Abstract
Cryo‐electron tomography and small‐angle X‐ray scattering were used to investigate the chromatin folding in metaphase chromosomes. The tomographic 3D reconstructions show that frozen‐hydrated chromatin emanated from chromosomes is planar and forms multilayered plates. The layer thickness was measured accounting for the contrast transfer function fringes at the plate edges, yielding a width of ~ 7.5 nm, which is compatible with the dimensions of a monolayer of nucleosomes slightly tilted with respect to the layer surface. Individual nucleosomes are visible decorating distorted plates, but typical plates are very dense and nucleosomes are not identifiable as individual units, indicating that they are tightly packed. Two layers in contact are ~ 13 nm thick, which is thinner than the sum of two independent layers, suggesting that nucleosomes in the layers interdigitate. X‐ray scattering of whole chromosomes shows a main scattering peak at ~ 6 nm, which can be correlated with the distance between layers and between interdigitating nucleosomes interacting through their faces. These observations support a model where compact chromosomes are composed of many chromatin layers stacked along the chromosome axis.
Keywords: chromatin higher‐order structure, cryo‐electron tomography, DNA packaging, metaphase chromosome structure, small‐angle X‐ray scattering
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; Structural Biology
Introduction
During mitosis in eukaryotic cells, the enormously long genomic DNA molecules are densely packed within metaphase chromosomes (Daban, 2003). Nucleosomes are the basic building blocks of chromatin and constitute the first level of DNA compaction in the cell nucleus. The nucleosome core is a flat cylindrical particle formed by 1.7 superhelical turns of DNA (146 bp) wrapped around the core histone octamer (Luger et al, 1997). In the chromatin filament, nucleosome cores are connected by variable lengths of linker DNA associated with histone H1. In vitro experiments showed that the compaction degree of the chromatin filament is extremely dependent on ionic conditions (Daban, 2011; Collepardo‐Guevara & Schlick, 2012; Grigoryev & Woodcock, 2012; Luger et al, 2012; Rippe, 2012; Boulé et al, 2015). The 30‐nm chromatin fiber is generally considered to be the second level of DNA compaction and, in particular, it is assumed that this fiber is the fundamental structural element for the packaging of DNA into chromosomes (Alberts et al, 2014). Several structural models have been proposed for the organization of chromatin in metaphase chromosomes. From early transmission electron microscopy (TEM) images obtained with histone‐depleted chromosomes, it was proposed that chromatin fibers form loops that are bound to a central protein scaffold (Paulson & Laemmli, 1977). However, subsequent chromosome stretching experiments in the presence of nucleases showed that chromosomes do not contain a continuous protein scaffold and it was suggested that chromatin fibers form an irregular network (Poirier & Marko, 2002). The analysis of chromosomes in different condensation stages indicated a hierarchical folding of fibers having diameters from 30 to 250 nm (Kireeva et al, 2004), and studies of chromosome conformation capture suggested that mitotic chromosomes are formed by a compact array of chromatin loops (Gibcus et al, 2018). In contrast to these models, the study of chromosome cryo‐sections and small‐angle X‐ray scattering (SAXS) experiments showed that condensed chromosomes in 5 mM Mg2+ do not have periodic structures larger than 11 nm (Eltsov et al, 2008; Nishino et al, 2012), indicating that chromatin is not folded as a 30‐nm fiber. Instead, the authors suggested that chromatin filaments in metaphase chromosomes are highly disordered and behave like a polymer melt.
Surprisingly, it was observed using conventional TEM that the incubation of metaphase chromosomes at 37°C produced the emanation of many multilayered plates (Caravaca et al, 2005; Gállego et al, 2009). This planar structure was confirmed with electron tomography (ET) of glutaraldehyde‐crosslinked metaphase chromatin adsorbed to the carbon substrate of typical TEM grids (Castro‐Hartmann et al, 2010), and with atomic force microscopy (AFM) of uncrosslinked chromatin in aqueous solution adsorbed to mica (Gállego et al, 2009). Although chromatin plates are very thin [each steep in a multilayered plate has an apparent thickness of 5–6 nm (Daban, 2011)], AFM‐based nanotribology and force spectroscopy showed that they are flexible and have good mechanical properties in the presence of structuring concentrations of Mg2+ (Gállego et al, 2009, 2010). However, incubation with solutions containing EDTA produced plate unfolding and emanation of chromatin fibers from plate edges (Gállego et al, 2009). Furthermore, it was found that fragments of chromatin fibers obtained from metaphase chromosomes digested with micrococcal nuclease associate spontaneously, forming multilaminar plates that have indistinguishable structure to plates emanated from chromosomes (Milla & Daban, 2012). These observations led to the proposal of the thin‐plate model, in which metaphase chromosomes are formed by many layers stacked along the chromosome axis (Gállego et al, 2009; Castro‐Hartmann et al, 2010). In this work, we used cryo‐electron tomography (cryo‐ET) to study the 3D structure of the plates from human metaphase chromosomes. In contrast to other microscopy techniques, the plates in these experiments were not oriented by adsorption to flat substrate surfaces; the uncrosslinked and unstained sample suspended in aqueous media containing 5 mM Mg2+ was immobilized in vitreous ice and imaged under cryogenic conditions. In order to achieve the maximum imaging resolution and contrast (Fernandez‐Leiro & Scheres, 2016), tomograms were acquired with a direct electron detector and a Volta phase plate (Danev et al, 2014). Furthermore, to study the internal structure of highly compacted chromosomes in the presence of the cation concentrations corresponding to metaphase [17 mM Mg2+ (Strick et al, 2001)], we took advantage of the high photon fluxes of third‐generation synchrotron radiation sources (García‐Gutiérrez & Rueda, 2009) to obtain SAXS data directly from whole chromosomes. Our cryo‐ET and SAXS results strengthen previous evidence indicating that chromatin in metaphase chromosomes is organized as stacked mononucleosome layers that are interdigitated with each other.
Results
Cryo‐tomograms of chromatin emanated from metaphase chromosomes
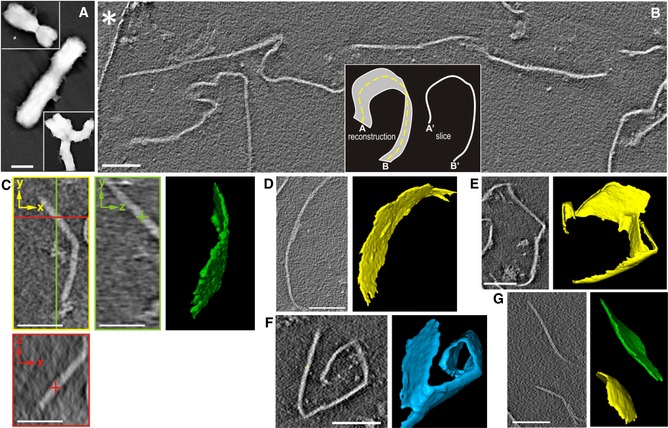
Chromosomes purified by centrifugation on sucrose step gradients in 5 mM Mg2+ (Fig 1A) were unfolded using the soft treatments described in the Materials and Methods. The resulting sample was deposited on EM grids coated with perforated carbon film, rapidly frozen and imaged using cryo‐ET. 2D slices through the 3D reconstructed tomographic volumes showed numerous lines of different shapes and orientations (Fig 1B). These lines in the x‐y plane persist along the z‐axis (Fig 1C), indicating that they correspond to slices of planar structures (see the inset in Fig 1B). Although we observed planar structures adopting a variety of orientations within the ice, the tomographic missing‐wedge causes anisotropic resolution (Lucic et al, 2005); planar structures approximately parallel to the x‐y plane are not well resolved, whereas those that are roughly perpendicular to the x‐y plane can be easily analyzed following the successive slices of the tomograms. Segmentation revealed the 3D architecture of the planar structures, which had a variety of dimensions and shapes (Fig 1C–G). We observed large plates spanning > 1 μm in the x‐y plane. The height (in the z‐axis) of many plates was 0.2–0.4 μm, which corresponds to the ice thickness in our preparations. In comparison with their large surface area, plates are very thin (~ 7.5 nm, see below), suggesting that the different sizes and shapes observed for 3D reconstructed plates are due to breakages and deformations produced during the preparation and deposition procedures. Thin plates emanated from chromosomes were observed previously using conventional TEM and AFM (Caravaca et al, 2005; Gállego et al, 2009; Castro‐Hartmann et al, 2010). Note that in these previous experiments, samples were deposited on carbon films and mica surfaces. In contrast, in our cryo‐ET experiments, the frozen‐hydrated chromatin plates were not adsorbed to a flat surface, but rather suspended in vitreous ice. Therefore, the cryo‐ET results show that chromatin emanated from chromosomes has an intrinsic planar geometry.
Figure 1. Examples of cryo‐tomograms containing plates emanated from metaphase chromosomes.
-
AWhole chromosomes imaged by conventional TEM.
-
BSlice from a large tomographic volume; part of the carbon film surrounding a hole with vitrified ice containing the plates is indicated with an asterisk; the inset illustrates that the slice of a plate corresponds to a line in the x‐y plane (perpendicular to the direction of the electron beam).
-
C–GSlices from different tomograms and the corresponding 3D segmentations showing plates with different sizes and shapes. In addition to a typical slice through the x‐y plane (yellow), two slices through the x‐z (red) and y‐z (green) planes (orthogonal to the x‐y plane) are shown in (C).
In addition to plates, we observed irregular aggregates and occasionally circular structures that have a diameter of ~ 30 nm (indicated with S in Fig 2A; see also the bottom‐left insets). As shown in previous studies performed with chromatin fragments (Bartolomé et al, 1994, 1995; Bermúdez et al, 1998; Daban & Bermúdez, 1998) and nucleosome arrays (Robinson et al, 2006), these circular structures likely correspond to cross‐sections of 30‐nm chromatin fibers folded as highly compact solenoids in which nucleosomes are tightly packed and cannot be distinguished as separate units. According to these studies, the dense annular zone of these structures is formed by nucleosomes of adjacent helical turns that are interdigitated (Daban & Bermúdez, 1998; Robinson et al, 2006). In our cryo‐tomograms, these fibers are short (their length in the z‐axis is 22–30 nm) and are probably formed by the helical folding of short chromatin fragments produced by mechanical breakage of the chromatin filament during the emanation of plates from soft‐denatured chromosomes. We did not observe long chromatin fibers in the tomograms.
Figure 2. Cryo‐tomograms of compact plates and distorted plates decorated with nucleosomes.
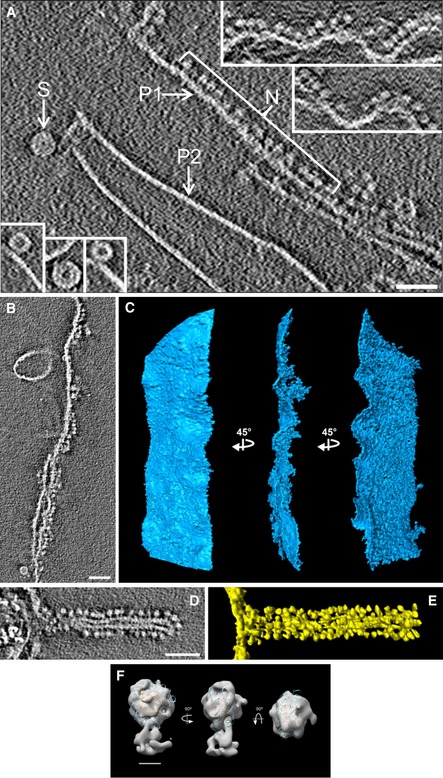
-
ANucleosomes (N) decorating a relaxed plate (P1); insets in the upper right show additional examples. Nucleosomes are not visible as individual units in typical compact plates (P2). Short compact interdigitated solenoids are shown in the main image (S) and in the bottom‐left insets.
-
B–ESlice (B) and segmentations in three different orientations (C) of a large relaxed plate decorated with many nucleosomes on its right side. Slice (D) and segmentation (E) of a relaxed plate forming a tube decorated with nucleosomes.
-
FStructure of the decorative particles (like those shown in N, panel A) after subtomogram averaging. The final density map was filtered to 25 Å and fitted with the molecular structure of the nucleosome core particle (Protein Data Bank code 2CV5).
Tight nucleosome packaging within the plates
Some chromatin plates were decorated by numerous small particles (indicated with N in Fig 2A; see also the upper‐right insets in Fig 2A and the slices and segmentations of plates decorated with many small particles shown in Fig 2B–E). The size of these particles (diameter ~ 9 nm; see Materials and Methods) suggests that they are nucleosomes. We performed reference‐free subtomogram averaging (Briggs, 2013) with classification for a set of 902 decorative particles; different views of the most relevant class (315 particles) are shown in Fig 2F (see also Fig EV1). The lower part of the reconstructed map is the link of the particle to the plate. Although the resolution of the average is low, likely due to both the small number of particles and the variety of orientations relative to the plate (see Materials and Methods), we observe that the dimensions of the average are consistent with the molecular structure of the nucleosome core particle. These results suggest that the particles decorating the plates shown in Fig 2 correspond to irregularly oriented nucleosomes, which were probably extruded from distorted plates.
Figure EV1. Subtomogram averaging of decorative nucleosomes.
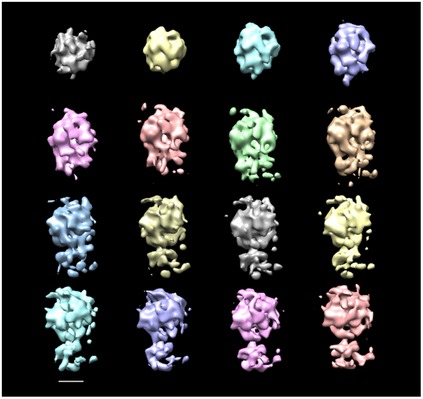
Initial random reference and averaged subvolumes of decorative nucleosomes like those shown in Fig 2A (region N) through 15 reference‐free alignment iterations; the averaged maps are shown without filtering. Scale bar: 5 nm.
The distorted plates (P1 in Fig 2A; see also the upper‐right insets) are relaxed and have less contrast than the typical compact plates observed in the cryo‐tomograms (P2 in Fig 2A; see also Figs 1 and 4), indicating that compact plates may have a higher nucleosome concentration. The thickness of these compact plates was ~ 7.5 nm (Table 1), measured directly from the tomographic slices while accounting for the contrast transfer function (CTF) fringes that border each plate (see Fig EV2). Compact plates do not show nucleosomes visible as separate units but, considering the dimensions of the nucleosome core particle (cylinder of 5.7 nm height and 11 nm diameter; Luger et al, 1997), the observed thickness suggests that each plate consists of a monolayer of nucleosomes aligned slightly tilted relative to the plate surface. These results indicate that compact plates are likely composed of tightly packed nucleosomes, which may have preferred orientations within the plate. In contrast, the relaxed plates have no well‐defined surfaces (compare P1 and P2 in Fig 2A), so they may contain loosely organized nucleosomes with more variable orientations.
Figure 4. Cryo‐tomograms of plates showing two‐layer contacts and edge‐to‐edge interactions.
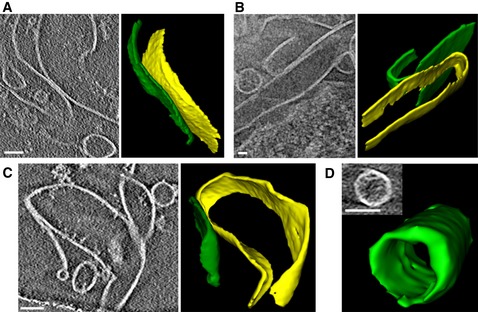
-
A–CSlices showing lateral association between two plates and the corresponding 3D segmentations.
-
DEdge‐to‐edge interactions form cylindrical structures.
Table 1.
Plate thickness
| Plate | Microscopea | Phase plate | Binning | Thicknessb (nm) |
|---|---|---|---|---|
| Monolayer | Polara | − | No | 7.2 ± 1.4 (n = 87) |
| Polara | − | 4× | 8.8 ± 2.0 (n = 85) | |
| Krios | + | 4× | 7.2 ± 1.0 (n = 366) | |
| Krios | − | 4× | 7.3 ± 0.9 (n = 39) | |
| Two layers in contact | Polara | − | No | 13.0 ± 2.0 (n = 26) |
| Krios | + | 4× | 12.4 ± 1.7 (n = 126) | |
| Krios | − | 4× | 13.0 ± 1.1 (n = 13) |
Tecnai Polara (27,500×); Titan Krios (33,000×).
Thickness measurements account for CTF fringes as described in Fig EV2. Values shown are means ± SD of the indicated number (n) of independent measurements.
Figure EV2. Measurement of plate thickness, adjusted for CTF fringes.
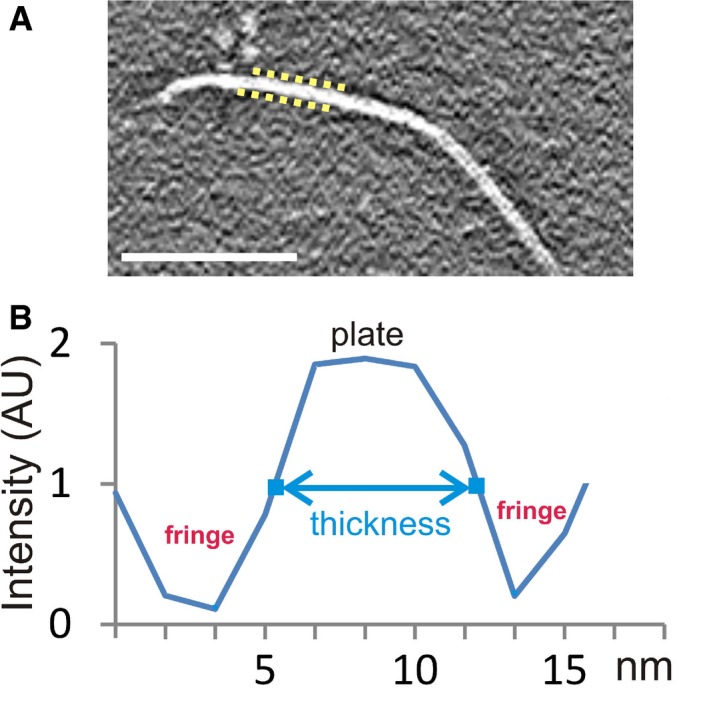
- The discontinuous yellow lines indicate the position of the CTF fringes on a select region of a plate. These fringes (black in reverse contrast) surround both sides of the plates in the tomograms. Scale bar: 100 nm.
- Example of an intensity profile along a vector perpendicular to a plate. Because of the CTF, the plate's intensity values do not have a hard edge, but rather gradually slope from the peak intensities of the plate to the valley of the surrounding CTF fringes. The plate thickness values presented in Table 1 correspond to the distance between the points (indicated in blue), approximately halfway between the peak and the valley. The intensities at these measurement points also roughly correspond to the background intensity of the tomogram. AU, arbitrary units.
Plates with many stacked layers
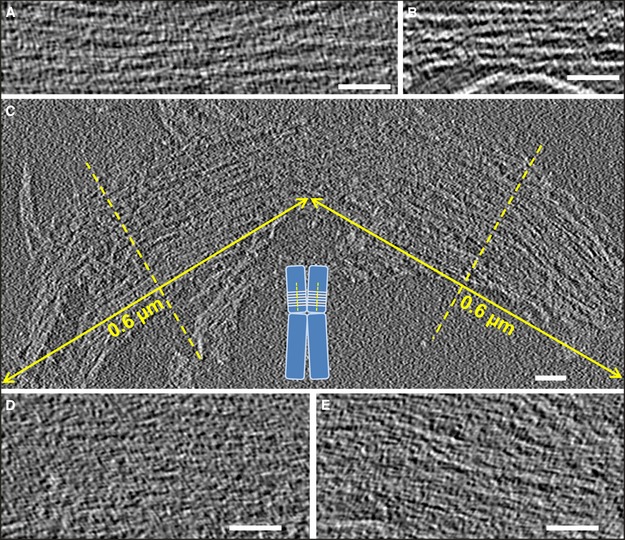
In the cryo‐tomograms, there are regions containing many parallel lines (see examples in Fig 3A and B). Each line persists in many consecutive slices along the z‐axis, indicating that these structures correspond to stacked plates oriented perpendicular to the x‐y plane. Our measurements of several structures containing parallel lines indicated lengths in the z‐axis ranging from 70 nm up to values approaching the ice thickness. The layers in the stacks have a thickness of ~ 7 nm (see Materials and Methods), which is equivalent to that observed for monolayer plates (Table 1).
Figure 3. Slices from tomographic volumes containing multilayered plates.
-
A, BPlates with several layers that are not closely appressed.
-
C–ELarge multilayer plates having the size of human metaphase chromatids [∼0.6 µm diameter (Daban, 2014)] (C); the inset schematically shows the perpendicular orientation of chromatin layers with respect to the chromatid axes proposed in the thin‐plate model (Gállego et al, 2009; Castro‐Hartmann et al, 2010). In other slices (D, E), the multilayer structures shown in (C) are more compact and the individual layers are not visible as separate elements.
Figure 3C shows a slice from a tomographic volume that contains a particularly large structure formed by many stacked layers. Figure EV3 shows additional examples of large multilayer structures. According to the thin‐plate model (Gállego et al, 2009; Castro‐Hartmann et al, 2010), chromosomes are formed by many stacked layers of chromatin oriented perpendicular to the axes of the chromatids (inset in Fig 3C); each chromatid of a human metaphase chromosome has a diameter of ~ 0.6 μm (Daban, 2014). Therefore, the dimensions of the structures in Fig EV3 suggest that they could be fragmented parts of chromatids. In the particular case of the multilayered structures in Fig 3C, since the left and right regions are apparently in contact, it is tempting to speculate that these two regions could correspond to stacked layers of two sister chromatids that broke apart during the preparation and deposition procedures. In the tomographic slice shown in this figure, parts of the layers can be seen separated from each other. However, in other slices of the same tomographic volume, the layers are much closer together and cannot be distinguished as separate units. For example, in the slices presented in Fig 3D and E (which correspond, respectively, to the left and right parts of the structure shown in Fig 3C), the layers are closely associated and, even at the high magnification used in these images, the structure is so compact that it is very difficult to distinguish individual layers.
Figure EV3. Additional examples of large multilayered plates.
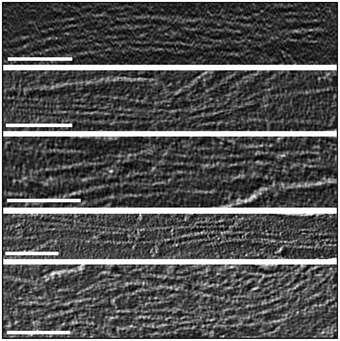
Slices from tomographic volumes containing large multilayered plates. Scale bars: 100 nm.
Layer interdigitation
In compact multilayered structures such as those presented in Fig 3D and E, it is not possible to analyze the structural details of the interaction between layers. Fortunately, in many tomographic volumes there are plates that interact with other plates in local regions that can be more readily analyzed. Figure 4A–C shows several examples of plates that make two‐layer contacts. Taking into account that the thickness of a monolayer plate is ~ 7.5 nm (Table 1), the expected thickness of two stacked layers should be ~ 15 nm. However, our measurements indicate that the thickness of two layers in close contact is ~ 13 nm (Table 1). These differences could be explained by a certain degree of interdigitation (~ 2 nm) between the two contacting layers. This structural solution was suggested previously from TEM and ET observations of dehydrated metaphase chromatin (Gállego et al, 2009; Castro‐Hartmann et al, 2010). Density profiles across interacting layers (Fig EV4) show that the layers are in such close contact that there is no empty space between them. Interdigitation would allow face‐to‐face interactions between nucleosomes in adjacent layers (see below).
Figure EV4. Contact between two plates.
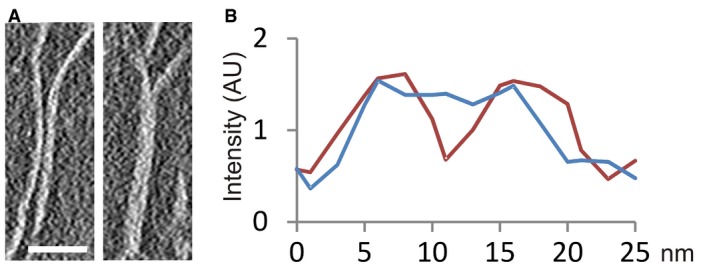
- Slice showing a region of a tomographic volume in which there are two independent plates (left); in another region, these two plates are in close contact (right). Scale bar: 50 nm.
- Intensity profile (red) along a vector perpendicular to the two independent plates shown in (A), and profile (blue) of the same plates in a region in which they are in contact. AU, arbitrary units.
In addition to lateral associations, plates can also interact through their edges to form closed structures. For instance, there are circular densities in the tomographic slices shown in Fig 4A–C. The 3D segmentation presented in Fig 4D shows that the observed circles correspond to slices of plates that form cylindrical structures. Additionally, Fig 2D and E shows a plate forming a tube decorated with nucleosomes. According to previous modeling studies (Daban, 2014), the lateral interaction between plates (inter‐layer association), as well as the edge‐to‐edge intra‐layer associations, can be interpreted considering that chromatin structures with nucleosomes exposed to the aqueous medium are less stable than structures in which there are more nucleosome‐nucleosome interactions. Both inter‐ and intra‐layer associations occur because there is a stabilization of the resulting structures due to the reduction of the number nucleosomes exposed to the medium.
SAXS analysis of condensed metaphase chromosomes
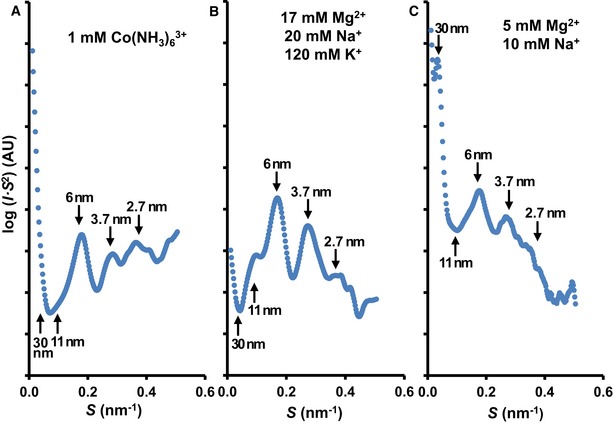
Whole metaphase chromosomes are large structures that are too dense and thick to observe chromatin organization using standard TEM (Fig 1A). To circumvent this problem, we analyzed the structure of thin chromatin plates emanated from soft‐denatured metaphase chromosomes using cryo‐ET (see the preceding sections), and we applied synchrotron SAXS to investigate the chromatin structure within whole intact chromosomes (Fig 5). The scattering study was performed using different divalent and trivalent cations that produce chromosome condensation (Strick et al, 2001; Poirier et al, 2002; Caravaca et al, 2005; Daban, 2011; Allahverdi et al, 2015; Maeshima et al, 2018). The peaks at ~ 3.7 and ~ 2.7 nm corresponding to the internal nucleosome structure (Widom & Klug, 1985) were observed in all samples with different intensities. The peak at ~ 30 nm, which is characteristic of side‐by‐side packaging of 30‐nm fibers (Widom, 1986), was only observed in the case of chromosomes prepared under relatively low Mg2+ concentration (Fig 5C). Nishino et al (2012) obtained a similar SAXS profile with metaphase chromosomes in 5 mM Mg2+, and these authors demonstrated that the 30‐nm peak can be eliminated by further chromosome purification. According to Strick et al (2001), in addition to physiological concentrations of K+ and Na+, metaphase chromosomes contain ~ 17 mM Mg2+ distributed homogeneously throughout the whole chromosome. Using these ionic conditions (Fig 5B), we observed that the 30‐nm peak was completely absent and the peak at ~ 6 nm was dominant. Similar results were obtained using the trivalent cation hexamminecobalt (Fig 5A).
Figure 5. SAXS profiles of metaphase chromosomes under different conditions.
-
A–CThe cation concentrations used are indicated for each experiment; AU, arbitrary units.
As can be seen in Fig 5A–C, in all the examined structuring conditions, the scattering intensity at ~ 11 nm is small compared to the prominent peak centered at ~ 6 nm. According to the X‐ray diffraction patterns observed for associated nucleosome cores that form columns (Mangenot et al, 2003; Bertin et al, 2007; Berezhnoy et al, 2016), the 6‐nm peak corresponds to the distance between nucleosome cores interacting face‐to‐face within the columns, and the 11‐nm peak is related to the distance between parallel columns (i.e., the distance corresponding to edge‐to‐edge nucleosome contacts). Narrow diffraction peaks were found in crystalline aggregates of nucleosome columns (Mangenot et al, 2003; Berezhnoy et al, 2016). Aggregates of nucleosome columns lacking the long‐range order of crystalline structures produce broad peaks (Mangenot et al, 2003). Therefore, in chromosomes formed by many stacked layers, the large number of face‐to‐face contacts that can be produced between nucleosomes in interdigitated layers can justify the broad peak at ~ 6 nm. Furthermore, this peak could also be related to the distance between stacked layers in condensed chromosomes (~ 6 nm; see Fig 6). This repeated distance strengthens the scattering peak at ~ 6 nm, and this can explain why the expected peak at ~ 11 nm corresponding to edge‐to‐edge contacts between nucleosomes in the compact multilayered structures has a low intensity in comparison with the 6‐nm peak. The scheme in Fig 6 integrates both the SAXS results and the main results obtained in the cryo‐ET experiments.
Figure 6. Schematic drawing showing the main structural elements of two layers in close contact.
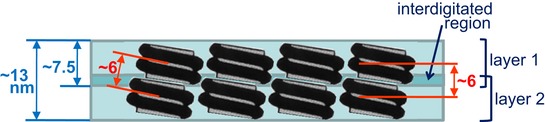
Only a few nucleosomes are shown to illustrate the main dimensions and the interdigitation of two layers. Our results suggest that the two turns of the nucleosomal DNA are oriented slightly tilted with respect to the axis normal to the plate surface, but they may have diverse orientations (not represented in this scheme) with respect to the other two axes of the plate. The thickness of single‐ and double‐layer plates (~ 7.5 and ~ 13 nm, respectively) obtained from cryo‐tomograms (Table 1) is indicated in blue. The main scattering peak at ~ 6 nm observed in SAXS experiments with condensed chromosomes under metaphase ionic conditions (Fig 5B) is probably due to the repetitive distances between nucleosomes (face‐to‐face interactions) and between stacked layers; these distances (~ 6 nm) are indicated in red.
Discussion
Our tomographic 3D reconstructions show that the chromatin filament in metaphase chromosomes is organized as a planar structure that forms many layers. This chromatin folding is completely different from the 30‐nm fiber proposed by many authors as the fundamental structural element of metaphase chromosomes. The chromatin emanated from soft‐denatured chromosomes forms monolayer plates (Fig 1B–G), plates containing two layers in close contact (Fig 4A–C), and multilayered plates (Figs 3 and EV3). Eltsov et al (2008) reported that 30‐nm fibers were not present in the cryo‐sections of chromosomes within mitotic cells. In principle, this observation is in agreement with our results, but these authors did not observe any higher‐order structure in their vitreous sections and concluded that the chromatin filament in metaphase chromosomes is completely disordered. Nevertheless, there is an alternative interpretation of these observations that is consistent with our findings: Chromatin plates cannot be distinguished as differentiated structural units in condensed chromosome cryo‐sections because, as can be seen in our cryo‐tomograms (Fig 3D and E), the individual layers are not visible in the interdigitated multilaminar plates. In these densely packed structures, space is completely filled by nucleosomes and consequently there is no visible higher‐order organization in the cryo‐sections of whole native chromosomes. Chromatin layers are only clearly visible in regions where chromosomes are distorted (Fig 3A–C).
Our SAXS results (Fig 5B) show that chromosomes prepared using the cation concentrations corresponding to metaphase conditions are not formed by densely packed 30‐nm fibers; this conclusion is in agreement with a previous SAXS study by Nishino et al (2012) with chromosomes in 5 mM Mg2+. The intense scattering at ~ 6 nm observed for native chromosomes under all the conditions analyzed (Fig 5A–C and Nishino et al, 2012) indicates that face‐to‐face association between nucleosomes (Fig 6) is a fundamental structural element of metaphase chromosomes. This is not surprising because face‐to‐face interactions with different degrees of overlap between nucleosomes were observed previously in different chromatin samples (purified nucleosome cores, chromatin fibers, and nucleosome arrays) using physicochemical methods (Tatchell & Van Holde, 1978), TEM (Finch et al, 1977; Dubochet & Noll, 1978; Bartolomé et al, 1994; Daban & Bermúdez, 1998; Robinson et al, 2006), cryo‐EM (Leforestier et al, 1999; Robinson et al, 2006; Scheffer et al, 2012; Song et al, 2014; Bilokapic et al, 2018), and X‐ray scattering (Mangenot et al, 2003; Bertin et al, 2007; Berezhnoy et al, 2016) and crystallography (Uberbacher & Bunick, 1985; Luger et al, 1997; Harp et al, 2000; White et al, 2001; Schalch et al, 2005; Ekundayo et al, 2017; Zhou et al, 2018); this interaction was also described in modeling studies (Stehr et al, 2010; Fan et al, 2013; Korolev et al, 2016, 2018; Saurabh et al, 2016; Ishida & Kono, 2017). The lateral association between two nucleosome cores involves an acidic surface formed by histones H2A and H2B and basic residues of the N‐terminal tail of histone H4 (Luger et al, 1997; Harp et al, 2000; White et al, 2001; Schalch et al, 2005). The diversity of nucleosome orientations observed in these lateral interactions (Mangenot et al, 2003; Ekundayo et al, 2017; Ishida & Kono, 2017; Bilokapic et al, 2018; Korolev et al, 2018; Zhou et al, 2018) may facilitate the association of nucleosomes in adjacent layers of chromatin plates.
The dense packaging observed in multilayered structures (Fig 3D and E), and the measurements (Table 1) showing that the thickness of two layers in close contact is smaller than the sum of two independent layers suggest that there is interdigitation between the stacked layers. This interdigitation allows face‐to‐face interactions of nucleosomes in adjacent layers (Fig 6). In vitro experiments and modeling studies demonstrated previously that interdigitation facilitates face‐to‐face nucleosome interactions that stabilize folded fibers with different conformations (Bartolomé et al, 1994; Daban & Bermúdez, 1998; Robinson et al, 2006; Wong et al, 2007; Depken & Schiessel, 2009; Rippe, 2012; Wu et al, 2016). The degree of interdigitation of the successive turns of compact solenoids is dependent on the orientation of the nucleosomes (Daban & Bermúdez, 1998). In agreement with this observation, our interpretation that nucleosomes are only slightly tilted within the plates explains why the observed degree of interdigitation in the multilayer plates is relatively low. Grigoryev (2004) and other authors (Eltsov et al, 2008; Castro‐Hartmann et al, 2010; Daban, 2011; Grigoryev & Woodcock, 2012; Luger et al, 2012; Collepardo‐Guevara & Schlick, 2014; Grigoryev et al, 2016; Maeshima et al, 2016; Bascom & Schlick, 2017) suggested that the lateral interdigitation of fibers can produce inter‐fiber associations. In vitro experiments performed with diluted chromatin fragments or nucleosome arrays in the presence of low cation concentrations showed that only intra‐fiber interactions between close neighbor nucleosomes can form (Eltsov et al, 2008; Daban, 2011); under these conditions, fibers folded as compact interdigitated solenoids (Daban & Bermúdez, 1998; Robinson et al, 2006) are the most complex structures that can be produced. In contrast, the high concentrations of chromatin (Daban, 2000) and cations (Strick et al, 2001) within condensed metaphase chromosomes allow the interaction between nucleosomes that are very distant in the single DNA molecule that is packed within each chromatid. All these observations strengthen the hypothesis that interdigitation combined with face‐to‐face interactions between nucleosomes in the successive layers stabilizes the multilaminar organization of planar chromatin in metaphase chromosomes.
We have performed an in vitro study using conditions that approach as much as possible the structuring ionic concentrations of metaphase cells, but future in vivo research will be required to validate the observed multilayered organization of chromatin. However, the functional role of this chromatin organization can be inferred from its structural and physical properties. The mechanical strength of planar chromatin (Gállego et al, 2010) and the stability of the stacked chromatin layers in metaphase chromosomes (see above) suggest that its primary biological role is the maintenance of the integrity of genomic DNA during mitosis. Furthermore, it was shown that this chromatin organization avoids topological entanglements of the chromatin filament (Milla & Daban, 2012) and can justify the elongated cylindrical structure of chromosomes as well as their outstanding mechanical properties (Poirier et al, 2000; Daban, 2014). It was also shown that if chromosomes consist of many stacked layers of planar chromatin, it is possible to explain many cytogenetic observations that were not previously understood (Daban, 2015). Presumably, the typical chromosome bands are produced by the preferential staining of several chromatin layers with different dyes, and the observed transverse orientation of the bands is due to the perpendicular orientation of the chromatin layers with respect to the chromosome axis. This also explains the splitting of broad bands (formed by several layers) observed in chromosome stretching experiments (Hliscs et al, 1997), and the maintenance of the orthogonal orientation of the split bands. According to the local concentration of DNA in metaphase chromosomes (~ 170 Mb/μm3; Daban, 2000, 2014), each chromatin layer of a human chromosome is formed by ~ 0.5 Mb of DNA, which justifies the existence of very thin bands containing < 1 Mb (International Human Genome Sequencing Consortium, 2001). The multilayered structure of chromatin in metaphase chromosomes is also compatible with the orthogonal orientation and planar structure of the connection surfaces seen in sister chromatid exchanges, and in the translocations observed in cancer cells. It has been argued (Daban, 2015) that the fibrillar models proposed by other authors (Paulson & Laemmli, 1977; Poirier & Marko, 2002; Kireeva et al, 2004; Eltsov et al, 2008; Naumova et al, 2013) require large quantities of DNA to cover the chromosome cross‐section and cannot justify the existence of very thin orthogonal bands and the orthogonal orientation of the connection surfaces in chromosome rearrangements.
There are several chromosome conformation capture methods capable of identifying contacts between distant regions of the chromatin filament via chemical crosslinking (Sajan & Hawkins, 2012; Bonev & Cavalli, 2016). In the genome‐wide Hi‐C method, the crosslinked contacting regions are identified by high‐throughput sequencing (Lieberman‐Aiden et al, 2009). Recently, Hi‐C results obtained with mitotic cells were modeled using polymer‐based simulations of chromatin structure, and it was proposed that chromatin in mitotic chromosomes is folded as a compact array of many loops having different sizes during mitosis (Gibcus et al, 2018). In the model proposed by these authors, the final compact chromosomes are formed by loops of ~ 0.5 Mb (consisting of ~ 400‐kb outer loops and ~ 80‐kb inner loops) and have a linear density of ~60 Mb/μm. For chromatids with a radius of ~ 0.36 μm (Gibcus et al, 2018), this linear density corresponds to ~ 150 Mb/μm3. This local DNA concentration is similar to the value considered above for multilayered chromosomes (~ 170 Mb/μm3; Daban, 2000, 2014) and is compatible with the high chromatin density observed for metaphase chromosomes by other authors (Eltsov et al, 2008; Ou et al, 2017). Obviously, to achieve this high density, the long chromatin filament in each 0.5‐Mb loop cannot be extended and must be tightly packed. We propose that the chromatin in the loops detected in the Hi‐C studies could be compacted into the multilayered plates observed in this work.
Materials and Methods
Preparation of metaphase chromosomes and chromatin plates
Chromosomes from HeLa cells blocked in metaphase with colcemid were prepared in TE buffer (15 mM triethanolamine‐HCl, pH 7.4, 2 mM EDTA, 0.5 mM EGTA, 20 mM NaCl, 80 mM KCl, 0.2 mM spermine, 0.5 mM spermidine, and 0.5% Triton X‐100) as described previously (Caravaca et al, 2005; Gállego et al, 2009); for SAXS experiments in the presence of 5 mM Mg2+, chromosomes were prepared in 10 mM PIPES (pH 7.2), 10 mM NaCl, 5 mM Mg2+, and 0.5% Triton X‐100. Chromosome suspensions were centrifuged at 4,000 g for 5 min. The resulting pellets were washed twice at 4°C with 10 mM PIPES (pH 7.2), 40% glycerol, and the concentrations of cations indicated in Fig 5. Finally, the samples were transferred to plastic capillaries (2 mm diameter, MiTeGen) and stored at −80°C for further SAXS measurements. For the preparation of chromatin plates, chromosomes in TE buffer (containing 1 mg/ml digitonin instead of Triton X‐100) were purified on a sucrose step gradient, with four layers (30, 40, 50, and 60% sucrose) containing 5 mM PIPES (pH 7.2), 5 mM NaCl, and 5 mM MgCl2 (PM buffer). Chromosomes were collected from the 40–50% and 50–60% sucrose interfaces; TEM images of these chromosomes were obtained following previously described procedures (Castro‐Hartmann et al, 2010). The chromosome suspension was diluted with four volumes of PM buffer without sucrose, passed several times through a 22‐gauge syringe needle, and finally dialyzed for 2.5 h at 37°C against the same buffer without sucrose. Each one of these treatments applied separately favors the emanation of chromatin plates from chromosomes (Gállego et al, 2009; Castro‐Hartmann et al, 2010), but we applied the three methods to obtain a high yield of plates. Note that in order to preserve the native chromatin structure as much as possible, the concentration of Mg2+ was maintained throughout these treatments. This sample was stored for 24 h in an ice bath and then used for the cryo‐preparations.
Cryo‐ET and image analysis
Perforated carbon films (Quantifoil R2/1) on 200‐mesh molybdenum grids were made hydrophilic by glow discharge before sample deposition. 200 μl of sample was pipetted onto the grid (placed in the cap of an inverted tube) and centrifuged at 1,500 g for 10 min. The grid was placed in a Vitrobot Mark III (set to 22°C and 95% humidity; blot offset −3 mm), 2 μl of gold particles (Aurion BSA tracers) in PM buffer were added, and the grid was blotted for 6 s from the reverse side and immediately plunged into a liquid ethane/propane mixture at liquid nitrogen temperature. Frozen‐hydrated preparations were stored in liquid nitrogen until used. Two sets of tomograms were obtained at the Instruct cryo‐ET platform at Max‐Planck‐Institute of Biochemistry (Martinsried). The first set (seven tomograms) was obtained with a Tecnai G2 Polara (FEI) microscope and the second set (25 tomograms) with a Titan Krios (FEI) microscope. Both instruments were equipped with field‐emission guns operated at 300 kV, post‐column energy filters (GIF 2002, Gatan), and K2 Summit (Gatan) direct electron‐detection cameras; in addition, in the case of Titan Krios most of the tomograms were obtained using a FEI Volta phase plate (Danev et al, 2014). Tilt series (range ±60°; increments of 2°) were collected under low‐dose conditions (total dose ~ 110 e−/Å2) using SerialEM software (Mastronarde, 2005) with −5 to −6 μm defocus at 27,500× magnification (image pixel size of 4.27 Å) in the Polara, and with −0.5 μm defocus at 33,000× magnification (image pixel size of 4.21 Å) in Titan Krios equipped with the phase plate. Image frames from the K2 direct detector were aligned using in‐house developed software based on the algorithm described in Li et al (2013). Alignment of the images of each tilt series was performed with IMOD software (Kremer et al, 1996) using fiducial gold particles, and tomographic 3D reconstruction was performed using the weighted back projection. Generally, to enhance contrast, tomograms were 4× binned, but in some cases (see Table 1) unbinned images were used for 3D reconstructions. Segmentations and the measurements of plate thickness and nucleosome dimensions were performed with Fiji‐ImageJ (NIH). These measurements accounted for the CTF fringes bordering the plates (caused by defocus and other modulating effects), as described in Fig EV2. The thickness of monolayers and two layers in close contact is presented in Table 1. The thickness of single layers in multilayered structures is 6.6 ± 1.5 (n = 205), and the diameter of nucleosomes decorating distorted plates is 8.7 ± 1.3 nm (n = 206). Unless otherwise indicated, measurements were made using slices from 4× binned tomograms obtained with the Titan Krios microscope, equipped with the phase plate. The slices from tomographic volumes are shown in reverse contrast.
Subtomogram averaging and classification
For a detailed analysis of the decorative particles (N in Fig 2A), a set of 902 particles was manually picked and extracted using the EMAN package (Tang et al, 2007). The subtomogram average was generated from data acquired on the Titan Krios microscope using the Volta phase plate and 0.5‐μm defocus. The 3D alignment and classification were performed using algorithms based on maximum likelihood (Scheres et al, 2009) included in the Xmipp package (de la Rosa‐Trevín et al, 2013). We did not apply a focused mask, only a standard spherical mask about the same size as the box to prevent hard box edges from influencing the alignment. We processed four classes (155, 192, 315, and 240 particles) using a 50 pixel box size and 15 iterations. Classes 1, 2, and 4 resulted in noisy versions of class 3, which was filtered to 25 Å (Fig 2F). The method is reference‐free; the process was started from a weighted average structure obtained from random orientations of all particles. Figure EV1 shows the evolution of the averaged subvolume at each iteration without filtering. Two parts can be distinguished in the reconstructed volume: the top region that has the size of a nucleosome core particle and the lower region corresponding to the link of the particle to the plate. Due to the large box size, part of the plate associated with decorative nucleosomes is also included in the subtomogram average. The limited number of particles available in cryo‐ET experiments did not allow to obtain a high‐resolution structure; the resolution estimated using the local resolution algorithm MonoRes (Vilas et al, 2018) is around 3 nm.
SAXS experiments
X‐ray scattering of condensed chromosomes was recorded at room temperature at the non‐crystalline diffraction (NCD) beamline of the ALBA Synchrotron (Cerdanyola del Vallès, Barcelona) using a SAXS Quantum 210r CCD detector from ADSC. Different zones of the chromosome pellets were exposed to X‐rays for 60 s. The wavelength λ was 1.29 Å and the sample‐to‐detector distance was 2.61 m. SAXS data are shown as plots of log (I·S 2) versus S, where I is the average intensity obtained after subtracting the buffer scattering and S = 2sinθ/λ is the scattering vector; 2θ is the scattering angle.
Author contributions
AC designed experiments, prepared chromatin plates, collected cryo‐ET and SAXS data, performed image processing, and analyzed data; EC designed experiments, prepared metaphase chromosomes, and collected and analyzed SAXS data; JO and RM processed cryo‐ET data and performed subtomogram averaging; BDE collected and analyzed cryo‐ET data, supervised the cryo‐ET experiments, and wrote the paper; J‐RD conceived and supervised the project, designed experiments, analyzed cryo‐ET and SAXS data, and wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Review Process File
Acknowledgements
The authors thank the staffs of the Serveis de Microscòpia and Cultius Cellulars (UAB), the cryo‐ET and image processing Instruct platforms, and the NCD‐BL11 staff from ALBA Synchrotron for their invaluable assistance. This work was supported in part by MINECO research grant BFU2010‐18939, European Union (EU) and Horizon 2020 through grant West‐Life (EINFRA‐2015‐1, Proposal: 675858), and a by a UAB‐PIF predoctoral fellowship to AC. Cryo‐ET experiments at Max‐Planck‐Institute of Biochemistry (Martinsried) and image processing at National Center of Biotechnology (Madrid) were funded by Instruct (PID250, PID2115), part of the European Strategy Forum on Research Infrastructures (ESFRI) and supported by national member subscriptions.
The EMBO Journal (2019) 38: e99769
See also: B Fierz (April 2019)
Data availability
The subtomogram average of nucleosome particles decorating unstructured plates and a cryo‐electron tomogram of chromatin plates emanated from metaphase chromosomes have been deposited in the Electron Microscopy Data Bank (EMDB; http://www.emdatabank.org) with accession numbers EMD‐0117 and EMD‐0119, respectively.
References
- Alberts B, Johnson A, Lewis J, Morgan D, Raff M, Roberts K, Walter P (2014) Molecular biology of the cell, pp 207–216. New York, NY: Garland Science; [Google Scholar]
- Allahverdi A, Chen Q, Korolev N, Nordenskiöld L (2015) Chromatin compaction under mixed salt conditions: opposite effects of sodium and potassium ions on nucleosome array folding. Sci Rep 5: 8512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomé S, Bermúdez A, Daban JR (1994) Internal structure of the 30 nm chromatin fiber. J Cell Sci 107: 2983–2992 [DOI] [PubMed] [Google Scholar]
- Bartolomé S, Bermúdez A, Daban JR (1995) Electrophoresis of chromatin on nondenaturing agarose gels containing Mg2+. Self‐assembly of small chromatin fragments and folding of the 30‐nm fiber. J Biol Chem 270: 22514–22521 [DOI] [PubMed] [Google Scholar]
- Bascom G, Schlick T (2017) Linking chromatin fibers to gene folding by hierarchical looping. Biophys J 112: 434–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezhnoy NV, Liu Y, Allahverdi A, Yang R, Su CJ, Liu CF, Korolev N, Nordenskiöld L (2016) The influence of ionic environment and histone tails on columnar order of nucleosome core particles. Biophys J 110: 1720–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermúdez A, Bartolomé S, Daban JR (1998) Partial denaturation of small chromatin fragments: direct evidence for the radial distribution of nucleosomes in folded chromatin fibers. J Cell Sci 111: 1707–1715 [DOI] [PubMed] [Google Scholar]
- Bertin A, Mangenot S, Renouard M, Durand D, Livolant F (2007) Structure and phase diagram of nucleosome core particles aggregated by multivalent cations. Biophys J 93: 3652–3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilokapic S, Strauss M, Halic M (2018) Cryo‐EM of nucleosome core particle interactions in trans. Sci Rep 8: 7046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Cavalli G (2016) Organization and function of the 3D genome. Nat Rev Genet 17: 661–678 [DOI] [PubMed] [Google Scholar]
- Boulé JB, Mozziconacci J, Lavelle C (2015) The polymorphisms of the chromatin fiber. J Phys Condens Matter 27: 033101 [DOI] [PubMed] [Google Scholar]
- Briggs JAG (2013) Structural biology in situ: the potential of subtomogram averaging. Curr Opin Struct Biol 23: 261–267 [DOI] [PubMed] [Google Scholar]
- Caravaca JM, Caño S, Gállego I, Daban JR (2005) Structural elements of bulk chromatin within metaphase chromosomes. Chromosome Res 13: 725–743 [DOI] [PubMed] [Google Scholar]
- Castro‐Hartmann P, Milla M, Daban JR (2010) Irregular orientation of nucleosomes in the well‐defined chromatin plates of metaphase chromosomes. Biochemistry 49: 4043–4050 [DOI] [PubMed] [Google Scholar]
- Collepardo‐Guevara R, Schlick T (2012) Crucial role of dynamic linker histone binding and divalent ions for DNA accessibility and gene regulation revealed by mesoscale modeling of oligonucleosomes. Nucleic Acids Res 18: 8803–8817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collepardo‐Guevara R, Schlick T (2014) Chromatin fiber polymorphism triggered by variations of DNA linker length. Proc Natl Acad Sci USA 111: 8061–8066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daban JR, Bermúdez A (1998) Interdigitated solenoid model for compact chromatin fibers. Biochemistry 37: 4299–4304 [DOI] [PubMed] [Google Scholar]
- Daban JR (2000) Physical constraints in the condensation of eukaryotic chromosomes. Local concentration of DNA versus linear packing ratio in higher order chromatin structures. Biochemistry 39: 3861–3866 [DOI] [PubMed] [Google Scholar]
- Daban JR (2003) High concentration of DNA in condensed chromatin. Biochem Cell Biol 81: 91–99 [DOI] [PubMed] [Google Scholar]
- Daban JR (2011) Electron microscopy and atomic force microscopy studies of chromatin and metaphase chromosome structure. Micron 42: 733–750 [DOI] [PubMed] [Google Scholar]
- Daban JR (2014) The energy components of stacked chromatin layers explain the morphology, dimensions and mechanical properties of metaphase chromosomes. J R Soc Interface 11: 20131043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daban JR (2015) Stacked thin layers of metaphase chromatin explain the geometry of chromosome rearrangements and banding. Sci Rep 5: 14891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danev R, Buijsse B, Khoshouei M, Plitzko JM, Baumeister W (2014) Volta potential phase plate for in‐focus phase contrast transmission electron microscopy. Proc Natl Acad Sci USA 111: 15635–15640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depken M, Schiessel H (2009) Nucleosome shape dictates chromatin fiber structure. Biophys J 96: 777–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubochet J, Noll M (1978) Nucleosome arcs and helices. Science 202: 280–286 [DOI] [PubMed] [Google Scholar]
- Ekundayo B, Richmond TJ, Schalch T (2017) Capturing structural heterogeneity in chromatin fibers. J Mol Biol 429: 3031–3042 [DOI] [PubMed] [Google Scholar]
- Eltsov M, MacLellan KM, Maeshima K, Frangakis AS, Dubochet J (2008) Analysis of cryo‐electron microscopy images does not support the existence of 30‐nm chromatin fibers in mitotic chromosomes in situ . Proc Natl Acad Sci USA 105: 19732–19737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Korolev N, Lyubartsev AP, Nordenskiöld L (2013) An advanced coarse‐grained nucleosome core particle model for computer simulations of nucleosome‐nucleosome interactions under varying ionic conditions. PLoS One 8: e54228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez‐Leiro R, Scheres SHW (2016) Unravelling biological macromolecules with cryo‐electron microscopy. Nature 537: 339–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch JT, Lutter LC, Rhodes D, Brown RS, Rushton B, Levitt M, Klug A (1977) Structure of nucleosome core particles of chromatin. Nature 269: 29–36 [DOI] [PubMed] [Google Scholar]
- Gállego I, Castro‐Hartmann P, Caravaca JM, Caño S, Daban JR (2009) Dense chromatin plates in metaphase chromosomes. Eur Biophys J 38: 503–522 [DOI] [PubMed] [Google Scholar]
- Gállego I, Oncins G, Sisquella X, Fernàndez‐Busquets X, Daban JR (2010) Nanotribology results show that DNA forms a mechanically resistant 2D network in metaphase chromatin plates. Biophys J 99: 3951–3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Gutiérrez MC, Rueda DR (2009) Bases of synchrotron radiation, light sources, and features of X‐ray scattering beamlines. Lect Notes Phys 776: 1–22 [Google Scholar]
- Gibcus JH, Samejima K, Goloborodko A, Samejima I, Naumova N, Nuebler J, Kanemaki MT, Xie L, Paulson JR, Earnshaw WC, Mirny LA, Dekker J (2018) A pathway for mitotic chromosome formation. Science 359: eaaao6135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryev SA (2004) Keeping fingers crossed: heterochromatin spreading through interdigitation of nucleosome arrays. FEBS Lett 564: 4–8 [DOI] [PubMed] [Google Scholar]
- Grigoryev SA, Woodcock CL (2012) Chromatin organization. The 30 nm fiber. Exp Cell Res 318: 1448–1455 [DOI] [PubMed] [Google Scholar]
- Grigoryev SA, Bascom G, Buckwalter JM, Schubert MB, Woodcock CL, Schlick T (2016) Hierarchical looping of zigzag nucleosome chains in metaphase chromosomes. Proc Natl Acad Sci USA 113: 1238–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harp JM, Hanson BL, Timm DE, Bunick GJ (2000) Asymmetries in the nucleosome core particle at 2.5 Å resolution. Acta Crystallogr D Biol Crystallogr 56: 1513–1534 [DOI] [PubMed] [Google Scholar]
- Hliscs R, Mühlig P, Claussen U (1997) The nature of G‐bands analyzed by chromosome stretching. Cytogenet Cell Genet 79: 162–166 [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Ishida H, Kono H (2017) H4 tails potentially produce the diversity in the orientation of two nucleosomes. Biophys J 113: 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva N, Lakonishok M, Kireev I, Hirano T, Belmont AS (2004) Visualization of early chromosome condensation: a hierarchical folding, axial glue model of chromosome structure. J Cell Biol 166: 775–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev N, Nordenskiöld L, Lyubartsev AP (2016) Multiscale coarse‐grained modelling of chromatin components: DNA and the nucleosome. Adv Colloid Interface Sci 232: 36–48 [DOI] [PubMed] [Google Scholar]
- Korolev N, Lyubartsev AP, Nordenskiöld L (2018) A systematic analysis of nucleosome core particle and nucleosome‐nucleosome stacking structure. Sci Rep 8: 1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR (1996) Computer visualization of three‐dimensional image data using IMOD. J Struct Biol 116: 71–76 [DOI] [PubMed] [Google Scholar]
- Leforestier A, Fudaley S, Livolant F (1999) Spermidine‐induced aggregation of nucleosome core particles: evidence for multiple liquid crystalline phases. J Mol Biol 290: 481–494 [DOI] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y (2013) Electron counting and beam‐induced motion correction enable near‐atomic‐resolution single‐particle cryo‐EM. Nat Methods 10: 584–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman‐Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J (2009) Comprehensive mapping of long‐range interactions reveals folding principles of the human genome. Science 326: 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucic V, Förster F, Baumeister W (2005) Structural studies by electron tomography: from cells to molecules. Annu Rev Biochem 74: 833–865 [DOI] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Luger K, Dechassa ML, Tremethick DJ (2012) New insight into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol 13: 436–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima K, Rogge R, Tamura S, Joti Y, Hikima T, Szerlong H, Krause C, Herman J, Seidel E, DeLuca J, Ishikawa T, Hansen JC (2016) Nucleosomal arrays self‐assemble into supramolecular globular structures lacking 30‐nm fibers. EMBO J 35: 1115–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima K, Matsuda T, Shindo Y, Imamura H, Tamura S, Imai R, Kawakami S, Nagashima R, Soga T, Noji H, Oka K, Nagai T (2018) A transient rise in free Mg2+ ions released from ATP‐Mg hydrolysis contributes to mitotic chromosome condensation. Curr Biol 28: 1–8 [DOI] [PubMed] [Google Scholar]
- Mangenot S, Leforestier A, Durand D, Livolant F (2003) X‐ray diffraction characterization of the dense phases formed by nucleosome core particles. Biophys J 84: 2570–2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (2005) Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152: 36–51 [DOI] [PubMed] [Google Scholar]
- Milla M, Daban JR (2012) Self‐assembly of thin plates from micrococcal nuclease‐digested chromatin of metaphase chromosomes. Biophys J 103: 567–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J (2013) Organization of the mitotic chromosome. Science 342: 948–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino Y, Eltsov M, Joti Y, Ito K, Takata H, Takahashi Y, Hihara S, Frangakis AS, Imamoto N, Ishikawa T, Maeshima K (2012) Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30‐nm chromatin structure. EMBO J 31: 1644–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou HD, Phan S, Deerinck TJ, Thor A, Ellisman MH, O'Shea CC (2017) ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357: eaag0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson JR, Laemmli UK (1977) The structure of histone‐depleted metaphase chromosomes. Cell 12: 817–828 [DOI] [PubMed] [Google Scholar]
- Poirier M, Eroglu S, Chatenay D, Marko JF (2000) Reversible and irreversible unfolding of mitotic newt chromosomes by applied force. Mol Biol Cell 11: 269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier MG, Marko JF (2002) Mitotic chromosomes are chromatin networks without a mechanically contiguous protein scaffold. Proc Natl Acad Sci USA 99: 15393–15397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier MG, Monhait T, Marko JF (2002) Reversible hypercondensation and decondensation of mitotic chromosomes studied using combined chemical‐micromechanical techniques. J Cell Biochem 85: 422–434 [DOI] [PubMed] [Google Scholar]
- Rippe K (2012) The folding of the nucleosome chain In Genome organization and function in the cell nucleus, Rippe K. (ed.), pp 139–167. Weinheim: Wiley‐VCH; [Google Scholar]
- Robinson PJJ, Fairall L, Huynh VAT, Rhodes D (2006) EM measurements define the dimensions of the “30‐nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc Natl Acad Sci USA 103: 6506–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Rosa‐Trevín JM, Otón J, Marabini R, Zaldívar A, Vargas J, Carazo JM, Sorzano COS (2013) Xmipp 3.0: an improved software suite for image processing in electron microscopy. J Struct Biol 184: 321–328 [DOI] [PubMed] [Google Scholar]
- Sajan SA, Hawkins RD (2012) Methods for identifying higher‐order chromatin structure. Annu Rev Genomics Hum Genet 13: 59–82 [DOI] [PubMed] [Google Scholar]
- Saurabh S, Glaser MA, Lansac Y, Maiti PK (2016) Atomistic simulation of stacked nucleosome core particles: tail bridging, the H4 tail, and effect of hydrophobic forces. J Phys Chem B 120: 3048–3060 [DOI] [PubMed] [Google Scholar]
- Schalch T, Duda S, Sargent DF, Richmond TJ (2005) X‐ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 436: 138–141 [DOI] [PubMed] [Google Scholar]
- Scheffer MP, Eltsov M, Bednar J, Frangakis AS (2012) Nucleosomes stacked with aligned dyad axes are found in native compact chromatin in vitro . J Struct Biol 178: 207–214 [DOI] [PubMed] [Google Scholar]
- Scheres SHW, Melero R, Valle M, Carazo JM (2009) Averaging of electron subtomograms and random conical tilt reconstructions through likelihood optimization. Structure 17: 1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F, Chen P, Sun D, Wang M, Dong L, Liang D, Xu RM, Zhu P, Li G (2014) Cryo‐EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science 344: 376–380 [DOI] [PubMed] [Google Scholar]
- Stehr R, Schöpflin R, Etting R, Kepper N, Rippe K, Wederman G (2010) Exploring the conformational space of chromatin fibers and their stability by numerical dynamic phase diagrams. Biophys J 98: 1028–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strick R, Strissel PL, Gavrilov K, Levi‐Setti R (2001) Cation‐chromatin binding as shown by ion microscopy is essential for the structural integrity of chromosomes. J Cell Biol 155: 899–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ (2007) EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157: 38–46 [DOI] [PubMed] [Google Scholar]
- Tatchell K, Van Holde KE (1978) Compact oligomers and nucleosome phasing. Proc Natl Acad Sci USA 75: 3583–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uberbacher EC, Bunick GJ (1985) X‐ray structure of the nucleosome core particle. J Biomol Struct Dyn 2: 1033–1055 [DOI] [PubMed] [Google Scholar]
- Vilas JL, Gómez‐Blanco J, Conesa P, Melero R, de la Rosa‐Trevín JM, Otón J, Cuenca J, Marabini R, Carazo JM, Vargas J, Sorzano COS (2018) MonoRes: automatic and accurate estimation of local resolution for electron microscopy maps. Structure 26: 1–8 [DOI] [PubMed] [Google Scholar]
- White CL, Suto RK, Luger K (2001) Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J 20: 5207–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widom J, Klug A (1985) Structure of the 300 Å chromatin filament: X‐ray diffraction from oriented samples. Cell 43: 207–213 [DOI] [PubMed] [Google Scholar]
- Widom J (1986) Physicochemical studies of the folding of the 100 Å nucleosome filament into the 300 Å filament. Cation dependence. J Mol Biol 190: 411–424 [DOI] [PubMed] [Google Scholar]
- Wong H, Victor JM, Mozziconacci J (2007) An all‐atom model of the chromatin fiber containing linker histones reveals a versatile structure tuned by the nucleosome repeat length. PLoS One 9: e877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, McGeehan JE, Travers A (2016) A metastable structure for the compact 30‐nm chromatin fibre. FEBS Lett 590: 935–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BR, Jiang J, Ghirlando R, Norouzi D, Yadav KNS, Feng H, Wang R, Zhang P, Zhurkin V, Bai Y (2018) Revisit of reconstituted 30‐nm nucleosome arrays reveals an ensemble of dynamic structures. J Mol Biol 430: 3093–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Review Process File
Data Availability Statement
The subtomogram average of nucleosome particles decorating unstructured plates and a cryo‐electron tomogram of chromatin plates emanated from metaphase chromosomes have been deposited in the Electron Microscopy Data Bank (EMDB; http://www.emdatabank.org) with accession numbers EMD‐0117 and EMD‐0119, respectively.


