Abstract
Kelch-like ECH-associated protein 1 (Keap1)/nuclear factor erythroid 2-related factor 2 (Nrf2) signaling has a protective effect on normal cells. A number of previous studies demonstrated that Keap1/Nrf2 signaling is associated with drug resistance in numerous tumors. The aim of the present study was to investigate the roles of Keap1 in renal cell carcinoma (RCC) and its effect on sensitivity to chemotherapy. Reverse transcription-quantitative polymerase chain reaction was used to detect the mRNA expression of Keap1 in 45 cases of RCC tumors and adjacent normal tissues. A total of five randomly selected patients with RCC, five RCC cell lines and normal renal tubular cells were examined to detect the protein and mRNA expressions of Keap1. The 5-year survival rate was analyzed by Kaplan-Meier analysis. The cell viability was assessed by a Cell Counting kit-8 assay. The cell apoptosis and reactive oxygen species (ROS) were determined by flow cytometry. The expressions of associated proteins were determined by western blot analysis. It was identified that in RCC tissues and RCC cell lines, the expression of Keap1 was downregulated, which was considered to be associated with poor prognosis. In total, 1 µM Axitinib significantly decreased cell viability, promoted ROS release and induced cell apoptosis in ACHN cells. Silencing Keap1 was able to reverse the inhibitory effect of Axitinib and enhance the protein expressions of Nrf2, NAD(P)H dehydrogenase [quinone] 1 and heme oxygenase 1. However, silencing Nrf2 increased the cell sensitivity to Axitinib. Under Axitinib condition, overexpressing Nrf2 was able to increase cell viability; however, overexpressing Keap1 resulted in an opposite effect. Keap1 serves as a tumor suppressor; its low expression was associated with poor prognosis and a decreased sensitivity of RCC cells to Axitinib. A possible mechanism underlying Axitinib resistance may involve Nrf2 overexpression.
Keywords: Kelch-like ECH-associated protein 1, nuclear factor erythroid 2-related factor 2, prognosis, Axitinib resistance, renal cell carcinoma
Introduction
Renal cell carcinoma (RCC) is a common malignant tumor in the urinary system. Among all the malignant tumors of the urinary system, the incidence rate of RCC is third following prostate cancer and bladder cancer, accounting for 2.7% of male malignant tumors worldwide in 2018 (1). Annually, there are >200,000 novel diagnosed cases of renal cancer and >100,000 mortalities occur worldwide. According to the latest statistics of the American Cancer Society, the morbidity and mortality of RCC in the last two years were among the leading 10 types of malignant tumors with highest morbidity and mortality (2,3). The morbidity and mortality of male patients with RCC are significantly higher compared with women (2,3). In recent years, with the emergence of molecular targeted drugs, the prognosis of patients with RCC has significantly improved; the clinical effect of molecular targeted drugs has been fully tested (4,5). Axitinib, a second generation vascular endothelial growth factor receptor inhibitor approved by the US Food and Drug Administration in 2012, is a small molecular tyrosine kinase inhibitor and a multi-target drug used for patients with advanced RCC that are not responding to other systemic therapies (6,7). Axitinib selectively acts on multiple targets, blocking the proliferation of tumor cells by blocking specific targets (8,9).
As a cytoplasmic protein, Kelch-like ECH-associated protein 1 (Keap1) is an important regulatory factor of the oxidative stress signaling pathway. It has five protein domains, which are in the N-terminal region (Broad complex, Tramtrack, Bric-a-Brac, linker intervening region and Kelch domain) and a C-terminal region (10). Numerous previous studies demonstrated that the expression of Keap1 in tumor tissues was significantly different compared with adjacent tissues. In particular, the Keap1 gene was inactivated or mutated at different structural sites in cancer tissues of patients with lung, ovarian and liver cancer (11-13). In a previous clinical analysis, it was identified that low expression of Keap1 was associated with a high mortality in patients with ovarian cancer (14).
The specific mechanism of Keap1 regulating tumorigenesis and development remains unclear. However, it was hypothesized that the progression of tumor is affected by three different ways; Keap1/nuclear factor erythroid 2-related factor 2 (Nrf2) signaling, Keap1/inhibitor of nuclear factor κ-B kinase subunit b/nuclear factor-κB signaling and Keap1/B cell lymphoma-2 (15-21). Keap1 forms a complex with cullin-3 (a ubiquitin ligase E3), which subsequently binds to a key transcription factor of cellular oxidative stress, Nrf2. It ubiquitinates and degrades Nrf2, thus regulating the Nrf2-antioxidant responsive element (ARE) signaling pathway and affecting anti-oxidation protein and phase II detoxification enzyme expression, which in turn affects tumor cell proliferation, apoptosis and sensitivity to chemotherapy drugs and radiation therapy (22-26). The aim of the present study was to investigate the effect of Keap1 on Axitinib resistance of RCC.
Patients and methods
Patients and tissue samples
All tissue specimens were collected from patients with RCC admitted to The Second Affiliated Hospital of Guangxi Medical University (Nanning, China) between May 2015 and April 2016. It was confirmed that all patients were diagnosed with RCC by pathology. All patients that were treated for the first time for renal tumor, clear cell RCC, papillary RCC, chromophobe RCC, multilocular cystic RCC and renal medullary carcinoma were included in the present study. All patients who had received preoperative radiotherapy, chemotherapy and cancer drug therapy were excluded from the present study. A portion of the RCC tissues and adjacent normal tissues were stored in 4% formaldehyde solution for pathological diagnosis routinely, and the rest were stored in a liquid nitrogen tank to perform reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blot analysis. In total, 45 cases of RCC and corresponding adjacent tissues were analyzed. The present study was approved by the Ethics Committee of The Second Affiliated Hospital of Guangxi Medical University. All tissue samples were obtained with written informed consent from the patients.
Cell culture
Normal human renal tubular epithelial cells (HK-2 cells) and five RCC cell lines (ACHN, 769-P, A-498, 786-0 and Caki-1) were obtained from China Union Medical University Basic Medical Cell Center (China). The cells were cultured in RPMI 1640 medium (HyClone; GE Healthcare Life Sciences, Logan, UT, USA) containing 10% fetal bovine serum (FBS; HyClone; GE Healthcare Life Sciences), 100 µ/ml penicillin and 100 µg/ml streptomycin, and placed in an incubator with 5% CO2 at 37°C. When confluence reached 70-80%, the cells were digested and subcultured.
Cell Counting kit-8 (CCK-8)
Cells were plated into 96-well plates at a density of 1×104 cells/well for 24 h. Subsequent to the cells being treated with different concentrations (0.1, 1, 10, 50 and 100 µM) of Axitinib (cat. no. PZ0193; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) or transfected at 37°C, 10 µl CCK-8 solution (Invitrogen; Thermo Fisher Scientific, Inc.) was added in each well and incubated for another 3 h at 37°C. Cell viability was determined by recording the optical density at a test wavelength of 450 nm using a microplate reader (Thermo Fisher Scientific, Inc.).
Cell transfection
ACHN cells were seeded in a 6-well plate (1×105) for 24 h prior to transfection. Small interfering (si)Keap1, siNrf2, overexpressing Keap1, overexpressing Nrf2 and empty control plasmids were purchased from Shanghai GeneChem Co., Ltd. (Shanghai, China). The sequence of siKeap1 was 5′-GAA TGA TCA CAG CAA TGA A-3′; the sequence of siNrf2 was 5′-GGT TGC CCA CAT TCC CAA ATC-3′; the sequence for overexpressing Keap1 was 5′-ATA CTC GAG ATG CAG CCA GAT CCC AGG CC-3′; the sequence for overexpressing Nrf2 was 5′-GTA CTA GTA TGA TGG ACT TGG AGT T-3′; and the NC sequence was 5′-GTT CTC CGA ACG TGT CAC GT-3′. Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used to perform the transient transfection, according to the manufacturer′s protocol. In total, 2 µg si/overexpressing RNA, negative control (NC) and Lipofectamine® 2000 were added to Opti-Minimum Essential Medium (Gibco; Thermo Fisher Scientific, Inc.) and incubated at 25°C for 20 min. Lipofectamine® 2000 was subsequently mixed into each well, which was cultured in Opti-MEM RPMI 1640. After 6 h of culturing, the fluid was replaced with RPMI 1640 medium containing 10% FBS.
Flow cytometry
The collected cells were stained with Annexin V-fluorescein isothiocyanate and propidium iodide (cat. no. CA1020; Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) at 4°C for 15 min. Within 1 h, a flow cytometer and analysis software (BD CellQuest™ Pro Software; version 1.2; BD Biosciences, San Jose, CA, USA) was used to assess cell apoptosis.
The reactive oxygen species (ROS) level was detected by 2,7-dichlorodi-hydrofluorescein diacetate (DCFH-DA; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Cells were treated with Axitinib or siKeap1 as previously described and 2×106 cells/ml were seeded into a 6-well plate. In total, 10 µM DCFH-DA with 1 ml PBS were added to cells at 37°C for 20 min. DCF fluorescence was determined using a flow cytometer and the data were analyzed using Summit Software v4.3 (Dako; Agilent Technologies, Inc., Santa Clara, CA, USA).
RT-qPCR
Total RNA from tissues or cultured cells were obtained using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RNA was reverse transcribed into cDNA using GoScript™ RT kit (Promega Corporation, Madison, WI, USA). The RT temperature protocol consisted of 37°C for 15 min and at 85°C for 5 sec. RT-qPCR was conducted using SYBR Fast qPCR Mix (Invitrogen; Thermo Fisher Scientific, Inc.) for Keap1 and GAPDH. The primer sequences used were as follows: Keap1: Sense, 5′-CTG GAG GAT CAT ACC AAG CAG G-3′ and antisense, 5′-GGA TAC CCT CAA TGG ACA CCA C-3′; Nrf2 sense, 5′-CAA CTC AGC ACC TTG T AT C-3′ and antisense, 5′-TTC TTA GTA TCT GGC TT C TT-3′; GAPDH sense, 5′-GAA GGT GAA GGT CGG AGT C-3′ and antisense, 5′-GAA GAT GGT GAT GGG ATT TC-3′. The cycling parameters were set as: 95°C for 5 min, followed by 40 cycles at 95°C for 15 sec, and 60°C for 30 sec. Subsequent to completing PCR amplification, comparative quantification was determined using the 2−ΔΔCq method (27). Primers were purchased from BioSune Biotechnology (Shanghai, China).
Western blot analysis
Total protein was extracted from tissues or cultured cells using radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Haimen, China). Protein concentrations were quantified using a bicinchoninic acid protein kit (Beyotime Institute of Biotechnology). Aliquots of protein (30 µg) were separated by 12% SDS-PAGE, and resolved proteins were transferred onto polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked in 5% milk PBS with 0.1% Triton X-100 at 4°C for 2 h, and incubated with different primary antibodies at 4°C overnight as follows: Anti-Keap1 antibody (cat. no. ab119403; 1:1,000; Abcam, Cambridge, UK), anti-Nrf2 antibody (cat. no. ab156883; 1:1,000), anti-Lamin B1 antibody (cat. no. ab133741; 1:1,000), anti-NAD(P)H dehydrogenase [quinone] 1 (NQO1) antibody (cat. no. ab28947; 1:1,000), anti-heme oxygenase 1 (HO1) antibody (cat. no. ab13243; 1:2,000), anti-phosphorylated (p)-extracellular signal-regulated kinase (ERK) antibody (cat. no. ab223500; 1:400), anti-ERK antibody (cat. no. ab17942; 1:1,000) and anti-GAPDH antibody (cat. no. ab9485; 1:2,500; all Abcam). The membranes were subsequently incubated with appropriate horseradish peroxidase-conjugated secondary antibodies (anti-rabbit; cat. no. ab205718; 1:2,000 and anti-mouse; cat. no. ab205719; 1:5,000) for 2 h at room temperature. Protein bands were detected with enhanced chemiluminescence (Thermo Fisher Scientific, Inc.) and visualized using Quantity One software, version 4.6.9 (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Statistical analysis
Statistical analysis was detected by Prism GraphPad version 6.0 software (GraphPad Software, Inc., La Jolla, CA, USA). The correlation between Keap1 expression and the survival was analyzed using the Kaplan-Meier method. All data are presented as the mean ± standard deviation from three independent experiments. χ2 test was used to analyze the discontinuous data. Differences were performed using one-way analysis of variance and Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Keap1 is downregulated in RCC tissues and is associated with a poor survival rate
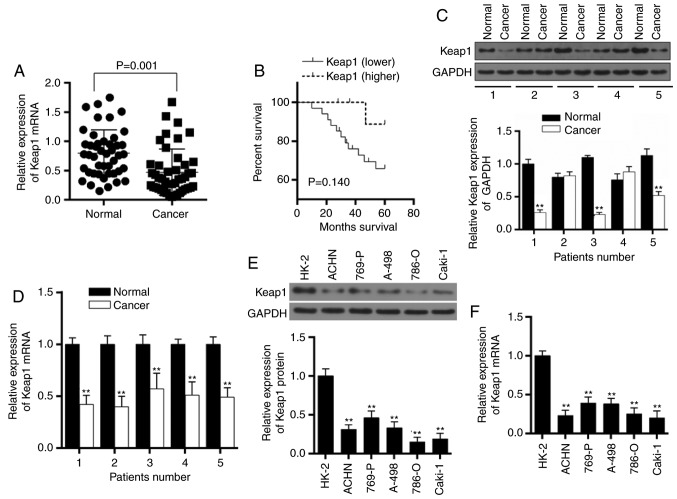
A total of 45 patients with RCC were recruited in the present study and the mRNA expressions of Keap1 in cancer and adjacent normal tissues were detected. The result demonstrated that Keap1 mRNA expressions were lower in cancer tissues compared with normal paired tissues (P=0.001; Fig. 1A). The association between Keap1 expression in RCC and patient survival was analyzed, and it was identified that low Keap1 expression was associated with a poor 5-year survival rate (P=0.140; Fig. 1B). A total of five patients with RCC were subsequently randomly selected to assess the expressions of Keap1 at the protein and mRNA expression level. The western blot analysis and RT-qPCR demonstrated that the expressions of Keap1 significantly decreased in three patients with cancer (P<0.01; Fig. 1C and D); however, an increased expression of Keap1 was detected in specific patients with cancer suggesting that the phenomenon may be affected by individual differences or different subtypes of RCC. The patients were divided into high and low Keap1 expression group based on the median value (0.36) of the relative Keap1 mRNA expression. The association between Keap1 expression and pathological classification data is presented in Table I. The low expression of Keap1 was identified to be associated with the recurrence of RCC.
Figure 1.
Low expression of Keap1 in RCC is associated with a poor survival rate. (A) In total, 45 cases of patients with RCC were analyzed to determine the mRNA expression of Keap1 in normal and cancer tissues (P=0.001). (B) Kaplan-Meier survival curve was determined to assess the association between the expression of Keap1 and survival rate (P=0.140). A total of five patients with RCC were randomly selected to assess the expression of Keap1 at the (C) protein and (D) mRNA level. GAPDH served as an internal control. Data are presented as the mean ± standard deviation from three independent experiments. **P<0.01 vs. respective normal RCC tissue. (E) Western blotting was used to detect the protein expression in normal human renal tubular epithelial cells (HK-2 cells) and five RCC cell lines (ACHN, 769-P, A-498, 786-0 and Caki-1). (F) Reverse transcription-quantitative polymerase chain reaction was used to detect the mRNA expression in normal human renal tubular epithelial cells and five RCC cell lines. GAPDH served as an internal control. Data are presented as the mean ± standard deviation from three independent experiments. **P<0.01 vs. HK-2 cells. Keap1, Kelch-like ECH-associated protein 1; RCC, renal cell carcinoma.
Table I.
Analysis between the clinical features and expression of Keap1 in patients with renal cell carcinoma.
| Characteristics | n | Keap1 expression
|
P-value | |
|---|---|---|---|---|
| Low group | High group | |||
| Age, years | 60±5.2 | 59±6.2 | 0.560 | |
| Sex | 0.556 | |||
| Male | 22 | 10 | 12 | |
| Female | 23 | 13 | 10 | |
| Location | 0.554 | |||
| Left | 24 | 11 | 13 | |
| Right | 21 | 12 | 9 | |
| T stage | 0.514 | |||
| T1 | 32 | 17 | 15 | |
| T2 | 12 | 5 | 7 | |
| T3 | 1 | 1 | 0 | |
| T4 | 0 | 0 | 0 | |
| Recurrence | 0.003 | |||
| No | 22 | 6 | 16 | |
| Yes | 23 | 17 | 6 | |
Keap1, Kelch-like ECH-associated protein 1.
Keap1 is downregulated in RCC cell lines
The expression of Keap1 in normal human renal tubular epithelial cells (HK-2 cells) and five RCC cell lines (ACHN, 769-P, A-498, 786-0 and Caki-1) was detected. All RCC cells demonstrated a downregulation of Keap1 protein (P<0.01; Fig. 1E) and mRNA expression (P<0.01; Fig. 1F), compared with normal HK-2 cells.
Inhibitory effect of Axitinib on RCC cell lines
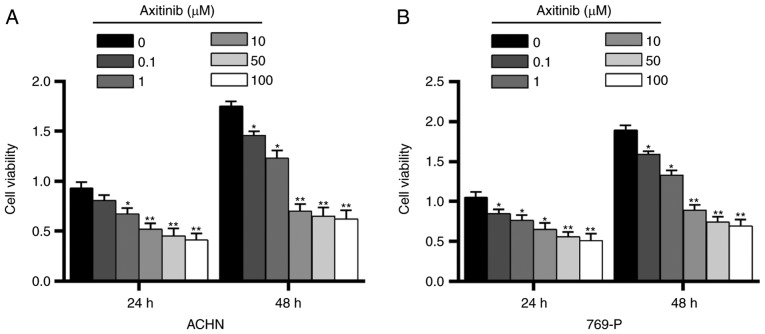
In order to test the effect of Axitinib on RCC cell lines, a CCK-8 assay was performed and different concentrations (0.1, 1, 10, 50 and 100 µM) of Axitinib were used to treat two of the five RCC cell lines, ACHN and 769-P cells, which were randomly selected, at 37°C for 24 and 48 h. In ACHN and 769-P cells, it was observed that Axitinib decreased the cell viability in a dose-dependent manner (Fig. 2). In ACHN cells, 1 µM Axitinib was able to significantly attenuate cell viability for 24 h of treatment (P<0.05; Fig. 2A) and 0.1 µM Axitinib treatment for 48 h was able to significantly decrease cell viability (P<0.05). In 769-P cells, treatment with 0.1 µM Axitinib for 24 and 48 h significantly decreased cell viability (P<0.05; Fig. 2B). The results suggested that Axitinib had a similar inhibitory effect in ACHN and 769-P cells. Therefore, one cell line, ACHN, was randomly selected and 1 µM Axitinib was selected for subsequent experimentation.
Figure 2.
Different concentrations of Axitinib are administered to renal cell carcinoma cell lines. Cell Counting kit-8 was used to detect the effect of different concentrations (0.1, 1, 10, 50 and 100 µM) of Axitinib on (A) ACHN and (B) 769-P cells for 24 and 48 h. Data are presented as the mean ± standard deviation from three independent experiments. *P<0.05, **P<0.01 vs. respective 0 µM Axitinib.
Transfection efficiencies of Keap1 and Nrf2 on ACHN cells
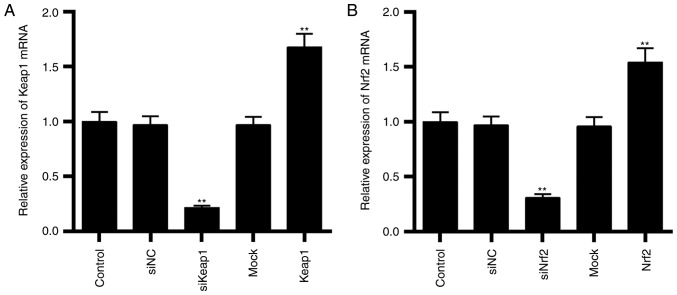
The transfection efficiencies of Keap1 and Nrf2 were assessed by RT-qPCR. It was demonstrated that the mRNA expressions of Keap1 (P<0.01; Fig. 3A) were downregulated by transfecting cells with siKeap1 and were upregulated by overexpressing Keap1 (P<0.01; Fig. 3A). As presented in Fig. 3B, the mRNA expressions of Nrf2 (P<0.01; Fig. 3B) were downregulated by transfecting cells with siNrf2 and were upregulated by overexpressing Nrf2 (P<0.01; Fig. 3B).
Figure 3.
Transfection efficiencies of Keap1 and Nrf2 in ACHN cells. (A) Keap1 was silenced and overexpressed in cells and the mRNA expression of Keap1 was determined by RT-qPCR. (B) Nrf2 was silenced and overexpressed in cells and the mRNA expression of Nrf2 was determined by RT-qPCR. Data are presented as the mean ± standard deviation from three independent experiments. **P<0.01 vs. control. Keap1, Kelch-like ECH-associated protein 1; Nrf2, nuclear factor erythroid 2-related factor 2; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; si, small interfering; NC, negative control.
Silencing Keap1 attenuates the inhibitory effect of Axitinib on ACHN cells
As presented in Fig. 4A, treatment with 1 µM Axitinib was able to significantly decrease cell viability, compared with the control (P<0.01; Fig. 4A); however, silencing Keap1 demonstrated an opposite result that cell viability was significantly increased compared with the Axitinib group (P<0.01). Cell viability was increased by siKeap1 alone compared with treatment with Axitinib alone (P<0.05). The result suggested that silencing Keap1 attenuated the inhibitory effect of Axitinib on ACHN cells.
Figure 4.
Effect of silencing Keap1 on the cell viability, ROS level and protein expression of Nrf2 and ERK signaling. ACHN cells were transfected with siKeap1 and divided into five groups, which were the control, Ax (1 µM Ax), NC+Ax, siKeap1+Ax and siKeap1 groups. (A) Cell viability was determined by a Cell Counting kit-8 assay. (B) ROS levels are presented in a bar graph. (C) Flow cytometry was used to detect the ROS level. (D) Nrf2 protein expression was detected by western blotting. Lamin B1 served as an internal control. (E) Keap1 protein expression was detected by western blotting. GAPDH served as an internal control. (F) NQO1, HO1, p-ERK and ERK protein expressions were detected by western blotting. GAPDH served as an internal control. Data are presented as the mean ± standard deviation from three independent experiments. *P<0.05, **P<0.01 vs. control; #P<0.05, ##P<0.01. Keap1, Kelch-like ECH-associated protein 1; ROS, reactive oxygen species; Nrf2, nuclear factor erythroid 2-related factor 2; ERK, extracellular signal-regulated kinase; Ax, Axitinib; NC, negative control; si, small interfering; NQO1, anti- NAD(P)H dehydrogenase [quinone] 1; HO1, heme oxygenase 1; p-, phosphorylated.
Silencing Keap1 decreases the ROS level induced by Axitinib in ACHN cells
Keap1 is known to be associated with oxidative stress (28). Therefore, the ROS level was detected by flow cytometry. The ROS level was significantly increased with treatment with Axitinib in ACHN cells compared with the control cells (P<0.01; Fig. 4B and C), suggesting that Axitinib had a damaging effect on RCC cells. However, Axitinib in combination with silencing Keap1 significantly decreased the ROS level compared with Axitinib treatment alone (P<0.01; Fig. 4B).
Silencing Keap1 enhances the expression of Nrf2 and promotes ERK signaling
In order to understand how siKeap1 affects ACHN cell sensitivity to Axitinib, the protein expression of Nrf2, and downstream target genes of Nrf2 and ERK signaling were detected. It was identified that treatment with Axitinib was able to promote the protein expression of Nrf2 (P<0.01; Fig. 4D) and downregulate the expression of Keap1 (P<0.01; Fig. 4E) compared with the control cells. Silencing Keap1 significantly promoted the effect of Axitinib by further upregulating the expression of Nrf2 protein (P<0.01; Fig. 4D). When silencing Keap1 was combined with treatment with Axitinib, the expression of Keap1 protein was further downregulated and was significantly decreased compared with treatment with Axitinib alone (P<0.05; Fig. 4E). The downstream target proteins of Nrf2 demonstrated a similar result to that of Nrf2 protein. As presented in Fig. 4F, treatment with Axitinib significantly increased the protein expressions of NQO1 and HO1 compared with the control (P<0.01). Furthermore, silencing Keap1 enhanced the effect of Axitinib in increasing NQO1 and HO1 protein (P<0.05; Fig. 4F). The protein expressions of p-ERK and ERK were detected. The result demonstrated that the expression of ERK had no marked differences among the control, Axitinib, NC+ Axitinib and siKeap1+ Axitinib groups, and groups that were treated with Axitinib demonstrated significant downregulated expressions of p-ERK/ERK, compared with the control (P<0.05; Fig. 4F). However, in the siKeap1 group, the protein expression of p-ERK/ERK demonstrated a significant increase compared with the treatment with Axitinib alone group (P<0.05).
Co-transfection of siKeap1 and siNrf2 affects the protein expression of Nrf2 and Keap1
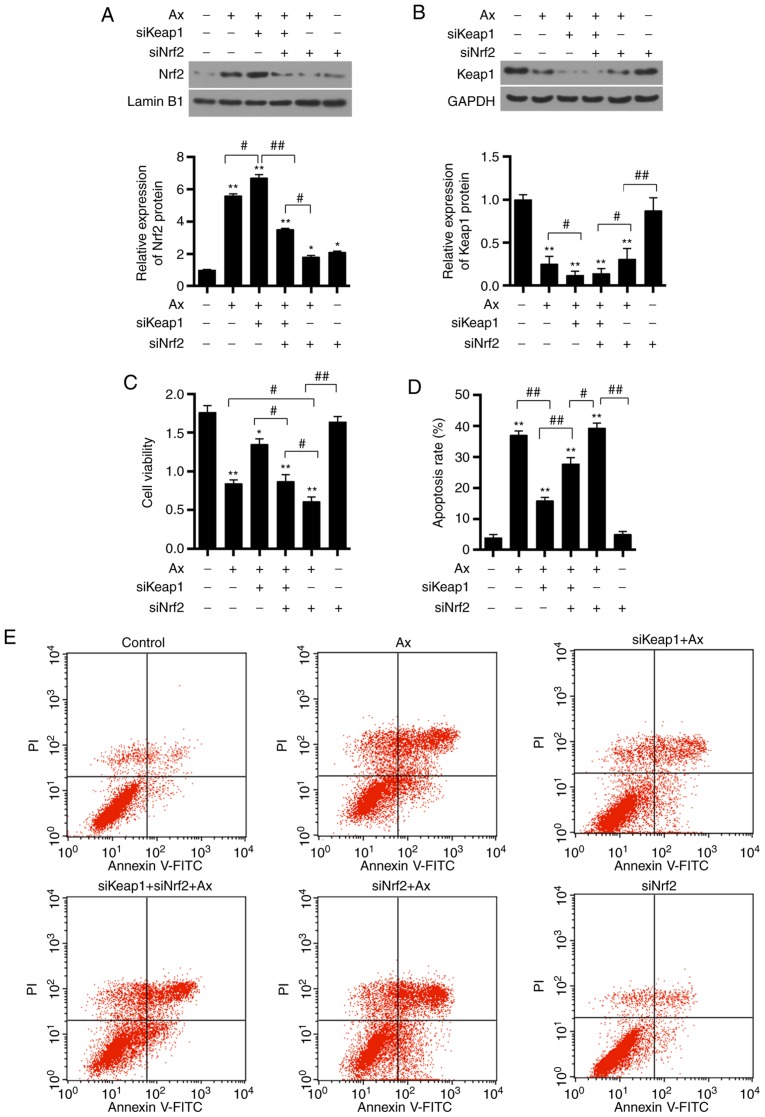
It was identified that transfection with siKeap1 and siNrf2 alone resulted in the downregulation of the respective mRNA and protein expressions (P<0.01; Fig. 3 and Fig. 5A). To further investigate the mechanism underlying the effect by Keap1, Keap1 and Nrf2 were silenced. Silencing Nrf2 in combination with Axitinib significantly decreased the protein expression of Nrf2, compared with co-transfection of siKeap1 and siNrf2, and treated with Axitinib (P<0.05). It was observed that co-transfection of siKeap1 and siNrf2 resulted in a similar effect on the Keap1 protein expression compared with siKeap1 alone (P>0.05; Fig. 5B). It was demonstrated that silencing Nrf2 only significantly increased the protein expression of Keap1 compared with the siNrf2+Axitinib group (P<0.01).
Figure 5.
Effect of co-transfection of siKeap1 and siNrf2 on protein expression of Nrf2 and Keap1, cell viability and apoptosis. ACHN were transfected with siKeap1 and siNrf2 and were divided into six groups, which were the control, Ax (1 µM Ax), siKeap1+Ax, siKeap1+siNrf2+Ax, siNrf2+Ax and siNrf2 groups. (A) Nrf2 protein expression was detected by western blotting. Lamin B1 served as an internal control. (B) Keap1 protein expression was detected by western blotting. GAPDH served as an internal control. (C) Cell viability was assessed by a Cell Counting kit-8 assay. (D) Apoptosis rate is presented as a bar graph. (E) Flow cytometry was used to detect the apoptosis level. Data are presented as the mean ± standard deviation from three independent experiments. *P<0.05, **P<0.01 vs. control; #P<0.05, ##P<0.01. Keap1, Kelch-like ECH-associated protein 1; Nrf2, nuclear factor erythroid 2-related factor 2; Ax, Axitinib; si, small interfering; PI, propidium iodide; FITC, fluorescein isothiocyanate.
Co-transfection of siKeap1 and siNrf2 affects the cell viability and cell apoptosis in ACHN cells
Co-transfection of siKeap1 and siNrf2 was able to decrease the increased cell viability that was induced by silencing Keap1+Axitinib (P<0.05; Fig. 5C). Furthermore, silencing Nrf2 enhanced the ACHN cell injury induced by Axitinib (P<0.05). However, silencing Nrf2 alone significantly increased the cell viability compared with the siNrf2+Axitinib group (P<0.01). It was additionally observed that treatment with Axitinib alone was able to significantly increase cell apoptosis, compared with the control (P<0.01; Fig. 5D and E) and that silencing Keap1 was able to significantly decrease the apoptosis induction by Axitinib (P<0.01). However, apoptosis was significantly increased by siNrf2 in cells transfected with siKeap1 (P<0.01).
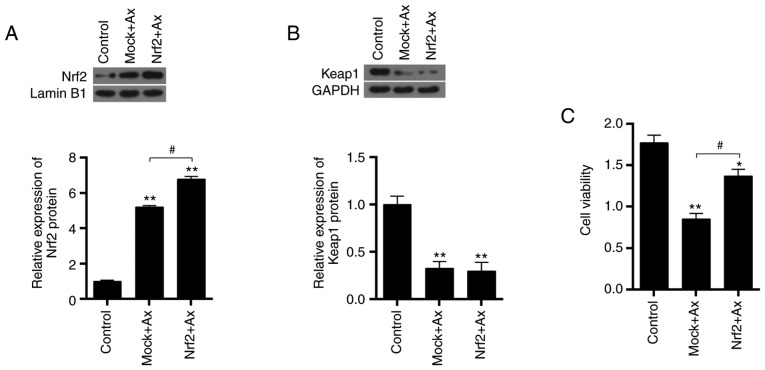
Overexpressing Nrf2 reverses the decrease of cell viability induced by Axitinib
The transfection efficiency of the overexpression plasmid of Nrf2 was assessed and it was demonstrated that the mRNA expression of Nrf2 was upregulated by overexpressing Nrf2 mRNA transfected in cells (P<0.01; Fig. 4B). Furthermore, the effect of overexpressing Nrf2 on ACHN cells was detected. Nrf2 overexpression was observed to significantly increase the Nrf2 protein expression induced by Axitinib (P<0.05; Fig. 6A). Under treatment with Axitinib, overexpressing Nrf2 did not influence the expression of Keap1 protein (P>0.05; Fig. 6B). Overexpressing Nrf2 in combination with Axitinib significantly increased cell viability, compared with the treatment with Axitinib alone (P<0.05; Fig. 6C).
Figure 6.
Effect of overexpressing Nrf2 on cell viability. Nrf2 was overexpressed in ACHN cells and were divided into three groups, which were the control, Mock+Ax and Nrf2+Ax groups. (A) Nrf2 protein expression was detected by western blotting. Lamin B1 served as an internal control. (B) Keap1 protein expression was detected by western blotting. GAPDH served as an internal control. (C) Cell viability was assessed by a Cell Counting kit-8 assay. Data are presented as the mean ± standard deviation from three independent experiments. *P<0.05, **P<0.01 vs. control; #P<0.05. Keap1, Kelch-like ECH-associated protein 1; Ax, Axitinib; Nrf2, nuclear factor erythroid 2-related factor 2.
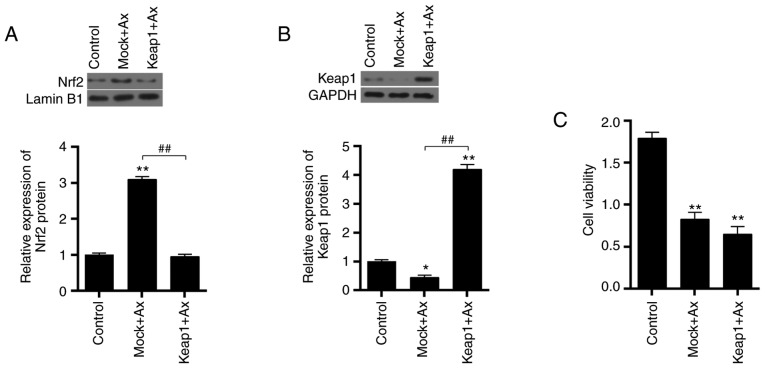
Overexpressing Keap1 enhances cell sensitivity to Axitinib
The transfection efficiency of the overexpression of Keap1 was assessed and it was demonstrated that the mRNA expression of Keap1 was upregulated by overexpressing Keap1 mRNA transfected in cells (P<0.01; Fig. 4A). As presented in Fig. 7A, treatment with Axitinib significantly increased the protein expression of Nrf2, compared with the control (P<0.01; Fig. 7A). Overexpressing Keap1 significantly decreased the expression of Nrf2 caused by Axitinib (P<0.01). Keap1 protein expression was significantly upregulated by overexpressing Keap1 (P<0.01; Fig. 7B). Furthermore, overexpressing Keap1 decreased the cell viability (P<0.01 vs. control; P>0.05 vs. Mock+Ax; Fig. 7C).
Figure 7.
Effect of overexpressing Keap1 on cell viability. Keap1 was overexpressed in ACHN cells and were divided into three groups, which were the control, Mock+Ax and Keap1+Ax groups. (A) Nrf2 protein expression was detected by western blotting. Lamin B1 served as an internal control. (B) Keap1 protein expression was detected by western blotting. GAPDH served as an internal control. (C) Cell viability was detected by a Cell Counting kit-8 assay. Data are presented as the mean ± standard deviation from three independent experiments. *P<0.05, **P<0.01 vs. control; ##P<0.01. Keap1, Kelch-like ECH-associated protein 1; Ax, Axitinib; Nrf2, nuclear factor erythroid 2-related factor 2.
Discussion
In the present study, Keap1 mRNA expression was lower in RCC tissues compared with adjacent normal tissues collected from 45 patients with RCC. This was additionally confirmed by the expressions of mRNA in five randomly selected patients with RCC. With regards to the protein expression level, it was identified that Keap1 protein expression increased in cancer tissues of two of the selected patients and this variation may be due to individual differences or different subtypes of RCC. A lower expression of Keap1 was associated with a poor 5-year survival analyzed by a Kaplan-Meier survival curve. In agreement with this result, a low expression of Keap1 is associated with various malignancies, including non-small cell lung cancer, colorectal cancer, liver cancer, gallbladder cancer, breast cancer, and head and neck squamous cell carcinoma (16,29-34). Solis et al (35) additionally observed that downregulated expression of Keap1 and high expression of Nrf2 were common abnormal phenomena in non-small cell lung carcinoma, and they were associated with a poor prognosis. The expression of Keap1 in normal human renal tubular epithelial cells and five RCC cell lines was further detected; as hypothesized, Keap1 expression was significantly decreased in RCC cell lines. As the protein expression of Keap1 was detected in five patients, the results may be limited as the Keap1 expression was not detected in the remaining patients. Furthermore, there were other limitations of the present study, including that the other two pathways involving NF-κB and Bcl-2 were not investigated.
Keap1 is not only associated with the poor prognosis of RCC; however, additionally serves an important role in chemotherapeutic resistance. It was previously demonstrated that Axitinib is effective in breast cancer, non-small-cell lung, pancreatic cancer and thyroid cancer (36-39). The present results additionally demonstrated that Axitinib had a similar inhibitory effect on RCC. In particular, it was able to inhibit RCC cell viability in a dose-dependent manner. In addition, treatment with Axitinib decreased cell viability, promoted ROS release and induced cell apoptosis. When Keap1 was silenced, the sensitivity of ACHN cells to Axitinib was decreased, specifically, cell viability was increased, the release of ROS was decreased and tumor cell apoptosis was suppressed by siKeap1. A previous study additionally observed that Keap1 mutations increased radio-resistance and was able to predict local tumor recurrence in patients with laryngeal squamous cell carcinoma subjected to radiotherapy (40). The present results demonstrated that siKeap1 decreased the ROS level and increased the cell viability.
The Keap1-Nrf2 signaling pathway has a protective effect on normal cells in addition to tumor cells (39,31). Numerous previous studies demonstrated that the signaling was able to induce drug resistance by reducing the sensitivity of tumor cells to chemotherapeutic drugs (41-44). Therefore, the effect of silencing Keap1 on the expression of Nrf2 and its effect on ERK signaling was investigated. The result demonstrated that treatment with Axitinib was able to decrease Keap1 expression and stimulate Nrf2 expression. Furthermore, the downstream proteins of Nrf2, NQO1 and HO1 were significantly enhanced under treatment with Axitinib. Silencing Keap1 increased the expression of Nrf2, NQO1 and HO1.
Nrf2 is a basic leucine Zipper structural transcription factor and cap 'n' collar family transcription factor (45). Human Nrf2 has 605 amino acid residues and forms conserved domains from Neh1 to Neh7 (46,47). Nrf2 has the function of activating the transcription and expression of the ARE gene, binding to Keap1, and regulating transcriptional activation and degradation (46,48). Nrf2 has been identified as one of the most important antioxidative regulators (49). Although a number of previous studies demonstrated that Nrf2 served an important role in tumor prevention (50,51), other previous studies observed that a high expression level of Nrf2 in tumor cells was additionally able to reduce its sensitivity to chemotherapeutic drugs and promote tumor growth (52-54). Stacy et al (55) identified that Nrf2 was highly expressed in head and neck squamous cell carcinoma, and that the high expression of Nrf2 was considered as one of the markers of tumor drug resistance. The Keap1-Nrf2 signaling pathway is activated in mammary cancer cells tolerant to tamoxifen, and the tolerance of tumor cells to tamoxifen may be altered subsequent to silencing Nrf2 with siRNA (42). Nrf2 may be a prognostic indicator of gastric cancer, and it may predict the efficacy of 5-Fu in patients with gastric cancer (56). Previous studies demonstrated that the target genes of Nrf2, including HO1, glutathione S-transferases, multidrug resistance associated protein and NQO1, were the core components of the drug resistance mechanism of tumors (57-59). HO1 has anti-oxidation and anti-apoptotic effects, whereas, Nrf2 may inhibit apoptosis of tumor cells and produce drug resistance by regulating the expression of HO1 (60). NQO1 is a flavozyme that catalyzes the redox reaction of cytotoxic substances to produce non-toxic or low-toxic substances (61). The present results suggest that decreased sensitivity to Axitinib in renal carcinoma cells induced by the downregulation of Keap1 is associated with the increase of Nrf2 expression.
Numerous previous studies demonstrated that activation of the mitogen-activated protein kinase/ERK signaling pathway contributed to resistance to receptor tyrosine kinase inhibitors (62-64). The protein expression of ERK signaling was assessed, and it was identified that the phosphorylation of ERK expression was decreased in Axitinib-treated cells. The activity of ERK signaling was not affected by siKeap1, suggesting that Axitinib resistance induced by silencing Keap1 was independent of the ERK pathway.
Silencing Keap1 and Nrf2, and overexpressing Nrf2 and Keap1 were performed to assess ACHN cell survival. The present results demonstrated that Axitinib decreased the cell viability and the knockdown of Keap1 increased the viability of Axitinib-treated cells, whereas, overexpression of Keap1 enhanced the effects of Axitinib. Taken together, it was concluded that the downregulation of Keap1 may contribute to the resistance of RCC cells to Axitinib.
In conclusion, the present study suggested that Keap1 serves as a tumor suppressor, and that its decreased expression is associated with a poor prognosis and a reduced Axitinib sensitivity of RCC cells. A possible mechanism involved Nrf2 overexpression. The present study suggested that regulation of Keap1 may be an attractive strategy to increase the therapeutic effect of cancer-drug resistance.
Acknowledgments
Not applicable.
Funding
The present study was supported by Natural Science Foundation of Guangxi Province (China; grant no. 2012GXNSFBA053087).
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Authors' contributions
HH wrote the manuscript. HH, YuW, WF, XW, LZ, XX, FH and YiW performed the experiments. HH, YuW and YiW designed the study. WF, XW, LZ, XX, FH and YiW performed data analysis. HH, YuW and YiW contributed to manuscript revisions and all the authors reviewed the manuscript.
Ethics approval and consent to participate
The project protocol was approved by the Ethics Committee of The Second Affiliated Hospital of Guangxi Medical University. All patients recruited in the present study provided written informed consent for the use of their tissue samples for clinical research.
Patient consent for publication
Written informed consent was obtained from all participants for the publication of their data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 4.Singer EA, Gupta GN, Srinivasan R. Targeted therapeutic strategies for the management of renal cell carcinoma. Curr Opin Oncol. 2012;24:284–290. doi: 10.1097/CCO.0b013e328351c646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Négrier S. Duration of targeted therapy for metastatic renal cell carcinoma: A review of current practices. Oncology. 2012;82:189–196. doi: 10.1159/000336888. [DOI] [PubMed] [Google Scholar]
- 6.Gross-Goupil M, Massard C, Ravaud A. Targeted therapies in metastatic renal cell carcinoma: Overview of the past year. Curr Urol Rep. 2012;13:16–23. doi: 10.1007/s11934-011-0232-y. [DOI] [PubMed] [Google Scholar]
- 7.Coppin C, Kollmannsberger C, Le L, Porzsolt F, Wilt TJ. Targeted therapy for advanced renal cell cancer (RCC): A Cochrane systematic review of published randomised trials. BJU Int. 2011;108:1556–1563. doi: 10.1111/j.1464-410X.2011.10629.x. [DOI] [PubMed] [Google Scholar]
- 8.Sonpavde G, Hutson TE, Rini BI. Axitinib for renal cell carcinoma. Expert Opin Investig Drugs. 2008;17:741–748. doi: 10.1517/13543784.17.5.741. [DOI] [PubMed] [Google Scholar]
- 9.Kelly RJ, Rixe O. Axitinib (AG-013736) Recent Results Cancer Res. 2010;184:33–44. doi: 10.1007/978-3-642-01222-8_3. [DOI] [PubMed] [Google Scholar]
- 10.Magesh S, Chen Y, Hu L. Small molecule modulators of Keap1-Nrf2-ARE pathway as potential preventive and therapeutic agents. Med Res Rev. 2012;32:687–726. doi: 10.1002/med.21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoo NJ, Kim HR, Kim YR, An CH, Lee SH. Somatic mutations of the KEAP1 gene in common solid cancers. Histopathology. 2012;60:943–952. doi: 10.1111/j.1365-2559.2012.04178.x. [DOI] [PubMed] [Google Scholar]
- 12.Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, Suzuki T, Kobayashi A, Yokota J, Sakiyama T, et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 13.Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 14.Wong TF, Yoshinaga K, Monma Y, Ito K, Niikura H, Nagase S, Yamamoto M, Yaegashi N. Association of keap1 and nrf2 genetic mutations and polymorphisms with endometrioid endometrial adenocarcinoma survival. Int J Gynecol Cancer. 2011;21:1428–1435. doi: 10.1097/IGC.0b013e31822d0eb2. [DOI] [PubMed] [Google Scholar]
- 15.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shibata T, Kokubu A, Gotoh M, Ojima H, Ohta T, Yamamoto M, Hirohashi S. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. 2008;135:1358–1368. 1368.e1–e4. doi: 10.1053/j.gastro.2008.06.082. [DOI] [PubMed] [Google Scholar]
- 17.Lister A, Nedjadi T, Kitteringham NR, Campbell F, Costello E, Lloyd B, Copple IM, Williams S, Owen A, Neoptolemos JP, et al. Nrf2 is overexpressed in pancreatic cancer: Implications for cell proliferation and therapy. Mol Cancer. 2011;10:37. doi: 10.1186/1476-4598-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee DF, Kuo HP, Liu M, Chou CK, Xia W, Du Y, Shen J, Chen CT, Huo L, Hsu MC, et al. KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol Cell. 2009;36:131–140. doi: 10.1016/j.molcel.2009.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee DF, Kuo HP, Chen CT, Hsu JM, Chou CK, Wei Y, Sun HL, Li LY, Ping B, Huang WC, et al. IKK beta suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway. Cell. 2007;130:440–455. doi: 10.1016/j.cell.2007.05.058. [DOI] [PubMed] [Google Scholar]
- 20.Niture SK, Jaiswal AK. Inhibitor of Nrf2 (INrf2 or Keap1) protein degrades Bcl-xL via phosphoglycerate mutase 5 and controls cellular apoptosis. J Biol Chem. 2011;286:44542–44556. doi: 10.1074/jbc.M111.275073. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Niture SK, Jaiswal AK. INrf2 (Keap1) targets Bcl-2 degradation and controls cellular apoptosis. Cell Death Differ. 2011;18:439–451. doi: 10.1038/cdd.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kansanen E, Jyrkkänen HK, Levonen AL. Activation of stress signaling pathways by electrophilic oxidized and nitrated lipids. Free Radic Biol Med. 2012;52:973–982. doi: 10.1016/j.freeradbiomed.2011.11.038. [DOI] [PubMed] [Google Scholar]
- 24.Galloway DC, Blake DG, Shepherd AG, McLellan LI. Regulation of human gamma-glutamylcysteine synthetase: Co-ordinate induction of the catalytic and regulatory subunits in HepG2 cells. Biochem J. 1997;328:99–104. doi: 10.1042/bj3280099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Traver RD, Horikoshi T, Danenberg KD, Stadlbauer TH, Danenberg PV, Ross D, Gibson NW. NAD(P)H:quinone oxidoreductase gene expression in human colon carcinoma cells: Characterization of a mutation which modulates DT-diaphorase activity and mitomycin sensitivity. Cancer Res. 1992;52:797–802. [PubMed] [Google Scholar]
- 26.Meijerman I, Beijnen JH, Schellens JH. Combined action and regulation of phase II enzymes and multidrug resistance proteins in multidrug resistance in cancer. Cancer Treat Rev. 2008;34:505–520. doi: 10.1016/j.ctrv.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Mizunoe Y, Kobayashi M, Sudo Y, Watanabe S, Yasukawa H, Natori D, Hoshino A, Negishi A, Okita N, Komatsu M, Higami Y. Trehalose protects against oxidative stress by regulating the Keap1-Nrf2 and autophagy pathways. Redox Biol. 2018;15:115–124. doi: 10.1016/j.redox.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frank R, Scheffler M, Merkelbach-Bruse S, Ihle MA, Kron A, Rauer M, Ueckeroth F, König K, Michels S, Fischer R, et al. Clinical and pathological characteristics of KEAP1- and NFE2L2-mutated non-small cell lung carcinoma (NSCLC) Clin Cancer Res. 2018;24:3087–3096. doi: 10.1158/1078-0432.CCR-17-3416. [DOI] [PubMed] [Google Scholar]
- 30.Wu B, Yang S, Sun H, Sun T, Ji F, Wang Y, Xu L, Zhou D. Keap1 inhibits metastatic properties of NSCLC cells by stabilizing architectures of F-actin and focal adhesions. Mol Cancer Res. 2018;16:508–516. doi: 10.1158/1541-7786.MCR-17-0544. [DOI] [PubMed] [Google Scholar]
- 31.Hanada N, Takahata T, Zhou Q, Ye X, Sun R, Itoh J, Ishiguro A, Kijima H, Mimura J, Itoh K, et al. Methylation of the KEAP1 gene promoter region in human colorectal cancer. BMC Cancer. 2012;12:66. doi: 10.1186/1471-2407-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartikainen JM, Tengström M, Winqvist R, Jukkola-Vuorinen A, Pylkäs K, Kosma VM, Soini Y, Mannermaa A. KEAP1 genetic polymorphisms associate with breast cancer risk and survival outcomes. Clin Cancer Res. 2015;21:1591–1601. doi: 10.1158/1078-0432.CCR-14-1887. [DOI] [PubMed] [Google Scholar]
- 33.Barbano R, Muscarella LA, Pasculli B, Valori VM, Fontana A, Coco M, la Torre A, Balsamo T, Poeta ML, Marangi GF, et al. Aberrant Keap1 methylation in breast cancer and association with clinicopathological features. Epigenetics. 2013;8:105–112. doi: 10.4161/epi.23319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klapproth E, Dickreuter E, Zakrzewski F, Seifert M, Petzold A, Dahl A, Schröck E, Klink B, Cordes N. Whole exome sequencing identifies mTOR and KEAP1 as potential targets for radiosensitization of HNSCC cells refractory to EGFR and β1 integrin inhibition. Oncotarget. 2018;9:18099–18114. doi: 10.18632/oncotarget.24266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Solis LM, Behrens C, Dong W, Suraokar M, Ozburn NC, Moran CA, Corvalan AH, Biswal S, Swisher SG, Bekele BN, et al. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res. 2010;16:3743–3753. doi: 10.1158/1078-0432.CCR-09-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma YH, Wang SY, Ren YP, Li J, Guo TJ, Lu W, Zhou TY. Antitumor effect of axitinib combined with dopamine and PK-PD modeling in the treatment of human breast cancer xenograft. Acta Pharmacol Sin. 2019;40:243–256. doi: 10.1038/s41401-018-0006-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bondarenko IM, Ingrosso A, Bycott P, Kim S, Cebotaru CL. Phase II study of axitinib with doublet chemotherapy in patients with advanced squamous non-small-cell lung cancer. BMC Cancer. 2015;15:339. doi: 10.1186/s12885-015-1350-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ioka T, Okusaka T, Ohkawa S, Boku N, Sawaki A, Fujii Y, Kamei Y, Takahashi S, Namazu K, Umeyama Y, et al. Efficacy and safety of axitinib in combination with gemcitabine in advanced pancreatic cancer: Subgroup analyses by region, including Japan, from the global randomized Phase III trial. Jpn J Clin Oncol. 2015;45:439–448. doi: 10.1093/jjco/hyv011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hui EP, Ma BBY, Loong HHF, Mo F, Li L, King AD, Wang K, Ahuja AT, Chan CML, Hui CWC, et al. Efficacy, safety, and pharmacokinetics of axitinib in nasopharyngeal carcinoma: A preclinical and phase II correlative study. Clin Cancer Res. 2018;24:1030–1037. doi: 10.1158/1078-0432.CCR-17-1667. [DOI] [PubMed] [Google Scholar]
- 40.Jeong Y, Hoang NT, Lovejoy A, Stehr H, Newman AM, Gentles AJ, Kong W, Truong D, Martin S, Chaudhuri A, et al. Role of KEAP1/NRF2 and TP53 mutations in lung squamous cell carcinoma development and radiotherapy response prediction. Cancer Discovery. 2017;7:86–101. doi: 10.1158/2159-8290.CD-16-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim HR, Kim S, Kim EJ, Park JH, Yang SH, Jeong ET, Park C, Youn MJ, So HS, Park R. Suppression of Nrf2-driven heme oxygenase-1 enhances the chemosensitivity of lung cancer A549 cells toward cisplatin. Lung Cancer. 2008;60:47–56. doi: 10.1016/j.lungcan.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 42.Kim SK, Yang JW, Kim MR, Roh SH, Kim HG, Lee KY, Jeong HG, Kang KW. Increased expression of Nrf2/ARE-dependent anti-oxidant proteins in tamoxifen-resistant breast cancer cells. Free Radic Biol Med. 2008;45:537–546. doi: 10.1016/j.freeradbiomed.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 43.Okawa H, Motohashi H, Kobayashi A, Aburatani H, Kensler TW, Yamamoto M. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem Biophys Res Commun. 2006;339:79–88. doi: 10.1016/j.bbrc.2005.10.185. [DOI] [PubMed] [Google Scholar]
- 44.Homma S, Ishii Y, Morishima Y, Yamadori T, Matsuno Y, Haraguchi N, Kikuchi N, Satoh H, Sakamoto T, Hizawa N, et al. Nrf2 enhances cell proliferation and resistance to anticancer drugs in human lung cancer. Clin Cancer Res. 2009;15:3423–3432. doi: 10.1158/1078-0432.CCR-08-2822. [DOI] [PubMed] [Google Scholar]
- 45.Vargas MR, Johnson JA. The Nrf2-ARE cytoprotective pathway in astrocytes. Expert Rev Mol Med. 2009;11:e17. doi: 10.1017/S1462399409001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taguchi K, Motohashi H, Yamamoto M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells. 2011;16:123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- 47.Wang H, Liu K, Geng M, Gao P, Wu X, Hai Y, Li Y, Li Y, Luo L, Hayes JD, et al. RXRα inhibits the NRF2-ARE signaling pathway through a direct interaction with the Neh7 domain of NRF2. Cancer Res. 2013;73:3097–3108. doi: 10.1158/0008-5472.CAN-12-3386. [DOI] [PubMed] [Google Scholar]
- 48.McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J Biol Chem. 2004;279:31556–31567. doi: 10.1074/jbc.M403061200. [DOI] [PubMed] [Google Scholar]
- 49.Rachakonda G, Xiong Y, Sekhar KR, Stamer SL, Liebler DC, Freeman ML. Covalent modification at Cys151 dissociates the electrophile sensor Keap1 from the ubiquitin ligase CUL3. Chem Res Toxicol. 2008;21:705–710. doi: 10.1021/tx700302s. [DOI] [PubMed] [Google Scholar]
- 50.Krajka-Kuźniak V, Paluszczak J, Baer-Dubowska W. The Nrf2-ARE signaling pathway: An update on its regulation and possible role in cancer prevention and treatment. Pharmacol Rep. 2017;69:393–402. doi: 10.1016/j.pharep.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 51.Moon EJ, Giaccia A. Dual roles of NRF2 in tumor prevention and progression: Possible implications in cancer treatment. Free Radic Biol Med. 2015;79:292–299. doi: 10.1016/j.freeradbiomed.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 53.Xu C, Huang MT, Shen G, Yuan X, Lin W, Khor TO, Conney AH, Kong AN. Inhibition of 7,12-dimethylbenz(a) anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res. 2006;66:8293–8296. doi: 10.1158/0008-5472.CAN-06-0300. [DOI] [PubMed] [Google Scholar]
- 54.auf dem Keller U, Huber M, Beyer TA, Kümin A, Siemes C, Braun S, Bugnon P, Mitropoulos V, Johnson DA, Johnson JA, et al. Nrf transcription factors in keratinocytes are essential for skin tumor prevention but not for wound healing. Mol Cell Biol. 2006;26:3773–3784. doi: 10.1128/MCB.26.10.3773-3784.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stacy DR, Ely K, Massion PP, Yarbrough WG, Hallahan DE, Sekhar KR, Freeman ML. Increased expression of nuclear factor E2 p45-related factor 2 (NRF2) in head and neck squamous cell carcinomas. Head Neck. 2006;28:813–818. doi: 10.1002/hed.20430. [DOI] [PubMed] [Google Scholar]
- 56.Hu XF, Yao J, Gao SG, Wang XS, Peng XQ, Yang YT, Feng XS. Nrf2 overexpression predicts prognosis and 5-FU resistance in gastric cancer. Asian Pac J Cancer Prev. 2013;14:5231–5235. doi: 10.7314/APJCP.2013.14.9.5231. [DOI] [PubMed] [Google Scholar]
- 57.Lau A, Villeneuve NF, Sun Z, Wong PK, Zhang DD. Dual roles of Nrf2 in cancer. Pharmacol Res. 2008;58:262–270. doi: 10.1016/j.phrs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lo HW, Ali-Osman F. Genetic polymorphism and function of glutathione S-transferases in tumor drug resistance. Curr Opin Pharmacol. 2007;7:367–374. doi: 10.1016/j.coph.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 59.Fiorillo M, Sotgia F, Sisci D, Cappello AR, Lisanti MP. Mitochondrial 'power' drives tamoxifen resistance: NQO1 and GCLC are new therapeutic targets in breast cancer. Oncotarget. 2017;8:20309–20327. doi: 10.18632/oncotarget.15852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanaka S, Akaike T, Fang J, Beppu T, Ogawa M, Tamura F, Miyamoto Y, Maeda H. Antiapoptotic effect of haem oxygenase-1 induced by nitric oxide in experimental solid tumour. Br J Cancer. 2003;88:902–909. doi: 10.1038/sj.bjc.6600830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oh ET, Park HJ. Implications of NQO1 in cancer therapy. BMB Rep. 2015;48:609–617. doi: 10.5483/BMBRep.2015.48.11.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Piscazzi A, Costantino E, Maddalena F, Natalicchio MI, Gerardi AM, Antonetti R, Cignarelli M, Landriscina M. Activation of the RAS/RAF/ERK signaling pathway contributes to resistance to sunitinib in thyroid carcinoma cell lines. J Clin Endocrinol Metab. 2012;97:E898–E906. doi: 10.1210/jc.2011-3269. [DOI] [PubMed] [Google Scholar]
- 63.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.