Abstract
Background
There have been few available prognostic biomarkers in gastric cancer. We rigorously assessed the clinical relevance of promoter DNA methylation of Cysteine dioxygenase type 1 (CDO1) gene, a cancer-specific aberration, in human gastric cancer.
Methods
Quantitative CDO1 methylation value (TaqMeth V) was initially calculated in 138 gastric cancer patients operated in 2005, and its clinical significance was elucidated. As a subsequent expanded set, 154 gastric cancer patients with pathological stage (pStage) II / III with no postoperative therapy were validated between 2000 and 2010.
Results
(1) Median TaqMeth V of CDO1 gene methylation of gastric cancer was 25.6, ranging from 0 to 120.9. As pStage progressed, CDO1 TaqMeth V became higher (p < 0.0001). (2) The optimal cut-off value was determined to be 32.6; gastric cancer patients with high CDO1 gene methylation showed a significantly worse prognosis than those with low CDO1 gene methylation (p < 0.0001). (3) A multivariate cox proportional hazards model identified high CDO1 gene methylation (p = 0.033) as an independent prognostic factor. (4) The results were recapitulated in the expanded set in pStage III, where high CDO1 gene methylation group had a significantly worse prognosis than low CDO1 gene methylation group (p = 0.0065). Hematogenous metastasis was unique in pStage III with high CDO1 gene methylation (p = 0.0075). (5) Anchorage independent growth was reduced in several gastric cancer cell lines due to forced expression of the CDO1 gene, suggesting that abnormal CDO1 gene expression may represent distant metastatic ability.
Conclusions
Promoter DNA hypermethylation of CDO1 gene was rigorously validated as an important prognostic biomarker in primary gastric cancer with specific stage.
Introduction
Gastric cancer is the fifth most common malignancy and the third leading cause of cancer-related death worldwide [1]. Advanced gastric cancer, defined as depth of muscularis propria or beyond, still exhibited poor prognosis by curative surgery even in combination with effective adjuvant chemotherapy [2, 3], and require prognostic factors reflecting their biology to enrich high-risk patients for recurrences. Although several prognostic biomarkers have been reported by immunohistochemistry [4, 5] or by mRNA quantification [6, 7], they have weak points; the former included issues of intra-tumoral heterogeneity and cut-off line between positive and negative cases, while the latter is instable and not appropriate for routine examination. Hence, stable and quantitative methods have been anticipated to develop like DNA markers [8].
Epigenetic gene silencing of the tumor suppresser genes (TSGs) through promoter DNA hypermethylation is a unique feature in human cancers, whereas such cancer specific methylation is rather a rare event [8, 9]. We had developed pharmacologic reversal of epigenetic silencing and uncovered a myriad of transcriptionally repressed genes in human cancers [10–13], and have finally identified outstanding candidate TSG with frequent promoter DNA methylation, a cysteine dioxygenase type 1 (CDO1) gene in human cancers including gastric cancer [14].
The CDO1 protein is a non-heme structured, iron-containing metalloenzyme involved in conversion of the cysteine to cysteine sulfinic acid (CSA), while it can promote apoptosis by increasing reactive oxygen species (ROS) through suppression of glutathione generation in breast cancer cells [15], thus suggesting that CDO1 is a TSG in the context of pseudo-inflammatory reaction during carcinogenesis. In addition to breast cancer [15, 16], promoter DNA hypermethylation of CDO1 gene has been reported to be highly specific to cancer cells [8], and exhibited prognostic relevance in specific cancers such as esophageal [17, 18], lung [19], colorectal [20], gallbladder [21], and kidney cancer [22].
Nevertheless, we have not found any reports on the clinical significance of CDO1 gene methylation in primary gastric cancer. In the present study, CDO1 gene promoter DNA methylation was for the first time examined and clarified for detailed clinicopathological factors in primary gastric cancer, and proved great clinical value in gastric cancer clinics.
Materials and methods
Patients and tissue samples
We recruited 140 primary gastric cancer patients who underwent curative gastrectomy at the Kitasato University Hospital in 2005. DNA was extracted from the formalin-fixed, paraffin embedded tumor tissues of the 138 patients who agreed to use pathological specimens. As an expanded set, 154 patients of pathological stage (pStage) II/III without postoperative adjuvant chemotherapy were collected from 1673 gastric cancer patients between 2000 and 2010 for validation. The median follow up term of the expanded one was 100.5 months, ranging from 2 to 148 months. pStage was used according to the Japanese Classification of Gastric Cancer staging system, 14th edition [23]. This study was approved by the Kitasato University Ethics Committee (number B17-251).
Cell lines
The 6 gastric cancer cell lines (MKN7, Kato III, SH-10-TC, KE-97, MKN74, and NUGC-4) were previously described [24]. All cell lines were grown in RPMI-1640 medium (GIBCO, Carlsbad, CA) supplemented with 10% FBS. The hepatocellular carcinoma cell line HepG2 and colorectal DLD-1 cells were used as positive and negative controls for methylation [25].
Genomic DNA extraction and bisulfite treatment
Genomic DNA was extracted from cell lines using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). Formalin-fixed paraffin-embedded tissue was cut into six slices of 10 μm thick before genomic DNA extraction using the QIAamp DNA FFPE Tissue Kit (Qiagen). Genomic DNA (2 μg) was bisulfite converted using the EZ DNA Methylation-Gold Kit (Zimo Research, Irvine, CA, USA).
Total RNA extraction and RT-PCR
Total RNA was extracted using an RNeasy Mini Kit (Qiagen). First strand cDNA was synthesized from RNA (2 μg) using SuperScript III reverse transcriptase (Invitrogen) and Oligo (dT) primers (Invitrogen). RT–PCR was carried out using Platinum Taq DNA Polymerase (Invitrogen). Primers sequences for CDO1 and β-actin were previously described [21].
Quantitative methylation-specific PCR (Q-MSP)
For Q-MSP of CDO1 gene, we performed real-time PCR using iQ Supermix (Bio-Rad, Hercules, CA) and CFX96 real-time systems and TaqMeth V was defined as previously described [21]. All reactions were performed in triplicate.
5-Aza-dC and TSA treatment
Cells were seeded in a 10 cm dishes, and were then treated every 24 h for 4 days with either 1 or 5 μM 5-Aza-dC (5-aza-20-deoxycytidine) dissolved in 50% acetic acid or were mock treated with PBS including the same amount of acetic acid. Trichostatin A (TSA; 300 nM; Sigma Aldrich, Inc, St Louis, MO, USA) was added to the medium for the final 24 h. On day 5, the cells were harvested and mRNA was extracted.
Immunostaining for CDO1 in primary gastric cancer tissues
FFPE tissue blocks were cut into thin sections (4 μm thick) and immunohistochemistry was performed as previously described [21]. The sections were incubated with primary rabbit anti-CDO1 polyclonal antibody (12589-1-AP) (proteintech, Rosemont, IL; 1:100). The secondary antibody reaction was performed using the Histofine Simple Stain MAX-PO (MULTI) kit (Nichirei, Tokyo, Japan). Mayer's Hematoxylin Solution was used to stain nuclei.
Plasmid construction for transfection into cell lines
Full-length CDO1 cDNA was inserted into the pcDNA3.1 myc-His C expression vector (Invitrogen) as previously described [20]. Cells were transfected using Lipofectamine 2000 (Invitrogen) in Opti-MEM (Invitrogen).
Western blotting analysis
Total cellular protein (60 μg) was loaded onto a NuPAGE 4–12% Bis-Tris gel (Invitrogen) and electrophoresis was performed, followed by electroblotting to a PVDF membrane (Invitrogen). The blots were incubated with anti-myc (Invitrogen) and anti β-Actin (Invitrogen) antibodies as previously described [26]. Signals were detected using the luminescent image analyzer ImageQuant LAS 4000 (GE Healthcare, CT, USA).
Cell proliferation assay
Cell proliferation was assayed using the CytoSelect water-soluble tetrazolium salt (WST-1) Cell Proliferation Assay Reagent (Cell Biolabs, San Diego, CA, USA). On day 1, the cells were cultured in a 96-well plate at a density of 1×104 cells per plate. On day 2, the cells were transiently transfected with CDO1. On day 3, cell proliferation was evaluated by measuring the optical density (OD) at 450 nm.
Anchorage-independent colony formation assay
The anchorage-independent colony formation assay was performed. In a six-well plate, 0.72% agarose (Bacto Agar; Becton, Dickinson and Company, Franklin Lakes, NJ) was placed on the bottom. Top agar was made with agarose mixed with 1 × 105 cells transfected with CDO1. After 3 weeks of culture, colonies with more than 100 cells were counted in 10 fields of view. The experiment was conducted twice.
Statistical analyses
All statistical analyses were performed using JMP 11 software (SAS Institute Inc., Cary, NC, USA). Continuous variables were evaluated by ANOVA, Student’s t test; categorical variables were evaluated by Fisher’s exact test or the Chi-square test, as appropriate. Overall survival (OS) was measured from the date of death or censored at the date of the last follow-up evaluation. Survival was estimated using the Kaplan-Meier method and compared by the log-rank test. Differences between results of comparative tests were considered significant if the two-sided P value was less than 0.05.
Results
Quantification of promoter DNA methylation of CDO1 gene in primary gastric cancer
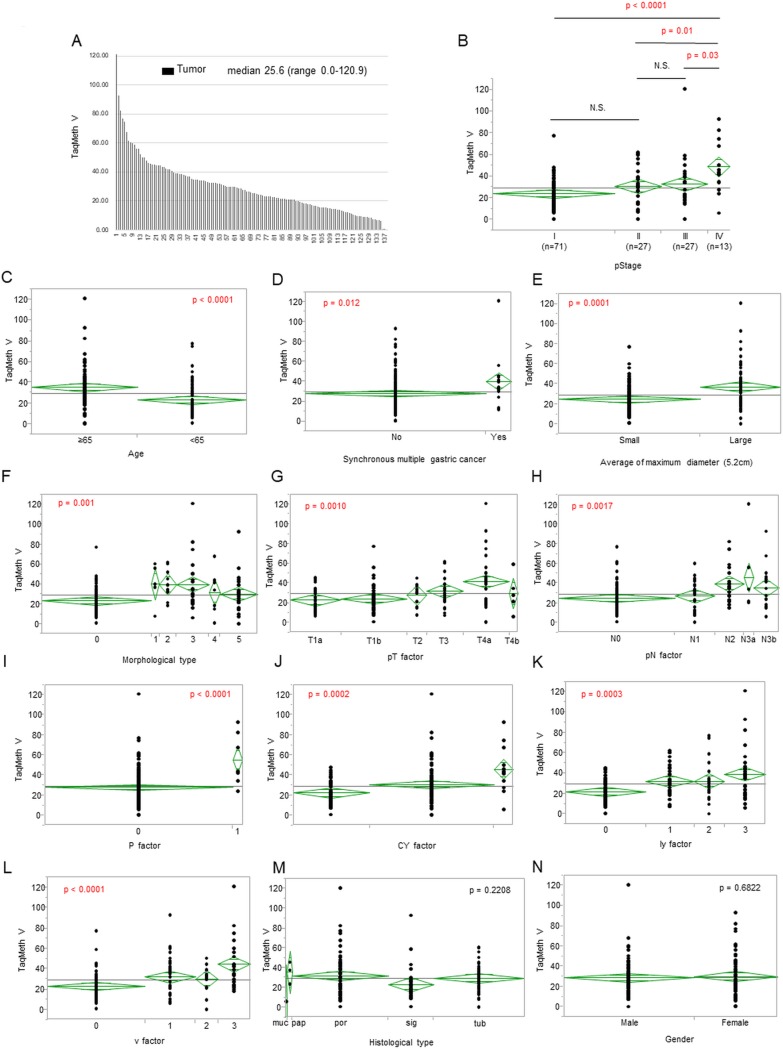
Q-MSP for CDO1 gene was initially performed in 138 primary gastric cancer. Median TaqMeth V of CDO1 gene was 25.6, ranging from 0 to 120.9 (Fig 1A). The CDO1 TaqMeth V tended to become higher as pStage progressed. There was a significant difference between pStage IV and pStage I / II / III (p < 0.0001 / p = 0.01 / p = 0.03, respectively)(Fig 1B). Age (Fig 1C, p < 0.0001), synchronous multiple gastric cancer (Fig 1D, p = 0.012), tumor size (divided by 5.2cm) (Fig 1E, p = 0.0001), morphological type (Fig 1F, p = 0.001), pT factor (Fig 1G, p = 0.001), pN factor (Fig 1H, p = 0.0017), P factor (Fig 1I, p < 0.0001), CY factor (p = 0.0002, Fig 1J), ly factor (Fig 1K, p = 0.0003), and v factor (Fig 1L, p < 0.0001) showed a significant difference. On the other hand, there was no significant difference between histological type (Fig 1M, p = 0.2208) and gender (Fig 1N, p = 0.6822).
Fig 1. CDO1 TaqMeth V in primary gastric cancer and its correlation with clinicopathological factor.
(A) CDO1 TaqMeth V distribution in the gastric cancer tissues. Median TaqMeth V of CDO1 gene was 25.6, ranging from 0 to 120.9. CDO1 TaqMeth V distribution in the gastric cancer tissues according to (B) pStage IV and pStage I / II / III (p < 0.0001 / p = 0.01 / p = 0.03), (C) Age (p < 0.0001), (D) synchronous multiple gastric cancer (p = 0.012), (E) average of maximum diameter (p = 0.0001), (F) morphological type (p = 0.001), (G) pT factor (p = 0.001), (H) pN factor (p = 0.0017), (I) P factor (p < 0.0001), (J) CY factor (p = 0.0002), (K) ly factor (p = 0.0003), (L) v factor (p < 0.0001), (M) histological type (p = 0.2208), (N) Gender (p = 0.6822).
Prognostic analysis according to CDO1 gene TaqMeth V in primary gastric cancer
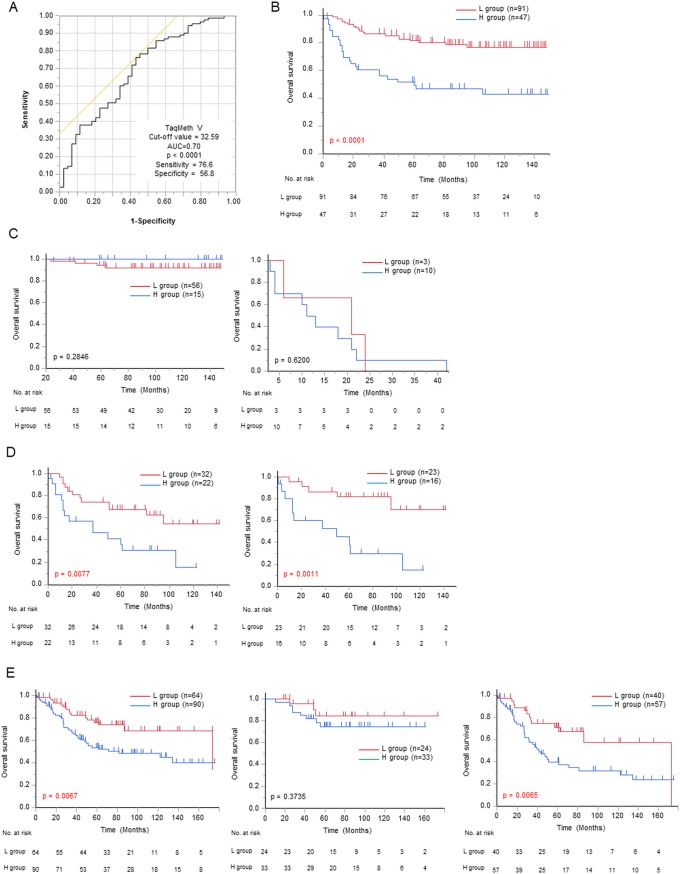
For prognostic analysis, the optimal cut-off value for OS was determined by ROC curve predicting death event. The most optimized TaqMeth V was determined to be 32.59 (AUC of 0.70, p < 0.0001, sensitivity 76.6%, specificity 56.8%)(Fig 2A). Gastric cancer patients were divided into two groups of H group (n = 47): high CDO1 TaqMeth V group (TaqMeth V ≥ 32.6) and L group (n = 91): low CDO1 TaqMeth V group (TaqMeth V <32.6). The H group had a significantly poorer prognosis (5-year OS 49.5%) than the L group (5-year OS 82.0%) (p < 0.0001) (Fig 2B). Age, procedures of gastrectomy, lymph node dissection, radical resection, tumor location, morphological type, pStage, and CDO1 TaqMeth V were significant (p < 0.05) prognostic factors in a univariate analysis. These univariate prognostic factors were applied to Cox proportional hazards model. As a result, pStage and CDO1 TaqMeth V (HR 2.28, CI 1.07–4.95, p = 0.033) were finally remnant independent prognostic factors in multivariate analysis (Table 1).
Fig 2. Prognostic analysis of CDO1 TaqMeth V in primary gastric cancer.
(A) ROC curve of the optimal cutoff value of CDO1TaqMeth V in death event. (B) Kaplan-Meier survival curves for OS comparing gastric cancer patients with CDO1 TaqMeth V below 32.6 and those with CDO1 TaqMeth V equal to or over 32.6 (p < 0.0001). In the same set of gastric cancer patients, survival curves are shown according to (Left panel of Fig 2C) pStage I (p = 0.2846), (Right panel of Fig 2C) pStage IV (p = 0.62), (Left panel of Fig 2D) pStage II / III (p = 0.0077). (Right panel of Fig 2D) Kaplan-Meier survival curves for OS comparing pStage II / III gastric cancer patients without postoperative adjuvant chemotherapy with CDO1 TaqMeth V below 32.6 and those with CDO1 TaqMeth V equal to or over 32.6 (p = 0.0011). In 154 patients as an expanded set of pStage II / III gastric cancer without postoperative adjuvant chemotherapy. (Left panel of Fig 2E) Kaplan-Meier survival curves for OS comparing gastric cancer patients with CDO1 TaqMeth V below 32.6 and those with CDO1 TaqMeth V equal to or over 32.6 (p = 0.0067). (Middle panel of Fig 2E) In expansion set of pStage II. Kaplan-Meier survival curves for OS comparing gastric cancer patients with CDO1 TaqMeth V below 32.6 and those with CDO1 TaqMeth V equal to or over 32.6 (p = 0.3735). (Right panel of Fig 2E) In expansion set of pStage III. Kaplan-Meier survival curves for OS comparing gastric cancer patients with CDO1 TaqMeth V below 32.6 and those with CDO1 TaqMeth V equal to or over 32.6 (p = 0.0065).
Table 1. Univariate and multivariate prognostic analysis for overall survival.
| Clinicopathological factor | Number (%) | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|---|
| 5-year OS | p-Value | Hazard ratio | 95% CI | p-Value | ||
| Age (years) | 0.0064 | 0.3479 | ||||
| <65 | 70 (50.7%) | 81.3% | Reference | |||
| ≥65 | 68 (49.3%) | 60.6% | 1.41 | 0.69–2.99 | ||
| Gender | 0.9844 | |||||
| Male | 84 (60.9%) | 70.5% | ||||
| Female | 54 (39.1%) | 71.9% | ||||
| Procedure of gastrectomy | 0.0005 | 0.5361 | ||||
| Distal gastrectomy | 75 (54.3%) | 83.8% | Reference | |||
| Total gastrectomy | 58 (42.0%) | 75.0% | 1.49 | 0.59–3.80 | ||
| Proximal gastrectomy | 5 (3.7%) | 54.0% | 2.00 | 0.10–13.08 | ||
| Field of lymph node dissection | 0.0315 | 0.1101 | ||||
| D1/1+ lymph node dissection | 86 (62.3%) | 77.5% | Reference | |||
| D2 lymph node dissection | 52 (37.7%) | 60.3% | 2.07 | 0.85–5.33 | ||
| Synchronous multiple gastric cancer | 0.6854 | |||||
| Absence | 121 (87.7%) | 75.0% | ||||
| Presence | 17 (12.3%) | 70.5% | ||||
| Resectability | < 0.0001 | 0.3759 | ||||
| R0 | 132 (95.7%) | 73.5% | Reference | |||
| R1-2 | 6 (4.3%) | 16.7% | 1.68 | 0.51–4.82 | ||
| Tumor location | 0.0315 | 0.0906 | ||||
| Lower | 27 (19.6%) | 76.7% | Reference | |||
| Middle | 99 (71.7%) | 73.4% | 1.37 | 0.49–3.51 | ||
| Upper | 12 (8.7%) | 35.0% | 2.11 | 0.58–8.11 | ||
| Histological type | 0.06 | |||||
| Differentiated type | 55 (39.9%) | 81.1% | ||||
| Undifferentiated type | 82 (60.1%) | 63.9% | ||||
| Morphological type | < 0.0001 | 0.383 | ||||
| Early cancer | 70 (50.7%) | 94.1% | Reference | |||
| Advanced cancer | 68 (49.3%) | 46.6% | 2.56 | 0.28–15.00 | ||
| Pathological stage (pStage) | < 0.0001 | < 0.0001 | ||||
| pStage I | 71 (51.4%) | 95.6% | Reference | |||
| pStage II | 27 (19.6%) | 72.2% | 10.52 | 1.12–80.28 | ||
| pStage III | 27 (19.6%) | 38.1% | 43.37 | 3.91–455.44 | ||
| pStage IV | 13 (9.4%) | 0% | 204.91 | 17.61–2128.60 | ||
| CDO1 methylation value (ROC 32.6) | < 0.0001 | 0.0326 | ||||
| <32.6 | 91 (65.9%) | 82.0% | Reference | |||
| ≥32.6 | 47 (34.1%) | 49.5% | 2.28 | 1.07–4.95 | ||
As the H group included higher pStage than the L group did (p = 0.0006, Table 2). prognosis was then compared in individual pStage. There was no significant difference between the H group and the L group in pStage I / IV (p = 0.2846/p = 0.62, respectively) (left and right panels of Fig 2C), while the H group (n = 22) exhibited significantly poor prognosis than the L Group (n = 32) in pStage II / III (p = 0.0077) (Fig 2D, left panel). In 2005, pathological stage II / III patients were recommended for postoperative adjuvant chemotherapy as a phase III clinical trial (ACTS-GC), the prognosis may have been modified by the adjuvant chemotherapy because the ACTS-GC proved prognostic efficacy [27]. We then restricted prognostic analysis to the 39 patients with pStage II / III gastric cancer without postoperative adjuvant chemotherapy, and still proved that the H group (n = 16) had a significantly poorer prognosis (5-year OS 37.7%) than the L group (n = 23, 5-year OS 81.7%) (p = 0.0011) (Fig 2D, right panel).
Table 2. Correlation of clinicopathologic characteristics and CDO1 methylation.
| CDO1 methylation value | |||||
|---|---|---|---|---|---|
| Low (<32.6) | High (≥32.6) | ||||
| n = 91 (65.9%) | n = 47 (34.1%) | ||||
| Clinicopathological factors | Number | Percentage (%) | Number | Percentage (%) | p-Value |
| Age (years) | 0.0004 | ||||
| <65 | 56 | 40.6% | 14 | 10.1% | |
| ≥65 | 35 | 25.4% | 33 | 23.9% | |
| Gender | NS | ||||
| Male | 57 | 41.3% | 27 | 19.6% | |
| Female | 34 | 24.6% | 20 | 14.5% | |
| Histological type | NS | ||||
| Differentiated carcinoma | 35 | 25.5% | 20 | 14.6% | |
| Undifferentiated carcinoma | 55 | 40.2% | 27 | 19.7% | |
| Pathological T factor (pT) | 0.0024 | ||||
| pT1a | 25 | 18.1% | 6 | 4.4% | |
| pT1b | 32 | 23.2% | 8 | 5.8% | |
| pT2 | 8 | 5.8% | 4 | 2.9% | |
| pT3 | 13 | 9.4% | 9 | 6.5% | |
| pT4a | 10 | 7.3% | 18 | 13.0% | |
| pT4b | 3 | 2.2% | 2 | 1.4% | |
| Pathological N factor (pN) | 0.0104 | ||||
| pN0 | 57 | 41.2% | 16 | 11.6% | |
| pN1 | 16 | 11.6% | 9 | 6.5% | |
| pN2 | 7 | 5.1% | 11 | 8.0% | |
| pN3a | 3 | 2.2% | 3 | 2.2% | |
| pN3b | 8 | 5.8% | 8 | 5.8% | |
| Peritoneal dissemination (P) | 0.0031 | ||||
| P0 | 90 | 65.2% | 41 | 29.7% | |
| P1 | 1 | 0.7% | 6 | 4.4% | |
| Cytorogy of peritoneal lavage (CY) | 0.0017 | ||||
| CY0 | 50 | 36.2% | 28 | 20.3% | |
| CY1 | 3 | 2.2% | 9 | 6.5% | |
| CYX | 38 | 27.5% | 10 | 7.3% | |
| Pathological stage (pStage) | 0.0006 | ||||
| pStage I | 56 | 40.6% | 15 | 10.9% | |
| pStage II | 17 | 12.3% | 10 | 7.2% | |
| pStage III | 15 | 10.9% | 12 | 8.7% | |
| pStage IV | 3 | 2.2% | 10 | 7.2% | |
| Lymph duct invasion (ly) | 0.001 | ||||
| ly0 | 46 | 33.6% | 9 | 6.6% | |
| ly1 | 18 | 13.1% | 15 | 10.9% | |
| ly2 | 14 | 10.2% | 7 | 5.1% | |
| ly3 | 12 | 8.8% | 16 | 11.7% | |
| Veinous invasion (v) | < 0.0001 | ||||
| v0 | 56 | 40.8% | 10 | 7.3% | |
| v1 | 18 | 13.1% | 16 | 11.7% | |
| v2 | 8 | 5.8% | 7 | 5.1% | |
| v3 | 8 | 5.8% | 14 | 10.2% | |
NS: not significant
In order to verify the results of prognostic significance in pStage II / III gastric cancer without adjuvant chemotherapy, prognostic analysis was performed on 154 patients in pStage II / III advanced gastric cancer without adjuvant chemotherapy who were collected between 2000 to 2010 as an expanded set. The result again proved similar prognosis with the learning set. CDO1 gene hypermethylation could predict poorer prognosis of pStage II / III gastric cancer without adjuvant chemotherapy than CDO1 gene hypomethylation (5-year OS 53.8% / 76.3% in the H / L group, respectively) (p = 0.0067) (Fig 2E, left panel). Statistical difference of OS was not recognized in pStage II (Fig 2E, middle panel), but in pStage III (Fig 2E, right panel); the 5-year OS of the H group was 40.1%, while 71.3% in the L group, and the prognosis was significantly poorer in the H group as compared with that in the L group (p = 0.0065).
The recurrence pattern of pStage II/III gastric cancer patients with no adjuvant chemotherapy in the expanded set was then clarified to explain the cause of poor prognosis by CDO1 gene hypermethylation. There were 8 recurrent cases (14.0%) in pStage II and 43 recurrent cases (44.3%) in pStage III, with significant more relapses in pStage III cases than in pStage II cases (p < 0.0001). There were 15 cases (9.6%) of initial recurrences at lymph nodes, 25 cases (15.9%) of initial recurrences at distant organ, 22 cases (14.0%) of initial recurrence at peritoneum, and 3 cases of initial recurrences at local location (1.9%). First of all, there were significantly more recurrences in the H group than in the L group among the pStage III gastric cancer patients (p = 0.0052), and the most outstanding features were characterized by more distant organ metastasis in the H group than in the L group in pStage III (p = 0.0075) (Table 3).
Table 3. Patterns of recurrence after gastrectomy.
| Pathological stage II | Pathological stage III | |||||||
|---|---|---|---|---|---|---|---|---|
| Recurrences | Low n = 24 |
High n = 33 |
Total n = 57 |
P-value | Low n = 40 |
High n = 57 |
Total n = 97 |
P-value |
| Total | 3 | 5 | 8 (14.0%) | NS | 11 | 32 | 43 (44.3%) | 0.0052 |
| Lymph node recurrence (n = 15, 9.6%) |
0 | 3 | 3 (5.3%) | NS | 4 | 8 | 12 (12.4%) | NS |
| Regional | 0 | 0 | 0 | 2 | 2 | 4 (4.1%) | ||
| Extra regional | 0 | 3 | 3 (5.3%) | 2 | 6 | 8 (8.2%) | ||
| Hematogeneous recurrence (n = 25, 15.9%) |
1 | 3 | 4 (7.0%) | NS | 3 | 18 | 21 (21.6%) | 0.0075 |
| Liver | 1 | 2 | 3 (5.3%) | 1 | 9 | 10 (10.3%) | ||
| Bone | 0 | 0 | 0 | 1 | 4 | 5 (5.2%) | ||
| Lung | 0 | 0 | 0 | 1 | 1 | 2 (2.1%) | ||
| Ovary | 0 | 0 | 0 | 0 | 2 | 2 (2.1%) | ||
| Brain | 0 | 0 | 0 | 0 | 1 | 1 (1.0%) | ||
| Colon | 0 | 1 | 1 (1.8%) | 0 | 0 | 0 | ||
| Liver+Lung | 0 | 0 | 0 | 0 | 1 | 1 (1.0%) | ||
| Peritoneal dissemination recurrence (n = 22, 14.0%) |
2 | 2 | 4 (7.0%) | NS | 6 | 12 | 18 (18.0%) | NS |
| Local recurrence (n = 3, 1.9%) |
0 | 0 | 0 | NS | 0 | 3 | 3 (3.0%) | NS |
NS: not significant
Functional assessment of CDO1 gene transfection on gastric cancer cells
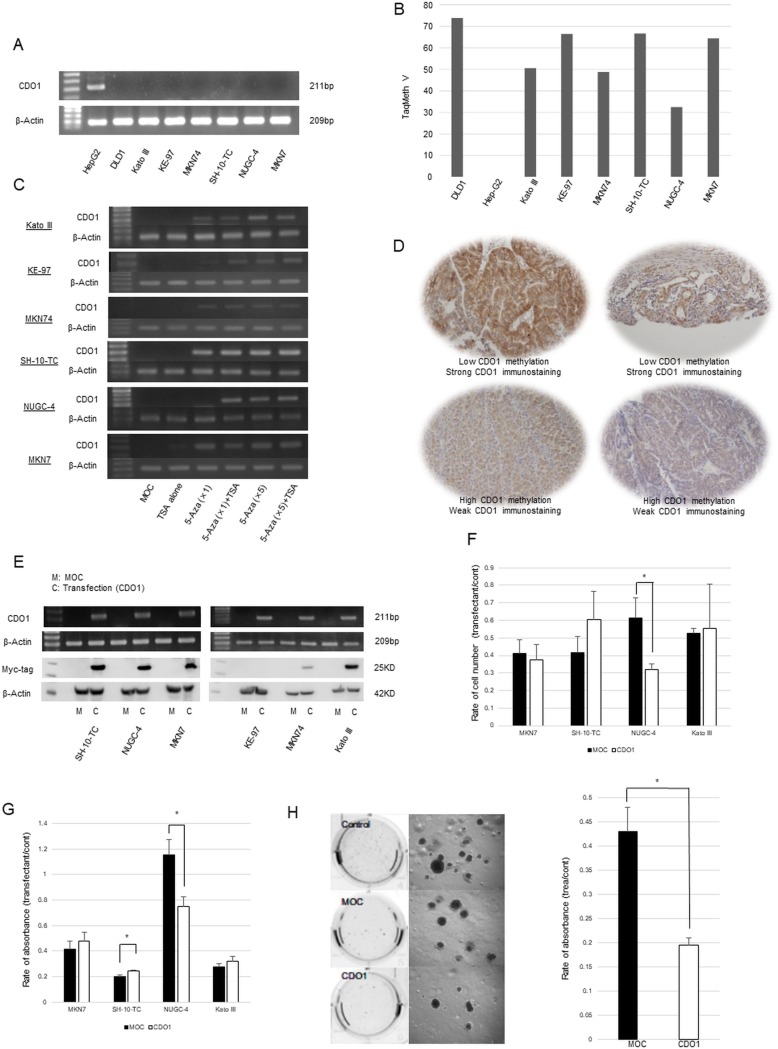
Expression of CDO1 gene was never observed in 6 gastric cancer cell lines, as compared with positive expression control of HepG2 cells at mRNA level (Fig 3A), where DNA hypermethylation was confirmed in all the 6 cell lines and DNA hypomethylation was seen in HepG2 cells (Fig 3B). Reactivation by demethylation treatments using 5-Aza-dC and Trichostatin A was confirmed in all 6 the cell lines (Fig 3C), suggesting that expression of CDO1 gene must be suppressed by epigenetic manners such as promoter DNA methylation. Immunostaining for CDO1 protein confirmed its localization in the cytoplasm of non-cancerous gastric mucosa gland cells (data not shown) or cancer cells harboring low value of CDO1 gene methylation (L group) (Fig 3D, upper panels). On the other hand, representative specimens of H group were weakly positive for CDO1 protein expression (Fig 3D, lower panels).
Fig 3.
(A) Expression of CDO1 gene in all 6cell lines in mRNA level. (B) DNA methylation levels in all 6 cell lines by using Q-MSP after bisulfite treatment. (C) Reactivation of CDO1 gene by demethylation. In Immunostaining chemistry, (Upper panel of Fig 3D) two specimens of H group were weakly positive, and (Lower panel of Fig 3D) two specimens of L group were strongly positive. (E) CDO1 gene was transfected to all 6 gastric cancer cell lines and the expression was confirmed by RT-PCR and Western blotting. (F) Cell count after gastric cancer cell lines was transfected CDO1 gene. (G) Cell viability after gastric cancer cell lines was transfected CDO1 gene (WST-1 assay). (H) Confirmation of colony formation on Soft agar. When the CDO1 gene was transfected, the number of colonies decreased compared to control and MOC. Anchorage independent growth ability significantly reduced the colony forming ability in CDO1-transfected cells in Kato III compared to MOC-transfected cells (p = 0.0245).
The CDO1 full length vector was transfected to all 6 gastric cancer cell lines and the expression was confirmed by RT-PCR and Western blotting (Fig 3E). Intense expression of CDO1 protein was confirmed in MKN7, SH-10-TC, NUGC-4 and Kato III, but not in MKN74 and KE-97 (Fig 3E). After CDO1 gene transfection, cell proliferation was suppressed in NUGC-4 by simple cell count (Fig 3F, p = 0.026) and WST-1 cell viability assays (Fig 3G, p = 0.03). Among gastric cancer cell lines tested in this study, only Kato III exhibited anchorage-independent colony formation. For demonstrating the tumor suppressive activity of CDO1 gene, only Kato III cells were considered to be appropriate. The CDO1-transfected cells showed suppressed capacity of anchorage independent growth compared to MOC-transfected cells (p = 0.0245) in Kato III cells (Fig 3H).
Discussion
Promoter DNA of the CDO1 gene is frequently hypermethylated in human cancers including gastric cancer [14], which showed the highest AUC (0.95) to differentiate tumor tissues from the corresponding non-cancerous tissues [8]. Hypermethylation in tumor tissues beyond 60% was designated as highly relevant methylation gene (HRMG), and CDO1 gene is the most common HRMG among human cancers [8]. Using the best optimized cut-off value of TaqMeth V to discriminate tumor from non-cancerous tissues, CDO1 gene hypermethylation is found in 72~91% in various cancers [14, 18, 21, 28]. These frequencies were determined, based on comparison of tumor tissues to the corresponding non-cancerous tissues, and affected by the methylation level of non-cancerous tissues. For example, corresponding non-cancerous tissues were relatively highly methylated for CDO1 gene in gallbladder cancer, and threshold cut-off value became high and the frequencies were underestimated [21]. Actual methylation frequencies are therefore considered higher than the report (~90% in almost human cancer). Anyway, CDO1 gene has an outstanding feature with regard to cancer-specific methylation in human cancer.
The current study is the first report describing clinicopathological relevance of CDO1 gene promoter DNA methylation status in primary gastric cancer. Recent literatures of the significant association between CDO1 methylation and poor prognosis have been reported in breast [15, 16], esophageal [17, 18], renal cells carcinoma [22], HPV associated malignancies [29], prostate cancer [30], gallbladder cancer [21], and colorectal cancer [20]. Nevertheless, there has never been reported with regard to prognostic relevance in primary gastric cancer.
In the present study, CDO1 TaqMeth V was rigorously validated as prognostic factor of primary gastric cancer. The most importantly, it could still show the prognostic relevance in pStage II/III gastric cancer patients without postoperative adjuvant chemotherapy. We have to clearly recognize the difference of patients with adjuvant chemotherapy and those without it from a prognostic point of view, because recent adjuvant chemotherapy is really effective to pStage II/III advanced gastric cancer [2, 3]. In order to know the biological role of CDO1 gene during natural clinical course, we had better not include gastric cancer with adjuvant chemotherapy. In our current study, CDO1 gene promoter methylation definitely accumulates as disease progressed, and it was significantly associated with the initial recurrences at distant organs in pStage III gastric cancer. CDO1 gene actually suppressed anchorage independent growth in Kato III cells, suggesting that it plays a functionally critical role in distant metastasis of gastric cancer.
The best optimized cut-off TaqMeth V with regard to prognosis was set as 32.6. This optimized cut-off value is always higher than those delineated tumor from non-cancerous tissues in various cancers [14, 17, 20, 21]. Moreover, cancer patients with CDO1 gene hypermethylation showed suppressed expression of CDO1 protein in immunohistochemistry [20, 21]. These findings suggested that higher promoter DNA methylation status represents strong suppression of CDO1 protein expression, which may be linked to tumor aggressiveness. Almost cancer cell lines including gastric cancer did not recognize CDO1 gene expression, and it is therefore difficult to conduct an experiment to suppress expression of CDO1 gene by RNA interference using siRNA. Only HepG2, a liver cancer cell line expressed CDO1 gene, and suppression of CDO1 gene by RNA interference resulted in invasive capacity as described by Brait M [14]. Reflected by this functional experiment, CDO1 gene promoter DNA methylation is an excellent prognostic marker, because it is DNA (that is stable in any environment), and it could be quantified by Q-MSP differently from immunohistochemistry.
Although there was no significant difference of the initial recurrences at peritoneum according to CDO1 gene TaqMeth V in gastric cancer, it can be used as a cancer detection marker, because it is highly specific to cancer cells [14]. We recently reported the usefulness of DNA diagnosis using the CDO1 gene methylation in DNA cytology test using the peritoneal lavage of gastric cancer [31]. DNA cytology of peritoneal lavage had higher diagnostic ability compared to the conventional cytology test of peritoneal lavage. Currently, prospective study is conducted to validate the clinical utility (UMIN000026191).
Cysteine biology has recently focused on cancer stem cell features [32]. CD44 variant interacts with xCT, a glutamate-cystine transporter, and permits intracellular increase of glutathione that protects stem cells, which is associated with inflammatory processes. Ablation of CD44 induced loss of xCT from the cell surface and suppressed tumor growth. CDO1 protein catalyzes the oxidation of cysteine to CSA [33], reducing intracellular cysteine concentration and subsequent reduction of glutathione. This molecular mechanism may be associated with tumor suppressive function of CDO1 gene [15].
Our DNA was extracted from the formalin-fixed, paraffin embedded (FFPE) tumor tissues, and not from noninvasive biopsy. Unlike fresh frozen samples, FFPE samples were demonstrated to exhibit deterioration in the quality of RNA [34], while verification of DNA methylation analysis using FFPE specimens has been done, and the usefulness has been confirmed [35]. For this reason, methylation analysis was also carried out using FFPE samples in this study.
In conclusion, CDO1 TaqMeth V was rigorously validated to be an important prognostic factor in primary gastric cancer. It was considered that the CDO1 methylation together with its aberrant expression may be causatively involved in the distant metastasis, resulting in poor prognosis of gastric cancer. If it is possible to predict distant metastasis, selection of patients requiring preoperative adjuvant chemotherapy can be effectively made, and can be highly expected for precision medicine.
Acknowledgments
The authors would like to thank FORTE Science Communications, Tokyo, Japan, for professional editorial assistance.
Data Availability
All relevant data are within the manuscript.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Rohrmann S, Linseisen J, Overvad K, Lund Wurtz AM, Roswall N, Tjonneland A, et al. Meat and fish consumption and the risk of renal cell carcinoma in the European prospective investigation into cancer and nutrition. Int J Cancer. 2015; 136(5):E423–31. https://onlinelibrary.wiley.com/doi/full/10.1002/ijc.29236 [DOI] [PubMed] [Google Scholar]
- 2.Sasako M, Sakuramoto S, Katai H, Kinoshita T, Furukawa H, Yamaguchi T, et al. Five-year outcomes of a randomized phase III trial comparing adjuvant chemotherapy with S-1 versus surgery alone in stage II or III gastric cancer. J Clin Oncol. 2011; 29(33):4387–93. http://ascopubs.org/doi/full/10.1200/JCO.2011.36.5908?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%3dpubmed [DOI] [PubMed] [Google Scholar]
- 3.Noh SH, Park SR, Yang HK, Chung HC, Chung IJ, Kim SW, et al. Adjuvant capecitabine plus oxaliplatin for gastric cancer after D2 gastrectomy (CLASSIC): 5-year follow-up of an open-label, randomised phase 3 trial. Lancet Oncol. 2014; 15(12):1389–96. https://www.sciencedirect.com/science/article/pii/S1470204514704735?via%3Dihub [DOI] [PubMed] [Google Scholar]
- 4.Terashima M, Kitada K, Ochiai A, Ichikawa W, Kurahashi I, Sakuramoto S, et al. Impact of expression of human epidermal growth factor receptors EGFR and ERBB2 on survival in stage II/III gastric cancer. Clin Cancer Res. 2012; 18(21):5992–6000. http://clincancerres.aacrjournals.org/content/18/21/5992.long [DOI] [PubMed] [Google Scholar]
- 5.Sasako M, Terashima M, Ichikawa W, Ochiai A, Kitada K, Kurahashi I, et al. Impact of the expression of thymidylate synthase and dihydropyrimidine dehydrogenase genes on survival in stage II/III gastric cancer. Gastric Cancer. 2015; 18(3):538–48. https://link.springer.com/article/10.1007%2Fs10120-014-0413-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ichikawa W, Terashima M, Ochiai A, Kitada K, Kurahashi I, Sakuramoto S, et al. Impact of insulin-like growth factor-1 receptor and amphiregulin expression on survival in patients with stage II/III gastric cancer enrolled in the Adjuvant Chemotherapy Trial of S-1 for Gastric Cancer. Gastric Cancer. 2017; 20(2):263–273. https://link.springer.com/article/10.1007%2Fs10120-016-0600-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheong JH, Yang HK, Kim H, Kim WH, Kim YW, Kook MC, et al. Predictive test for chemotherapy response in resectable gastric cancer: a multi-cohort, retrospective analysis. Lancet Oncol. 2018; 19(5):629–638. https://www.sciencedirect.com/science/article/pii/S1470204518301086?via%3Dihub [DOI] [PubMed] [Google Scholar]
- 8.Yamashita K, Hosoda K, Nishizawa N, Katoh H, Watanabe M. Epigenetic Biomarkers of Promoter DNA Methylation in the New Era of Cancer Treatment. Cancer Sci. 2018. https://onlinelibrary.wiley.com/doi/full/10.1111/cas.13812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ooki A, Yamashita K, Kikuchi S, Sakuramoto S, Katada N, Kokubo K, et al. Potential utility of HOP homeobox gene promoter methylation as a marker of tumor aggressiveness in gastric cancer. Oncogene. 2010; 29(22):3263–75. https://www.nature.com/articles/onc201076 [DOI] [PubMed] [Google Scholar]
- 10.Yamashita K, Upadhyay S, Osada M, Hoque MO, Xiao Y, Mori M, et al. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell. 2002; 2(6):485–95. https://www.sciencedirect.com/science/article/pii/S1535610802002155?via%3Dihub [DOI] [PubMed] [Google Scholar]
- 11.Kim MS, Yamashita K, Baek JH, Park HL, Carvalho AL, Osada M, et al. N-methyl-D-aspartate receptor type 2B is epigenetically inactivated and exhibits tumor-suppressive activity in human esophageal cancer. Cancer Res. 2006; 66(7):3409–18. http://cancerres.aacrjournals.org/content/66/7/3409.long [DOI] [PubMed] [Google Scholar]
- 12.Yamashita K, Park HL, Kim MS, Osada M, Tokumaru Y, Inoue H, et al. PGP9.5 methylation in diffuse-type gastric cancer. Cancer Res. 2006; 66(7):3921–7. http://cancerres.aacrjournals.org/content/66/7/3921.long [DOI] [PubMed] [Google Scholar]
- 13.Kim MS, Chang X, Yamashita K, Nagpal JK, Baek JH, Wu G, et al. Aberrant promoter methylation and tumor suppressive activity of the DFNA5 gene in colorectal carcinoma. Oncogene. 2008; 27(25):3624–34. https://www.nature.com/articles/1211021 [DOI] [PubMed] [Google Scholar]
- 14.Brait M, Ling S, Nagpal JK, Chang X, Park HL, Lee J, et al. Cysteine dioxygenase 1 is a tumor suppressor gene silenced by promoter methylation in multiple human cancers. PLoS One. 2012; 7(9):e44951 https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0044951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeschke J, O'Hagan HM, Zhang W, Vatapalli R, Calmon MF, Danilova L, et al. Frequent inactivation of cysteine dioxygenase type 1 contributes to survival of breast cancer cells and resistance to anthracyclines. Clin Cancer Res. 2013; 19(12):3201–11. http://clincancerres.aacrjournals.org/content/19/12/3201.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minatani N, Waraya M, Yamashita K, Kikuchi M, Ushiku H, Kojo K, et al. Prognostic Significance of Promoter DNA Hypermethylation of cysteine dioxygenase 1 (CDO1) Gene in Primary Breast Cancer. PLoS One. 2016; 11(1):e0144862 https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0144862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ushiku H, Yamashita K, Katoh H, Ema A, Minatani N, Kikuchi M, et al. Promoter DNA methylation of CDO1 gene and its clinical significance in esophageal squamous cell carcinoma. Dis Esophagus. 2017; 30(2):1–9. https://academic.oup.com/dote/article-abstract/30/2/1/2725542?redirectedFrom=fulltext [DOI] [PubMed] [Google Scholar]
- 18.Kojima K, Yamashita K, Ushiku H, Katoh H, Ishii S, Tanaka T, et al. The clinical significance of cysteine dioxygenase type 1 methylation in Barrett esophagus adenocarcinoma. Dis Esophagus. 2017; 30(3):1–9. https://academic.oup.com/dote/article-abstract/30/3/1/2981971?redirectedFrom=fulltext [DOI] [PubMed] [Google Scholar]
- 19.Ooki A, Maleki Z, Tsay JJ, Goparaju C, Brait M, Turaga N, et al. A Panel of Novel Detection and Prognostic Methylated DNA Markers in Primary Non-Small Cell Lung Cancer and Serum DNA. Clin Cancer Res. 2017; 23(22):7141–7152. http://clincancerres.aacrjournals.org/content/23/22/7141.long [DOI] [PubMed] [Google Scholar]
- 20.Kojima K, Nakamura T, Ohbu M, Katoh H, Ooizumi Y, Igarashi K, et al. Cysteine dioxygenase type 1 (CDO1) gene promoter methylation during the adenoma-carcinoma sequence in colorectal cancer. PLoS One. 2018; 13(5):e0194785 https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0194785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Igarashi K, Yamashita K, Katoh H, Kojima K, Ooizumi Y, Nishizawa N, et al. Prognostic significance of promoter DNA hypermethylation of the cysteine dioxygenase 1 (CDO1) gene in primary gallbladder cancer and gallbladder disease. PLoS One. 2017; 12(11):e0188178 https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0188178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deckers IA, Schouten LJ, Van Neste L, van Vlodrop IJ, Soetekouw PM, Baldewijns MM, et al. Promoter Methylation of CDO1 Identifies Clear-Cell Renal Cell Cancer Patients with Poor Survival Outcome. Clin Cancer Res. 2015; 21(15):3492–500. http://clincancerres.aacrjournals.org/content/21/15/3492.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Japanese Gastric Cancer Association. Japanese classification of gastric carcinoma: 3rd English edition. Gastric Cancer. 2011; 14(2):101–12. https://link.springer.com/article/10.1007%2Fs10120-011-0041-5 [DOI] [PubMed] [Google Scholar]
- 24.Ooki A, Yamashita K, Kikuchi S, Sakuramoto S, Katada N, Waraya M, et al. Therapeutic potential of PRL-3 targeting and clinical significance of PRL-3 genomic amplification in gastric cancer. BMC Cancer. 2011; 11:122 https://bmccancer.biomedcentral.com/articles/10.1186/1471-2407-11-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamashita K, Waraya M, Kim MS, Sidransky D, Katada N, Sato T, et al. Detection of methylated CDO1 in plasma of colorectal cancer; a PCR study. PLoS One. 2014; 9(12):e113546 https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0113546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katoh H, Yamashita K, Waraya M, Margalit O, Ooki A, Tamaki H, et al. Epigenetic silencing of HOPX promotes cancer progression in colorectal cancer. Neoplasia. 2012; 14(7):559–71. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3421953/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakuramoto S, Sasako M, Yamaguchi T, Kinoshita T, Fujii M, Nashimoto A, et al. Adjuvant chemotherapy for gastric cancer with S-1, an oral fluoropyrimidine. N Engl J Med. 2007; 357(18):1810–20. https://www.nejm.org/doi/10.1056/NEJMoa072252?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%3dwww.ncbi.nlm.nih.gov [DOI] [PubMed] [Google Scholar]
- 28.Vedeld HM, Andresen K, Eilertsen IA, Nesbakken A, Seruca R, Gladhaug IP, et al. The novel colorectal cancer biomarkers CDO1, ZSCAN18 and ZNF331 are frequently methylated across gastrointestinal cancers. Int J Cancer. 2015; 136(4):844–53. https://onlinelibrary.wiley.com/doi/full/10.1002/ijc.29039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feber A, Arya M, de Winter P, Saqib M, Nigam R, Malone PR, et al. Epigenetics markers of metastasis and HPV-induced tumorigenesis in penile cancer. Clin Cancer Res. 2015; 21(5):1196–206. http://clincancerres.aacrjournals.org/content/21/5/1196.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meller S, Zipfel L, Gevensleben H, Dietrich J, Ellinger J, Majores M, et al. CDO1 promoter methylation is associated with gene silencing and is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients. Epigenetics. 2016; 11(12):871–880. https://www.tandfonline.com/doi/full/10.1080/15592294.2016.1241931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ushiku H, Yamashita K, Ema A, Minatani N, Kikuchi M, Kojo K, et al. DNA diagnosis of peritoneal fluid cytology test by CDO1 promoter DNA hypermethylation in gastric cancer. Gastric Cancer. 2017; 20(5):784–792. https://link.springer.com/article/10.1007%2Fs10120-017-0697-6 [DOI] [PubMed] [Google Scholar]
- 32.Ishimoto T, Nagano O, Yae T, Tamada M, Motohara T, Oshima H, et al. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell. 2011; 19(3):387–400. https://www.cell.com/cancer-cell/fulltext/S1535-6108(11)00050-X [DOI] [PubMed] [Google Scholar]
- 33.Prabhu A, Sarcar B, Kahali S, Yuan Z, Johnson JJ, Adam KP, et al. Cysteine catabolism: a novel metabolic pathway contributing to glioblastoma growth. Cancer Res. 2014; 74(3):787–96. http://cancerres.aacrjournals.org/content/74/3/787.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Ahlfen S, Missel A, Bendrat K, Schlumpberger M. Determinants of RNA quality from FFPE samples. PLoS One. 2007; 2(12):e1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalmár A, Péterfia B, Hollósi P, Wichmann B, Bodor A, Patai ÁV, et al. Bisulfite-Based DNA Methylation Analysis from Recent and Archived Formalin-Fixed, Paraffin Embedded Colorectal Tissue Samples. Pathol Oncol Res. 2015; 21(4):1149–56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the manuscript.