Abstract
The hundreds of cichlid fish species in Lake Malawi constitute the most extensive recent vertebrate adaptive radiation. Here we characterize its genomic diversity by sequencing 134 individuals covering 73 species across all major lineages. Average sequence divergence between species pairs is only 0.1-0.25%. These divergence values overlap diversity within species, with 82% of heterozygosity shared between species. Phylogenetic analyses suggest that diversification initially proceeded by serial branching from a generalist Astatotilapia-like ancestor. However, no single species tree adequately represents all species relationships, with evidence for substantial gene flow at multiple times. Common signatures of selection on visual and oxygen transport genes shared by distantly related deep water species point to both adaptive introgression and independent selection. These findings enhance our understanding of genomic processes underlying rapid species diversification, and provide a platform for future genetic analysis of the Malawi radiation.
The formation of every lake or island represents a fresh opportunity for colonization, proliferation and diversification of living forms. In some cases, the ecological opportunities presented by underutilized habitats facilitate adaptive radiation - rapid and extensive diversification of the descendants of the colonizing lineages1–3. Adaptive radiations are thus exquisite examples of the power of natural selection, as seen for example in Darwin’s finches in the Galapagos4,5, Anolis lizards of the Caribbean6 and in East African cichlid fishes7,8.
Cichlids are one of the most species-rich and diverse families of vertebrates, and nowhere are their radiations more spectacular than in the Great Lakes of East Africa: Malawi, Tanganyika, and Victoria2, each of which contains several hundred endemic species, with the largest number in Lake Malawi9. Molecular genetic studies have made major contributions to reconstructing the evolutionary histories of these adaptive radiations, especially in terms of the relationships between the lakes10,11, between some major lineages in Lake Tanganyika12, and in describing the role of hybridization in the origins of the Lake Victoria radiation13. However, the task of reconstructing within-lake relationships remains challenging due both to retention of large amounts of ancestral genetic polymorphism (i.e. incomplete lineage sorting) and gene flow between taxa12,14–18.
Initial genome assemblies of cichlids from East Africa suggest that an increased rate of gene duplication together with accelerated evolution of some regulatory elements and protein coding genes may have contributed to the radiations11. However, understanding of the genomic mechanisms contributing to adaptive radiations is still in its infancy3.
Here we provide an overview of and insights into the genomic signatures of the haplochromine cichlid radiation of Lake Malawi. The species that comprise the radiation can be divided into seven groups with differing ecology and morphology (see Supplementary Note): 1) the rock-dwelling ‘mbuna’; 2) Rhamphochromis - typically midwater pelagic piscivores; 3) Diplotaxodon - typically deep-water pelagic zooplanktivores and piscivores; 4) deep-water and twilight feeding benthic species; 5) ‘utaka’ feeding on zooplankton in the water column but breeding on or near the lake bottom (here utaka corresponds to the genus Copadichromis); 6) a diverse group of benthic species, mainly found in shallow non-rocky habitats. In addition, Astatotilapia calliptera is a closely related generalist that inhabits shallow weedy margins of Lake Malawi, and other lakes and rivers in the catchment, as well as river systems to the east and south of the Lake Malawi catchment. This division into seven groups has been partially supported by previous molecular phylogenies based on mtDNA and amplified fragment length polymorphism (AFLP) data18–20. However, published phylogenies show numerous inconsistencies and, in particular, the question of whether the groups are genetically separate remained unanswered.
To characterize the genetic diversity, species relationships, and signatures of selection across the whole radiation, we obtained Illumina whole-genome sequence data from 134 individuals of 73 species distributed broadly across the seven groups (Fig. 1a; Supplementary Note). This includes 102 individuals at ~15× coverage and 32 additional individuals at ~6× (Supplementary Table 1).
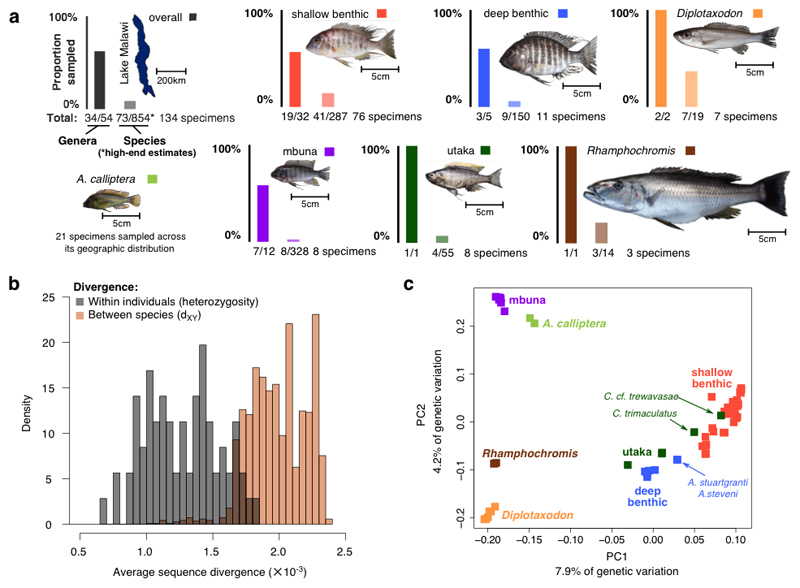
Fig. 1. The Lake Malawi haplochromine cichlid radiation.
a, The sampling coverage of this study: overall and for each of the seven main eco-morphological groups within the radiation. A representative specimen is shown for each group (Diplotaxodon: D. limnothrissa; shallow benthic: Lethrinops albus; deep benthic: Lethrinops gossei; mbuna: Metriaclima zebra; utaka: Copadichromis virginalis; Rhamphochromis: R. woodi). Numbers of species and genera are based on ref. 29. b, The distributions of genomic sequence diversity within individuals (heterozygosity; π) and of divergence between species (dXY). c, Principal component analysis (PCA) of whole genome variation data.
Results
Low genetic diversity and species divergence
Sequence data were aligned to and variants called against a Metriaclima zebra reference genome11. Average divergence from the reference was 0.19% to 0.27% (Supplementary Fig. 1). After filtering and variant refinement, we obtained 30.6 million variants of which 27.1 million were single nucleotide polymorphisms (SNPs) and the rest were short insertions and deletions. All the following analyses are based on biallelic SNPs.
To estimate nucleotide diversity (π) within the species, we measured the frequency of heterozygous sites in each individual. The estimates are distributed within a relatively narrow range between 0.7 and 1.8×10-3 per bp (Fig. 1b). The mean π estimate of 1.2×10-3 per bp is at the low end of values found in other animals21. There does not appear to be a relationship between π and the rate of speciation: individuals in the species-rich mbuna and shallow benthic groups show levels of π comparable to the relatively species-poor utaka, Diplotaxodon, and Rhamphochromis (Supplementary Fig. 1).
Despite their extensive phenotypic differentiation, species within the Lake Malawi radiation are genetically closely related22,23. However, genome-wide genetic divergence has never been quantified. We calculated the average pairwise sequence differences (dXY) between species and compared dXY against heterozygosity, finding that the two distributions partially overlap (Fig. 1b). Thus, the sequence divergence within a single diploid individual is sometimes higher than the divergence between two distinct species. The average dXY is 2.0×10-3 with a range between 1.0 and 2.4×10-3 per bp. The maximum dXY is therefore approximately one fifth of the divergence between human and chimpanzee24. In addition to the low ratio of divergence to diversity, most genetic variation is shared between species. On average both alleles are observed in other species for 82% of heterozygous sites within individuals, consistent with the expected and previously observed high levels of incomplete lineage sorting (ILS)23. Supplementary Fig. 2 shows dXY and FST values for comparisons between the seven eco-morphological groups and Supplementary Fig. 3 shows patterns of linkage disequilibrium across the radiation, within groups, and within individual species.
Low per-generation mutation rate
It has been suggested that the species richness and morphological diversity of teleosts in general and of cichlids in particular might be explained by elevated mutation rates compared to other vertebrates25,26. To obtain a direct estimate of the per-generation mutation rate, we reared offspring of three species from three different Lake Malawi groups (A. calliptera, Aulonocara stuartgranti and Lethrinops lethrinus). We sequenced both parents and one offspring of each to high coverage (40x), applied stringent quality filtering, and counted variants present in each offspring but absent in both its parents (Supplementary Fig. 4). There was no evidence for significant difference in mutation rates between species. The overall mutation rate (μ) was estimated at 3.5×10-9 (95%CI: 1.6×10-9 to 4.6×10-9) per bp per generation, approximately three to four times lower than in human27, although, given much shorter mean generation times, the per-year rate is still expected to be higher in cichlids than in humans. We note that Recknagel et al.26 obtained a much higher mutation rate estimate (6.6×10-8 per bp per generation) in Midas cichlids, but from relatively low depth RADseq data that may have made accurate verification more difficult. We also note that our per generation rate estimate, although low, is still higher than the lowest μ estimate in vertebrates: 2×10-9 per bp per generation recently reported for Atlantic herring28. By combining our mutation rate with nucleotide diversity (π) values, we estimate the long term effective population sizes (Ne) to be in the range of approximately 50,000 to 130,000 breeding individuals (with Ne = π/4μ).
Genome data support for eco-morphological groupings
Principal component analysis (PCA) of the whole-genome genotype data generally separates the major eco-morphological groups (Fig. 1c). The most notable exceptions to this are (1) the utaka, for which some species cluster more closely with deep benthics and others with shallow benthics, and (2) two species of the genus Aulonacara, A. stuartgranti and A. steveni, which are located between the shallow and deep benthic groups. Although these have enlarged lateral line sensory apparatus like many deep benthic species including other Aulonocara, they are typically found in shallower water29. Another interesting pattern in the PCA plot is that the utaka and benthic samples are often spread along principal component (PC) axes (Fig. 1c, Supplementary Fig. 5), a pattern typical for admixed populations (e.g. ref. 30). Along the two main PCs, the deeper water benthic species extend towards the deep water Diplotaxodon, an observation we will return to in the context of gene flow and shared mechanisms of depth adaptation.
To further verify the consistency of group assignments, we tested whether pairs of species from the same group always share more derived alleles with each other than with any species from other groups. Group assignments were again supported, except for the four species also highlighted in the PCA: the two shallow-living Aulonocara are closer to shallow benthics than to deep benthics in 71% and 82% of tests respectively when comparing these alternatives, and Copadichromis trimaculatus is closer to shallow benthics than to utaka in 58% of the comparisons. Copadichromis cf. trewavasae always clustered with shallow benthics; therefore, we treat it as a member of the shallow benthic group throughout the remainder of this manuscript. With the three intermediate samples removed and C. cf. trewavasae reassigned, all other species showed 100% consistency with their group assignment.
Allele sharing inconsistent with tree-like relationships
The above observations suggest that some species may be genetic intermediates between well-defined groups, consistent with previous studies which have suggested that hybridization and introgression subsequent to initial separation of species may have played a significant role in cichlid radiations, including in Lakes Tanganyika12,14–16 and Malawi18,20. Where this happens, there is no single tree relating the species.
To assess the overall extent of violation of tree-like species relationships, we calculated Patterson’s D statistic (ABBA-BABA test)31,32 for all possible trios of Lake Malawi species, without assuming any a priori knowledge of their relationships. N. brichardi from Lake Tanganyika was always used as the outgroup. The test statistic Dmin is the minimum absolute value of Patterson’s D for each trio, across all possible tree topologies. Therefore, a significantly positive Dmin score signifies that the sharing of derived alleles between the three species is inconsistent with a single species tree relating them, even in the presence of incomplete lineage sorting.
Overall, 62% of trios (75,616 out of 121,485) have significantly positive Dmin score (Holm-Bonferroni FWER < 0.01). The Dmin values are not independent: for example, a single gene flow event between ancestral lineages can affect multiple contemporary species and thus more trios than a more recent gene flow event would. However, tree violations are numerous and pervasive throughout the dataset, within all the major groups and also between groups (Fig. 2a), revealing reticulate evolution at multiple levels. Therefore, phylogenetic trees alone cannot fully describe the evolutionary relationships of Lake Malawi cichlids.
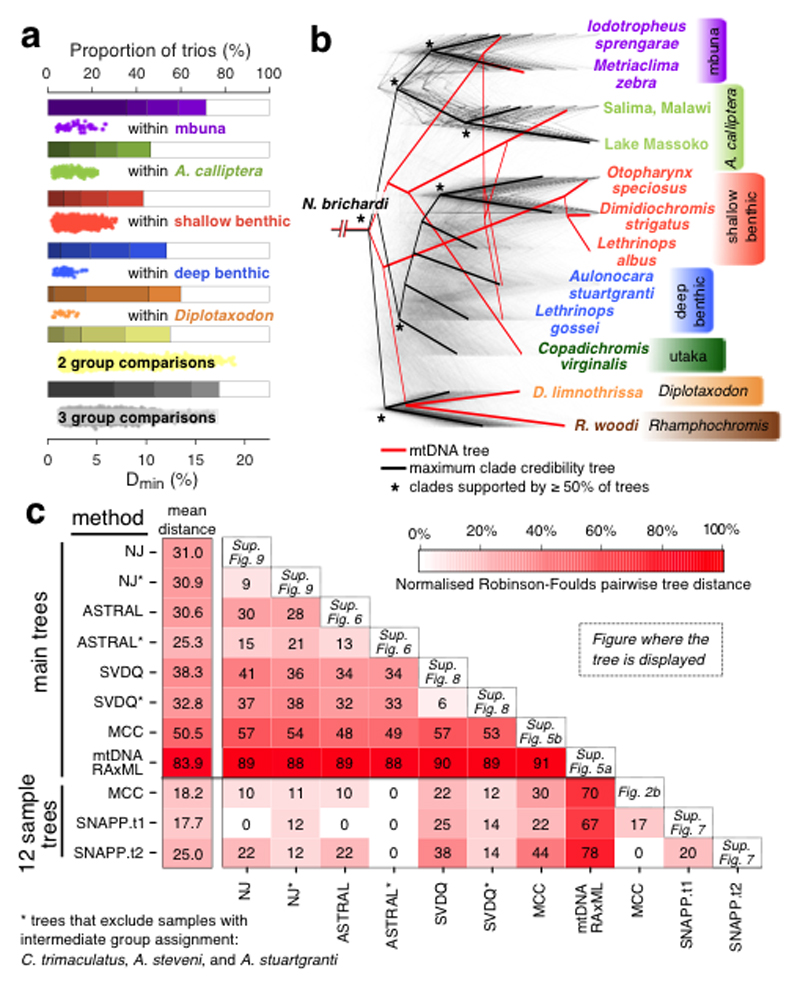
Fig. 2. Excess allele sharing and patterns of species relatedness.
a, Derived allele sharing reveals non-tree-like relationships among trios of species. The bars show the proportion of significantly elevated Dmin scores (see main text). Shading corresponds to FWER q values of (from light to dark) 10-2, 10-4, 10-8, 10-14. The scatterplots show the Dmin scores that were significant at FWER<0.01. Results are shown separately for comparisons where all three species in the trio are from the same group, and for cases where the species come from two or three different groups. Rhamphochromis and utaka within-group comparisons are not shown due to the low number of data points. b, A set of 2543 Maximum Likelihood (ML) phylogenetic trees for non-overlapping regions along the genome. Branch lengths were scaled for visualization so that the total height of each tree is the same. The local trees were built with 71 species and then subsampled for display to 12 individuals representing the eco-morphological groups. The maximum clade credibility tree shown here was built from the subsampled local trees. A ML mitochondrial phylogeny is shown for comparison. c, A summary of all phylogenies from this study and the normalised Robinson-Foulds distances between them, reflecting the topological distance between pairs of trees on the scale from zero to 100%. The least controversial 12 sample tree is SNAPP.t1, with an average distance to other trees of 17.7%, while ASTRAL* is the least controversial among the ‘main trees’ (mean distance of 25.3%). To compare trees with differing sets of taxa, the trees were downsampled so that only matching taxa were present. The position of the outgroup/root was considered in all comparisons.
Phylogenetic framework
Despite no tree giving a complete and accurate picture of the relationships between species, standard phylogenetic approaches are useful to provide a framework for discussion. To obtain an initial picture we divided the genome into 2543 non-overlapping windows, each comprising 8000 SNPs (average size: 274kb) and constructed a Maximum Likelihood (ML) phylogeny separately for the full sequences within each window, obtaining trees with 2542 different topologies. We also calculated the maximum clade credibility (MCC) summary tree33 and an ML phylogeny based on the full mtDNA genome (Fig. 1c and Supplementary Fig. 6).
We next applied a range of further phylogenomic methods which are known to be robust to incomplete lineage sorting. These included three multispecies coalescent methods34,35: the Bayesian SNAPP36 (with a subset of 48,922 unlinked SNPs in 12 individuals representing the eco-morphological groups), the algebraic method SVDquartets37,38, which allows for site-specific rate variation and is robust to gene-flow between sister taxa39, and the summary method ASTRAL40,41, using the 2543 local ML trees that were described above as input. We also built a whole genome Neighbour-Joining (NJ) tree using the Dasarathy et al. algorithm, which has been shown to be a statistically consistent and accurate species tree estimator under ILS42,43. The above methods have also been applied to datasets where the individuals that are genetically intermediate between eco-morphological groups (C. trimaculatus, A. stuartgranti, and A. steveni) have been removed, thus likely reducing the extent of violation of the multispecies coalescent model.
Despite extensive variation among the 2543 individual ML trees (at least in part attributable to ILS), and, to a lesser extent, variation between the different genome-wide phylogenetic methods, there is some general consensus (Fig. 2c and Supplementary Figs. 6-10). Except for the three previously identified intermediate species, individuals from within each of the previously identified eco-morphological groups cluster together in all the whole genome phylogenies, forming well supported reciprocally monophyletic groups. The pelagic Diplotaxodon and Rhamphochromis together form a sister group to the rest of the radiation, except in the all-sample MCC and SVDquartets phylogenies. Perhaps surprisingly, all the methods place the generalist A. calliptera as the sister taxon to the specialized rocky-shore mbuna group in a position that is nested within the Lake Malawi radiation. On a finer scale, many similarities between the resulting phylogenies reflect features of previous taxonomic assignment, but some currently-recognized genera are always polyphyletic, including Placidochromis, Lethrinops, and Mylochromis.
The mtDNA phylogeny is an outlier, substantially different from all the whole-genome phylogenies and also from the majority of the local ML trees (Fig. 2b,c and Supplementary Figs. 6 and 11). Discordances between mtDNA and nuclear phylogenies in Lake Malawi have been reported previously and interpreted as a signature of past hybridization events18,20. However, as we discuss below, some of these previously suggested hybridization events are not reflected in the whole genome data. Indeed, large discrepancies between mitochondrial and nuclear phylogenies have been shown in many other systems, reflecting both that mtDNA as a single locus is not expected to reflect the consensus under ILS, and high incidence of mitochondrial selection44–46. This underlines the importance of evaluating species relationships in the Lake Malawi radiation from a genome-wide perspective.
Specific signals of introgression
We applied a variety of methods to identify the species and groups whose relationships violate the framework trees described in the previous section. First, we contrasted the pairwise genetic distances used to produce the NJ tree against the distances between samples along the tree branches, calculating the residuals (Supplementary Fig. 12). If the tree captured all the genetic relationships in our sample perfectly, the residuals would all be zero. However, as expected in the light of the Dmin analysis above, we found numerous differences, affecting both groups of species and individual species, with some standout cases. Among the strongest signals on individual species, in addition to the previously discussed C. trimaculatus, we can see that 1) Placidochromis cf. longimanus is genetically closer to the deep benthic clade and to a subset of the shallow benthic (mainly Lethrinops species) than the tree suggests; and 2) our sample of Otopharynx tetrastigma (from Lake Ilamba) is much closer to A. calliptera (especially to the sample from Lake Kingiri, only 3.2km away) than expected from the tree.
Second, the sharing of long haplotypes between otherwise distantly related species is an indication of recent admixture or introgression. To investigate this type of gene flow signature, we used the chromopainter software package47 and calculated the ‘coancestry matrix’ of all species - a summary of nearest neighbour (therefore recent) haplotype relationships. The Lake Ilamba O. tetrastigma and Lake Kingiri A. calliptera also stand out in this analysis, showing a strong signature of recent gene flow between individual species from distinct eco-morphological groups (Supplementary Fig. 13). The other tree-violation signatures described above are also visible on the haplotype sharing level but are less pronounced, consistent with being older events involving the common ancestors of multiple present-day species. However, the chromopainter results indicate additional recent introgression events (e.g. the utaka C. virginalis with Diplotaxodon; more highlighted in Supplementary Fig. 13). Furthermore, the clustering based on recent co-ancestry is different from all phylogenetic trees: in particular a number of shallow benthics, including P. cf. longimanus, cluster next to the deep benthics.
Third, we used the f4 admixture ratio31,32,48 (f statistic; closely related to Patterson's D), computing f(A,B;C,O) for all groups of species that fit the relationships ((A, B), C) in the ASTRAL* tree (Supplementary Fig. 7), with the outgroup fixed as N. brichardi. When elevated due to introgression, the f statistic is expected to be linear in relation to the proportion of introgressed material. The ASTRAL* tree has the lowest mean topological distance to all the other trees, and excludes the three species with intermediate group assignment, a choice made here because we were interested in identifying additional signals beyond the admixed status of A. stuartgranti, A. steveni, and C. trimaculatus. Out of the 164,320 computed f statistics, 97,889 were significant at FWER < 0.001.
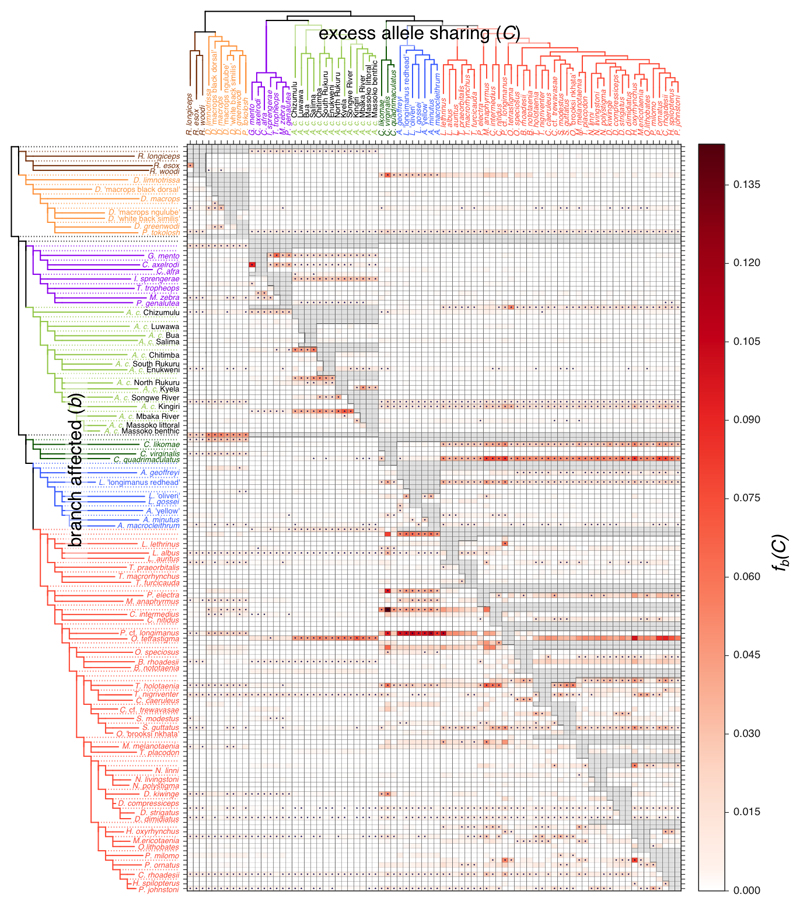
As in the case of Dmin, a single gene flow event can lead to multiple significant f statistics. Noting that the values for different combinations of ((A, B), C) groups are not independent as soon as they share branches on the tree, we sought to obtain branch-specific estimates of excess allele sharing that would be less correlated. Building on the logic employed to understand correlated gene flow signals in ref. 49, we developed “f-branch” or fb(C): a summary of f scores that, on a given tree, captures excess allele sharing between a species C and a branch b compared to the sister branch of b (Methods). Therefore, an fb(C) score is specific to the branch b (on the y-axis in Fig. 3), but a single introgression event can still lead to significant fb(C)’s across multiple related C. There were 11,158 fb(C) scores of which 1,421 were significantly elevated at FWER<0.001 (Supplementary Fig. 14), and 238 scores were larger than 3% (the value inferred for human-Neanderthal introgression in ref. 31). The majority of nodes in the tree are affected: 92 of the 158 branches in the phylogeny show significant excess allele sharing with at least one other species C (Fig. 3).
Fig. 3. Identifying tree violating branches and possible gene flow events.
The branch-specific statistic fb(C) identifies excess sharing of derived alleles between the branch of the tree on the y-axis and the species C on the x-axis (see Supplementary Note). The ASTRAL* tree was used as a basis for the branch statistic and grey data points in the matrix correspond to tests that are not consistent with the phylogeny. Colours correspond to eco-morphological groups as in Fig. 1. The * sign denotes block jack-knifing significance at |Z|>3.17 (Holm-Bonferroni FWER<0.001).
Overall, the highest fb(C) (14.2%) is between the ancestor of the two sampled Ctenopharynx species from the shallow benthic group and the utaka Copadichromis virginalis (Fig. 3). Notably, Ctenopharynx species, particularly C. intermedius and C. pictus have very large numbers of long slender gill rakers, a feature shared with Copadichromis species, and believed to be related to a diet of small invertebrates50. Several other benthic lineages also share excess alleles with C. virginalis at a lower level. Next, the significantly elevated fb(C) scores between the shallow and the deep benthic lineages suggest that genetic exchanges between these two groups go beyond the clearly admixed shallow-living Aulonacara (not included in this analysis). The f-branch signals between O. tetrastigma and A. calliptera Kingiri are observed in both directions – A. calliptera Kingiri with shallow benthics (and most strongly O. tetrastigma) and O. tetrastigma with A. calliptera (most strongly A. calliptera Kingiri), suggesting bi-directional introgression.
At the level of the major eco-morphological groups, the strongest signal indicates that the ancestral lineage of benthics and utaka shares excess derived alleles with Diplotaxodon and, to a lesser degree, Rhamphochromis, as previously suggested by the PCA plot (Fig 1c). Furthermore, there is evidence for additional ancestry from the pelagic groups in utaka, which could be explained either by an additional, more recent, gene flow event or by differential fixation of introgressed material, possibly due to selection. Reciprocally, Diplotaxodon shares excess derived alleles (relative to Rhamphochromis) with utaka and deep benthics, as does Rhamphochromis with mbuna and A. calliptera. Furthermore, mbuna show excess allele sharing (relative to A. calliptera) with Diplotaxodon and Rhamphochromis. (Fig. 3) On the other hand, while ref. 18 suggested gene flow between the deep benthic and mbuna groups on the basis of a discrepancy between mtDNA and nuclear phylogenies, our genome-wide analysis did not find any signal of substantial genetic exchange between these groups.
The f statistic tests are robust to the occurrence of incomplete lineage sorting, in the sense that ILS alone cannot generate a significant test result32. We note, however, that pronounced population structure within ancestral species, coupled with rapid succession of speciation events, can also substantially violate the assumptions of a strictly bifurcating species tree and lead to significantly elevated f scores32,51. This needs to be taken into account when interpreting non-treelike relationships, for example among major groups early in the radiation. However, in cases of excess allele sharing between ‘distant’ lineages that are separated by multiple speciation events, ancestral population structure would have needed to segregate through these speciation events without affecting sister lineages, a scenario that is not credible in general. Therefore, we suggest that there is strong evidence for multiple cross-species gene flow events. Additionally, simulations suggest that, compared with treemix52, fb(C) is robust to misspecification of the initial tree (Supplementary Note).
Overall, the NJ tree residuals, the haplotype sharing patterns, and the many elevated fb(C) scores paint a consistent picture. They confirm the extensive violations of the bifurcating species tree model initially revealed by the Dmin analysis, and suggest many independent gene flow events at different times during the evolutionary history of the adaptive radiation.
Origins of the radiation
The generalist Astatotilapia calliptera has been referred to as the 'prototype' for the endemic Lake Malawi cichlids29,53, and discussions concerning the origin of the radiation often centre on ascertaining its relationship to the Malawi species20,54. Previous phylogenetic analyses, using mtDNA and small numbers of nuclear markers, showed inconsistencies in this respect18,20,54. In contrast, our whole genome data indicated a clear and consistent position of the Lake Malawi catchment A. calliptera as a sister group to the mbuna, in agreement with the nuclear DNA phylogeny in ref. 18. While it is not certain whether the 320 remaining mbuna species form a monophyletic group with the eight species we used here, the eight species represent the majority of the genera of mbuna and therefore are likely to be representative of much of the genetic diversity within the group.
To explore the origins of the Lake Malawi radiation in greater detail, we obtained 24 additional Astatotilapia whole genome sequences from outside of Lake Malawi: five A. calliptera from Indian Ocean catchments (IOC), thus covering most of its geographical distribution, and 19 individuals from seven other Astatotilapia species (Supplementary Table 2). We generated new variant calls (Supplementary Methods) and first constructed a NJ tree, finding that all the A. calliptera (including IOC) cluster as a single group nested at the same place within the radiation, whereas the other Astatotilapia species branched off well before the lake radiation (Fig. 4a,b,c). All A. calliptera individuals cluster by geography (Fig. 4b,c), except for the specimen from crater Lake Kingiri, whose position in the tree is likely a result of the admixture signals with O. tetrastigma. Indeed, a NJ tree built only with A. calliptera samples (Supplementary Fig. 17) places the Kingiri individual according to geography with the specimens from the nearby crater Lake Massoko and Mbaka River.
Fig. 4. Origins of the radiation and the role of A. calliptera.
a, An NJ phylogeny showing the Lake Malawi radiation in the context of other East African Astatotilapia taxa. b, A Lake Malawi NJ phylogeny with expanded view of A. calliptera, with all other groups collapsed. c, Approximate A. calliptera sampling locations shown on a map of the broader Lake Malawi region. Black lines correspond to present day level 3 catchment boundaries from the US Geological Survey’s HYDRO1k dataset. d, Strong f4 admixture ratio signal showing that Malawi catchment A. calliptera are closer to mbuna than their Indian Ocean catchment counterparts. e, PCA of body shape variation of Lake Malawi endemics, A. calliptera and other Astatotilapia taxa, obtained from geometric morphometric analysis. f, A phylogeny with the same topology as in panel (b) but displayed with a straight line between the ancestor and A. calliptera. For each branch off this lineage, we show mean sequence divergence (dXY) minus mean heterozygosity, and translation of this value into a mean divergence time estimate with 95% CI reflecting the statistical uncertainty in mutation rate. Dashed lines with arrows indicate likely instances of gene flow between major groups; their true timings are uncertain.
Applying the same logic as above, we tested whether the position of the A. calliptera group in the NJ tree changes when the tree is built without mbuna (as would be expected if A. calliptera were affected by hybridization with mbuna). We found that the position of A. calliptera is unaffected (Supplementary Fig. 18), suggesting that the nested position is not due to later hybridization. The f statistics in Fig. 3 further support this, because the signals involving the whole mbuna or A. calliptera groups are modest and do not suggest erroneous placement of these groups in all phylogenetic analyses. Furthermore, the nested position of A. calliptera is also supported by the vast majority of the genome. Searching for the basal branch in a set of 2638 local ML phylogenies, we found results that agree with the whole genome ASTRAL, SNAPP and NJ trees: the most common basal branches are the pelagic groups Rhamphochromis and Diplotaxodon (in 42.12% of the genomic windows). In comparison, A. calliptera (including IOC samples) were found to be basal only in 5.99% of the windows (Supplementary Fig. 19).
Joyce et al.20 reported that the mtDNA haplogroup of Indian Ocean catchment (IOC) A. calliptera clustered with mbuna (as we confirm in Supplementary Fig. 15) and suggested that there had been repeated colonization of Lake Malawi by two independent Astatotilapia lineages with different mitochondrial haplogroups: the first founding the entire species flock, and the second, with the IOC mtDNA haplogroup, introgressing into the Malawi radiation and contributing strongly to the mbuna. This hypothesis predicts that, compared with the Malawi catchment A. calliptera, the IOC A. calliptera should be closer to mbuna. However, across the nuclear genome we found a strong signal in the opposite direction, with 30% excess allele sharing between Malawi catchment A. calliptera and mbuna (Fig. 4d). Therefore, the Joyce et al. hypothesis that the mbuna, the most species rich group within the radiation, may be a hybrid lineage formed from independent invasions is not supported by genome-wide data.
It has been repeatedly suggested that A. calliptera may be the direct descendant of the riverine-generalist lineage that seeded the Lake Malawi radiation7,50,53,54. Our interpretation of this argument is that the ancestor was likely a riverine generalist that was ecologically and phenotypically similar to A. calliptera and other Astatotilapia. This hypothesis is lent further support by geometric morphometric analysis. Using 17 homologous body shape landmarks we established that, despite the relatively large genetic divergence, A. calliptera is nested within the morphospace of the other more distantly related but ecologically similar Astatotilapia species (Fig. 4a,e), and these together have a central position within the morphological space of the Lake Malawi radiation (Fig. 4e and Supplementary Fig. 16).
To reconcile the nested phylogenetic position of A. calliptera with its generalist ‘prototype’ phenotype, we propose a model where the Lake Malawi species flock consists of three separate radiations splitting off from the lineage leading to A. calliptera. The relationships between the major groups supported by the ASTRAL, SNAPP and NJ methods suggest that the pelagic radiation was seeded first, then the benthic + utaka, and finally the rock-dwelling mbuna, all in a relatively quick succession, followed by subsequent gene flow as described above (Fig. 4f; the pelagic vs. utaka + benthic branching order is swapped in SVDquartets tree in Supplementary Fig. 9b). Applying our per-generation mutation rate to observed genomic divergences we obtained mean divergence time estimates between these lineages between 460 thousand years ago (ka) [95%CI: (350ka to 990ka)] and 390ka [95%CI: (300ka to 860ka)] (Fig. 4f), assuming three years per generation as in ref. 55. The point estimates all fall within the second most recent prolonged deep lake phase inferred from the Lake Malawi paleoecological record56 while the upper ends of the confidence intervals cover the third deep lake phase at ~800ky. Considering that our split time estimates from sequence divergence are likely to be reduced by subsequent gene-flow, leading to underestimates, the data are consistent with a previous report based on fossil time calibration which put the origin of the Lake Malawi radiation at 700-800ka12.
The fact that the common ancestor of all the A. calliptera appears younger than the Malawi radiation suggests that the Lake Malawi A. calliptera population has been a reservoir that has repopulated the river systems and more transient lakes following dry-wet transitions in East African hydroclimate56,57. Our results do not fully resolve whether the lineage leading from the common ancestor to A. calliptera retained its riverine generalist phenotype throughout or whether a lacustrine species evolved at some point (e.g. the common ancestor of A. calliptera and mbuna) and later de-specialized again to recolonize the rivers. However, while it is a possibility, we suggest it is unlikely that the many strong phenotypic affinities of A. calliptera to the basal Astatotilapia (Fig. 4e; refs. 58,59) would be reinvented from a lacustrine species.
Signatures and consequences of selection on coding sequences
To gain insight into the functional basis of diversification and adaptation in Lake Malawi cichlids, we next turned our attention to protein coding genes. We compared the between-species levels of non-synonymous variation to synonymous variation in 20,664 genes and calculated the difference between these two values Overall, coding sequence exhibits signatures of purifying selection: the average between-species was 54% lower than in a random matching set of non-coding regions. Interestingly, the average between-species synonymous variation in genes was 13% higher than in non-coding control regions (p <2.2 × 10−16, one tailed Mann-Whitney test). One possible explanation of this observation would be if intergenic regions were homogenized by gene-flow, whereas protein coding genes were more resistant to this.
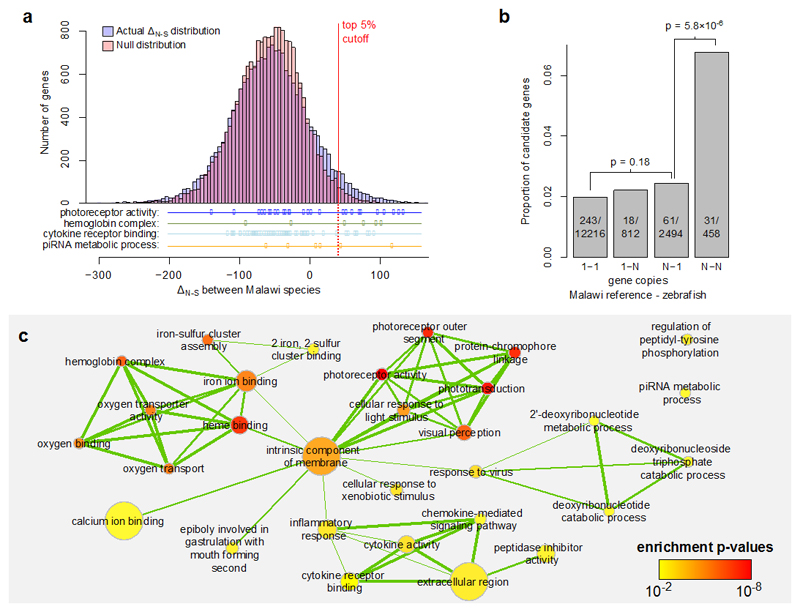
To control for statistical effects of variation in gene length and sequence composition we normalized the δN−S values per gene by taking into account the variance across all pairwise sequence comparisons for each gene, deriving the non-synonymous excess score ΔN−S (see Methods). Values at the upper tail of the distribution of ΔN−S are substantially overrepresented in the actual data when compared to a null model based on random sampling of codons (Fig. 5a). We focus below on the top 5% of the distribution (ΔN−S > 40.2, 1034 candidate genes). Genes with elevated ΔN−S are expected to have been under positive selection at multiple non-synonymous sites, either recently repeatedly within multiple species or ancestrally. Therefore, the statistic reveals only a limited subset of positive selection events from the history of the radiation (e.g. a selection event on a single amino acid would not be detected). Furthermore, to minimise any effect of gene prediction errors, all the following analyses focus on the 15980 (77.3% of total) genes for which zebrafish homologs were found in ref. 11; selection scores of genes without homologs are briefly discussed in a Supplementary Note.
Fig. 5. Gene selection scores, copy numbers, and ontology enrichment.
a, The distribution of the non-synonymous variation excess scores (ΔN−S) highlighting the top 5% cutoff, compared against a null model. The null was derived by calculating the statistic on randomly sampled combinations of codons. We also show the distributions of genes in selected Gene Ontology (GO) categories which are overrepresented in the top 5%. b, The relationship between the probability of ΔN−S being in the top 5% and the relative copy numbers of genes in the Lake Malawi reference (M. zebra) and zebrafish. The p-values are based on χ2 tests of independence. Genes existing in two or more copies in both zebrafish and Malawi cichlids are disproportionately represented among candidate selected genes. c, An enrichment map for significantly enriched GO terms (cutoff at p ≤ 0.01). The level of overlap between GO enriched terms is indicated by the thickness of the edge between them. The colour of each node indicates the p-value for the term and the size of the node is proportional to the number of genes annotated with that GO category.
Cichlids have an unexpectedly large number of gene duplicates, which possibly contributed to their extensive adaptive radiations3,11. To investigate the extent of divergent selection on gene duplicates, we examined how the ΔN−S scores are related to gene copy numbers in the reference genomes. Focusing on homologous genes annotated both in the Malawi reference (M. zebra) and in the zebrafish genome, we found that the highest proportion of candidate genes was among genes with two or more copies in both genomes (N - N). The relative enrichment in this category is both substantial and highly significant (Fig. 5b). On the other hand, the increase in proportion of candidate genes in the N - 1 category (multiple copies in the M. zebra genome but only one copy in zebrafish) is of a much lesser magnitude and is not significant (χ2 test p = 0.18), suggesting that selection is occurring more often within ancient multi-copy gene families, rather than on genes with cichlid-specific duplications.
We used Gene Ontology (GO) annotation of zebrafish homologs to test whether candidate genes are enriched for particular functional categories (Methods). We found significant enrichment for 30 GO terms (range: 1.6×10-8 < p < 0.01, weigh algorithm60; Supplementary Table 3): 10 in the Molecular Function, 4 in the Cellular Component and 16 in Biological Process category. Combining all the results in a network (connecting terms that share many genes) revealed clear clusters of enriched terms related to (i) haemoglobin function and oxygen transport; (ii) phototransduction and visual perception; and (iii) the immune system, especially inflammatory response and cytokine activity (Fig. 5c). That evolution of genes in these functional categories has contributed to cichlid radiations has been suggested previously (see below); it is nevertheless interesting that these categories stand out in a genome-wide analysis.
Shared mechanisms of depth adaptation
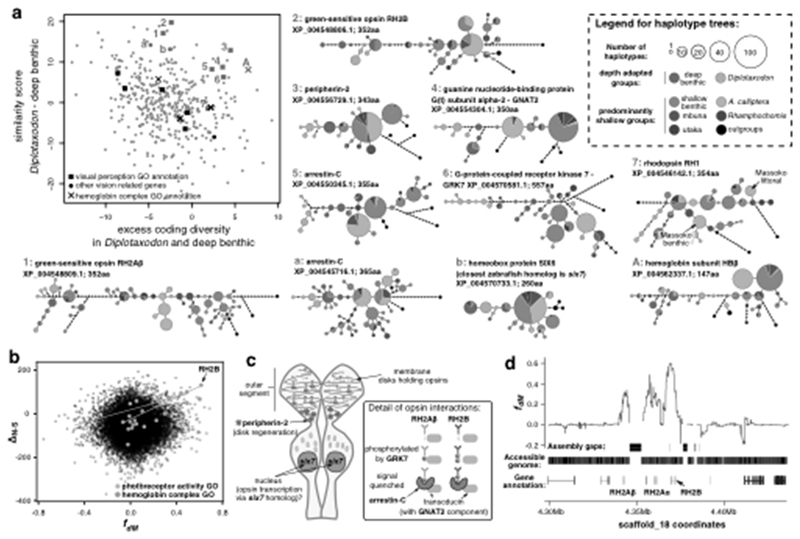
To gain insight into the distribution of adaptive alleles across the radiation, we built maximum likelihood trees from amino acid sequences of candidate genes, thus summarising potentially complex haplotype genealogy networks. Focusing on the significantly enriched GO categories, many haplotype trees have features that are unusual in the broader dataset: the haplotypes from the deep benthic group and the deep-water pelagic Diplotaxodon tend to group together (despite these two groups being distant in whole-genome phylogenies and monophyletic in only two out of 2638 local ML trees) and also tend to be disproportionally diverse when compared with the rest of the radiation. We quantified both excess similarity and diversity, and found that both measures are elevated for candidate genes in the ‘visual perception’ category (Fig. 6a; Mann-Whitney tests: p=0.007 for similarity, p=0.08 for shared diversity, and p=0.003 when the scores are added) and also for the ‘haemoglobin complex’ category (p values not significant due to the small number of genes).
Fig. 6. Shared selection between the deep water adapted groups Diplotaxodon and deep benthic.
a, The scatterplot shows the distribution of genes with high ΔN−S scores (candidates for positive selection) along axes reflecting shared selection signatures. Only genes with zebrafish homologs are shown. Amino acid haplotype trees, shown for genes as indicated by the red symbols and numbers, indicate that Diplotaxodon and deep benthic species are often divergent from other taxa, but similar to each other. Outgroups include Oreochromis niloticus, Neolamprologus brichardi, Astatotilapia burtoni, and Pundamilia nyererei. b, Selection scores plotted against fdM (mbuna, deep benthic, Diplotaxodon, N. brichardi), a measure of local excess allele sharing between deep benthic and Diplotaxodon. Overall there is no correlation between ΔN−S and fdM. However, the strong correlation between ΔN−S and fdM in the highlighted GO categories suggests that positively selected alleles in those categories tend to be subject to introgression or convergent selection between Diplotaxodon and the deep benthic group. c, A schematic drawing of a double cone photoreceptor expressing the green sensitive opsins and illustrating the functions of other genes with signatures of shared selection. d, fdM calculated in sliding windows of 100 SNPs around the green opsin cluster, revealing that excess allele sharing between deep benthic and Diplotaxodon extends far beyond the coding sequences.
Sharply decreasing levels of dissolved oxygen and low light intensities with narrow short wavelength spectra are the hallmarks of the habitats at below ~50 meters to which the deep benthic and Diplotaxodon groups have both adapted, either convergently or in parallel61. Shared signatures of selection in genes involved in vision and in oxygen transport therefore point to shared molecular mechanisms underlying this ecological parallelism. Further evidence of shared mechanisms of adaptation is that, for genes annotated with ‘photoreceptor activity’ and ‘haemoglobin complex’ GO terms, the ΔN-S selection score is strongly correlated with the local levels of excess allele sharing between the two depth-adapted groups (Fig. 6b; ρS = 0.63 and 0.81, p = 0.001 and p = 0.051, respectively).
Vision genes with high similarity and diversity scores for the deep benthic and Diplotaxodon groups include three opsins: the green sensitive RH2Aβ and RH2B, and rhodopsin (Fig. 6a and Supplementary Fig. 20). The specific residues that distinguish the deep adapted groups from the rest of the radiation differ between the two RH2 copies, with only one shared mutation out of a possible fourteen (Supplementary Fig. 20). RH2Aβ and RH2B are located less than 40kb apart on the same chromosome (Fig. 6c); a third paralog, RH2Aα, is located between them, but does not show signatures of shared depth adaptation (Supplementary Fig. 21), consistent with reports of functional divergence between RH2Aα and RH2Aβ62,63. A similar, albeit weaker signature of shared depth-related selection is apparent in rhodopsin, which is known to play a role in deep-water adaptation in cichlids64. Previously, we discussed the role of coding variants in rhodopsin in the early stages of speciation of A. calliptera in the crater Lake Massoko55. The haplotype tree presented here for the broader radiation shows that the Massoko alleles did not originate by mutation in that lake but were selected out of ancestral variation (Fig. 6a). The remaining opsin genes are less likely to be involved in shared depth adaptation (Supplementary Note).
There have been many studies of selection on opsin genes in fish65–67, including selection associated with depth preference, but having whole genome coverage allows us to investigate other components of primary visual perception in an unbiased fashion. We found shared patterns of selection between deep benthics and Diplotaxodon in six other vision associated candidate genes (Fig. 6a). The functions of these genes, together with the fact that RH2Aβ and RH2B are expressed exclusively in double-cone photoreceptors, suggest a prominent role of cone cell vision in depth adaptation. The wavelength of maximum absorbance in cone cells expressing a mixture of RH2Aβ with RH2B (λmax = 498nm) corresponds to the part of light spectrum that transmits the best into deep water in Lake Malawi67.
Figure 6c illustrates interactions of the vision genes with shared selection patterns in the cichlid double-cone photoreceptor. The homeobox protein six7 governs the expression of RH2 opsins and is essential for the development of green cones in zebrafish68 (specific mutations are highlighted in Supplementary Fig. 20). The kinase GRK7 and the retinal cone arrestin-C have complementary roles in photoresponse recovery: arrestin produces the final shutoff of the cone pigment following phosphorylation by GRK7, thus determining the temporal resolution of motion vision69. Bases near to the C-terminus in RH2Aβ mutated away from serine (S290Y and S292G), thus reducing the number of residues that can be modified by GRK7 (Supplementary Fig. 20). The transducin subunit GNAT2 is located exclusively in the cone receptors and is a key component of the pathway which converts light stimulus into electrical response in these cells70. Finally, peripherin-2 is essential to the development and renewal of the membrane system that holds the opsin pigments in both rod and cone cells71.
Haemoglobin genes in teleost fish are found in two separate chromosomal locations: the minor ‘LA’ cluster and the major ‘MN’ cluster72. The region around the LA cluster has been highlighted by selection scans among four Diplotaxodon species by Hahn et al.73, who also noted the similarity of the haemoglobin subunit beta (HBβ) haplotypes between Diplotaxodon and deep benthic species. We confirmed signatures of selection in the two annotated LA cluster haemoglobins. In addition, we found that four haemoglobin subunits (HBβ1, HBβ2, HBα2, HBα3) from the MN cluster are also among the genes with high selection scores (Supplementary Fig. 22). The shared patterns of depth selection may be particular to the β-globin genes (Supplementary Fig. 22), although this hypothesis remains tentative, because the repetitive nature of the MN cluster precludes us from confidently examining all haemoglobin genes.
A key question concerns the mechanism leading to the similarity of haplotypes in Diplotaxodon and deep benthics. Possibilities include parallel selection on variation segregating in both groups due to common ancestry, selection on the gene flow that we described in a previous section, or independent selection on new mutations. From considering the haplotype trees and local patterns of excess allele sharing (fdM statistics55), there is evidence for each of these processes acting on different genes. The haplotype trees for rhodopsin and HBβ have outgroup taxa (and also A. calliptera) appearing at multiple locations on their haplotype networks (Fig. 6a), suggesting that the haplotype diversity of these genes may reflect ancestral variation. In contrast, trees for the green cone genes show the Malawi radiation all being derived with respect to outgroups and we found substantially elevated fdM scores extending for around 40kb around the RH2 cluster (Fig. 6d), consistent with adaptive introgression in a pattern reminiscent of mimicry loci in Heliconius butterflies74. Finally, the peaks in fdM around peripherin-2 and one of the arrestin-C genes are narrow, ending at the gene boundaries, and fdM scores are elevated only for non-synonymous variants; synonymous variants do not show excess allele sharing (Supplementary Fig. 23). Due to the close proximity of non-synonymous and synonymous sites within the same gene, this suggests that for these two genes there may have been independent selection on the same de novo mutations.
Discussion
Variation in genome sequences forms the substrate for evolution. Here we described genome variation at the full sequence level across the Lake Malawi haplochromine cichlid radiation. We focused on ecomorphological diversity, representing more than half the genera from each major group, rather than obtaining deep coverage of species within any particular group. Therefore, we have more samples from the morphologically highly diverse benthic lineages than, for example, the mbuna where there are relatively fewer genera and many species are largely recognised by colour differences.
The observation that cichlids within an African Great Lake radiation are genetically very similar is not new75, but we now quantify the relationship of this to within-species variation, and the consequences for variation in local phylogeny across the genome. The fact that between-species divergence is generally only slightly higher than within species diversity, is likely the result of the young age of the radiation, the relatively low mutation rate, and of gene flow between taxa. Within-species diversity itself is relatively low for vertebrates, at around 0.1%, suggesting that low genome-wide nucleotide diversity levels do not necessarily limit rapid adaptation and speciation. This conclusion appears in contrast for example with a recent report that high diversity levels may have been important for rapid adaptation in Atlantic killifish76. One possibility is that in cichlids repeated selection has maintained diversity in adaptive alleles for a range of traits that support ecological diversification, as we have concluded for rhodopsin and HBβ and appears to be the case for some adaptive variants in sticklebacks77.
We provide evidence that gene flow during the radiation, although not ubiquitous, has certainly been extensive. Overall, the numerous violations of the bifurcating species tree model suggest that full resolution of interspecies relationships in this system will require network approaches (see e.g. ref. 35; section 6.2) and population genomic analyses within the framework of the structured coalescent with gene flow. The majority of the signals affect groups of species, suggesting events involving their common ancestors, or are between closely-related species within the major ecological groups. The only strong and clear example of recent gene flow between individual distantly-related species is not within Lake Malawi itself, but between Otopharynx tetrastigma from crater Lake Ilamba and local A. calliptera. Lake Ilamba is very turbid and the scenario is reminiscent of cichlid admixture in low visibility conditions in Lake Victoria78. It is possible that some of the earlier signals of gene flow between lineages we observed in Lake Malawi may have happened during low lake level periods when the water is known to have been more turbid56.
Our model of the early stages of radiation in Lake Malawi (Fig. 4f) is broadly consistent with the model of initial separation by major habitat divergence23, although we propose a refinement in which there were three relatively closely-spaced separations from a generalist Astatotilapia type lineage, initially of pelagic genera Rhamphochromis and Diplotaxodon, then of shallow- and deep-water benthics and utaka (this includes Kocher’s sand dwellers23,29), and finally of mbuna. Thus, we suggest that Lake Malawi contains three separate haplochromine cichlid radiations stemming from the generalist lineage, interconnected by subsequent gene flow.
The finding that cichlid-specific gene duplicates do not tend to diverge particularly strongly in coding sequences (Fig. 5b) suggests that other mechanisms of diversification following gene duplications may be more important. Divergence via changes in expression patterns has previously been illustrated and discussed11, and future studies addressing structural variation between cichlid genomes will assess the contribution of differential retention of duplicated genes.
The evidence concerning shared adaptation of the visual and oxygen-transport systems to deep-water environments between deep benthics and Diplotaxodon suggests different evolutionary mechanisms acting on different genes, even within the same cellular system. It will be interesting to see whether the same genes or even specific mutations underlie depth adaptation in Lake Tanganyika, which harbours specialist deep water species in least two different tribes79 and has a similar light attenuation profile but a steeper oxygen gradient than Lake Malawi61.
Over the last few decades, East African cichlids have emerged as a model for studying rapid vertebrate evolution11,23. Taking advantage of recently assembled reference genomes11, our data and results provide unprecedented information about patterns of sequence sharing and adaptation across the Lake Malawi radiation, with insights into mechanisms of rapid phenotypic diversification. The data sets are openly available (see Acknowledgements) and will underpin further studies on specific taxa and molecular systems. For example, we envisage that our results, clarifying the relationships between all the main lineages and many individual species, will facilitate speciation studies, which require investigation of taxon pairs at varying stages on the speciation continuum80,81, and studies on the role of adaptive gene flow in speciation.
Methods
Samples
Ethanol preserved fin clips were collected by M.J. Genner and G.F. Turner between 2004 and 2014 from Tanzania and Malawi, in collaboration with the Tanzania Fisheries Research Institute (the MolEcoFish Project) and with the Fisheries Research Unit of the Government of Malawi (various collaborative projects). Samples were collected and exported with the permission of the Tanzania Commission for Science and Technology, the Tanzania Fisheries Research Institute, and the Fisheries Research Unit of the Government of Malawi
From sequencing to a variant callset
The analyses presented above are based on SNPs obtained from Illumina short (100bp-125bp) reads, aligned to the Metriaclima zebra reference assembly version 1.111 with bwa-mem82, followed by GATK haplotype caller83 and samtools/bcftools84 variant calling restricted to 653Mb of ‘accessible genome’ where variants can be determined confidently with short reads, filtering, genotype refinement, imputation, and phasing in BEAGLE85 and further haplotype phasing with shapeit v286, including the use of phase-informative reads87. For details please see Supplementary Methods.
Linkage disequilibrium (LD) calculations
Haplotype r2 between pairs of SNPs was calculated along the phased scaffolds 0 to 201, using vcftools v0.1.12b with the options --hap-r2 --ld-window-bp 50000. To reduce the computational burden, we used a random subsample of 10% of SNPs. We binned the r2 values according to the distance between SNPs into 1kb or 100bp windows and plotted the average values in each bin.
To estimate background LD, we calculated haplotype r2 between variants mapping to different linkage groups (LG) in the Oreochromis niloticus genome assembly. First, we used the chain files generated by the whole genome alignment pipeline88 (see Supplementary Methods) and the UCSC liftOver tool to translate the genomic coordinates of all SNPs to the O. niloticus coordinates. Then we calculated LD between variants mapping to LG1 and LG2.
De novo mutation rate estimation
In each trio we looked for mutations in the child that were not present in either of its parents. Because the results of this analysis are very sensitive to false positives and false negative rates, we used higher coverage sequencing (~40x average) and applied more stringent genome masks than in the population genomic work. Increased coverage supports clean separation of sequencing errors and somatic mutations from true heterozygous calls in the offspring, and improved ability to distinguish single copy vs. multi-copy sequence on a per-individual basis.
First we determined the “Accessible Genome” (i.e. the regions of the genome where we can confidently call de novo mutations) for each trio by excluding:
Genomic regions where mapped read depth in any member of a trio is ≤25× or >50×
Bases where either of the parents has a mapped read that does not match the reference (the specific bases where any read has non-reference alleles in the parents were masked)
Sequences where indels were called in any sample (we also excluded +/- 3bp of sequence surrounding the indel)
Sites which were called as multiallelic among the nine samples in the overall trios dataset
Known segregating variable sites - i.e. sites with alternative alleles found in four and more copies in the overall Lake Malawi dataset
Sites in the reference where less than 90% of overlapping 50-mers (sub-sequences of length 50) could be matched back uniquely and without 1-difference. For this we used Heng Li’s SNPable tool (http://lh3lh3.users.sourceforge.net/snpable.shtml), dividing the reference genome into overlapping k-mers (sequences of length k – we used k=50), and then aligning the extracted k-mers back to the genome (we used bwa aln -R 1000000 -O 3 -E 3).
After excluding sites in the categories above, we were left with an “Accessible Genome” of 516.6Mb in the A. calliptera trio, 459Mb in the A. stuartgranti trio and 404Mb in the L. lethrinus trio. Because any observed de novo mutation could have occurred either on the chromosome inherited from the mother or on the chromosome inherited from the father, the point estimate of the per generation per basepair mutation rate is: μ = nMutations/(2×AccessibleGenome).
Next we set out to search for de novo mutations: i.e. heterozygous sites in the offspring within the Accessible Genome. Under random sampling there is an equal probability of seeing a read with either of the two alleles at a heterozygous site. Therefore, Na - the number of reads supporting the alternative allele is distributed as: ~Binomial(ReadDepth, 0.5). We filtered out variants with observed Na below 2.5th or above 97.5th percentiles of this distribution, thus accepting a false negative rate of 5%. We also filtered out sites where the offspring call had Read Position or Base Quality rank sum test Z-score > 99.5th percentile of standard normal distribution or where the Strand Bias phred scaled p-value was ≥ 20 or where the phred scaled GQ (genotype quality) in either mother, father, or offspring was ≤ 30. For simplicity, assuming these filters are independent they are expected to introduce a false negative rate of 7.17%. The mutation rate estimate was adjusted to account for this.
After filtering, we found nine de novo mutations across the three offspring. For each mutation we double-checked the alignment in the IGV genome browser and found all of them were single base mutations supported by high number of reads (>8) in the offspring. The 95% confidence intervals for the number of observed mutations were calculated using the “exact” method relating chi-squared and Poisson distributions89,90. If N is the number of observed mutations, the lower (ciNL) and upper (ciNU) limits are:
where 2N and 2(N+1) are degrees of freedom of the corresponding chi-squared distributions.
Principal Component Analysis
SNPs with minor allele frequency >= 0.05 were selected using the bcftools (v1.2) view option --min-af 0.05:minor. The program vcftools v0.1.12b was then used to export that data into PLINK format91. Next, the variants were LD-pruned to obtain a set of variants in approximate linkage equilibrium (unlinked sites) using the --indep-pairwise 50 5 0.2 option in PLINK v1.0.7. Principal Component Analysis on the resulting set of variants was performed using the smartpca program from the eigensoft v5.0.2 software package92 with default parameters.
Genome-wide FST calculations
In addition to performing PCA, the smartpca program from the eigensoft v5.0.2 software package also calculates genome-wide FST for all pairs of populations specified by the sixth column in the .pedind file. For the calculation, it uses the Hudson estimator, as defined by Bhatia, Patterson et al.93 in equation 10, and the ‘ratio of averages’ is used to combine estimates of FST across multiple variants, as recommended in that manuscript. We used all SNPs (no minor allele frequency filtering).
Allele sharing test for group assignment
We tested if two individuals who come from the same group always share more derived alleles with each other than with any individuals from other groups. Technically, we implemented this using the D statistic (ABBA-BABA tests) framework31,32, by calculating D (A, G1, G2, O) for all permutations of individuals, where G1 and G2 come from the same eco-morphological group and A from a different group. The outgroup O was always N. brichardi from Lake Tanganyika. Note that this is an unusual use of the D statistics and our aim here was not to look for gene flow but to test if allele sharing is greater within eco-morphological groups (G1 with G2) compared to across groups (A with G2), in which case D (A, G1, G2, O) > 0. All results were statistically significant, which was assessed using block jackknife31 on windows of 60k SNPs.
Dmin statistic
Here we calculated the D statistic for each trio of species (A,B,C) and for all possible tree topologies (the outgroup again fixed as N. brichardi). Therefore, Dmin=min(|D(A,B,C,O)|, |D(A,C,B,O)|, |D(C,B,A,O)|. If this is significantly elevated, then allele sharing within the trio of species is inconsistent with any simple tree topology. Note that this approach is conservative in the sense that the Dmin score for each trio is considered in isolation and we ignore ‘higher-order’ inconsistencies where different Dmin trio topologies are inconsistent with each other. Statistical significance was assessed using block jackknife31 on windows of 60k SNPs and FWER was calculated following the Holm-Bonferroni method.
Sample selection for demographic analyses
To prevent potential confounding effects of uneven sequencing depth, we limited these analyses to one high coverage (15x) individual per species. Species without a high coverage sample (P. subocularis, F. rostratus and L. trewavasae) were not included.
Outgroup sequences/alleles
Outgroup (Supplementary Table 5) sequences in M. zebra genomic coordinates were obtained based on pairwise whole-genome alignments (Supplementary Methods). Insertions in the outgroup were ignored and deletions filled by N characters.
Local phylogenetic trees and maximum clade credibility
To generate a multiple alignment input in fasta format we used the getWGSeq subprogram of evo We set the window size in terms of the numbers of variants rather than physical length (8000 variants; --split 8000 option) aiming for the local regions to have similar strengths of phylogenetic signal. Small windows at the ends of scaffolds were discarded. We limited the sequence output to the accessible genome using the --accessibleGenomeBED option. The N. brichardi outgroup sequence in M. zebra genomic coordinates was added via the --incl-Pn option.
Maximum likelihood phylogenies were inferred using RAxML v7.7.894 under the GTRGAMMA model. The best tree for each region was selected out of twenty alternative runs on distinct starting maximum parsimony trees (the -N 20 option).
The maximum clade credibility (MCC) trees were calculated in TreeAnnotator v.2.4.2, a part of the BEAST2 platform95. Clade credibility is the frequency with which a clade appears in the tree set; the MCC tree is the tree (from among the trees in the set) that maximizes the product of the frequencies of all its clades33. The node heights for the MCC trees are derived as a summary from the heights of each clade in the whole tree set via the Common Ancestor heights option.
Mitochondrial (mtDNA) phylogenies
The mtDNA sequence corresponds to scaffolds 747 and 2036 in the Metriaclima zebra reference. Variants from these scaffolds were subjected to the same filtering as in the rest of the genome except for the depth filter because the mapped read depth was much higher (approximately 300-400x per sample). Because of the greater sequence diversity in the mtDNA genome, we found that more than 10% of variants were multiallelic. Therefore, we separated SNPs from indels at multiallelic sites using bcftools norm with the --multiallelics - option, then removed indels and the merged multiallelic SNPs back together with the --multiallelics + option. Sequences in the fasta format were generated using the bcftools consensus command, and missing genotypes in the VCF replaced by the N character with the --mask option. N. brichardi outgroup sequence in M. zebra genomic coordinates was added to the fasta files.
A maximum likelihood tree was inferred using RAxML v7.7.894 under the GTRGAMMA model. The best tree was selected out of twenty alternative runs on distinct starting maximum parsimony trees (using the -N 20 option) and two hundred bootstrap replicates were obtained using RAxML’s rapid bootstrapping algorithm96 satisfying the -N autoFC frequency-based bootstrap stopping criterion. Bipartition bootstrap support was drawn on the maximum likelihood tree using the RAxML -f b option.
Neighbour-joining trees and the residuals
For the Neighbour-Joining (NJ)97 trees we calculated the average numbers of single nucleotide differences between haplotypes for each pair of species. This simple pairwise difference matrix was divided by the accessible genome size to obtain pairwise differences per bp, which are equivalent to of Dasarathy et al.42. Then we followed equation 8 from Dasarathy et al. and calculated their corrected measure of dissimilarity:
The values were then used as input into the nj() tree-building function implemented in the APE package98 in R language.
We measured the distances between all pairs of species in the reconstructed NJ tree (i.e. the lengths of branches) using the get_distance() method implemented in the ETE3 toolkit for phylogenetic trees99. Our first measure of ‘tree violation’ is the difference between these distances and the distances between samples in the original matrix that was used to build the NJ tree.
Multispecies coalescent methods
We applied three different methods that attempt to reconstruct the species tree under the multispecies coalescent model. For a brief discussion of these approaches see Supplementary Methods.
For SNAPP36 we used a random subset of ~0.5% of genome-wide SNPs (48,922 SNPs) for 12 individuals representing the eco-morphological groups and the Lake Victoria outgroup P. nyererei whose alleles were filled in based on the whole genome alignment. The P. nyererei alleles were assigned as ‘ancestral’ (0 in the nexus input file). The ‘forward‘ and ‘backward‘ mutation rate parameters u and v were calculated directly from the data by SNAPP (the Calc mutation rates option). The default value 10 was used for the Coalescent rate parameter and the value of the parameter was sampled (estimated in the MCMC chain). We used uninformative priors as we don’t assume strong a priori knowledge about the parameters. The prior for ancestral population sizes was chosen to be a relatively broad gamma distribution with parameters α = 4 and β = 20. The tree height prior λ was set to the initial value of 100 but sampled in the MCMC chain with an uninformative uniform hyperprior on the interval [0,50000]. We ran three independent MCMC chains with the same starting parameters, each on 30 threads with a total runtime of over 10 CPU years. The first one million steps from each MCMC chain was discarded as burn-in. In total, more than 30 million MCMC steps were sampled in the three runs. For the MCMC traces for each run see Supplementary Fig. 24.
Next we used SVDquartets37,38 as implemented in PAUP* [v4.0a (build 159)]100. We prepared the data into the NEXUS ‘dna’ format, using evo with the getWGSeq --whole-genome --makeSVDinput -r options. This command outputs for each individual the DNA base at each variable site, randomly sampling one of the two alleles at heterozygous sites, and ignoring sites that become monomorphic due to this random sampling of alleles. The final dataset contained 17,833,187 SNPs. Then we ran SVDquartets in PAUP* setting outgroup to N. brichardi and then executing svdq evalq=all; specifying that all quartets should be evaluated (not just a random subset). In the final step, PAUP* version of the QFM algorithm101 is used to search for the overall tree that minimizes the number of quartets that are inconsistent with it.
Finally we used ASTRAL40 (v.5.6.1) with default parameters and the full set of 2543 local trees generated by RAxML (see above) as input.
Tree comparisons
To summarise the degree of (dis)agreement between the topologies of trees produced by different phylogenetic methods (Fig. 2c), we calculated the normalised Robinson-Foulds distances between pairs of trees102 using the RF.dist function from the phangorn103 package in R with the option normalize=TRUE.
Chromopainter and fineSTRUCTURE
Singleton SNPs were excluded using the bcftools v.1.1 -c 2:minor option, before exporting the remaining variants in the PLINK format91. The chromopainter v0.0.4 software47 was then run for the 201 largest genomic scaffolds on shapeit phased SNPs. Briefly, we created a uniform recombination map using the makeuniformrecfile.pl script, then estimated the effective population size (Ne) for a subsample of 20 individuals using the chromopainter inbuilt expectation-maximization procedure47, averaged over the 20 Ne values using the provided neaverage.pl script. The chromopainter program was then run for each scaffold independently, with the -a 0 0 option to run all individuals against all others. Results for individual scaffolds were combined using the chromocombine tool before running fineSTRUCTURE v0.0.5 with 1,000,000 burn in iterations, and 200,000 sample iterations, recording a sample every 1,000 iterations (options -x 1000000 -y 200000 -z 1000). Finally, the sample relationship tree was built with fineSTRUCTURE using the -m T option and 20,000 iterations.
The f-branch statistic
The f4-admixture ratio (f statistic) statistic was developed to estimate the proportion of introgressed material in an admixed population [see SOM18 in ref. 31, and fG in ref. 48]. However, when calculated for different subsets of samples within the same phylogeny, there are a very large number of highly correlated f values that are hard to interpret. To make the interpretation easier, we developed “f-branch” or fb(C): fb (C) = medianA [minB [f(A,B,C,O) ] ], where B are samples descending from branch b, and A are samples descending from the sister branch of b. The outgroup O was always N. brichardi. The fb(C) score provides for each branch b of a given phylogeny and each sample C a summary of excess allele sharing of branch b with sample C (Fig 3, Supplementary Figure 26). Each fb(C) score was also assigned an associated z-score to assess statistical significance Zb (C) = medianA [minB [Z(A,B,C,O) ] ]. Additional information on the f and fb(C) statistics, including detailed reasoning behind the design of fb(C), are in Supplementary Methods.
Geometric morphometric analyses
A total of 168 photographs were used to compare the gross body morphology of Astatotilapia calliptera to that of endemic Lake Malawi species and other East African Astatotilapia lineages (Supplementary Table 7). Coordinates for 17 homologous landmarks [following ref. 104] were collected using tpsDig2 v2.26105. After landmark digitization, analysis of shape variation was carried out in R (v3.3.2) using the package GeoMorph v3.0.2106. First a General Procrustes Analysis was applied to remove non-shape variation and shape data were corrected for allometric size effects by performing a regression of Procrustes coordinates (10,000 iterations). The resulting allometry corrected residuals were used in PCA.
Maps
Present day catchment boundary maps are based on ‘level 3’ detail of Hydro1K dataset from the US Geological Survey. We downloaded the watershed boundary data from the United Nations Environment Programme website (http://ede.grid.unep.ch) and processed it using the QGIS geographic information system software (http://www.qgis.org/en/site/).
Protein-coding gene annotations
We used the BROADMZ2 annotation generated by the cichlid genome project11 and removed overlapping transcripts using Jim Kent’s genePredSingleCover program. Genes whose annotated length in nucleotides was not divisible by three were discarded, as they typically had inaccuracies in annotation that would require manual curation (2495 out of 23698 genes). We also used the cichlid genome project11 assignment of homologs between the M. zebra genome reference and zebrafish (Danio rerio).
Coding sequence positive selection scan
We used evo with the getCodingSeq -H b --no-stats options to obtain the coding sequences for each allele and each gene. The excess of non-synonymous variation (δN−S) and the non-synonymous variation excess score (ΔN-S) were calculated on a per-gene basis as follows. Let NTS be the number of possible non-synonymous transitions and NTV the number of possible non-synonymous transversion between two sequences; analogously STS and STV represent possible synonymous differences. We do not specify the ancestral allele, and therefore consider it equally likely that allele i mutated into allele j or that allele j mutated to allele i. Then let Nd be the number of observed non-synonymous mutations and Sd the number of observed synonymous mutations. If there are more than one difference within a codon, all “mutation pathways” (i.e. the different orders in which mutations could have happened) have equal probabilities. When a particular allele contained a premature stop codon, the remainder of the sequence after the stop was excluded from the calculations.
Because the transition:transversion ratio in the Lake Malawi dataset was 1.73, and hence (because there are two possible transversions for each possible transition) the prior probability of each transition is 3.46 times that of each transversion, we account for the unequal probabilities of transitions and transversions in calculating the proportions of non-synonymous (pN) and of synonymous differences (pS) as follows:
The excess of non-synonymous variation (δN−S) is the average of pN − pS over pairwise sequence comparisons. Only between-species sequence comparisons are considered for the Lake Malawi dataset. We normalized the δN−S values in order to take into account the effect on the variance of this statistic introduced by differences in gene length and by sequence composition. To achieve this, we used the leave-one-out jackknife procedure across different pairwise comparisons for each gene, estimating the standard error. The non-synonymous variation excess score (ΔN-S) is then:
Note that because the sequences are related by a genealogy, there is a correlation structure between the pairwise comparisons. Therefore, the jackknife approach substantially underestimates the true standard error of δN−S and is used here simply as a normalization factor.
The null model shown in Fig. 5a was derived by splitting all the coding sequence into its constituent codons, and then randomly sampling these codons with replacement to build new sequences that matched the actual coding genes in their numbers and the length distribution. Then we calculated the ΔN−S scores, as we did for the actual genes and compared the two distributions. High positive values at the upper tail of the distribution are substantially overrepresented in the actual data when compared to a null model.
We also calculated the above statistics for random non-coding regions, matching the gene sequences in length. We used the bedtools v2.26.0107 shuffle command to permute the locations of exons along the chromosomes. Of the total length of all the permuted sequences, 98.4% was within the ‘accessible genome’ and outside of coding sequences (we required at least 95% in any of the permuted locations). The specific command was:
bedtools shuffle -chrom -I exons.bed -excl InaccessibleGenome_andExons.bed -f 0.05 -g chrom.sizes
Gene Ontology enrichment
Zebrafish has the most extensive functional gene annotation of any fish species, providing a basis for Gene Ontology (GO)108 term enrichment analysis. Gene Ontology (GO) enrichment for the genes that were candidates for being under positive selection (the top 5% of ΔN−S values) was calculated in R using the topGO v2.26.0 package109 from the Bioconductor project110. The GO hierarchical structure was obtained from the GO.db v3.4.0 annotation and linking zebrafish gene identifiers to GO terms was accomplished using the org.Dr.eg.db v3.4.0 annotation package. Genome-wide, between 9024 and 9353 genes had a GO annotation that could be used by topGO, the exact number depending on the GO category being assessed. The nodeSize parameter was set to 5 to remove GO terms which have fewer than 5 annotated genes, as suggested in the topGO manual.
There is often an overlap between gene sets annotated with different GO terms, in part because the terms are related to each other in a hierarchical structure108. This is partly accounted for by our use in topGO of the weight algorithm that accounts for the GO graph structure by down-weighing genes in the GO terms that are neighbours of the locally most significant terms in the GO graph60. All the p-values we report are from the weight algorithm, which the authors suggest should be reported without multiple testing correction109.
Some interdependency between significant GO terms remains after using the weight algorithm. Therefore, we used the Enrichment Map111 app for Cytoscape (http://www.cytoscape.org) to organize all the significantly enriched terms into networks where terms are connected if they have a high overlap, i.e. if they share many genes.
Diplotaxodon and deep benthic convergence
To obtain a quantitative measure of the similarity between and the extent of excess diversity in the Diplotaxodon and deep benthic amino acid sequences, we calculated simple statistics based on the proportions of non-synonymous differences (pN scores). Intuitively, the similarity score is high if Diplotaxodon and deep benthic jointly have higher pN then all the others, but are not very different from each other relative to how much diversity there is within Diplotaxodon and deep benthic.
Specifically, the similarity score s is calculated as follows:
and
where is the mean pN between Diplotaxodon jointly with deep benthic and all the other Lake Malawi species, is the mean pN between Diplotaxodon and deep benthic, and is the mean pN within Diplotaxodon and deep benthic. The jackknife normalization is analogous to the one used for ΔN-S and the mean is subtracted to center the statistic at zero.
The excess diversity score is high when the mean pN scores within Diplotaxodon and within deep benthic are high relative to the mean pN in the rest of the radiation. Specifically, the excess score ex is defined as:
where is the mean pN within Diplotaxodon, is the mean pN within deep benthic, and is the mean pN within the rest of the radiation.
Haplotype trees
To view the relationship between haplotypes for genes of interest, we translated nucleotide sequences to amino acid sequences and loaded these into Haplotype Viewer (http://www.cibiv.at/~greg/haploviewer). This software requires that a tree is loaded together with the sequences. Therefore, we inferred gene trees using RAxML v7.7.894 with the PROTGAMMADAYHOFFF model of substitution.
Local excess allele sharing between Diplotaxodon and deep benthic
We used an extension of the fd statistic48; this extension is referred to as fdM55. fdM is a conservative version of the f statistic that is particularly suited for analysis of small genomic windows48,55. For the gene scores shown in Fig. 6b, we calculated fdM (mbuna, deep benthic, Diplotaxodon, N. brichardi) for each gene in window from the transcription start site (TSS) to 10 kb into the gene. For the ‘along the genome’ plots, as shown in Fig. 6d and Supplementary Fig. 23, we used a product of two fdM statistics [fdM(shallow benthic, deep benthic, Diplotaxodon, N. brichardi) x fdM(Rhamphochromis, Diplotaxodon, deep benthic, N. brichardi)], an approach which we found to increase the local resolution. This score was calculated in sliding windows of 100 SNPs across a region of +- 100kb around the genes. Finally, we also calculated fdM (mbuna, deep benthic, Diplotaxodon, N. brichardi) separately for synonymous and non-synonymous mutations in each gene.
Supplementary Material
Acknowledgments
This work was supported by the Wellcome Trust (097677/Z/11/Z to MM, WT206194 and WT207492 for RD and HS), the Royal Society-Leverhulme Trust Africa Awards (AA100023 and AA130107 to MG and GFT) and the European Molecular Biology Organization (ALTF 456-2016 to MM). We want to thank the Sanger Institute sequencing core for DNA sequencing, Mingliu Du for DNA extractions, David Swofford and Michael Matschiner for advice on phylogenomic analyses, Walter Salzburger and Ian Wilson for comments on the manuscript. We also thank the Tanzania Fisheries Research Institute, and the Fisheries Research Unit of the Government of Malawi for their assistance and support.
Footnotes
Reporting Summary. Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Code availability. The majority of the custom code used in this project is available on Github as a part of the evo package (https://github.com/millanek/evo). All other custom codes are available from the authors upon request.
Data availability. All raw sequencing reads have been deposited to the NCBI Short Read Archive: (BioProjects PRJEB1254 and PRJEB15289). Sample accessions are listed in Supplementary Table 4. In addition, we are making whole-genome variant calls in the Variant Call Format (VCF), phylogenetic trees and protein coding sequence alignments, and tables with f4 statistics available through the Dryad Digital Repository (http://dx.doi.org/10.5061/dryad.7rj8k6c).
Author contributions:
EM, GFT, MJG, MM and RD devised the study. GFT and MJG collected the samples. AMT bred parent-offspring trios and performed geometric morphometric analyses. MM performed the DNA extractions. HS and MM analysed the genomic data. All authors participated in interpretation of the results. MM, HS and RD drafted the manuscript, and all others commented.
Competing interests
RD declares that he owns stock in Illumina from previous consulting. The authors declare no other competing interests.
References
- 1.Losos JB, Ricklefs RE. Adaptation and diversification on islands. Nature. 2009;457:830–836. doi: 10.1038/nature07893. [DOI] [PubMed] [Google Scholar]
- 2.Wagner CE, Harmon LJ, Seehausen O. Ecological opportunity and sexual selection together predict adaptive radiation. Nature. 2012;487:366–369. doi: 10.1038/nature11144. [DOI] [PubMed] [Google Scholar]
- 3.Berner D, Salzburger W. The genomics of organismal diversification illuminated by adaptive radiations. Trends Genet. 2015;31:491–499. doi: 10.1016/j.tig.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Darwin C. On the Origin of Species. OUP; Oxford: 2008. [Google Scholar]
- 5.Lamichhaney S, et al. Evolution of Darwin's finches and their beaks revealed by genome sequencing. Nature. 2015;518:371–375. doi: 10.1038/nature14181. [DOI] [PubMed] [Google Scholar]
- 6.Losos J, Jackman T, Larson A, Queiroz K, Rodriguez-Schettino L. Contingency and determinism in replicated adaptive radiations of island lizards. Science. 1998;279:2115–2118. doi: 10.1126/science.279.5359.2115. [DOI] [PubMed] [Google Scholar]
- 7.Fryer G, Iles TD. The cichlid fishes of the great lakes of Africa: their biology and evolution. Oliver and Boyd; 1972. [Google Scholar]
- 8.Salzburger W, Van Bocxlaer B, Cohen AS. Ecology and Evolution of the African Great Lakes and Their Faunas. Annu Rev Ecol Evol Syst. 2014;45:519–545. [Google Scholar]
- 9.Genner MJ, et al. How does the taxonomic status of allopatric populations influence species richness within African cichlid fish assemblages? Journal of Biogeography. 2004;31:93–102. [Google Scholar]
- 10.Meyer A. Phylogenetic relationships and evolutionary processes in East African cichlid fishes. Trends Ecol Evol. 1993;8:279–284. doi: 10.1016/0169-5347(93)90255-N. [DOI] [PubMed] [Google Scholar]
- 11.Brawand D, et al. The genomic substrate for adaptive radiation in African cichlid fish. Nature. 2014;513:375–381. doi: 10.1038/nature13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer BS, Matschiner M, Salzburger W. Disentangling incomplete lineage sorting and introgression to refine species-tree estimates for Lake Tanganyika cichlid fishes. Syst Biol. 2016 doi: 10.1093/sysbio/syw069. [DOI] [PubMed] [Google Scholar]
- 13.Meier JI, et al. Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat Commun. 2017;8 doi: 10.1038/ncomms14363. 14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koblmüller S, Egger B, Sturmbauer C, Sefc KM. Rapid radiation, ancient incomplete lineage sorting and ancient hybridization in the endemic Lake Tanganyika cichlid tribe Tropheini. Mol Phylogenet Evol. 2010;55:318–334. doi: 10.1016/j.ympev.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 15.Weiss JD, Cotterill FPD, Schliewen UK. Lake Tanganyika—A ‘Melting Pot’ of Ancient and Young Cichlid Lineages (Teleostei: Cichlidae)? PLoS ONE. 2015;10:e0125043. doi: 10.1371/journal.pone.0125043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gante HF, et al. Genomics of speciation and introgression in Princess cichlid fishes from Lake Tanganyika. Mol Ecol. 2016 doi: 10.1111/mec.13767. [DOI] [PubMed] [Google Scholar]
- 17.Wagner CE, et al. Genome-wide RAD sequence data provide unprecedented resolution of species boundaries and relationships in the Lake Victoria cichlid adaptive radiation. Mol Ecol. 2012;22:787–798. doi: 10.1111/mec.12023. [DOI] [PubMed] [Google Scholar]
- 18.Genner MJ, Turner GF. Ancient hybridization and phenotypic novelty within Lake Malawi's cichlid fish radiation. Mol Biol Evol. 2012;29:195–206. doi: 10.1093/molbev/msr183. [DOI] [PubMed] [Google Scholar]
- 19.Moran P, Kornfield I, Reinthal PN. Molecular Systematics and Radiation of the Haplochromine Cichlids (Teleostei: Perciformes) of Lake Malawi. Copeia. 1994;1994:274. [Google Scholar]
- 20.Joyce DA, et al. Repeated colonization and hybridization in Lake Malawi cichlids. 2011;21:R108–9. doi: 10.1016/j.cub.2010.11.029. [DOI] [PubMed] [Google Scholar]
- 21.Leffler EM, et al. Revisiting an old riddle: what determines genetic diversity levels within species? PLoS Biol. 2012;10:e1001388. doi: 10.1371/journal.pbio.1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Albertson RC, Markert JA, Danley PD, Kocher TD. Phylogeny of a rapidly evolving clade: the cichlid fishes of Lake Malawi, East Africa. Proc Natl Acad Sci USA. 1999;96:5107–5110. doi: 10.1073/pnas.96.9.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kocher TD. Adaptive evolution and explosive speciation: the cichlid fish model. Nat Rev Genet. 2004;5:288–298. doi: 10.1038/nrg1316. [DOI] [PubMed] [Google Scholar]
- 24.Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 25.Ravi V, Venkatesh B. Rapidly evolving fish genomes and teleost diversity. Curr Opin Genet Dev. 2008;18:544–550. doi: 10.1016/j.gde.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Recknagel H, Elmer KR, Meyer A. A hybrid genetic linkage map of two ecologically and morphologically divergent Midas cichlid fishes (Amphilophus spp.) obtained by massively parallel DNA sequencing (ddRADSeq) G3 (Bethesda) 2013;3:65–74. doi: 10.1534/g3.112.003897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ségurel L, Wyman MJ, Przeworski M. Determinants of mutation rate variation in the human germline. Annu Rev Genomics Hum Genet. 2014;15:47–70. doi: 10.1146/annurev-genom-031714-125740. [DOI] [PubMed] [Google Scholar]
- 28.Feng C, et al. Moderate nucleotide diversity in the Atlantic herring is associated with a low mutation rate. Elife. 2017;6:e23907. doi: 10.7554/eLife.23907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konings A. Malaŵi Cichlids in Their Natural Habitat. Cichlid Press; 2007. [Google Scholar]
- 30.1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durand EY, Patterson N, Reich D, Slatkin M. Testing for ancient admixture between closely related populations. Mol Biol Evol. 2011;28:2239–2252. doi: 10.1093/molbev/msr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heled J, Bouckaert RR. Looking for trees in the forest: summary tree from posterior samples. BMC Evol Biol. 2013;13:221. doi: 10.1186/1471-2148-13-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edwards SV. Is a new and general theory of molecular systematics emerging? Evolution. 2009;63:1–19. doi: 10.1111/j.1558-5646.2008.00549.x. [DOI] [PubMed] [Google Scholar]
- 35.Edwards SV, et al. Implementing and testing the multispecies coalescent model: A valuable paradigm for phylogenomics. Mol Phylogenet Evol. 2016;94:447–462. doi: 10.1016/j.ympev.2015.10.027. [DOI] [PubMed] [Google Scholar]
- 36.Bryant D, Bouckaert R, Felsenstein J, Rosenberg NA, RoyChoudhury A. Inferring species trees directly from biallelic genetic markers: bypassing gene trees in a full coalescent analysis. Mol Biol Evol. 2012;29:1917–1932. doi: 10.1093/molbev/mss086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chifman J, Kubatko L. Quartet Inference from SNP Data Under the Coalescent Model. Bioinformatics. 2014;30:3317–3324. doi: 10.1093/bioinformatics/btu530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chifman J, Kubatko L. Identifiability of the unrooted species tree topology under the coalescent model with time-reversible substitution processes, site-specific rate variation, and invariable sites. J Theor Biol. 2015;374:35–47. doi: 10.1016/j.jtbi.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 39.Long C, Kubatko L. The Effect of Gene Flow on Coalescent-Based Species-Tree Inference. arXiv.org. 2017:1–19. doi: 10.1093/sysbio/syy020. [DOI] [PubMed] [Google Scholar]
- 40.Mirarab S, et al. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics. 2014;30:i541–8. doi: 10.1093/bioinformatics/btu462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang C, Rabiee M, Sayyari E, Mirarab S. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics. 2018;19:153. doi: 10.1186/s12859-018-2129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dasarathy G, Nowak R, Roch S. Data Requirement for Phylogenetic Inference from Multiple Loci: A New Distance Method. IEEE/ACM Trans Comput Biol Bioinform. 2015;12:422–432. doi: 10.1109/TCBB.2014.2361685. [DOI] [PubMed] [Google Scholar]
- 43.Rusinko J, McPartlon M. Species tree estimation using Neighbor Joining. J Theor Biol. 2017;414:5–7. doi: 10.1016/j.jtbi.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 44.Ballard JWO, Whitlock MC. The incomplete natural history of mitochondria. Mol Ecol. 2004;13:729–744. doi: 10.1046/j.1365-294x.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- 45.Toews DPL, Brelsford A. The biogeography of mitochondrial and nuclear discordance in animals. Mol Ecol. 2012;21:3907–3930. doi: 10.1111/j.1365-294X.2012.05664.x. [DOI] [PubMed] [Google Scholar]
- 46.Consuegra S, John E, Verspoor E, de Leaniz CG. Patterns of natural selection acting on the mitochondrial genome of a locally adapted fish species. Genet Sel Evol. 2015;47:58. doi: 10.1186/s12711-015-0138-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lawson DJ, Hellenthal G, Myers S, Falush D. Inference of Population Structure using Dense Haplotype Data. PLoS Genet. 2012;8:e1002453. doi: 10.1371/journal.pgen.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin SH, Davey JW, Jiggins CD. Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol Biol Evol. 2015;32:244–257. doi: 10.1093/molbev/msu269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin SH, et al. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 2013;23:1817–1828. doi: 10.1101/gr.159426.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eccles DH, Trewavas E. Malawian Cichlid Fishes. Lake Fish Movies; 1989. [Google Scholar]
- 51.Eriksson A, Manica A. Effect of ancient population structure on the degree of polymorphism shared between modern human populations and ancient hominins. Proc Natl Acad Sci USA. 2012;109:13956–13960. doi: 10.1073/pnas.1200567109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peterson EN, Cline ME, Moore EC, Roberts NB, Roberts RB. Genetic sex determination in Astatotilapia calliptera, a prototype species for the Lake Malawi cichlid radiation. Naturwissenschaften. 2017;104:41. doi: 10.1007/s00114-017-1462-8. [DOI] [PubMed] [Google Scholar]
- 54.Genner MJ, Ngatunga BP, Mzighani S, Smith A, Turner GF. Geographical ancestry of Lake Malawi's cichlid fish diversity. Biol Lett. 2015;11 doi: 10.1098/rsbl.2015.0232. 20150232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Malinsky M, et al. Genomic islands of speciation separate cichlid ecomorphs in an East African crater lake. Science. 2015;350:1493–1498. doi: 10.1126/science.aac9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ivory SJ, et al. Environmental change explains cichlid adaptive radiation at Lake Malawi over the past 1.2 million years. Proceedings of the National Academy of Sciences. 2016;113:11895–11900. doi: 10.1073/pnas.1611028113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lyons RP, et al. Continuous 1.3-million-year record of East African hydroclimate, and implications for patterns of evolution and biodiversity. Proc Natl Acad Sci USA. 2015;112:15568–15573. doi: 10.1073/pnas.1512864112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Greenwood PH. Towards a phyletic classification of the ‘genus’ Haplochromis (Pisces, Cichlidae) and related taxa. Part 1. 1979;35:265–322. [Google Scholar]
- 59.Lippitsch E. A phyletic study on lacustrine haplochromine fishes (Perciformes, Cichlidae) of East Africa, based on scale and squamation characters. Journal of Fish Biology. 1993;42:903–946. [Google Scholar]
- 60.Alexa A, Rahnenführer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 61.Van Bocxlaer B, SCHULTHEIß R, Plisnier P-D, Albrecht C. Does the decline of gastropods in deep water herald ecosystem change in Lakes Malawi and Tanganyika? Freshwater Biology. 2012;57:1733–1744. [Google Scholar]
- 62.Spady TC, et al. Evolution of the cichlid visual palette through ontogenetic subfunctionalization of the opsin gene arrays. Mol Biol Evol. 2006;23:1538–1547. doi: 10.1093/molbev/msl014. [DOI] [PubMed] [Google Scholar]
- 63.Weadick CJ, Chang BSW. Complex patterns of divergence among green-sensitive (RH2a) African cichlid opsins revealed by Clade model analyses. BMC Evol Biol. 2012;12:206. doi: 10.1186/1471-2148-12-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sugawara T, et al. Parallelism of amino acid changes at the RH1 affecting spectral sensitivity among deep-water cichlids from Lakes Tanganyika and Malawi. Proc Natl Acad Sci USA. 2005;102:5448–5453. doi: 10.1073/pnas.0405302102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bowmaker JK, Hunt DM. Evolution of vertebrate visual pigments. Current Biology. 2006;16:R484–R489. doi: 10.1016/j.cub.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 66.Davies WIL, Collin SP, Hunt DM. Molecular ecology and adaptation of visual photopigments in craniates. Mol Ecol. 2012;21:3121–3158. doi: 10.1111/j.1365-294X.2012.05617.x. [DOI] [PubMed] [Google Scholar]
- 67.Carleton KL, Dalton BE, Escobar-Camacho D, Nandamuri SP. Proximate and ultimate causes of variable visual sensitivities: Insights from cichlid fish radiations. Genesis. 2016;54:299–325. doi: 10.1002/dvg.22940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ogawa Y, Shiraki T, Kojima D, Fukada Y. Homeobox transcription factor Six7 governs expression of green opsin genes in zebrafish. Proceedings of the Royal Society B: Biological Sciences. 2015;282 doi: 10.1098/rspb.2015.0659. 20150659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Renninger SL, Gesemann M, Neuhauss SCF. Cone arrestin confers cone vision of high temporal resolution in zebrafish larvae. Eur J Neurosci. 2011;33:658–667. doi: 10.1111/j.1460-9568.2010.07574.x. [DOI] [PubMed] [Google Scholar]
- 70.Brockerhoff SE, et al. Light stimulates a transducin-independent increase of cytoplasmic Ca2+ and suppression of current in cones from the zebrafish mutant nof. J Neurosci. 2003;23:470–480. doi: 10.1523/JNEUROSCI.23-02-00470.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boesze-Battaglia K, Goldberg AFX. Photoreceptor renewal: a role for peripherin/rds. Int Rev Cytol. 2002;217:183–225. doi: 10.1016/s0074-7696(02)17015-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Opazo JC, Butts GT, Nery MF, Storz JF, Hoffmann FG. Whole-genome duplication and the functional diversification of teleost fish hemoglobins. Mol Biol Evol. 2013;30:140–153. doi: 10.1093/molbev/mss212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hahn C, Genner MJ, Turner GF, Joyce DA. The genomic basis of cichlid fish adaptation within the deepwater ‘twilight zone’ of Lake Malawi. Evolution Letters. 2017;1:184–198. doi: 10.1002/evl3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heliconius Genome Consortium. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487:94–98. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meyer A, Kocher TD, Basasibwaki P, Wilson AC. Monophyletic origin of Lake Victoria cichlid fishes suggested by mitochondrial DNA sequences. Nature. 1990;347:550–553. doi: 10.1038/347550a0. [DOI] [PubMed] [Google Scholar]
- 76.Reid NM, et al. The genomic landscape of rapid repeated evolutionary adaptation to toxic pollution in wild fish. Science. 2016;354:1305–1308. doi: 10.1126/science.aah4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jones FC, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seehausen O. Cichlid Fish Diversity Threatened by Eutrophication That Curbs Sexual Selection. Science. 1997;277:1808–1811. [Google Scholar]
- 79.Konings A. Tanganyika cichlids in their natural habitat. Cichlid Press; 2015. [Google Scholar]
- 80.Coyne JA, Orr HA. Speciation. 2004.
- 81.Feder JL, Egan SP, Nosil P. The genomics of speciation-with-gene-flow. Trends in Genetics. 2012;28:342–350. doi: 10.1016/j.tig.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 82.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv.org. 2013 q-bio.GN. [Google Scholar]
- 83.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 87.Delaneau O, Howie B, Cox AJ, Zagury J-F, Marchini J. Haplotype estimation using sequencing reads. Am J Hum Genet. 2013;93:687–696. doi: 10.1016/j.ajhg.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miller W, et al. 28-way vertebrate alignment and conservation track in the UCSC Genome Browser. Genome Res. 2007;17:1797–1808. doi: 10.1101/gr.6761107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ulm K. A simple method to calculate the confidence interval of a standardized mortality ratio (SMR) Am J Epidemiol. 1990;131:373–375. doi: 10.1093/oxfordjournals.aje.a115507. [DOI] [PubMed] [Google Scholar]
- 90.Dobson AJ, Kuulasmaa K, Eberle E, Scherer J. Confidence intervals for weighted sums of Poisson parameters. Stat Med. 1991;10:457–462. doi: 10.1002/sim.4780100317. [DOI] [PubMed] [Google Scholar]
- 91.Purcell S, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. The American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190–e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bhatia G, Patterson N, Sankararaman S, Price AL. Estimating and interpreting FST: The impact of rare variants. Genome Res. 2013;23:1514–1521. doi: 10.1101/gr.154831.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 95.Bouckaert R, et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 97.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 98.Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 99.Huerta-Cepas J, Serra F, Bork P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol Biol Evol. 2016;33:1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Swofford DL. PAUP*: Phylogenetic analysis using parsimony (and other methods) Sinauer Associates; 2002. [Google Scholar]
- 101.Reaz R, Bayzid MS, Rahman MS. Accurate Phylogenetic Tree Reconstruction from Quartets: A Heuristic Approach. PLoS ONE. 9:e104008. doi: 10.1371/journal.pone.0104008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Robinson DF, Foulds LR. Comparison of phylogenetic trees. Mathematical Biosciences. 1981 [Google Scholar]
- 103.Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Theis A, Ronco F, Indermaur A, Salzburger W, Egger B. Adaptive divergence between lake and stream populations of an East African cichlid fish. Mol Ecol. 2014;23:5304–5322. doi: 10.1111/mec.12939. [DOI] [PubMed] [Google Scholar]
- 105.Rohlf FJ. tpsDig2
- 106.Adams DC, Castillo EO. geomorph: an R package for the collection and analysis of geometric morphometric shape data. Methods in Ecology and …. 2013 [Google Scholar]
- 107.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Alexa A, Rahnenfuhrer J. topGO: enrichment analysis for gene ontology. R package version. 2010 [Google Scholar]
- 110.Huber W, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS ONE. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.