Abstract
Upon viral infection, the 2′, 5′-oligoadenylate synthetase (OAS)-ribonuclease L (RNaseL) system works to cleave viral RNA, thereby blocking viral replication. However, it is unclear whether OAS proteins have a role in regulating gene expression. Here, we show that OAS1 and OAS3 act as negative regulators of the expression of chemokines and interferon-responsive genes in human macrophages. Clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein-9 nuclease (Cas9) technology was used to engineer human myeloid cell lines in which the OAS1 or OAS3 gene was deleted. Neither OAS1 nor OAS3 was exclusively responsible for the degradation of rRNA in macrophages stimulated with poly(I:C), a synthetic surrogate for viral double-stranded (ds)RNA. An mRNA sequencing analysis revealed that genes related to type I interferon signaling and chemokine activity were increased in OAS1−/− and OAS3−/− macrophages treated with intracellular poly(I:C). Indeed, retinoic-acid-inducible gene (RIG)-I- and interferon-induced helicase C domain-containing protein (IFIH1 or MDA5)-mediated induction of chemokines and interferon-stimulated genes was regulated by OAS3, but Toll-like receptor 3 (TLR3)- and TLR4-mediated induction of those genes was modulated by OAS1 in macrophages. However, stimulation of these cells with type I interferons had no effect on OAS1- or OAS3-mediated chemokine secretion. These data suggest that OAS1 and OAS3 negatively regulate the expression of chemokines and interferon-responsive genes in human macrophages.
Keywords: Macrophage, Negative regulation, OAS, poly(I:C), RNA degradation
INTRODUCTION
During infection with RNA viruses, germline-encoded pattern-recognition receptors (PRRs), such as Toll-like receptors (TLRs) and retinoic-acid-inducible gene (RIG)-I-like receptors (RLRs), recognize viral RNA and trigger various signaling pathways that lead to the production of type I interferons (IFNs) and proinflammatory cytokines (1). It is well known that the IFN system plays an important role in the response to viral infection and virus clearance. IFN-α and IFN-β induce the expression of IFN-stimulated genes (ISGs), which encode transcription factors and antiviral effectors, including the 2′, 5′-oligoadenylate synthetase (OAS) proteins (2).
The OASs are a family of IFN- and virus-induced proteins. The human OAS family consists of four genes, OAS1 (p40/p46; the short form), OAS2 (p69/p71; the intermediate form), OAS3 (p100; the long form), and OASL (OAS-like protein; p59), which are located on chromosome 12. Because OAS1, OAS2, and OAS3 can synthesize 2′, 5′-oligoadenylates (2-5A) from ATP upon binding to double-stranded (ds)RNA, it was thought that these three enzymes are involved in the activation of ribonuclease L (RNaseL) during viral infection (3). During infection with RNA viruses, viral dsRNA directly binds to and activates OAS1-3, resulting in the production of 2-5A from ATP (4); 2-5A leads to the dimerization and activation of RNaseL, which cleaves viral and host RNA, thereby blocking viral replication and inhibiting viral protein synthesis (5).
OAS1 is the most well-studied member of the OAS family and plays an important role in resistance to flaviviruses, such as the West Nile virus and dengue virus (6, 7). Polymorphism at the OAS1 locus has also been implicated in susceptibility to enteroviruses and respiratory syncytial virus (8, 9). Structural and biochemical studies show that human OAS1 undergoes a conformational change to recognize dsRNA, resulting in the production of 2-5A (10). However, OAS1 produces short 2-5A, which is much less able to activate RNaseL than are the forms of 2-5A generated by OAS2 and OAS3 (11). Furthermore, enzymatically inactive OAS1 can inhibit viral replication in vitro, suggesting that OAS1 can play various roles in the regulation of signaling pathways during viral infection (3). However, the functions of OAS1 in immune cells have yet to be investigated.
OAS3 is superior to OAS1 in its ability to synthesize 2-5A (11). A lower concentration of dsRNA is needed for OAS3 activation than for OAS1 activation, and 2-5A synthesized by OAS3 is longer than the 2-5A synthesized by OAS1 (11). A structural study of human OAS3 revealed that it is specialized for binding long dsRNA (12). Moreover, consistent with these properties of OAS3, it was recently reported that OAS3 is mainly responsible for activating RNaseL during viral infection of human cell lines (the lung carcinoma cell line A549 and the fibrosarcoma cell line HT1080) (13). However, there are large differences in the basal expression of OAS proteins among different cell types (14), which results in cell-type-specific effects. Activation of the OAS/RNaseL pathway increased IFN production in fibroblasts, which have low levels of OASs, but suppressed it in macrophages, which have high basal levels of OASs (15). Although OAS3 is a dominant factor for RNaseL activation in fibroblasts, it is possible that other OASs might play this role in macrophages, and/or that OAS3 itself plays a different role in macrophages. In addition, because OAS3 has a nuclear import sequence and is mainly localized to the cytoplasm and nucleus, OAS3 proteins may have a role in gene regulation during viral infection (16). Therefore, investigating OAS3-induced genes during viral infection in the major viral target-cell type, human macrophages, is required to understand the antiviral immune response.
To investigate the roles of OAS1 and OAS3 in human myeloid cells, we generated THP-1 (human monocytic leukemia) cell lines individually that lacked OAS1 or OAS3 by using clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein-9 nuclease (Cas9) gene-editing technology (17). Unexpectedly, RNaseL activation was independent on OAS3 in differentiated THP-1 (dTHP-1) cells, but both OAS1 and OAS3 negatively regulated the expression of chemokines in response to specific pathogen-associated molecular patterns (PAMPs). Thus, we report for the first time that OAS1 and OAS3 are negative regulators of cytokine expression, rather than RNaseL activation, during viral infection in human macrophages.
RESULTS
OAS1 and OAS3 are not required for poly(I:C)-induced rRNA degradation in THP-1 cells
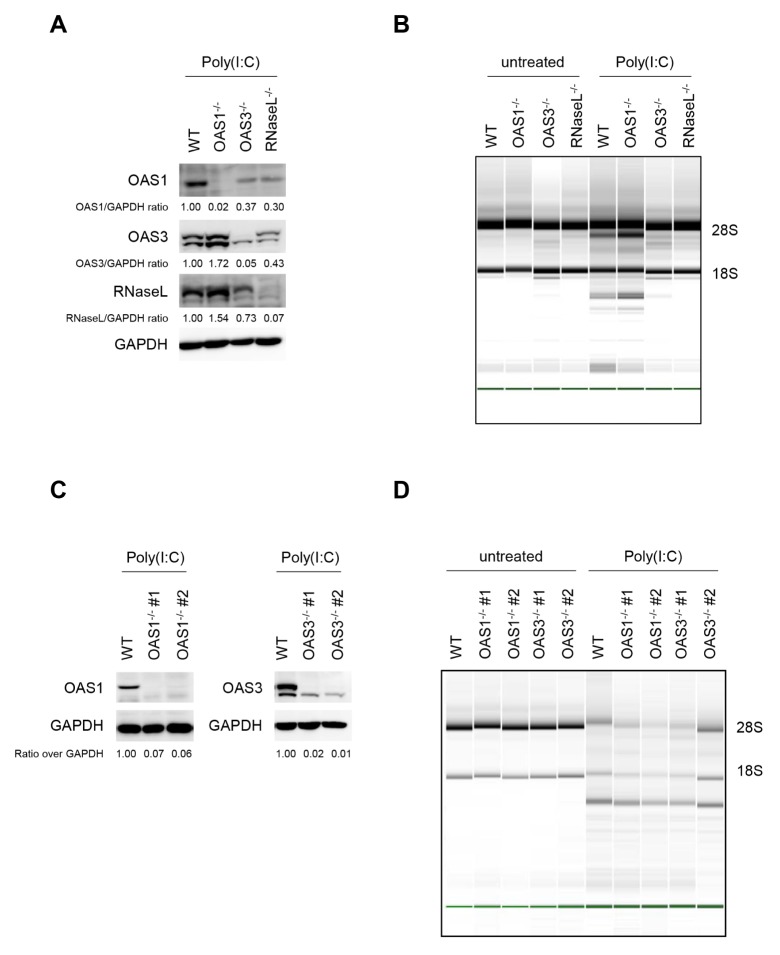
To investigate the antiviral responses of OAS species, OAS1, OAS3, or RNaseL was knocked out in HT1080 cells, a human fibrosarcoma-derived cell line, using the CRISPR-Cas9 system (Fig. 1A). Each knock-out cell line was transfected with poly(I:C) for 6 h. As expected, the level of rRNA degradation in cells lacking OAS1 was similar to that in wild-type (WT) cells, whereas rRNA remained intact in cells lacking OAS3 or RNaseL (Fig. 1B). These results confirmed that OAS3 was required for the RNaseL-mediated degradation of rRNA in response to poly(I:C) in HT1080 cells.
Fig. 1.
Degradation of RNA in response to poly(I:C) in dTHP-1 cells does not require OAS1 or OAS3. (A) Knock-out of OAS1, OAS3, or RNaseL in HT1080 cells that were stimulated with 10 μg/ml poly(I:C) overnight was confirmed by immunoblot. (B) WT and KO HT1080 cells were transfected with 1 μg/ml poly(I:C) for 6 h, and then lysed. RNA integrity was assessed by Bioanalyzer. The positions of 18S and 28S rRNA are indicated. (C) OAS1−/− and OAS3−/− dTHP-1 cells were treated with 10 μg/ml poly(I:C) overnight. Cells were lysed, and expression of OAS1 and OAS3 was analyzed by immunoblot. (D) WT and KO dTHP-1 cells were transfected with 1 μg/ml poly(I:C) for 6 h, and RNA integrity was analyzed. *P < 0.05 (two-tailed unpaired Student’s t-test).
Next, we examined whether OAS3 affected RNaseL activity in THP-1 cells, which can differentiate into macrophage-like cells. First, OAS1 or OAS3 knock-out THP-1 cells were generated (Fig. 1C), and then the dTHP-1 cells were treated intracellularly with poly(I:C). Interestingly, rRNA degradation was observed not only in WT and OAS1−/− cells, but also in OAS3−/− THP-1 cells (Fig. 1D). These results indicate that neither OAS1 nor OAS3 is exclusively responsible for the degradation of rRNA in response to poly(I:C).
OAS1 and OAS3 are negative regulators of intracellular poly(I:C)-induced chemokines and type I IFN signaling genes
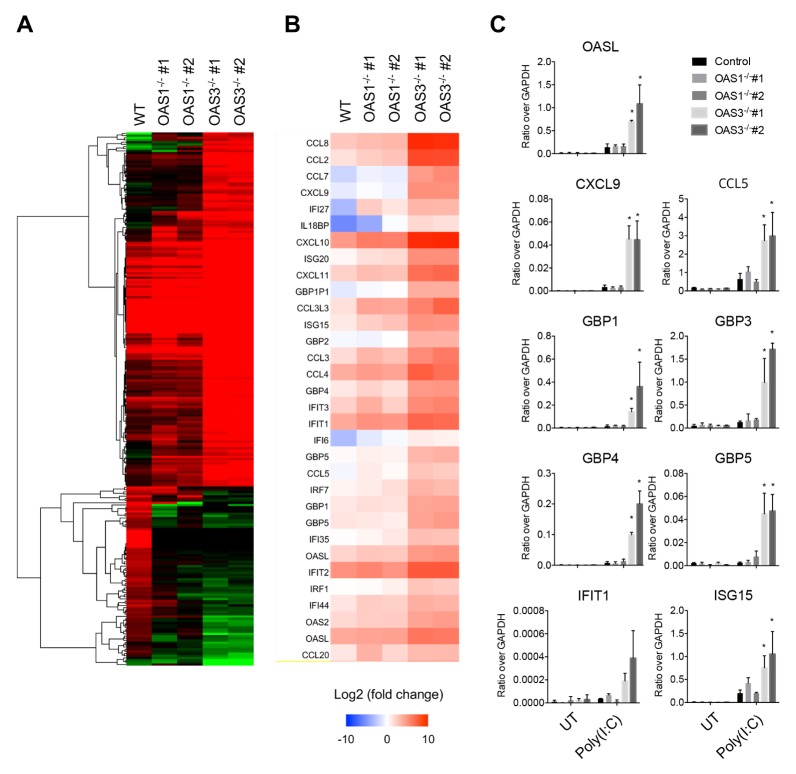
To investigate the role of OAS1 and OAS3 on gene expression induced by dsRNA, we did mRNA sequencing analysis of WT, OAS1−/−, and OAS3−/− dTHP-1 cells transfected with poly(I:C). OAS3−/− cells stimulated with intracellular poly(I:C) showed strong transcriptional changes (more than four-fold) in 244 genes, mostly enriched in the innate immune-response ontology group (Fig. 2A). In general, OAS1−/− dTHP-1 cells transfected with poly(I:C) showed a transcription profile similar to that of OAS3−/− cells. However, several genes were significantly increased or decreased by intracellular poly(I:C) in OAS3−/− cells but not in WT or OAS1−/− cells. The genes that were upregulated in both OAS1−/− and OAS3−/− cells were predominantly involved in type I IFN signaling, viral defense responses, and chemokine activity, whereas the genes that were downregulated were mostly involved in transforming growth factor-β (TGF-β) signaling, nuclear envelope organization, and negative regulation of intrinsic apoptosis signaling (Fig. 2A and Supplementary Table 1). Focusing on the genes related to type I IFN signaling, transcripts encoding IFN regulatory factor 7 (IRF7), ISG15, ISG20, IFN-induced protein with tetratricopeptide repeats 1 (IFIT1), guanine nucleotide-binding protein 1 (GBP1), and OASL were all upregulated in OAS1−/− and OAS3−/− cells treated with intracellular poly(I:C) compared with WT cells (Fig. 2B). In addition, many chemokines, including CCL2, CCL3, CCL4, CCL8, CXCL9, and CXCL10, showed abundant changes in transcription levels in poly(I:C)-transfected OAS1−/− and OAS3−/− cells (Fig. 2B). However, the magnitude of these changes was much greater in OAS3−/− cells than in OAS1−/− cells. These sequencing results suggest that OAS1 and OAS3 are both negative regulators of dsRNA-dependent antiviral responses, but that OAS3 modulates these responses more strongly than OAS1.
Fig. 2.
OAS3 regulates intracellular poly(I:C)-induced expression of chemokines and interferon-stimulated genes in THP-1 cells. (A) Changes in gene expression in WT, OAS1−/−, and OAS3−/− dTHP-1 cells transfected with 1 μg/ml poly(I:C) for 6 h. The heat map represents genes that were increased (a, 162 genes) or decreased (b, 82 genes) by more than twice in OAS3−/− cells compared with WT cells. (B) Heat map depicting the expression of genes related to type I interferon signaling and chemokine activity in WT, OAS1−/−, and OAS3−/− dTHP-1 cells stimulated with intracellular poly(I:C). Relative gene expression compared with WT cells is shown for each independent sample. Over-expression is shown in red, under-expression in blue. (C) WT, OAS1−/−, and OAS3−/− dTHP-1 cells were transfected with 1 μg/ml poly(I:C) for 6 h, and the mRNA levels of the indicated genes were quantified by qRT-PCR. Data are representative of at least three independent experiments. *P < 0.05 (two-tailed unpaired Student’s t-test).
Next, the intracellular poly(I:C)-induced expression of type I IFN signaling genes and chemokine family genes was confirmed by qRT-PCR. The qRT-PCR results were consistent with the mRNA sequencing analyses, demonstrating that stimulation of OAS3−/− dTHP-1 cells with intracellular poly(I:C) leads to a significant upregulation of type I IFN signaling genes, including OASL, GBP1, GBP3, GBP4, GBP5, IFIT1, and ISG15, and chemokine family genes, including CCL5 and CXCL9 (Fig. 2C). However, OAS1−/− dTHP-1 cells showed only a modest increase in the expression of these genes, which is also consistent with the mRNA sequencing results. These results suggested that OAS3 strongly suppresses the intracellular poly(I:C)-induced expression of type I IFN signaling genes and chemokine family genes.
OAS3 negatively regulates RIG-I- and MDA5-dependent antiviral responses in THP-1 cells
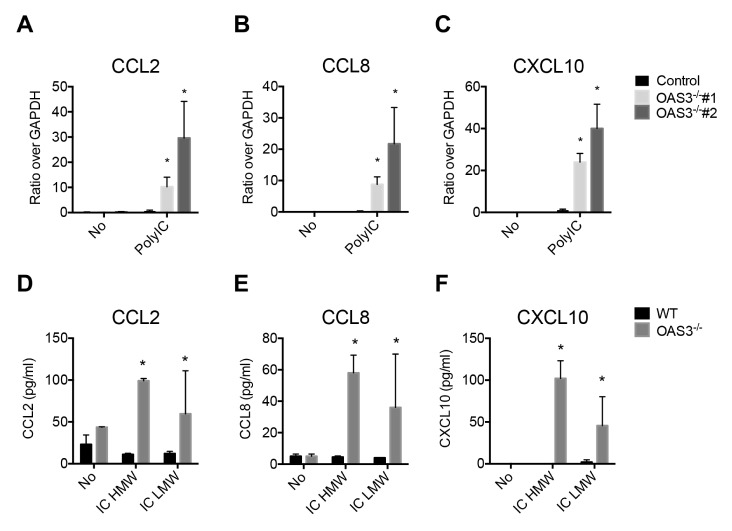
In macrophages, intracellular poly(I:C) is recognized mainly by RIG-I and MDA5 (18). Intracellular high molecular weight (HMW) poly(I:C) is a ligand for MDA5, whereas RIG-I preferentially binds to intracellular low molecular weight (LMW) poly(I:C) (19, 20). Thus, we examined whether OAS3 regulates the expression of RIG-I- or MDA5-induced chemokines. OAS3-deficient dTHP-1 cells showed a higher induction of the chemokines CCL2, CCL8, and CXCL10 in response to intracellular poly(I:C) than did WT cells (Fig. 3A–C). To analyze whether OAS3-mediated chemokine expression is dependent on MDA5 or RIG-I, we transfected HMW poly(I:C) and LMW poly(I:C) separately into WT and OAS3−/− dTHP-1 cells. The secretion of CCL2, CCL8, and CXCL10 was increased in OAS3−/− cells in response to both HMW poly(I:C) and LMW poly(I:C) treatment, compared with WT dTHP-1 macrophages (Fig. 3D–F). These data demonstrate that RIG-I- and MDA5-mediated chemokine induction is regulated by OAS3.
Fig. 3.
The levels of the chemokines CCL2, CCL8, and CXCL10 are increased in OAS3−/− dTHP-1 cells stimulated intracellularly with poly(I:C). (A–C) WT and OAS3−/− dTHP-1 cells were transfected with 1 μg/ml poly(I:C) for 6 h, and the mRNA levels of (A) CCL2, (B) CCL8, and (C) CXCL10 were quantified by qRT-PCR. (D–F) WT and OAS3−/− dTHP-1 cells were transfected with 1 μg/ml HMW or LMW poly(I:C) for 6 h. The secretion of (D) CCL2, (E) CCL8, and (F) CXCL10 was measured by ELISA. Data are representative of at least three independent experiments. *P < 0.05 (two-tailed unpaired Student’s t-test).
OAS1 regulates poly(I:C)- and LPS-mediated chemokine induction in dTHP-1 cells
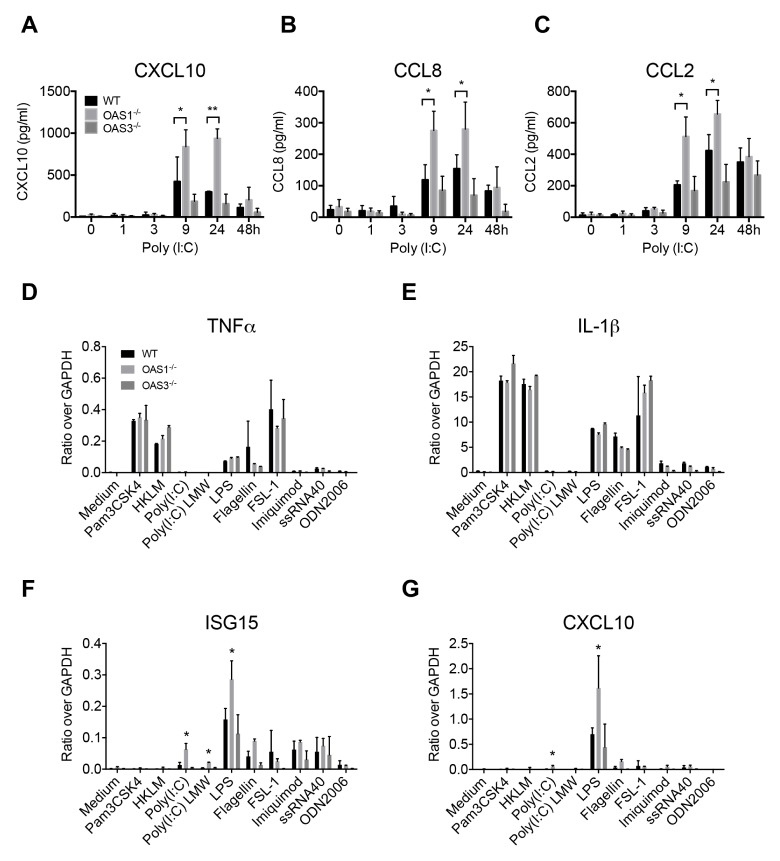
Stimulation of macrophages with extracellular poly(I:C) activates TLR3 (18). OAS1 and OAS3 are highly induced at the mRNA level by extracellular poly(I:C) (21). Thus, we analyzed the kinetic expression of the chemokines CCL2, CCL8, and CXCL10 in WT, OAS1−/−, and OAS3−/− dTHP-1 cells after poly(I:C) treatment. Unexpectedly, OAS1−/− dTHP-1 cells showed high levels of expression of these chemokines at 9 and 24 h after poly(I:C) stimulation, compared with the levels in WT or OAS3−/− cells (Fig. 4A–C). These findings suggest that OAS1 negatively regulates the induction of chemokines in response to TLR3 stimulation in macrophages.
Fig. 4.
Expression of cytokines and chemokines after treatment of OAS1−/− or OAS3−/− dTHP-1 cells with TLR agonists. (A–C) WT, OAS1−/−, and OAS3−/− dTHP-1 cells were stimulated with poly(I:C) (10 μg/ml) for the indicated times, and mRNA levels of (A) CXCL10, (B) CCL8, and (C) CCL2 were quantified by qRT-PCR. (E–G) WT, OAS1−/−, and OAS3−/− dTHP-1 cells were cultured for 16 h in the absence or presence of Pam3CSK4 (TLR1/2 agonist; 1 μg/ml), HKLM (TLR2 agonist; 108 cells/ml), poly(I:C) (TLR3 agonist; 10 μg/ml), poly(I:C) LMW (TLR3 agonist; 10 μg/ml), LPS (TLR4 agonist; 1 μg/ml), FLA (TLR5 agonist; 1 μg/ml), FSL-1 (TLR2/6 agonist; 100 ng/ml), imiquimod (TLR7 agonist; 1 μg/ml), ssRNA40 (TLR8 agonist; 1 μg/ml), or ODN2006 (TLR9 agonist; 5 μM), and the mRNA levels of (D) TNF-α, (E) IL-1β, (F) ISG15, and (G) CXCL10 were quantified by qRT-PCR. Data are representative of at least three independent experiments. *P < 0.05, **P < 0.01 (two-tailed unpaired Student’s t-test).
Next, to investigate whether OAS1 or OAS3 regulates the expression of cytokines or chemokines in response to TLR signaling, we examined the induction of TNF-α, IL-1β, ISG15, and CXCL10 after stimulation with various human TLR ligands. As expected, since THP-1 cells express all TLR family members except for TLR9 (22), stimulation with a TLR9 agonist (CpG ODN2006) had no effect on cytokine/chemokine production (Fig. 4D–G). TNF-α and IL-1β were induced at the mRNA level in WT, OAS1−/−, and OAS3−/− cells after stimulation with agonists of TLR1/2 (Pam3CSK4), TLR2 [heat-killed Listeria monocytogenes (HKLM)], TLR4 [lipopolysaccharide (LPS)], TLR5 [flagellin (FLA)], and TLR6/2 (the synthetic diacylated lipoprotein FSL-1) (Fig. 4D–E). However, interestingly, OAS1−/− dTHP-1 cells showed much higher induction of ISG15 and CXCL10 after treatment with TLR3 and TLR4 agonists than did WT and OAS3−/− cells (Fig. 4F–G and Supplementary Fig. 1A–B). These findings suggest that OAS1 negatively regulates the induction of chemokines and ISGs in response to TLR3 and TLR4 signaling in macrophages.
Type I IFNs are not involved in OAS1- or OAS3-mediated chemokine expression
TLR3, RIG-I, and MDA5 are critical in triggering the production of type I IFNs, which can induce chemokines such as CCL2, CCL8, and CXCL10 (23). For this reason, we evaluated the induction of IFN-β in response to intracellular poly(I:C) in WT, OAS1−/−, and OAS3−/− dTHP-1 cells, but found no significant differences between the cell types (Supplementary Fig. 2). Next, we evaluated CCL2, CCL8, and CXCL10 secretion in response to type I IFNs, and the levels of these chemokines were also similar among WT, OAS1−/−, and OAS3−/− dTHP-1 cells after stimulation with IFN-α (Supplementary Fig. 3A–C) or IFN-β (Supplementary Fig. 3D–F). These results suggest that OAS1- and OAS3-dependent chemokine expression is not dependent on the type I IFN pathway.
DISCUSSION
It has previously been demonstrated that both OAS1 and OAS3 have antiviral activity in human and mouse model systems (13, 24, 25). However, no attempt has been made to generate knock-out cell lines of these genes in human macrophages. In this study, we demonstrate that OAS1 and OAS3 have a role in gene regulation in human THP-1 macrophages. First, the expression of OAS1 and OAS3 is not required for RNaseL activation in dTHP-1 cells. Furthermore, OAS3 suppresses the expression of type I IFN signaling-related genes in response to stimulation by RIG-I and MDA5, whereas OAS1 suppresses the expression of chemokines after stimulation via TLR3 and TLR4.
The observation that intracellular poly(I:C) induced rRNA degradation in dTHP-1 cells despite the absence of OAS1 or OAS3 suggested that OAS1 and OAS3 might be able to compensate for each other’s loss in macrophages. These results are not consistent with those of a previous study in which OAS3 knock-out human lung carcinoma and fibrosarcoma cells had deficient RNaseL activation because of disrupted 2-5A synthesis (13). However, OAS1, OAS2, and OAS3 can all synthesize 2-5A in vitro in human cell lines, and their basal expression is higher in macrophages than in fibroblasts (11, 15). Thus, the ability of each OAS family member to induce 2-5A synthesis may differ by cell type. In addition, the use of double or triple knock-outs of OAS genes in dTHP-1 cells is needed to evaluate compensation for OAS protein loss.
OAS3-deficient dTHP-1 cells showed upregulation of genes related to type I IFN signaling and chemokine activity in response to stimulation by RIG-I and MDA5 (Figs. 2 and 3). However, the mechanism of OAS3-mediated negative regulation of these genes was not clearly demonstrated in our experiments. Because RIG-I and MDA5 activate mitochondrial antiviral signaling protein (MAVS) on the mitochondrial membrane (26), one possibility is that OAS3 may inhibit the ability of MAVS to induce innate immunity. Interestingly, a proteomic analysis of the human innate immunity interactome indicated that OAS3 interacts with MAVS (27). However, these findings do not provide sufficient evidence for OAS3-mediated MAVS inhibition, and further studies will be needed to find out whether OAS3 directly inhibits MAVS, thereby triggering negative regulation of antiviral gene expression.
In summary, our study provides the first evidence that OAS1 and OAS3 are involved in the negative regulation of the innate antiviral response in human macrophages. OAS1 regulates the expression of chemokines and IFN-responsive genes following stimulation via TLR3 and TLR4, whereas OAS3 regulates the expression of these genes following stimulation via RIG-I- and MDA5. These findings suggest that OAS proteins regulate host innate immune signaling in macrophages, which has broad implications for the treatment of viral diseases.
MATERIALS AND METHODS
Cell lines
Human HT-1080 cells were cultured in Dulbecco’s modified Eagle medium (DMEM; ThermoFisher Scientific) supplemented with 10% fetal bovine serum (FBS; Gibco/ThermoFisher Scientific), 50 U/ml penicillin, and 50 mg/ml streptomycin. Human monocytic THP-1 cells were maintained in culture in Roswell Park Memorial Institute culture medium (RPMI 1640; Invitrogen/ThermoFisher Scientific) containing 10% FBS, 50 U/ml penicillin, and 50 mg/ml streptomycin. THP-1 monocytes were differentiated into macrophages by 48 h incubation with 50 nM phorbol 12-myristate 13-acetate (PMA, Sigma, P8139).
CRISPR-Cas9-dependent gene knock-out in HT-1080 and THP-1 cells
The oligonucleotide sequences (Supplementary Table 2) used for the generation of small guide RNAs (sgRNAs) were designed using the site http://crispr.mit.edu. Two sgRNA sequences were selected for each OAS gene knock-out, and one was selected for knock-out of the RNaseL gene. The synthesized single-strand guide RNAs (gRNAs) (listed in Table 2) were annealed to form gRNA oligo duplexes and ligated into digested pLentiCRISPR v2 vectors (Addgene plasmid #52961; a gift from Feng Zhang) according to the manufacturer’s instructions.
For packaging of pseudolentiviruses, HEK 293FT cells were transfected with lentiCRISPR v2, psPAX2 (packaging vector), and pMD2.G (envelope-expressing vector). After 48 h, the supernatants were harvested and stored at −80°C. The 48 h supernatants were used for further knock-out experiments.
To construct HT-1080 and THP-1 knock-out cells using the pLentiCRISPRv2 vector, HT-1080 or THP-1 cells were transduced with pseudolentiviruses and polybrene (8 μg/ml, Sigma). At 48 h post transduction, cells were cultured in medium containing puromycin (2 μg/ml) for 3 d. Puromycin-resistant cells were cloned by limiting dilution and further selected by plating in 96-well plates. Gene knock-out colonies were validated by immunoblot.
Statistical analysis
Statistical analysis was performed with Prism 6.0 software (GraphPad). An unpaired two-tailed t-test with a 95% confidence interval was used for calculation of P values. Group sizes, reproducibility, and P values for each experiment are given in the figure legends.
SUPPLEMENTARY INFORMATION
ACKNOWLEDGEMENTS
This research was supported by the Basic Science Research Program of the NRF, funded by the Ministry of Science, ICT & Future Planning (NRF-2017R1D1A1B06029547 to J.S.K-H), by the Collaborative Genome Program for Fostering New Post-Genome Industry of the National Research Foundation of Korea (NRF), which is funded by the Ministry of Science, Communications Technology (ICT), and Future Planning (grant number: 2016M3C9A4921712, to Y.J.K.), and by an intramural grant (2Z05610, to W.-B.L.) from the Korea Institute of Science and Technology (KIST), Republic of Korea.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Justesen J, Hartmann R, Kjeldgaard NO. Gene structure and function of the 2′-5′-oligoadenylate synthetase family. Cell Mol Life Sci. 2000;57:1593–1612. doi: 10.1007/PL00000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kristiansen H, Gad HH, Eskildsen-Larsen S, Despres P, Hartmann R. The oligoadenylate synthetase family: an ancient protein family with multiple antiviral activities. J Interferon Cytokine Res. 2011;31:41–47. doi: 10.1089/jir.2010.0107. [DOI] [PubMed] [Google Scholar]
- 4.Dong B, Silverman RH. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J Biol Chem. 1995;270:4133–4137. doi: 10.1074/jbc.270.8.4133. [DOI] [PubMed] [Google Scholar]
- 5.Hovanessian AG, Justesen J. The human 2′-5′oligoadenylate synthetase family: unique interferon-inducible enzymes catalyzing 2′-5′ instead of 3′-5′ phosphodiester bond formation. Biochimie. 2007;89:779–788. doi: 10.1016/j.biochi.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Alagarasu K, Honap T, Damle IM, Mulay AP, Shah PS, Cecilia D. Polymorphisms in the oligoadenylate synthetase gene cluster and its association with clinical outcomes of dengue virus infection. Infect Genet Evol. 2013;14:390–395. doi: 10.1016/j.meegid.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 7.Lim JK, Lisco A, McDermott DH, et al. Genetic variation in OAS1 is a risk factor for initial infection with West Nile virus in man. PLoS Pathog. 2009;5:e1000321. doi: 10.1371/journal.ppat.1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behera AK, Kumar M, Lockey RF, Mohapatra SS. 2′-5′ Oligoadenylate synthetase plays a critical role in interferon-gamma inhibition of respiratory syncytial virus infection of human epithelial cells. J Biol Chem. 2002;277:25601–25608. doi: 10.1074/jbc.M200211200. [DOI] [PubMed] [Google Scholar]
- 9.Cai Y, Chen Q, Zhou W, et al. Association analysis of polymorphisms in OAS1 with susceptibility and severity of hand, foot and mouth disease. Int J Immunogenet. 2014;41:384–392. doi: 10.1111/iji.12134. [DOI] [PubMed] [Google Scholar]
- 10.Donovan J, Dufner M, Korennykh A. Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proc Natl Acad Sci U S A. 2013;110:1652–1657. doi: 10.1073/pnas.1218528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibsen MS, Gad HH, Thavachelvam K, Boesen T, Despres P, Hartmann R. The 2′-5′-oligoadenylate synthetase 3 enzyme potently synthesizes the 2′-5′-oligoadenylates required for RNase L activation. J Virol. 2014;88:14222–14231. doi: 10.1128/JVI.01763-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donovan J, Whitney G, Rath S, Korennykh A. Structural mechanism of sensing long dsRNA via a noncatalytic domain in human oligoadenylate synthetase 3. Proc Natl Acad Sci U S A. 2015;112:3949–3954. doi: 10.1073/pnas.1419409112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Banerjee S, Wang Y, et al. Activation of RNase L is dependent on OAS3 expression during infection with diverse human viruses. Proc Natl Acad Sci U S A. 2016;113:2241–2246. doi: 10.1073/pnas.1519657113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao L, Birdwell LD, Wu A, et al. Cell-type-specific activation of the oligoadenylate synthetase-RNase L pathway by a murine coronavirus. J Virol. 2013;87:8408–8418. doi: 10.1128/JVI.00769-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banerjee S, Chakrabarti A, Jha BK, Weiss SR, Silverman RH. Cell-type-specific effects of RNase L on viral induction of beta interferon. MBio. 2014;5:e00856–00814. doi: 10.1128/mBio.00856-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malaguarnera L, Nunnari G, Di Rosa M. Nuclear import sequence identification in hOAS3 protein. Inflamm Res. 2016;65:895–904. doi: 10.1007/s00011-016-0972-8. [DOI] [PubMed] [Google Scholar]
- 17.Shen S, Loh TJ, Shen H, Zheng X, Shen H. CRISPR as a strong gene editing tool. BMB Rep. 2017;50:20–24. doi: 10.5483/BMBRep.2017.50.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takeuchi O, Akira S. MDA5/RIG-I and virus recognition. Curr Opin Immunol. 2008;20:17–22. doi: 10.1016/j.coi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 19.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 20.Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 21.Das A, Chai JC, Kim SH, et al. Transcriptome sequencing of microglial cells stimulated with TLR3 and TLR4 ligands. BMC Genomics. 2015;16:517. doi: 10.1186/s12864-015-1728-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeshita F, Suzuki K, Sasaki S, Ishii N, Klinman DM, Ishii KJ. Transcriptional regulation of the human TLR9 gene. J Immunol. 2004;173:2552–2561. doi: 10.4049/jimmunol.173.4.2552. [DOI] [PubMed] [Google Scholar]
- 23.Mantovani A, Bonecchi R, Locati M. Tuning inflammation and immunity by chemokine sequestration: decoys and more. Nat Rev Immunol. 2006;6:907–918. doi: 10.1038/nri1964. [DOI] [PubMed] [Google Scholar]
- 24.Samuel MA, Whitby K, Keller BC, et al. PKR and RNase L contribute to protection against lethal West Nile Virus infection by controlling early viral spread in the periphery and replication in neurons. J Virol. 2006;80:7009–7019. doi: 10.1128/JVI.00489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kristiansen H, Scherer CA, McVean M, et al. Extracellular 2′-5′ oligoadenylate synthetase stimulates RNase L-independent antiviral activity: a novel mechanism of virus-induced innate immunity. J Virol. 2010;84:11898–11904. doi: 10.1128/JVI.01003-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim TH, Lee HK. Innate immune recognition of respiratory syncytial virus infection. BMB Rep. 2014;47:184–191. doi: 10.5483/BMBRep.2014.47.4.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li S, Wang L, Berman M, Kong YY, Dorf ME. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity. 2011;35:426–440. doi: 10.1016/j.immuni.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.