemia levels) of the animal models included in the proteomic analysis. Statistical significance between the diseased (Ldlr−/−, n = 5; Ldlr−/−STZ, n = 5) and control (WT, n = 5) animals was determined using 2-tailed Mann-Whitney U tests (*P < 0·05, **P < 0·01). (b) Principal component analysis of the thoracic aorta proteome was performed in an unsupervised fashion. The log2 transformed values (prior adding one) of the total protein identifications for each animal were used in this analysis. The scatter plot illustrates the first three principal component scores in a three-dimensional space. Three distinct clusters are inferred indicating the absence of outlying samples. (c) The differential expression analysis results are graphically represented with volcano plots; (i) Volcano plot of log2(fold change: (Ldlr−/−)/WT) versus –log10(Mann-Whitney p-value) and (ii) Volcano plot of log2(fold change: (Ldlr−/−STZ)/WT) versus –log10(Mann-Whitney p-value). Proteins uniquely expressed per model were filtered out. Red data points indicate 385 differentially expressed proteins when comparing Ldlr−/− with WT animals and 489 proteins between Ldlr−/−STZ and WT that were detected based on at least two peptides identification, had a fold change of >1·5 (or < 1/1·5) and a p-value of <0·05. Among these, (d) comparative proteomic analysis identified 284 proteins that were common in both group comparisons. (e) The heatmap illustrates the expression level changes of the 284 differentially expressed proteins among the WT, Ldlr−/− and Ldlr−/−STZ animals. (f) Correlation of the levels of the 284 differentially expressed proteins was analyzed using the 2-tailed Pearson correlation coefficient (P < 0·0001) after log2 transformation of the values. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To assess the quality of the proteomics data in an unsupervised fashion, Principal Component Analysis (PCA) was performed. As shown in Fig. 1b, there were no significant outliers and all of the three animal groups were well separated. The LC-MS/MS analysis resulted in a total of 2,365 proteins identified based on at least 2 peptides (Supplementary Table 2). Only proteins fulfilling the following criteria were considered as differentially expressed: a) were statistically significant different (P ≤ 0.05) and b) had a fold change of at least 1.5 (|fold change| ≥1.5) in Ldlr−/− or Ldlr−/−STZ versus WT. This led to the identification of 385 and 489 protein changes in Ldlr−/− and Ldlr−/−STZ, compared to WT, respectively (Fig. 1c). Of these, 284 proteins were common (Fig. 1d) and consistently changed (e.g. with same trend of expression) in disease in both comparisons (Fig. 1e, Supplementary Table 2). A strong correlation between the abundance of these 284 proteins was observed (Fig. 1f). This finding further suggests that these changes may reflect common disease manifestations regardless of the presence of diabetes.
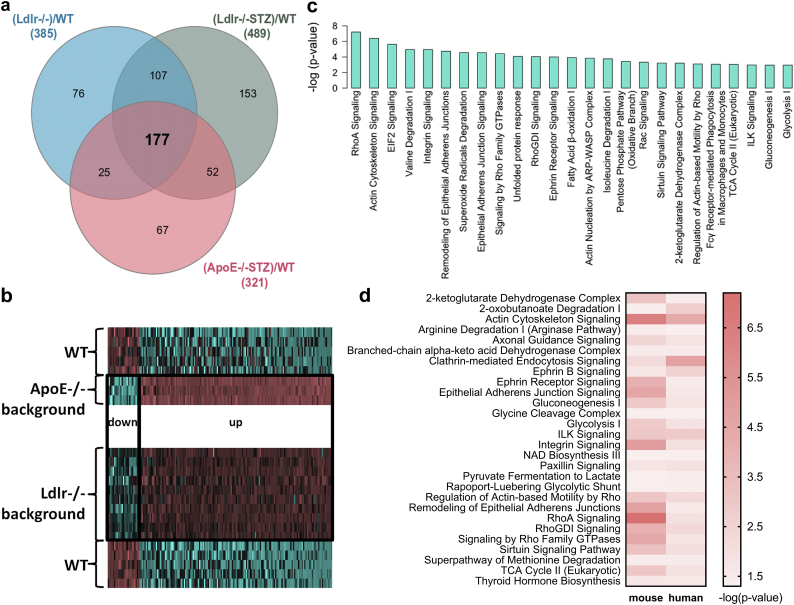
To exclude protein changes that may be specifically related to the Ldlr−/− background, we subsequently performed proteomic analysis of thoracic aortas from another atherosclerotic model characterized by the ApoE−/− background and the presence of diabetes (ApoE−/−STZ, n = 3). Phenotypic and biochemical characterization prior to the proteomic analysis confirmed the presence of diabetic atherosclerosis in these animals (Supplementary Fig. 1a, Supplementary Table 1). PCA exploratory analysis of the proteomics findings indicated the separation of the experimental groups and the absence of outliers (Supplementary Fig. 1b). Following comparative proteomics analysis, 321 differentially expressed proteins were identified, having a statistical significant fold change of >1.5 in ApoE−/−STZ versus WT animals (Supplementary Fig. 1c and 1d, Supplementary Table 3). Comparison of the 284 differentially expressed proteins of the Ldlr−/− and the 321 proteins of the ApoE−/− analysis revealed an overlap of 177 proteins (Fig. 2a, Supplementary Table 4). These had the same trend of expression (Fig. 2b) in all animal models in comparison to controls, suggesting their association with the development of atherosclerosis regardless the specific disease aetiology (e.g. genetic background or presence of diabetes, as reflected in the employed models).
Fig. 2.
Comparative proteomic analysis and functional annotation. (a) Comparative analysis of the proteomic findings led to the identification of 177 differentially expressed proteins that are common in all disease models (Ldlr−/−, Ldlr−/−STZ, ApoE−/−STZ) when compared to WT controls. (b) The expression changes of these 177 proteins in disease are illustrated in the heatmap for each group, indicating the similar trend of expression of these proteins in all cases. (c) Ingenuity Pathway Analysis (IPA) mapping of the representative molecular pathways by the 177 differentially expressed proteins. (d) The common molecular pathways between mouse and human proteomic findings based on IPA.
3.2. Assessment of the biological relevance of the proteomic findings
Analyzing the function (based on UniProt database) of the top 15 most significantly deregulated proteins in each of the three models (Table 1, Supplementary Table 4), we observed that these proteins were associated with processes such as the mitochondrial respiratory chain (e.g. Letm1, Ndufa10, Afg1l), cell migration (e.g. Stk25), adhesion (e.g. Kif26b), cytoskeleton dynamics (e.g. Cfl1), gene expression (e.g. Xrn1, Ice1), biosynthesis of hormonal steroids (e.g. Hsd3b4) and signal transduction (e.g. Ndrg2). Moreover, proteins associated with histone modifications such as Kdm5d and Hpf1, were also included (Table 1, Supplementary Table 4). Further mapping of all common 177 differentially expressed proteins into molecular pathways (Supplementary Table 5, representative shown on Fig. 2c) based on the Ingenuity Pathway Analysis (IPA) software revealed that these proteins participate in metabolic processes including Fatty Acid β-oxidation I, Pentose Phosphate Pathway, 2-ketoglutarate dehydrogenase complex, tricarboxylic acid (TCA) cycle, glycolysis, gluconeogenesis and metabolism of amino acids and derivatives (e.g. Valine and Isoleucine degradation). Additionally, disease-associated changes in pathways related to detoxification of ROS (Superoxide radicals degradation, NRF2-mediated oxidative stress response), immune system (Fcγ Receptor-mediated phagocytocis in macrophages and monocytes, fMLP signaling in neutrophils, CD28 signaling in T Helper cells, chemokine and leukocyte extravasation signaling), translation (EIF2 signaling), cytoskeleton remodeling and cell adhesion (actin cytoskeleton signaling, integrin signaling, remodeling of epithelial adherens junctions) as well as in Endoplasmic Reticulum Stress pathway and unfolded protein response were predicted (Supplementary Table 5). These pathways, as well as the enriched Rho and ILK signaling pathways (Fig. 2c), have been previously described to be involved in the atherosclerotic process [[19], [20], [21], [22], [23], [24], [25], [26], [27], [28]], supporting the biological relevance of the proteomic findings.
3.3. KDM5D abundance changes in human CVD
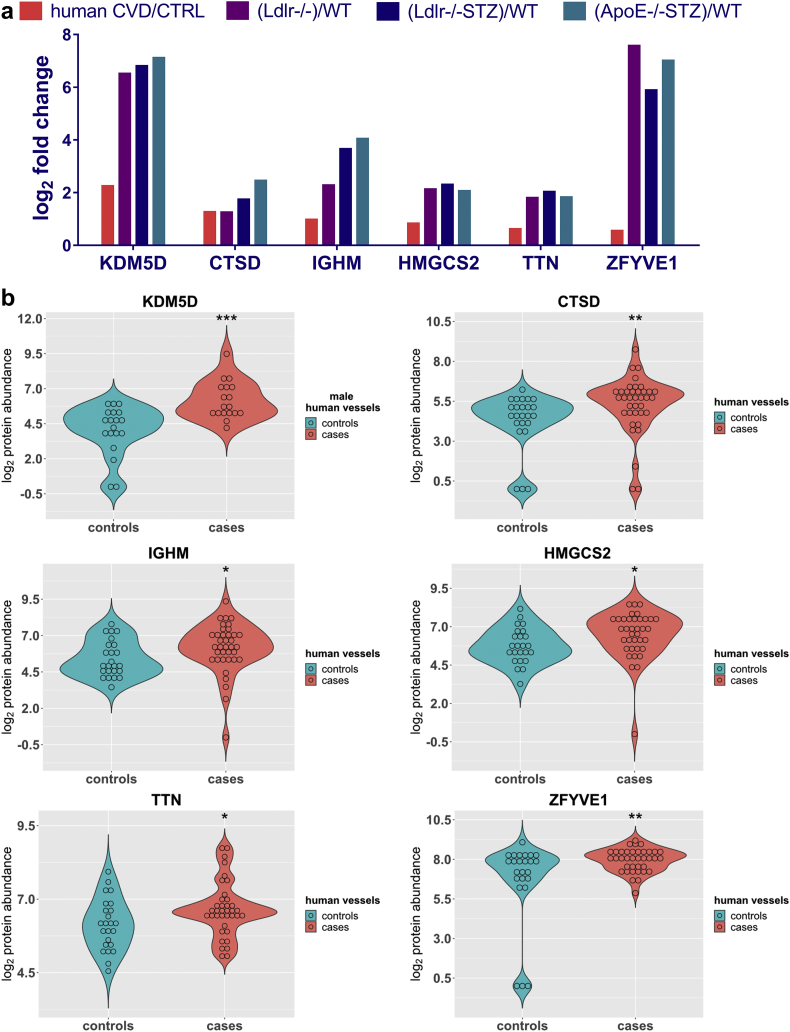
To investigate transferability of the animal model findings to human disease, the list of 177 differentially expressed proteins in the atherosclerotic animal models, was compared to protein changes in human vascular tissues from patients with CVD (cases; n = 37 samples from 30 patients; Table 2) in comparison to vessels from organ donors with no evidence of CVD (controls, n = 24 samples from 20 subjects; Table 2), as revealed following application of a similar LC-MS/MS approach. In this analysis of human vascular tissue, 94 proteins were found to be differentially expressed. Similar to the animal models, the proteins represented pathways associated with metabolism (e.g. 2-ketoglutarate dehydrogenase complex, TCA cycle, glycolysis, and metabolism of amino acids), cytoskeleton remodeling and cell adhesion (e.g. actin cytoskeleton signaling, integrin signaling, remodeling of epithelial adherens junctions) as well as pathways known to be implicated in atherosclerosis including the Rho signaling pathways (RhoA signaling, signaling by Rho family GTPases, RhoGDI signaling), the ILK signaling, Ephrin B signaling and the Sirtuin signaling pathway (Fig. 2d). Following a cross-species comparison of the two protein lists (177 mouse proteins versus 94 human proteins) based on orthologues, an overlap of six proteins could be observed (KDM5D, CTSD, IGHM, HMGCS2, TTN, ZFYVE1), (Fig. 3a). Interestingly, of these, the male specific demethylase KDM5D was found at significantly increased abundance levels in all atherosclerotic models (Table 1, Fig. 3a), being also fairly abundant in the human tissues of male subjects with CVD (n = 18 vessels from 14 patients, and n = 19 vessels from 16 controls; Supplementary Table 6), having the most pronounced differences in CVD in comparison to the other five proteins (Fig. 3b, Supplementary Table 7). This difference was also observed even when considering only male samples for the analysis of the expression changes of all six proteins (Supplementary Fig. 2). Of note, among the tested vascular samples, 8 originated from patients with diabetes (cases, n = 6) and when compared to the 19 vascular tissues from non-diabetic healthy individuals (controls, n = 16) pronounced differences in the KDM5D levels were also observed, again in agreement to the animal model data (Supplementary Fig. 3).
Table 2.
Demographic, physical and clinical characteristics of the participants in this study.
| Variable | Control group (n = 20) | Case group (n = 30) |
|---|---|---|
| Demographics | ||
| Sex [n (%)] | 16 (80%) M, 4 (20%) F | 14 (46·7%) M, 16 (53·3%) F |
| Age [years] | 46·6 ± 18·5 | 62·8 ± 13 |
| Physical characteristics | ||
| Weight [kg] | 80·1 ± 14·2 | 75·7 ± 12·3 |
| Height [m] | 1·8 ± 0·1 | 1·7 ± 0·1 |
| Body mass index [kg/m2] | 26·1 ± 3·7 | 26·2 ± 3·1 |
| Clinical information | ||
| Dyslipoproteinemia [n (%)] | 4 (20%) | 5 (16·7%) |
| Obesity [n (%)] | 3 (15%) | 4 (13·3%) |
| Diabetes [n (%)] | 0 (0%) | 8 (26·7%) |
| Vascular disease [n (%)] | 0 (0%) | 30 (100%) |
| Left ventricular hypertrophy [n (%)] | 0 (0%) | 12 (40%) |
| Angina pectoris [n (%)] | 0 (0%) | 2 (6·7%) |
| Claudicatio intermittens [n (%)] | 0 (0%) | 10 (33·3%) |
| Myocardial infarction [n (%)] | 0 (0%) | 3 (10%) |
M: Males, F: Females; Dyslipoproteinemia was characterized by total cholesterol ≥ 5·2 mmol/l (≥ 200 mg/dl), low-density lipoprotein (LDL) cholesterol ≥ 3·4 mmol/l (≥ 130 mg/dl), high-density lipoproteins (HDL) cholesterol ≤ 1·0 mmol/l (≤ 40 mg/dl), and triglycerides ≥ 1·7 mmol/l (≥ 150 mg/dl); Obesity was defined by a body mass index > 30 kg/m2.
Fig. 3.
The expression levels of the six -common with mouse- proteins in human vascular tissues. (a) Bar graph illustrating the fold change difference of the six proteins that were identified in all mouse and human proteomic datasets, and at statistically significant levels when comparing cases and controls. (b) Violin plots showing the protein abundance of each one of the six proteins in human vascular tissues from patients with CVD (cases, n = 37) compared to organ donors without CVD background (controls, n = 24) based on LC-MS/MS analysis. The male specific KDM5D protein was detected only in male human vascular tissues (controls, n = 19 and cases, n = 18). The expression levels are expressed as log2(x + 1) values with x representing the raw data from the LC-MS/MS analysis. Statistical significance was determined using 2-tailed Mann-Whitney U tests (*P < 0·05, **P < 0·01, ***P < 0·001).
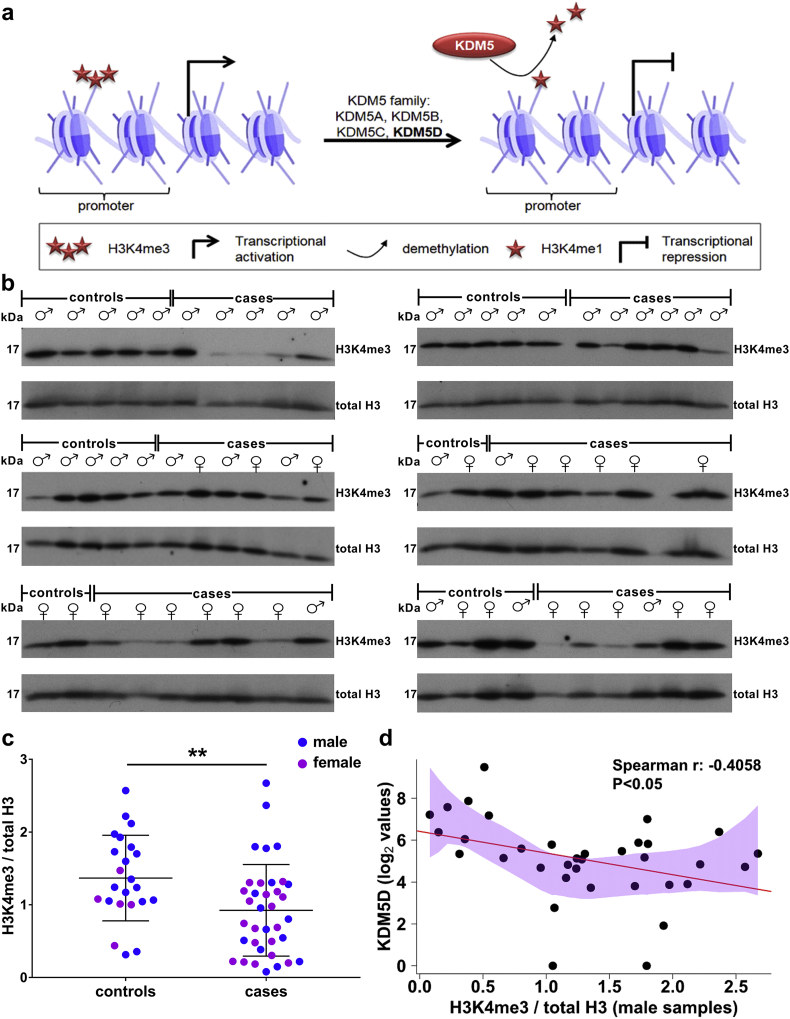
To further verify this observation, we next examined whether the increased levels of KDM5D were associated with a reduction of its substrate. As depicted in Fig. 4a, H3K4me3 is the main substrate of KDM5D in males as well as of all members of KDM5 family of histone demethylases in both sexes [29]. Western blot analysis for H3K4me3 of all available human vessel samples (Table 2) indicated a significant decrease in the levels of the protein in CVD in comparison to controls (Fig. 4b and 4c). In male patients where KDM5D expression levels were evaluated (Fig. 3b), Spearman correlation analysis revealed a statistical significant negative correlation between the expression levels of KDM5D and the abundance of H3K4me3 (Fig. 4d), further suggesting that KDM5D overexpression was associated with increased activity. H3K4me3 also appeared at overall decreased levels at the available samples from females affected by CVD, indicating increased activity of further members of the KDM5 family in CVD.
Fig. 4.
Alteration in the expression levels of the KDM5 substrate, H3K4me3, in CVD. (a) KDM5D is a member of the KDM5 family of histone demethylases that specifically demethylate trimethylated and dimethylated lysine 4 of histone H3. KDM5 are transcriptional regulators that may act as transcriptional repressors by demethylating H3K4me3 at promoters of transcribed genes. (b) Western blot (WB) analysis of H3K4me3 in all available human vascular tissues from organ donors without a CVD background (controls; male: 18 vascular tissues from 15 individuals and female: 5 vascular tissues from 4 individuals) and patients with CVD (cases; male: 17 vascular tissues from 14 patients and female: 19 vascular tissues from 16 patients). Total histone H3 was used as loading control. (c) Quantification of the WB data following normalization to total H3. Statistical significance was determined using 2-tailed Mann-Whitney U tests (**P < 0·01). (d) Nonparametric regression analysis to evaluate association of the enzyme (KDM5D) and substrate (H3K4me3) levels in the male vascular tissues. The polygon represents the 95% confidence interval of local polynomial regression (LOESS function in R statistical package). Spearman correlation analysis between the enzyme and the substrate indicates a negative correlation between the two variables which is graphically represented with the linear model regression line.
3.4. The impact of KDM5 inhibition on H3K4me3 levels and on proliferation, migration and tube-forming ability of HUVEC
An increasing number of studies support an important role of epigenetic mechanisms including post-translational modifications of histones in the regulation of the endothelial function. More specifically, the methylation status of H3K4 has been proven essential for endothelial cell sprouting and migration and therefore for angiogenesis [30]. Based on the literature, the KDM5 family is expected to affect a wide variety of molecular processes including proliferation, migration and angiogenesis [31]. Since all these are processes involved in the development of atherosclerosis, we investigated the possible impact of KDM5 inhibition on these processes using the KDM5-C70 inhibitor, a KDM5-family inhibitor [32,33], and HUVEC as in vitro model [34].
HUVEC expressing KDM5D (Supplementary Fig. 4) were subjected to treatment with KDM5-C70 at different concentrations (10 μΜ, and 20 μΜ). After 3 days of treatment, abundance of H3K4me3 was significantly increased in the cells, as evaluated by western blot analysis, demonstrating efficient inhibition of KDM5 demethylase activity by KDM5-C70 (Fig. 5a). Absence of non-specific toxic effects was demonstrated by flow cytometry analysis for apoptotic (Annexin-V) and necrotic (7-AAD) cells (Supplementary Fig. 5).
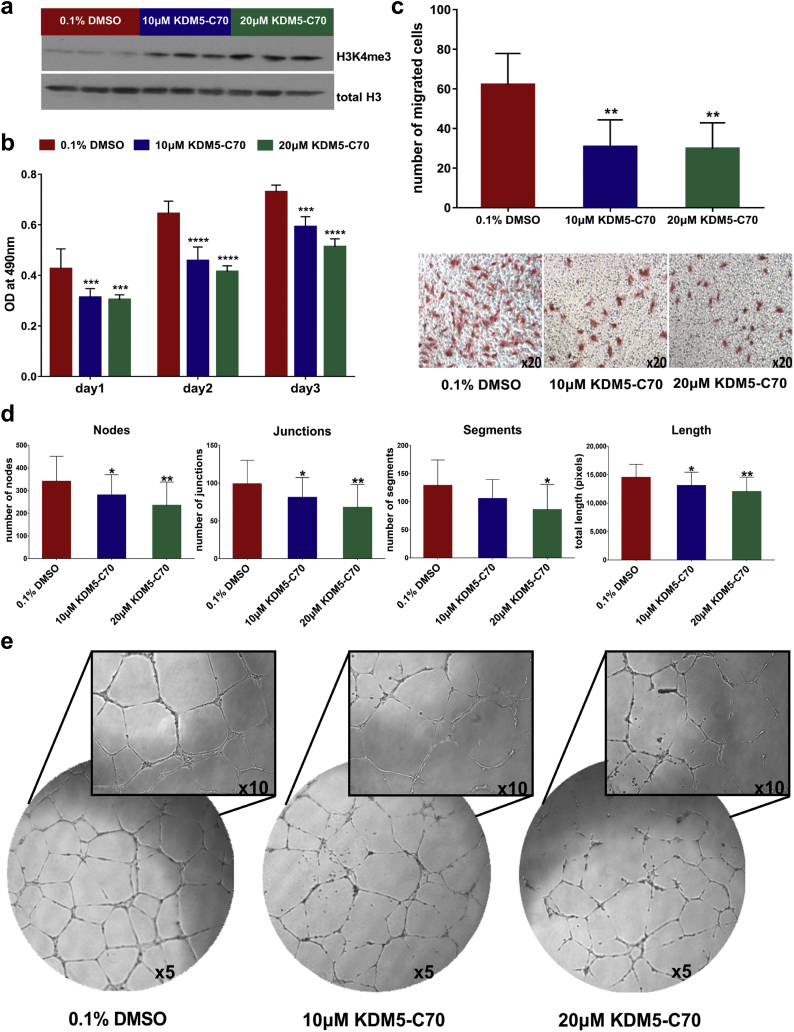
Fig. 5.
The effect of KDM5 inhibition on the endothelial properties of HUVEC. (a) Western blot analysis of H3K4me3 of HUVEC treated with 0·1% DMSO, 10 μM or 20 μM KDM5-C70 for 3 days. Total histone H3 was used as loading control. (b) MTS cell proliferation assay. Bar graph representing the absorbance (OD) at 490 nm of HUVEC at three different time points (day 1, day 2, day 3). The values represent the means ± SD from three independent experiments performed in five replicates. Statistical significance was determined using the two-way ANOVA test (***P < 0·001, ****P < 0·0001). (c) The graph illustrates the number of migrated cells towards complete medium with representative images displayed underneath. Magnification: 20×. The values represent the means ± SD from three independent experiments performed in duplicate. Statistical significance was determined using the one-way ANOVA test (**P < 0·01). (d) Graphs illustrate the impact of the inhibitor on the number of nodes, junctions and segments as well as on the total length of segments, isolated elements and branches formed by HUVEC. The values represent the means ± SD from three independent experiments performed in duplicate. Statistical significance was determined using the one-way ANOVA test (*P < 0·05, **P < 0·01). (e) KDM5-C70 treatment has a significant effect on the angiogenic properties of HUVEC as illustrated by the representative images of the tube formation assay. Magnification: 5× and 10×.
The impact on proliferation was assessed by MTS assay. Treatment with 10 μΜ and 20 μΜ of the inhibitor significantly reduced the proliferation rate of HUVEC in comparison to the DMSO treated cells (Fig. 5b). KDM5 inhibition also resulted in a statistically significant reduction of the migratory capacity of HUVEC, as assessed using the transwell system (Fig. 5c). As shown in Fig. 5d, treatment with the inhibitor significantly reduced the number of nodes, junctions, and segments, as well as the total length of segments, isolated elements and branches formed by HUVEC. Collectively, these results indicate that KDM5 inhibition attenuated the tube-forming ability of HUVEC (Fig. 5e).
4. Discussion
With this work, we aimed to characterize protein changes associated with CVD using high resolution proteome analysis. These changes may have a potential to serve as biomarkers for disease, or even as possible therapeutic targets. We used three widely employed disease models representing different disease aetiologies (Ldlr−/−, Ldlr−/−STZ, ApoE−/−STZ) thus allowing for the definition of as possible, ‘common’ atherosclerosis-associated proteomic alterations [13]. Of note, a good overlap was also observed when comparing the common proteins of the aforementioned three animal models with changes observed in ApoE−/− versus WT mice (111 common proteins with the same expression trend at statistical significant levels – Supplementary Table 8). Male mice were selected for this study since they develop more pronounced atherosclerosis than the females [35]. It should be noted that protein changes unique per model are also observed (as may be seen in Fig. 2a and Supplementary Tables 2 and 3) which even though not investigated in this study, may merit further investigation.
To the best of our knowledge this is the first study that describes common proteomic alterations in these three atherosclerotic mouse models. An in-depth analysis of these protein changes suggests that they reflect different well-characterized atherosclerosis-associated mechanisms such as inflammation [19], metabolism [20], oxidative stress [21], and extracellular matrix remodeling [23]. Further comparison of the proteomics findings from the animal models with their orthologues from the proteomic profiling of human vascular tissues from patients affected by CVD, revealed a high degree of overlap at the pathway level, although a small overlap at the individual molecule level. This has been also observed in studies comparing the overlap of human genes in coronary artery disease (CAD) identified by genome-wide association studies (GWAS) with those from atherosclerotic mouse models [36]. In these studies, although an overlap of only 18.4% of human CAD genes with mouse orthologs was detected, their relevance at pathways was significantly higher (over than 50% being consistent between the two species) [36]. In our case, the observed relatively small overlap at the individual molecule level may be additionally attributed to the use of different vessel types (central in mice versus peripheral and central in humans). Nevertheless and despite this variability, six common proteins were identified. A more detailed investigation of the expression of these proteins in a gender-specific manner is required to exclude with high confidence an impact of gender on our findings. Of note, among them was cathepsin D (CTSD) whose increased expression levels have been previously associated with atherosclerosis [37] and the ketogenic enzyme hydroxymethylglutaryl-CoA synthase, mitochondrial (HMGCS2) whose upregulation may be implicated in type 1 diabetes induced cardiac dysfunction [38]. Furthermore, proteins with a possible role in CVD were also included such as the immunoglobulin heavy chain mu (IGHM) which was significantly increased in B cells in the artery tertiary lymphoid organs (ATLOs) with the latter associated with B cell responses in the atherosclerotic aortas [39] and titin (TTN), a highly abundant protein in striated muscle whose truncated variants have been associated with several cardiomyopathies [40]. Proteins without a known/previous implication in CVD such as the zinc finger FYVE domain-containing protein 1 (ZFYVE1) which is a protein participating to autophagosome formation [41], and KDM5D were also included with the latter being highlighted as a predominant CVD-associated change in both species. KDM5D was also found upregulated (fold change = 50.8 in cases versus controls) yet did not reach statistical significance in the ApoE−/− mice (in the absence of diabetes) when comparing with WT mice (data not shown).
KDM5D (also known as JARID1D or SMCY) is encoded on the Y chromosome and until now it has been implicated in prostate cancer invasion and metastasis [42], spermatogenesis [43], and sex-specific tissue transplantation rejection [44]. KDM5D acts as a direct epigenetic modulator, and as one of the four members of the KDM5 family of histone demethylases, specifically demethylates trimethylated and dimethylated Lys-4 of histone H3 [29]. The elevated expression of KDM5D in the human tissues of patients with CVD was accompanied by a reduction of its substrate (H3K4me3) implying increased activity of KDM5D.
Alteration in the trimethylated state of H3K4 has been previously related to the development of heart failure in humans and rat models [45] and also associated with the physiological function of cardiomyocytes [46]. Mutations in genes involved in the H3K4me pathway (production, removal or reading of H3K4me), including KDM5A and KDM5B, have been implicated in the pathogenesis of congenital heart disease (CHD) [47]. Given this published evidence with respect to H3K4 and the observed increased KDM5D and concomitant decreased H3K4me3 levels in human tissues of patients with CVD, we investigated the impact of the KDM5-C70 inhibitor on HUVEC. This compound targets all four members of the KDM5 family, is a cell-permeable prodrug with no reported cytotoxicity, and shows selectivity for KDM5 family members compared to KDM6 and KDM4 [32,33]. Considering that endothelial cell dysfunction is an important contributor in atherosclerosis [1] we employed HUVEC. Inhibition of KDM5 activity increased the H3K4me3 levels and significantly attenuated the proliferation rate, the migratory capacity and the tube-forming ability of the endothelial cells in vitro. These findings are in line with another study that describes impaired angiogenic properties of the endothelial cells after inhibition of KDM5B and gene silencing with shRNA, linked to induced expression of the antiangiogenic transcription factor HOXA5 [48].
Interpretation of our findings in the context of atherosclerosis suggests that treatment with a KDM5 inhibitor may be beneficial, considering that the atherosclerotic plaques are characterized by increased neovascularization that contributes to intraplaque hemorrhage and plaque progression [49]. In response to hypoxia and to the VEGF gradient in the atherosclerotic lesion, the endothelial cells of the vasa vasorum switch from a quiescent phenotype to a highly proliferative and migratory state to start sprouting and forming the new vessels [49]. Since KDM5 inhibition significantly attenuated growth and function of endothelial cells in vitro, it may have therapeutic potential in vivo- a working hypothesis warranting further investigation.
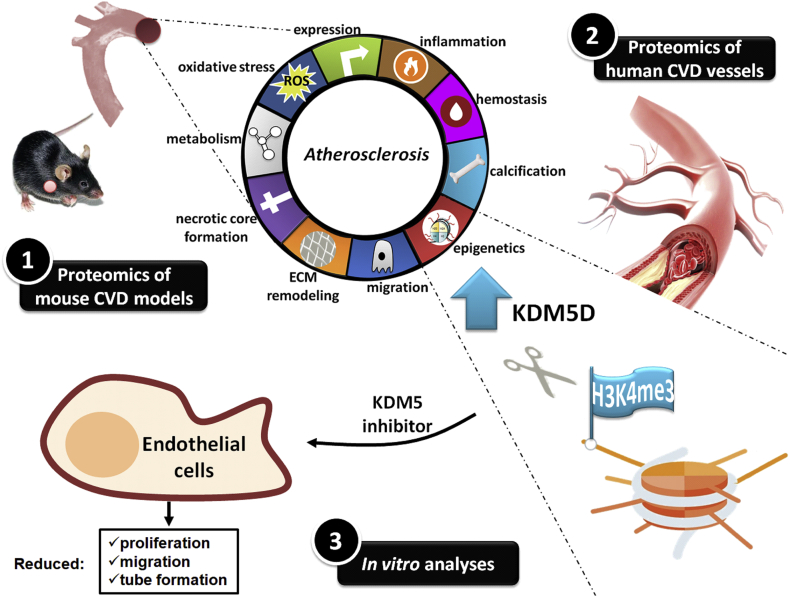
Collectively, in this study, by applying a systematic approach that included high-throughput proteomics of mouse models and human vascular tissues as well as in vitro assays, a role for KDM5 histone demethylases in CVD, likely via affecting H3K4 methylation, is suggested. The key findings of this study are summarized in Fig. 6. In addition to multiple high confidence proteomic changes reflecting common molecular manifestations of atherosclerosis, further underlining the validity of the approach, other yet unknown proteomic changes were identified, which may be of significant value in further systems biology approaches and model selection for pre-clinical studies.
Fig. 6.
Graphical illustration of the workflow and key findings of the study.
Limitations of the study
Our proteomics based approach using mouse and human vascular tissues with CVD indicates KDM5D as a novel candidate pharmacological target associated with CVD progression. However, there are several shortcomings of this study with regards to the role of KDM5D in the context of CVD. Given the fact that the cellular composition of the aortas especially in the areas of the lesions is quite heterogeneous, further experiments and efforts to identify which cells contribute to the elevated expression of KDM5D appear well justified. Along the same lines, in this study we focused on the impact of KDM5 inhibition on endothelial cells, given that endothelial dysfunction is a crucial event in atherosclerosis and impact of H3K4 methylation on endothelial migration and proliferation has been reported. Nevertheless, studying the impact of KDM5 inhibition on other cell types involved in atherosclerotic plaque formation including smooth muscle cells and inflammatory cells is well justified, especially given the reported impact of H3K4 methylation on inflammation [50]. Additionally, characterization of the genes affected upon the KDM5D-mediated alteration of the methylation status of H3K4 may be valuable to improve our understanding of the mechanism of KDM5 inhibition. In parallel, the impact of inhibition of KDM5 in vivo on CVD preferably in a gender-specific manner is needed. However this is currently limited due to the lack of well characterized inhibitors applicable for in vivo studies. Although limitations of the current study, these are multiple research avenues to be further explored, for which the presented study forms a solid starting point.
Acknowledgments
Acknowledgements
All authors have read and approved the article.
Funding sources
This work was supported by the European Commission via funding to the Systems Biology to Identify Molecular Targets for Vascular Disease Treatment [SysVasc, HEALTH-2013 603288]. Part of the study was financed by the ERA-CVD PROACT project (numbers ANR-17-ECVD-0006 for J.K, J.S.S.B and J.P.S and 01KL1805 via the BMBF for H.M), and by a grant from the “Fondation pour la Recherche Médicale” (grant number DEQ20170336759) for J.K, J.S.S.B and J.P.S. The funders had no role in study design, data collection, data analysis, data interpretation, patient recruitment, or writing of the manuscript. The authors have not been paid to write this article by a pharmaceutical company or other agency. The corresponding author had full access to all the data in the study and had the final responsibility for the decision to submit for publication.
Declaration of interests
Harald Mischak is the co-founder and co-owner of Mosaiques Diagnostics.
Author contributions
A.V, J.P.S, H.M, B.P, M.G.R, J.Z conceived and designed the research; M.M*, J.K, M.M, V.B, W.M, J.L.B, J.S.S.B, M.S acquired the data; M.M* performed statistical analysis of the data; M.M*, A.V, J.P.S, J.K drafted the manuscript. All co-authors have read the manuscript, revised it and agree with the submitted version.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.02.040.
Contributor Information
Joost P. Schanstra, Email: joost-peter.schanstra@inserm.fr.
Antonia Vlahou, Email: vlahoua@bioacademy.gr.
Appendix A. Supplementary data
Supplementary Table 1. Biochemical characteristics of the animal models included in this study.
Supplementary Table 2. LC-MS/MS data corresponding to the thoracic aorta proteins detected in the Ldlr−/−, Ldlr−/−STZ and WT animals.
Supplementary Table 3. LC-MS/MS data corresponding to the thoracic aorta proteins detected in the ApoE−/−STZ and WT animals.
Supplementary Table 4. The 177 differentially expressed proteins that are common in all disease models (Ldlr−/−, Ldlr−/−STZ, ApoE−/−STZ) when compared to WT controls.
Supplementary Table 5. Mapping of the 177 (common in all models) differentially expressed proteins into molecular pathways based on the Ingenuity Pathway Analysis (IPA) software.
Supplementary Table 7. LC-MS/MS quantification data of KDM5D, CTSD, IGHM, HMGCS2, TTN, and ZFYVE1 in human vascular tissues.
Supplemental material includes Supplementary Figures 1-5 and Supplementary Tables 6 and 8.
References
- 1.Gimbrone M.A., Jr., Garcia-Cardena G. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ Res. 2016;118(4):620–636. doi: 10.1161/CIRCRESAHA.115.306301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Libby P., Ridker P.M., Hansson G.K. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473(7347):317–325. doi: 10.1038/nature10146. [DOI] [PubMed] [Google Scholar]
- 3.Dominiczak A.F., Herget-Rosenthal S., Delles C. Systems biology to battle vascular disease. Nephrology, dialysis, transplantation : official publication of the European Dialysis and transplant association. European Renal Association. 2010;25(4):1019–1022. doi: 10.1093/ndt/gfq025. [DOI] [PubMed] [Google Scholar]
- 4.Shalhoub J., Sikkel M.B., Davies K.J., Vorkas P.A., Want E.J., Davies A.H. Systems biology of human atherosclerosis. Vasc Endovascular Surg. 2014;48(1):5–17. doi: 10.1177/1538574413510628. [DOI] [PubMed] [Google Scholar]
- 5.Mokou M., Lygirou V., Vlahou A., Mischak H. Proteomics in cardiovascular disease: recent progress and clinical implication and implementation. Expert Review of Proteomics. 2017;14(2):117–136. doi: 10.1080/14789450.2017.1274653. [DOI] [PubMed] [Google Scholar]
- 6.Martin-Lorenzo M., Gonzalez-Calero L., Maroto A.S. Cytoskeleton deregulation and impairment in amino acids and energy metabolism in early atherosclerosis at aortic tissue with reflection in plasma. Biochim Biophys Acta. 2016;1862(4):725–732. doi: 10.1016/j.bbadis.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 7.Martin-Lorenzo M., Zubiri I., Maroto A.S. KLK1 and ZG16B proteins and arginine-proline metabolism identified as novel targets to monitor atherosclerosis, acute coronary syndrome and recovery. Metabolomics. 2015;11(5):1056–1067. doi: 10.1007/s11306-014-0761-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herrington D.M., Mao C., Parker S.J. Proteomic architecture of human coronary and aortic atherosclerosis. Circulation. 2018;137(25):2741–2756. doi: 10.1161/CIRCULATIONAHA.118.034365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emini Veseli B., Perrotta P., De Meyer G.R.A. Animal models of atherosclerosis. Eur J Pharmacol. 2017;816:3–13. doi: 10.1016/j.ejphar.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Saulnier-Blache J.S., Wilson R., Klavins K. Ldlr−/− and ApoE−/− mice better mimic the human metabolite signature of increased carotid intima media thickness compared to other animal models of cardiovascular disease. Atherosclerosis. 2018;276:140–147. doi: 10.1016/j.atherosclerosis.2018.07.024. [DOI] [PubMed] [Google Scholar]
- 11.Beckman J.A., Creager M.A. Vascular complications of diabetes. Circ Res. 2016;118(11):1771–1785. doi: 10.1161/CIRCRESAHA.115.306884. [DOI] [PubMed] [Google Scholar]
- 12.Shen X., Bornfeldt K.E. Mouse models for studies of cardiovascular complications of type 1 diabetes. Ann N Y Acad Sci. 2007;1103:202–217. doi: 10.1196/annals.1394.004. [DOI] [PubMed] [Google Scholar]
- 13.Getz G.S., Reardon C.A. Do the Apoe−/− and Ldlr−/− mice yield the same insight on Atherogenesis? Arteriosclerosis, Thrombosis, and Vascular Biology. 2016;36(9):1734–1741. doi: 10.1161/ATVBAHA.116.306874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lygirou V., Latosinska A., Makridakis M. Plasma proteomic analysis reveals altered protein abundances in cardiovascular disease. J Transl Med. 2018;16(1):104. doi: 10.1186/s12967-018-1476-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makridakis M., Vlahou A. GeLC-MS: a sample preparation method for proteomics analysis of minimal amount of tissue. Methods Mol Biol. 2018;1788:165–175. doi: 10.1007/7651_2017_76. [DOI] [PubMed] [Google Scholar]
- 16.Latosinska A., Mokou M., Makridakis M. Proteomics analysis of bladder cancer invasion: targeting EIF3D for therapeutic intervention. Oncotarget. 2017;8(41):69435–69455. doi: 10.18632/oncotarget.17279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Babicki S., Arndt D., Marcu A. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44(W1):W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kramer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30(4):523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geovanini G.R., Libby P. Atherosclerosis and inflammation: overview and updates. Clin Sci. 2018;132(12):1243–1252. doi: 10.1042/CS20180306. [DOI] [PubMed] [Google Scholar]
- 20.Koelwyn G.J., Corr E.M., Erbay E., Moore K.J. Regulation of macrophage immunometabolism in atherosclerosis. Nat Immunol. 2018;19(6):526–537. doi: 10.1038/s41590-018-0113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forstermann U., Xia N., Li H. Roles of vascular oxidative stress and nitric oxide in the pathogenesis of atherosclerosis. Circ Res. 2017;120(4):713–735. doi: 10.1161/CIRCRESAHA.116.309326. [DOI] [PubMed] [Google Scholar]
- 22.Borissoff J.I., Spronk H.M., ten Cate H. The hemostatic system as a modulator of atherosclerosis. N Engl J Med. 2011;364(18):1746–1760. doi: 10.1056/NEJMra1011670. [DOI] [PubMed] [Google Scholar]
- 23.Yurdagul A., Jr., Finney A.C., Woolard M.D., Orr A.W. The arterial microenvironment: the where and why of atherosclerosis. Biochem J. 2016;473(10):1281–1295. doi: 10.1042/BJ20150844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khyzha N., Alizada A., Wilson M.D., Fish J.E. Epigenetics of atherosclerosis: emerging mechanisms and methods. Trends Mol Med. 2017;23(4):332–347. doi: 10.1016/j.molmed.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 25.Ivanova E.A., Orekhov A.N. The role of endoplasmic reticulum stress and unfolded protein response in atherosclerosis. Int J Mol Sci. 2016;17(2) doi: 10.3390/ijms17020193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai A., Zhou Y., Li L. Rho-GTPase and atherosclerosis: pleiotropic effects of statins. J Am Heart Assoc. 2015;4(7) doi: 10.1161/JAHA.115.002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rolfe B.E., Worth N.F., World C.J., Campbell J.H., Campbell G.R. Rho and vascular disease. Atherosclerosis. 2005;183(1):1–16. doi: 10.1016/j.atherosclerosis.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 28.Finney A.C., Stokes K.Y., Pattillo C.B., Orr A.W. Integrin signaling in atherosclerosis. Cellular and molecular life sciences. CMLS. 2017;74(12):2263–2282. doi: 10.1007/s00018-017-2490-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee M.G., Norman J., Shilatifard A., Shiekhattar R. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell. 2007;128(5):877–887. doi: 10.1016/j.cell.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Fraineau S., Palii C.G., Allan D.S., Brand M. Epigenetic regulation of endothelial-cell-mediated vascular repair. FEBS J. 2015;282(9):1605–1629. doi: 10.1111/febs.13183. [DOI] [PubMed] [Google Scholar]
- 31.Harmeyer K.M., Facompre N.D., Herlyn M., Basu D. JARID1 histone demethylases: emerging targets in cancer. Trends in Cancer. 2017;3(10):713–725. doi: 10.1016/j.trecan.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horton J.R., Liu X., Gale M. Structural basis for KDM5A histone lysine Demethylase inhibition by diverse compounds. Cell Chemical Biology. 2016;23(7):769–781. doi: 10.1016/j.chembiol.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johansson C., Velupillai S., Tumber A. Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nat Chem Biol. 2016;12(7):539–545. doi: 10.1038/nchembio.2087. [DOI] [PubMed] [Google Scholar]
- 34.Goodwin A.M. In vitro assays of angiogenesis for assessment of angiogenic and anti-angiogenic agents. Microvasc Res. 2007;74(2–3):172–183. doi: 10.1016/j.mvr.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Getz G.S., Reardon C.A. Animal models of atherosclerosis. Arteriosclerosis, Thrombosis, and Vascular Biology. 2012;32(5):1104–1115. doi: 10.1161/ATVBAHA.111.237693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Scheidt M., Zhao Y., Kurt Z. Applications and limitations of mouse models for understanding human atherosclerosis. Cell Metab. 2017;25(2):248–261. doi: 10.1016/j.cmet.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao C.F., Herrington D.M. The function of cathepsins B, D, and X in atherosclerosis. American Journal of Cardiovascular Disease. 2016;6(4):163–170. [PMC free article] [PubMed] [Google Scholar]
- 38.Shukla S.K., Liu W., Sikder K. HMGCS2 is a key ketogenic enzyme potentially involved in type 1 diabetes with high cardiovascular risk. Sci Rep. 2017;7(1):4590. doi: 10.1038/s41598-017-04469-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srikakulapu P., Hu D., Yin C. Artery tertiary lymphoid organs control multilayered territorialized atherosclerosis B-cell responses in aged ApoE−/− mice. Arterioscler Thromb Vasc Biol. 2016;36(6):1174–1185. doi: 10.1161/ATVBAHA.115.306983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang C., Zhang H., Wu G. Titin-truncating variants increase the risk of cardiovascular death in patients with hypertrophic cardiomyopathy. Can J Cardiol. 2017;33(10):1292–1297. doi: 10.1016/j.cjca.2017.05.020. [DOI] [PubMed] [Google Scholar]
- 41.Gatica D., Chiong M., Lavandero S., Klionsky D.J. Molecular mechanisms of autophagy in the cardiovascular system. Circ Res. 2015;116(3):456–467. doi: 10.1161/CIRCRESAHA.114.303788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li N., Dhar S.S., Chen T.Y. JARID1D is a suppressor and prognostic marker of prostate cancer invasion and metastasis. Cancer Res. 2016;76(4):831–843. doi: 10.1158/0008-5472.CAN-15-0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akimoto C., Kitagawa H., Matsumoto T., Kato S. Spermatogenesis-specific association of SMCY and MSH5. Genes to Cells. 2008;13(6):623–633. doi: 10.1111/j.1365-2443.2008.01193.x. [DOI] [PubMed] [Google Scholar]
- 44.Wang W., Meadows L.R., den Haan J.M. Human H-Y: a male-specific histocompatibility antigen derived from the SMCY protein. Science. 1995;269(5230):1588–1590. doi: 10.1126/science.7667640. [DOI] [PubMed] [Google Scholar]
- 45.Kaneda R., Takada S., Yamashita Y. Genome-wide histone methylation profile for heart failure. Genes to Cells. 2009;14(1):69–77. doi: 10.1111/j.1365-2443.2008.01252.x. [DOI] [PubMed] [Google Scholar]
- 46.Stein A.B., Jones T.A., Herron T.J. Loss of H3K4 methylation destabilizes gene expression patterns and physiological functions in adult murine cardiomyocytes. J Clin Invest. 2011;121(7):2641–2650. doi: 10.1172/JCI44641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zaidi S., Choi M., Wakimoto H. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498(7453):220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fork C., Gu L., Hitzel J. Epigenetic regulation of angiogenesis by JARID1B-induced repression of HOXA5. Arteriosclerosis, Thrombosis, and Vascular Biology. 2015;35(7):1645–1652. doi: 10.1161/ATVBAHA.115.305561. [DOI] [PubMed] [Google Scholar]
- 49.de Vries M.R., Quax P.H. Plaque angiogenesis and its relation to inflammation and atherosclerotic plaque destabilization. Curr Opin Lipidol. 2016;27(5):499–506. doi: 10.1097/MOL.0000000000000339. [DOI] [PubMed] [Google Scholar]
- 50.Zhao S., Zhong Y., Fu X. H3K4 methylation regulates LPS-induced proinflammatory cytokine expression and release in macrophages. Shock. 2019;51(3):401–406. doi: 10.1097/SHK.0000000000001141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Biochemical characteristics of the animal models included in this study.
Supplementary Table 2. LC-MS/MS data corresponding to the thoracic aorta proteins detected in the Ldlr−/−, Ldlr−/−STZ and WT animals.
Supplementary Table 3. LC-MS/MS data corresponding to the thoracic aorta proteins detected in the ApoE−/−STZ and WT animals.
Supplementary Table 4. The 177 differentially expressed proteins that are common in all disease models (Ldlr−/−, Ldlr−/−STZ, ApoE−/−STZ) when compared to WT controls.
Supplementary Table 5. Mapping of the 177 (common in all models) differentially expressed proteins into molecular pathways based on the Ingenuity Pathway Analysis (IPA) software.
Supplementary Table 7. LC-MS/MS quantification data of KDM5D, CTSD, IGHM, HMGCS2, TTN, and ZFYVE1 in human vascular tissues.
Supplemental material includes Supplementary Figures 1-5 and Supplementary Tables 6 and 8.