Abstract
The tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) initiates the extrinsic apoptotic pathway through formation of the death-inducing signaling complex (DISC), followed by activation of effector caspases. TRAIL receptors are composed of death receptors (DR4 and DR5), decoy receptors (DcR1 and DcR2), and osteoprotegerin. Among them, only DRs activate apoptotic signaling by TRAIL. Since the levels of DR expressions are higher in cancer cells than in normal cells, TRAIL selectively activates apoptotic signaling pathway in cancer cells. However, multiple mechanisms, including down-regulation of DR expression and pro-apoptotic proteins, and up-regulation of anti-apoptotic proteins, make cancer cells TRAIL-resistant. Therefore, many researchers have investigated strategies to overcome TRAIL resistance. In this review, we focus on protein regulation in relation to extrinsic apoptotic signaling pathways via ubiquitination. The ubiquitin proteasome system (UPS) is an important process in control of protein degradation and stabilization, and regulates proliferation and apoptosis in cancer cells. The level of ubiquitination of proteins is determined by the balance of E3 ubiquitin ligases and deubiquitinases (DUBs), which determine protein stability. Regulation of the UPS may be an attractive target for enhancement of TRAIL-induced apoptosis. Our review provides insight to increasing sensitivity to TRAIL-mediated apoptosis through control of post-translational protein expression.
Keywords: Deubiquitinase, DISC, E3 ligase, Extrinsic pathway, TRAIL
INTRODUCTION
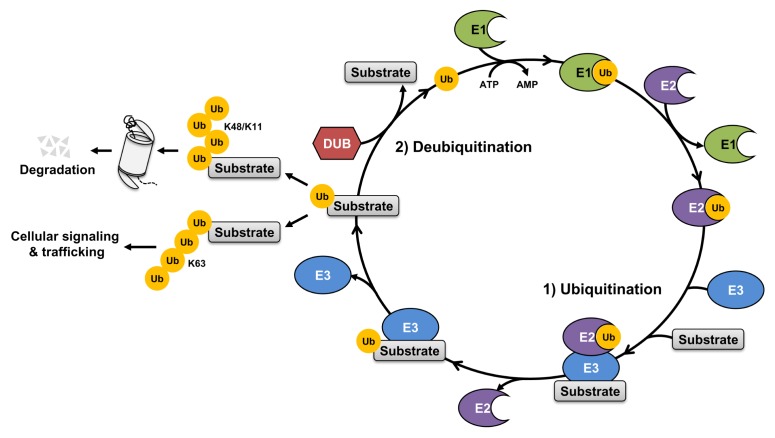
The ubiquitin-proteasome system (UPS) affects various intracellular processes and substrates. Ubiquitination is induced by covalent binding of ubiquitin to lysine residues of target proteins and needs to be catalyzed by an enzymatic cascade that includes E1 activating, E2 conjugating and E3 ligase enzymes (1–4). E1 (ubiquitin-activating enzymes) starts the ubiquitination process, and transfers ubiquitin to E2. E3 ubiquitin ligases are recruited by E2 conjugating enzymes and transfer ubiquitin from E2 to the target protein, resulting in the formation of polyubiquitin chains (5, 6). Proteins are monoubiquitylated by covalent attachment of one ubiquitin, but ubiquitin itself induces polyubiquitination of target proteins through isopeptide linkages. Many proteins are the targets of ubiquitinations, with many functions. Lysine 48 (K48)-linked ubiquitin chains are known primarily to induce proteasomal degradation of target proteins, whereas Lysine 63 (K63)-linked ubiquitin chains have multiple functions, such as DNA repair, cellular signaling, trafficking events and lysosomal degradation of target proteins (7–9) (Fig. 1). Moreover, the UPS is involved in cancer development and therapy, because of their roles in cell cycle regulation, proliferation and apoptosis of cancer cells (10–12).
Fig. 1.
The process of the ubiquitin proteasome system (UPS). 1) Ubiquitin (Ub) is activated by E1 activating enzymes in an ATP-dependent manner, then transferred to E2 conjugating enzymes. E2 conjugating enzymes can recruit E3 ligases enzymes with target substrates. E3 ligase enzymes directly catalyze transfer of activated ubiquitin from E2 conjugating enzymes to substrates, leading to the formation of polyubiquitin chains on target substrates. This process is called ubiquitination. Lys48- and Lys11-linked polyubiquitin chains usually induce degradation of target substrate through proteasome activation. Conversely, Lys63-linked polyubiquitin chains regulate cellular signaling and trafficking. 2) Ubiquitination of target substrate is reversed by deubiquitinases (DUBs). DUBs are critical roles for regulating the function of ubiquitinated proteins by removal of polyubiquitin chains. This process is called deubiquitination. In addition, ubiquitin released from substrates by DUBs can be recycled for activation of ubiquitination.
Ubiquitination is reversed by activation of deubiquitinases (DUBs) that depolymerize ubiquitin in polyubiquitin chains and cleave isopeptide bonds between ubiquitin and the target protein (13, 14) (Fig. 1). In mammals, there are approximately 100 DUBs, and they are classified into six classes depending on the catalytic domain; five cysteine proteases and one metalloprotease. Cysteine proteases include ubiquitin specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), ovarian tumor proteases (OTUs), Machado-joseph disease proteases (MJDs) and MIU-containing novel DUB family (MINDY) protease (15–18). Metalloproteases include Jab1/MPN/MOV34 metalloenzymes (JAMMs). The ubiquitin E3 ligases and DUBs regulate protein stability and are strategic targets for drug discovery (19–21). Therefore, the modulation of protein expression through ubiquitination by E3 ubiquitin ligases and DUBs may enhance the efficiency of cancer therapies.
Tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL), one of the TNF ligand family members, selectively induces apoptosis in tumor cells, but not normal cells (22–24). TRAIL can interact with the complex receptor of death receptors (DR) and decoy receptors (DcR), such as DR4, DR5, DcR1, DcR2, and osteoprotegrin (24). The two DRs initiate apoptosis signaling by binding TRAIL, whereas the three DcRs inhibit TRAIL-mediated apoptosis by competing with the DRs (25, 26). TRAIL-mediated signaling is classified into extrinsic and intrinsic pathways, and results in activation of effector-caspases and induction of apoptosis (27). The extrinsic pathway is initiated by binding of TRAIL to DRs, which induces recruitment of FAS-associated protein with death domain (FADD) and pro-caspase-8, leading to death-inducing signal complex (DISC) formation. Activated caspase-8 by DISC formation directly activates caspase-3 and caspase-7, and eventually induces apoptosis (28). In the intrinsic pathway, truncation of Bid through activated caspase-8 is translocated to the mitochondria, causing cytochrome c release from mitochondria into the cytosol through mitochondria membrane permeabilization, which results in induction of apoptosis via activation of caspase-9 (29). However, many cancer cells exhibit down-regulation of DRs and acquire TRAIL resistance (30–32).
In this article, we review the modulatory mechanisms of the TRAIL-mediated extrinsic pathway through ubiquitination by E3 ubiquitin ligases and DUBs.
REGULATION OF DEATH RECEPTORS BY E3 LIGASE AND DUB
Various E3 ubiquitin ligases and DUBs are involved in the regulation of DR expressions. The E3 ligase c-Casitas B-lineage lymphoma (c-Cbl) directly binds to DRs, following induction of mono-ubiquitination of DRs. Interestingly, mono-ubiquitination of DR4/5 by c-Cbl is degraded in a lysosome-dependent manner, resulting in the increase of early phase TRAIL resistance (33). Moreover, knockdown of c-Cbl by small hairpin RNA (shRNA)-expressing adenovirus has been shown to enhance sensitivity to TRAIL-induced apoptosis in vivo and in vitro through the induction of DR4/5 expression (34).
Van de Kooij et al. reported that membrane associated RING-CH (MARCH)-8 induced down-regulation of DR4 expression on the cell surface, but not DR5 expression (35). MARCH-8 increased polyubiquitination of DR4 on a lysine residue 273 in the C-terminus, followed by degradation of DR4 protein through the lysosomal pathway. The authors reported that overexpression of MARCH-8 reduced sensitivity to TRAIL-mediated apoptosis, and the RING mutant form of MARCH-8 had no effect on resistance to TRAIL-induced apoptosis (35). Furthermore, lysosome inhibitor (bafilomycin A1) reversed DR4 degradation, whereas proteasome inhibitor (MG132) failed to recover DR4 expression. Therefore, the authors suggested that DR4 is degraded by lysosomes at steady-state. In addition, a previous study reported that proteasome or lysosomal inhibitors sensitized TRAIL-induced apoptosis through E3 ligase-mediated up-regulation of DR, overcoming TRAIL resistance in cancer cells (36). Unlike the function of E3 ligase in DR regulation, the modulatory mechanisms of DR expression by DUB are largely unclear.
b-AP15 is a novel inhibitor of proteasome-associated DUBs (USP14 and UCHL5) in 19S proteasome regulatory particles, and both DUBs mediate removal of ubiquitin from the distal end of polyubiquitinated proteins. Therefore, b-AP15 induces accumulation of highly polyubiquitinated proteins through inhibition of proteasome function (37, 38). DR5 is one of the proteins accumulated by b-AP15, and stabilization of DR5 by inhibition of proteasome activity increases TRAIL sensitivity (39). In addition, Oh et al. reported that monocyte chemotactic protein-induced protein-1 (MCPIP1) directly induced deubiquitination of DR5 through its DUB function (40). Ectopic expression of MCPIP1 induced degradation of DR5 in a lysosome-dependent manner, and b-AP15 reversed MCPIP1-mediated DR5 degradation (40). Taken together, the regulation of E3 ligases and DUBs is associated with upregulation of DR expressions, resulting in an increased sensitivity to TRAIL.
REGULATION OF CASPASE-8 BY E3 LIGASE AND DUB
When TRAIL binds DRs, DISC is formed by recruitment of pro-caspase-8 and FADD, following the activation of effect caspases, such as caspase-3 and -7 (28). Several studies have reported that ubiquitination is involved in activation and stabilization of caspase-8. Jin et al. reported that stimulation with TRAIL induced ubiquitination of caspase-8, and cullin3-based E3 ligase (CUL3) was associated with this ubiquitination (41). Neddylation of CUL3 by TRAIL treatment is augmented by cullin-RING ubiquitin ligases (42, 43), and interacts with pro-caspase-8 at DISC. CUL3 promotes K48- and K63-linked polyubiquitination of caspase-8 and polyubiquitinated caspase-8 translocates from the DISC to the ubiquitin-rich foci in a p62-dependent manner. Localization of caspase-8 with p62 in ubiquitin-rich foci drives full activation via autocatalytic processes of caspase-8, followed by induction of apoptosis (41). Moreover, knockdown of CUL3 inhibits caspase-8 polyubiquitination and activation, resulting in reduction of TRAIL-induced apoptosis (41). In contrast, these results were reversed by activation of A20 as a DUB, which removes the ubiquitin chains from pro-capsase-8 (41). Therefore, this evidence suggests that ubiquitin-mediated regulation of caspase-8 activity needs a balance between E3 ligase and DUB. The E3 ligase TNF receptor-associated factor 2 (TRAF2) directly interacts with caspase-8 at DISC and induces K48-linked polyubiquitination of caspase-8, leading to proteasomal degradation of activated caspase-8, and depletion of TRAF2 overcomes resistance to TRAIL through prevention of K48-linked polyubiquitination of caspase-8 (44, 45). CUL3-induced caspase-8 ubiquitination is on K461 in the p10 region of caspase-8 (41), whereas TRAF2-mediated ubiquitination sites are K224, 229, and 231 of the p10 region of caspase-8 (44).
Whereas CUL3 and TRAF2 promote K48-linked polyubiquitination of caspase-8, another E3 ligase (HECTD3) increases ubiquitination of caspase-8 through K63-linked polyubiquitin chain (46). Caspase-8 ubiquitination by the induction of HECTD3 is associated with inactivation of caspase-8, but not degradation. Moreover, HECTD3 is overexpressed in breast carcinoma and inhibits TRAIL-induced caspase-8 cleavage in an E3 ligase activity-dependent manner (46). In addition, several other E3 ligases, WWP1, Siah2, and POSH, do not regulate ubiquitination of caspase-8. However, inhibition of WWP1 increases recruitment of caspase-8 into DISC, and silencing of Siah2 and POSH enhances caspase-8 activity, ultimately sensitizing TRAIL-mediated apoptosis (47, 48). Taken together, the various E3 ligases can regulate caspase-8 activity through ubiquitination of caspase-8, and determine sensitivity to TRAIL in cancer. But caspase-8 degradation and TRAIL sensitivity may depend on action of E3 ligases. Therefore, it remains to be identified how modifications of caspase-8 by E3 ligases differ from other molecular mechanisms, in order to promote cancer cell death.
REGULATION OF FADD BY E3 LIGASE AND DUB
Upon TRAIL stimulation, FADD binds to the death domain of DRs and recruits components of DISC, including caspase-8 and receptor-interacting protein kinase 1 (RIPK1) (28, 49, 50). Lee et al. reported that E3 ligase makorin ring finger protein 1 (MKRN1) down-regulated FADD protein levels through proteasomal degradation (51). MKRN1 directly interacts with FADD, and exogenous MKRN1 induces FADD ubiquitination and destabilization. Depletion of MKRN1 rapidly promotes DISC formation and caspase-8 activation, ultimately increasing TRAIL-induced cell death in vitro and in vivo (51). In addition, knockdown of MKRN1 facilitates necroptosis through increase of necrosome formation upon caspase-8 inhibition. However, DUB-mediated FADD regulation is still unknown.
REGULATION OF RIPK1 BY E3 LIGASE AND DUB
RIPK1 is involved in both complex I and complex II TRAIL signaling through FADD-caspase-8-dependent recruitment to DISC. The C-terminal death domain (DD) of RIPK1 can interact with other DD-containing proteins (52, 53). Therefore, RIPK1 has emerged as a central controller, downstream of DR signaling, that determines cell fate (54, 55). Interestingly, A20 has two domains that are an N-terminal OTU domain of DUB and a C-terminal Zinc finger domain of E3 ligase (56). Several reports have suggested that the function of A20 as an E3 ligase is to inhibit TRAIL-induced apoptosis through ubiquitination of RIPK1 (57, 58). A20 increases K63-linked polyubiquitin chain-mediated RIPK1 ubiquitination, and ubiquitination of RIPK1 by A20 binds to caspase-8 protease domains, followed by protection against TRAIL-induced apoptosis via inhibition of capsase-8 dimerization (57). In addition, silencing of A20 expression increases RIPK1 cleavage-dependent TRAIL sensitivity (58). Therefore, A20 has a dual function as both E3 ligase and DUB, and differentially regulated according to substrate target.
Recently, Lafont et al. reported that RIPK1 was a substrate of the linear ubiquitin chain assembly complex (LUBAC), upon TRAIL treatment (59). The HOIL-1L interacting protein (HOIP), a catalytic component of LUBAC, acts as a E3 ligase, which generates the linear ubiquitin linkages (60). HOIP is recruited to FADD-dependent DRs-associated complex I, resulting in cleavage of complex I. HOIP induces linear ubiquitination of RIPK1, and knockdown of HOIP decreases linear ubiquitination and sensitizes TRAIL-induced apoptosis. Moreover, depletion of cIAP1/2 by SMAC mimetics, significantly reduces LUBAC recruitment to complex I and RIPK1 ubiquitination, following enhancement of TRAIL-induced cell death (59).
REGULATION OF c-FLIP BY E3 LIGASE AND DUB
The long and short isoforms of cellular FLICE-inhibitory protein (c-FLIP) are major inhibitors of TRAIL-mediated apoptosis, and interfere with caspase-8-mediated DISC formation through competition with caspase-8 (24, 61, 62). Many cancer cells have high levels of c-FLIP expression (63–65), and overexpression of c-FLIP is associated with resistance to TRAIL-mediated apoptosis and correlation with a poor prognosis (66–68). Several studies reported E3 ligase Itch regulates proteasomal degradation of c-FLIP(L) through its K48-linked ubiquitination (69, 70). Itch interacts with c-FLIP(L) and induces its degradation, but Itch does not interact with c-FLIP(S) (69). Down-regulation of c-FLIP(L) by Itch is associated with increased TRAIL sensitivity, and knockdown of Itch induces c-FLIP(L) up-regulation and inhibition of TRAIL sensitization (70). Seo et al. recently reported that increased E3 ligase Cbl expression down-regulated c-FLIP(L) expression in a p53-dependent manner, and down-regulation of Cbl by shRNA blocked the c-FLIP(L) down-regulation and sensitivity to TRAIL (71). In addition, Cbl regulated c-FLIP(S) stabilization, and silencing of Cbl abrogated the reduction of c-FLIP(S) expression (72). Recently, Hsu et al. identified E3 ligase deltex1 as a novel regulator of c-FLIP expression (73). Deltex1 bound c-FLIP(L) and promoted down-regulation of c-FLIP(L) through the endosome-lysosomal degradation pathway, but not the proteasome pathway. Overexpression of deltex1 increased sensitivity to TRAIL-induced apoptosis, whereas knockdown of deltex1 attenuated apoptosis by TRAIL (73).
Unlike the ability of E3 ligase to regulate c-FLIP expression, the underlying mechanisms of DUB-dependent stabilization of c-FLIP are unclear. Previous studies have demonstrated that two DUBs, USP2 and USP8, can indirectly regulate c-FLIP(S) expression through deubiquitination of Itch (74, 75). Overexpression of USP2 accumulates Itch by its deubiquitination, resulting in degradation of c-FLIP(S) (74). Moreover, upregulation of USP8 by regulation of PTEN-Akt signaling decreases c-FLIP(S) steady state levels and induces down-regulation of c-FLIP(S) through Itch-mediated c-FLIP(S) ubiquitination, followed by induction of TRAIL sensitivity (75). Recently, Jeong et al. demonstrated that USP8 directly interacted with the caspase-like domain in c-FLIP(L) and induced deubiquitination and stabilization of c-FLIP(L), but not c-FLIP(S). Depletion of USP8 destabilized c-FLIP(L) through ubiquitin-proteasome pathway leading to sensitization of TRAIL-induced apoptosis in vitro and in a xenograft model (76).
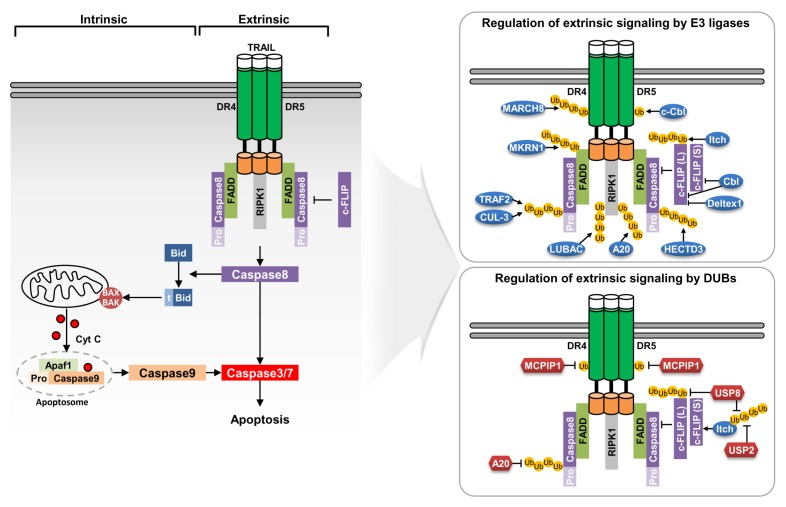
Although E3 ligase-dependent degradation and stabilization of c-FLIP has been demonstrated, studies on the DUB-dependent regulation of c-FLIP are lacking. Therefore, further study is needed to identify the correct mechanism of direct c-FLIP regulation through DUBs. We summarized the molecular mechanisms of E3 ligases and DUBs that regulate expression and activation of extrinsic pathway-related proteins (Fig. 2 and Table 1).
Fig. 2.
Modulatory mechanisms of TRAIL extrinsic signaling pathway by E3 ligases and deubiquitinases. TRAIL-induced apoptosis is executed by the death receptor-mediate extrinsic pathway and mitochondria-mediated intrinsic pathway through activation of effector caspases. Binding of TRAIL to death receptors triggers the formation of DISC by recruiting FADD and caspase-8. Various E3 ubiquitin ligases and deubiquitinases (DUBs) modulate the expression and function of proteins involved in the extrinsic signaling pathway.
Table 1.
E3 ligases and deubiquitinases as regulators in TRAIL extrinsic signaling
| Enzyme | Target | Mechanisms | TRAIL-induced cell death | References |
|---|---|---|---|---|
| E3 ligase | ||||
| c-Cbl | DR4, DR5 | Mono-Ub/Proteasome | Inhibition | (33, 34) |
| MARCH-8 | DR4 | Poly-Ub/Lysosome | Inhibition | (35) |
| CUL-3 | Pro-caspase-8 | Poly-Ub (K48) | Sensitization | (41) |
| TRAF-2 | Caspase-8 | Poly-Ub (K48)/Proteasome | Sensitization | (44, 45) |
| HECTD3 | Caspase-8 | Poly-Ub (K63) | Inhibition | (46) |
| WWP1 | Caspase-8 | Inhibition of recruitmentto DISC | Inhibition | (47) |
| Siah2, POSH | Caspase-8 | Inhibition of activity | Inhibition | (48) |
| MKRN1 | FADD | Ub/destabilization | Inhibition | (51) |
| A20 | RIPK1 | Poly-Ub (K63) | Inhibition | (57, 58) |
| LUBAC | RIPK1 | Linear ubiquitination | Inhibition | (59) |
| Itch | c-FLIP(L) | Poly-Ub (K48)/Proteasome | Sensitization | (69, 70) |
| Cbl | c-FLIP(L) | Degradation/Proteasome | Sensitization | (71) |
| c-FLIP(S) | Stabilization | Sensitization | (72) | |
| Deltex1 | c-FLIP(L) | Degradation by endosome-lysosome | Sensitization | (73) |
| Deubiquitinase | ||||
| MCPIP1 | DR5 | Deubiquitination | Inhibition | (40) |
| USP14, UCHL5 | DR5 | Indirect degradation by proteasome | Inhibition | (39) |
| A20 | Pro-caspase-8 | Removing the ubiquitin chains | Inhibition | (41) |
| USP2, USP8 | c-FLIP(S) | Indirect degradation by DUB of Itch | Sensitization | (74, 75) |
| USP8 | c-FLIP(L) | DUB and stabilization | Inhibition | (76) |
CONCLUSION
Because cancer cells preferentially have TRAIL receptor expression on their cell surface, DRs-mediated TRAIL signaling can provide a therapeutic target for cancer treatment. Therefore, various ways to improve TRAIL-mediated apoptosis signals through DRs regulation have been investigated. The UPS is a key modulator of cellular physiological processes in cancer, such as cell cycle, proliferation and apoptosis. In addition, the controlled activation and degradation of TRAIL signaling regulators by ubiquitination affect TRAIL-induced apoptosis in many cancer cells. Many studies have demonstrated that UPS-mediated regulation of DRs, as well as DISC components, modulates sensitivity and resistance to TRAIL-mediated apoptosis. Here, we describe the regulatory molecular mechanisms of the TRAIL extrinsic pathway through E3 ligases and DUBs (Fig. 2). Since TRAIL treatment alone is inefficient in treating cancer and preventing its recurrence, targeting E3 ligases and DUBs for regulation of TRAIL signaling could provide management for development of new TRAIL adjuvants.
ACKNOWLEDGEMENTS
This work was supported by an NRF grant funded by the Korea Government (MSIP) (2014R1A5A2010008, NRF-2016R1A2B 2013393 and NRF-2018R1D1A3B07049596).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 3.Finley D, Ciechanover A, Varshavsky A. Ubiquitin as a central cellular regulator. Cell. 2004;116:S29–32. doi: 10.1016/S0092-8674(03)00971-1. 2 p following S32. [DOI] [PubMed] [Google Scholar]
- 4.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 6.Joazeiro CA, Weissman AM. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/S0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 7.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kulathu Y, Komander D. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol. 2012;13:508–523. doi: 10.1038/nrm3394. [DOI] [PubMed] [Google Scholar]
- 9.Olzmann JA, Chin LS. Parkin-mediated K63-linked polyubiquitination: a signal for targeting misfolded proteins to the aggresome-autophagy pathway. Autophagy. 2008;4:85–87. doi: 10.4161/auto.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen M, Schmitt S, Buac D, Dou QP. Targeting the ubiquitin-proteasome system for cancer therapy. Expert Opin Ther Targets. 2013;17:1091–1108. doi: 10.1517/14728222.2013.815728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding F, Xiao H, Wang M, Xie X, Hu F. The role of the ubiquitin-proteasome pathway in cancer development and treatment. Front Biosci (Landmark Ed) 2014;19:886–895. doi: 10.2741/4254. [DOI] [PubMed] [Google Scholar]
- 12.Orlowski RZ. The role of the ubiquitin-proteasome pathway in apoptosis. Cell Death Differ. 1999;6:303–313. doi: 10.1038/sj.cdd.4400505. [DOI] [PubMed] [Google Scholar]
- 13.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leznicki P, Kulathu Y. Mechanisms of regulation and diversification of deubiquitylating enzyme function. J Cell Sci. 2017;130:1997–2006. doi: 10.1242/jcs.201855. [DOI] [PubMed] [Google Scholar]
- 15.Nijman SM, Luna-Vargas MP, Velds A, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Clague MJ, Barsukov I, Coulson JM, Liu H, Rigden DJ, Urbe S. Deubiquitylases from genes to organism. Physiol Rev. 2013;93:1289–1315. doi: 10.1152/physrev.00002.2013. [DOI] [PubMed] [Google Scholar]
- 17.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 18.Abdul Rehman SA, Kristariyanto YA, Choi SY, et al. MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol Cell. 2016;63:146–155. doi: 10.1016/j.molcel.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bielskiene K, Bagdoniene L, Mozuraitiene J, Kazbariene B, Janulionis E. E3 ubiquitin ligases as drug targets and prognostic biomarkers in melanoma. Medicina (Kaunas) 2015;51:1–9. doi: 10.1016/j.medici.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 20.D’Arcy P, Wang X, Linder S. Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol Ther. 2015;147:32–54. doi: 10.1016/j.pharmthera.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Nicholson B, Marblestone JG, Butt TR, Mattern MR. Deubiquitinating enzymes as novel anticancer targets. Future Oncol. 2007;3:191–199. doi: 10.2217/14796694.3.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashkenazi A, Pai RC, Fong S, et al. Safety and antitumor activity of recombinant soluble Apo2 ligand. J Clin Invest. 1999;104:155–162. doi: 10.1172/JCI6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walczak H, Miller RE, Ariail K, et al. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat Med. 1999;5:157–163. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
- 24.Wang S, El-Deiry WS. TRAIL and apoptosis induction by TNF-family death receptors. Oncogene. 2003;22:8628–8633. doi: 10.1038/sj.onc.1207232. [DOI] [PubMed] [Google Scholar]
- 25.Pan G, O’Rourke K, Chinnaiyan AM, et al. The receptor for the cytotoxic ligand TRAIL. Science. 1997;276:111–113. doi: 10.1126/science.276.5309.111. [DOI] [PubMed] [Google Scholar]
- 26.Sheridan JP, Marsters SA, Pitti RM, et al. Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science. 1997;277:818–821. doi: 10.1126/science.277.5327.818. [DOI] [PubMed] [Google Scholar]
- 27.Srivastava RK. TRAIL/Apo-2L: mechanisms and clinical applications in cancer. Neoplasia. 2001;3:535–546. doi: 10.1038/sj.neo.7900203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kischkel FC, Lawrence DA, Chuntharapai A, Schow P, Kim KJ, Ashkenazi A. Apo2L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity. 2000;12:611–620. doi: 10.1016/S1074-7613(00)80212-5. [DOI] [PubMed] [Google Scholar]
- 29.Kantari C, Walczak H. Caspase-8 and bid: caught in the act between death receptors and mitochondria. Biochim Biophys Acta. 20111813:558–563. doi: 10.1016/j.bbamcr.2011.01.026. [DOI] [PubMed] [Google Scholar]
- 30.Zhang L, Fang B. Mechanisms of resistance to TRAIL-induced apoptosis in cancer. Cancer Gene Ther. 2005;12:228–237. doi: 10.1038/sj.cgt.7700792. [DOI] [PubMed] [Google Scholar]
- 31.Jin Z, McDonald ER, 3rd, Dicker DT, El-Deiry WS. Deficient tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) death receptor transport to the cell surface in human colon cancer cells selected for resistance to TRAIL-induced apoptosis. J Biol Chem. 2004;279:35829–35839. doi: 10.1074/jbc.M405538200. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Zhang B. TRAIL resistance of breast cancer cells is associated with constitutive endocytosis of death receptors 4 and 5. Mol Cancer Res. 2008;6:1861–1871. doi: 10.1158/1541-7786.MCR-08-0313. [DOI] [PubMed] [Google Scholar]
- 33.Song JJ, Szczepanski MJ, Kim SY, et al. c-Cbl-mediated degradation of TRAIL receptors is responsible for the development of the early phase of TRAIL resistance. Cell Signal. 2010;22:553–563. doi: 10.1016/j.cellsig.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SY, Kim JH, Song JJ. c-Cbl shRNA-expressing adenovirus sensitizes TRAIL-induced apoptosis in prostate cancer DU-145 through increases of DR4/5. Cancer Gene Ther. 2013;20:82–87. doi: 10.1038/cgt.2012.88. [DOI] [PubMed] [Google Scholar]
- 35.van de Kooij B, Verbrugge I, de Vries E, et al. Ubiquitination by the membrane-associated RING-CH-8 (MARCH-8) ligase controls steady-state cell surface expression of tumor necrosis factor-related apoptosis inducing ligand (TRAIL) receptor 1. J Biol Chem. 2013;288:6617–6628. doi: 10.1074/jbc.M112.448209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park EJ, Min KJ, Choi KS, et al. Chloroquine enhances TRAIL-mediated apoptosis through up-regulation of DR5 by stabilization of mRNA and protein in cancer cells. Sci Rep. 2016;6:22921. doi: 10.1038/srep22921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D’Arcy P, Linder S. Proteasome deubiquitinases as novel targets for cancer therapy. Int J Biochem Cell Biol. 2012;44:1729–1738. doi: 10.1016/j.biocel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 38.D’Arcy P, Brnjic S, Olofsson MH, et al. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat Med. 2011;17:1636–1640. doi: 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
- 39.Oh YT, Deng L, Deng J, Sun SY. The proteasome deubiquitinase inhibitor b-AP15 enhances DR5 activation-induced apoptosis through stabilizing DR5. Sci Rep. 2017;7:8027. doi: 10.1038/s41598-017-08424-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oh YT, Qian G, Deng J, Sun SY. Monocyte chemotactic protein-induced protein-1 enhances DR5 degradation and negatively regulates DR5 activation-induced apoptosis through its deubiquitinase function. Oncogene. 2018;37:3415–3425. doi: 10.1038/s41388-018-0200-9. [DOI] [PubMed] [Google Scholar]
- 41.Jin Z, Li Y, Pitti R, et al. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–735. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 42.Bosu DR, Kipreos ET. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 2008;3:7. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 44.Gonzalvez F, Lawrence D, Yang B, et al. TRAF2 Sets a threshold for extrinsic apoptosis by tagging caspase-8 with a ubiquitin shutoff timer. Mol Cell. 2012;48:888–899. doi: 10.1016/j.molcel.2012.09.031. [DOI] [PubMed] [Google Scholar]
- 45.Xu L, Zhang Y, Qu X, et al. DR5-Cbl-b/c-Cbl-TRAF2 complex inhibits TRAIL-induced apoptosis by promoting TRAF2-mediated polyubiquitination of caspase-8 in gastric cancer cells. Mol Oncol. 2017;11:1733–1751. doi: 10.1002/1878-0261.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Kong Y, Zhou Z, et al. The HECTD3 E3 ubiquitin ligase facilitates cancer cell survival by promoting K63-linked polyubiquitination of caspase-8. Cell Death Dis. 2013;4:e935. doi: 10.1038/cddis.2013.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Z, Liu R, Chen C. The WWP1 ubiquitin E3 ligase increases TRAIL resistance in breast cancer. Int J Cancer. 2012;130:1504–1510. doi: 10.1002/ijc.26122. [DOI] [PubMed] [Google Scholar]
- 48.Christian PA, Fiandalo MV, Schwarze SR. Possible role of death receptor-mediated apoptosis by the E3 ubiquitin ligases Siah2 and POSH. Mol Cancer. 2011;10:57. doi: 10.1186/1476-4598-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bodmer JL, Holler N, Reynard S, et al. TRAIL receptor-2 signals apoptosis through FADD and caspase-8. Nat Cell Biol. 2000;2:241–243. doi: 10.1038/35008667. [DOI] [PubMed] [Google Scholar]
- 50.Sprick MR, Weigand MA, Rieser E, et al. FADD/MORT1 and caspase-8 are recruited to TRAIL receptors 1 and 2 and are essential for apoptosis mediated by TRAIL receptor 2. Immunity. 2000;12:599–609. doi: 10.1016/S1074-7613(00)80211-3. [DOI] [PubMed] [Google Scholar]
- 51.Lee EW, Kim JH, Ahn YH, et al. Ubiquitination and degradation of the FADD adaptor protein regulate death receptor-mediated apoptosis and necroptosis. Nat Commun. 2012;3:978. doi: 10.1038/ncomms1981. [DOI] [PubMed] [Google Scholar]
- 52.Chaudhary PM, Eby M, Jasmin A, Bookwalter A, Murray J, Hood L. Death receptor 5, a new member of the TNFR family, and DR4 induce FADD-dependent apoptosis and activate the NF-kappaB pathway. Immunity. 1997;7:821–830. doi: 10.1016/S1074-7613(00)80400-8. [DOI] [PubMed] [Google Scholar]
- 53.Hsu H, Shu HB, Pan MG, Goeddel DV. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84:299–308. doi: 10.1016/S0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 54.Sessler T, Healy S, Samali A, Szegezdi E. Structural determinants of DISC function: new insights into death receptor-mediated apoptosis signalling. Pharmacol Ther. 2013;140:186–199. doi: 10.1016/j.pharmthera.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 55.Christofferson DE, Li Y, Yuan J. Control of life-or-death decisions by RIP1 kinase. Annu Rev Physiol. 2014;76:129–150. doi: 10.1146/annurev-physiol-021113-170259. [DOI] [PubMed] [Google Scholar]
- 56.Wertz IE, O’Rourke KM, Zhou H, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 57.Bellail AC, Olson JJ, Yang X, Chen ZJ, Hao C. A20 ubiquitin ligase-mediated polyubiquitination of RIP1 inhibits caspase-8 cleavage and TRAIL-induced apoptosis in glioblastoma. Cancer Discov. 2012;2:140–155. doi: 10.1158/2159-8290.CD-11-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dong B, Lv G, Wang Q, et al. Targeting A20 enhances TRAIL-induced apoptosis in hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2012;418:433–438. doi: 10.1016/j.bbrc.2012.01.056. [DOI] [PubMed] [Google Scholar]
- 59.Lafont E, Kantari-Mimoun C, Draber P, et al. The linear ubiquitin chain assembly complex regulates TRAIL-induced gene activation and cell death. EMBO J. 2017;36:1147–1166. doi: 10.15252/embj.201695699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kirisako T, Kamei K, Murata S, et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006;25:4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roth W, Reed JC. FLIP protein and TRAIL-induced apoptosis. Vitam Horm. 2004;67:189–206. doi: 10.1016/S0083-6729(04)67011-7. [DOI] [PubMed] [Google Scholar]
- 62.Shirley S, Micheau O. Targeting c-FLIP in cancer. Cancer Lett. 2013;332:141–150. doi: 10.1016/j.canlet.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 63.Griffith TS, Chin WA, Jackson GC, Lynch DH, Kubin MZ. Intracellular regulation of TRAIL-induced apoptosis in human melanoma cells. J Immunol. 1998;161:2833–2840. [PubMed] [Google Scholar]
- 64.Xiao CW, Yan X, Li Y, Reddy SA, Tsang BK. Resistance of human ovarian cancer cells to tumor necrosis factor alpha is a consequence of nuclear factor kappaB-mediated induction of Fas-associated death domain-like interleukin-1beta-converting enzyme-like inhibitory protein. Endocrinology. 2003;144:623–630. doi: 10.1210/en.2001-211024. [DOI] [PubMed] [Google Scholar]
- 65.Zhang X, Jin TG, Yang H, DeWolf WC, Khosravi-Far R, Olumi AF. Persistent c-FLIP(L) expression is necessary and sufficient to maintain resistance to tumor necrosis factor-related apoptosis-inducing ligand-mediated apoptosis in prostate cancer. Cancer Res. 2004;64:7086–7091. doi: 10.1158/0008-5472.CAN-04-1498. [DOI] [PubMed] [Google Scholar]
- 66.Valnet-Rabier MB, Challier B, Thiebault S, et al. c-Flip protein expression in Burkitt’s lymphomas is associated with a poor clinical outcome. Br J Haematol. 2005;128:767–773. doi: 10.1111/j.1365-2141.2005.05378.x. [DOI] [PubMed] [Google Scholar]
- 67.Valente G, Manfroi F, Peracchio C, et al. cFLIP expression correlates with tumour progression and patient outcome in non-Hodgkin lymphomas of low grade of malignancy. Br J Haematol. 2006;132:560–570. doi: 10.1111/j.1365-2141.2005.05898.x. [DOI] [PubMed] [Google Scholar]
- 68.Ullenhag GJ, Mukherjee A, Watson NF, Al-Attar AH, Scholefield JH, Durrant LG. Overexpression of FLIPL is an independent marker of poor prognosis in colorectal cancer patients. Clin Cancer Res. 2007;13:5070–5075. doi: 10.1158/1078-0432.CCR-06-2547. [DOI] [PubMed] [Google Scholar]
- 69.Chang L, Kamata H, Solinas G, et al. The E3 ubiquitin ligase itch couples JNK activation to TNFalpha-induced cell death by inducing c-FLIP(L) turnover. Cell. 2006;124:601–613. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 70.Yang F, Tay KH, Dong L, et al. Cystatin B inhibition of TRAIL-induced apoptosis is associated with the protection of FLIP(L) from degradation by the E3 ligase itch in human melanoma cells. Cell Death Differ. 2010;17:1354–1367. doi: 10.1038/cdd.2010.29. [DOI] [PubMed] [Google Scholar]
- 71.Seo BR, Min KJ, Woo SM, et al. Inhibition of cathepsin s induces mitochondrial ros that sensitizes trail-mediated apoptosis through p53-Mediated Downregulation of Bcl-2 and c-FLIP. Antioxid Redox Signal. 2017;27:215–233. doi: 10.1089/ars.2016.6749. [DOI] [PubMed] [Google Scholar]
- 72.Zhao L, Yue P, Khuri FR, Sun SY. mTOR complex 2 is involved in regulation of Cbl-dependent c-FLIP degradation and sensitivity of TRAIL-induced apoptosis. Cancer Res. 2013;73:1946–1957. doi: 10.1158/0008-5472.CAN-12-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hsu TS, Mo ST, Hsu PN, Lai MZ. c-FLIP is a target of the E3 ligase deltex1 in gastric cancer. Cell Death Dis. 2018;9:135. doi: 10.1038/s41419-017-0165-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haimerl F, Erhardt A, Sass G, Tiegs G. Down-regulation of the de-ubiquitinating enzyme ubiquitin-specific protease 2 contributes to tumor necrosis factor-alpha-induced hepatocyte survival. J Biol Chem. 2009;284:495–504. doi: 10.1074/jbc.M803533200. [DOI] [PubMed] [Google Scholar]
- 75.Panner A, Crane CA, Weng C, et al. Ubiquitin-specific protease 8 links the PTEN-Akt-AIP4 pathway to the control of FLIPS stability and TRAIL sensitivity in glioblastoma multiforme. Cancer Res. 2010;70:5046–5053. doi: 10.1158/0008-5472.CAN-09-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jeong M, Lee EW, Seong D, et al. USP8 suppresses death receptor-mediated apoptosis by enhancing FLIPL stability. Oncogene. 2017;36:458–470. doi: 10.1038/onc.2016.215. [DOI] [PubMed] [Google Scholar]