Abstract
We investigated the effect of aging on the multi-dimensional characteristics and heterogeneity of human peripheral CD8+ T cells defined by the expression of a set of molecules at the single cell level using the recently developed mass cytometry or Cytometry by Time-Of-Flight (CyTOF) and computational algorithms. CD8+ T cells of young and older adults had differential expression of molecules, especially those related to cell activation and migration, permitting the clustering of young and older adults through an unbiased approach. The changes in the expression of individual molecules were collectively reflected in the altered high-dimensional profiles of CD8+ T cells in older adults as visualized by the dimensionality reduction analysis tools principal component analysis (PCA) and t-distributed stochastic neighbor embedding (t-SNE). A combination of PhenoGraph clustering and t-SNE analysis revealed heterogeneous subsets of CD8+ T cells that altered with aging. Furthermore, intermolecular quantitative relationships in CD8+ T cells appeared to change with age as determined by the computational algorithm conditional-Density Resampled Estimate of Mutual Information (DREMI). The results of our study showed that heterogeneity, multidimensional characteristics, and intermolecular quantitative relationships in human CD8+ T cells altered with age, distinctively clustering young and older adults through an unbiased approach.
Keywords: Aging, Human CD8+ T cells, Mass cytometry, High-dimensional analysis, Heterogeneity
1. Introduction
Age-associated changes occur in the immune system [1–6], T cells play a major role in host defense and inflammatory diseases. In human T cells, probably the most prominent change with age is found in the proportions of naive and memory CD8+ T cells. With age, the frequency of naive CD8+ T cells decreases while the frequency of memory CD8+ T cells increases in human peripheral blood [7–9]. Other age-associated alterations in CD8+ T cells include impaired functions like cell proliferation, increased expression of the senescent marker CD57, and a decrease in the expression of the lymphoid tissue homing chemokine receptor CCR7 [10, 11]. Also, CD8+ T cells had decreased gene expression of the naive cell marker CD27 and the cell adhesion molecule Sell (CD62L) in humans with age as determined by gene expression array [12].
Mass cytometry or Cytometry by Time-Of-Flight (CyTOF), which uses heavy metal ions and mass spectrometry as labels and a readout, respectively, is a recently developed single-cell cytometry technique that can determine high-dimensional cellular profiles [13–15]. CyTOF can discriminate isotopes of different atomic weights with high accuracy, which allows for the measuring of 40+ molecules in a single tube [16–18]. The complexity of data generated by CyTOF requires high-dimensional data analytic approaches since multi-parameter data sets containing more than 20 molecules can generate several hundreds of two-dimensional dot plots [14]. Indeed, CyTOF data can be analyzed to show high-dimensional relationships of individual molecules by applying computational methods such as t-distributed Stochastic Neighbor Embedding (t-SNE) and Principal Component Analysis (PCA) [13, 14]. In combination with the PhenoGraph clustering analysis, the t-SNE can robustly identify cell populations with distinct traits [19, 20]. In addition to dimensionality reduction tools, the strength of molecular relationship can be quantified and visualized by conditional-Density Resampled Estimate of Mutual Information (DREMI) and conditional-Density Rescaled Visualization (DREVI), respectively [21], In conjunction with CyTOF, these data analytic tools enhance our ability to evaluate complex cellular traits in human immune cells.
Here we investigated whether aging could affect high dimensional profiles of human peripheral CD8+ T cells as defined by the expression of a set of molecules at the single cell level in young and older adults using CyTOF and algorithmic data analytic tools including t-SNE, PC A, PhenoGraph clustering and DREMI. The results of our study showed that heterogeneity, multidimensional characteristics, and intermolecular quantitative relationships in human CD8+ T cells altered with age, permitting the clustering of young and older adults distinctively through an unbiased approach.
2. Material and methods
2.1. Human Subjects
Healthy young subjects 35 years of age or young (n = 17) and healthy older subjects 65 years of age (n = 11) or older and were recruited for this study (mean age ± SD, 25.8 years ± 2.2 and 73.4 years ± 5.8, respectively, Table S1 supplement). The gender distribution was not different between the two groups (F:M, 9:8 and 4:7, respectively for young and older adult groups, P = 0.4601 by Fisher’s exact test). Individuals on immunosuppressive drugs or had a medical condition potentially affecting the immune system, including cancer and autoimmunity were excluded [22–27]. Informed consent was obtained from all subjects. This work was approved by the institutional review committee of Yale University.
2.2. CyTOF analysis
All mass cytometry reagents were purchased from Fluidigm, Inc (South San Francisco, CA) unless otherwise stated. Peripheral blood mononuclear cells (PBMCs) were prepared from blood on FicollPAQUE gradients. PBMCs (2 × 106) were stained with a panel of metal-tagged antibodies (Table S2 supplement) followed by Cisplatin staining. After fixing cells with Maxpar Fix 1 buffer, stained cells were washed and kept overnight in the MaxPar Fix & Perm Buffer containing intercalator-Ir. Cells were resuspended with MaxPar Water containing EQ Four Element Calibration Beads and acquired on a CyTOF system Helios (Fluidigm). All FCS files were normalized and analyzed using the analytic tool CYT, an open source analytic tool for CyTOF data, and FlowJo software (FlowJo, LLC) (Fig. S1 and S2) [19, 28]. The FCS files were transformed using an inverse hyperbolic sine (arcsinh) function with a cofactor of 5 and pre-gated manually to exclude EQ beads, cell debris, cell doublets and dead cells before additional analysis [18]. t-SNE, PCA, PhenoGraph, DREMI and DREMI were performed on gated cells (3,500 cells) [20, 21, 29]. We set the number of neighbors to use (k parameter) in PhenoGraph clustering at 30, the default number in the CYT program. For metaclustering, the k parameter was set at 10. The numbers of cell subsets clustered at different k parameters supported the relatively stable clustering patterns at different k parameters (data not shown). Unbiased hierarchical clustering analysis was done on z-score converted geometric mean mental intensities (GMMI) of individual molecules, frequencies of metaclusters (MC) and DREMI scores using R software.
2.3. Statistical analysis.
The unpaired t-test was used to compare GMMI, frequency of metaclusters, and DREMI scores between young and older adults. Multiple comparisons were controlled with the two-stage step-up method of Benjamini, Krieger and Yekutieli, with a false discovery rate (Q) of 5% [30]. The statistical analysis was completed using GraphPad Prism 7.01.
3. Results
3.1. CD8+ T cells of young and older adults have differential expression of a set of molecules related to cell activation and migration.
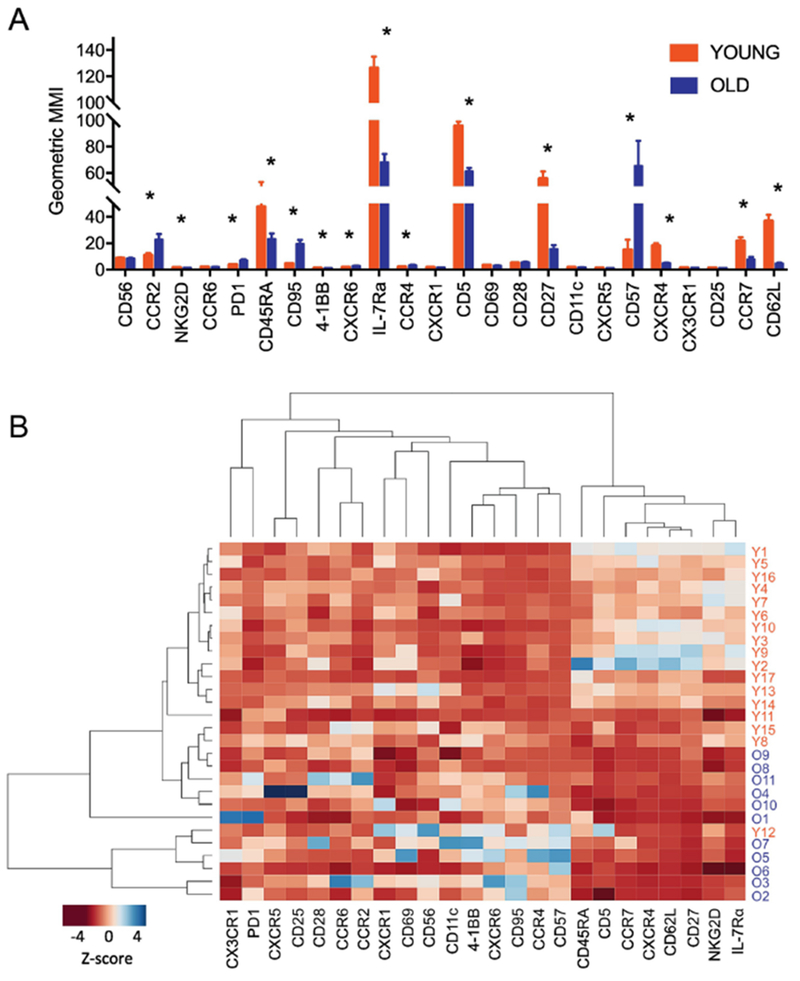
We analyzed the expression of a group of 24 molecules by CD8+ T cells related to cell activation and migration in young (age ≤ 35) and older (age ≥ 65) adults using CyTOF as any differential expression of such molecules could affect cell function and subsequent immune response. Also, some of these molecules can be utilized to define naïve and memory T cell subsets. The expressional intensities of individual molecules by CD8+ T cells were diverse as determined by geometric mean metal intensity (GMMI, Fig. 1A ) because metals could have different channel sensitivities like in fluorescent flow cytometry. Of note, the GMMI of some molecules were different between young and older adults, indicating altered expression of such molecules by human CD8+ T cells with age. These molecules include CD45RA, CCR7, CD62L and CD27, which are markers for naive and memory CD8+ T cells, as well as the replication senescent marker CD57, the death receptor CD95 (Fas receptor), the NK cell receptor NKG2D, and the pro-survival cytokine receptor IL-7Rα. Indeed, these findings are consistent with the results of previous studies on human CD8+ T cells in young and older adults using flow cytometry[10–12, 22, 31, 32], Furthermore, we first noticed differential expression of the immune checkpoint molecule PD-1 and the T cell receptor (TCR) signaling regulator CD5 as well as the chemokine receptors CCR2, CXCR6, CCR4 and CXCR4 by human CD8+ T cells at the protein level in young and older adults. The unbiased hierarchical clustering analysis showed the grouping of the measured molecules based on the expression levels in young and older adults (Fig. 1B). For instance, IL-7Rα, NKG2D, CD27, CD62L, CXCR4, CCR7, CD5 and CD45RA were clustered together with higher expression levels in young adults than in older adults. Also, the differential expression of such analyzed molecules by CD8+ T cells permitted clustering of young and older adults in an unbiased manner (Fig. 1B).
Fig.1. A set of molecules expressed by CD8+ T cells alter with age, resulting in the clustering of young and older adults by an unbiased analysis.
PBMCs of young and older adults were stained with antibodies to a set of molecules (Table S2) and run on a Helios CyTOF instrument. The acquired data were normalized and analyzed using FlowJo software. (A) Geometric mean metal expression intensities (GMMI) of individual molecules in CD8+ T cells of young and older adults (n = 17 and 11, respectively). *q < 0.05 by the t-test with multiple comparison control (false discovery rate or FDR, 5%). Bars and error bars indicate mean and standard error of mean (SEM). (B) Heatmap showing the results of an unbiased hierarchical clustering analysis of molecule and studied subjects based on the z-scores of GMMI of individual molecules.
3.2. High-dimensional analysis reveals heterogeneous subsets of CD8+ T cells that alter with aging.
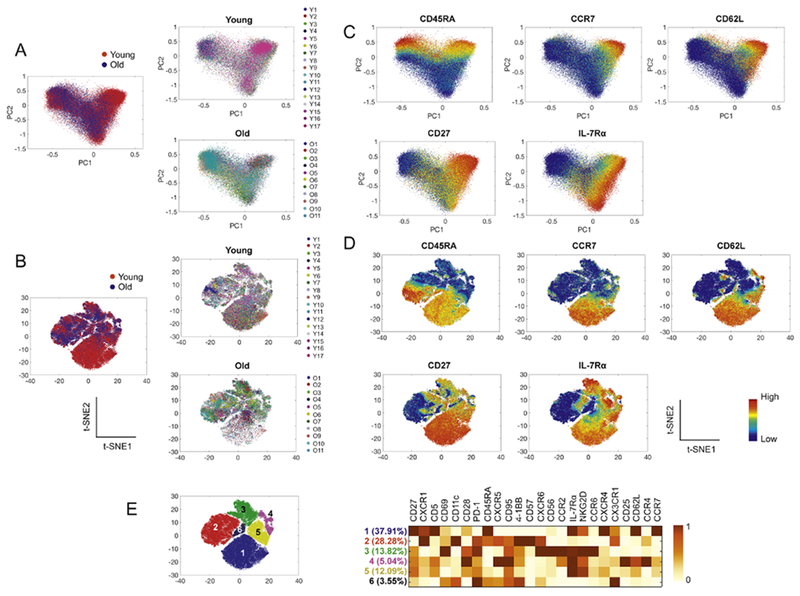
We explored whether the multiple molecules analyzed in our study could define the heterogeneity of CD8+ T cells that alters with aging using PCA, t-SNE and PhenoGraph algorithms. PCA is a linear dimensionality reduction technique while t-SNE is a nonlinear dimensionality-reduction tool. PCA and t-SNE can visualize relationships in multidimensional data [13, 14], We first applied both algorithms to the CyTOF data of 17 young and 11 older adults based on the expression of 24 molecules listed in Fig. 1A. The landscapes of intercellular relationships of CD8+ T cells visualized by both algorithms appear different between the two groups (Fig. 2A, B), suggesting an alteration in the characteristics of CD8+ T cells with aging as defined by the 24 analyzed molecules. The expression levels of some molecules by distinct regions on the PCA and t-SNE plots were different (Fig. 2C, D). Of note, the lowest region referred to as region 1 on the t-SNE plot in Fig. 2E contained fewer cells with age (Fig. 2B). This population expressed high levels of CCR7, CD62L and CD45RA, indicating the characteristics of naïve CD8+ T cells while cells in region 3 expressed low levels of these molecules, which were consistent with CD45RA− effector memory CD8+ T cells (Fig. 2E). Region 2 expressed low levels of CCR7 and CD62L but high levels of CD45RA, which were CD45RA+ effector memory CD8+ T cells. Regions 4 and 5 contained cells possessing characteristics of central memory CD8+ T cells with high levels of CCR7 and CD62 but low levels of CD45RA. The results of our dimensionality reduction analysis on CD8+ T cells, as visualized by the landscape patterns of these cells on PCA and t-SNE plots, support that multidimensional characteristics of CD8+ T cells alter with age.
Fig. 2. Multidimensional characteristics of CD8+ T cells alter with age as visualized by the dimensionality reduction analysis tools PCA and t-SNE.
CD8+ T cells gated from the acquired CyTOF data in Fig 1 were analyzed using PCA and t-SNE. (A-B) The relationships of CD8+ T cells from 17 young and 11 older adults were visualized on PCA (A) and t-SNE (B) plots based on the expression of 24 molecules (see Fig. 1A). (C-D) The expression of the indicated molecules on PCA and t-SNE plots is shown. (E) Heatmap shows the mean expression levels of individual molecules in the cell subsets indicated by the numbers in the t-SNE plot.
3.3. A combination of PhenoGraph and metaclustering approaches showed CD8+ T cell subsets that altered with aging.
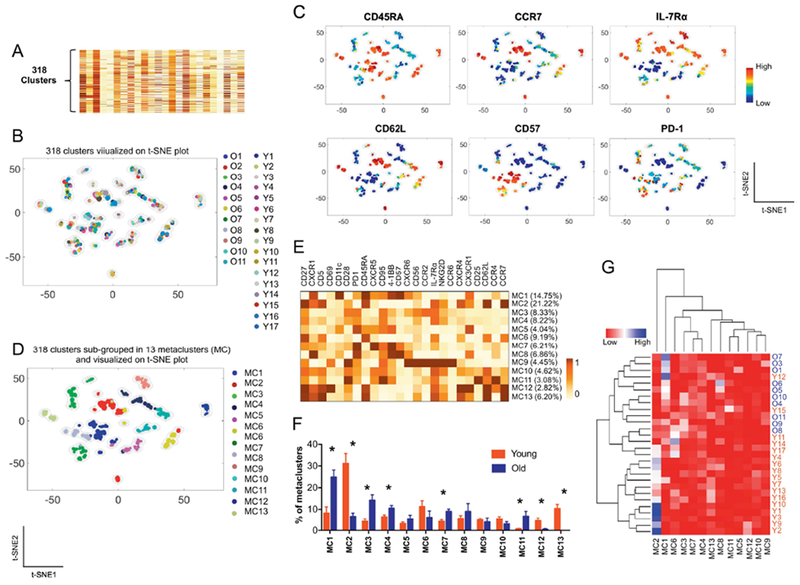
The PhenoGraph clustering analysis can robustly identify subsets of cells with distinct characteristics which can be visualized on a t-SNE plot [19, 20]. Using the PhenoGraph clustering, we interrogated the characteristics of CD8+ T cell populations that altered with aging. A total of 318 clusters were identified by the PhenoGraph clustering on CD8+ T cells of young and older adults based on the expression levels of the 24 analyzed molecules (Fig. 3A). The 318 PhenoGraph clusters were landscaped on a global t-SNE plot (Fig. 3B) where each subset and its size were indicated by a single point scaled to represent its proportion of the total 318 subsets. Although the donors of these cell subsets were different, some subsets were closely located on the global t-SNE plot, supporting the similarities of such subsets. Indeed, the closely located subsets had similar expression levels of the analyzed molecules (Fig. 3C).
Fig. 3. A combination of PhenoGraph and metaclustering analysis shows CD8+ T cell subsets that altered with aging.
(A-E) The PhenoGraph clustering was performed on CD8+ T cells from 17 young and older adults based on the expression of 24 molecules (Fig 1A), dividing them into 318 subpopulations (A). Subsequently, metaclustering or secondary clustering analysis was done on the 318 subpopulations (D). t-SNE plots showing a landscape of the 318 subpopulations and their relationships to (B) individual subjects, (C) expression levels of the indicated molecules and (D) metaclusters (MC). (E) Heatmap showing the percentages of metaclusters (MC) (Y-axis) and mean expression levels of 24 molecules by individual metaclusters (X-axis). (F) The frequency of different metaclusters (MC) between young and older adults. *q < 0.05 by the t-test with multiple comparison control (false discovery rate, 5%). (G) Heatmap showing the results of an unbiased hierarchical clustering analysis of metaclusters and studied subjects based on the frequency of metaclusters in individual subjects.
We employed a metaclustering approach to merge the 318 subsets of CD8+ T cells from the young and older subjects into a set of secondary clusters or metaclusters to compare the characteristics of the 318 PhenoGraph clusters of CD8+T cells from young and older adults [20, 33], Thirteen metaclusters were identified and visualized on a global t-SNE plot (Fig. 3D). The expression levels of analyzed molecules were different among the metaclusters (Fig. 3E). The proportions of individual metaclusters in CD8+ T cells varied among the studied subjects (Fig. S3 supplement). The proportions of some metaclusters were different between young and older adults (Fig. 3F). Metacluster 1, which was higher in older adults, contained cells with low levels of CD27, CD28, CCR7, CD62L and IL-7Rα but high levels of CD45RA, CD57 and 4-1BB. This indicates that CD8+ T cells in metacluster 1 were those with the characteristics of CD45RA+ effector memory CD8+ T cells with low levels of IL-7Rα (Fig. 3E). These cells also expressed high levels of the chemokine receptors CXCR1 and CX3CR1 as well as the integrin subunit CD1 lc which was reported to be expressed by activated human CD8+ T cells [34], Metaclusters 3 and 4 also expanded in older adults. The expression patterns of the effector memory cell markers CCR7 and CD62L were similar in metaclusters 1, 3 and 4 although the latter two had increased IL-7Rα expression and decreased CD45RA expression compared to metacluster 1. Metacluster 7 had memory CD8+ T cells with low levels of CCR7 and CD62L and high levels of PD-1 expanded in older adults. In contrast, young adults had a higher frequency of metacluster 2, which had high levels of CD27, CCR7, CD62L and IL-7Rα but low levels of 4-1BB and CD57, possess the characteristics of naïve CD8+ T cells. Also, metacluster 2 had high level of CD5 expression. Metaclusters 12 and 13 expanded in young adults. The expression profiles of metaclusters 12 and 13 are largely like those of metacluster 1 except for the expression of CD69 at high levels in metacluster 12 and CD56 at moderate levels in metacluster 13. We next performed cluster analysis based on the frequency of individual metaclusters in young and older adults, showing the clustering of most young and older adults (Fig. 3G). Overall, these findings indicate that heterogeneous subsets of human CD8+ T cells can be identified by high-dimensional CyTOF data analysis and that the frequency of some cell subsets alter with age.
3.4. The quantitative relationships of molecules in human CD8+ T cells alter with aging as determined by the computational algorithm DREMI.
We explored whether aging affected quantitative relationships between molecules at the single cell level using the computational algorithms DREMI and DREVI [21]. DREMI computes mutual information of how the state (e.g. abundance) of Y changes with different states of X [21]. The strength of the statistical dependency between two molecules is shown by DREMI scores. The function underlying this molecular interaction can be visualized by DREVI [21] as shown in Fig. 4A where the expression of CD27 increased proportionately with the expression of CD5. We computed DREMI scores among the 24 molecules we measured, which yielded scores for a total of 552 possible molecular combinations in individual subjects. We next performed an unbiased clustering analysis on the DREMI scores of these combinations, which showed clustering of young and older adults based on the DREMI scores (Fig. 4B). This finding suggests the possible alteration in the expressional relationship of molecules in human CD8+ T cells with age as determined by the DREMI algorithm. Of the 552 combinations, we selected 187 molecular combinations that had mean DREMI scores of 0.1 or greater, which we considered meaningful, in young and older adults. Indeed, the DREMI scores of some of the 187 molecular combinations were lower or higher in older adults compared to young adults (Fig. 4C, q < 0.05, false discovery rate or FDR = 5%). Most molecules involved in the DREMI combinations that changed in older adults included those with altered expression by CD8+ T cells with aging (Fig. 1A). Although the biological significance and potential mechanism of these findings still need to be elucidated, our observations indicate the effect of aging on the intermolecular quantitative relationships in human CD8+ T cells.
Fig. 4. The intermolecular quantitative relationships in human CD8+ T cells alter with age as determined by the computational algorithm conditional-Density Resampled Estimate of Mutual Information (DREMI).
Pre-gated CD8+ T cells from CyTOF data of young (Y, n = 17) and older adults (O, n = 11) were analyzed using DREMI algorithm. (A) Representative data for the quantitative relationships of the indicated molecules as determined and visualized by DREMI and conditional-Density Rescaled Visualization (DREVI), respectively. The strength of the statistical dependency between two molecules (CD5-CD27; CD57-CD27) is shown by DREMI scores. (B) DREMI scores for 552 possible combinations of two molecules from the 24 analyzed molecules (Fig 1A) were subjected to unbiased hierarchal clustering using R program. (C) The molecular combinations with DREMI score ≥0.1 and q < 0.05 (t-test with multiple comparison, false discovery rate 5%) between young and older adults are shown. Bars and error bars indicate mean and SEM.
4. Discussion
Here we investigated whether aging could affect multi-dimensional characteristics and the heterogeneity of CD8+ T cells as defined by the expression of a set of molecules at the single cell level in young and older adults using CyTOF and algorithmic data analytic tools. We found that CD8+ T cells of young and older adults had differential expression of molecules, especially ones related to cell activation and migration, permitting the clustering of young and older adults by an unbiased approach. The changes in the expression of individual molecules by CD8+ T cells with age were collectively reflected in the altered multidimensional characteristics of CD8+ T cells in older adults as visualized by the dimensionality reduction analysis tools PCA and t-SNE. A combination of high-dimensional PhenoGraph clustering and t-SNE analysis revealed heterogeneous subsets of CD8+ T cells that altered with aging. Furthermore, intermolecular quantitative relationships in CD8+ T cells appeared to change with age as determined by the computational algorithm DREMI. Overall, the results of our study show that heterogeneity, multidimensional characteristics, and intermolecular quantitative relationships in human CD8+ T cells alter with age.
We analyzed the expression of a group of 24 molecules related to cell activation and migration by CD8+ T cells in young and older adults using CyTOF. In comparison to the expression levels of individual molecules between the two groups, older adults had altered expression levels of molecules, including CD45RA, CCR7, CD62L and CD27, by CD8+ T cells as determined by GMMI. This finding is consistent with the results of previous studies reporting an alteration in the frequency of naïve and memory CD8+ T cells with age. Indeed, our PhenoGraph clustering analysis showed the expansion of CD8+ T cell subsets with the memory cell phenotype in older adults while CD8+ T cell subsets with the naive cell phenotype decreased. Of note, we noticed differential expression of the immune checkpoint molecule PD-1 and the TCR signaling regulator CD5 as well as the chemokine receptors CCR2, CXCR6, CCR4 and CXCR4 by human CD8+ T cells between young and older adults. These findings suggest an age-associated alteration in CD8+ T cell activation and migration through these molecules. CD8+ T cell subsets with high levels of CCR2, CXCR6 and CCR4 had the memory phenotype as shown in Fig 3E. However, the subsets expressing these chemokine receptors were largely distinct as shown by our metaclustering approach, which further supports the heterogeneity of memory CD8+ T cells.
Age-associated alterations in the expression of molecules by CD8+ T cells are well known. However, it is unknown whether aging affects the intermolecular quantitative relationship in human CD8+ T cells. Our DREMI analysis on CD8+ T cells showed that the quantitative relationships of molecules, especially ones related to cell activation, survival, and migration altered with age. The mechanism underlying this phenomenon and its biological significance are yet to be demonstrated. A possible explanation could be an age-associated alteration in an upstream mechanism regulating cell differentiation and expression of multiple molecules. Our unbiased hierarchical clustering approach revealed that the expression levels of the 24 analyzed molecules in CD8+ T cells as determined by GMMI could cluster young and older subjects. By taking a similar approach, we also showed clustering of young and older subjects based on the frequency of metaclusters as well as the intermolecular DREMI scores. Our approach of combining high-dimensional CyTOF data analysis with unbiased hierarchical clustering provides an illustration which can be utilized in analyzing and comparing high-dimensional characteristics and heterogeneity of cells in different groups.
In summary, we investigated the effect of aging on the high-dimensional cellular characteristics of CD8+ T cells at the single cell level in young and older adults using CyTOF and algorithmic data analytic tools including t-SNE, PCA, PhenoGraph and DREMI along with an unbiased hierarchical clustering approach. To the best of our knowledge, no similar analytic approach was taken to investigate the cellular characteristics of CD8+ T cells in young and older adults. This unique approach robustly identified age-associated changes in heterogeneity, multidimensional characteristics, and intermolecular quantitative relationships in human CD8+ T cells, which permitted the distinct clustering of studied subjects into young and older adults.
Supplementary Material
Highlights.
High-dimensional profiles of CD8+ T cells alter with age.
Intermolecular quantitative relationships in CD8+ T cells change with age.
Such changes permit clustering of young and older adults by an unbiased approach.
Acknowledgments
The authors thank Dr. Ala Nassar and Ms. Shelly Ren of the Yale CyTOF Core.
Funding information
This work was supported in part by grants from the National Institutes of Health (2R56AG0280691, 1R01AG056728, and R21AI126604 to IK; K24 AG042489 to ACS; and R01 AG055362 to IK and ACS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare no competing financial interests.
References
- [1].Shaw AC, Joshi S, Greenwood H, Panda A, Lord JM, Aging of the innate immune system, Curr Opin Immunol, 22 (2010) 507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Dorshkind K, Montecino-Rodriguez E, Signer RA, The ageing immune system: is it ever too old to become young again?, Nat Rev Immunol, 9 (2009) 57–62. [DOI] [PubMed] [Google Scholar]
- [3].Nikolich-Zugich J, Aging of the T Cell Compartment in Mice and Humans: From No Naïve Expectations to Foggy Memories, J Immunol, 193 (2014) 2622–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Nikolich-Zugich J, The twilight of immunity: emerging concepts in aging of the immune system, Nat Immunol, 19 (2018) 10–19. [DOI] [PubMed] [Google Scholar]
- [5].Haberthur K, Engelman F, Barron A, Messaoudi I, Immune senescence in aged nonhuman primates, Exp Gerontol, 45 (2010) 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Goronzy JJ, Weyand CM, Successful and Maladaptive T Cell Aging, Immunity, 46 (2017) 364–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Callahan JE, Kappler JW, Marrack P, Unexpected expansions of CD8-bearing cells in old mice, J Immunol, 151 (1993) 6657–6669. [PubMed] [Google Scholar]
- [8].Posnett DN, Sinha R, Kabak S, Russo C, Clonal populations of T cells in normal elderly humans: the T cell equivalent to “benign monoclonal gammapathy” [published erratum appears in J Exp Med 1994 Mar 1;179(3):1077], J Exp Med, 179 (1994) 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hong MS, Dan JM, Choi JY, Kang I, Age-associated changes in the frequency of naive, memory and effector CD8+ T cells, Mech Ageing Dev, 125 (2004) 615–618. [DOI] [PubMed] [Google Scholar]
- [10].Effros RB, Cai Z, Linton PJ, CD8 T cells and aging, Crit Rev Immunol, 23 (2003) 45–64. [DOI] [PubMed] [Google Scholar]
- [11].Clambey ET, van Dyk LF, Kappler JW, Marrack P, Non-malignant clonal expansions of CD8+ memory T cells in aged individuals, Immunol Rev, 205 (2005) 170–189. [DOI] [PubMed] [Google Scholar]
- [12].Cao JN, Gollapudi S, Sharman EH, Jia Z, Gupta S, Age-related alterations of gene expression patterns in human CD8+ T cells, Aging Cell, 9 (2010) 19–31. [DOI] [PubMed] [Google Scholar]
- [13].Saeys Y, Gassen SV, Lambrecht BN, Computational flow cytometry: helping to make sense of high-dimensional immunology data, Nat Rev Immunol, 16 (2016) 449–462. [DOI] [PubMed] [Google Scholar]
- [14].Chester C, Maecker HT, Algorithmic Tools for Mining High-Dimensional Cytometry Data, J Immunol, 195 (2015) 773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Spitzer MH, Nolan GP, Mass Cytometry: Single Cells, Many Features, Cell, 165 (2016) 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, Pavlov S, Vorobiev S, Dick JE, Tanner SD, Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry, Anal Chem, 81 (2009) 6813–6822. [DOI] [PubMed] [Google Scholar]
- [17].Ornatsky O, Bandura D, Baranov V, Nitz M, Winnik MA, Tanner S, Highly multiparametric analysis by mass cytometry, J Immunol Methods, 361 (2010) 1–20. [DOI] [PubMed] [Google Scholar]
- [18].Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, Balderas RS, Plevritis SK, Sachs K, Pe’er D, Tanner SD, Nolan GP, Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum, Science, 332 (2011) 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Amir el AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP, Pe’er D, viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia, Nat Biotechnol, 31 (2013) 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, Finck R, Gedman AL, Radtke I, Downing JR, Pe’er D, Nolan GP, Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis, Cell, 162 (2015) 184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Krishnaswamy S, Spitzer MH, Mingueneau M, Bendall SC, Litvin O, Stone E, Pe’er D, Nolan GP, Systems biology. Conditional density-based analysis of T cell signaling in single-cell data, Science, 346 (2014) 1250689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kim HR, Hong MS, Dan JM, Kang I, Altered IL-7R{alpha} expression with aging and the potential implications of IL-7 therapy on CD8+ T-cell immune responses, Blood, 107 (2006) 2855–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kang I, Hong MS, Nolasco H, Park SH, Dan JM, Choi JY, Craft J, Age-associated change in the frequency of memory CD4+ T cells impairs long term CD4+ T cell responses to influenza vaccine, J Immunol, 173 (2004) 673–681. [DOI] [PubMed] [Google Scholar]
- [24].Hwang KA, Kim HR, Kang I, Aging and human CD4(+) regulatory T cells, Mech Ageing Dev, 130 (2009) 509–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lee JS, Lee WW, Kim SH, Kang Y, Lee N, Shin MS, Kang SW, Kang I, Age-associated alteration in naive and memory Th17 cell response in humans, Clin Immunol, 140 (2011) 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lee WW, Shin MS, Kang Y, Lee N, Jeon S, Kang I, The relationship of cytomegalovirus (CMV) infection with circulatory IFN-alpha levels and IL-7 receptor alpha expression on CD8(+) T cells in human aging, Cytokine, (2012) [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Panda A, Qian F, Mohanty S, van Duin D, Newman FK, Zhang L, Chen S, Towle V, Belshe RB, Fikrig E, Allore HG, Montgomery RR, Shaw AC, Age-associated decrease in TLR function in primary human dendritic cells predicts influenza vaccine response, J Immunol, 184 (2010) 2518–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Finck R, Simonds EF, Jager A, Krishnaswamy S, Sachs K, Fantl W, Pe’er D, Nolan GP, Bendall SC, Normalization of mass cytometry data with bead standards, Cytometry A, 83 (2013) 483–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bendall SC, Davis KL, Amir el AD, Tadmor MD, Simonds EF, Chen TJ, Shenfeld DK, Nolan GP, Pe’er D, Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development, Cell, 157 (2014) 714–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Benjamini Y, Krieger AM, Yekutieli D, Adaptive linear step-up procedures that control the false discovery rate, Biometrika, 93 (2006) 491–507. [Google Scholar]
- [31].Almeida-Oliveira A, Smith-Carvalho M, Porto LC, Cardoso-Oliveira J, Ribeiro Ados S, Falcao RR, Abdelhay E, Bouzas LF, Thuler LC, Ornellas MH, Diamond HR, Age-related changes in natural killer cell receptors from childhood through old age, Hum Immunol, 72 (2011) 319–329. [DOI] [PubMed] [Google Scholar]
- [32].Hsu HC, Scott DK, Zhang P, Zhou J, Yang P, Wu Q, Schroeder HW Jr., Gerald LB, Ravussin E, Jazwinski SM, Mountz JD, S. Louisiana Healthy Aging, CD8 T-cell immune phenotype of successful aging, Mech Ageing Dev, 127 (2006) 231–239. [DOI] [PubMed] [Google Scholar]
- [33].Pyne S, Hu X, Wang K, Rossin E, Lin TI, Maier LM, Baecher-Allan C, McLachlan GJ, Tamayo P, Hafler DA, De Jager PL, Mesirov JP, Automated high-dimensional flow cytometric data analysis, Proc Natl Acad Sci U S A, 106 (2009) 8519–8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Qualai J, Li LX, Cantero J, Tarrats A, Fernandez MA, Sumoy L, Rodolosse A, McSorley SJ, Genesca M, Expression of CD11c Is Associated with Unconventional Activated T Cell Subsets with High Migratory Potential, PLoS One, 11 (2016) e0154253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.