Abstract
Most genetic risk for psychiatric disease lies in regulatory regions, implicating pathogenic dysregulation of gene expression and splicing. However, comprehensive assessments of transcriptomic organization in disease brain are limited. Here, we integrate genotype and RNA-sequencing in brain samples from 1695 subjects with autism, schizophrenia, bipolar disorder and controls. Over 25% of the transcriptome exhibits differential splicing or expression, with isoform-level changes capturing the largest disease effects and genetic enrichments. co-expression networks isolate disease-specific neuronal alterations, as well as microglial, astrocyte, and interferon response modules defining novel neural-immune mechanisms. We prioritize disease loci likely mediated by cis-effects on brain expression via transcriptome-wide association analysis. This transcriptome-wide characterization of the molecular pathology across three major psychiatric disorders provides a comprehensive resource for mechanistic insight and therapeutic development.
Graphical Abstract
Introduction
Developing more effective treatments for autism (ASD), schizophrenia (SCZ), and bipolar disorder (BD), three common psychiatric disorders that confer lifelong disability, is a major international public health priority (3). Studies have identified hundreds of causal genetic variants robustly associated with these disorders, and thousands more that likely contribute to their pathogenesis (4). However, the neurobiological mechanisms through which genetic variation imparts risk, both individually and in aggregate, are still largely unknown (4–6).
The majority of disease-associated genetic variation lies in non-coding regions (7) enriched for non-coding RNAs and cis regulatory elements that regulate gene expression and splicing of their cognate coding gene targets (8, 9). Such regulatory relationships show substantial heterogeneity across human cell types, tissues, and developmental stages (10), and are often highly species-specific (11). Recognizing the importance of understanding transcriptional regulation and non-coding genome function, several consortia (10, 12–14) have undertaken large-scale efforts to provide maps of the transcriptome and its genetic and epigenetic regulation across human tissues. Although some have included CNS tissues, a more comprehensive analysis focusing on the brain in both healthy and disease states is necessary to accelerate our understanding of the molecular mechanisms of these disorders (1, 15–17).
We present results of the analysis of RNA-sequencing (RNA-Seq) data from the PsychENCODE Consortium (17), integrating genetic and genomic data from over 2000 well-curated, high-quality post-mortem brain samples from individuals with SCZ, BD, ASD, and controls (18). We provide a comprehensive resource of disease-relevant gene expression changes and transcriptional networks in the postnatal human brain (see Resource.PsychENCODE.org for raw data and annotations). Data was generated across eight studies (2, 19, 20), uniformly processed, and combined through a consolidated genomic data processing pipeline ((21); Fig S1), yielding a total of 2188 samples passing quality control (QC) for this analysis, representing frontal and temporal cerebral cortex from 1695 unique subjects across the lifespan, including 279 technical replicates (Fig S2). Extensive quality control steps were taken within and across individual studies resulting in the detection of 16,541 protein-coding and 9,233 non-coding genes with the Gencode v19 annotations ((21); Fig S3). There was substantial heterogeneity in RNA-Seq methodologies across cohorts, which was accounted for by including 28 surrogate variables and aggregate sequencing metrics as covariates in downstream analyses of differential expression (DE) at gene, isoform, and local splicing levels (21). Differential expression did not overlap with experimentally defined RNA degradation metrics in brain, indicating that results were not driven by RNA-quality confounds (Fig S4) (22).
To provide a comprehensive view of the genomic architecture of these disorders, we characterize several levels of transcriptomic organization – gene-level, transcript isoform, local splicing, and co-expression networks – for protein-coding and non-coding gene biotypes. We integrate results with common genetic variation and disease GWAS to identify putative regulatory targets of genetic risk variants. Although each level provides important disease-specific and shared molecular pathology, we find that isoform-level changes show the largest effects in disease brain, are most reflective of genetic risk, and provide the greatest disease specificity when assembled into co-expression networks.
We recognize that these analyses involve a variety of steps and data types and are necessarily multifaceted and complex. We therefore organize results into two major sections. The first is at the level of individual genes and gene products, starting with gene level transcriptomic analyses, isoform and splicing analyses, followed by identification of potential genetic drivers. The second section is anchored in gene network analysis, where we identify co-expression modules at both gene and isoform levels and assess their relationship to genetic risk. As these networks reveal many layers of biology, we provide an interactive website to permit their in depth exploration (Resource.PsychENCODE.org).
Gene and Isoform Expression Alterations in Disease
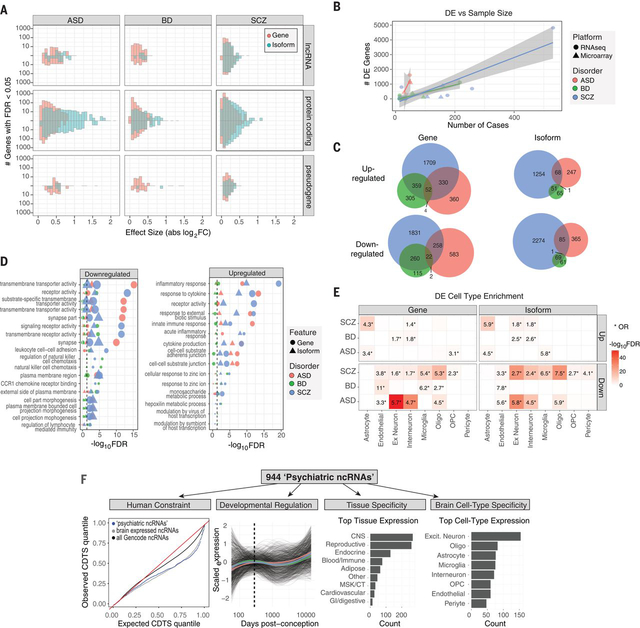
RNA-Seq based quantifications enabled assessment of coding and non-coding genes and transcript isoforms, imputed using RSEM guided by Gencode v19 annotations (21, 23). In accordance with previous results (1), we observed pervasive differential gene expression (DGE) in ASD, SCZ, and BD (n=1611, 4821, and 1119 genes at FDR<0.05, respectively; Fig 1A; Table S1). There was substantial cross-disorder sharing of this DE signal and a gradient of transcriptomic severity with the largest changes in ASD compared with SCZ or BD (ASD vs SCZ, mean |log2FC| 0.26 vs 0.10, P<2×10−16, Kolmogorov-Smirnov (K-S) test; ASD vs BD, mean |log2FC| 0.26 vs 0.15, P<2×10−16, K-S test), as observed previously (1). Altogether, over a quarter of the brain transcriptome was affected in at least one disorder (Fig 1A-C; complete gene list, Table S1).
Figure 1. Gene and isoform expression dysregulation in psychiatric brain.
A) Differential expression effect size (|log2FC|) histograms are shown for protein-coding, lncRNA, and pseudogene biotypes up or downregulated (FDR<0.05) in disease. Isoform-level changes (DTE; blue) show larger effect sizes than at gene level (DGE; red), particularly for protein-coding biotypes in ASD and SCZ. B) A literature-based comparison shows that the number of DE genes detected is dependent on study sample size for each disorder. C) Venn diagrams depict overlap among up or downregulated genes and isoforms across disorders. D) Gene ontology enrichments are shown for differentially expressed genes or isoforms. The top 5 pathways are shown for each disorder. E) Heatmap depicting cell type specificity of enrichment signals. Differentially expressed features show substantial enrichment for known CNS cell type markers, defined at the gene level from single cell RNA-Seq. F) Annotation of 944 unique non-coding RNAs DE in at least one disorder. From left to right: Sequence-based characterization of ncRNAs for measures of human selective constraint; brain developmental expression trajectories are similar across each disorder (colored lines represent mean trajectory across disorders); tissue, and CNS cell type expression patterns.
DGE results were concordant with previously published datasets for all three disorders (Fig S4), although some had overlapping samples. We observed significant concordance of DGE effect sizes with those from a microarray meta-analysis of each disorder (ASD: ⍴=0.8, SCZ: ⍴=0.78, BD: ⍴=0.64, Spearman ⍴ of log2FC, all P’s<10−16, (1)) and with previous RNA-Seq studies of individual disorders (ASD: ⍴=0.96, ref (19); SCZ ⍴=0.78, ref (2); SCZ ⍴=0.80, ref (24); BD ⍴=0.85, ref (1); Spearman ⍴ of log2FC, all P’s<10−16). These DE genes exhibited substantial enrichment for known pathways and cell type specific markers derived from single nucleus RNA-Seq in human brain (Fig 1D-E) (21), consistent with previously observed patterns (1, 19).
Expanding these analyses to the transcript isoform-level, we observe widespread differential transcript expression (DTE) across ASD, SCZ, and BD (n=767, 3803, and 248 isoforms at FDR<0.05, respectively; Table S1). Notably, at the DTE level, the cross-disorder overlap was significantly attenuated (Fig 1C), suggesting that alternative transcript usage and/or splicing confers a substantial portion of disease specificity. In addition, isoform-level alterations in disease exhibited substantially larger effect sizes compared with gene-level changes (mean |log2FC| 0.25 vs 0.14, P<2×10−16, K-S test), particularly for protein coding biotypes (Fig 1A), consistent with recent work demonstrating the importance of splicing dysregulation in disease pathogenesis (25). Furthermore, although isoform and gene-level changes exhibited similar pathway and cell type enrichments (e.g. Fig 1D-E), isoform-level analysis identified DE transcripts that did not show DGE (‘isoform-only DE’), including 811 in SCZ, 294 in ASD, and 60 in BD. These isoform-only DE genes were more likely to be downregulated than upregulated in disease (one sample t-test, P<10−16), exhibited greatest overlap with excitatory neuron clusters (OR’s> 4, Fisher’s exact test, FDR’s<10−10), and showed significant enrichment for neuron projection development, mRNA metabolism, and synaptic pathways (FDR<3×10−3; Table S1). To validate DTE results, we performed PCR on several selected transcripts in a subset of ASD, SCZ and control samples (21), and find significant concordance in fold-changes compared with those from RNA-Seq data (Fig S5A-B). Together, these results suggest that isoform-level changes are most reflective of neuronal and synaptic dysfunction characteristic of each disorder.
Differential Expression of the Non-coding Transcriptome
Non-coding RNAs (ncRNAs) represent the largest class of transcripts in the human genome and have increasingly been associated with complex phenotypes (26). However, most have limited functional annotation, particularly in human brain, and have been only minimally characterized in the context of psychiatric disease. Based on Gencode annotations, we identify 944 ncRNAs exhibiting gene- or isoform-level DE in at least one disorder (herein referred to as ‘neuropsychiatric (NP) ncRNAs’ (21)), 693 of which were DE in SCZ, 178 in ASD, and 174 in BD, of which 208, 60, and 52 are annotated as intergenic long non-coding RNAs (lincRNAs), in each disorder, respectively. To place these NPncRNAs within a functional context, we examined expression patterns across human tissues, cell types, and developmental time periods, as well as sequence characteristics including evolutionary conservation, selection, and constraint. We highlight several noncoding genes exhibiting DE across multiple disorders (Fig S6) and provide comprehensive annotations for each NPncRNA (Table S2), including cell type specificity, developmental trajectory, and constraint, to begin to elucidate a functional context in human brain.
As a class, NPncRNAs were under greater selective constraint compared to all Gencode annotated ncRNAs (Fig 1F), consistent with the observed increased purifying selection in brain-expressed genes (27). We identify 74 NPncRNAs (~8%) under purifying selection in humans, with average exon-level context-dependent tolerance scores (CDTS) below the 10th percentile (21). Over 200 NPncRNAs exhibited broad and non-specific expression patterns across cell types, whereas 66 were expressed within a specific cell type class (Table S2). Notable examples are: LINC00996, which is downregulated in SCZ (log2FC −0.71, FDR<5×10−11) and BD (log2FC −0.45, FDR=0.02) and restricted to microglia in brain (Fig S6); LINC00343, expressed in excitatory neurons, and downregulated in BD (log2FC −0.33, FDR=0.012) with a trend in SCZ (log2FC −0.15, FDR 0.065); and LINC00634, an unstudied brain enriched lincRNA downregulated in SCZ (log2FC −0.06, FDR 0.027) with a genome-wide significant SCZ TWAS association as described below.
Local Splicing Dysregulation in Disease
Isoform-level diversity is achieved by combinatorial use of alternative transcription start sites, polyadenylation, and splicing (28). We used LeafCutter (29) to assess local differential splicing (DS) differences in ASD, SCZ and BD using de novo aligned RNA-seq reads, controlling for the same covariates as DGE/DTE (Fig S7). This approach complements DTE by considering aggregate changes in intron usage affecting exons that may be shared by multiple transcripts and is consequently not restricted to the specified genome annotation (21). Previous studies have identified alterations in local splicing events in ASD (19, 30) and in smaller cohorts in SCZ (2, 24) and BD (31).
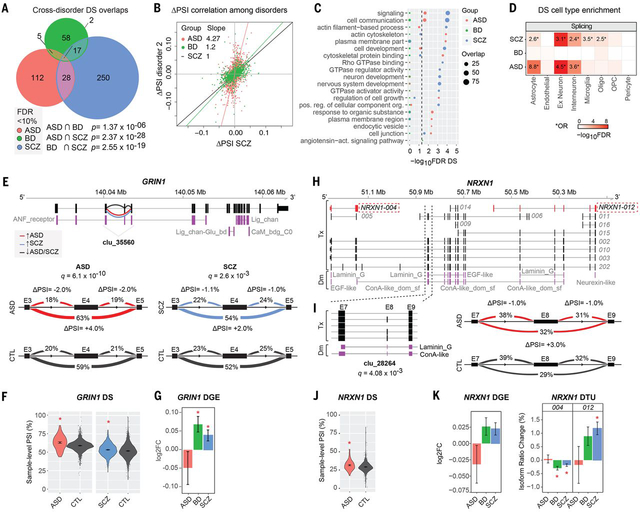
We identified 515 DS intron clusters in 472 genes across all disorders (FDR<0.1), 117 of which (25%) contained one or more novel exons (Table S3; Fig 2A). Validation of DS changes for 9 genes in a subset of cases and controls (n= 5–10 in each group) by semiquantitative RT-PCR showed percent spliced-in (PSI) changes consistent with those reported by LeafCutter (Fig S5C-E). The most commonly observed local splicing change was exon skipping (41–60%), followed by alternative 5’ exon inclusion (e.g. due to alternative promoter usage; 11–21%) and alternative 3’ splice site usage (5–18%) (Table S3; Fig S8A). DS genes overlapped significantly with DTE results for ASD and SCZ (Fig S8B), but not BD, which likely still remains underpowered. There was significant cross-disorder correlation in PSI changes (Spearman’s ⍴=0.59 SCZ-BD, ⍴=0.52 SCZ-ASD, all P<10−4) and subsequently, overlap among DS genes (Fig 2A-B), although the majority of splicing changes still are disorder specific. Only two genes, DTNA and AHCYL1, were significantly DS in all three disorders (Fig S9). DS genes showed significant (FDR<0.05) enrichment for signaling, cell communication, actin cytoskeleton, synapse, and neuronal development pathways across disorders (Figs 2C, S8C), and were relatively broadly expressed across cell types (Fig 2D). Disorder specific pathways implicated by splicing dysfunction include plasma membrane receptor complex, endocytic vesicle, regulation of cell growth and cytoskeletal protein binding in ASD; angiotensin receptor signaling in BD; and GTPase receptor activity, neuron development and actin cytoskeleton in SCZ. We also find significant enrichment of splicing changes in targets of two RNA binding proteins that regulate synaptic transmission and whose targets are implicated in both ASD and SCZ, the neuronal splicing regulator RBFOX1 (FDR=5.16×10−11) (32) and the fragile X mental retardation protein (FMRP) (FDR=3.10×10−21) (33). Notably, 48 DS genes (10%; FDR=8.8×10−4) encode RNA binding proteins or splicing factors (34), with at least six splicing factors also showing DTE in ASD (MATR3), SCZ (QKI, RBM3, SRRM2, U2AF1) or both (SRSF11).
Figure 2. Aberrant local splicing and isoform usage in ASD, SCZ and BD.
A) Venn diagram showing cross-disorder overlap for 472 genes with significant differentially spliced (DS) intron clusters (FDR< 10%) identified by LeafCutter. P values for hypergeometric tests of pairwise overlaps between each disorder are shown at the bottom. B) Scatter plots comparing percent spliced-in (PSI) changes for all 1,287 introns in 515 significant DS clusters in at least one disorder, for significant disease pairs SCZ vs ASD and SCZ vs BD (Spearman’s ⍴=0.52 and ⍴=0.59, respectively). Principal component regression lines are shown in red, with regressions slopes for ASD and BD delta PSI compared to SCZ in the top-left corner. C) Top 10 gene ontology (GO) enrichments for DS genes in each disorder (see also Fig S8C). D) Significant enrichment for neuronal and astrocyte markers (ASD and SCZ), as well as oligodendrocyte and microglia (SCZ) cell type markers in DS genes. *Odds Ratio (OR) is given only for FDR< 5% and OR> 1. Oligo - oligodendrocytes; OPC - oligodendrocyte progenitor cells. E) A significant DS intron cluster in GRIN1 (clu_35560; chr9:140,040,354–140,043,461) showing increased exon 4 (E4) skipping in both ASD and SCZ. Increased or decreased intron usage in ASD/SCZ cases compared to controls are highlighted in red and blue, respectively. Protein domains are annotated as ANF_receptor - Extracellular receptor family ligand binding domain; Lig_chan - Ionotropic glutamate receptor; Lig_chan-Glu_bd - Ligated ion channel L-glutamate- and glycine-binding site; CaM_bdg_C0 - Calmodulin-binding domain C0 of NMDA receptor NR1 subunit. Visualization of splicing events in cluster clu_35560 with the change in PSI (ΔPSI) for ASD (left) and SCZ (right) group comparisons. FDR-corrected p-values (q) are indicated for each comparison. Covariate-adjusted average PSI levels in ASD or SCZ (red) vs CTL (blue) are indicated at each intron. F) Violin-plots with the distribution of covariate-adjusted PSI per sample for the intron skipping E4 are shown for each disease group comparison. G) DGE for GRIN1 in each disorder (*FDR< 5%). H) Whole-gene view of NRXN1 highlighting (dashed lines) the intron cluster with significant DS in ASD (clu_28264; chr2:50,847,321–50,850,452), as well as transcripts NRXN1–004 and NRXN1–012 that show significant DTU in SCZ and/or BD. Protein domain mappings are shown in purple. DM - Protein domains; Tx - Transcripts. ConA-like_dom_sf - Concanavalin A-like lectin/glucanase domain. EGF-like - Epidermal growth factor-like domain; Laminin_G - Laminin G domain; Neurexin-like - Neurexin/syndecan/glycophorin C domain. I) Left: close-up of exons and protein domains mapped onto the DS cluster, and FDR-corrected p-value (q). Right: visualization of introns in cluster clu_28264 with their change in percent spliced in (ΔPSI). Covariate-adjusted average PSI levels in ASD (red) vs CTL (blue) are indicated for each intron. J) Violin-plots with the distribution of covariate-adjusted PSI per sample for the largest intron skipping exon 8 (E8). K) Bar plots for changes in gene expression and transcript usage for NRXN1–004 and NRXN1–012 (*FDR< 5%).
Many differential splicing events show predictable functional consequences on protein isoforms. Notable examples include GRIN1 and NRXN1, which are known risk loci for neurodevelopmental disorders (35, 36). GRIN1 encodes the obligatory subunit of the NMDA-type glutamate ionotropic receptors, is upregulated in SCZ and BD and shows increased skipping of exon 4 in both ASD and SCZ that impacts its extracellular ligand-binding domain (Fig 2E-G). NRXN1 is a heterotypic, presynaptic cell adhesion molecule that undergoes extensive alternative splicing and plays a key role in the maturation and function of synapses (35, 37). We observed various DS and/or differential transcript usage (DTU) changes in NRXN1 in ASD, SCZ and/or BD (Fig 2H-K). An exon skipping event in ASD disrupts a laminin domain in NRXN1 (Fig 2I-J); changes which are predicted to have major effects on its function (Fig 2H). Another example is CADPS, which is located within an ASD GWAS risk locus and supported by Hi-C defined chromatin interactions as a putative target gene (38) and manifests multiple isoform and splice alterations in ASD (Fig S9; Tables S1 and S3).
We found significant overlap (42%, P=3.42×10−27; Fisher’s exact test) of the ASD DS intron clusters and splicing changes identified in a previous study (19) that used a different method and only a subset of the samples in our ASD and control cohorts (Table S3). Overall, this examination of local splicing across three major neuropsychiatric disorders, coupled with the analysis of isoform-level regulation, emphasizes the need to understand the regulation and function of transcript isoforms at a cell type specific level in the human nervous system.
Identifying Drivers of Transcriptome Dysregulation
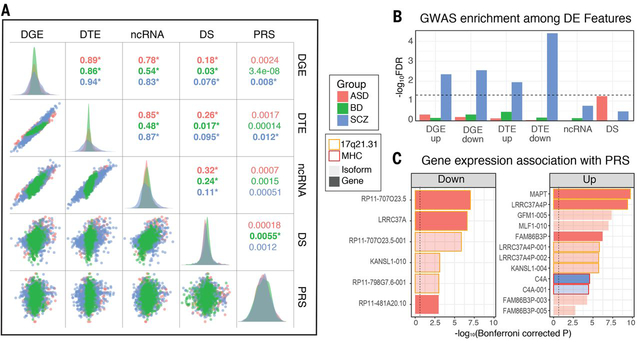
We next set to determine whether changes observed across levels of transcriptomic organization in psychiatric disease brain are reflective of the same, or distinct, underlying biological processes. Further, transcriptomic changes may represent a causal pathophysiology or may be a consequence of disease. To begin to address this, we assessed the relationships among transcriptomic features and with polygenic risk scores (PRS) for disease, which provide a directional, genetic anchor (Fig 3A). Across all three disorders, there was strong concordance among differential gene, isoform, and ncRNA signals, as summarized by their first principal component (Fig 3A). Notably, DS exhibited greatest overlap with the ncRNA signal, suggesting a role for non-coding genes in regulating local splicing events.
Figure 3. Overlaps and genetic enrichment among dysregulated transcriptomic features.
A) Scatterplots demonstrate overlap among dysregulated transcriptomic features, summarized by their first principle component across subjects (R2 values; *P<0.05). Polygenic risk scores (PRS) show greatest association with differential transcript signal in SCZ. B) SNP-heritability in SCZ is enriched among multiple differentially expressed transcriptomic features, with downregulated isoforms showing most substantial association via stratified LD-score regression. C) Several individual genes and isoforms exhibit genome-wide significant associations with disease PRS. Plots are split by direction of association with increasing PRS. In ASD, most associations localize to the 17q21.31 locus, harboring a common inversion polymorphism. In SCZ, a significant association as observed with C4A in the MHC locus.
Significant associations with PRS were observed for DGE and DTE signal in SCZ, with greater polygenic association at the isoform level in accordance with the larger transcript isoform effect sizes observed. Transcript-level differential expression also showed the greatest enrichment for SCZ SNP-heritability, as measured by stratified LD score regression (21, 39) (Fig 3B). The overall magnitude of genetic enrichment was modest, however, suggesting that most observed transcriptomic alterations are less a proximal effect of genetic variation and more likely the consequence of a downstream cascade of biological events following earlier acting genetic risk factors.
We were also interested to determine the degree to which genes showed increases in the magnitude of DE over the duration of illness, as a positive relationship would be expected if age-related cumulative exposures (e.g. drugs, smoking) were driving these changes. To assess this, we fit local regression models to case and control sample-level expression measurements as a function of age and computed age-specific DE effect-sizes (Fig S10). Of 4821 DE genes in SCZ, only 143 showed even nominal association between effect size magnitude and age. Similar associations were seen in 29 of 1119 DE genes in BD and 85 of 1611 DE genes in ASD. Consequently, this would not support substantial age-related environmental exposures as the mechanism for the vast majority of differentially expressed genes.
Using gene expression data from animal models, we investigated whether exposure to commonly used psychiatric medications could recapitulate observed gene expression changes in disease (Fig S11). Overall, with the exception of lithium, chronic exposure to medications including antipsychotics (clozapine, haloperidol), mood stabilizers (lamotrigine), and SSRI antidepressants (fluoxetine) had a small effect on the transcriptome, in many cases with no differentially expressed genes at traditional FDR thresholds (21). Even at more liberal thresholds, the overlap between medication-driven and disease signal remains sparse. One notable exception was a module that reflects major components of a well-described (40) neural activity-dependent gene expression program, whose disease relationships are refined in the network analysis section below. Finally, we note that other unmeasured factors could potentially contribute to gene expression variation in post-mortem tissue, including agonal events or smoking (22, 41, 42) in addition to those measured and used as covariates, such as RNA integrity and post mortem interval (PMI). We used surrogate variable correction in our analyses to account for such unmeasured confounders (43), which is a standard approach (44).
Transcriptome-wide Association
We next sought to leverage this transcriptomic dataset to prioritize candidate disease risk genes with predicted genetically-driven effects on expression in brain. We identified 18 genes or isoforms whose expression was significantly associated with PRS ((21); Bonferroni-corrected P < 0.05), 16 in ASD, 2 in SCZ, with none in BD (Fig 3C; Table S4). In ASD, the majority of associations map to 17q21.31, which harbors a common inversion polymorphism and rare deleterious structural variants associated with intellectual disability (45). Additional associations for ASD included two poorly annotated pseudogenes, FAM86B3P and RP11–481A20.10. In SCZ, PRS shows the genome-wide significant association with upregulation of the established risk gene C4A (5). Concordantly, we find a strong positive correlation between C4A expression and genetically imputed C4A copy number (R=0.36, P=6×10−21) and imputed number of C4-HERV elements (R=0.35, P=4×10−20), but a slight negative association with C4B copy number (R=−0.087, P=0.03; ref (21)). At less stringent thresholds (FDR-corrected P<0.05), we identify BD PRS associations with isoforms of the neuronal calcium sensor NCALD and SNF8, an endosomal sorting protein, as well as several additional associations in the MHC region in SCZ, which harbors the largest GWAS peak comprised of multiple independent signals (5), but is difficult to parse due to complex patterns of LD. These included two lncRNAs, HCG17 and HCG23, as well as the MHC class I heavy chain receptor HLA-C. However, expression of all three was also significantly (P<0.05) correlated with imputed C4A copy number, suggesting pleiotropic effects.
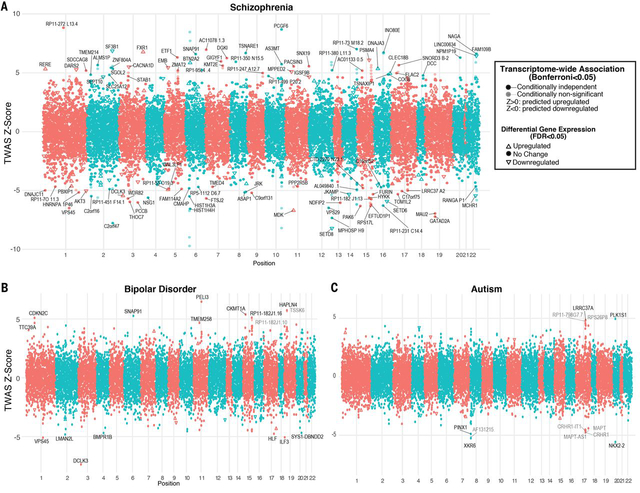
Taking an orthogonal approach, we performed a formal transcriptome-wide association study (TWAS; (46)) to directly identify those genes whose cis-regulated expression is associated with disease (21). TWAS and related methods have the advantage of aggregating the effects of multiple SNPs onto specific genes, reducing multiple comparisons and increasing power for association testing, although results can still be influenced by LD and pleiotropy (46, 47). Further, by imputing the cis-regulated heritable component of brain gene expression into the association cohort, TWAS enables direct prediction of the transcriptomic effects of disease-associated genetic variation, identifying potential mechanisms through which variants may impart risk. However, the limited size of brain eQTL datasets to date has necessitated use of non-CNS tissues to define TWAS weights (46). Given the enrichment of psychiatric GWAS signal within CNS expressed regulatory elements (39), we reasoned that our dataset would provide substantial power and specificity. Indeed, we identify 14,750 genes with heritable cis-regulated expression in brain in the PsychENCODE cohort, enabling increased transcriptomic coverage for detection of association signal (Fig 4). In BD, TWAS prioritizes 17 genes across 14 distinct loci (Bonferroni-corrected P<0.05; Fig 4; Table S4), none of which exhibited DE. At loci with multiple hits, we applied conditional analyses to further finemap these regions (21). For orthogonal validation, we conducted summary based mendelian randomization (SMR), a complementary method that tests for pleiotropic associations in the cis window with an accompanying HEIDI test to distinguish linkage from pleiotropy (48). Eight genes showed consistent association (21) across multiple analyses -- BMPR1B, DCLK3, HAPLN4, HLF, LMAN2L, MCHR1, UBE2Q2L, SNAP91, TTC39A, TMEM258, and VPS45 (Table S4). The two isoforms with PRS associations in BD (NCALD, SNF8) were non-significant in TWAS, perhaps due to lack of a nearby genome-wide significant locus or isoform-specific regulation, suggesting those expression changes may be driven by trans-acting factors.
Figure 4. Transcriptome-wide association.
Results from TWAS prioritize genes whose cis-regulated expression in brain is associated with disease. Plots show conditionally-independent TWAS prioritized genes, with lighter shade depicting marginal associations. The sign of TWAS Z-scores indicates predicted direction of effect. Genes significantly up or downregulated in disease brain are shown with arrows, indicating directionality. A) In SCZ, 193 genes (164 outside of MHC) are prioritized by TWAS at Bonferroni-corrected P<0.05, including 107 genes with conditionally independent signals. Of these, 23 are also differentially expressed in SCZ brain with 11 in the same direction as predicted. B) Seventeen genes are prioritized by TWAS in BD, of which 15 are conditionally independent. C) In ASD, TWAS prioritizes 12 genes, of which 5 are conditionally independent.
In ASD, TWAS prioritizes 12 genes across 3 genomic loci (Bonferroni-corrected P<0.05; Fig 4). This includes the 17q21.31 region, which showed multiple PRS associations as described above, but did not reach genome-wide significance in the largest GWAS to date (38). Of the seven TWAS-significant genes at this locus, conditional analysis prioritizes one – LRRC37A, which is further supported by SMR and Hi-C interaction in fetal brain (38). LRRC37A is intriguing due to its primate-specific evolutionary expansion, loss-of-function intolerance, and expression patterns in brain and testis (45). However, common variants in GWAS are also likely tagging the common inversion and other recurrent structural variants present at this locus (45). TWAS additionally prioritizes genes on chromosomes 8 and 20 (Fig 4). Altogether, five genes showed consistent associations with ASD across multiple methods: LRRC37A, FAM86B3P, PINX1, XKR6, RP11–481A20.10 (Table S4; (21)).
In SCZ, TWAS identifies 193 genes of which 107 remain significant following conditional analysis at each gene within multi-hit loci. Excluding the MHC region, there remained 164 significant genes representing 78 genome-wide significant GWAS loci (Fig 4; Table S4). A previous TWAS study in SCZ primarily based on non-neural tissue prioritized 157 genes, 37 of which are identified here, a significant overlap (OR 61, p<10−42, Fisher’s exact test). Moreover, 60 TWAS prioritized genes overlapped with the list of 321 ‘high confidence’ SCZ risk genes identified in the companion manuscript (18), identified using gene regulatory networks and a deep learning approach (OR 34.7, p<10−60, Fisher’s exact test). Of the 107 conditionally significant genes prioritized by TWAS, 62 were further supported by SMR (PSMR<0.05, PHEIDI>0.05) and 11 were also concordantly DE in SCZ brain in the same direction as predicted by TWAS. Altogether, 64 genes were consistently prioritized across multiple methods, including 10 ncRNAs (Table S4; (21)). These included a number of intriguing novel candidates for SCZ: two downregulated lysine methyltransferayjec9vses (SETD6, SETD8); RERE, a downregulated, mutationally intolerant nuclear receptor co-regulator of retinoic acid signaling associated with a rare neurodevelopmental genetic syndrome; LINC00634, a downregulated poorly annotated brain-enriched lincRNA; and SLC12A5, encoding a mitochondrial Ca2+ binding aspartate/glutamate carrier protein, associated with a recessive epileptic encephalopathy. Most genes identified in this analysis show disease-specific effects, as only four genes (MCHR1, VPS45, SNAP91, DCLK3) showed overlap between SCZ and BD TWAS, and none overlapped with ASD. Overall, this analysis provides a core set of strong candidate genes implicated by risk loci, and provides a mechanistic basis for the composite activity of disease risk variants.
Networks Refine Shared Cross-Disorder Signals
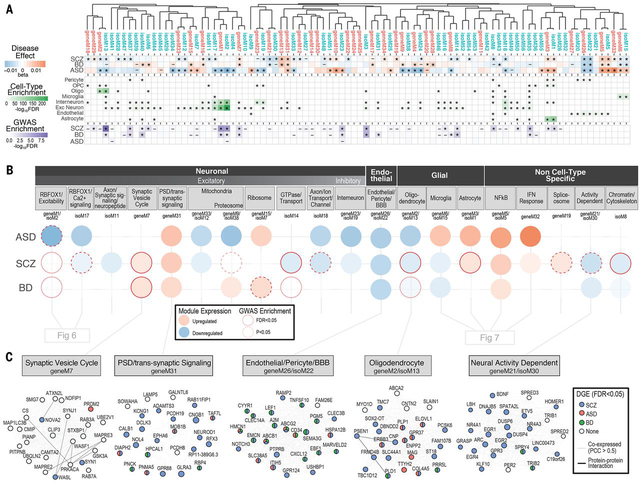
To place transcriptomic changes within a systems-level context and more fully interrogate the specific molecular neuropathology of these disorders, we performed weighted correlation network analysis (WGCNA) to create independent gene and isoform-level networks (15, 49, 50), which we then assessed for disease association and GWAS enrichment using stratified LD score regression ((21); see Resource.PsychENCODE.org for interactive visualization). Although calculated separately, gene and isoform-level networks generally reflect equivalent biological processes, as demonstrated by hierarchical clustering (Fig 5A). However, the isoform-level networks captured greater detail and a larger proportion were associated with disease GWAS than gene-level networks (61% vs 41% with nominal GWAS enrichment, P=0.07, χ2; Fig 5A). Consistent with expectations, modules showed enrichment for gene ontology pathways and we identified modules strongly and selectively enriched for markers of all major CNS cell types (Fig 5A-B; Fig S12), facilitating computational deconvolution of cell type specific signatures (15, 49, 51). For ease of subsequent presentation, we group gene-isoform module pairs that co-cluster, have overlapping parent genes, and represent equivalent biological processes.
Figure 5. Gene and isoform co-expression networks capture shared and disease-specific cellular processes and interactions.
A) Gene and isoform co-expression networks demonstrate pervasive dysregulation across psychiatric disorders. Hierarchical clustering shows that separate gene- and isoform-based networks are highly overlapping, with greater specificity conferred at the isoform level. Disease associations are shown for each module (linear regression β value, * FDR<0.05, – P<0.05). Module cell type enrichments (*FDR<0.05) are shown for major CNS cell types defined from PsychENCODE UMI single cell clusters. Enrichments are shown for GWAS results from SCZ (59), BD (90), and ASD (38), using stratified LD score regression (* FDR<0.05, – P<0.05). B) Co-expression modules capture specific cellular identities and biological pathways. Colored circles represent module differential expression effect size in disease, with red outline representing GWAS enrichment in that disorder. Modules are organized and labeled based on CNS cell type and top gene ontology enrichments. C) Examples of specific modules dysregulated across disorders, with top 25 hub genes shown. Edges represent co-expression (Pearson correlation > 0.5) and known protein-protein interactions. Nodes are colored to represent disorders in which that gene is differentially expressed (*FDR<0.05).
The large sample sizes, coupled with the specificity of isoform-level quantifications, enabled refinement of previously identified gene networks related to ASD, BD and SCZ (1, 2, 15, 16, 19, 52). Of a combined 90 modules, including 34 gene- (geneM) and 56 isoform-level (isoM) modules, 61 (68%) showed significant association with at least one disorder, demonstrating the pervasive nature of transcriptome dysregulation in psychiatric disease. Five modules are shared across all three disorders, 3 up and two downregulated; 22 modules are shared by 2 of the 3 disorders, and 36 demonstrate more specific patterns of dysregulation in either ASD, SCZ or BD (Fig 5; Table S5). It is notable that of these 61 co-expression modules with a disease-association, 41 demonstrate cell type enrichments, consistent with the strong cell type disease-related signal observed using both supervised and unsupervised methods in our companion paper (18). This demonstrates the importance of cell type specific changes in the molecular pathology of these major psychiatric disorders; the cell type relationships defined by the disease modules substantially enhance our knowledge of these processes, as we outline below.
The five modules shared between ASD, BD and SCZ can be summarized to represent 3 distinct biological processes. Two of these processes are upregulated, including an inflammatory NFkB signaling module pair (geneM5/isoM5; further discussed in neural-immune section below), and a module (geneM31) enriched primarily for genes with roles in the postsynaptic density, dendritic compartments, and receptor mediated presynaptic signaling that are expressed in excitatory neurons, and to a lesser extent, inhibitory neurons (Fig 5C). Remarkably, DCLK3, one of the hubs of geneM31, is a genome-wide significant TWAS hit in both SCZ and BD. The third biological process, geneM26/isoM22 (Fig 5C), is downregulated, and enriched for endothelial and pericyte genes, with hubs that represent markers of the blood-brain barrier, including ITIH5, SLC38A5, ABCB1, and GPR124, a critical regulator of brain-specific angiogenesis (53, 54). This highlights specific, shared alterations in neuronal-glial-endothelial interactions across these neuropsychiatric disorders.
In contrast to individual genes or isoforms, no modules were significantly associated with PRS scores after multiple-testing correction. However, 19 modules were significantly (FDR < 0.05) enriched for SNP-heritability based on published GWAS ((21); Fig 5A; Fig S13). A notable example is geneM2/isoM13, which is enriched for oligodendrocyte markers and neuron projection developmental pathways and is downregulated in ASD and SCZ, with a trend in BD (Fig 5C). isoM13 showed the greatest overall significance of enrichment for SCZ and educational attainment GWAS, and was also enriched in BD GWAS to a lesser degree, Further, this module is enriched for genes harboring ultra-rare variants identified in SCZ (55) (Fig S13). Finally, we also observe pervasive and distinct enrichments for syndromic genes and rare variants identified through whole exome sequencing in individuals with neurodevelopmental disorders (Table S5; Fig S13).
Neuronal Isoform Networks Capture Disease Specificity
Multiple neuronal and synaptic signaling pathways have been previously demonstrated to be downregulated in a diminishing gradient across ASD, SCZ, and BD brains without identification of clear disease-specific signals for these neuronal-synaptic gene sets (1, 2, 16, 19, 56, 57). We do observe neuronal modules broadly dysregulated across multiple disorders, including a neuronal/synaptic module (isoM18) with multiple isoforms of the known ASD risk gene, ANK2, as hubs. However, the large sample size, coupled with the specificity of isoform-level qualifications, enabled us to identify synaptic modules containing unique isoforms with distinct disease associations and to separate signals from excitatory and inhibitory neurons (Fig 5B).
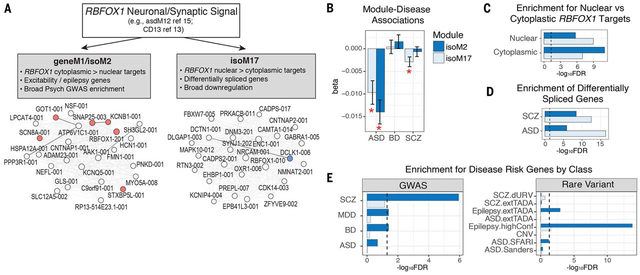
A salient example of differential module membership and disease association of transcript isoforms is RBFOX1, a major neuronal splicing regulator implicated across multiple neurodevelopmental and psychiatric disorders (16, 32, 58, 59). Previous work has identified downregulated neuronal modules in ASD and SCZ containing RBFOX1 as a hub (1, 16). Here, we identify two neuronal modules with distinct RBFOX1 isoforms as hub genes (Fig 6A). The module pair geneM1/isoM2, downregulated only in ASD (Fig 6B), contains the predominant brain-expressed RBFOX1 isoform and includes several cation channels (e.g., HCN1, SCN8A). The second most abundant RBFOX1 isoform is in another module, isoM17, which is downregulated in both ASD and SCZ (Fig 6B). Experiments in mouse indicate that RBFOX1 has distinct nuclear and cytoplasmic isoforms with differing functions, the nuclear isoform primarily regulating pre-mRNA alternative splicing, and the cytoplasmic isoform binding to the 3’ UTR to stabilize target transcripts involved in regulation of neuronal excitability (28, 32, 58, 60). Here, we find that isoM17 shows greater enrichment for nuclear RBFOX1 targets (Fig 6C), whereas isoM2 shows stronger overlap with cytoplasmic targets (32). Consistent with a predicted splicing-regulatory effect, isoM17 shows greater enrichment for genes exhibiting DS in ASD and SCZ (Fig 6D). In accordance with a predicted role in regulating excitability, isoM2 shows strong enrichment for epilepsy risk genes (Fig 6E). Moreover, the two modules show differential association with common genetic risk (Fig 6E), with isoM2 exhibiting GWAS enrichment across SCZ, BD, and MDD. This widespread enrichment of neurodevelopmental and psychiatric disease risk factors -- from rare variants in epilepsy to common variants in BD, SCZ, and MDD – is consistent with a model where broad neuropsychiatric liability emanates from myriad forms of dysregulation in neuronal excitability, all linked via RBFOX1. These results highlight the importance of further studies focused on understanding the relationship between human RBFOX1 transcript diversity and functional divergence, as most of what is known is based on mouse, and the human shows far greater transcript diversity (32, 58, 61).
Figure 6. Two RBFOX1 isoform modules capture distinct biological and disease associations.
A) Previous studies have identified RBFOX1 as a critical hub of neuronal and synaptic modules downregulated across multiple psychiatric disorders (1, 16, 19, 32). Here, we identify two pairs of modules with distinct RBFOX1 isoforms as hub genes. Plots show the top 25 hub genes of modules isoM2 and isoM17, following the same coloring scheme as Fig 5C. B) Distinct module-eigengene trait associations are observed for isoM2 (downregulated in ASD only) compared with isoM17, which is downregulated in ASD and SCZ. C) Modules show distinct enrichments for nuclear and cytoplasmic RBFOX1 targets, defined experimentally in mouse (32). D) Genes harboring differential splicing events observed in ASD and SCZ show greater overlap with isoM17, consistent with its association with nuclear RBFOX1 targets. E) Modules show distinct patterns of genetic association. isoM2 exhibits broad enrichment for GWAS signal in SCZ, BD, and MDD, as well as for epilepsy risk genes, whereas isoM17 shows no apparent genetic enrichment. GWAS enrichments show FDR-corrected P-values calculated using stratified-LDSC, and rare-variant associations were calculated using logistic regression, controlling for gene length and GC content (21).
Previous transcriptional networks related to ASD, BD and SCZ did not separate inhibitory and excitatory neuron signals (1). The increased resolution here allowed us to identify several modules enriched in inhibitory interneuron markers (Fig 5B), including geneM23/isoM19, which is downregulated in ASD and SCZ, with a trend toward downregulation observed in BD; downsampling in the SCZ dataset suggests that the lack of significance in BD may be due to smaller sample size (Fig S14). This module pair contained as hubs the two major GABA synthesizing enzymes (GAD1, GAD2), multiple GABA transporters (SLC6A1, SLC24A3), many other known interneuron markers (RELN, VIP), as well as DLX1 and the lncRNA DLX6-AS1, both critical known regulators of inhibitory neuron development (62). This inhibitory neuron-related module is not enriched for common or rare genetic disease-associated variation, although other studies have found enrichment for SCZ GWAS signal among interneuron markers defined in other ways (63).
Several neuronal modules that distinguish between the disorders differentiate BD and SCZ from ASD, including the module pair geneM21/isoM30 (Fig 5C), which captures known elements of activity-dependent neuronal gene regulation, whose hubs include classic early-response (ARC, EGR1, NPAS4, NR4A1) and late-response genes (BDNF, HOMER1) (40). Although these modules were not significantly downregulated in ASD, subsampling indicates that the differences between disorders could be driven by sample size (Fig S14). These genes play critical roles in regulating synaptic plasticity and the balance of excitatory and inhibitory synapses (40). Remarkably, a nearly identical module was recently identified as a sex-specific transcriptional signature of major depression and stress susceptibility (64). Since psychiatric drug use is more prevalent in SCZ and BD than ASD, and the geneM21/isoM30 module pair are altered more substantially in these disorders, we explored whether these modules may be affected by medication exposure. Indeed, geneM21/isoM30 was associated with genes downregulated by chronic high-doses (but not low-doses) of haloperidol, as well as genes upregulated by the antidepressant fluoxetine (Fig S11A). Furthermore, geneM21/isoM30 expression was negatively correlated with the degree of lifetime antipsychotic exposure in the subset of patients for whom these data were available (P=0.001, Pearson; Fig S11B). As such, it will be worthwhile to determine whether this module is a core driver of the therapeutic response, as has been suggested (65). Other neuronal modules distinguished SCZ and BD from ASD (Fig 5B), including geneM7, enriched for synaptic and metabolic processes with the splicing regulator NOVA2 (Fig 5C). This neuronal module was significantly enriched for both BD and SCZ GWAS signals, supporting a causal role for this module.
Distinct Trajectories of Neural-Immune Dysregulation
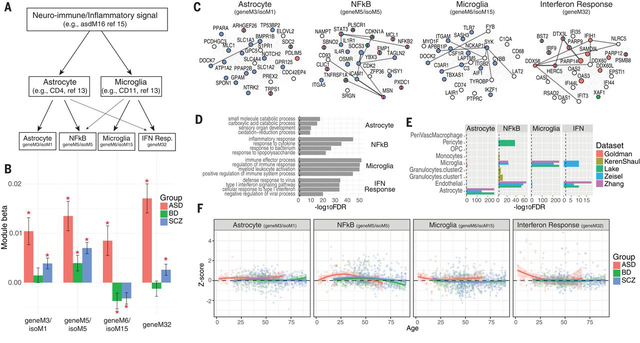
Previous work has identified differential activation of glial and neural-immune processes in brain from patients with psychiatric disorders (16, 52, 57, 66–69), including upregulation of astrocytes in SCZ and BD (1, 57) and both microglia and astrocytes in ASD (19, 70). Evidence supports hyperactive complement-mediated synaptic pruning in SCZ pathophysiology, presumably through microglia (5), although post-mortem microglial upregulation was observed only in ASD (1, 19, 70). We examined whether our large cohort including ~1000 control brains, capturing an age range from birth to 90 years, would enable refinement of the nature and timing of this neuroinflammatory signal and potential relationship to disease pathogenesis (Fig 7A). Four modules were directly related to neural-immune processes (Fig 7A-C), two of which are gene/isoform module pairs that correspond clearly to cell type specific gene expression; one representing microglia (geneM6/isoM15) and the other astrocytes (geneM3/isoM1), as they are strongly and selectively enriched for canonical cell type specific marker genes (Fig 7C-E). Two additional immune-related modules appear to represent more broadly expressed signaling pathways: interferon response (geneM32) and NFkB (geneM5/isoM5). The interferon response module (geneM32) contains critical components of the IFN-stimulated gene factor 3 (ISGF3) complex that activates the transcription of downstream interferon-stimulated genes (ISGs), which comprise a striking 59 of the 61 genes in this module (71). The NFkB module pair (geneM5/isoM5) includes four out of five of the NFkB family members (NFkB1, NFkB2, REL, RELA), as well as many downstream transcription factor targets and upstream activators of this pathway.
Figure 7. Distinct neural-immune trajectories in disease.
A) Co-expression networks provide substantial refinement of the neuro-immune/inflammatory processes upregulated in ASD, SCZ, and BD. Previous work has identified specific contributions to this signal from astrocyte and microglial populations (1, 19). Here, we further identify additional, distinct interferon (IFN)-response and NFkB signaling modules. B) Eigengene-disease associations are shown for each of 4 identified neural-immune module pairs. The astrocyte and IFN-response modules are upregulated in ASD and SCZ. NFkB signaling is elevated across all three disorders. The microglial module is upregulated in ASD and downregulated in SCZ and BD. C) Top hub genes for each module are shown, along with edges supported by co-expression (light grey; Pearson correlation>0.5) and known protein-protein interactions (dark lines). Nodes follow same coloring scheme as in Fig 5C. Hubs in the astrocyte module (geneM3/isoM1) include several canonical, specific astrocyte markers, including SOX9, GJA1, SPON1, and NOTCH2. Microglial module hub genes include canonical, specific microglial markers, including AIF1, CSF1R, TYROBP, TMEM119. The NFkB module includes many known downstream transcription factor targets (JAK3, STAT3, JUNB, FOS) and upstream activators (IL1R1, 9 TNF receptor superfamily members) of this pathway. D) The top 4 GO enrichments are shown for each module. E) Module enrichment for known cell type-specific marker genes, collated from sequencing studies of neural-immune cell types (91–95). F) Module eigengene expression across age demonstrates distinct and dynamic neural-immune trajectories for each disorder.
The dynamic trajectories of these processes in cases with respect to controls reveal distinct patterns across disorders (Fig 7F). The IFN-response and microglial modules are most strongly upregulated in ASD, peaking during early development, coincident with clinical onset. In contrast, in SCZ and BD, the microglial module is actually downregulated and driven by a later dynamic decrease, dropping below controls after age 30. The NFkB module, which is upregulated across all three disorders, maximally diverges from controls during early adulthood, coincident with typical disease onset in SCZ and BD (~25). Accordingly, this NFkB module contained C4A – the top GWAS-supported, and strongly upregulated, risk gene for SCZ (5). This pattern is clearly distinct from ASD, which shows a dynamic trajectory, but remains upregulated throughout (Fig 7F).
Non-coding Modules and lncRNA Regulatory Relationships
Given that many lncRNAs are predicted to have transcriptional regulatory roles, we next assessed whether mRNA-based co-expression networks could provide additional functional annotation for ncRNAs. As a subset of lncRNAs are thought to function by repressing mRNA targets (72), we applied csuWGCNA (73) to identify potential regulatory relationships (21). We identified 39 modules (csuM) using csuWGCNA, all preserved in the signed networks with strong cell type and GWAS enrichments, which captured 7186 negatively correlated lncRNA-mRNA pairs within the same module (Fig S15). We provide a table of putative mRNA targets for these brain expressed lncRNAs, including 209 exhibiting DE in ASD, 122 in BD and 241 in SCZ (Table S6).
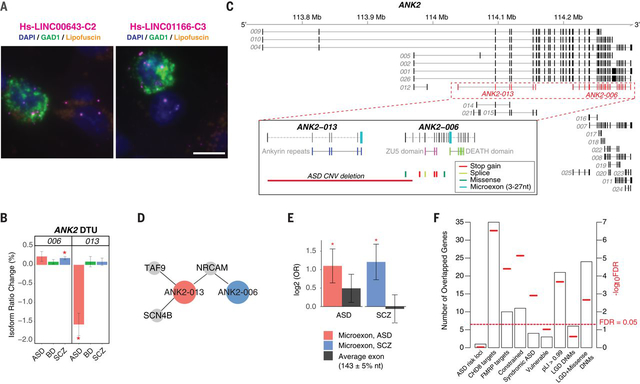
A salient example of the power of this approach for functional annotation is LINC00473, a hub of the neuronal activity dependent gene regulation module (geneM21/isoM30; Fig 5C). Expressed in excitatory neurons and downregulated in SCZ (log2FC −0.16, FDR<0.002), LINC00473 is regulated by synaptic activity and downregulates immediate early gene expression (74), consistent with its hub status in this module. Similarly, we identify the lncRNA DLX6-AS1, a known development regulator of interneuron specification (62), as the most central hub gene in the interneuron module (geneM23/isoM19), which is downregulated in ASD and SCZ. This interneuron module also contains LINC00643 and LINC01166, two poorly annotated, brain enriched lncRNAs. LINC00643 is downregulated in SCZ (log2FC −0.06, FDR=0.04) whereas LINC01166 is significantly downregulated in BD (log2FC −0.17, FDR<0.05) with trends in ASD and SCZ (FDR’s < 0.1). Our data suggest a role for these lncRNAs in interneuron development, making them intriguing candidates for follow-up studies. Using fluorescence in situ hybridization (FISH), we confirmed that both LINC00643 and LINC1166 are expressed in GAD1+ GABAergic neurons in area 9 of adult brain, present both in the cell nucleus and cytoplasm (Fig 8A; Fig S16), although expression was also detected in other non GAD1+ neurons as well.
Figure 8. LncRNA annotation, ANK2 isoform switching & microexon enrichment.
A) FISH images demonstrate interneuron expression for two poorly annotated lincRNAs – LINC00643 and LINC01166 – in area 9 of adult human prefrontal cortex. Sections were labeled with GAD1 probe (green) to indicate GABAergic neurons and lncRNA (magenta) probes for LINC00643 (left) or for LINC01166 (right). All sections were counterstained with DAPI (blue) to reveal cell nuclei. Lipofuscin autofluorescence is visible in both the green and red channels and appears yellow/orange. Scale bar, 10 μm. FISH was repeated at least twice on independent samples (Table S9 (21)) with similar results (see also Fig S16). B) ANK2 isoforms ANK2–006 and ANK2–013 show significant DTU in SCZ and ASD, respectively (*FDR<0.05). C) Exon structure of ANK2 highlighting (dashed lines) the ANK2–006 and ANK2–013 isoforms. Inset, these isoforms have different protein domains and carry different microexons. ANK2–006 is hit by multiple ASD DNMs while ANK2–013 could be entirely eliminated by a de novo CNV deletion in ASD. D) Disease-specific coexpressed PPI network. Both ANK2–006 and ANK2–013 interact with NRCAM. The ASD-associated isoform ANK2–013 has two additional interacting partners, SCN4B and TAF9. E) As a class, switch isoforms are significantly enriched in microexon(s). In contrast, exons of average length are not enriched among switch isoforms. Y-axis displays odds ratio on log2 scale. P-values are calculated using logistic regression and corrected for multiple comparisons. F) Enrichment of 64 genes with switch isoforms in: ASD risk loci (81); CHD8 targets (96); FMRP targets (33); Mutationally constraint genes (97); Syndromic and highly ranked (1 and 2) genes from SFARI Gene database; Vulnerable ASD genes (98); Genes with probability of loss-of-function intolerance (pLI) > 0.99 as reported by the Exome Aggregation Consortium (99); Genes with likely-gene-disruption (LGD) or LGD plus missense de novo mutations (DNMs) found in patients with neurodevelopmental disorders (21).
Multiple ncRNAs including SOX2-OT, MIAT, and MEG3 are enriched in oligodendrocyte modules (geneM2/isoM13/csuM1; Fig 5C) that are downregulated in both SCZ and ASD. SOX2-OT is a heavily spliced, evolutionarily-conserved lncRNA exhibiting predominant brain expression and a hub of these oligodendrocyte modules, without previous mechanistic links to myelination (75, 76). The lncRNAs MIAT and MEG3 are negatively correlated with most of the hubs in this module, including SOX2-OT (Fig S15). MIAT is also known to interact with QKI, an established regulator of oligodendrocyte-gene splicing also located in this module (77, 78). These analyses predict critical roles for these or these often overlooked non-coding genes in oligodendrocyte function (77, 78) and potentially in psychiatric conditions.
Isoform Network Specificity and Switching
To more comprehensively assess whether aspects of disease specificity are conferred by alternative transcript usage or splicing, versus DE, we surveyed genes exhibiting DTU across disorders (21). We identified 134 such ‘switch isoforms’, corresponding to 64 genes displaying different DTU between ASD and SCZ (Table S7). As an example, isoforms of SMARCA2, a member of the BAF-complex strongly implicated in several neurodevelopmental disorders including ASD (79), are up and downregulated in ASD and SCZ, respectively (Fig S17). Conversely, the isoforms of NIPBL, a gene associated with Cornelia de Lange Syndrome (80) are down and upregulated in ASD and SCZ, respectively (Fig S17). Such opposing changes in isoform expression of various genes may represent differences in disease progression or symptom manifestation in diseases as ASD and SCZ, mediated by genetic risk variants that create subtle differences in isoforms within the same gene that exhibit distinct biological effects in each disorder. A remarkable example is the ASD risk gene ANK2 (81), whose two alternatively spliced isoforms, ANK2–006 and ANK2–013, are differentially regulated in SCZ and ASD (Fig 8B). These switch isoforms show markedly different expression patterns, belonging to different co-expression modules, geneM3/isoM1 (Fig 7C) and isoM18, which are enriched in astrocyte and neuronal cell types, respectively (Fig 5A; Fig S12). The protein domain structure of these transcripts is also non-overlapping, with ANK2–006 carrying exclusively ZU5 and DEATH domains, and ANK2–013 carrying exclusively ankyrin repeat domains (Fig 8C). Both isoforms are impacted by a de novo ASD CNV, and ANK-006 also carries de novo mutations from neurodevelopmental disorders. Both isoforms bind to the neuronal cell adhesion molecular NRCAM, but ANK2–013 has two additional, unique partners – TAF9 and SCN4B (Fig 8D), likely cell type specific interactions that suggest distinct functions of the isoforms of this gene in different neural cell types and diseases.
Finally, several studies have demonstrated that genes carrying microexons are preferentially expressed in brain and their splicing is dysregulated in ASD (30, 82, 83). This PsychENCODE sample provided the opportunity to assess the role of microexons in a far larger cohort and across several disorders. Indeed, we find that switch isoforms with microexons (3–27 bp) are significantly enriched in both ASD (FDR=0.03) and SCZ (FDR=0.03, logistic regression) (Fig 8E; (21)). Genes with switch isoforms are also enriched for the regulatory targets of two ASD risk genes, CHD8 and FMRP, as well as highly mutationally constrained genes (pLI>0.99), syndromic ASD genes, and in genes with de novo exonic mutations in ASD, SCZ and BD (Fig 8F; Table S7; (21)). These data confirm the importance of microexon regulation in neuropsychiatric disorders beyond ASD, and their potential role in distinguishing among biological pathways differentially affected across conditions. This role for microexons further highlights local splicing regulation as a potential mechanism conferring key aspects of disease specificity, extending the larger disease signal observed at the isoform-level in co-expression and differential expression analyses.
Discussion
We present a large-scale RNA-Seq analysis of the cerebral cortex across three major psychiatric disorders, including extensive analyses of the non-coding and alternatively spliced transcriptome, as well as gene- and isoform-level co-expression networks. The scope and complexity of these data do not immediately lend themselves to simple mechanistic reduction. Nevertheless, at each level of analysis, we present concrete examples that provide proofs-of-principle and starting points for investigations targeting shared and distinct disease mechanisms to connect causal disease drivers with brain-level perturbations.
Broadly, we find that isoform-level changes exhibit the largest effect sizes in disease brain, are most enriched for genetic risk, and provide the greatest disease specificity when assembled into co-expression networks. Remarkably, disturbances in the expression of distinct isoforms of more than 50 genes are differentially observed in SCZ and ASD, which in the case of the ASD risk gene ANK2, is predicted to affect different cell types in each disorder. Moreover, we observe disease-associated changes in the splicing of dozens of RNA-binding proteins and splicing factors, most of whose targets and functions are unknown. Similarly, nearly 1000 ncRNAs are dysregulated in at least one disorder and most of these ncRNAs show significant CNS enrichment, but until now, have limited functional annotation.
This work highlights isoform-level dysregulation as a critical, and relatively underexplored, proximal mechanism linking genetic risk factors with psychiatric disease pathophysiology. In contrast to local splicing changes, isoform-level quantifications require imputation from short-read RNA-Seq data guided by existing genomic annotations. Consequently, the accuracy of these estimates is hindered by incomplete annotations, as well as by limitations of short-read sequencing, coverage, and genomic biases like GC content (84, 85). This may be particularly problematic in brain where alternative splicing patterns are more distinct than in other organ systems (82). We present experimental validations for several specific isoforms, but try to focus on the class of dysregulated isoforms, and the modules and biologically processes they represent, rather than individual cases which may be more susceptible to bias. Longer read sequencing, which provides a more precise means for isoform quantification, will be of great utility as it becomes more feasible at scale.
Several broad shared patterns of gene expression dysregulation have been observed in post mortem brain in previous studies, most prominently, a gradient of downregulation of neuronal and synaptic signaling genes, and upregulation of glial-immune or neuroinflammatory signals. Here, we refine these signals by distinguishing both up and downregulated neuron-related processes that are differentially altered across these three disorders. Furthermore, we extend previous work that identified broad neuroinflammatory dysregulation in SCZ, ASD, and BD, by identifying specific pathways involving IFN-response, NFkB, astrocytes and microglia that manifest distinct temporal patterns across conditions. A module enriched for microglial-associated genes, for example, shows a clear distinction between disorders, with strong upregulation observed on ASD and significant downregulation in SCZ and BD. Overall, these results provide increased specificity to the observations that ASD, BD, and SCZ are associated with elevated neuroinflammatory processes (69, 86–88).
By integrating transcriptomic data with genetic variation, we identify multiple disease-associated co-expression modules enriched for causal variation, as well as new mechanisms potentially underlying specific disease loci in each of the diseases. In parallel, by performing a well-powered brain-relevant TWAS in SCZ, and to a lesser extent in BD and ASD, we are further able to elucidate candidate molecular mechanisms through which disease-associated variants may act. TWAS prioritizes dozens of new candidate disease genes, including many dysregulated in disease brain. Similar to the eQTLs identified in the companion paper (18), the majority of these new loci do not overlap with disease GWAS association signals. Rather, most are outside of the LD block and quite distal to the original association signal, highlighting the importance of orthogonal functional data types, such as transcriptome or epigenetic data (17, 47, 82, 89), in deciphering the underlying mechanisms of disease-associated genetic effects.
As with any case/control association study, multiple potential factors contribute to gene expression changes in post-mortem human brain, many of which may represent reactive processes. At each step of analysis, we have attempted to mitigate the contribution of these factors through known and hidden covariate correction, assessment of age trajectories, and via enrichment for causal genetic variation. Supporting the generalizability of our findings, we find significant correlations of the log2FC between randomly split halves of the data (Fig S3). This likely varies by transcript class, and some of the modest correlations are likely due to low abundance genes, such as ncRNAs, which we prefer to include, while recognizing the inherent tension between expression level and measurement accuracy. We provide access to this extensive resource, both in terms of raw and processed data and as browsable network modules (Resource.PsychENCODE.org).
A large proportion of disease-associated co-expression modules are enriched for cell type specific markers, as is overall disease DE signal, indicating that transcriptomic alterations in disease are likely driven substantially by (even subtle) shifts in cell type proportions, or cell type specific pathways, consistent with our previous observations (1) and those in the companion PsychENCODE manuscript (18). Functional genomic studies often remove such cell type-specific signals, through use of large numbers of expression-derived principle components or surrogate variables as covariates, to remove unwanted sources of variation and maximize detection of cis eQTLs (44). We retain the cell type-specific signals as much as possible, reasoning that cell type-related alterations may directly inform the molecular pathology of disease in psychiatric disorders, in which there is no known microscopic or macroscopic pathology. This rationale is supported by the consistent observation of the dynamic and disease-specific microglial upregulation observed in ASD, and the shared astrocyte upregulation in SCZ and ASD. This approach, however, reduces the ability to detect genetic enrichment from GWAS, as current methods predominately capture cis-acting regulatory effects. The modesty of genetic enrichments among disease-associated transcriptomic alterations may also indicate that gene expression changes reflect an indirect cascade of molecular events triggered by environmental as well as genetic factors, or that genetic factors may act earlier such as during development.
Finally, these data, while providing a unique, large-scale resource for the field, also suggest that profiling additional brains, especially from other implicated brain regions from patients will continue to be informative. Similarly, these data suggest that isoform level analyses including the identification of isoform-specific PPI and cell type specificity, while posing major challenges for high-throughput studies, are likely to add substantial value to understanding brain function and neuropsychiatric disorders. Finally, as GWAS studies in ASD and BD increase in size and subsequently in power, their continued integration with these transcriptome data will likely prove critical in identifying the functional impact of disease-associated genetic variation.
Supplementary Material
Acknowledgements
We would like to acknowledge the National Institute of Mental Health (NIMH) for funding. Also, we acknowledge program staff, in particular T. Lehner, L. Bingaman, D. Panchision, A. Arguello and G. Senthil, for providing institutional support and guidance for this project. The authors also acknowledge A Silva, M López-Aranda, B Pasanuic, N Mancuso, C Giambartolomei for helpful discussions. Brain tissue for the study was obtained from the following brain bank collections: the Mount Sinai NIH Brain and Tissue Repository, the University of Pennsylvania Alzheimer’s Disease Core Center, the University of Pittsburgh NeuroBioBank and Brain and Tissue Repositories, the Banner Sun Health Research Institute, the Harvard Brain Bank as part of the Autism Tissue Project (ATP), the Stanley Medical Research Institute, and the NIMH Human Brain Collection Core. Tissue for the RNAscope experiments was obtained from the Mount Sinai Neuropathology Research Core and Brain Bank.
Funding: The work is funded by the U.S. National Institute of Mental Health (NIMH) (grants P50-MH106438, D.H.G.; R01-MH094714, D.H.G.; U01-MH103339, D.H.G.; R01-MH110927, D.H.G.; R01-MH100027, D.H.G.; R01-MH110920, C.L.; U01-MH103340, C.L.; MH109885, LMI; MH104766, LMI; MH105524, LMI; MH108528, LMI; R21-MH105881, D.P.; R01-MH110555, D.P.; R01-MH109715, D.P.), the Simons Foundation for Autism Research Initiative (SFARI grant 206733, D.H.G.; SFARI grant 345469, LMI; SFARI Bridge to Independence Award, M.J.G.; Simons Foundation Grant FA 345922, P.R.H.), the National Natural Science Foundation of China (grants 81401114, CC; 31571312, CC), the National Key Plan for Scientific Research and Development of China (2016YFC1306000, CC), the Innovation-Driven Project of Central South University (No. 2015CXS034, CC; 2018CX033, CC), and the Icahn School of Medicine at Mount Sinai computational resources by the Office of Research Infrastructure of the NIH (award number S10OD018522, D.P.). Data were generated as part of the PsychENCODE Consortium, supported by grants U01MH103339, U01MH103365, U01MH103392, U01MH103340,U01MH103346, R01MH105472, R01MH094714, R01MH105898,R21MH102791, R21MH105881, R21MH103877, and P50MH106934 awarded to Schahram Akbarian (Icahn School of Medicine at Mount Sinai), Gregory Crawford (Duke), Stella Dracheva (Icahn School of Medicine at Mount Sinai), Peggy Farnham (USC), Mark Gerstein (Yale), Daniel Geschwind (UCLA), Thomas M. Hyde (LIBD), Andrew Jaffe (LIBD), James A. Knowles (USC), Chunyu Liu (SUNY Upstate), Dalila Pinto (Icahn School of Medicine at Mount Sinai), Nenad Sestan (Yale), Pamela Sklar (Icahn School of Medicine at Mount Sinai), Matthew State (UCSF), Patrick Sullivan (UNC), Flora Vaccarino (Yale), Sherman Weissman(Yale), Kevin White (UChicago), and Peter Zandi (JHU). RNA-Seq data from the CommonMind Consortium used in this study (Synapse accession no. syn2759792) was supported by funding from Takeda Pharmaceuticals Company, F. Hoffman-La Roche and NIH grants R01MH085542, R01MH093725, P50MH066392, P50MH080405, R01MH097276, R01MH075916, P50M096891, P50MH084053S1, R37MH057881, R37MH057881S1, HHSN271201300031C, AG02219, AG05138, and MH06692.
Footnotes
Competing Interests: K.P.W. is associated with Tempus Labs. The other authors declare no direct competing interests.
Data and Materials Availability: All data are available at doi.org/10.7303/syn1208024 (100). Summary data are available on our companion website at Resource.PsychENCODE.org.
References and Notes
- 1.Gandal MJ et al. , Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science. 359, 693–697 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fromer M et al. , Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci 19, 1442–1453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whiteford HA, Ferrari AJ, Degenhardt L, Feigin V, Vos T, The global burden of mental, neurological and substance use disorders: an analysis from the Global Burden of Disease Study 2010. PLoS One. 10, e0116820 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gandal MJ, Leppa V, Won H, Parikshak NN, Geschwind DH, The road to precision psychiatry: translating genetics into disease mechanisms. Nat. Neurosci 19, 1397–1407 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sekar A et al. , Schizophrenia risk from complex variation of complement component 4. Nature. 530, 177–183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanders SJ, First glimpses of the neurobiology of autism spectrum disorder. Curr. Opin. Genet. Dev 33, 80–92 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Maurano MT et al. , Systematic localization of common disease-associated variation in regulatory DNA. Science. 337, 1190–1195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ward LD, Kellis M, Interpreting noncoding genetic variation in complex traits and human disease. Nat. Biotechnol 30, 1095–1106 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Visel A, Rubin EM, Pennacchio LA, Genomic views of distant-acting enhancers. Nature. 461, 199–205 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.GTEx Consortium, Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 348, 648–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reilly SK et al. , Evolutionary genomics. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science. 347, 1155–1159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The ENCODE Project Consortium, Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 447, 799 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roadmap Epigenomics Consortium et al. , Integrative analysis of 111 reference human epigenomes. Nature. 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andersson R et al. , An atlas of active enhancers across human cell types and tissues. Nature. 507, 455–461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parikshak NN, Gandal MJ, Geschwind DH, Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat. Rev. Genet 16, 441–458 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voineagu I et al. , Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 474, 380–384 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.PsychENCODE Consortium et al. , The PsychENCODE project. Nat. Neurosci 18, 1707–1712 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Daifeng, Liu Shuang, Warrell Jonathan, Won Hyejung, Shi Xu, Navarro Fabio C.P., Clarke Declan, Gu Mengting, Emani Prashant, Yang Yucheng T., Xu Min, Gandal Michael J., Lou Shaoke, Zhang Jing, Park Jonathan J., Yan Chengfei, Rhie Suhn Kyong, Manakongtreecheep Kasidet, Zhou Holly, Nathan Aparna, Peters Mette, Mattei Eugenio, Fitzgerald Dominic, Brunetti Tonya, Moore Jill, Jiang Yan, Girdhar Kiran, Hoffman Gabriel E., Kalayci Selim, Gümüş Zeynep H., Crawford Gregory E., PsychENCODE Consortium, Roussos Panos, Akbarian Schahram,, Jaffe Andrew E., White Kevin P., Weng Zhiping, Sestan Nenad, Geschwind Daniel H., Knowles James A., Gerstein Mark B., Comprehensive functional genomic resource and integrative model for the human brain. Science. Under revision (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parikshak NN et al. , Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature. 540, 423–427 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang HJ et al. , Spatio-temporal transcriptome of the human brain. Nature. 478, 483–489 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.See supplemental methods.
- 22.Jaffe AE et al. , qSVA framework for RNA quality correction in differential expression analysis. Proc. Natl. Acad. Sci. U. S. A 114, 7130–7135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li B, Dewey CN, RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaffe AE et al. , Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat. Neurosci 21, 1117–1125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li YI et al. , RNA splicing is a primary link between genetic variation and disease. Science. 352, 600–604 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Batista PJ, Chang HY, Long noncoding RNAs: cellular address codes in development and disease. Cell. 152, 1298–1307 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.di Iulio J et al. , The human noncoding genome defined by genetic diversity. Nat. Genet 50, 333–337 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Vuong CK, Black DL, Zheng S, The neurogenetics of alternative splicing. Nat. Rev. Neurosci 17, 265–281 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li YI et al. , Annotation-free quantification of RNA splicing using LeafCutter. Nat. Genet 50, 151–158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irimia M et al. , A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 159, 1511–1523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akula N et al. , RNA-sequencing of the brain transcriptome implicates dysregulation of neuroplasticity, circadian rhythms and GTPase binding in bipolar disorder. Mol. Psychiatry 19, 1179–1185 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J-A et al. , Cytoplasmic Rbfox1 Regulates the Expression of Synaptic and Autism-Related Genes. Neuron. 89, 113–128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darnell JC et al. , FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 146, 247–261 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sebestyén E et al. , Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks. Genome Res. 26, 732–744 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gauthier J et al. , Truncating mutations in NRXN2 and NRXN1 in autism spectrum disorders and schizophrenia. Hum. Genet. 130, 563–573 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamdan FF et al. , Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am. J. Hum. Genet 88, 306–316 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Treutlein B, Gokce O, Quake SR, Südhof TC, Cartography of neurexin alternative splicing mapped by single-molecule long-read mRNA sequencing. Proc. Natl. Acad. Sci. U. S. A 111, E1291–E1299 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grove J et al. , Common risk variants identified in autism spectrum disorder. bioRxiv (2017), p. 224774. [Google Scholar]
- 39.Finucane HK et al. , Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet 47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.West AE, Greenberg ME, Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb. Perspect. Biol 3 (2011), doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu Y, Wang L, Yin Y, Yang E, Systematic analysis of gene expression patterns associated with postmortem interval in human tissues. Sci. Rep 7, 5435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li JZ et al. , Systematic changes in gene expression in postmortem human brains associated with tissue pH and terminal medical conditions. Hum. Mol. Genet 13, 609–616 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Leek JT, Storey JD, Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 3, 1724–1735 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.GTEx Consortium, Genetic effects on gene expression across human tissues. Nature. 550, 204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zody MC et al. , Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat. Genet 40, 1076–1083 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gusev A et al. , Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet 48, 245–252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gamazon ER et al. , A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet 47, 1091–1098 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu Z et al. , Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet 48, 481–487 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Oldham MC et al. , Functional organization of the transcriptome in human brain. Nat. Neurosci 11, 1271–1282 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang B, Horvath S, A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol 4, Article17 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Miller JA, Oldham MC, Geschwind DH, A systems level analysis of transcriptional changes in Alzheimer’s disease and normal aging. J. Neurosci 28, 1410–1420 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen C et al. , Two gene co-expression modules differentiate psychotics and controls. Mol. Psychiatry 18, 1308–1314 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Daneman R et al. , The mouse blood-brain barrier transcriptome: a new resource for understanding the development and function of brain endothelial cells. PLoS One. 5, e13741 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Obermeier B, Daneman R, Ransohoff RM, Development, maintenance and disruption of the blood-brain barrier. Nat. Med 19, 1584–1596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Genovese G et al. , Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nat. Neurosci 19, 1433–1441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ellis SE, Panitch R, West AB, Arking DE, Transcriptome analysis of cortical tissue reveals shared sets of downregulated genes in autism and schizophrenia. Transl. Psychiatry 6, e817 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramaker RC et al. , Post-mortem molecular profiling of three psychiatric disorders. Genome Med. 9, 72 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vuong CK et al. , Rbfox1 Regulates Synaptic Transmission through the Inhibitory Neuron-Specific vSNARE Vamp1. Neuron. 98, 127–141.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pardiñas AF et al. , Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet 50, 381–389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gehman LT et al. , The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat. Genet 43, 706–711 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fogel BL et al. , RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum. Mol. Genet 21, 4171–4186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bond AM et al. , Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci 12, 1020–1027 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Skene NG et al. , Genetic identification of brain cell types underlying schizophrenia. Nat. Genet 50, 825–833 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Labonté B et al. , Sex-specific transcriptional signatures in human depression. Nat. Med 23, 1102–1111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Bartolomeis A et al. , Immediate-Early Genes Modulation by Antipsychotics: Translational Implications for a Putative Gateway to Drug-Induced Long-Term Brain Changes. Front. Behav. Neurosci 11, 240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Birnbaum R et al. , Investigating the neuroimmunogenic architecture of schizophrenia. Mol. Psychiatry 23, 1251–1260 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Fillman SG et al. , Increased inflammatory markers identified in the dorsolateral prefrontal cortex of individuals with schizophrenia. Mol. Psychiatry 18, 206–214 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Pacifico R, Davis RL, Transcriptome sequencing implicates dorsal striatum-specific gene network, immune response and energy metabolism pathways in bipolar disorder. Mol. Psychiatry 22, 441–449 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Rao JS, Harry GJ, Rapoport SI, Kim HW, Increased excitotoxicity and neuroinflammatory markers in postmortem frontal cortex from bipolar disorder patients. Mol. Psychiatry 15, 384–392 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gupta S et al. , Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nat. Commun 5, 5748 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rusinova I et al. , Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 41, D1040–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Necsulea A et al. , The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 505, 635–640 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Dai R, Xia Y, Liu C, Chen C, csuWGCNA: a combination of signed and unsigned WGCNA to capture negative correlations. bioRxiv (2018), p. 288225. [Google Scholar]
- 74.Pruunsild P, Bengtson CP, Bading H, Networks of Cultured iPSC-Derived Neurons Reveal the Human Synaptic Activity-Regulated Adaptive Gene Program. Cell Rep. 18, 122–135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Amaral PP et al. , Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA. 15, 2013–2027 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS, Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. U. S. A 105, 716–721 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aberg K, Saetre P, Jareborg N, Jazin E, Human QKI, a potential regulator of mRNA expression of human oligodendrocyte-related genes involved in schizophrenia. Proc. Natl. Acad. Sci. U. S. A 103, 7482–7487 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barry G et al. , The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 19, 486–494 (2014). [DOI] [PubMed] [Google Scholar]
- 79.Koga M et al. , Involvement of SMARCA2/BRM in the SWI/SNF chromatin-remodeling complex in schizophrenia. Hum. Mol. Genet 18, 2483–2494 (2009). [DOI] [PubMed] [Google Scholar]
- 80.Krantz ID et al. , Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat. Genet 36, 631–635 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sanders SJ et al. , Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 87, 1215–1233 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Melé M et al. , Human genomics. The human transcriptome across tissues and individuals. Science. 348, 660–665 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Quesnel-Vallières M et al. , Misregulation of an Activity-Dependent Splicing Network as a Common Mechanism Underlying Autism Spectrum Disorders. Mol. Cell 64, 1023–1034 (2016). [DOI] [PubMed] [Google Scholar]
- 84.Steijger T et al. , Assessment of transcript reconstruction methods for RNA-seq. Nat. Methods 10, 1177–1184 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Love MI, Hogenesch JB, Irizarry RA, Modeling of RNA-seq fragment sequence bias reduces systematic errors in transcript abundance estimation. Nat. Biotechnol 34, 1287–1291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Estes ML, McAllister AK, Immune mediators in the brain and peripheral tissues in autism spectrum disorder. Nat. Rev. Neurosci 16, 469–486 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meyer U, Feldon J, Dammann O, Schizophrenia and autism: both shared and disorder-specific pathogenesis via perinatal inflammation? Pediatr. Res 69, 26R–33R (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rosenblat JD et al. , Inflammation as a neurobiological substrate of cognitive impairment in bipolar disorder: Evidence, pathophysiology and treatment implications. J. Affect. Disord 188, 149–159 (2015). [DOI] [PubMed] [Google Scholar]
- 89.Lappalainen T, Functional genomics bridges the gap between quantitative genetics and molecular biology. Genome Res. 25, 1427–1431 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ruderfer DM et al. , Genomic Dissection of Bipolar Disorder and Schizophrenia, Including 28 Subphenotypes. Cell. 173, 1705–1715.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lake BB et al. , Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat. Biotechnol. 36, 70–80 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goldmann T et al. , Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat. Immunol. 17, 797–805 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Keren-Shaul H et al. , A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell. 169, 1276–1290.e17 (2017). [DOI] [PubMed] [Google Scholar]
- 94.Zeisel A et al. , Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 347, 1138–1142 (2015). [DOI] [PubMed] [Google Scholar]
- 95.Zhang Y et al. , Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron. 89, 37–53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wilkinson B et al. , The autism-associated gene chromodomain helicase DNA-binding protein 8 (CHD8) regulates noncoding RNAs and autism-related genes. Transl. Psychiatry. 5, e568 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Samocha KE et al. , A framework for the interpretation of de novo mutation in human disease. Nat. Genet 46, 944–950 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Iossifov I et al. , Low load for disruptive mutations in autism genes and their biased transmission. Proc. Natl. Acad. Sci. U. S. A 112, E5600–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Karczewski KJ et al. , The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Res. 45, D840–D845 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.PsychENCODE Capstone Data Collection, doi: 10.7303/syn12080241. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.