Abstract
The thoracic ossification of the posterior longitudinal ligament (T-OPLL) can cause thoracic spinal stenosis, which results in intractable myelopathy and radiculopathy. Our previous whole-genome sequencing study first reported rs199772854 in the interleukin 17 receptor C (IL17RC) gene as a potentially pathogenic loci for T-OPLL. The aim of the present study was to examine the effects of the IL17RC gene rs199772854A site mutation on osteogenesis by establishing a model of osteogenic differentiation. IL17RC gene mutation site and wild-type site mouse embryonic osteoblast (3T3-E1) models were constructed in order to induce the differentiation of the cells into osteoblasts. Whether the mutation site causes the abnormal expression of the IL17RC gene and osteogenic markers was analyzed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blot analysis. The IL17RC gene rs199772854A site mutation was demonstrated to play a biological role through the overexpression of its own gene, and also to significantly increase the expression levels of osteogenic markers. Furthermore, the mutation upregulated the expression of the key proteins, tumor necrosis factor receptor (TNFR)-associated factor 6 (TRAF6) and nuclear factor (NF)-κB, in the interleukin (IL)-17 signaling axis. On the whole, the findings of this study suggest that the IL17RC gene rs199772854A loci mutation propels mouse embryonic osteoblasts towards osteogenic differentiation and may play an important role in the pathogenesis of T-OPLL. The IL17RC gene may promote osteogenesis through the IL-17 signaling pathway and may thus be involved in the process of ectopic osteogenesis in T-OPLL.
Keywords: IL17RC, gene, ossification of the posterior longitudinal ligament, thoracic
Introduction
The thoracic ossification of the posterior longitudinal ligament (T-OPLL) can cause thoracic spinal stenosis, which results in intractable myelopathy and radiculopathy. T-OPLL is a rare disease of the spine; however, it has a high disability rate and is progressively aggravating. The disability rate of patients with T-OPLL is much higher than that of patients with cervical spinal cord disease caused by the cervical ossification of the posterior longitudinal ligament (C-OPLL). The prevalence of T-OPLL in individuals of Japanese ethnicity has been shown to be 1.6-1.9%. Among patients with T-OPLL, the most frequently encountered type is the liner type, the most common level is T3-T4, and the mean age of onset is >40 years (1,2).
Surgery is the only effective treatment for T-OPLL; however, surgery is complex and the risk level is high. Although a variety of surgical treatment methods have been reported, there is currently no standard method available for the effective treatment of T-OPLL. Furthermore, the post-operative complication rate is 9.6-40.8% (3–5), and the cause of neurological deterioration in the early post-operative period is unclear. Therefore, numerous studies have focused on elucidating the pathogenesis of T-OPLL. Although the pathogenesis of T-OPLL remains unclear, studies on C-OPLL have suggested that genetic factors play an important role in the pathogenesis of OPLL (6,7). Previous studies have reported >10 susceptibility genes/loci that are linked to OPLL susceptibility, which include R-spondin 2 (8,9), bone morphogenetic protein (BMP)2 (10,11), BMP4 (12), BMP9 (13), runt-related transcription factor 2 (14,15), collagen (COL) type VI α 1 chain (16–18), COL type XVII α 1 chain (19), transforming growth factor β1 (TGF-β1) (20), Toll-like receptor 5 (21), nucleotide pyrophosphatase/phosphodiesterase 1 (22) and interleukin (IL)15 receptor α (23).
Due to the limited movement of the thoracic spine, the stability of the thoracic vertebrae is higher than that of the cervical vertebrae. Furthermore, the local mechanical stress that affects degenerative disease of the spine is small. Therefore, we hypothesized that genetic factors may play a greater role in the pathogenesis of T-OPLL than environmental factors. Our recent whole-genome sequencing study identified that rs199772854 in the IL17 receptor C (IL17RC) gene is a potentially pathogenic loci for T-OPLL (24). In addition, our case-controlled association study confirmed that this mutation was potentially associated with susceptibility to T-OPLL in individuals of Chinese Han ethnicity (25).
The IL17RC gene is primarily responsible for encoding type I transmembrane proteins (26). The majority of studies on the pathogenic role of the IL17RC gene have demonstrated that it mainly functions through the IL-17 signaling axis. The dysfunction of the IL-17 signaling axis has been implicated in numerous human diseases (27,28). The IL-17 signaling axis plays a central role in the regulation of inflammation (29). Recent studies have demonstrated that the IL-17 signaling axis also affects bone formation and remodeling, and can protect bone mass in the case of bone loss due to infection or hormone imbalance. IL17RC serves an indispensable role in the development of osteoblasts, which accelerates the differentiation of osteoblasts (30,31). OPLL results in increased bone formation in the ligament tissues, and there is also an association between OPLL disease and increased bone mineral density throughout the body (7). These studies indicate the potential role of IL17RC in the pathogenesis and progression of osteogenic diseases.
In the present study, mouse embryonic osteoblast cells were induced to differentiate into osteoblasts by transfection of the IL17RC gene carrying the rs199772854A site lentivirus, rs199772854C site lentivirus and empty lentivirus into 3T3-E1 mouse embryonic osteoblasts. Furthermore, whether the mutation loci causes the abnormal expression of the IL17RC gene was analyzed and differences in the ability to induce osteogenesis were detected in order to provide an experimental basis for the potential role of the IL17RC gene in the pathogenesis of T-OPLL.
Materials and methods
Cell lines and cell culture
The 3T3-E1 mouse embryonic osteoblast cell line (American Type Culture Collection, Manassas, VA, USA) was cultured in minimum Essential medium (MEM)-α with ribonucleosides, deoxyribonucleosides, 2 mM L-glutamine and 1 mM sodium pyruvate, and without ascorbic acid (cat no. A1049001; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Cells were incubated at 37°C with 5% CO2 in a 95% humidified atmosphere, and the culture medium was replaced every 2 days. 293 cells (American Type Culture Collection, Manassas, VA, USA) were cultured in Dulbecco's modified Eagle medium (DMEM; cat. no. 10564-037; Life Technologies; Thermo Fisher Scientific, Inc.). The complete growth medium was prepared by the addition of fetal bovine serum (FBS) to the base medium to reach a final concentration of 10% under 37°C and a humidified atmosphere containing 5% CO2, and the culture medium was replaced every 2 days.
Construction of the IL17RC gene vector
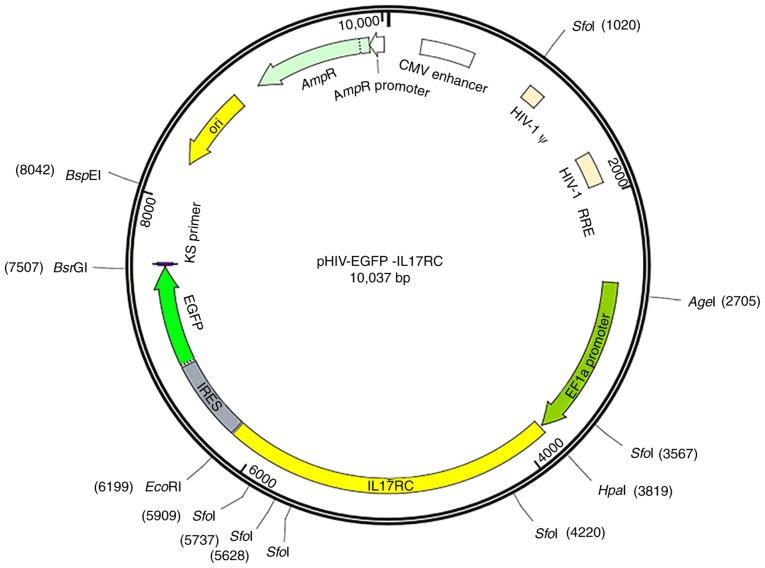
The primers were designed and synthesized according to the human IL17RC sequence information. The upstream HpaI restriction site and the downstream EcoRI restriction site were added, and the primer sequences are presented in Table I. The pHIV-EGFP vector contained the EF1a-MCS-IRES-EGFP component sequence (Addgene, Watertown, MA, USA), HpaI and EcoRI enzyme cleavage sites and the following control insert: TAT TCT AGG GAT CCA ACC CTC GAG TGA CCC GTC TAG AGG GCT AG. The vector map is presented in Fig. 1. GFP is located downstream of the target gene separated by the IRES sequence. The positive bacterial Sanger sequencing primer sequences are shown in Table II. Mutant primers were designed based on the human IL17RC gene sequence information, and wild-type plasmids were used as templates to design mutant primers according to the primer design principles of the Site-Directed Mutagenesis kits (cat. no. 210514; Agilent Technologies, Inc., Santa Clara, CA, USA). The mutant and wild-type IL17RC gene primer sequences are presented in Table III. The IL17RC gene wild-type and mutant plasmids were subjected to endotoxin plasmid extraction and extracted using Plasmid DNA Purification kits (cat. no. K210006; Thermo Fisher Scientific, Inc.).
Table I.
IL17RC gene primer sequences.
| Gene | Primer sequence | Length (bp) |
|---|---|---|
| IL17RC-F | 5′-ATCGGTTAACATGAGGGCGGCCCGTGCTCTGCT-3′ | 2,376 |
| IL17RC-R | 5′-CGGAATTCTTAGCCCAGCGCCACCTTCCTGGA-3′ |
IL17RC, interleukin 17 receptor C.
Figure 1.
pHIV-EGFP lentiviral vector information. The component sequence is EF1a-MCS-IRES-EGFP and the enzyme cleavage sites are HpaI and EcoRI.
Table II.
Sanger sequencing primer sequences.
| Gene | Primer sequence | Length (bp) |
|---|---|---|
| IL17RC-F | 5′-AAGCCTCAGACAGTGGTTCAAAG-3′ | 24 |
| IL17RC-R | 5′-CAAGCGGCTTCGGCCAGTAACGT-3′ | 25 |
IL17RC, interleukin 17 receptor C.
Table III.
Mutant and wild-type gene primer sequences.
| Gene | Primer sequence | Length (bp) |
|---|---|---|
| IL17RC-rs199772854 C-F | 5′-gtgtcccgggcccttcagccagccctggat-3′ | 2,376 |
| IL17RC-rs199772854 C-R | 5′-ggctggctgaagggcccgggacacttgctc-3′ | |
| IL17RC-rs199772854 A-F | 5′-gtgtcccgggccattcagccagccctggat-3′ | 2,376 |
| IL17RC-rs199772854 A-R | 5′-ggctggctgaatggcccgggacacttgctc-3′ |
IL17RC, interleukin 17 receptor C.
Transfection of the pHIV-IL17RC-rs199772854A, pHIV-IL17RC-rs199772854C and pHIV-GFP expression vectors into the 3T3-E1 cells
A total of 5 ml poly-D-lysine (PDL; cat. no. P7280; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was pre-added to a 10-cm dish, which was incubated at 37°C for 30 min. The PDL was aspirated, and the dish was washed twice with phosphate-buffered saline (cat. no. AM9625; Thermo Fisher Scientific, Inc.) and stored in a 37°C incubator for 7 days. 293 cells within 20 passages were adjusted to the logarithmic growth phase, and the cells were divided into the 10-cm dishes to a density of ~70% following trypsin digestion. The cells were cultured in DMEM at 37°C overnight, and then the medium was discarded and a small amount of Opti-MEM medium (cat. no. 51985034; Thermo Fisher Scientific, Inc.) was added to rinse the dish. Subsequently, 6 ml Opti-MEM medium were added to the dish and it was placed in an incubator for use. The helper plasmid pMD2G, pSPAX2 and the wild-type or mutant expression plasmid pHIV-EGFP-IL17RC (Addgene) were added to 500 μl Opti-MEM medium at a ratio of 3:6:9 μg, and 15 μl PLUS reagent (cat. no. 11514015; Thermo Fisher Scientific, Inc.) was added to the mixture. In another tube containing 500 μl Opti-MEM medium, 30 μl Lipofectin Transfection Reagent (cat. no. 18292037; Thermo Fisher Scientific, Inc.) was added and mixed well. After being allowed to stand for 5 min at room temperature, the two tubes were mixed in a volume of 1 ml. The liposomes were mixed with the plasmid and then coated at room temperature for 30 min. The liposome and plasmid mixture was added to the 3T3-E1 cells that had been added to the medium containing Opti-MEM, and DMEM complete medium containing 10% FBS was replaced after 6-8 h. The cells were cultured for 48 h at 37°C in a 5% CO2 incubator. The virus solution was concentrated and purified, and the cells were infected with the high quality virus solution of pHIV-IL17RC-rs199772854C (titer: 6.4×108 TU/ml) and pHIV-IL17RC-rs199772854A (titer: 7.0×108 TU/ml) for 24 h. The viral supernatant was aspirated and fresh DMEM was added. The 3T3-E1 cells were plated in 6-well plates at a density of ~30% and after 12 h, the concentrated virus solution was added to infect the cells, at an MOI of 50 for each group infection. GFP and target gene expression were monitored at nearly the same time using a fluorescence microscope 48 h after exposure. The cells with GFP expression were sorted using a CytoFLEX flow cytometer (Beckman Coulter, Brea, CA, USA) 72 h after exposure. The GFP-positive cells sorted by FACS and part of the sorted cells were examine by western blot analysis and cultured for further analysis. Once the cells had grown to a confluence of 80-90%, they were subcultured.
Differentiation of 3T3-E1 cells into osteoblasts
pHIV-I L17RC-rs199772854C (wild-type group), pHIV-IL17RC-rs199772854A (mutation group), pHIV-GFP infected 3T3-E1 cells (empty lentivirus control group) and the 3T3-E1 cells without adding any vector (mock group). 3T3-E1 cells were cultured to the logarithmic growth phase and then divided into 6-well plates at a density of ~70%. The plates were incubated at 37°C for 16-18 h. When the cells were close to fusion, osteogenic induction medium (low-glucose DMEM (cat. no. 11885-084), 10% FBS (cat no. 10099141; both Gibco; Thermo Fisher Scientific, Inc.), 10 mmol/l β-glycerophosphate (cat. no. G9422), 100 nmol/l dexamethasone (cat. no. D4902), 50 μmol/l ascorbyl phosphate (cat. no. A4544; all Sigma-Aldrich, Inc.), 100 U/ml penicillin and 0.1 mg/ml streptomycin (cat no. 15140-122; Gibco; Thermo Fisher Scientific, Inc.) was added. The cells were harvested at 21 days following osteogenic induction.
Alkaline phosphatase (ALP) activity assay and Alizarin red staining
After the cells were seeded in 24-well plates at a density of 1×105 cells/well and cultured in osteogenic medium for 21 days, osteogenic identification was performed using an ALP activity staining kit (cat. no. GMS80033.1; Genmed Scientific, Shanghai, China) as follows: i) The culture solution was carefully removed from the 24-well plates; ii) this was followed by the addition of 400 μl cleaning solution (Reagent A) to clean the surface of the cells, and this was then removed; iii) 400 μl fixative (Reagent B) was then added followed by incubation for 2 min at room temperature, and removal; iv) 400 μl cleaning solution (Reagent A) was then added for washing and this was then removed; this process was repeated 2 times to wash the fixative residue; v) 400 μl staining working solution was then added followed by incubation for 5-15 min in the dark at room temperature, and the staining solution was then removed; vi) 400 μl cleaning solution (Reagent A) was then added followed by incubation for 5 min at room temperature for washing which was then removed; this was repeated 2 times to wash the stain residue; vii) 200 μl cleaning solution (Reagent A) or PBS were then added to cover the cell surface, and observe the cell staining by the naked eye.
In addition, Mineralization was assessed using an Alizarin Red S kit (cat. no. GMS80046.3; Genmed Scientific) as follows: i) The culture solution was carefully removed the from the 24-well plates; ii) 400 μl cleaning solution (Reagent A) were then added to clean the surface of the cells, and this was then removed; iii) 400 μl fixative (Reagent B) was then added followed by incubation for 10 min at room temperature, and this was then removed; iv) 400 μl cleaning solution (Reagent A) were added for washing and this was then removed; this process was repeated 2 times to wash the fixative residue; v) 400 μl staining solution (Reagent C) were then added followed by incubation at room temperature until orange-red color became visible, drain, and drying at room temperature; vi) 400 μl cleaning solution (Reagent A) were added for washing and this was then removed; this was repeated 2 times to wash the stain residue; vii) 200 μl cleaning solution (Reagent A) or PBS were then added to cover the cell surface, and observe the cell staining by the naked eye.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from the cells using TRIzol reagent (Thermo Fisher Scientific, Inc.), and reverse transcription was performed using a M-MLV Reverse Transcriptase kit (Promega Corp., Madison, WI, USA) according to the manufacturer's instructions. qPCR was applied to quantify the mRNA levels of IL17RC, ALP and GAPDH using SYBR-Green Real-Time PCR Master mix on the LightCycler 480 Real-Time System (Roche Diagnostics, Basel, Switzerland) under 2-Step Cycling (95°C for 10 min hold, 40 cycles of 95°C for 15 sec and 60°C for 60 sec). All experiments were performed in triplicate and normalized to GAPDH, and the relative gene expression was calculated based on 2-ΔΔCq method (29). The primers are listed in Table IV.
Table IV.
Primer sequences for reverse transcription-quantitative PCR.
| Gene | Primer sequence | Length (bp) |
|---|---|---|
| IL17RC-RT-F | 5′-CTGCCCTTGTGCAGTTTGG-3′ | 85 |
| IL17RC-RT-R | 5′-CAGATTCGTACCTCACTCCCTA-3′ | |
| ALP-RT-F | 5′-GTGAACCGCAACTGGTACTC-3′ | 81 |
| ALP-RT-R | 5′-GAGCTGCGTAGCGATGTCC-3′ | |
| GAPDH-RT-F | 5′-TGGGTGTGAACCATGAGAAGT-3′ | 126 |
| GAPDH-RT-R | 5′-TGAGTCCTTCCACGATACCAA-3′ |
IL17RC, interleukin 17 receptor C.
Western blot analysis
Cell lysates were obtained using ice-cold RIPA lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China) containing 10 mM PMSF as a protease inhibitor. The total concentration of the extracted proteins was determined using an Enhanced BCA Protein Assay kit (Beyotime Institute of Biotechnology). A total of 30 μg protein was subjected to 12% SDS-PAGE and transferred onto a nitrocellulose membrane. The membrane was incubated in 1% bovine serum albumin containing primary rabbit anti-human polyclonal antibodies at 4°C overnight. Subsequently, the membrane was incubated with horseradish peroxidase-conjugated goat anti-rabbit antibody at room temperature for 1 h and the protein was detected using electro-chemiluminescence (Merck Millipore, Darmstadt, Germany). The following corresponding primary and secondary antibodies were used: Anti-IL17RC (1:1,000; cat. no. ab69673; Abcam, Cambridge, MA, USA); anti-tumor necrosis factor receptor (TNFR)-associated factor 6 (TRAF6; 1:2,000; cat. no. ab33915; Abcam); anti-nuclear factor (NF)-κB p65 (1:1,000; cat. no. 8242; Cell Signaling Technology, Inc., Danvers, MA, USA); anti-IKKe (1:2,500; cat. no. ab210927); anti-GFP (1:10,000; cat. no. ab183734); and anti-GAPDH (1:2,500; cat. no. ab9485; all Abcam) antibodies; and goat anti-mouse antibody (1:2,500; cat. no. CW0102M; Kangwei Biotech Co. Ltd., Beijing, China); and goat anti-rabbit antibody (1:2,500; cat. no. CW0103M; Kangwei Biotech Co. Ltd.). The blots were detected using a Kodak film developer (Fujifilm, Tokyo, Japan). The protein levels were quantified by densitometric analysis using Image-Pro Plus software version 6.0 (Media Cybernetics, Inc., Rockville, MD, USA). GAPDH was used as the endogenous control.
Statistical analysis
All statistical analyses were performed using SPSS software version 17.0 (SPSS, Inc., Chicago, IL, USA). Descriptive data for continuous variables are presented as the means ± standard deviation. The Student's t-test was used to compare the means between 2 groups. For the comparisons with the control group, statistical analyses were performed using one-way analysis of variance (ANOVA) with the post hoc Dunnett's test. A value of P<0.05 was considered to indicate a statistically significant difference. All experiments were performed 3 times.
Results
Wild-type and mutant IL17RC gene bacteria Sanger sequencing
The strains that were identified as positive using the electrophoresis of the PCR products were sequenced to obtain the wild-type and mutant IL17RC gene clones (Fig. 2), indicating that the wild-type and mutant IL17RC gene lentiviral packaging vectors were successfully constructed.
Figure 2.
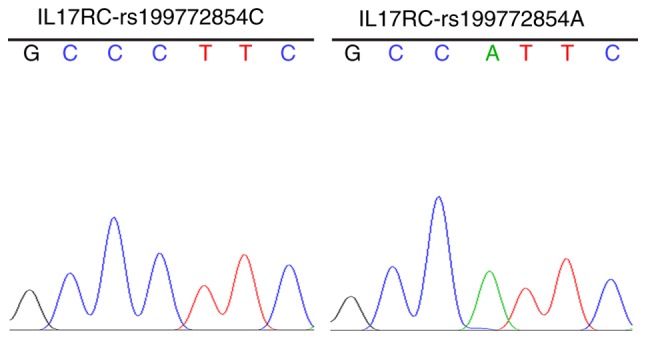
Wild-type and mutant IL17RC gene bacteria Sanger sequencing. Sanger sequencing of the strains that were identified as positive using electrophoresis of the PCR products. A allele, green line; T allele, red line; C allele, blue line; and G allele, black line. IL17RC, interleukin 17 receptor C.
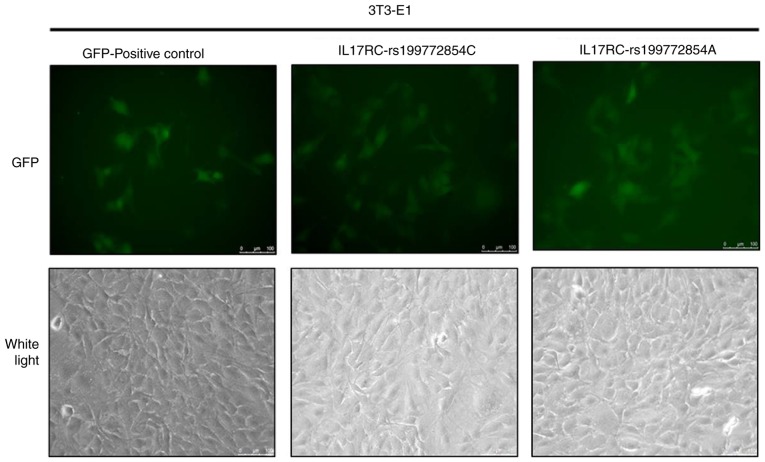
Lentivirus infection of the 3T3-E1 cells
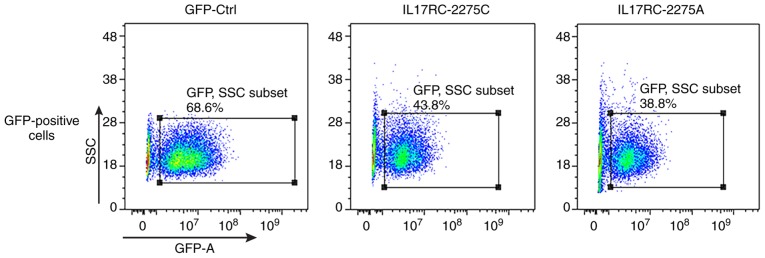
Significant levels of GFP expression were observed using fluorescence microscopy at 48 h after cell exposure (Fig. 3), which indicated that the 3T3-E1 cells were successfully infected. The 3T3-E1 cells were trypsinized at 72 h after they were infected, and the GFP-positive cells were sorted by flow cytometry, as shown in Fig. 4. The positive cells were plated and cultured, and then subjected to the subsequent osteogenesis induction experiments. Cell lines expressing the wild-type and mutant IL17RC gene were therefore obtained.
Figure 3.
Lentivirus infection of 3T3-E1 cells. 3T3-E1 cells were infected with pHIV-IL17RC-rs199772854C and pHIV-IL17RC-rs199772854A lentiviruses and significant GFP expression was identified. IL17RC, interleukin 17 receptor C.
Figure 4.
Flow cytometric sorting of the GFP-positive cells. After the 3T3-E1 cells were infected with the lentivirus, flow cytometry was used to detect the GFP-positive cells.
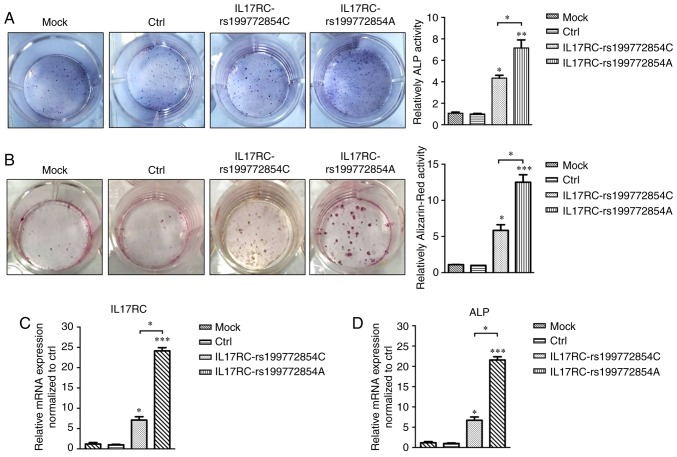
IL17RC gene rs199772854A site mutation induces the osteogenic differentiation of 3T3-E1 cells
Following 21 days of osteogenic induction, the ALP content of the IL17RC gene mutation group or the wild-type group was significantly higher than that of the mock group and the empty lentivirus control group. The ALP content of the IL17RC gene mutation group was significantly higher than that of the wild-type group (Fig. 5A). The Alizarin red content in the IL17RC gene mutation group or the wild-type group was significantly higher than that of the mock group and the empty lentivirus control group. The Alizarin red content of the IL17RC gene mutation group was significantly higher than that of the wild-type group (Fig. 5B). The results of RT-qPCR revealed that the mRNA expression of IL17RC in the rs199772854A transgenic IL17RC gene 3T3-E1 cell group was 18-, 24.1- and 3.4-fold higher than that of the mock group and the empty lentivirus control group along with wild-type group, respectively. The expression in the wild-type group was 6- and 7.1-fold higher than that in the mock group and the empty lentivirus control group, respectively (Fig. 5C). The mRNA expression of ALP in the rs199772854A transgenic IL17RC gene 3T3-E1 cell group was 20-, 21.6- and 3.2-fold higher than that of the mock group and the empty lentivirus control group along with wild-type group, respectively. The expression in the wild-type group was 5.6- and 7.1-fold higher than that in the mock group and the empty lentivirus control group, respectively (Fig. 5D). At the protein level, the expression of exogenous IL17RC in the cells was confirmed by western blot analysis of GFP tags. The expression levels of IL17RC, ALP, TRAF6, NF-κB and p-IKKe in the rs199772854A transgenic IL17RC gene 3T3-E1 cell group were higher than those in the mock group and the empty lentivirus control group along with rs199772854C transgenic IL17RC gene 3T3-E1 cell group, respectively. The protein expression of IKKe in the rs199772854A transgenic IL17RC gene 3T3-E1 cell group was higher than that in the mock group and control group, although no significance was observed between the mutation group and the wild-type group (Fig. 6). The expression of IKKe was enhanced in the mutation group and the wild-type group, whereas the expression of p-IKKe was further enhanced in the mutant group. As the lentivirus (MOI) was added to the cells in the different groups and the lentivirus-infected cells used in our study for ALP, mRNA and protein detection were collected after FACS sorting, the expression levels of ALP, mRNA and protein were not affected by MOI. These results suggest that the IL17RC gene mutation promotes the osteogenic differentiation of 3T3-E1 cells and may increase IL17RC gene expression by regulating the IL17 family signaling pathway.
Figure 5.
IL17RC gene rs199772854A site mutation induces osteogenic differentiation of 3T3-E1 cells. (A) After 21 days of osteogenic induction, the ALP content of the IL17RC gene mutation group and the non-mutation group was significantly higher than that of the control group. **P<0.01 and *P<0.05, vs. mock group and control group. The ALP content of the IL17RC gene mutation group was significantly higher than that of the non-mutation group. *P<0.05, compared with the non-mutation group. (B) The content of Alizarin red in the IL17RC gene mutation group and the non-mutation group was significantly higher than that of the mock group and control group. ***P<0.001 and *P<0.05, vs. mock group and control group. The Alizarin red content of the IL17RC gene mutation group was significantly higher than that of the non-mutation group. *P<0.05, compared with the non-mutation group. The mRNA expression levels of (C) IL17RC and (D) ALP in the IL17RC gene mutation group and non-mutation group normalized to the control group. ***P<0.001 and *P<0.05, vs. mock group and control group; *P<0.05, compared with the non-mutation group. IL17RC, interleukin 17 receptor C; ALP, alkaline phosphatase.
Figure 6.
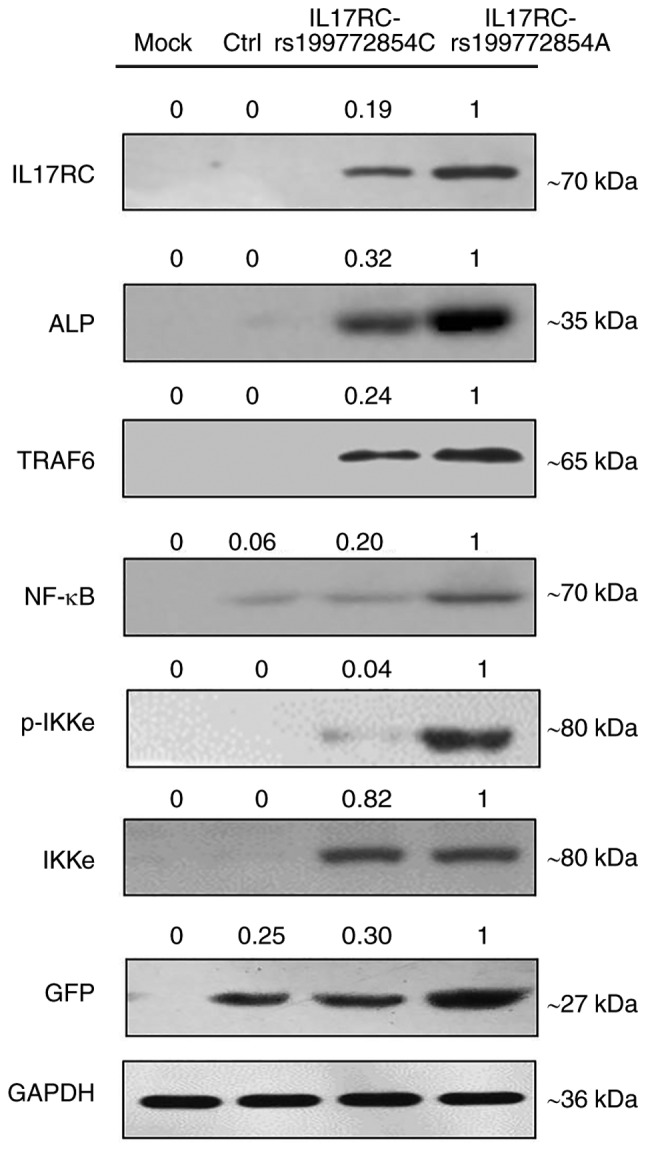
The expression levels of GFP-tagged IL17RC, ALP, TRAF6, NF-κB, p-IKKe and IKKe proteins detected by western blot analysis. IL17RC, interleukin 17 receptor C; ALP, alkaline phosphatase; TRAF6, tumor necrosis factor receptor (TNFR)-associated factor 6; NF-κB, nuclear factor-κB.
Discussion
The pathological process of T-OPLL involves the differentiation of posterior longitudinal ligament fibroblasts into osteoblasts. To verify the pathogenic potential of a mutation site that was identified in our previous study (24), IL17RC-induced mouse embryonic osteoblasts (3T3-E1) cells were established in the present study. In the present study, in the osteogenic differentiation model, the IL17RC gene carrying the rs199772854A and rs199772854C sites was transfected into mouse embryonic osteoblast (3T3-E1) cells, and osteogenic identification, western blot analysis and RT-qPCR were conducted at 21 days following osteogenic induction.
The rs199772854 locus of the IL17RC gene is located in the promoter region. SNPs of the promoter region can aggravate or attenuate transcriptional activity by affecting the binding efficiency of transcription factors and various action elements, consequently interfering with gene expression and potentially causing disease. The sequence of IL17RC mRNA exhibits no difference between the wild-type and mutant. Thus, RT-qPCR and the western blot analysis cannot distinguish between the wild-type and mutant IL17RC. The mutation in the promoter region just enhances the expression level of the IL17RC gene mRNA which further translates more IL17RC protein.
The results of the present study demonstrated that the IL17RC gene rs199772854A mutation significantly increased the expression of its own gene. Therefore, these results suggest that this mutation site exerts its biological functions through the overexpression of its own genes and the different influences of downstream signaling were enhanced by the mutation of IL17RC.
Previous studies have suggested that IL17RC plays a major role in disease pathogenesis through its function in the IL-17 signaling axis (32,33). IL17RC plays an indispensable role in the development of osteoblasts and accelerates the differentiation of osteoblasts (34). OPLL results in increased bone formation in the ligament tissue, and there is also an association between OPLL and increased bone mineral density in the body. Thus, IL17RC plays a potential role in the pathogenesis and progression of osteogenic diseases. IL17RC may act directly in the disease pathology or may have an indirect effect due to its function as a receptor for IL-17F and as a subunit of the IL-17R complex. Therefore, IL17RC is a potential therapeutic target in IL-17-dependent diseases (35).
TRAF6 is involved in the regulation of multiple signaling pathways as a key upstream regulatory molecule, including the IL-17 family signaling pathway (36). The overexpression of IL17RC can affect TRAF6 and increase the expression of NF-κB (37,38). The transcription factor NF-κB is a key factor in the expression of various genes that are regulated by the immune system and inflammatory response, proliferation, tumorigenesis and survival (39,40). The current study demonstrated that the ratio of phosphorylated IKKe versus total protein in the IL17RC gene mutation group was 18.5-fold higher than that of the wild-type group. IKKe is required for the activation of NF-κB through the phosphorylation and further degradation of IKKe. The inadequate regulation of NF-κB is associated with cancer, inflammation and autoimmune diseases, septic shock, viral infection and immune dysfunction. Therefore, NF-κB is a key signaling molecule between bone and the immune response. It has previously been demonstrated that environmental factors can cause PDGA-BB and TGF-β1 in ligament cells to stimulate NF-κB, which then influences the differentiation of undifferentiated mesenchymal cells into osteoblasts (41).
In the present study, mouse embryonic osteoblast (3T3-E1) cells were induced to differentiate into osteoblasts by transfecting the IL17RC gene carrying the rs199772854A site mutation into these cells. The potential mechanism of the IL17RC gene in T-OPLL was further investigated to confirm our hypothesis that the IL17RC gene regulates the development of T-OPLL through the IL17 family signaling pathway. Western blot analysis and RT-qPCR were used to analyze the in vitro model of osteogenesis. The results demonstrated that the expression levels of TRAF6 and NF-κB in the mouse embryonic osteoblast (3T3-E1) cells that were carrying the IL17RC gene rs199772854A site mutation were higher than those in the unmutated group.
There were some limitations to the present study. Due to the fact that the surgical removal of the posterior longitudinal ligament of the thoracic spine is complex and the risk level is high, the resected non-ossified posterior longitudinal ligament is too small. Hence, we were not able to carry out the primary cell culture of thoracic posterior longitudinal ligament during the current period. Although the classic cellular line utilized in osteogenic differentiation was the 3T3-E1 by its great osteogenic differentiation potential (42–44), using mouse cell lines may have some differences in the activation of the signaling pathway. Therefore, we will try to use posterior longitudinal ligament cell as target cell studying T-OPLL in the future. For the lentivirus system utilized in the establishment of wild-type and mutant cellular lines, the random insertion of the target is also a limitation of this study. In further studies, other systems will be considered for use in our research, such as AAV system. Furthermore, to the best of our knowledge, there are currently no reports available on the IL17RC gene related to C-OPLL. To ac complish this issue, we are currently collecting data from patients with C-OPLL and aim to verify whether the IL17RC gene is associated with C-OPLL in the future.
In conclusion, the present study suggests that the IL17RC gene rs199772854A site mutation causes significantly higher levels of IL17RC gene expression and TRAF6 and NF-κB protein expression compared to the rs199772854C site, indicating that the IL17RC gene rs199772854A loci mutation induces mouse embryonic osteoblasts towards osteogenic differentiation and may play a role in the pathogenesis of T-OPLL, and the IL17 signal axis may be involved in the regulation of T-OPLL disease. Therefore, the present study provides a theoretical basis for the early detection and diagnosis of T-OPLL diseases and the investigation of treatments other than surgery.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81672201).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
PW, SL and XiaogL conceived the study and designed the experiments. PW, YM, CK, and CL performed the experiments. ZT, LY, XiaoL and GH analyzed the data. PW wrote the manuscript. All authors reviewed the manuscript and all authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Fujimori T, Watabe T, Iwamoto Y, Hamada S, Iwasaki M, Oda T. Prevalence, concomitance, and distribution of ossification of the spinal ligaments: Results of whole spine CT scans in 1500 Japanese patients. Spine (Phila Pa 1976) 2016;41:1668–1676. doi: 10.1097/BRS.0000000000001643. [DOI] [PubMed] [Google Scholar]
- 2.Mori K, Imai S, Kasahara T, Nishizawa K, Mimura T, Matsusue Y. Prevalence, distribution, and morphology of thoracic ossification of the posterior longitudinal ligament in Japanese: Results of CT-based cross-sectional study. Spine (Phila Pa 1976) 2014;39:394–399. doi: 10.1097/BRS.0000000000000153. [DOI] [PubMed] [Google Scholar]
- 3.Yu LJ, Li WJ, Guo SG, Zhao Y. Transforaminal thoracic interbody fusion: Treatment of thoracic myelopathy caused by anterior compression. Orthopade. 2018;47:985–991. doi: 10.1007/s00132-018-3588-6. [DOI] [PubMed] [Google Scholar]
- 4.Imagama S, Ando K, Takeuchi K, Kato S, Murakami H, Aizawa T, Ozawa H, Hasegawa T, Matsuyama Y, Koda M, et al. Perioperative complications after surgery for thoracic ossification of posterior longitudinal ligament: Nationwide multicenter prospective study. Spine (Phila Pa 1976) 2018;43:E1389–E1397. doi: 10.1097/BRS.0000000000002703. [DOI] [PubMed] [Google Scholar]
- 5.Hu P, Yu M, Liu X, Liu Z, Jiang L. A circumferential decompression- based surgical strategy for multilevel ossification of thoracic posterior longitudinal ligament. Spine J. 2015;15:2484–2492. doi: 10.1016/j.spinee.2015.08.060. [DOI] [PubMed] [Google Scholar]
- 6.Ikegawa S. Genetics of ossification of the posterior longitudinal ligament of the spine: A mini review. J Bone Meta. 2014;21:127–132. doi: 10.11005/jbm.2014.21.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikegawa S. Genomic study of ossification of the posterior longitudinal ligament of the spine. Proc Jpn Acad Ser B Phys Biol Sci. 2014;90:405–412. doi: 10.2183/pjab.90.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakajima M, Kou I, Ohashi H, Genetic Study Group of the Investigation Committee on the Ossification of Spinal Ligaments. Ikegawa S. Identification and functional characterization of RSPO2 as a susceptibility gene for ossification of the posterior longitudinal ligament of the spine. Am J Hum Genet. 2016;99:202–207. doi: 10.1016/j.ajhg.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakajima M, Takahashi A, Tsuji T, Karasugi T, Baba H, Uchida K, Kawabata S, Okawa A, Shindo S, Takeuchi K, et al. A genome- wide association study identifies susceptibility loci for ossification of the posterior longitudinal ligament of the spine. Nat Genet. 2014;46:1012–1016. doi: 10.1038/ng.3045. [DOI] [PubMed] [Google Scholar]
- 10.Chen X, Guo J, Cai T, Zhang F, Pan S, Zhang L, Wang S, Zhou F, Diao Y, Zhao Y, et al. Targeted next- generation sequencing reveals multiple deleterious variants in OPLL- associated genes. Sci Rep. 2016;6:26962. doi: 10.1038/srep26962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Liu D, Yang Z, Tian B, Li J, Meng X, Wang Z, Yang H, Lin X. Association of bone morphogenetic protein- 2 gene polymorphisms with susceptibility to ossification of the posterior longitudinal ligament of the spine and its severity in Chinese patients. Eur Spine J. 2008;17:956–964. doi: 10.1007/s00586-008-0651-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren Y, Feng J, Liu Z, Wan H, Li J, Lin X. A new haplotype in BMP4 implicated in ossification of the posterior longitudinal ligament (OPLL) in a Chinese population. J Orhop Res. 2012;30:748–756. doi: 10.1002/jor.21586. [DOI] [PubMed] [Google Scholar]
- 13.Ren Y, Liu ZZ, Feng J, Wan H, Li JH, Wang H, Lin X. Association of a BMP9 haplotype with ossification of the posterior longitudinal ligament (OPLL) in a Chinese population. PLoS One. 2012;7:e40587. doi: 10.1371/journal.pone.0040587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang H, Shi L, Shi G, Guo Y, Chen D, Chen D, Shi J. Connexin 43 affects osteogenic differentiation of the posterior longitudinal ligament cells via regulation of ERK activity by stabilizing Runx2 in ossification. Cell Physiol Biochem. 2016;38:237–247. doi: 10.1159/000438625. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Zhao Y, Chen Y, Shi G, Yuan W. RUNX2 polymorphisms associated with OPLL and OLF in the Han population. Clin Orthop Relat Res. 2010;468:3333–3341. doi: 10.1007/s11999-010-1511-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu C, Chen Y, Zhang H, Chen Y, Shen X, Shi C, Liu Y, Yuan W. Integrated microRNA- mRNA analyses reveal OPLL specific microRNA regulatory network using high- throughput sequencing. Sci Rep. 2016;6:21580. doi: 10.1038/srep21580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong Q, Ma X, Li F, Guo Z, Qi Q, Li W, Yuan H, Wang Z, Chen Z. COL6A1 polymorphisms associated with ossification of the ligamentum flavum and ossification of the posterior longitudinal ligament. Spine (Phila Pa 1976) 2007;32:2834–2838. doi: 10.1097/BRS.0b013e31815b761c. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka T, Ikari K, Furushima K, Okada A, Tanaka H, Furukawa K, Yoshida K, Ikeda T, Ikegawa S, Hunt SC, et al. Genomewide linkage and linkage disequilibrium analyses identify COL6A1, on chromosome 21, as the locus for ossification of the posterior longitudinal ligament of the spine. Am J Hum Genet. 2003;73:812–822. doi: 10.1086/378593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei W, He HL, Chen CY, Zhao Y, Jiang HL, Liu WT, Du ZF, Chen XL, Shi SY, Zhang XN. Whole exome sequencing implicates PTCH1 and COL17A1 genes in ossification of the posterior longitudinal ligament of the cervical spine in Chinese patients. Genet Mol Res. 2014;13:1794–1804. doi: 10.4238/2014.March.17.7. [DOI] [PubMed] [Google Scholar]
- 20.Kamiya M, Harada A, Mizuno M, Iwata H, Yamada Y. Association between a polymorphism of the transforming growth factor-beta1 gene and genetic susceptibility to ossification of the posterior longitudinal ligament in Japanese patients. Spine (Phila Pa 1976) 2001;26:1266–1267. doi: 10.1097/00007632-200106010-00017. [DOI] [PubMed] [Google Scholar]
- 21.Chung WS, Nam DH, Jo DJ, Lee JH. Association of toll- like receptor 5 gene polymorphism with susceptibility to ossification of the posterior longitudinal ligament of the spine in Korean population. J Korean Neurosurg Soc. 2011;49:8–12. doi: 10.3340/jkns.2011.49.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He Z, Zhu H, Ding L, Xiao H, Chen D, Xue F. Association of NPP1 polymorphism with postoperative progression of ossification of the posterior longitudinal ligament in Chinese patients. Genet Mol Res. 2013;12:4648–4655. doi: 10.4238/2013.October.18.3. [DOI] [PubMed] [Google Scholar]
- 23.Guo Q, Lv SZ, Wu SW, Tian X, Li ZY. Association between single nucleotide polymorphism of IL15RA gene with susceptibility to ossification of the posterior longitudinal ligament of the spine. J Orthop Surg Res. 2014;9:103. doi: 10.1186/s13018-014-0103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang P, Liu X, Zhu B, Ma Y, Yong L, Teng Z, Wang Y, Liang C, He G, Liu X. Identification of susceptibility loci for thoracic ossification of the posterior longitudinal ligament by whole- genome sequencing. Mol Med Rep. 2018;17:2557–2564. doi: 10.3892/mmr.2017.8171. [DOI] [PubMed] [Google Scholar]
- 25.Wang P, Liu X, Zhu B, Ma Y, Yong L, Teng Z, Liang C, He G, Liu X. Association of IL17RC and COL6A1 genetic polymorphisms with susceptibility to ossification of the thoracic posterior longitudinal ligament in Chinese patients. J Orthop Surg Res. 2018;13:109. doi: 10.1186/s13018-018-0817-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho AW, Gaffen SL. IL- 17RC: A partner in IL- 17 signaling and beyond. Semin Immunopathol. 2010;32:33–42. doi: 10.1007/s00281-009-0185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dhaouadi T, Chahbi M, Haouami Y, Sfar I, Abdelmoula L, Ben Abdallah T, Gorgi Y. IL- 17A, IL- 17RC polymorphisms and IL17 plasma levels in Tunisian patients with rheumatoid arthritis. PLoS One. 2018;13:e0194883. doi: 10.1371/journal.pone.0194883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roos AB, Mori M, Gura HK, Lorentz A, Bjermer L, Hoffmann HJ, Erjefält JS, Stampfli MR. Increased IL- 17RA and IL- 17RC in end- stage COPD and the contribution to mast cell secretion of FGF- 2 and VEGF. Respir Res. 2017;18:48. doi: 10.1186/s12931-017-0534-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Righetti RF, Dos Santos TM, Camargo LDN, Aristóteles LRCRB, Fukuzaki S, de Souza FCR, Santana FPR, de Agrela MVR, Cruz MM, Alonso-Vale MIC, et al. Protective effects of anti- IL17 on acute lung injury induced by LPS in mice. Front Pharmacol. 2018;9:1021. doi: 10.3389/fphar.2018.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato K, Suematsu A, Okamoto K, Yamaguchi A, Morishita Y, Kadono Y, Tanaka S, Kodama T, Akira S, Iwakura Y, et al. Th17 functions as an osteoclastogenic helper T cell subset that links T cell activation and bone destruction. J Exp Med. 2006;203:2673–2682. doi: 10.1084/jem.20061775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kotake S, Udagawa N, Takahashi N, Matsuzaki K, Itoh K, Ishiyama S, Saito S, Inoue K, Kamatani N, Gillespie MT, et al. IL- 17 in synovial fluids from patients with rheumatoid arthritis is a potent stimulator of osteoclastogenesis. J Clin Invest. 1999;103:1345–1352. doi: 10.1172/JCI5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campfield BT, Eddens T, Henkel M, Majewski M, Horne W, Chaly Y, Gaffen SL, Hirsch R, Kolls JK. Follistatin- like protein 1 modulates IL- 17 signaling via IL- 17RC regulation in stromal cells. Immunol Cell Biol. 2017;95:656–665. doi: 10.1038/icb.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Luca A, Pariano M, Cellini B, Costantini C, Villella VR, Jose SS, Palmieri M, Borghi M, Galosi C, Paolicelli G, et al. The IL- 17F/IL- 17RC axis promotes respiratory allergy in the proximal airways. Cell Rep. 2017;20:1667–1680. doi: 10.1016/j.celrep.2017.07.063. [DOI] [PubMed] [Google Scholar]
- 34.Zhou S, Qiu XS, Zhu ZZ, Wu WF, Liu Z, Qiu Y. A single- nucleotide polymorphism rs708567 in the IL- 17RC gene is associated with a susceptibility to and the curve severity of adolescent idiopathic scoliosis in a Chinese Han population: A case- control study. BMC Musculoskelet Disord. 2012;13:181. doi: 10.1186/1471-2474-13-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang H, Kim HJ, Chang EJ, Lee ZH, Hwang SJ, Kim HM, Lee Y, Kim HH. IL- 17 stimulates the proliferation and differentiation of human mesenchymal stem cells: Implications for bone remodeling. Cell Death Differ. 2009;16:1332–1343. doi: 10.1038/cdd.2009.74. [DOI] [PubMed] [Google Scholar]
- 36.Yin Y, Li F, Shi J, Li S, Cai J, Jiang Y. MiR- 146a regulates inflammatory infiltration by macrophages in polymyositis/dermatomyositis by targeting TRAF6 and affecting IL- 17/ICAM-1 pathway. Cell Physiol Biochem. 2016;40:486–498. doi: 10.1159/000452563. [DOI] [PubMed] [Google Scholar]
- 37.Qu F, Gao H, Zhu S, Shi P, Zhang Y, Liu Y, Jallal B, Yao Y, Shi Y, Qian Y. TRAF6- dependent Act1 phosphorylation by the IκB kinase- related kinases suppresses interleukin- 17- induced NF-κB activation. Mol Cell Biol. 2012;32:3925–3937. doi: 10.1128/MCB.00268-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang SH, Dong C. Signaling of interleukin- 17 family cytokines in immunity and inflammation. Cell Signal. 2011;23:1069–1075. doi: 10.1016/j.cellsig.2010.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C, Petriello MC, Zhu B, Hennig B. PCB 126 induces monocyte/macrophage polarization and inflammation through AhR and NF-κB pathways. Toxicol Appl Pharmacol. 2019;367:71–81. doi: 10.1016/j.taap.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng Y, Bi R, Guo H, Yang J, Du Y, Wang C, Wei W. Andrographolide enhances TRAIL- induced apoptosis via p53- mediated death receptors up- regulation and suppression of the NF-κB pathway in bladder cancer cells. Int J Biol Sci. 2019;15:688–700. doi: 10.7150/ijbs.30847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kosaka T, Imakiire A, Mizuno F, Yamamoto K. Activation of nuclear factor kappaB at the onset of ossification of the spinal ligaments. J Orthop Sci. 2000;5:572–578. doi: 10.1007/s007760070008. [DOI] [PubMed] [Google Scholar]
- 42.Damsongsang P, Chaikiawkeaw D, Phoolcharoen W, Rattanapisit K, Kaewpungsup P, Pavasant P, Hoven VP. Surface- immobilized plant- derived osteopontin as an effective platform to promote osteoblast adhesion and differentiation. Colloids Surf B Biointerfaces. 2019;173:816–824. doi: 10.1016/j.colsurfb.2018.10.032. [DOI] [PubMed] [Google Scholar]
- 43.Ye M, Shi B. Zirconia nanoparticles- induced toxic effects in osteoblast- like 3T3- E1 cells. Nanoscale Res Lett. 2018;13:353. doi: 10.1186/s11671-018-2747-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee YH, Bhattarai G, Park IS, Kim GR, Kim GE, Lee MH, Yi HK. Bone regeneration around N- acetyl cysteine- loaded nano-tube titanium dental implant in rat mandible. Biomaterials. 2013;34:10199–10208. doi: 10.1016/j.biomaterials.2013.08.080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.