Abstract
OBJECTIVE:
To investigate associations between genetic mutations and qualitative as well as quantitative features on MRI in rectal adenocarcinoma at primary staging.
METHODS:
In this retrospective study, patients with rectal adenocarcinoma, genome sequencing, and pretreatment rectal MRI were included. Statistical analysis was performed to evaluate associations between qualitative features obtained from subjective evaluation of rectal MRI and gene mutations as well as between quantitative textural features and gene mutations. For the qualitative evaluation, Fisher’s Exact test was used to analyze categorical associations and Wilcoxon Rank Sum test was used for continuous clinical variables. For the quantitative evaluation, we performed manual segmentation of T2-weighted images for radiomics-based quantitative image analysis. Thirty-four texture features consisting of first order intensity histogram-based features (n=4), second order Haralick textures (n=5), and Gabor-edge based Haralick textures were computed at two different orientations. Consensus clustering was performed with 34 computed texture features using the K-means algorithm with Euclidean distance between the texture features. The clusters resulting from the algorithm were then used to enumerate the prevalence of gene mutations in those clusters.
RESULTS:
In 65 patients, 45 genes were mutated in more than 3/65 patients (5%) and were included in the statistical analysis. Regarding qualitative imaging features, on univariate analysis, tumor location was significantly associated with APC (p=0.032) and RASA1 mutation (p=0.032); CRM status was significantly associated with ATM mutation (p=0.021); and lymph node metastasis was significantly associated with BRCA2 (p=0.046) mutation. However, these associations were not significant after adjusting for multiple comparisons. Regarding quantitative imaging features, Cluster C1 had tumors with higher mean Gabor edge intensity compared with cluster C2 (θ=0°, p=0.018; θ=45°, p=0.047; θ=90°, p=0.037; cluster C3 (θ=0°, p=0.18; θ=45°, p=0.1; θ=90°, p=0.052), and cluster C4 (θ=0°, p=0.016; θ=45°, p=0.033; θ=90°, p=0.014) suggesting that the cluster C1 had tumors with more distinct edges or heterogeneous appearance compared with other clusters.
CONCLUSIONS:
Although this preliminary study showed promising associations between quantitative features and genetic mutations, it did not show any correlation between qualitative features and genetic mutations. Further studies with larger sample size are warranted to validate our preliminary data.
Keywords: Rectal Neoplasms, Magnetic Resonance Imaging, Genomics, Precision Medicine
INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer worldwide and the second-leading cause of cancer death. Genetic alterations are common in CRC and are the driving force of tumorigenesis [1, 2]. Each tumor has unique genetic alterations that may serve to guide treatment and monitor disease. In rectal cancer, most investigative efforts related to genetic alterations have focused on the identification of response predictors, given that neoadjuvant chemoradiation therapy is widely used for locally advanced rectal cancer, and for the prediction of lymph node metastasis. The preliminary results have been encouraging and some studies have demonstrated different outcomes depending on the type of mutation.
Rectal magnetic resonance imaging (MRI) plays an important role in local staging and in identifying risk factors for local and distant recurrence, thus helping to tailor patient treatment. As the current standard reference for treatment planning is moving towards genetic level assessment and personalized oncology, additional information obtained from imaging must be further explored and refined. Considering the high cost and limited availability of genetic evaluation, the concept of radiogenomics has emerged. Radiogenomics refers to the relationship between imaging features of a disease and its gene expression [3, 4].
Radiogenomics has been evaluated in several types of cancer, including glioblastoma, breast cancer, non-small cell lung cancer, neuroblastoma, clear cell renal carcinoma, and high-grade serous carcinoma, with promising results [5–11]. In rectal cancer, Hong et al. investigated the correlations between parameters of dynamic contrast-enhanced MRI with molecular prognostic factors, including mutation status of the KRAS oncogene and microsatellite instability; however, no significant correlation between qualitative imaging parameters and KRAS mutation or microsatellite instability was found [12]. Radiogenomics is not equivalent to radiomics. Quantitative texture analyses referred to as radiomics aims to extract large amounts of quantitative radiological data from imaging (texture features), using data characterization algorithms to obtain predictive or prognostic information [13]. Radiomics can be used to predict genetic signatures of tumors [14]. To the best of our knowledge, there is no previous study in the literature that exploits radiogenomics in rectal cancer based on the evaluation of qualitative diagnostic and quantitative texture imaging features on rectal MRI.
In this context, the purpose of this study is to investigate possible associations between genetic mutations and rectal MRI qualitative diagnostic imaging features and radiomics-based quantitative texture features in patients with rectal adenocarcinoma at primary staging.
MATERIALS AND METHODS
Study population
The institutional review board approved this retrospective study, which was compliant with the Health Insurance Portability and Accountability Act, and waived the requirement for informed consent. We searched our retrospectively maintained database for consecutive patients who underwent rectal biopsy with a diagnosis of adenocarcinoma at our institution from January 2009 to March 2016. The inclusion criteria were patients with rectal adenocarcinoma, genetic analysis, and pretreatment (baseline) rectal MRI. The exclusion criteria were (a) patients without baseline rectal MRI, (b) recurrent disease, (c) patients with colon cancer, (d) no visible tumor on MRI, and (e) poor image quality. Patient accrual for this study is summarized in Figure 1.
Figure 1.
Flowchart demonstrating patient accrual.
Histopathologic analyses
All histopathologic analyses were performed by specialized gastrointestinal pathologists with at least 10 years of experience, and pathologic results were reported in a standardized manner. We performed a retrospective chart review of pathologic results and no additional pathologic analysis was done for our study.
MR imaging protocol
MRI scans were acquired on either a GE Healthcare System 1.5T or 3.0T platform (GE Discovery MR750, GE Optima MR450w, GE Signa EXCITE, GE Signa HDxt, and GE Signa HDxt; GE Healthcare, Waukesha, WI) using a phase-array coil. The minimum sequence required was high spatial resolution axial oblique T2-weighted image (WI) through the tumor. Diffusion weighted imaging (DWI) and dynamic contrast-enhanced (DCE) were used when available. The oblique axial T2WI sequence was obtained perpendicular to the long axis of the rectal tumor. MR imaging parameters at our institution are summarized in Table 1. Rectal MRI scans were included if fulfilled the minimum standards agreed upon by the investigators. The minimum standards were the presence of high spatial resolution axial oblique T2WI through the tumor with a slice thickness of 3 mm and an FOV of 180 mm.
Table 1.
MR imaging parameters at our institution.
| Parameter | Seq | TR/TE (ms) | Matrix | FOV (mm) |
ST/SG (mm) | BW (kHz)/ FA | b Value (s/mm2) |
|---|---|---|---|---|---|---|---|
| ObliqueT2WI | |||||||
| 1.5T | FSE | 4000–6000/120 | 320×224 | 180 | 3/1 | 31/160 | - |
| 3.0T | 4000–6000/120 | 320×320 | 180 | 3/1 | 41/110 | - | |
| DWI | |||||||
| 1.5T | DWI | 6000/minimum | 128×128 | 240 | 5/1 | 250/90 | 0,800 |
| 3.0T | 6000/minimum | 128×128 | 240 | 5/1 | 250/90 | 0,800 | |
| DCE | |||||||
| 1.5T | 3D FSPGR | minimum | 256×160 | 240 | 5/0 | 62/12 | - |
| 3.0T | minimum | 256×160 | 240 | 5/0 | 62/12 | - |
BW, bandwidth; TE, echo time; FA, flip angle; FOV, field of view; FSE, fast spin echo; FSPGR, fast spoiled gradient echo; TR, repetition time; Seq, sequence; SG, section gap; ST, slice thickness; T2WI, T2-weighted image.
Selected gene sequencing and identification of mutations
Genomic profiling was performed using a sequencing-based molecular profiling platform employing solution-phase exon capture and massively parallel next-generation sequencing developed at MSK (MSK-IMPACT) [15]. This platform included all exons and selected introns of 410 oncogenes and tumor suppressor genes. Several samples were sequenced with an earlier version of the assay that sequenced the exomes of 341 genes, and the rest were sequenced via the 410-gene panel. Mutation calling and copy number aberrations were called according to the standard assay pipeline.
Qualitative MRI evaluation
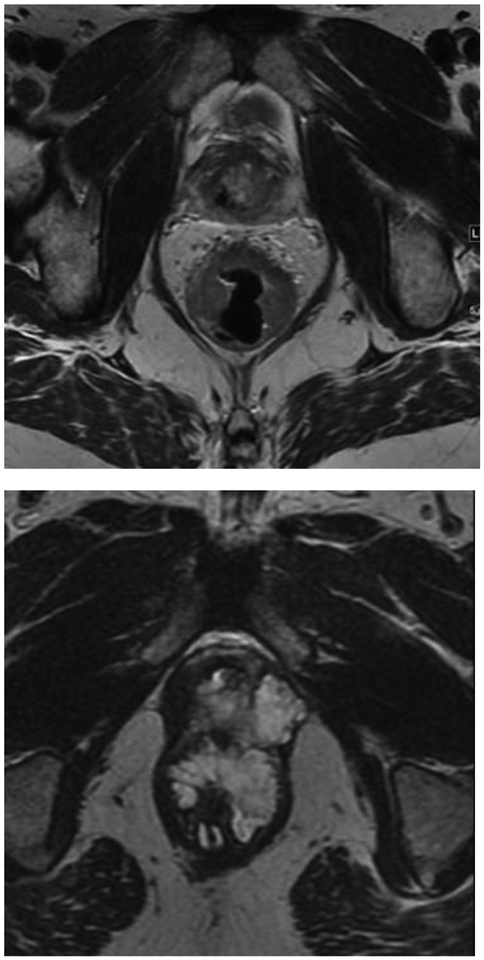
Qualitative MRI data for our study replicated the standardized reports for rectal MRI at our institution which we used in our everyday clinical practice. An abdominal radiologist with 5 years of experience in rectal MRI reviewed all the MRI images and assessed the following qualitative imaging features: (a) tumor location, (b) tumor length, (c) depth of infiltration beyond muscularis propria, (d) mucin content, (e) circumferential resection margin status, (f) presence of extramural vascular invasion, (g) diffusion weighted imaging restriction, (h) early perfusion on dynamic contrast-enhanced imaging, and (i) presence of suspicious lymph nodes (Figure 2). The imaging features were then compared with those described in the standardized report, and disagreements were resolved by a second radiologist with 9 years of experience in rectal MRI.
Figure 2.
Magnetic resonance imaging (MRI) qualitative features evaluated in this study. Nonmucinous tumor (a); mucinous tumor (b); positive circumferential resection margin, due to infiltration of mesorectal fascia (arrow, c); positive mesorectal lymph nodes, one measuring > 9 mm (dashed arrow, d) and the other measuring 7 mm and with round shape and irregular signal intensity (arrowhead, d); restriction on diffusion weighted imaging (asterisk, E); and extramural vascular invasion (curved arrow, f).
The tumor location was defined as low (0–5 cm from anal verge), middle (5.1–10 cm from anal verge), or high (10.1–15 cm from anal verge) [16]. Mucin content was deemed present if tumor contained high signal intensity on T2WI. CRM was obtained by measuring the shortest distance between the outermost part of the tumor beyond the muscularis propria and the mesorectal fascia. Extramural vascular invasion was defined as the presence of (a) vessel wall expansion or irregularity, (b) loss of normal vascular flow void, and (c) intraluminal intermediate signal intensity of the tumor within a vessel continuous with the tumor. For nodal evaluation, we used size and morphological criteria to determine if a node was suspicious for malignancy. The morphological criteria were irregular borders, heterogeneous signal intensity, and round shape. If the node measured <5 mm in short axis, we deemed it as suspicious if three morphological criteria were present; if the node measured between 5 and 8 mm, we deemed it as suspicious if two morphological criteria were present; and if the node measured >9 mm, we considered it suspicious based on size alone [17].
Quantitative MR texture analysis
Image segmentation
Two abdominal radiologists (N.H. and M.C.F.P.T.J.) with 5 and 3 years of experience in rectal MRI, respectively, reviewed all images and reached a consensus on the tumor location. One of the radiologists (M.C.F.P.T.J.) then manually segmented the tumor in all slices on the high spatial resolution axial oblique T2WI using a free open-source software package (ITK-SNAP, version 3.4.0; http://itksnap.org), to provide the volume of interest of the tumor for radiomics-based quantitative image analysis.
Quantitative texture analyses
Thirty-four texture features consisting of first order intensity histogram-based features (n=4), second order Haralick textures (n=5), and Gabor-edge based Haralick textures were computed at two different orientations as in Horvat et al [18]. The texture features were then used to compute clusters that separated the patients by their texture features using consensus clustering.
Gabor filters are edge detectors that detect edges at specific orientations (or angles) and scales (or pixel width). A Gabor filter is composed of a Gaussian envelope function superimposed on a sinusoidal wave. This approach of edge detection is particularly well suited for texture representation and discrimination [19].
Consensus clustering is a technique to automatically extract the appropriate number of clusters that would lead to stable partitioning of the data regardless of random perturbations in the data samples [20]. Concretely, consensus clustering tries to extract clusters that are produced stably regardless of how the data is partitioned by computing an ensemble of clustering using multiple partitions of the data. The central idea of the technique is that if sub-populations of data are representative of the whole data, then samples drawn through random bootstrap resamples of the data should produce the same set of clusters. Therefore, stable clusters are generated by extracting multiple sets of clusters through bootstrapping (or resampling of the data) across multiple runs from which an agreement or consensus matrix is computed. The consensus matrix is an NxN (where N is the number of data samples) that records the number of times a pair of samples occur in the same cluster divided by the number of times those two samples were selected together in a bootstrapping run. A perfect consensus cluster should lead to a block diagonal consensus matrix with ones for sample pairs that always co-occur in the clusters and zeros for sample pairs that will never co-occur in a cluster. Any clustering method such as agglomerative or K-means clustering can be applied to the consensus matrix to extract the individual clusters.
Statistical analysis
Patient characteristics were summarized using medians and ranges for continuous variables and frequencies and percent for categorical variables. Our study comprised 65 patients tested for somatic mutations across 241 genes. Of the 241 genes, 196 genes were mutated in fewer than 3/65 (5%) patients; these genes were not included in the statistical analysis for this study. The remaining 45 genes were mutated in more than 3/65 (>5%) patients and were included in the statistical analysis to evaluate their association with clinical characteristics with respect to their mutation status (wild type vs mutant) as well as with qualitative features obtained from subjective evaluation of rectal MRI and quantitative textural features. Fisher’s Exact test was used to analyze categorical associations and Wilcoxon Rank Sum test was used for continuous clinical variables. The False Discovery Rate method was used to correct for Type 1 error when conducting statistical tests for multiple genes. A P value of >0.05 was considered statistically significant. For the quantitative evaluation, we used the ConsensusClusterPlus package available in R.
RESULTS
Patient characteristics
Among the 65 patients included in our study, there were 38 (58.5%) men and 27 (41.5%) women. The median age was 57 years (33–88): 57 years in men (38–80), and 56 years in women (33–88).
Selected gene sequencing and identification of mutations
The median number of gene mutations was 5 (interquartile range: 4–8). The five most frequent gene mutations were APC (n=57, 87.7%), TP53 (n=45, 69.2%), KRAS (n=30, 46.2%), PIK3CA (n=13, 20%), and SOX9 (n=12, 18.5%). Table 2 demonstrates the 45 genes that were included in the statistical analysis.
Table 2.
List of genes in our population.
| Wild Type (n%) | Mutation (n%) | |
|---|---|---|
| APC | 8 (12.31) | 57 (87.69) |
| TP53 | 20 (30.77) | 45 (69.23) |
| KRAS | 35 (53.85) | 30 (46.15) |
| FBXW7 | 54 (83.08) | 11 (16.92) |
| PIK3CA | 52 (80) | 13 (20) |
| SOX9 | 53 (81.54) | 12 (18.46) |
| AR | 54 (83.08) | 11 (16.92) |
| AMER1 | 61 (93.85) | 4 (6.15) |
| PTPRT | 60 (92.31) | 5 (7.69) |
| NRAS | 62 (95.38 | 3 (4.62) |
| ARID1A | 60 (92.31) | 5 (7.69) |
| MLL2 | 62 (95.38) | 3 (4.62) |
| FAT1 | 59 (90.77) | 6 (9.23) |
| ERBB3 | 59 (90.77) | 6 (9.23) |
| CIC | 60 (92.31) | 5 (7.69) |
| ATM | 60 (92.31) | 5 (7.69) |
| PTPRS | 61 (93.85) | 4 (6.15) |
| MLL3 | 60 (92.31) | 5 (7.69) |
| FLT4 | 62 (95.38) | 3 (4.62) |
| R0S1 | 61 (93.85) | 4 (6.15) |
| BRCA2 | 61 (93.85) | 4 (6.15) |
| ASXL2 | 60 (92.31) | 5 (7.69) |
| KDR | 62 (95.38) | 3 (4.62) |
| NSD1 | 61 (93.85) | 4 (6.15) |
| RECQL4 | 62 (95.38) | 3 (4.62) |
| RNF43 | 61 (93.85) | 4 (6.15) |
| NCOR1 | 62 (95.38) | 3 (4.62) |
| SOX17 | 60 (92.31) | 5 (7.69) |
| LATS2 | 62 (95.38) | 3 (4.62) |
| NOTCH1 | 62 (95.38) | 3 (4.62) |
| MLH1 | 61 (93.85) | 4 (6.15) |
| SPEN | 62 (95.38) | 3 (4.62) |
| PTCH1 | 62 (95.38) | 3 (4.62) |
| PBRM1 | 62 (95.38) | 3 (4.62) |
| REL | 62 (95.38) | 3 (4.62) |
| RASA1 | 62 (95.38) | 3 (4.62) |
| EPHA3 | 62 (95.38) | 3 (4.62) |
| EPHB1 | 62 (95.38) | 3 (4.62) |
| BRD4 | 62 (95.38) | 3 (4.62) |
| RBM10 | 62 (95.38) | 3 (4.62) |
| DNMT1 | 62 (95.38) | 3 (4.62) |
| RET | 62 (95.38) | 3 (4.62) |
| PAK7 | 62 (95.38) | 3 (4.62) |
| TSC1 | 62 (95.38) | 3 (4.62) |
| KDM5C | 62 (95.38) | 3 (4.62) |
Qualitative MRI evaluation
The qualitative evaluation was performed in all 65 patients; however, DWI imaging was not available in 5 patients. The median tumor length was 4.2 cm (range, 1.6–10.3). Patients tended to have tumor in the middle rectum (31/65, 47.7%), without mucin content (52/65, 80%), with negative CRM (42/65, 64.6%), without extramural vascular invasion (51/65, 78.5%), with DWI restriction (56/60, 93.3%), and with suspicious lymph nodes (34/65, 52.3%). Table 3 summarizes the qualitative rectal MRI imaging features that were assessed.
Table 3.
Imaging features on rectal MRI.
| Imaging Features | |||
|---|---|---|---|
| Tumor Localization | Lower | Middle | Upper |
| n (%) | 16 (24.62) | 31 (47.69) | 18 (27.69) |
| Tumor Length (cm) | |||
| median (range) | 4.2 (1.6–10.3) | ||
| Mucin Content | No | Yes | |
| n (%) | 52 (80) | 13 (20) | |
| CRM distance (mm) | |||
| Median (range) | 7 (0–27) | ||
| CRM status | Negative | Positive | NA |
| n (%) | 42 (64.62) | 17 (26.15) | 6 (9.23) |
| Extramural vascular invasion | Absent | Present | |
| n (%) | 51 (78.46) | 14 (21.54) | |
| DWI restriction | No | Yes | NA |
| n (%) | 4 (6.15) | 56 (86.15) | 5 (7.69) |
| Early perfusion on DCE | No | Yes | NA |
| n (%) | 15 (23.08) | 42 (64.62) | 8 (12.3) |
| Metastatic lymph nodes | No | Yes | |
| n (%) | 31 (47.69) | 34 (52.31) | |
CRM, circumferential resection margin; DCE, dynamic contrast-enhances; DWI, diffusion weighted imaging
After comparing the imaging features with mutational profiles, CRM status was associated with ATM mutant status (p=0.021, adjusted p=0.715). Tumor location was associated with APC mutation (p=0.032, adjusted p=0.723) and RASA1 mutation (p=0.032, adjusted p=0.723). Suspicious lymph nodes were associated with and BRCA2 (p=0.046, adjusted p=1.0) mutation. Furthermore, tumor length in patients with a mutation of FLT4 was higher than in patients with wild type FLT4 (p=0.049, adjusted p=0.513). However, none of the associations was significant after adjusting for multiple comparisons (adjusted p>0.05). Table 4 summarizes the genes that were associated with rectal MRI imaging features.
Table 4.
Genes that were associated with rectal MRI imaging features.
| P VALUE (UNADJUSTED) | P VALUE (ADJUSTED) | ||||
|---|---|---|---|---|---|
| TUMOR LOCATION | |||||
| Lower (n=16) | Middle (n=31) | Upper (n=18) | |||
| APC | 0.032 | 0.723 | |||
| WILD TYPE | 5 (31%) | 2 (6%) | 1 (6%) | ||
| MUTATED | 11 (69%) | 29 (94%) | 17 (94%) | ||
| RASA1 | 0.032 | 0.723 | |||
| WILD TYPE | 16 (100%) | 31 (100%) | 15 (83%) | ||
| MUTATED | 0 (0%) | 0 (0%) | 3 (17%) | ||
| CRM STATUS | |||||
| Negative (n=42) | Positive (n=17) | Not applicable (n=6) | |||
| ATM | 0.021 | 0.715 | |||
| WILD TYPE | 41 (98%) | 13 (76%) | 6 (100%) | ||
| MUTATED | 1 (2%) | 4 (24%) | 0 (0%) | ||
| METASTATIC LYMPH NODES | |||||
| Negative (n=31) | Positive (n=34) | ||||
| BRCA2 | 0.046 | 1 | |||
| WILD TYPE | 27 (87%) | 34 (100%) | |||
| MUTATED | 4 (13%) | 0 (0%) | |||
| TUMOR LENGTH | |||||
| Number | Median (IQR) cm | 0.049 | 0.513 | ||
| FLT4 | |||||
| WILD TYPE | 62 | 4.2 (1.6–10.3) | |||
| MUTATED | 3 | 6.3 (6.2–6.3) | |||
CRM, circumferential resection margin; IQR, interquartile range; MRI, magnetic resonance imaging
Quantitative texture analyses
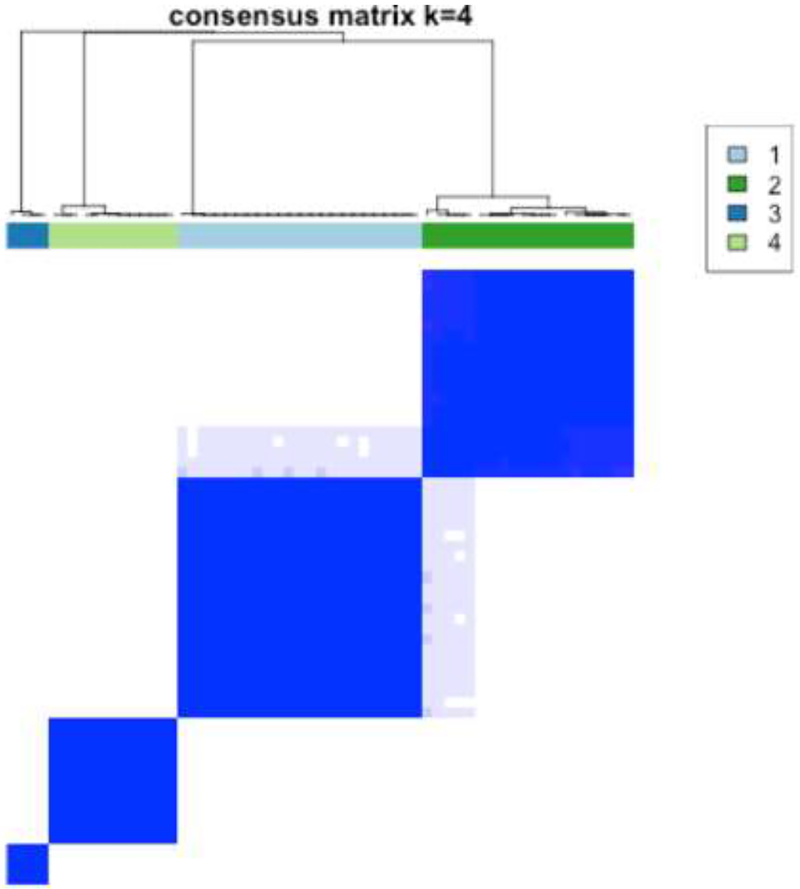
The quantitative evaluation was performed in 59 patients with high spatial resolution T2 images; in 6 cases, there was insufficient tumor coverage. Consensus clustering was performed using all the computed texture features (n=34) using the K-means algorithm with Euclidean distance between the texture features. Consensus clustering was evaluated for clusters ranging from 2–10 and 4 was chosen as the best cluster as it produced the best split in the data. Distribution of conditional density function (CDF) for those clusters by varying consensus index were assessed and the most stable CDF across all consensus indices was K=4 (Figure 3).
Figure 3.
Consensus clustering results using K=4 (a). Distribution of conditional density function (CDF) for the various clusters by varying consensus index. At K=4, the CDF is most stable across all consensus indices (b).
The clusters resulting from the algorithm were then used to enumerate the prevalence of gene mutations in those clusters. The clusters sizes were as follows: C1: 23, C2: 20, C3: 4, and C4: 12. Cluster C1 had tumors with higher mean Gabor edge intensity compared with cluster C2 (θ = 0°, p = 0.018; θ = 45°, p=0.047; θ = 90°, p = 0.037), cluster C3 (θ = 0°, p = 0.18; θ = 45°, p = 0.1; θ = 90°, p=0.052), and cluster C4 (θ = 0°, p = 0.016; θ = 45°, p=0.033; θ = 90°, p = 0.014) suggesting that the cluster C1 had tumors with more distinct edges or heterogeneous appearance compared with other clusters. In fact, the Gabor intensities progressively decreased from cluster C1 to C4 (see Supplementary Figure 1). The heatmap of the mutations is shown in Figure 4. As demonstrated, no significant associations were detected between the clusters and gene mutations, except for PTPRT (p=0.01); however, this was not significant after adjusting for multiple comparisons.
Figure 4.
Heatmap of mutations. Rows demonstrate the genes, columns the samples and black stripe means a mutation was seen for a sample for that particular gene. Light blue bar shows cluster 1, dark green cluster 2, dark blue cluster 3, and light green cluster 4.
DISCUSSION
In our study population, the three most common mutations were APC (87.7%), TP53 (69.2%), and KRAS (46.2%). The frequency of mutations in our study are in line with the literature regarding the three most common genes involved in the tumorigenesis of CRC [1, 2]. APC is a tumor suppressor gene located on chromosome 5q21 and its mutation is responsible for familial adenomatous polyposis and around 85% of sporadic CRC [21]. TP53 is a tumor suppressor gene that regulates apoptosis, and nearly 50% of all CRC demonstrate TP53 mutation with frequencies are even higher in distal colon and rectal cancers [22]. The RAS family (HRAS, KRAS, and NRAS) is important in the control of cell proliferation, and KRAS is present in up to 50% of CRC [23].
Rectal cancer biology has evolved over the last decade as gene mutations and protein expression have been demonstrated to correlate with clinical outcome, e.g., chemoradiation therapy response and lymph node metastases. Patients with locally advanced rectal cancer who have KRAS and combined KRAS/TP53 mutations have been shown to have decreased response to neoadjuvant therapy [24]. Wild type TP53 has been associated with pathological complete response and good response to chemoradiation therapy [25]. The investigation of individual associations between diagnostic imaging features and mutations is considered the critical first step of radiogenomics of rectal cancer. Chen et al. showed in their study with PETCT that patients with KRAS mutation had different PET-CT imaging aspects. Another study investigated the correlations between parameters of dynamic contrast-enhanced MRI with molecular prognostic factors, including mutation status of the KRAS oncogene and microsatellite instability; however, no significant correlations were found between qualitative imaging parameters and KRAS mutation or microsatellite instability [12]. In our study, patients with positive CRM showed a higher frequency of ATM mutation, patients with negative lymph node showed a higher frequency BRCA2 mutation, the length of tumor was associated with FLT4 mutation, and tumor location was associated with APC and RASA1 mutations. Although we found associations in this pilot study between qualitative diagnostic MRI imaging features and genetic mutations, after statistical adjustments for multiple comparisons, no significant correlation was maintained. We believe this could be due to the small sample size and further studies with larger datasets are needed to evaluate radiogenomics in rectal cancer. This study was primarily executed as a preliminary, discovery-phase pilot analysis of radiogenomics MRI associations in rectal cancer.
Promising associations were observed when quantitative features were correlated with genetic mutations. For example, unsupervised clustering of MRI radiomic features of tumor heterogeneity showed that the cluster with least heterogeneity (texture entropy) was associated with protein tyrosine phosphatase T (PTPRT); however, this was not significant after adjusting for multiple comparisons. In addition, cluster C1 had tumors with higher mean Gabor edge intensity compared with cluster C2, suggesting that the cluster C1 had tumors with more distinct edges or heterogeneous appearance compared with that of other clusters. Higher values of Gabor edges that quantify the edges in images indicate larger heterogeneity within the tumors. However, these results are preliminary and analysis on larger cohorts is necessary for validation. Radiomics extracts large amounts of quantitative data from imaging and may complement visual assessment, may reflect information related to tumor heterogeneity and microenvironment, and is less susceptible to intra and interobserver variability [26]. Therefore, radiomics might detect differences beyond the capability of human vision. In the new era of precision medicine, knowing the genetic profile of the rectal adenocarcinoma in advance based on imaging modalities may guide the multidisciplinary team in choosing the best treatment for the patient [27], without adding the cost of genetic analysis and potentially improving patient outcomes [28, 29].
Our pilot study had limitations. First, we had a small sample size of patients with both primary rectal MRI and genetic analysis. We might have introduced selection bias by not including patients without rectal MRI and/or genetic analyses. Given the small sample size of our population, we did not evaluate the possible differences in textural features between scans performed on 1.5T and 3T units; however, we addressed that issue in a previous study and no statistically significant difference were detected among the textural features obtained from 1.5T and 3T MRI units [30]. Second, the image segmentation and qualitative assessment were done after consensus; therefore, inter reader agreement was not evaluated. Third, the volume of interest of the tumor was manually obtained, which is a time-consuming process. It is vital to develop a robust user-friendly tool that requires minimal operator input to encourage the use of radiomic measures in daily clinical practice. Finally, we did not perform external validation of our results; thus, generalization is limited. Further studies are therefore needed to overcome these limitations and to provide a better, more comprehensive evaluation of radiogenomics in rectal cancer. Furthermore, studies using textural features obtained from other sequences besides T2WI, such as DWI and dynamic contrast-enhanced imaging techniques, may be explored in future. However, as there is no consensus regarding these sequences in the literature and among different institutions, it may be difficult to obtain large datasets.
In conclusion, this preliminary study showed no significant associations between qualitative MRI imaging features and genetic mutations after adjusting for multiple comparisons. However, promising correlations were observed between quantitative radiomic-based texture features and genetic mutations. Further studies with larger sample size are warranted for additional evaluation and to validate our preliminary data.
Supplementary Material
HIGHLIGHTS.
Tumor location was associated with APC and RASA1 mutation
Circumferential resection margin status was associated with ATM mutation
Lymph node metastasis was associated with BRCA2 mutation
These correlations were not significant after adjusting for multiple comparisons
PTPRT was enriched in radiomic cluster C4
ACKNOWLEDGMENTS
We thank Joanne Chin and Joseph Singer for their editorial support on this manuscript.
Funding Sources: This study has received funding by the NIH/NCI P30 CA008748 Cancer Center Support Grant and the Colorectal Cancer Research Center CC50367 at Memorial Sloan Kettering Cancer Center. The funding sources had no involvement in the study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
ABBREVIATIONS:
- CDF
conditional density function
- CRC
colorectal cancer
- CRM
circumferential resection margin
- DWI
diffusion weighted imaging
- MRI
Magnetic resonance imaging
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest: The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
REFERENCES
- [1].Armaghany T, Wilson JD, Chu Q, Mills G, Genetic alterations in colorectal cancer, Gastrointest Cancer Res 5(1) (2012) 19–27. [PMC free article] [PubMed] [Google Scholar]
- [2].Ungerbäck J, Belenki D, Jawad ul-Hassan A, Fredrikson M, Fransén K, Elander N, Verma D, Söderkvist P, Genetic variation and alterations of genes involved in NFκB/TNFAIP3- and NLRP3-inflammasome signaling affect susceptibility and outcome of colorectal cancer, Carcinogenesis 33(11) (2012) 2126–34. [DOI] [PubMed] [Google Scholar]
- [3].Pinker K, Shitano F, Sala E, Do RK, Young RJ, Wibmer AG, Hricak H, Sutton EJ, Morris EA, Background, current role, and potential applications of radiogenomics, Journal of magnetic resonance imaging : JMRI 47(3) (2018) 604–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sala E, Mema E, Himoto Y, Veeraraghavan H, Brenton JD, Snyder A, Weigelt B, Vargas HA, Unravelling tumour heterogeneity using next-generation imaging: radiomics, radiogenomics, and habitat imaging, Clin Radiol 72(1) (2017) 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Brisse HJ, Blanc T, Schleiermacher G, Mosseri V, Philippe-Chomette P, Janoueix-Lerosey I, Pierron G, Lapouble E, Peuchmaur M, Fréneaux P, Galmiche L, Algret N, Peycelon M, Michon J, Delattre O, Sarnacki S, Radiogenomics of neuroblastomas: Relationships between imaging phenotypes, tumor genomic profile and survival, PLoS One 12(9) (2017) e0185190.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Grimm LJ, Breast MRI radiogenomics: Current status and research implications, J Magn Reson Imaging 43(6) (2016) 1269–78. [DOI] [PubMed] [Google Scholar]
- [7].Karlo CA, Di Paolo PL, Chaim J, Hakimi AA, Ostrovnaya I, Russo P, Hricak H, Motzer R, Hsieh JJ, Akin O, Radiogenomics of clear cell renal cell carcinoma: Associations between CT imaging features and mutations, Radiology 270(2) (2014) 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liu X, Mangla R, Tian W, Qiu X, Li D, Walter KA, Ekholm S, Johnson MD, The preliminary radiogenomics association between MR perfusion imaging parameters and genomic biomarkers, and their predictive performance of overall survival in patients with glioblastoma, J Neurooncol (2017). [DOI] [PubMed] [Google Scholar]
- [9].Vargas HA, Huang EP, Lakhman Y, Ippolito JE, Bhosale P, Mellnick V, Shinagare AB, Anello M, Kirby J, Fevrier-Sullivan B, Freymann J, Jaffe CC, Sala E, Radiogenomics of High-Grade Serous Ovarian Cancer: Multireader Multi-Institutional Study from the Cancer Genome Atlas Ovarian Cancer Imaging Research Group, Radiology 285(2) (2017) 482–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhou M, Leung A, Echegaray S, Gentles A, Shrager JB, Jensen KC, Berry GJ, Plevritis SK, Rubin DL, Napel S, Gevaert O, Non-Small Cell Lung Cancer Radiogenomics Map Identifies Relationships between Molecular and Imaging Phenotypes with Prognostic Implications, Radiology (2017) 161845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Woodard GA, Ray KM, Joe BN, Price ER, Qualitative Radiogenomics: Association between Oncotype DX Test Recurrence Score and BI-RADS Mammographic and Breast MR Imaging Features, Radiology 286(1) (2018) 60–70. [DOI] [PubMed] [Google Scholar]
- [12].Hong HS, Kim SH, Park HJ, Park MS, Kim KW, Kim WH, Kim NK, Lee JM, Cho HJ, Correlations of dynamic contrast-enhanced magnetic resonance imaging with morphologic, angiogenic, and molecular prognostic factors in rectal cancer, Yonsei Med J 54(1) (2013) 123–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gillies RJ, Kinahan PE, Hricak H, Radiomics: Images Are More than Pictures, They Are Data, Radiology 278(2) (2016) 563–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Napel S, Giger M, Special Section Guest Editorial:Radiomics and Imaging Genomics: Quantitative Imaging for Precision Medicine, J Med Imaging (Bellingham) 2(4) (2015) 041001.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, Chandramohan R, Liu ZY, Won HH, Scott SN, Brannon AR, O’Reilly C, Sadowska J, Casanova J, Yannes A, Hechtman JF, Yao J, Song W, Ross DS, Oultache A, Dogan S, Borsu L, Hameed M, Nafa K, Arcila ME, Ladanyi M, Berger MF, Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology, J Mol Diagn 17(3) (2015) 251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nougaret S, Reinhold C, Mikhael HW, Rouanet P, Bibeau F, Brown G, The use of MR imaging in treatment planning for patients with rectal carcinoma: have you checked the “DISTANCE”?, Radiology 268(2) (2013) 330–44. [DOI] [PubMed] [Google Scholar]
- [17].Beets-Tan RGH, Lambregts DMJ, Maas M, Bipat S, Barbaro B, Curvo-Semedo L, Fenlon HM, Gollub MJ, Gourtsoyianni S, Halligan S, Hoeffel C, Kim SH, Laghi A, Maier A, Rafaelsen SR, Stoker J, Taylor SA, Torkzad MR, Blomqvist L, Correction to: Magnetic resonance imaging for clinical management of rectal cancer: Updated recommendations from the 2016 European Society of Gastrointestinal and Abdominal Radiology (ESGAR) consensus meeting, Eur Radiol 28(6) (2018) 2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Horvat N, Veeraraghavan H, Khan M, Blazic I, Zheng J, Capanu M, Sala E, Garcia-Aguilar J, Gollub MJ, Petkovska I, MR Imaging of Rectal Cancer: Radiomics Analysis to Assess Treatment Response after Neoadjuvant Therapy, Radiology (2018) 172300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Daugman JG, Uncertainty relation for resolution in space, spatial frequency, and orientation optimized by two-dimensional visual cortical filters, J Opt Soc Am A 2(7) (1985) 1160–9. [DOI] [PubMed] [Google Scholar]
- [20].T. P Monti S, Mesirov J, Golub T, Consensus Clustering: A Resampling-Based Method for Class Discovery and Visualization of Gene Expression Microarray Data, Machine Learning 52 (2003) 91–118. [Google Scholar]
- [21].Kinzler KW, Vogelstein B, Lessons from hereditary colorectal cancer, Cell 87(2) (1996) 159–70. [DOI] [PubMed] [Google Scholar]
- [22].Iacopetta B, TP53 mutation in colorectal cancer, Hum Mutat 21(3) (2003) 271–6. [DOI] [PubMed] [Google Scholar]
- [23].Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N, Beranek M, Jandik P, Benamouzig R, Jullian E, Laurent-Puig P, Olschwang S, Muller O, Hoffmann I, Rabes HM, Zietz C, Troungos C, Valavanis C, Yuen ST, Ho JW, Croke CT, O’Donoghue DP, Giaretti W, Rapallo A, Russo A, Bazan V, Tanaka M, Omura K, Azuma T, Ohkusa T, Fujimori T, Ono Y, Pauly M, Faber C, Glaesener R, de Goeij AF, Arends JW, Andersen SN, Lövig T, Breivik J, Gaudernack G, Clausen OP, De Angelis PD, Meling GI, Rognum TO, Smith R, Goh HS, Font A, Rosell R, Sun XF, Zhang H, Benhattar J, Losi L, Lee JQ, Wang ST, Clarke PA, Bell S, Quirke P, Bubb VJ, Piris J, Cruickshank NR, Morton D, Fox JC, Al-Mulla F, Lees N, Hall CN, Snary D, Wilkinson K, Dillon D, Costa J, Pricolo VE, Finkelstein SD, Thebo JS, Senagore AJ, Halter SA, Wadler S, Malik S, Krtolica K, Urosevic N, Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study, Br J Cancer 85(5) (2001) 692–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chow OS, Kuk D, Keskin M, Smith JJ, Camacho N, Pelossof R, Chen CT, Chen Z, Avila K, Weiser MR, Berger MF, Patil S, Bergsland E, Garcia-Aguilar J, KRAS and Combined KRAS/TP53 Mutations in Locally Advanced Rectal Cancer are Independently Associated with Decreased Response to Neoadjuvant Therapy, Ann Surg Oncol 23(8) (2016) 2548–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Karagkounis G, Kalady MF, Molecular Biology: Are We Getting Any Closer to Providing Clinically Useful Information?, Clin Colon Rectal Surg 30(5) (2017) 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lubner MG, Smith AD, Sandrasegaran K, Sahani DV, Pickhardt PJ, CT Texture Analysis: Definitions, Applications, Biologic Correlates, and Challenges, Radiographics 37(5) (2017) 1483–1503. [DOI] [PubMed] [Google Scholar]
- [27].Rosenstein BS, Radiogenomics: Identification of Genomic Predictors for Radiation Toxicity, Semin Radiat Oncol 27(4) (2017) 300–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yang L, Dong D, Fang M, Zhu Y, Zang Y, Liu Z, Zhang H, Ying J, Zhao X, Tian J, Can CT-based radiomics signature predict KRAS/NRAS/BRAF mutations in colorectal cancer?, Eur Radiol 28(5) (2018) 2058–2067. [DOI] [PubMed] [Google Scholar]
- [29].Pinker K, Shitano F, Sala E, Do RK, Young RJ, Wibmer AG, Hricak H, Sutton EJ, Morris EA, Background, current role, and potential applications of radiogenomics, J Magn Reson Imaging (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Horvat N, Veeraraghavan H, Khan M, Blazic I, Zheng J, Capanu M, Sala E, Garcia-Aguilar J, Gollub MJ, Petkovska I, MR Imaging of Rectal Cancer: Radiomics Analysis to Assess Treatment Response after Neoadjuvant Therapy, Radiology 287(3) (2018) 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.