Abstract
Cells must fine-tune their gene expression programs for optimal cellular activities in their natural growth conditions. Transcriptional memory, a unique transcriptional response, plays a pivotal role in faster reactivation of genes upon environmental changes, and is facilitated if genes were previously in an active state. Hyper-activation of gene expression by transcriptional memory is critical for cellular differentiation, development, and adaptation. TREM (Transcriptional REpression Memory), a distinct type of transcriptional memory, promoting hyper-repression of unnecessary genes, upon environmental changes has been recently reported. These two transcriptional responses may optimize specific gene expression patterns, in rapidly changing environments. Emerging evidence suggests that they are also critical for immune responses. In addition to memory B and T cells, innate immune cells are transcriptionally hyperactivated by restimulation, with the same or different pathogens known as trained immunity. In this review, we briefly summarize recent progress in chromatin-based regulation of transcriptional memory, and its potential role in immune responses.
Keywords: H3K4me3, Rpd3L HDAC, Trained Immunity, Transcriptional memory, Transcriptional Repression Memory (TREM)
INTRODUCTION
All organisms must accordingly respond to stressful stimuli resulting from environmental changes, including nutrient starvation, accumulation of byproduct, and other cellular damage (1). Establishment of optimal gene expression patterns can support cellular differentiation, development, and adaptation. In addition, cells are often re-exposed to the same or different environmental changes, and they can more rapidly and strongly induce genes that support cellular functions. This transcriptional response is known as ‘transcriptional memory’ that increases the kinetics of reactivation (2). In yeast, transcriptional memory involving inducible GAL genes and INO1 has been extensively studied, and chromatin factors and cytoplasmic proteins involved have been identified (3–5). Transcriptional memory of GAL genes is positively regulated by the SWI/SNF chromatin remodeling complex, and the Htz1 histone variant (3, 6). In addition, the Gal1 and Gal3 metabolic proteins, and the nuclear pore complex, are also required for GAL memory (6–8).
Turning-off unnecessary genes in a given condition, is also crucial for cells to save cellular resources. We have recently reported that ~540 yeast genes are more strongly repressed, if they were in an inactive state during carbon sources shifts (9). This novel transcriptional response has been named ‘transcriptional repression memory’ (TREM) (9). Modulation of gene expression dynamics by transcriptional memory, and TREM are likely critical for optimized cellular functions, in rapidly changing environments.
Although immune memory is known as a specific response of T or B cells, increasing evidence suggests transcriptional/epigenetic memory is a vital mechanism that boosts innate immune response. Trained immunity, a transcriptional memory response in non-lymphoid cells including macrophage and innate lymphoid cells (ILCs) plays a crucial role in innate immune responses (10). Hyper-activation and -repression of interferon-γ (INF-γ) response genes upon restimulation is also observed in human macrophages (11–13). Furthermore, papain-stimulated ILCs can enhance lung inflammation upon restimulation with IL-33 (10).
Eukaryotic gene expression is regulated by post-translational modifications, including acetylation, methylation, phosphorylation, and ubiquitination of histone tails, and by chromatin remodeling factors that directly affect chromatin structure (14, 15). Although these factors do not strongly affect global gene expression in steady-state conditions, they play central roles in regulating the kinetics of transcriptional responses during cellular development, differentiation, or adaptation to environmental changes (16–18). In this review, we summarize recent findings on molecular mechanisms, of two distinct transcriptional memories and their possible roles in immune memory.
TRANSCRIPTIONAL MEMORY OF GAL GENES IN YEAST
GAL genes including GAL1, GAL10, and GAL7 are involved in galactose metabolism and strongly induced in media containing galactose. For example, GAL1 encoding the galactokinase is transcriptionally induced by ~1,000-fold when cells are exposed to galactose (19, 20). Transcription of GAL genes is controlled by multiple regulatory factors. A key regulator of GAL genes is the Gal4 activator that directly binds to the upstream activating sequence (UAS) of these genes. The Gal4 activator becomes activated in the presence of galactose to promote transcription of target genes. In contrast, the activation domain of this protein is masked by the Gal80 repressor in media, containing neutral carbon sources including raffinose, sucrose, or glycerol, and thus the Gal4 activator fails to activate GAL genes under these conditions (19, 21). GAL genes are strongly repressed, when glucose is present in media known as ‘glucose repression’ (22, 23). Sequence-specific transcriptional repressors, Mig1 and Nrg1, and a general corepressor complex, Ssn6-Tup1, directly bind to upstream regions of GAL genes to repress transcription (20, 24).
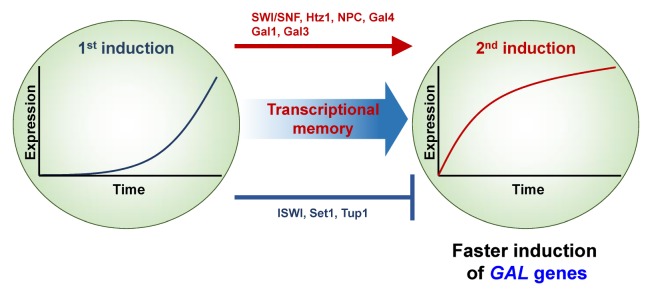
GAL genes have been extensively used to study transcriptional memory in yeast. GAL1 is induced when cells are transferred to media containing galactose, and maximum levels of GAL1 transcripts are observed after approximately one hour incubation in galactose media. However, when cells are re-exposed to galactose after a short period growth in the presence of glucose, reactivation of GAL1 transcription occurs rapidly and peaks within 10 minutes of the second galactose exposure (3) (Fig. 1). Therefore, GAL1 gene remembers its previous active state to be hyper-activated, upon re-stimulation with galactose. This response is known as transcriptional memory, that increases the kinetics of reactivation. Two distinct types of GAL transcriptional memory have been proposed. Whereas short-term memory persists for 1–2 generations in absence of galactose, long-term memory continues for over six cell divisions (3, 6, 7). These findings suggest that transcriptional memory is epigenetically inherited to daughter cells.
Fig. 1.
Transcriptional memory of GAL genes. When yeast cells are grown in media containing galactose, the GAL genes necessary for galactose metabolism are induced (1st induction). Transcriptional memory upon re-exposure to galactose after a short period growth in glucose media significantly increases the rate of activation (2nd induction). Chromatin regulators including SWI/SNF and Htz1, Nuclear pore complex, and Gal4 activator positively regulate transcriptional memory of GAL genes. Furthermore, the Gal1 metabolic enzyme and the Gal3 protein regulating the function of the Gal4 activator are specifically required for long-term memory of GAL genes. Several factors including ISWI chromatin remodeling complex, Set1 HMT, and Tup1 negatively affect GAL memory.
FACTORS THAT MODULATE TRANSCRIPTIONAL MEMORY OF GAL GENES
Epigenetic inheritance of GAL memory is positively and negatively regulated by multiple factors. An ATP-dependent chromatin remodeling complex, SWI/SNF, promotes transcriptional memory of GAL genes (3). Histone H2A variant, Htz1 incorporated into the recently inactivated promoter regions, is also critical for faster reactivation of GAL genes (6) (Fig. 1). In addition, the nuclear pore complex that targets active genes to the nuclear periphery, positively regulates transcriptional memory of GAL genes (5) (Fig. 1). However, how these factors functionally interact each other, to modulate GAL memory remains elusive. Interestingly, a metabolic enzyme, Gal1 galactokinase, is required for rapid reactivation of GAL genes (2, 4, 7). In addition Gal3, a cytoplasmic protein negatively regulating Gal80 repressor, also promotes GAL memory (4) (Fig. 1). These factors differentially affect short-term or long-term memory of GAL genes. Some chromatin regulators are specifically required for short-term memory. For example, SWI/SNF complex is involved in short-term memory, but not in long-term memory of GAL genes. In contrast, Gal1 and Gal3 function to promote long-term memory, but not short-term memory (Fig. 1).
Transcriptional memory of GAL genes is also negatively regulated by several chromatin regulators. ISWI (Imitation SWitch) chromatin remodeling complex, antagonizes the positive effect of SWI/SNF on GAL memory (3) (Fig. 1). In addition, H3K4 methylation by Set1 also has a negative role in GAL memory. A recent study has proposed that H3K4 methylation remaining at recently transcribed GAL gene, delays reactivation by targeting the ISWI remodeling complex (25) (Fig. 1).
TRANSCRIPTIONAL MEMORY AT GENOME-WIDE LEVELS
Several inducible genes stimulated by specific stimulus, have been also used to study transcriptional memory in different organisms. In yeast, INO1 induced by inositol, PHO5 activated by phosphate starvation, and heat-shock response gene HSP26, have been used to understand molecular mechanisms of their transcriptional memory responses (25, 26). These genes are also positively or negatively regulated by the factors affecting GAL memory. In human cells, cytokine-response genes such as genes activated by IFN-γ (Interferon-γ) have been studied (11, 12). However, a specific stimulus often reprograms global gene expression patterns. For example, heat-shock or H2O2 treatment in yeast induce or repress > 500 genes (1, 27). IFN-γ (Interferon-γ) treatment in human cells also can increase expression of ~2,000 genes (11). Therefore, it is important to understand how many target genes show memory of previous stimulation, and if all memory genes share a common mechanism for transcriptional memory.
Although galactose exposure has been used to study transcriptional memory of GAL genes, it also changes expression of ~1,000 genes that are not directly involved in galactose metabolism (28, 29). The key regulator, Gal4 binds to ~10 genes in yeast genome (30). Molecular mechanisms for GAL memory involving the Gal4 activator may not be applicable to other galactose response genes. To better understand how transcriptional memory by a specific stimulus affect cellular functions, a detailed analysis of transcriptional memory response at genome-wide levels will be required.
TRANSCRIPTIONAL REPRESSION MEMORY: TREM
Hyperactivation of genes that support cellular functions via transcriptional memory is important for optimal cellular adaptation to environmental changes. Studies on transcriptional memory have focused on how genes were strongly induced upon restimulation. However, it is also critical for cells to efficiently suppress transcription of unnecessary ones in a given growth condition. We recently revealed that ~1000 genes dynamically and distinctly induced or repressed, during carbon-source shifts (raffinose → galactose → glucose → galactose) (9, 28, 29). Among the genes repressed during galactose incubation, approximately 540 yeast genes exhibited ‘memory’ of their prior inactive states, during carbon-source shifts (Fig. 2A). They were slightly down-regulated during the first galactose pulse. However, transcriptional repression of these genes was more robust and rapid, once cells were shifted to media containing galactose after a short period growth in glucose media. This novel phenomenon has been termed ‘Transcriptional Repression Memory’ (TREM) (9) (Fig. 2A). TREM is biologically important, since a rapid and strong repression of unnecessary genes by TREM prevents loss of cellular energy and resources that can be eventually used for hyperactivation of genes critical for cell survival and function.
Fig. 2.
Models for regulation of TREM by Rpd3L HDAC. (A) Upon environmental changes, unnecessary genes are repressed by a stimulus (1st repression). TREM upon re-exposure to the same stimulus promotes optimal repression of unnecessary genes to save cellular resources (2nd repression). Rpd3L HDAC bound to active promoters facilitates TREM but the factors that antagonize the effect of Rpd3L need to be determined. (B) Set1 complex (Set1C) is targeted to active genes via the interaction with elongating RNA Pol II and/or nascent RNA transcripts and deposits H3K4me3 at active promoters. This methyl mark is recognized by the Pho23 PHD finger and instructs Rpd3L to deacetylate histones. Hypoacetylation by the H3K4me3-Rpd3L pathway results in optimal repression of unnecessary genes by TREM (wild type). In contrast, hyperacetylation by the loss of Rpd3L function may result in sustained expression of unnecessary ones (mutants for Rpd3L).
MOLECULAR MECHANISM OF TREM
TREM is likely facilitated by transcriptional repressors/corepressors that bind to active promoters. Although Rpd3 histone deacetylase functions as a transcriptional corepressor, its binding to promoters significantly correlated with transcription frequency (31–33). However, its function at active genes remained elusive. Interestingly, loss of Rpd3 significantly delayed gene repression specifically during the second galactose incubation, indicating that Rpd3 functions as a key regulator of TREM. Rpd3 is the catalytic subunit of two distinct HDACs, Rpd3 large (Rpd3L) and Rpd3 small (Rpd3S) (34, 35). We have revealed that ~45% of TREM genes are directly regulated by Rpd3L, but not Rpd3S. Two subunits, Pho23 and Cti6 of Rpd3L have a PHD (Plant Homeodomain) finger that preferentially binds to trimethylated K4 of histone H3 (H3K4me3). In particular, the interaction between H3K4me3 and Pho23 PHD finger facilitates histone deacetylation, and TREM by Rpd3L HDAC (9) (Fig. 2B).
H3K4me3 is a well-known active mark, and has been considered as a memory mark of previous transcriptional activity (36). Set1/COMPASS interacts with elongating RNA Polymerase II (RNA Pol II) and/or nascent RNA transcripts to deposit H3K4me3 at promoter regions, followed by H3K4me2 and H3K4me1 in 5′ and 3′ transcribed regions, respectively (37–39). In particular, H3K4me3 level is strongly correlated with transcription frequency. Some factors including SAGA (Spt-Ada-Gcn5-Acetyltransferase), NuA3 (Nucleosomal Acetyl-transferase of histone H3), and NuA4 (Nucleosomal Acetyl-transferase of histone H4) HATs that positively affect transcription directly bind to H3K4me3 (40–44). However, loss of this modification had no strong effect on global transcription in a steady-state growth condition (16). Interestingly, factors that negatively regulate transcription also bind to this mark. For example, ING2 (INhibitor of Growth 2) directly binds to H3K4me3 via its PHD finger and functionally interacts with Sin3-HDAC1 complex for active gene repression (45). Loss of Set1 also negatively affects kinetics of gene induction (17). These findings suggest that H3K4me3 may have dual functions: one for gene activation, and the other, for active gene repression and TREM. Based on findings, we proposed that H3K4me3 instructed Rpd3L to promote TREM by deacetylating histones at active promoters.
EPIGENETIC MEMORY IN IMMUNE CELLS
Immune responses are divided into innate immune responses and adaptive (acquired) immune responses. Initially, innate immunity was considered a primitive form of immune response, and adaptive immunity was considered a more evolved immune response. The major difference between the two was explained by if they could make a memory response, upon previously encountered immunological challenges. However, even if adaptive immune cells such as T cells and B cells play a more specialized and elaborate memory response, innate immune cells also undergo a memory response (46–49). From a molecular point of view, if memory response of immune cell is rapid and efficient expression of related genes upon the repeated immune stimulation, epigenetic regulation is preceded before expression of those genes. In this section, we will look at these epigenetic changes that are made, for rapid response to repeatedly encountered stimuli in innate immune cells, rather than those in adaptive immune cells.
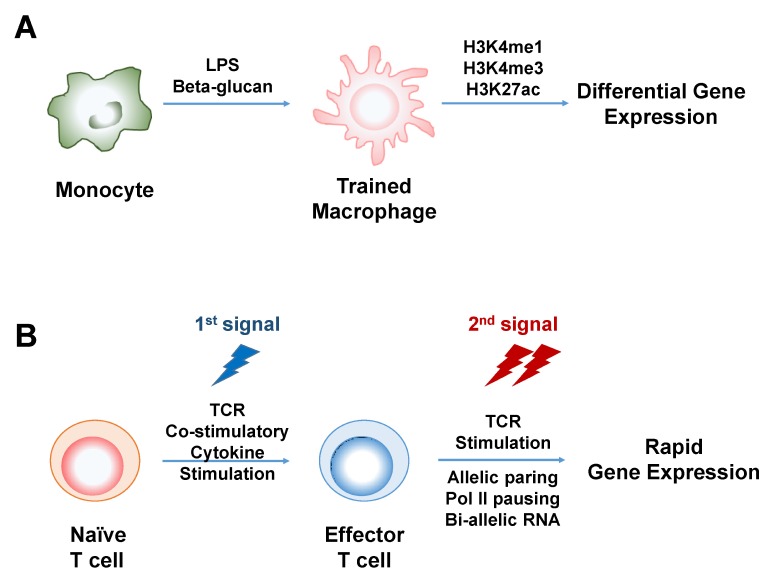
There are a variety of innate immune cells such as monocytes, macrophages, granulocytes, innate lymphoid cells (ILCs) and natural killer (NK) cells. Monocytes are differentiated into macrophages (Fig. 3A). If immune response can be remembered, monocytes that have undergone immune stimulation, will have a trace of memory, even after differentiation into macrophages. To prove this, monocytes were pre-treated with beta-glucan or lipopolysaccharide (50). Macrophages that were experienced with immune stimulus revealed different genome-wide levels of H3K4me3, H3K4me1 and H3K27ac and expression of related genes compared to unexperienced macrophages (50) (Fig. 3A). H3K9me2 and ATF7 were also reported to be important histone modification and transcription factor respectively, in macrophage immunological memory response (51). During viral infection, a few thousands of interferon stimulated genes (ISGs) were expressed in fibroblasts, as well as macrophages. This experience was inherited through multiple cell divisions, and led to the rapid expression of at least parts of ISGs (11). At this time, RNA polymerase II (pol II) was rapidly recruited, and coincided with H3K36me3 and histone H3.3 acquisition (11).
Fig. 3.
Transcriptional memory in immune cells. (A) Monocyte exposed to immune stimulation such as lipopolysaccharide (LPS) or beta-glucan exhibits different levels of H3K4me1, H3K4me3 and H3K27ac globally even after differentiation into macrophage. (B) Effector T cell exhibits transcriptional memory upon 2nd stimulation. At that time, the chromosomal allelic pairing and Pol II pausing are occurred at locus of the related genes, which lead to bi-allelic RNA expression and finally rapid gene expression.
T cells are clearly one of the adaptive immune cells. In particular, memory T cells play a highly sophisticated role in immunological memory response. However, memory T cells are survivors of effector T cells. Most effector T cells decrease after immune response is over. Interestingly, effector T cells also reveal transcriptionally rapid and efficient expression of the related genes, upon repeated immune challenges (Fig. 3B). Tumor necrosis factor-α (TNF-α) is one of pro-inflammatory cytokines expressed at a very early time point in most of the immune cells. When naïve T cells are differentiated into effector T cells, TNF-α is also expressed. If stimulation occurs again in effector T cells, TNF-α is re-expressed in much more rapid and efficient manner (Fig. 3B). At this time, each chromosome locus containing TNF-α makes a chromosomal allelic pairing by nuclear motor protein Myosin VI, that leads to bi-allelic RNA expression (52). Interestingly, RNA pol II is already bound on the promoter of TNF-α, that may make it possible rapid expression (52). Such a phenomenon is not limited to TNF-α, but applied to many of rapidly induced immune related genes and transcription factors such as Tbx21 that is a master transcription factor of T helper 1 effector T cell (52). Despite such recent discoveries, there is a lack of research, on epigenetic memory responses in immune cells. Much more research is needed from a more diverse perspective.
CONCLUSION
Transcriptional memory is a unique transcriptional response, that directs faster induction of genes upon restimulation and is important, for cellular adaptation to environmental changes. However, once the stimulus is removed, it is also important for cells to effectively suppress hyper-activated genes. This may require TREM to promote optimal repression of unnecessary genes. Transcriptional memory and TREM likely function together to optimize gene expression dynamics in rapidly changing environments. In particular, these transcriptional memories may play important roles, in innate immune responses. Innate immune cells can be hyper-activated by re-exposure, to the same or different allergens. This step requires transcriptional memory for hyper-activation of genes, supporting innate immune response. Once the allergen has been removed, a small portion of trained cells will become naïve cells, stimulated via repression of hyper-active genes by TREM.
A stimulus can often reprogram genome-wide gene expression patterns. However, it is unclear if all memory genes activated by a stimulus share a common mechanism. To better understand how transcriptional memory affects cellular functions, a more detailed analysis of transcriptome dynamics or transcriptional memory at transcriptome levels is required.
Although transcriptional memory is important for optimal induction or repression of gene expression, some genes may require a mechanism that prevents hyper-activation or -repression by transcriptional memory. Circadian clock genes or cell cycle regulators must be induced and then repressed, with the exact same kinetics. If not, mis-regulation of these genes by transcriptional memory may cause defects in cell cycle progression. It is important to understand the mechanism that erases, or removes, transcriptional memory of these genes.
ACKNOWLEDGEMENTS
We thank the Kim Laboratory members for helpful discussion on the manuscript. This work was supported by grants from the National Research Foundation [NRF-2017M3A9B5060887 and NRF-2012R1A5A1048236] to T.K.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Guan Q, Haroon S, Bravo DG, Will JL, Gasch AP. Cellular memory of acquired stress resistance in Saccharomyces cerevisiae. Genetics. 2012;192:495–505. doi: 10.1534/genetics.112.143016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brickner JH. Transcriptional memory at the nuclear periphery. Curr Opin Cell Biol. 2009;21:127–133. doi: 10.1016/j.ceb.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kundu S, Horn PJ, Peterson CL. SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev. 2007;21:997–1004. doi: 10.1101/gad.1506607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kundu S, Peterson CL. Dominant role for signal transduction in the transcriptional memory of yeast GAL genes. Mol Cell Biol. 2010;30:2330–2340. doi: 10.1128/MCB.01675-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Light WH, Freaney J, Sood V, et al. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol. 2013;11:e1001524. doi: 10.1371/journal.pbio.1001524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brickner DG, Cajigas I, Fondufe-Mittendorf Y, et al. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zacharioudakis I, Gligoris T, Tzamarias D. A yeast catabolic enzyme controls transcriptional memory. Curr Biol. 2007;17:2041–2046. doi: 10.1016/j.cub.2007.10.044. [DOI] [PubMed] [Google Scholar]
- 8.Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee BB, Choi A, Kim JH, et al. Rpd3L HDAC links H3K4me3 to transcriptional repression memory. Nucleic Acids Res. 2018;46:8261–8274. doi: 10.1093/nar/gky573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez-Gonzalez I, Matha L, Steer CA, Ghaedi M, Poon GF, Takei F. Allergen-Experienced Group 2 Innate Lymphoid Cells Acquire Memory-like Properties and Enhance Allergic Lung Inflammation. Immunity. 2016;45:198–208. doi: 10.1016/j.immuni.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 11.Kamada R, Yang W, Zhang Y, et al. Interferon stimulation creates chromatin marks and establishes transcriptional memory. Proc Natl Acad Sci U S A. 2018;115:E9162–E9171. doi: 10.1073/pnas.1720930115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gialitakis M, Arampatzi P, Makatounakis T, Papamatheakis J. Gamma interferon-dependent transcriptional memory via relocalization of a gene locus to PML nuclear bodies. Mol Cell Biol. 2010;30:2046–2056. doi: 10.1128/MCB.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boehm U, Klamp T, Groot M, Howard JC. Cellular responses to interferon-gamma. Annu Rev Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 14.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 16.Lenstra TL, Benschop JJ, Kim T, et al. The specificity and topology of chromatin interaction pathways in yeast. Mol Cell. 2011;42:536–549. doi: 10.1016/j.molcel.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiner A, Chen HV, Liu CL, et al. Systematic dissection of roles for chromatin regulators in a yeast stress response. PLoS Biol. 2012;10:e1001369. doi: 10.1371/journal.pbio.1001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woo H, Dam Ha S, Lee SB, Buratowski S, Kim T. Modulation of gene expression dynamics by co-transcriptional histone methylations. Exp Mol Med. 2017;49:e326. doi: 10.1038/emm.2017.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhat PJ, Murthy TV. Transcriptional control of the GAL/MEL regulon of yeast Saccharomyces cerevisiae: mechanism of galactose-mediated signal transduction. Mol Microbiol. 2001;40:1059–1066. doi: 10.1046/j.1365-2958.2001.02421.x. [DOI] [PubMed] [Google Scholar]
- 20.Frolova E, Johnston M, Majors J. Binding of the glucose-dependent Mig1p repressor to the GAL1 and GAL4 promoters in vivo: regulationby glucose and chromatin structure. Nucleic Acids Res. 1999;27:1350–1358. doi: 10.1093/nar/27.5.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang F, Frey BR, Evans ML, Friel JC, Hopper JE. Gene activation by dissociation of an inhibitor from a transcriptional activation domain. Mol Cell Biol. 2009;29:5604–5610. doi: 10.1128/MCB.00632-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston M, Flick JS, Pexton T. Multiple mechanisms provide rapid and stringent glucose repression of GAL gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3834–3841. doi: 10.1128/MCB.14.6.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlson M. Regulation of glucose utilization in yeast. Curr Opin Genet Dev. 1998;8:560–564. doi: 10.1016/S0959-437X(98)80011-7. [DOI] [PubMed] [Google Scholar]
- 24.Zhou H, Winston F. NRG1 is required for glucose repression of the SUC2 and GAL genes of Saccharomyces cerevisiae. BMC Genet. 2001;2:5. doi: 10.1186/1471-2156-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou BO, Zhou JQ. Recent transcription-induced histone H3 lysine 4 (H3K4) methylation inhibits gene reactivation. J Biol Chem. 2011;286:34770–34776. doi: 10.1074/jbc.M111.273128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shivaswamy S, Iyer VR. Stress-dependent dynamics of global chromatin remodeling in yeast: dual role for SWI/SNF in the heat shock stress response. Mol Cell Biol. 2008;28:2221–2234. doi: 10.1128/MCB.01659-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruiz-Roig C, Vieitez C, Posas F, de Nadal E. The Rpd3L HDAC complex is essential for the heat stress response in yeast. Mol Microbiol. 2010;76:1049–1062. doi: 10.1111/j.1365-2958.2010.07167.x. [DOI] [PubMed] [Google Scholar]
- 28.Kim T, Xu Z, Clauder-Munster S, Steinmetz LM, Buratowski S. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell. 2012;150:1158–1169. doi: 10.1016/j.cell.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JH, Lee BB, Oh YM, et al. Modulation of mRNA and lncRNA expression dynamics by the Set2-Rpd3S pathway. Nat Commun. 2016;7:13534. doi: 10.1038/ncomms13534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren B, Robert F, Wyrick JJ, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 31.Kurdistani SK, Robyr D, Tavazoie S, Grunstein M. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat Genet. 2002;31:248–254. doi: 10.1038/ng907. [DOI] [PubMed] [Google Scholar]
- 32.Robyr D, Suka Y, Xenarios I, et al. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell. 2002;109:437–446. doi: 10.1016/S0092-8674(02)00746-8. [DOI] [PubMed] [Google Scholar]
- 33.Drouin S, Laramee L, Jacques PE, Forest A, Bergeron M, Robert F. DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLoS Genet. 2010;6:e1001173. doi: 10.1371/journal.pgen.1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carrozza MJ, Li B, Florens L, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 35.Keogh MC, Kurdistani SK, Morris SA, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 36.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/S1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 37.Liu CL, Kaplan T, Kim M, et al. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pokholok DK, Harbison CT, Levine S, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 39.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Shi X, Kachirskaia I, Walter KL, et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem. 2007;282:2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taverna SD, Ilin S, Rogers RS, et al. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bian C, Xu C, Ruan J, et al. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J. 2011;30:2829–2842. doi: 10.1038/emboj.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin DG, Baetz K, Shi X, et al. The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol Cell Biol. 2006;26:7871–7879. doi: 10.1128/MCB.00573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choy JS, Kron SJ. NuA4 subunit Yng2 function in intra-S-phase DNA damage response. Mol Cell Biol. 2002;22:8215–8225. doi: 10.1128/MCB.22.23.8215-8225.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi X, Hong T, Walter KL, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Netea MG, Latz E, Mills KH, O’Neill LA. Innate immune memory: a paradigm shift in understanding host defense. Nat Immunol. 2015;16:675–679. doi: 10.1038/ni.3178. [DOI] [PubMed] [Google Scholar]
- 47.Netea MG, Joosten LA, Latz E, et al. Trained immunity: A program of innate immune memory in health and disease. Science. 2016;352:aaf1098. doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Logie C, Stunnenberg HG. Epigenetic memory: A macrophage perspective. Semin Immunol. 2016;28:359–367. doi: 10.1016/j.smim.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 49.Woodworth AM, Holloway AF. The Role of Epigenetic Regulation in Transcriptional Memory in the Immune System. Adv Protein Chem Struct Biol. 2017;106:43–69. doi: 10.1016/bs.apcsb.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 50.Saeed S, Quintin J, Kerstens HH, et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014;345:1251086. doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshida K, Maekawa T, Zhu Y, et al. The transcription factor ATF7 mediates lipopolysaccharide-induced epigenetic changes in macrophages involved in innate immunological memory. Nat Immunol. 2015;16:1034–1043. doi: 10.1038/ni.3257. [DOI] [PubMed] [Google Scholar]
- 52.Zorca CE, Kim LK, Kim YJ, et al. Myosin VI regulates gene pairing and transcriptional pause release in T cells. Proc Natl Acad Sci U S A. 2015;112:E1587–1593. doi: 10.1073/pnas.1502461112. [DOI] [PMC free article] [PubMed] [Google Scholar]