Abstract
Growth substrates that maximize energy yield are widely thought to be utilized preferentially by microorganisms. However, observed distributions of microorganisms and their activities often deviate from predictions based solely on thermodynamic considerations of substrate energy supply. Here we present observations of the bioenergetics and growth yields of a metabolically flexible, thermophilic strain of the archaeon Acidianus when grown autotrophically on minimal medium with hydrogen (H2) or elemental sulfur (S°) as an electron donor, and S° or ferric iron (Fe3+) as an electron acceptor. Thermodynamic calculations indicate that S°/Fe3+ and H2/Fe3+ yield three- and four-fold more energy per mol electron transferred, respectively, than the H2/S° couple. However, biomass yields in Acidianus cultures provided with H2/S° were eight-fold greater than when provided S°/Fe3+ or H2/Fe3+, indicating the H2/S° redox couple is preferred. Indeed, cells provided with all three growth substrates (H2, Fe3+, and S°) grew preferentially by reduction of S° with H2. We conclude that substrate preference is dictated by differences in the energy demand of electron transfer reactions in Acidianus when grown with different substrates, rather than substrate energy supply.
Microorganisms and their activities drive global biogeochemical cycles and interconnect ecosystem processes affecting plant, animal, and human health 1, 2. Consequently, a central goal in microbial ecology is to predict the distributions and activities of microorganisms in natural environments 3. The application of thermodynamic calculations to geochemical data makes it possible to predict and infer the variety and abundance of chemical energy available to microbial populations 4, 5. Such approaches have been widely used to rationalize the observed distribution of microbial populations and their activities in a variety of environments 6, 7, 8.
Thermodynamic-based approaches rank assemblages of redox reactions according to the amount of energy that can be dissipated, with the distribution of microorganisms and their activities presumed to follow this sequence of reactions due to competition for energy-rich substrates 6, 7, 9, 10. However, thermodynamic-based predictions often fall short of accurately describing the distribution of microorganisms and metabolic processes in natural 11, 12, 13, 14 and engineered systems 15. To provide insight into why these predictions often fail to play out in natural systems and to improve our ability to accurately predict patterns of substrate usage and their effects on biogeochemical cycles, we aimed to quantify the bioenergetics and biomass yields of a metabolically versatile organism. To limit the influence of confounding variables in our determination of the efficiency of biomass synthesis, we sought to identify an autotrophic strain with the ability to grow on defined mineral medium with minimal biosynthetic deficiencies (minimal auxotrophy) and that exhibits versatility in its energy metabolism.
The thermoacidophilic strain Acidianus DS80, within the crenarchaeal order Sulfolobales, was isolated under autotrophic conditions on defined mineral medium (supplemented with vitamins) from Dragon Spring (pH 3.1, 78°C), Yellowstone National Park, Wyoming, U.S.A. Strain DS80 displays versatility in lithotrophic energy metabolism involving hydrogen (H2) or elemental sulfur (S°) as electron donors and S° or ferric iron (Fe3+) as electron acceptors (Supp. Fig. 1) when grown at pH 3.0 and 80°C. Moreover, the ability of strain DS80 to couple the iron, sulfur, and hydrogen cycles in overlapping ways, in addition to the broader metabolic flexibility and global distribution of Acidianus in thermal environments16, 17, 18, makes strain DS80 a good model system to examine factors that influence substrate preference.
Growth yields and substrate transformation kinetics.
Generation times (Tn) of autotrophically grown DS80 cells provided with the redox couples H2/S°, S°/Fe3+, or H2/Fe3+ were statistically indistinguishable (Table 1), indicating that measurements of Tn are not sensitive to processes that dictate substrate preference. The amount of CO2 assimilated per cell (Table 1, Supp. Table 1–2) when cultures were provided with H2/S° or S°/Fe3+ was also statistically indistinguishable (P = 0.59). This is consistent with similar cellular morphologies (Fig. 1; Supp. Fig. 2) and cell sizes (1038 ± 68 nm and 1128 ± 38 nm in diameter, respectively) when grown under these conditions (Table 1). In contrast, the amount of carbon assimilated from CO2 per cell when grown with H2/Fe3+ was 2-fold lower than when cells were grown with H2/S° or S°/Fe3+ (Table 1), which was reflected by the smaller size (826 ± 55 nm diameter) and the 2-fold smaller calculated volume of cells grown with H2/Fe3+. Since the amount of CO2 assimilated per unit cell volume (Table 1) was not significantly different (P > 0.33 for all pairwise comparisons) among cells grown under these three conditions, these observations suggest that the differences in the electron transfer pathways attending utilization of the substrates involved in the cells energy metabolism and their thermodynamic efficiencies (energy conserved as chemical bonds or ionic gradients as opposed to energy lost as heat) 19 lead to an observed phenotype at the morphological level.
Table 1.
Generation times (Tn), lag phase times, yields of carbon assimilated per cell, measured cell diameters, yields of carbon assimilated per unit cell volume, and yields of carbon assimilated per μJ of energy released in cultures of Acidianus sp. DS80 grown at 80°C in base salts medium (pH 3.0) with CO2 as sole carbon source with the electron donors and acceptors specified. The average and standard deviation of three replicate cultures is presented. In the case of cell diameter determinations, the average and standard deviation of measurements of >10 cells, based on TEM, are presented.
| Redox reaction | Tn (hrs) | Lag phase (hrs) | fmol CO2 /cell | Cell diameter (nm) | fmol CO2 /m3 b | fmol CO2 /μJ |
|---|---|---|---|---|---|---|
| H2(g) + S°(s) → H2S(aq) | 35 ± 17 | 24 | 3.00 ± 0.34 | 1038 ± 68 | 4.41 ± 3.24 | 267.4 ± 36.2 |
| S°(s) + 6Fe3+(aq) + 4H2O(aq) → 6Fe2+(aq) + HSO4−(aq) + 7H+ | 33 ± 19 | 96 | 3.44 ± 1.90 | 1128 ± 38 | 4.39 ± 2.92 | 31.7 ± 1.7 |
| H2(g) + 2Fe3+(aq) → 2Fe2+(aq) + 2H+ | 27 ± 4 | 120 | 1.64 ± 0.23 | 826 ± 55 | 4.85 ± 2.87 | 34.2 ± 5.0 |
| Competition Experiment (H2 / S° / Fe3+)a | 31 ± 2 | 24 | 3.32 ± 0.25 | 1017 ± 29 | 5.87 ± 1.83 | N/D |
Redox reaction is not defined since multiple potential electron donors and acceptors are present in the cultivation medium.
Calculated assuming a spherical (coccoid) morphology (Fig. 1).
Abbreviations: N/D, not determined.
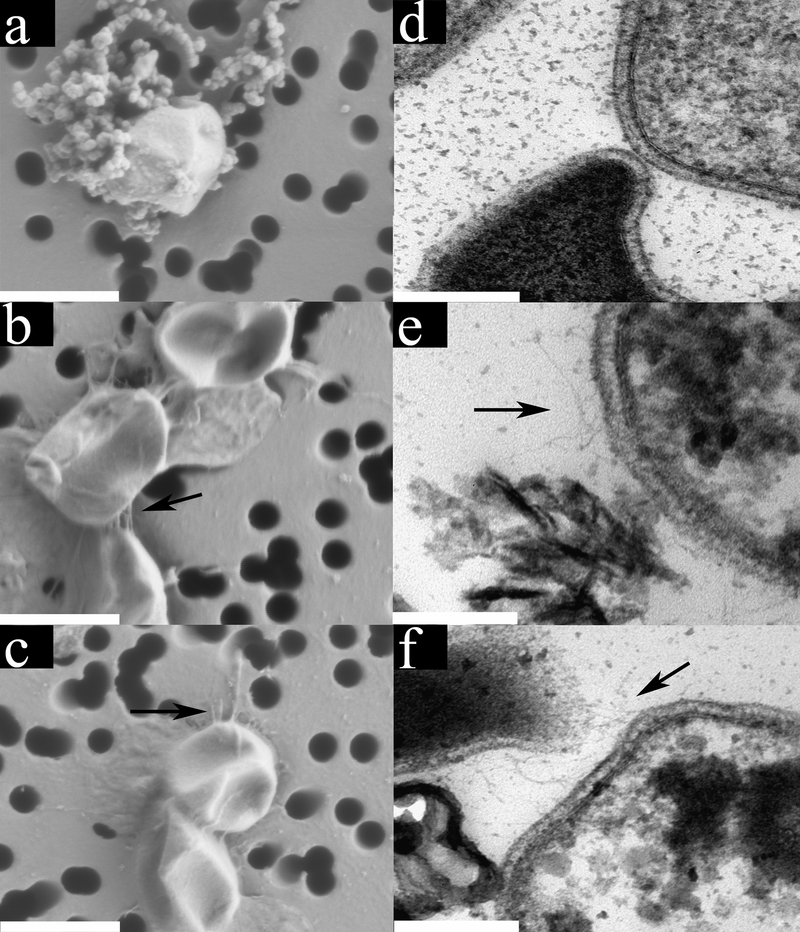
Fig. 1. Electron micrographs of DS80 cells grown with different electron donor/acceptor pairs reveal differences in morphology.
Field emission scanning electron micrographs (FE SEMs) of H2/S° (a), S°/Fe3+ (b), and H2/Fe3+ (c) grown cells with arrows denoting pili-like structures where present. Globules outside cells in panel a correspond to nanoparticles of S° 20. FE SEM scale bars represent 1000 nm. Thin section transmission electron micrographs (TEMs) of cells grown with H2/S° (d), S°/Fe3+ (e), and H2/Fe3+ (f) with arrows denoting pilin-like structures where present. Insoluble precipitates of unknown composition are visible in panel e and f. TEM scale bars represent 200 nm.
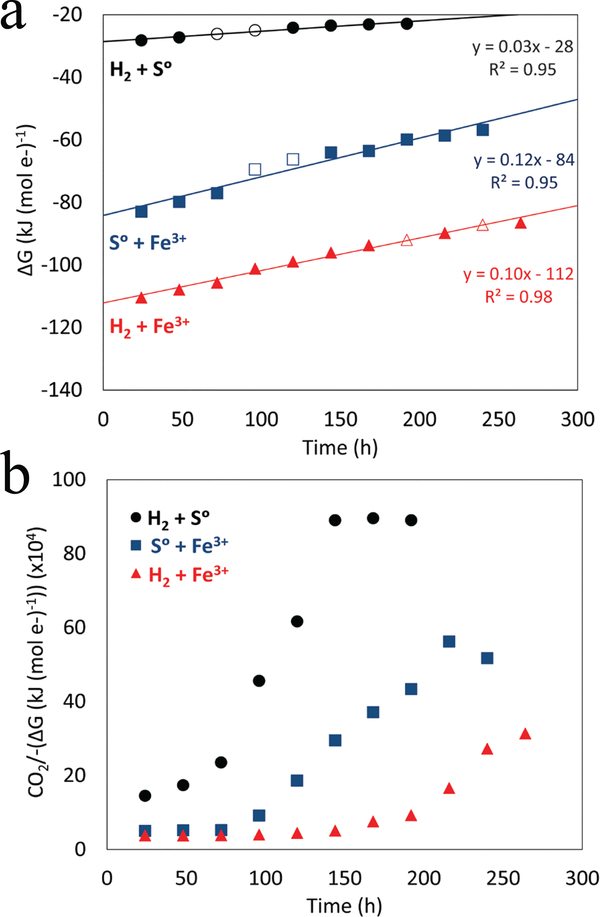
To further explore this possibility, measured rates of substrate transformation were used to calculate the amount of energy released during log phase growth assuming stoichiometric conversion of reactants to products as depicted by reactions listed in Table 1. The amount of CO2 assimilated per μJ of energy released in log phase cells grown with H2/S° was ~ 8-fold greater than in cells grown with S°/Fe3+ or H2/Fe3+ (Table 1). This indicates that the electron transfer pathways used to conserve energy in cells growing with H2/S° are more efficient than those operating in cells grown under iron reducing conditions, allowing for greater biomass synthesis. Furthermore, a plot of the change in the overall Gibbs free energy per mol of electron transfer as a function of culture incubation time (Fig. 2a) revealed different slopes among the three growth conditions. The growth condition with the shallowest slope was H2/S° which is consistent with a lower energy demand for biomass synthesis in cells respiring S° than those respiring Fe3+ (Fig. 2b).
Fig. 2. Available Gibbs free energy (ΔG) per mol e- transferred in DS80 cultures provided with H2/S°, S°/Fe3+, and H2/Fe3+.
Trends in the data are illustrated with the least-squares best-fit regressions. The standard deviations of the slope of the regression for H2/S°, S°/Fe3+, and H2/Fe3+ grown cultures are 0.002, 0.010, 0.004, respectively. Empty circles represent logarithmic growth and are the points where generation times and growth yields were calculated (a). Carbon assimilated (fmol) divided by the negative value of (ΔG) per mol e- transferred in cultures of DS80 provided with H2/S°, S°/Fe3+, and H2/Fe3+ as a function of incubation time (b).
The energy demand for biomass synthesis in cells respiring S° is likely lower than that reported here. This is due to the observation that S° reducing thermoacidophilic Archaea reduce nanoparticulate S° that is generated when hydrogen sulfide (H2S; from respiration of S°) reacts with bulk S° to form polysulfides (Sx2−) 20. Under acidic conditions (pH <6.0), Sx2− rapidly disproportionate to yield H2S and nanoparticulate S°, the latter of which is thought to be the electron acceptor in thermoacidophiles reducing S° 20. However, the involvement of other intermediates in the reduction of S° by thermoacidophiles cannot be excluded. The activity of nanoparticulate S° in previous growth experiments 20 and in the growth experiments reported here is not known, but is likely to be far lower than the activity of 1.0 specified for bulk S° in thermodynamic calculations considering previously determined low concentrations of S° (478 nM at 80°C in equilibrium with excess S° solid phase) in abiotic aqueous solutions under controlled experimental conditions 21. Furthermore, in the case of Fe3+ reduction, the chemical equilibrium models that were used took aqueous Fe3+ complexes into account and thus thermodynamic calculations were based on the concentration of free Fe3+ only, which was quite low (0.19 mM) when compared to total Fe3+ (17.2 mM). Taken together, these observations suggest that cells grown in the presence of H2/S°/Fe3+ will preferentially utilize the H2/S° couple, despite this couple being energetically less favorable per mol e- transferred than the H2/Fe3+ and S°/Fe3+ couples (Fig. 2).
Preferential use of redox couples.
We tested the hypothesis that DS80 will preferentially utilize the H2/S° couple by growing cells in the presence of all three growth substrates (H2/S°/Fe3+), simultaneously. Concentrations of soluble Fe3+, Fe2+, SO42-, and H2S were determined to track the use of available electron donors and acceptors (Supp. Table 4). In addition, the number of cells produced, the amount of CO2 assimilated per cell, and cell size were quantified. Gibbs free energies for each of the three individual redox couples (H2/S°, S°/Fe3+, H2/Fe3+) at the start of the competition experiment varied by < 1% of the Gibbs free energies computed at the onset of growth experiments prepared with only a single possible redox couple. During growth in the presence of all three growth substrates the concentration of SO42- in biological controls never exceeded that of abiological controls. Since oxidation products of S° (i.e., thiosulfate and sulfite) are instable in aqueous solutions with pH <4.0, in particular in the presence of Fe3+ 22, and ultimately degrade to SO42- in acid solutions 23, this observation indicates that cells were not oxidizing S° and thus not growing with the S°/Fe3+ couple (i.e., oxidizing S°).
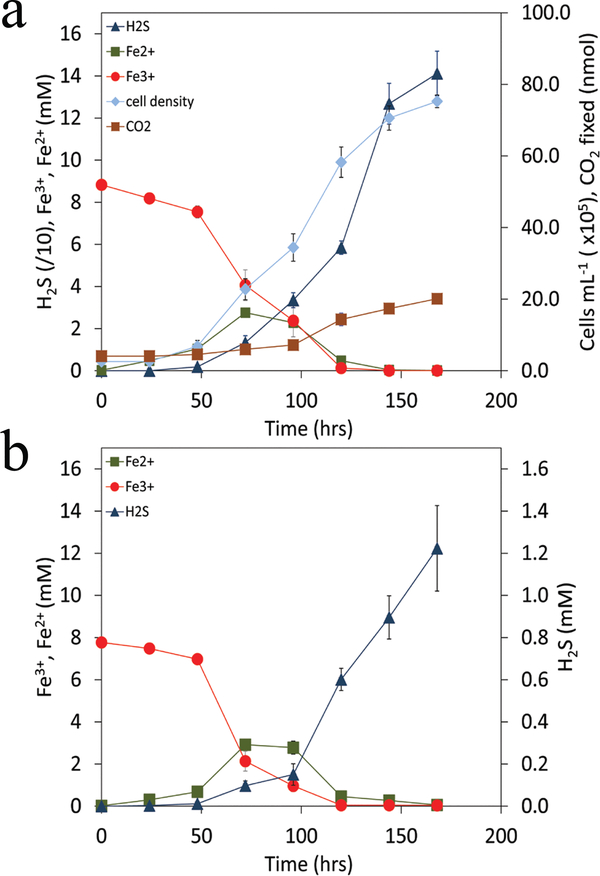
Within the first day of growth the concentration of Fe2+ increased with a near stoichiometric decrease in the concentration of soluble Fe3+ (Fig. 3a). This differed from cells grown with H2/Fe3+ only, where detectable Fe2+ was not observed until 6 days of growth (Supp. Fig. 1). This suggests that the Fe2+ produced in the H2/S°/Fe3+ cultivation was due to abiological reduction of Fe3+ by H2S produced from the biological reduction of S°. Indeed, following depletion of soluble Fe3+ after 48 hrs, the concentration of H2S increased rapidly as did concentrations of cells and amounts of CO2 assimilated, consistent with growth with the H2/S° couple. These observations are independent of whether cells used as inoculum were grown with H2/S°, S°/Fe3+ or H2/Fe3+ (Supp. Fig. 3) indicating that preference for H2/S° is not due to a “priming effect” or a difference in the need to acclimate to new cultivation conditions. Further evidence that DS80 cells prefer to utilize H2/S° when provided with H2/S°/Fe3+ comes from the size of cells in cultures, which were statistically indistinguishable (P = 0.47) from those provided with H2/S° but which were significantly larger (P < 0.01) than cells grown with H2/Fe3+. Likewise, the amount of CO2 assimilated per cell in cultures provided with H2/S°/Fe3+ was statistically indistinguishable (P = 0.62) from measurements made on cells provided with H2/S° and the lag phase corresponded with cultures grown with H2/S° (Table 1).
Fig. 3. Growth and activities of DS80.
Cells were provided with H2/S°/Fe3+ with CO2 as the sole carbon source when a culture growing on H2/S° was used as inoculum (a). Cultures innoculated with S°/Fe3+ or H2/Fe3+ grown cells (Supp. Fig. 3), exhibited indistinguishable growth kinetics and activities when compared to those presented. Titration of anoxic and sterile cultivation medium containing H2/S°/Fe3+ with H2S (b) reveals a similar activity profile when compared to the biological control (a). Depletion in the concentration of soluble Fe2+ in both abiotic and biotic assays was due to formation of insoluble iron sulfides (Supp. Fig. 4).
To assess whether Fe3+ reduction observed early in cultures provided with H2/S°/Fe3+ was due to abiological reduction by H2S, sterile medium containing H2/S°/Fe3+ was titrated with H2S (Fig. 3b) at rates that corresponded with those determined in cultures grown with H2/S° only (Supp. Fig. 1). Production of Fe2+ and consumption of soluble Fe3+ were observed at similar rates and over similar time frames in these abiotic experiments (Fig. 3b) when compared to biotic assays (Fig. 3a). The amount of soluble Fe2+ detected could not account for the amount of Fe3+ reduction that had taken place. Evidence that this was due to formation of insoluble iron sulfides comes from depletion in the concentration of soluble Fe2+ in both abiotic and biotic assays (Fig. 3) concomitant with the formation of black precipitates in cultures (Supp. Fig. 4), which are likely iron mono- or di-sulfides 24.
Energy demand of redox reactions dictates substrate preference.
The results of these experiments raise questions about why the energy demands differ in cells supported by these reactions, and where and how electron transfer takes place in cells that grow preferentially with H2/S° over S°/Fe3+ or H2/Fe3+. Respiration of S° with H2 in Acidianus is mediated by a short electron transfer chain comprising a membrane-associated [NiFe]-hydrogenase and a membrane-associated sulfur reductase complex, linked by a quinone cycle, with S reduction thought to take place on the outer side of the membrane 25. Respiration of Fe3+ with H2 or S° in crenarchaeotes, including Acidianus, is not well understood but in DS80 likely involves the same [NiFe]-hydrogenase, since there is a single hydrogenase encoded in its genome, and a sulfur-oxygenase reductase (SOR) complex 26 which is also encoded in the draft genome (IMG submission identifier 92883; estimated to be 99% complete; data not shown). Despite the absence of a characterized Fe3+ reduction pathway, DS80 cells grown with Fe3+ as the electron acceptor had numerous hair-like protrusions extending from the plasma membrane (Fig. 1; Supp. Fig. 5) that resembled “nanowires” 27, 28. Such structures were observed when cells were grown with “soluble” Fe3+ [i.e., Fe2(SO4)3] (Figs. 1e & 1f) or insoluble ferric oxyhydroxides such as ferrihydrite or hematite (Supp. Fig. 6) but not with S° as oxidant (Fig. 1d), suggesting that this is a generalized physiological response to growth via Fe3+ reduction. Intriguingly, these structures were often observed in association with mineral precipitates of unknown composition in Fe3+ grown cultures (Figs. 1; Supp. Figs. 5a & 5b). Such structures were not observed when cells were provided with H2/S°/Fe3+ (data not shown), which is consistent with these cells growing preferentially via the H2/S° redox couple under this condition.
C-type multiheme cytochromes (cMHC) have been suggested to be involved in iron reduction in both Bacteria 29, 30 and Archaea 31, 32, and have been suggested to be the electrically conductive component of nanowires in Shewanella 33. However, many iron reducers, including a number of archaea from the Sulfolobales and Thermoproteales 31, 34, lack detectable c-type cytochromes 35 or the genes encoding for c-type cytochromes. Consistent with this observation, BLASTp searches failed to detect conventional (CXXCH) 36, 37 and unconventional heme binding motifs (CXXXH and CXXXXH) 37 in the DS80 and the closely related Acidianus copahuensis genome 16, suggesting that they also do not encode for cMHCs. Collectively, these observations point to a different mechanism of Fe3+ reduction in members of the Sulfolobales that is independent of c-type cytochromes.
Although the physical properties conferring conductivity to microbial nanowires has been controversial, it has been recently suggested that overlap of the pi-pi orbitals of aromatic amino acids in the structural pilin protein PilA in Geobacter imparts metallic-like conductivity to the structures, independent of the presence of cMHC 38, 39. The primary structural components of nanowires (type IV pili) include an assembly ATPase, a transmembrane protein, a pilin protein, and a prepilin peptidase 40, 41. BLASTp analyses of proteins encoded in the DS80 genome revealed the presence of the ATPase, a transmembrane protein, and a prepilin peptidase in an apparent operon while screens for proteins with class III signal peptides thought to be specific for type IV pili using the FlaFind (ver. 1.2) server 42, 43 revealed several type IV pilin-like proteins in the DS80 genome. Pilin proteins that have been shown to be involved in electron transfer are enriched in aromatic amino acids, with the more conductive pilins averaging an aromatic density (number of aromatic rings in amino acids divided by the total number of amino acids) of 0.15 44. Intriguingly, one of the pilin proteins encoded in the DS80 genome is predicted to have an aromatic density of 0.12, which may point to its potential involvement in Fe3+ reduction.
While the role of pili-like structures in Fe3+ reduction in strain DS80 is not clear, their synthesis would involve an additional energetic cost to cells grown with Fe3+ but not with S°. In addition, if these structures were involved in Fe3+ reduction, it is possible that energy is lost as heat during successive electron transfers along the array of electrically conductive components (e.g., aromatic amino acids) leading to a lower thermodynamic efficiency per electron transported. Energy loss through either of these mechanisms would represent additional energetic costs to cells, which could otherwise contribute to biomass synthesis and more efficient cellular yields. The longer lag times associated with growth via Fe3+ reduction when compared to S° reduction (Table 1; Supp. Fig. 1) may point to the influence of these mechanisms on the inefficiency of converting energy captured to biomass synthesis. Alternatively, the short lag associated with the H2/S° versus the H2/Fe3+ and S°/Fe3+ couples may suggest that the biochemical machinery necessary to process the H2/S° couple is the energetically less costly to assemble when compared to the machinery necessary to process the other two redox couples.
Collectively, these observations indicate that the preferential use of electron donors and acceptors in metabolically versatile strains, such as DS80, cannot be understood in terms of energetic supplies based on thermodynamic analysis or on measurements of Tn alone. Rather, these data show that combining thermodynamic calculations with measurements of biomass yields provides a metric to describe the physiology of cells in a way that more accurately captures the physiological nuances experienced by cells and that dictate differences in energy demands attending substrate utilization. We suggest that a combined approach, as outlined above, could be used to develop more accurate models for describing the distribution of microorganisms and their activities in natural and engineered environments.
Online-Only Methods Section
METHODS
Data Availability.
The data that support the findings of this study are available from the corresponding author upon request. The draft genome sequence obtained for strain DS80 has been deposited in the Joint Genome Institute IMG database, IMG submission identifier 92883, genome ID 2690315630.
Sample collection and enrichment of Acidianus strain DS80.
Samples for enrichment and isolation of a chemolithotrophic strain capable of metabolizing substrates that can be easily tracked were collected from the source of ‘Dragon Spring’, an acidic hot spring (78°C; pH 3.1; determined with a YSI combination pH and temperature probe) located in the Hundred Springs Plain area of Norris Geyser Basin in YNP (44°43′55″ N, 110°42′39″ W). The source of ‘Dragon Spring’ has clear depositional zones of sulfur and iron oxides and samples were collected from this area (see images in 45, 46). Detailed reports describing the aqueous- and solid-phase chemistry of ‘Dragon Spring’ have been published previously 45, 46 and thus only basic chemical measurements will be reported here. Ferrous iron (Fe2+) and total sulfide (S2-total) were quantified using Hach ferrozine pillows and Hach sulfide reagents 1 and 2, respectively, and a Hach DR/890 spectrophotometer (Hach Company, Loveland, CO). The concentration of Fe2+ in source water was 55 μM and the concentration of S2-total was 46 μM.
Flocculent elemental sulfur (S°) precipitate and spring water were sampled aseptically using a sterile syringe and this precipitate slurry was immediately injected into a sterile, N2-purged serum bottle. The serum bottle was placed in an insulated thermos bottle containing spring water to keep the samples at ambient spring temperature during the 120-min. transit to the lab where they were immediately used to inoculate enrichments. Roughly 250 μL (~100 mg of dry solids) of the S° precipitate slurry was used to inoculate a base salts medium containing NH4Cl (0.33 g L−1), KCl (0.33 g L−1), CaCl2 • 2H2O (0.33 g L−1), MgCl2 • 6H2O (0.33 g L−1), and KH2PO4 (0.33 g L−1) 47. The pH of the base salts medium was adjusted to 3.0 with concentrated hydrochloric acid. Eighteen mL of base salts medium were dispensed into 70 mL serum bottles and bottles and their contents were subjected to autoclave sterilization. Following autoclave sterilization, filter-sterilized Wolfe’s vitamins (1 mL L−1 final concentration), filter sterilized SL-10 trace metals (1 mL L−1 final concentration), and S° (sulfur precipitated powder, EMD Millipore, USA; 5 g L−1 final concentration; sterilized by baking at 100°C for 24 h) were added, and the bottles and contents were deoxygenated by purging with sterile nitrogen gas passed over heated (210°C) and hydrogen reduced copper shavings. The serum bottles were sealed with butyl rubber stoppers and heated to 80°C prior to the replacement of the headspace with H2 and CO2 (80%:20%) or N2 and CO2 (80%:20%). The serum bottles were then inoculated with the sulfur precipitates slurry recovered from the source of ‘Dragon Spring’ and were incubated at 80°C.
Progress of enrichments was monitored by filtering cells onto 0.22 μm black polycarbonate filters (Millipore, Billerica, MA), staining cells with SYBR Gold Nucleic Acid Gel Stain (Life Technologies, Inc., Grand Island, NY) at a final concentration of 1/4000 (vol/vol), followed by epifluorescence microscopy using an Axioskop 2 Plus microscope (Zeiss, Thornwood, NY). Following 4 rounds of dilution to extinction, a single morphotype was observed, which was designated as strain DS80. The purity of the culture was further confirmed by PCR. Briefly, PCR amplification of 16S rRNA gene was performed according to previously described protocols 48, 49 using archaeal primers 344F (5’-ACGGGGYGCAGCAGGCGCGA-3’) and 915R (5’-GTGCTCCCCCGCCAATTCCT-3’) and bacterial primers 1100F (5’-YAACGAGCGCAACCC-3’) and 1492R (5’-GGTTACCTTGTTACGACTT-3’). Bacterial 16S rRNA gene amplicons were not detected. Archaeal 16S rRNA gene amplicons were purified, cloned, sequenced, assembled, and analyzed using previously published protocols 47. A single 16S rRNA gene phylotype was recovered and this was 100% identical to the 16S rRNA gene from Acidianus hospitalis. Nucleotide sequence of the 16S rRNA gene of strain DS80 have been deposited in the GenBank database under the accession number KX608545. Genomic DNA was subjected to Illumina MiSeq sequencing, assembly, and annotation as described previously 50. The assembled genome (IMG submission identifier 92883), which is estimated to be 99% complete (data not shown).
Physiological characterization.
The temperature range for growth of strain DS80 was tested between 40 to 100°C, at pH 3.0. The pH range for growth was tested between 1.0 and 6.0, at 80°C. Optimal growth was observed at 80°C and pH 3.0 and thus all experiments were conducted under these conditions. The ability of strain DS80 to utilize H2 or S° as electron donors and Fe3+ or S° as electron acceptors was tested in anoxic base salts medium amended with trace elements and vitamins as previously described 47. Medium was supplemented with S° (5 g L−1) or Fe2(SO4)3 (3.7 g liter −1). Fe2(SO4)3 was prepared at a pH 1.9 (fully dissolved) and was added to medium with a pH of 7.5 (final pH when mixed was 3.0). A headspace gas mixture of 80%:20% N2-CO2 was used when S° was the electron donor and Fe3+ the electron acceptor. A headspace gas mixture of 80%:20% H2-CO2 was used when S° or Fe3+ were supplied as the electron acceptor. All cultures were incubated at 80°C. Cultures were prepared in triplicate for both biological and abiological (un-inoculated) assays as described above.
Preferential substrate use determination.
Cells of strain DS80 were grown in the presence of H2, S°, and Fe2(SO4)3 • H2O to test for the preferential use of H2/S°, H2/Fe3+, S°/Fe3+ as a redox couple. Anoxic base salts medium amended with trace elements and vitamins was used, as previously described 47. Medium was supplemented with S° (5 g L−1) and Fe2(SO4)3 • H2O (3.7 g liter −1), using a headspace gas mixture of 80%:20% H2-CO2. Cultures grown with H2/S°, S°/Fe3+ or H2/Fe3+ were separately used as inoculum to rule out any “priming effect” or carryover effect from previous growth experiments. Abiotic experiments were performed by titration of sterile medium containing H2/S°/Fe3+ with Na2S at rates that corresponded with those determined in cultures grown with H2/S°. Cultures were prepared in triplicate for both biological and abiological (un-inoculated) assays and incubated at 80°C as described above.
Evaluation of growth and activity.
Growth of strain DS80 with different electron donors and acceptors was quantified in terms of cell density, dissolved inorganic carbon (DIC) assimilation (described below), sulfur or iron reduction activity, or sulfur oxidation activity. Cell density was determined by staining with SYBR Gold and enumeration via epifluorescence microscopy, as described above. A subsample (0.9 mL) of culture was briefly centrifuged (14,000 x g, 1 min.) to remove particulate iron before activity measurements. Dissolved sulfide concentrations were determined with the methylene blue reduction method 51. Total sulfide production (dissolved and gaseous) was calculated using standard gas-phase equilibrium calculations as described previously 47. The concentration of total and ferrous iron was determined using the Ferrozine method; the concentration of ferric iron was calculated as the difference between total (obtained after reduction of all iron with hydroxylamine hydrochloride) and ferrous iron measurements 52. The concentration of sulfate was determined after precipitation with barium chloride, as previously described 53. The addition of Fe2(SO4)3 • H2O was considered in the measurement of sulfate production.
Carbon assimilation.
The amount of CO2 assimilated into cellular biomass in cultures of DS80 was determined using a radioisotope tracer method as described previously 50. Briefly, a 160 mL serum bottle containing 55 mL of base salts medium (described above) with a 20% CO2 (V/V) headspace (balance was H2 or N2, depending on experimental condition) was amended with 14C-labeled sodium bicarbonate (American Radiolabeled Chemical) to a final concentration of 27.5 μCi. Given that the pKa for bicarbonate/CO2(aq) at 80°C is ~6.4 54, we assumed that the added bicarbonate completely dissociated to CO2 when added to medium (pH 3.0) and equilibrated equally among the unlabeled DIC pools. The amount of 14C assimilated into cellular biomass was quantified by removal of 2 mL subsamples of culture from each serum bottle (followed by replacement of the gas phase) with sterile gas. Concentrated hydrochloric acid was added to subsamples to achieve a final pH of less than 2.0 in order to volatilize unreacted DIC. Following 2 hrs of equilibration and degassing, subsamples were filtered onto 0.22 μm white polycarbonate membranes, washed with sterile base salts medium, and dried at 80°C for 24 hrs. Dried filters were transferred to 20 mL liquid scintillation vials containing 10 mL CytoScint ES™ liquid scintillation fluid (MP Biomedicals, USA) and radioactivity associated with filtered biomass was measured on a Beckman LS 6500 liquid scintillation counter (LSC) (Beckman Coulter, Inc., Indianapolis, IN). Rates of CO2 assimilation were calculated from LSC counts using previously described methods 50. The amount of C assimilated per cell (fmol/cell) during autotrophic growth was calculated by dividing the total amount of C assimilated from CO2 during exponential growth by the number of new cells produced over this time.
Energetics.
The total amount of energy that can be obtained by an organism catalyzing a redox reaction in a non-standard state displaced from equilibrium is given by equation 1:
| (1) |
where ΔG represents the Gibbs free energy of reaction, ΔG° represents the standard Gibbs free energy of reaction (J mol−1), R designates the ideal gas constant (~8.314 J mol−1 K−1), T is the temperature in Kelvin (K), and Q is the activity product. The activity (α) of each reactant and product can be calculated from its aqueous concentration and the ionic strength of the aqueous solution in which the reaction proceeds using the extended Debye-Hückel equation 55.
As was previously described by Urschel et al. 50, the activities of reactant and product at each time point during growth of strain DS80 on H2/S°, S°/Fe3+ and H2/Fe3+ were calculated based on the ionic strength and composition of the base salts medium, as well as the measured or estimated concentration of each reactant and product, using the freeware chemical equilibrium modeling application Visual MINTEQ 3.0. Since we did not quantify differences in H2 concentration during the growth of strain DS80, we assumed a constant aqueous H2 concentration equal to the saturation concentration of H2 in H2O at 80°C and 1 atmosphere of partial pressure (~768 μM). Reactant and product activities were then used to calculate logQ based on the following chemical reactions (number in parenthesis indicates equation number):
| (2) |
| (3) |
| (4) |
ΔG° of the different reactions was calculated using the SUPCRT online program provided by the “Group Exploring Organic Processes in Geochemistry” from Arizona State University.
Calculated log Q and ΔG° values at each time point during exponential phase were then used to calculate ΔG with equation (1) above, and the resulting ΔG value was divided by the total number of electrons transferred per mol substrate transformed to determine the energy available per mol electron transferred kJ (mol e-)−1 at each time point during exponential phase. Because of the differences in the stoichiometry of the chemical reactions 2, 3 and 4, the biomass yield per μJ of energy was calculated in order to be able to directly compare the efficiency of coupling energy conservation to biomass synthesis under these three growth conditions. The difference in the amount of C assimilated between two sampling points during exponential growth was then divided by the difference in total energy dissipated (Fig. 2, Supp. Tables 1–3) between these time points to compute the amount of C assimilated per unit of energy dissipated (fmol CO2 μJ−1). These calculations were conducted using measurements made during exponential phase growth (Supp. Fig. 1).
Transmission electron microscopy.
Cells were fixed on ice with 2% paraformaldehyde and 1.25% glutaraldehyde and washed with Hepes buffer as previously described 56. Cells were negatively stained with sodium phosphotungstate and imaged on a Leo912AB transmission electron microscope (TEM) equipped with a 2K X 2K CCD camera, operated at 100-kV accelerating voltage. The diameters of negative stained cells were measured from the TEM micrographs. Cells from all growth conditions were prepared for TEM analysis using the same protocol in order to account for modification of cell size associated with the preparation technique.
Field emission scanning electron microscopy.
Cells were fixed with 2% paraformaldehyde and 1.25% glutaraldehyde overnight at room temperature. Cells were washed four times with distilled water. Washed cells were subjected to a series of dehydration steps with 25% ethanol for 20 min, 50% ethanol for 20 min, 75% ethanol for 20 min, 95% ethanol for 30 min. and three final dehydrations using 100% ethanol for 60 min each. Dehydrated cells were then subjected to critical point drying using a Tousimis Samdri®−795 critical point dryer. Dried cells were coated with iridium and imaged on a Zeiss SUPRA 55VP Field Emission Scanning Electron Microscope.
Supplementary Material
Acknowledgements
This work was supported by National Science Foundation grant EAR - 1123689 to ESB and EAR-1123649 to ELS and NASA Exobiology and Evolutionary Biology Grant (NNX13AI11G) to ESB. The NASA Astrobiology Institute is supported by grant NNA13AA94A to EER and ESB and grant NNA15BB02A to ELS and ESB. MJA acknowledges support from the CONICYT Becas-Chile Scholarship program.
Footnotes
Competing Financial Interests The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
REFERENCES.
- 1.Falkowski PG, Fenchel T & Delong EF The microbial engines that drive Earth’s biogeochemical cycles. Science. 320, 1034–1039 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Lin W, Wang Y, Gorby Y, Nealson K & Pan Y Integrating niche-based process and spatial process in biogeography of magnetotactic bacteria. Sci. Rep 3, 1643 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atlas RM & Bartha R Microbial Ecology: Fundamentals and Applications. Benjamin-Cummings Pub. Co.: California, 1986. [Google Scholar]
- 4.Amend JP, Rogers KL, Shock EL, Gurrieri S, & Inguaggiato S Energetics of chemolithoautotrophy in the hydrothermal system of Vulcano Island, southern Italy. Geobiology. 1, 37–58 (2003). [Google Scholar]
- 5.Shock EL et al. Quantifying inorganic sources of geochemical energy in hydrothermal ecosystems, Yellowstone National Park, USA. Geochim. Cosmochim. Acta 74, 4005–4043 (2010). [Google Scholar]
- 6.Lovley DR & Klug MJ Sulfate reducers can outcompete methanogens at freshwater sulfate concentrations. Appl. Environ. Microbiol 45, 187–192 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nealson KH & Stahl DA Microorganisms and biogeochemical cycles; what can we learn from layered microbial communities? Rev. Mineral. Geochem. 35, 5–34 (1997). [Google Scholar]
- 8.Spear JR, Walker JJ, McCollom TM & Pace NR Hydrogen and bioenergetics in the Yellowstone geothermal ecosystem. Proc. Nat. Acad. Sci. U.S.A 102, 2555–2560 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Froelich PN et al. Early oxidation of organic matter in pelagic sediments of the eastern equatorial Atlantic: suboxic diagenesis. Geochim. Cosmochim. Acta 43, 1075–1090 (1979). [Google Scholar]
- 10.Roden EE & Wetzel RG Competition between Fe(III)-reducing and methanogenic bacteria for acetate in iron-rich freshwater sediments. Microb. Ecol 45, 252–258 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Boyd ES, Skidmore M, Mitchell AC, Bakermans C & Peters JW Methanogenesis in subglacial sediments. Environ. Microbiol. Rep 2, 685–692 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Canfield DE & Des Marais DJ Aerobic sulfate reduction in microbial mats. Science. 251, 1471–1473 (1991). [DOI] [PubMed] [Google Scholar]
- 13.D’Hondt S et al. Distributions of microbial activities in deep subseafloor sediments. Science. 306, 2216–2221 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Hansel CM et al. Dominance of sulfur-fueled iron oxide reduction in low-sulfate freshwater sediments. ISME J. 9, 2400–2412 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marounek M, Fliegrova K & Bartos S Metabolism and some characteristics of ruminal strains of Megasphaera elsdenii. Appl. Environ. Microbiol. 55, 1570–1573 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urbieta MS et al. Draft genome sequence of the novel thermoacidophilic archaeon Acidianus copahuensis strain ALE1, isolated from the Copahue volcanic area in Neuquén, Argentina. Genome Announc. 2, e00259–00214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Segerer A, Neuner A, Kristjansson JK & Stetter KO Acidianus infernus gen. nov., sp. nov., and Acidianus brierleyi Comb. nov.: Facultatively aerobic, extremely acidophilic thermophilic sulfur-metabolizing Archaebacteria. Int. J. Syst. Bacteriol 36, 559–564 (1986). [Google Scholar]
- 18.Plumb JJ, Haddad CM, Gibson JAE & Franzmann PD Acidianus sulfidivorans sp. nov., an extremely acidophilic, thermophilic archaeon isolated from a solfatara on Lihir Island, Papua New Guinea, and emendation of the genus description. Int. J. Syst. Evol. Microbiol 57, 1418–1423 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Thauer RK, Jungermann K & Decker K Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev 41, 100–180 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyd ES & Druschel GK Involvement of intermediate sulfur species in biological reduction of elemental sulfur under acidic, hydrothermal conditions. Appl. Environ. Microbiol. 79, 2061–2068 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamyshny A Solubility of cyclooctasulfur in pure water and sea water at different temperatures. Geochim. Cosmochim. Acta. 73, 6022–6028 (2009). [Google Scholar]
- 22.Xu Y & Schoonen MAA The stability of thiosulfate in the presence of pyrite in low-temperature aqueous solutions. Geochim Cosmochim Acta. 59, 4605–4622 (1995). [Google Scholar]
- 23.Nordstrom DK, Ball JW & McClesky RB Ground water to surface water: chemistry of thermal outflows in Yellowstone National Park In: Inskeep WP, McDermott TR (eds). Geothermal biology and geochemistry in Yellowstone National Park. Montana State University: Bozeman, 2005, pp 143–162. [Google Scholar]
- 24.Rickard D & Luther GW Chemistry of iron sulfides. Chem. Rev 107, 514–562 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Laska S, Lottspeich F & Kletzin A Membrane-bound hydrogenase and sulfur reductase of the hyperthermophilic and acidophilic archaeon Acidianus ambivalens. Microbiology. 149, 2357–2371 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Veith A et al. Substrate pathways and mechanisms of inhibition in the sulfur oxygenase reductase of Acidianus ambivalens. Front. Microbiol 2, 37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorby YA et al. Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc. Nat. Acad. Sci. U.S.A 103, 11358–11363 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reguera G et al. Extracellular electron transfer via microbial nanowires. Nature. 435, 1098–1101 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Leang C, Coppi MV & Lovley DR OmcB, a c-type polyheme cytochrome, involved in Fe(III) reduction in Geobacter sulfurreducens. J. Bacteriol 185, 2096–2103 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myers CR & Myers JM Cloning and sequence of cymA a gene encoding a tetraheme cytochrome c required for reduction of iron(III), fumarate, and nitrate by Shewanella putrefaciens MR-1. J. Bacteriol 179, 1143–1152 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kletzin A et al. Cytochromes c in Archaea: distribution, maturation, cell architecture, and the special case of Ignicoccus hospitalis. Frontiers in microbiology. 6, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mardanov AV et al. Complete genome sequence of strain 1860, a crenarchaeon of the genus Pyrobaculum able to grow with various electron acceptors. J. Bacteriol 194, 727–728 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pirbadian S et al. Shewanella oneidensis MR-1 nanowires are outer membrane and periplasmic extensions of the extracellular electron transport components. Proc. Nat. Acad. Sci. U.S.A 111, 12883–12888 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feinberg LF, Srikanth R, Vachet RW & Holden JF Constraints on anaerobic respiration in the hyperthermophilic archaea Pyrobaculum islandicum and Pyrobaculum aerophilum. Appl. Environ. Microbiol 74, 396–402 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lovley DR, Phillips EJP, Lonergan DJ & Widman PK Fe(III) and S0 Reduction by Pelobacter carbinolicus. Appl. Environ. Microbiol 61, 2132–2138 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thony-Meyer L Haem-polypeptide interactions during cytochrome c maturation. Biochim. Biophys. Acta-Bioenerg 1459, 316–324 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Stevens JM, Daltrop O, Allen JWA & Ferguson SJ C-type cytochrome formation: Chemical and biological enigmas. Accounts Chem. Res 37, 999–1007 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Malvankar NS, Yalcin SE, Tuominen MT & Lovley DR Visualization of charge propagation along individual pili proteins using ambient electrostatic force microscopy. Nat. Nanotechnol 9, 1012–1017 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Malvankar NS et al. Structural basis for metallic-like conductivity in microbial nanowires. mBio. 6, e00084–00015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lassak K, Ghosh A & Albers SV Diversity, assembly and regulation of archaeal type IV pili-like and non-type-IV pili-like surface structures. Res. Microbiol 163, 630–644 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Makarova KS, Koonin EV & Albers SV Diversity and evolution of type IV pili systems in Archaea. Front. Microbiol 7, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szabo Z et al. Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J. Bacteriol 189, 772–778 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Esquivel RN, Xu R & Pohlschroder M Novel archaeal adhesion pilins with a conserved N terminus. J. Bacteriol 195, 3808–3818 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan Y et al. Expressing the Geobacter metallireducens PilA in Geobacter sulfurreducens yields pili with exceptional conductivity. mBio. 8, e02203–02216 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langner HW, Jackson CR, Mcdermott TR & Inskeep WP Rapid oxidation of arsenite in a hot spring ecosystem, Yellowstone National Park. Environ. Sci. Technol 35, 3302–3309 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Colman DR et al. Ecological differentiation in planktonic and sediment-associated chemotrophic microbial populations in Yellowstone hot springs. FEMS Microbiol. Ecol 92, fiw137 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Boyd ES et al. Isolation, characterization, and ecology of sulfur-respiring Crenarchaea inhabiting acid-sulfate-chloride-containing geothermal springs in yellowstone national park. Appl. Environ. Microbiol 73, 6669–6677 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boyd ES, Cummings DE & Geesey GG Mineralogy influences structure and diversity of bacterial communities associated with geological substrata in a pristine aquifer. Microb. Ecol 54, 170–182 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Hamilton TL, Peters JW, Skidmore ML & Boyd ES Molecular evidence for an active endogenous microbiome beneath glacial ice. ISME J. 7, 1402–1412 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urschel MR, Hamilton TL, Roden EE & Boyd ES Substrate preference, uptake kinetics and bioenergetics in a facultatively autotrophic, thermoacidophilic crenarchaeote. FEMS Microbiol. Ecol 92, fiw069 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Fogo JK & Popowsky M Spectrophotometric determination of hydrogen sulfide - methylene blue method. Anal. Chem 21, 732–734 (1949). [Google Scholar]
- 52.Viollier E, Inglett PW, Hunter K, Roychoudhury AN & Van Cappellen P The ferrozine method revisited: Fe(II)/Fe(III) determination in natural waters. Appl. Geochem 15, 785–790 (2000). [Google Scholar]
- 53.Cypionka H & Pfennig N Growth yields of Desulfotomaculum orientis with hydrogen in chemostat culture. Arch. Microbiol 143, 396–399 (1986). [Google Scholar]
- 54.Amend JP & Shock EL Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and bacteria. FEMS Microbiol. Rev 25, 175–243 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Helgeson HC Prediction of the thermodynamic properties of electrolytes at high pressures and temperatures. Phys. Chem. Earth 13, 133–177 (1981). [Google Scholar]
- 56.McGlynn SE, Chadwick GL, Kempes CP & Orphan VJ Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature. 526, 531–535 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request. The draft genome sequence obtained for strain DS80 has been deposited in the Joint Genome Institute IMG database, IMG submission identifier 92883, genome ID 2690315630.