Abstract
Breast cancer (BRC) is the most invasive cancer in women. Although the survival rate of BRC is gradually increasing due to improved screening systems, development of novel therapeutic targets for inhibition of BRC proliferation, metastasis and recurrence have been constantly needed. Thus, in this study, we identified overexpression of SETDB1 (SET Domain Bifurcated 1), a histone methyltransferase, in RNA-seq data of BRC derived from TCGA portal. In Gene Ontology (GO) analysis, cell migration-related GO terms were enriched, and we confirmed down-regulation of cell migration/invasion and alteration of EMT/MET markers after knockdown of SETDB1. Moreover, gene network analysis showed that SMAD7 expression is regulated by SETDB1 levels, indicating that up-regulation of SMAD7 by SETDB1 knockdown inhibited BRC metastasis. Therefore, development of SETDB1 inhibitors and functional studies may help develop more effective clinical guidelines for BRC treatment.
Keywords: Breast cancer, Metastasis, SETDB1
INTRODUCTION
Triple-negative breast cancer (TNBC) has a high rate of recurrence and metastasis and a poor survival rate. As TNBC is an ER/PR-negative type, TNBC does not use estrogen and progesterone for cancer cell growth and metastasis, Thus, TNBC does not respond to hormone therapy, such as tamoxifen. In addition, as TNBC has high levels of HER2 receptors, antibody therapy, such as Herceptin, has a slight effect, suggesting that development of therapeutic targets for TNBC treatment is urgently needed (1–3). Recently, among various therapeutic targets for TNBC treatment, epigenetic modifiers, such as histone methyltransferase/demethylase, have been highlighted in studies on inhibition of TNBC growth and metastasis (4, 5).
SET domain bifurcated 1 (SETDB1) is a histone methyltransferase, which mainly catalyzes histone H3 lysine 9 tri-methylation. In suppression of gene expression, histone H3K9 tri-methylation plays an important role in the structural change from euchromatin (open chromatin) to heterochromatin (silent chromatin) (6). In epigenetic regulation for cancer development, repressive histone markers, such as H3K9 and H3K27, are commonly found on tumor suppressor genes for suppression of gene expression at the transcriptional level (7–9). Thus, epigenetic modifiers such as SETDB1 are regarded as a promising therapeutic targets for cancer treatment. Recently, several studies have indicated that SETDB1 is overexpressed in several types of cancer, such as hepatocellular cancer (HCC), prostate cancer, melanoma, glioma, colon cancer and breast cancer (BRC), and overexpressed SETDB1 was associated with cancer proliferation and metastasis progression (10–15). However, the molecular mechanisms of SETDB1-related proliferation and metastasis in BRC are not fully understood.
Therefore, in this study, we identified SETDB1 overexpression in BRC samples compared to normal samples derived from TCGA portal and Oncomine data. In Gene Ontology (GO) analysis using RNA-seq data of SETDB1 knockdown, cell migration-related GO terms were found, and cell migration and invasion were reduced by SETDB1 knockdown. Finally, SMAD7 was selected as a target gene for SETDB1-related BRC metastasis via IPA analysis. Thus, we suggest that SETDB1 is a potential metastasis target in BRC, especially TNBC metastasis.
RESULTS
SETDB1 was overexpressed in BRC
SETDB1 (KMT1E) is a histone methyltransferase responsible for histone H3 lysine 9 tri-methylation and is involved in gene silencing via formation of heterochromatin. SETDB1 is overexpressed in colorectal cancer and lung cancer; moreover, SETDB1 is associated with meiosis/mitosis in development (11, 16, 17). Additionally, in tissue-wide expression profile analysis in GENT2 (Gene Expression patterns across Normal and Tumor tissues, http://gene2.appex.kr/gent2/), we clearly observed SETDB1 overexpression in several types of cancer compared to normal tissues (Supplementary Fig. 1A). To verify expression levels of SETDB1 in BRC, we used RNA-seq data (normal; 113, BRC; 1,098) derived from TCGA portal and found overexpression of SETDB1 in BRC compared to normal breast (Supplementary Fig. 1B, Supplementary Table 1). Additionally, in Oncomine data, the expression level of SETDB1 was increased in breast carcinoma and invasive breast carcinoma as shown in Supplementary Fig. 1A and B (Supplementary Fig. 1C). Therefore, we suggest that SETDB1 overexpression may be involved in BRC proliferation or metastasis.
Knockdown of SETDB1 suppressed cell migration and invasion in MDA-MB-231 cells
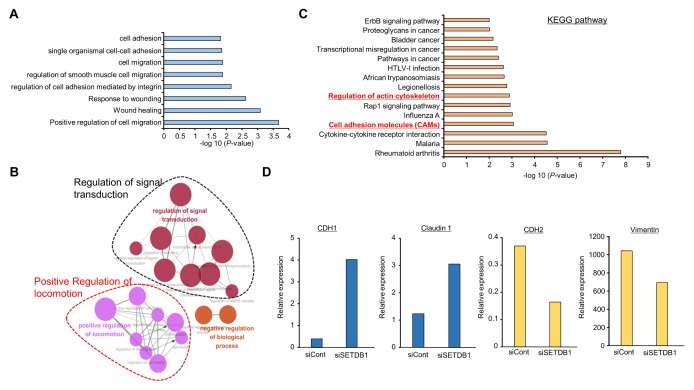
To identify SETDB1-related cellular functions in BRC, we treated MB-231 cells with SETDB1-specific siRNA and control siRNA for knockdown of SETDB1 expression and performed RNA-seq analysis. Fig. 1A shows that SETDB1-related genes were enriched in cell adhesion, cell migration, wound healing and positive regulation of cell migration via GO analysis using DAVID (version 6.8). Moreover, networking analysis using the ClueGO plug-in Cytoscape showed that SETDB1 is involved in regulation of signal transduction and positive regulation of locomotion (Fig. 1B), and regulation of actin cytoskeleton and cell adhesion molecules pathways was observed after SETDB1 knockdown through KEGG pathway analysis, implying that SETDB1 was associated with BRC metastasis (Fig. 1C). RNA-seq analysis showed that after knockdown of SETDB1, epithelial-mesenchymal transition (EMT) markers (CDH1 and Claudin1) were up-regulated and mesenchymal-epithelial transition (MET) markers (CDH2 and Vimentin) were down-regulated. Thus SETDB1 was associated with cell migration and was a metastasis regulator in BRC (Fig. 1D).
Fig. 1.
RNA-seq analysis of SETDB1 knockdown MB-231 cells. (A) DAVID-based gene ontology analysis of RNA-seq results from the siSETDB1 and siCont groups. Enriched terms are shown. Raw transcriptome sequence data (RNA-seq) were deposited in the Short Read Archive (SRA) of NCBI under the accession number PRJNA506342. (B) GO pathway term enrichment networks. GO pathway term networks in the SETDB1 knockdown and control groups were functionally grouped by ClueGO. (C) KEGG pathway analysis of RNA-seq results from the siSETDB1 and siCont groups. (D) RNA-seq results from the siSETDB1 and siCont groups. Up-regulation of epithelial cell markers (CDH1 and Claudin 1) and down-regulation of mesenchymal cell markers (CDH2 and Vimentin).
SETDB1 induced the EMT process in BRC
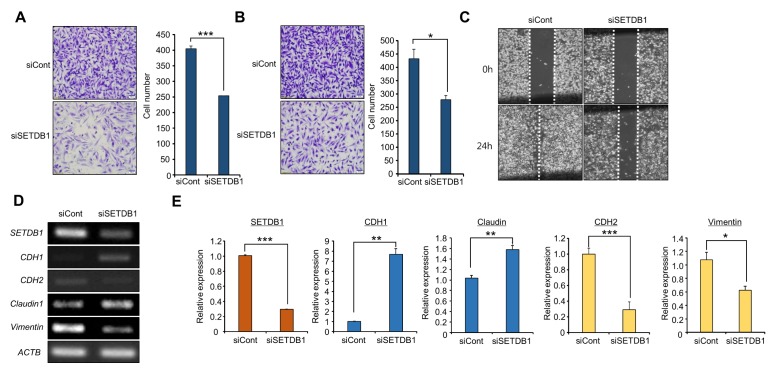
SETDB1 is mainly associated with cell proliferation, migration and invasion in prostate cancer and hepatocellular carcinoma, according to suppression of cell growth and metastasis by SETDB1 knockdown, indicating that SETDB1 is a potential therapeutic target in cancer (14, 18). In this study, we verified the role of this protein in cell growth by SETDB1 knockdown; we assessed the metastatic functions of SETDB1. After treatment with SETDB1 siRNA, cell migration and invasion assays indicated that the numbers of migrating and invading cells were dramatically reduced in the SETDB1 knockdown group compared to the control group, as shown in Fig. 2A and B. Additionally, in wound healing assays, the wound closure process was slowed in the SETDB1 knockdown group (Fig. 2C). Thus, SETDB1 controlled cell migration and invasion, but not proliferation (Supplementary Fig. 2), in BRC cell lines.
Fig. 2.
SETDB1 knockdown changed EMT/MET markers. (A, B) Migration and invasion assays after SETDB1 knockdown. Cell migration and invasion assays were performed after 36 h. Migrated/invaded cells were stained with crystal violet (left); P values were calculated using Student’s t-tests (***P < 0.001, *P < 0.05) (right). (C) Wound healing assay. After 24 h of SETDB1 knockdown, scratch assays were performed. After another 24 h, wound closure was measured. (D, E) RT-PCR (D) and qRT-PCR (E) analysis of EMT markers (CDH1, CDH2, Claudin 1, and Vimentin). Actin (ACTB) was used as an internal control.
In metastasis, EMT and MET markers indicate the status of BRC cells; therefore, we examined the alterations of EMT/MET markers by SETDB1 knockdown using RT-PCR and qPCR analyses. As shown to Fig. 1D, epithelial markers (CDH1 and Claudin1) were up-regulated, while mesenchymal markers (CDH2, Vimentin) were down-regulated. Taken together, these findings suggested that SETDB1 is an important regulator in cell migration and invasion of BRC (Fig. 2D and E).
SETDB1 was involved in the regulation of SMAD7 for BRC metastasis
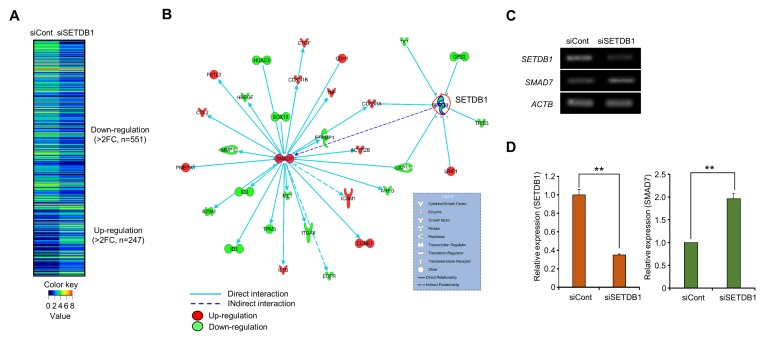
In RNA-seq results, we identified 247 up-regulated genes and 551 down-regulated genes after SETDB1 knockdown (Fig. 3A). Using the SETDB1-related gene set for identification of SETDB1 target genes responding to BRC metastasis, we performed gene-network analysis using IPA software and found a connection between SMAD7 up-regulation and SETDB1 down-regulation (Fig. 3B). The qRT-PCR and RT-PCR results showed that SMAD7 was induced by SETDB1 knockdown (Fig. 3C and D).
Fig. 3.
Up-regulation of SMAD7 by SETDB1 knockdown. (A) Heat map of gene expression related to SETDB1 knockdown, sorted by fold change in the siCont/siSETDB1 FPKM value. In the heat map, red indicates up-regulation, while green indicates down-regulation. (B) Gene-to-gene networks of SETDB1 and SMAD7. Up- and down-regulated genes are indicated in red and green, respectively. Each line and arrow represents functional and physical interactions between the genes and the direction of regulation reported in the literature. (C, D) Up-regulation of SMAD7 after the knockdown of SETDB1. RT-PCR (C) and qRT-PCR analysis (D), P values were calculated using Student’s t-tests (**P < 0.01).
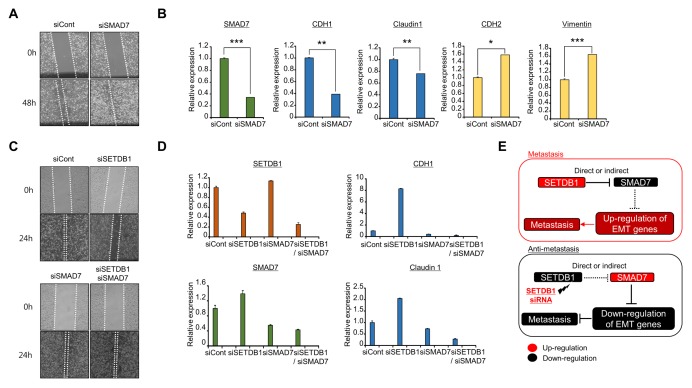
SMAD7 is an antagonist of the TGF-beta signaling pathway, which is involved in immune response, inflammation and fibrosis (19). Moreover, the regulation of SMAD7 expression is closely associated with EMT and cancer cell migration and invasion in BRC, glioma and colorectal cancer (20–23). Therefore, to confirm the relationship between SMAD7 and BRC metastasis, we performed wound healing analysis following SMAD7 knockdown and observed faster wound closure in the SMAD7 knockdown group than the control group (Fig. 4A). Additionally, down-regulation of EMT markers and up-regulation of MET markers were detected by qRT-PCR analysis after SMAD7 knockdown (Fig. 4B). Next, to verify the link between SMAD7 and SETDB1 in more detail, we performed rescue analysis by co-transfection with SETDB1 and SMAD7 siRNAs. In wound healing analysis, the rate of wound closure in the SETDB1 knockdown group was slower than that in the control group; however, co-transfection of SETDB1 and SMAD7 rescued the wound closure rate compared to SETDB1 knockdown only (Fig. 4C). Equally, EMT markers up-regulated by SETDB1 knockdown were down-regulated by double knockdown of SETDB1 and SMAD7, as shown by qRT-PCR analysis (Fig. 4D). Thus, knockdown of SETDB1 reduced migration and invasion of BRC cells via up-regulation of SMAD7 expression. Therefore, we hypothesized that regulation or inactivation of SETDB1 prevented the spread of BRC metastasis.
Fig. 4.
Down-regulation of SMAD7 recovered SETDB1-induced migration and invasion. (A) Wound healing assay. After 24 h of SMAD7 knockdown, scratch assays were performed. After another 48 h, wound closure was measured. (B) qRT-PCR analysis of EMT markers (CDH1, CDH2, Claudin 1, and Vimentin). P values were calculated using Student’s t-tests (***P < 0.001, **P < 0.01, *P < 0.05). (C) Wound healing assay. After treatment with siCont, siSETDB1, siSMAD7 and siSETDB1/siSMAD7 for 24 h, wound closure was measured after another 24 h. (D) qRT-PCR analysis of EMT markers (CDH1 and Claudin 1) after treatment with siCont, siSETDB1, siSMAD7 and siSETDB1/siSMAD7 for 48 h. (E) Schematic summary of the effects of SETDB1 on BRC metastasis.
DISCUSSION
For effective BRC treatment, BRC has been divided four subtypes, such as ER-positive (luminal A and B) and ER-negative (HER2 positive and basal-like) cancers. In targeted molecular therapy for BRC, tamoxifen, as a specific inhibitor for the estrogen receptor, is used as an ER-positive BRC treatment (24). Moreover, in the ER-negative subtype, Herceptin (Trastuzumab), a specific antibody for the HER2 receptor, is used for HER2-overexpressing BRC (25). Among BRC subtypes, TNBC shows HER2 overexpression and lacks the progesterone/estrogen receptor. Compared to other subtypes of BRC, TNBC shows more invasive/metastatic characteristics, higher recurrence and a poor survival rate (1). Thus, the treatment of TNBC and inhibition of TNBC metastasis have been recognized as an important issue. Therefore, in this study, we used the MDA-MB-231 cell line as a TNBC cell line for a functional study of SETDB1 in BRC.
Interestingly, knockdown of SETDB1 in MB231 cells inhibited migration and invasion via alteration of EMT/MET markers (Fig. 2). However, Regina et al. demonstrated that knockdown of SETDB1 decreased the number of colonies formed in the MCF7 cell line (26). The MCF7 cell line was classified as ER positive (positive ER, positive PR, negative HER2) and p53 wild-type. In contrast, the MB-231 cell line is a TNBC cell line and p53 mutant. Additionally, MCF7 has epithelial characteristics, and MB-231 shows a mesenchymal phenotype (27, 28). Therefore, we hypothesized that SETDB1 was clearly associated with cell proliferation in ER-positive BRC cell lines, such as MCF7, and in more invasive BRC cell lines, such as MB-231, SETDB1 could function as a metastasis regulator, implying that development of a SETDB1-specific inhibitor could have an important role in effective BRC therapy.
In RNA-seq results, the expression level of SMAD7 was up-regulated by SETDB1 knockdown, and we confirmed the high expression of SMAD7 using RT-PCR and qRT-PCR (Fig. 3). SETDB1 mainly catalyzes histone H3 lysine 9 tri-methylation for suppression of gene expression at the transcriptional level via heterochromatin formation. Thus, we focused to up-regulated genes after SETDB1 knockdown and finally selected SMAD7 genes via IPA analysis (Fig. 3A). From an epigenetic point of view, we hypothesized that the status of histone H3K9 trimethylation in the SMAD7 promoter region is significantly decreased by SETDB1 knockdown; accordingly, closed chromatin regions dramatically change to open chromatin regions. Finally, up-regulation of SMAD7 expression may occur. However, direct regulation between SMAD7 and SETDB1 should be confirmed in further studies.
TGF-beta signaling induces cancer cell metastasis via induction of cancer stemness (29). Many specific inhibitors have been developed to block TGF-beta signaling. In addition, SMAD7, an antagonist of the TGF-beta signaling pathway, has been recognized as a metastasis therapeutic target in several types of cancer. The inhibition of TGF-beta signaling by treatment with halofuginone or melatonin and deregulation of miR-21 and OTUD1 expression leads to suppression of metastasis in lung cancer, glioma, BRC and HCC (19–22, 29). Therefore, inhibition of BRC metastasis by co-treatment with specific inhibitors of SETDB1 and TFG-beta may lead to more efficient metastasis therapy in BRC.
In summary, overexpression of SETDB1 in TNBC may promote metastasis by down-regulation of SMAD7. Thus, development of SETDB1-specific inhibitors may contribute to the management of TNBC metastasis and recurrent cases (Fig. 4E).
MATERIALS AND METHODS
Cell culture
The human breast cancer cell line MDA-MB-231 was purchased from the Korean Cell Line Bank (Seoul, South Korea) and cultured at 37°C in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin in a humidified atmosphere with 5% CO2 (30).
siRNA transfection
siRNA duplexes against SETDB1 (siSETDB1; 5′-GACCU AUCAGGAAUGAGCA-3′, 5′-UGCUCAUUCCUCAUAGGUC-3′) were purchased from Sigma-Aldrich (Milwaukee, Wis., USA). siRNA duplexes against SMAD7 (siSMAD7; 5′-CGACACUG UUCUUAGCUCA-3′, 5′-UGAGCUAAGAACAG UGUCG-3′ ) were purchased from Bioneer (Daejeon, South Korea). Negative control siRNA (siCont; 5′-AUGAACGUGA AUUGCUCAATT-3′, 5′-UUGAGCAAUUCACGUUCACTT-3′) was used for control treatments. The siRNAs (100 nM) were transfected into cancer cell lines using RNAiMax (Invitrogen, Carlsbad, CA) for 48 h (31).
Semi-quantitative reverse transcription PCR and quantitative real-time PCR
Total RNA was isolated from the indicated cell lines using a Qiagen RNeasy Mini Kit according to the manufacturer’s instructions. RNA aliquots of 1 μg were then reverse transcribed using the iScriptTM cDNA synthesis kit (Bio-Rad, Hercules, CA), according to the standard protocols. For semi-quantitative RT-PCR, cDNA was used as a template for PCR using AccuPower® HotStart PCR PreMix (Bioneer, Daejeon, South Korea). For quantitative RT-PCR, PCR reactions were performed using the SimpliAmp Thermal Cycler (Applied Biosystems) following the manufacturer’s instructions. Quantitative real-time PCR was performed on cDNA samples using Brilliant III Ultra-Fast SYBR® Green QPCR Master Mix (Agilent Technologies), and the signal was detected by AriaMx Real-time PCR System (Agilent Technologies). The fluorescence threshold value was calculated using Agilent Aria 1.6 software (32–34). PCR Primers (SETDB1 F-5′-AGGAACTTCGGCATTTCATCG-3′, R-5′-TGTCCCGGTATTGTAGTCCCA-3′; SMAD7 F-5′-GGACA GCTCAATTCGGACAAC-3′, R-5′-GTACACCCACACACCATCC AC-3′; CDH1 F-5′-CGAGAGCTACACGTTCACGG-3′, R-5′-GGGTGTCGAGGGAAAAATAGG-3′; CDH2 F-5′-AGCCAACC TTAACTGAGGA GT-3′, R-5′-GGCAAGTTGATTGGAGGGAT G-3′; Claudin1 F-5′-TGGTCAGGCTCTCTTC ACTG-3′, R-5′-TTGGATAGGGCCTTGGTGTT-3′; Vimentin F-5′-CCCTCACCT GTGAAG TGGAT-3′, R-5′-TGACGAGCCATTTCCTCCTT-3′; A CTB F-5′-ACTCTTCCAGCC TTCCTTCC-3′, R-5′-CAATGCCAG GGTACATGGTG-3′;).
Wound healing assay
Cells were seeded in 6-well plates and wounded by scratching with sterile plastic 10 μl micropipette tips after 24 h of siRNA infection. Then cells were cleaned with PBS and added fresh serum medium. The cells were photographed 0 h, 24 h and 48 h after the wounding by the CELENATM S Digital Imaging System (Logos Biosystems). Cell migration distance was observed in the photographs.
Migration and invasion assays
Transwell inserts were coated with a 2% gelatin solution and incubated at room temperature for 4 h for the migration assay. The gelatin-coated transwell inserts (353097, BD Falcon, Bedford, MA) and invasion chambers (354480, Corning, Corning, NY) were rehydrated in serum-free medium. Complete medium with 20% FBS (700 μl) served as a chemoattractant in the bottom chamber. Approximately 1 × 105 cells/well were incubated in the plates for 36 h at 37°C with 5% CO2. At the end of the incubation period, the migrated and invaded cells were fixed with methanol for 5 min and stained with 0.1% crystal violet.
Supplementary Information
ACKNOWLEDGEMENTS
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (2016M3A9C4953144, 2017R1A2B4003757, 2018M3A9H3023077) and the KRIBB Research Initiative Program. The funders had no role in the study design, data collection or analysis, decision to publish, or preparation of the manuscript.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Dent R, Trudeau M, Pritchard KI, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 2.Lee A, Djamgoz MBA. Triple negative breast cancer: Emerging therapeutic modalities and novel combination therapies. Cancer Treat Rev. 2017;62:110–122. doi: 10.1016/j.ctrv.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Park J, Jang JH, Park GS, Chung Y, You HJ, Kim JH. BLT2, a leukotriene B4 receptor 2, as a novel prognostic biomarker of triple-negative breast cancer. BMB Rep. 2018;51:373–377. doi: 10.5483/BMBRep.2018.51.8.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim K, Son MY, Jung CR, Kim DS, Cho HS. EHMT2 is a metastasis regulator in breast cancer. Biochem Biophys Res Commun. 2018;496:758–762. doi: 10.1016/j.bbrc.2018.01.074. [DOI] [PubMed] [Google Scholar]
- 5.Song H, Li D, Wu T, et al. MicroRNA-301b promotes cell proliferation and apoptosis resistance in triple-negative breast cancer by targeting CYLD. BMB Rep. 2018;51:602–607. doi: 10.5483/BMBRep.2018.51.11.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kazanets A, Shorstova T, Hilmi K, Marques M, Witcher M. Epigenetic silencing of tumor suppressor genes: Paradigms, puzzles, and potential. Biochim Biophys Acta. 20161865:275–288. doi: 10.1016/j.bbcan.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Kondo Y, Shen L, Issa JP. Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer. Mol Cell Biol. 2003;23:206–215. doi: 10.1128/MCB.23.1.206-215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han TS, Ban HS, Hur K, Cho HS. The epigenetic regulation of HCC metastasis. Int J Mol Sci. 2018;19:3978. doi: 10.3390/ijms19123978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ceol CJ, Houvras Y, Jane-Valbuena J, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen K, Zhang F, Ding J, et al. Histone Methyltransferase SETDB1 promotes the progression of colorectal cancer by inhibiting the expression of TP53. J Cancer. 2017;8:3318–3330. doi: 10.7150/jca.20482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho YJ, Lin YM, Huang YC, et al. Significance of histone methyltransferase SETDB1 expression in colon adenocarcinoma. APMIS. 2017;125:985–995. doi: 10.1111/apm.12745. [DOI] [PubMed] [Google Scholar]
- 13.Spyropoulou A, Gargalionis A, Dalagiorgou G, et al. Role of histone lysine methyltransferases SUV39H1 and SETDB1 in gliomagenesis: modulation of cell proliferation, migration, and colony formation. Neuromolecular Med. 2014;16:70–82. doi: 10.1007/s12017-013-8254-x. [DOI] [PubMed] [Google Scholar]
- 14.Wong CM, Wei L, Law CT, et al. Up-regulation of histone methyltransferase SETDB1 by multiple mechanisms in hepatocellular carcinoma promotes cancer metastasis. Hepatology. 2016;63:474–487. doi: 10.1002/hep.28304. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, Cai K, Wang J, et al. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells. 2014;32:2858–2868. doi: 10.1002/stem.1795. [DOI] [PubMed] [Google Scholar]
- 16.Eymery A, Liu Z, Ozonov EA, Stadler MB, Peters AH. The methyltransferase Setdb1 is essential for meiosis and mitosis in mouse oocytes and early embryos. Development. 2016;143:2767–2779. doi: 10.1242/dev.132746. [DOI] [PubMed] [Google Scholar]
- 17.Na HH, Kim KC. SETDB1-mediated FosB regulation via ERK2 is associated with an increase in cell invasiveness during anticancer drug treatment of A549 human lung cancer cells. Biochem Biophys Res Commun. 2018;495:512–518. doi: 10.1016/j.bbrc.2017.10.176. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Huang J, Li Q, et al. Histone methyltransferase SETDB1 promotes cells proliferation and migration by interacting withTiam1 in hepatocellular carcinoma. BMC Cancer. 2018;18:539. doi: 10.1186/s12885-018-4464-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyazawa K, Miyazono K. Regulation of TGF-beta family signaling by inhibitory smads. Cold Spring Harb Perspect Biol. 2017;9 doi: 10.1101/cshperspect.a022095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Wang Z, Ma H, et al. Melatonin attenuates hypoxia-induced epithelial-mesenchymal transition and cell aggressive via Smad7/CCL20 in glioma. Oncotarget. 2017;8:93580–93592. doi: 10.18632/oncotarget.20525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juarez P, Fournier PGJ, Mohammad KS, et al. Halofuginone inhibits TGF-beta/BMP signaling and in combination with zoledronic acid enhances inhibition of breast cancer bone metastasis. Oncotarget. 2017;8:86447–86462. doi: 10.18632/oncotarget.21200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Nie L, Wu L, Liu Q, Guo X. NR2F2 inhibits Smad7 expression and promotes TGF-beta-dependent epithelial-mesenchymal transition of CRC via transactivation of miR-21. Biochem Biophys Res Commun. 2017;485:181–188. doi: 10.1016/j.bbrc.2017.02.049. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z, Fan Y, Xie F, et al. Breast cancer metastasis suppressor OTUD1 deubiquitinates SMAD7. Nat Commun. 2017;8:2116. doi: 10.1038/s41467-017-02029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sauter ER. Breast cancer prevention: current approaches and future directions. Eur J Breast Health. 2018;14:64–71. doi: 10.5152/ejbh.2018.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross JS, Fletcher JA. The HER-2/neu oncogene in breast cancer: prognostic factor, predictive factor, and target for therapy. Stem Cells. 1998;16:413–428. doi: 10.1002/stem.160413. [DOI] [PubMed] [Google Scholar]
- 26.Regina C, Compagnone M, Peschiaroli A, et al. Setdb1, a novel interactor of DeltaNp63, is involved in breast tumorigenesis. Oncotarget. 2016;7:28836–28848. doi: 10.18632/oncotarget.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Comsa S, Cimpean AM, Raica M. The Story of MCF-7 Breast Cancer Cell Line: 40 years of Experience in Research. Anticancer Res. 2015;35:3147–3154. [PubMed] [Google Scholar]
- 28.Gest C, Joimel U, Huang L, et al. Rac3 induces a molecular pathway triggering breast cancer cell aggressiveness: differences in MDA-MB-231 and MCF-7 breast cancer cell lines. BMC Cancer. 2013;13:63. doi: 10.1186/1471-2407-13-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellomo C, Caja L, Moustakas A. Transforming growth factor beta as regulator of cancer stemness and metastasis. Br J Cancer. 2016;115:761–769. doi: 10.1038/bjc.2016.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim SK, Kim K, Ryu JW, et al. The novel prognostic marker, EHMT2, is involved in cell proliferation via HSPD1 regulation in breast cancer. Int J Oncol. 2019;54:65–76. doi: 10.3892/ijo.2018.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryu JW, Kim SK, Son MY, et al. Novel prognostic marker PRMT1 regulates cell growth via downregulation of CDKN1A in HCC. Oncotarget. 2017;8:115444–115455. doi: 10.18632/oncotarget.23296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung KB, Lee H, Son YS, et al. In vitro and in vivo imaging and tracking of intestinal organoids from human induced pluripotent stem cells. FASEB J. 2018;32:111–122. doi: 10.1096/fj.201700504R. [DOI] [PubMed] [Google Scholar]
- 33.Kim DS, Ryu JW, Son MY, et al. A liver-specific gene expression panel predicts the differentiation status of in vitro hepatocyte models. Hepatology. 2017;66:1662–1674. doi: 10.1002/hep.29324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Son MY, Jung CR, Kim DS, Cho HS. Comparative in silico profiling of epigenetic modifiers in human tissues. Mol Biol Rep. 2018;45:309–314. doi: 10.1007/s11033-018-4163-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.