Abstract
We examined the association between genetic risk score (GRS) for blood pressure (BP), based on SNPs identified in previous BP genome-wide association study meta-analyses, and salt- and potassium-sensitivity of BP among participants of the Genetic Epidemiology Network of Salt-Sensitivity (GenSalt) study. The GenSalt study was conducted among 1,906 participants who underwent a 7-day low-sodium (51.3 mmol sodium/day), 7-day high-sodium (307.8 mmol sodium/day), and 7-day high-sodium plus potassium (60 mmol potassium/day) intervention. BP was measured nine times at baseline and at the end of each intervention period using a random-zero sphygmomanometer. Associations between systolic BP (SBP), diastolic BP (DBP) and mean arterial pressure (MAP) GRS and respective SBP, DBP and MAP responses to the dietary interventions were assessed using mixed linear regression models that accounted for familial dependencies and adjusted for age, gender, field center, body mass index and baseline BP. As expected, baseline SBP, DBP, and MAP significantly increased per quartile increase in GRS (P=2.7×10−8, 9.8×10−8, and 6.4×10−6 respectively). In contrast, increasing GRS quartile conferred smaller SBP, DBP, and MAP responses to the low-sodium intervention (P=1.4×10−3, 0.02, and 0.06, respectively) and smaller SBP responses to the high sodium and potassium interventions (P=0.10 and 0.05). In addition, overall findings were similar when examining GRS as a continuous measure. Contrary to our initial hypothesis, we identified an inverse relationship between BP GRS and salt- and potassium-sensitivity of BP. These data may provide novel implications regarding the relationship between BP responses to dietary sodium and potassium and hypertension.
Keywords: blood pressure, hypertension, genetics, sodium, diet
Introduction
Elevated blood pressure (BP) is a leading risk factor for morbidity and mortality globally (1). The substantial public health toll arising from hypertension can be attributed to its high prevalence and associated risk of cardiovascular disease (CVD) (2, 3). While several rare monogenic BP disorders have been identified, mostly involving genes that influence renal sodium handling (4) the vast majority of hypertension cases result from interactions of multiple genetic and environmental factors (5). Numerous genetic loci for the complex BP and hypertension phenotypes have been identified through genome-wide association study (GWAS) meta-analyses (6–12). Among environmental factors, dietary sodium and potassium intake have been established as important determinants of BP (13, 14). While BP responses to dietary sodium and potassium intake are known as moderately heritable phenotypes (4, 15–17), the cumulative effects of robustly identified BP loci on salt- and potassium-sensitivity has never been explored. Associations between BP GRS and salt- and -potassium-sensitivity would implicate these phenotypes as mediators on the causal pathway between genomic susceptibility and hypertension, which would add to the observational evidence linking salt-sensitivity to this complex condition (18, 19). Furthermore, identification of a relation between BP GRS and BP salt- and -potassium-sensitivity could eventually identify individuals who might receive enhanced BP-lowering benefits from dietary sodium and potassium interventions, and hence, provide targeted strategies for reducing BP and ultimately preventing hypertension in high-risk subgroups of the population.
The purpose of our study was to examine the relationship between BP genetic risk score (GRS), a score comprised of loci identified by previous BP GWAS meta-analyses, and BP responses to dietary sodium and potassium intake. To explore this research question, our study leveraged genome-wide genotype and unique salt- and potassium-sensitivity phenotype data from 1,906 Han Chinese participants of a carefully conducted dietary sodium and potassium feeding study.
Methods
Study Population
The GenSalt study is a family feeding study that was conducted from 2003 to 2005 among Han Chinese residents of rural north China. A community-based BP screening was conducted among persons aged 18–60 years in the study villages to identify potential probands and their families. Detailed eligibility criteria for the probands and siblings/spouses/offspring have been presented elsewhere (20). Briefly, probands with untreated prehypertension or stage-1 hypertension and their spouses, siblings, and offspring were recruited as volunteers for the dietary intervention study. Those who had stage-2 hypertension, secondary hypertension, a history of clinical CVD or diabetes, used antihypertensive medications, or were pregnant, heavy alcohol drinkers, or currently on a low-sodium diet were excluded from the study. Among the 1,906 eligible participants from 633 families, 1,850 (98.4%), 1,840 (97.8%) and 1,836 (97.6%) participants with GWAS data completed the low-sodium, high-sodium, and potassium-supplementation interventions, respectively (21), and were included in the current analysis. Institutional review boards or ethnic committees at all participating institutions approved the GenSalt study. Written informed consent was obtained from each GenSalt study participant before baseline data collection and intervention.
Dietary Intervention
The dietary intervention consisted of three successive 7-day interventions, including a low-sodium diet (3-gram salt or 51.3 mmol sodium/day), a high-sodium diet (18 gram salt or 307.8 mmol sodium/day), and a high-sodium diet plus potassium-supplementation (60 mmol potassium/day). During the period of sodium intervention, dietary potassium intake remained unchanged. Total energy intake was varied according to each participant’s baseline energy intake. All study foods were cooked without salt, and prepackaged salt was added to the individual study participant’s meal when it was served by the study staff. To ensure study participants’ compliance with the intervention program, they were required to have their breakfast, lunch, and dinner at the study kitchen under supervision of the study staff during the entire study period. The study participants were instructed to avoid consuming any foods that were not provided by study personnel. Three timed urinary specimens (one 24-hour and two overnights) were collected at baseline and at the end of each intervention phase (days 5, 6, and 7) to monitor each participant’s compliance to the dietary interventions. The overnight urinary excretions of sodium and potassium were converted to 24-hour values based on formulas developed from a random subsample of 238 participants who had collected overnight and 24-hour measures on the same days at baseline and during each phase of intervention.
Phenotype Measurement
A standard questionnaire was administered by trained staff at the baseline examination to collect information on family structure, demographic characteristics, personal and family medical history, and lifestyle risk factors. Body weight and height were also measured twice with participants wearing light indoor clothing without shoes during the baseline examination. Body mass index (BMI) was calculated as kilograms per meters squared (kg/m2). Three BP measurements were recorded at the same time each morning, according to a standard protocol recommended by the American Heart Association (22), during each of the 3-days of baseline observation, and during days five, six, and seven of each intervention period. All BP measurements were obtained by trained and certified observers using a random zero sphygmomanometer after five minutes of rest, with the participants in the sitting position and their arm placed at the level of their heart. Participants were advised to avoid alcohol, cigarette smoking, coffee/tea, and exercise for at least 30 minutes prior to their BP measurements. All observers were blinded to the dietary intervention stages.
Salt sensitivity phenotypes were defined as the absolute changes in SBP, DBP, and MAP when switching from baseline to low-sodium and low-sodium to high-sodium interventions, while potassium sensitivity was defined as the absolute changes in SBP, DBP and MAP when switching from high-sodium to high-sodium plus potassium supplementation intervention. Mean BP response to low-sodium was calculated as the mean of 9 BP measures on days 5, 6, and 7 of the low-sodium intervention minus the mean of 9 BP measures at baseline. Mean BP response to high-sodium was calculated as the mean of 9 BP measures on days 5, 6, and 7 of the high-sodium intervention minus the mean of 9 BP measures during the low-sodium intervention. Mean BP response to potassium was calculated as the mean of 9 BP measures on days 5, 6, and 7 of the high-sodium plus potassium intervention minus the mean of 9 BP measures during the high-sodium intervention.
Genotyping, SNP Selection, and Genetic Risk Score (GRS)
Lymphocyte DNA samples were obtained from study participants. A total of 868,158 autosomal SNPs were genotyped using the Affymetrix Genomewide Human SNP array 6.0 (Affymetrix, Inc, Santa Clara, CA). After removal of monomorphic SNPs, SNPs with deviations from Hardy-Weinberg equilibrium (P<1×10−6), missing rate >25% and a minor allele frequency <1%, 820,017 SNPs remained. The overall non-Mendelian inheritance error rate was 0.019%. Non-Mendelian inheritance errors were identified and corrected using PLINK (23) and PedCheck (24). We used MiniMac (25, 26) software to impute variants from the 1000 Genomes Phase 3 All Ancestry Panel. Imputation results were summarized as “dosage” scores (fractional values ranging from 0 to 2, representing the expected number of copies of the coded allele at each SNP).
Variants comprising the GRSs for SBP, DBP, and MAP included those robustly identified in individuals of East Asian ancestry. The GWAS catalogue (27) was searched to identify BP GWAS meta-analyses conducted in at least 10,000 participants of East Asian ancestry in the replication or combined sample. The variants comprising each GRS from the eight studies identified (6–8, 28–32) included independent SNPs (r2<0.8) achieving genome-wide significance (P<5×10−8) in the combined sample, and at least nominal significance (P<0.05) and same effect direction in the replication sample. In cases where multiple lead SNPs were reported within one linkage disequilibrium (LD) block by different studies, the SNP identified by the study with the largest replication sample size was selected. Using effect sizes from the largest replication sample should yield the most precise beta estimate for GRS weighting while avoiding bias due to the winner’s curse. The three GRSs utilized in this study were comprised of 43, 42, and 24 independent SNPs for SBP, DBP, and MAP, respectively, with imputation quality r2 >0.5. In addition, a sensitivity analysis was conducted using all loci identified from previous BP GWAS meta-analyses (6–12, 28–35), regardless of ancestry. These combined ancestry GRSs (caGRSs) were comprised of 94, 93, and 38 independent SNPs for SBP, DBP, and MAP, respectively (Table S1, please see http://hyper.ahajournals.org). To limit the analysis to only variants that may influence BP through pathways underlying BP responses to sodium and potassium, another sensitivity analysis was conducted using only loci within 1Mb of genes previously implicated in BP salt- and potassium sensitivity or those thought to influence BP via renal sodium or potassium handling (4). These pathway restricted GRSs (prGRSs) comprised of 9, 7, and 4 independent SNPs for SBP, DBP, and MAP, respectively (Table S6, please see http://hyper.ahajournals.org).
Statistical Analysis
Weighted GRSs for SBP, DBP, and MAP were calculated for each participant, as the sum of the products of the participant’s dosage score for each SNP and the SNP’s estimated effect size. Participants were then categorized into quartiles of risk for each GRS.
The means and standard deviations or frequency and percent of baseline characteristics were calculated for each quartile of the SBP GRS. P values for linear trends in baseline characteristics across quartiles were estimated. A mixed linear regression model was used to assess changes in baseline BP and BP response to each dietary intervention per quartile increase in the categorized GRS and per standard deviation increase in the quantitative GRS. Familial correlations were accounted for using a sandwich estimator and compound symmetry correlation matrix, which assumes the same degree of dependency among family members. All models were adjusted for age, gender, field center, and baseline BMI. The three intervention models were also adjusted for baseline BP. Non-linear trends were assessed by adding a quadratic term to the models. Post-hoc analyses were conducted to explore the roles of baseline urinary sodium and potassium excretions on the relation between GRS and BP/BP response phenotypes. All analyses were conducted using SAS software (version 9.3; SAS Institute, Inc., Cary, North Carolina).
Results
Baseline characteristics of GenSalt participants, separated by quartile of SBP GRS, are presented in Table 1. Genetic burden for elevated blood pressure, represented by SBP GRS, ranged from 24.37 to 54.79. Neither age nor gender was associated with GRS (P=0.12 and 0.11 respectively). Mean BMI ranged from 23.13 in the first quartile to 23.61 in the fourth quartile (P=0.01). As expected, mean baseline SBP increased significantly from 114.51 in the first quartile to 118.58 in the fourth quartile (P=9.9×10−7); DBP from 72.41 in the first quartile to 74.90 in the third quartile (P=1.0×10−3); and MAP from 86.44 in the first quartile to 89.42 in the third quartile (P=2.5×10−5).
Table 1.
Characteristics of GenSalt participants according to SBP GRS quartile.
| Characteristic | Quartile 1 | Quartile 2 | Quartile 3 | Quartile 4 | P-value |
|---|---|---|---|---|---|
| GRS, median (IQR) | 33.00 (30.93, 34.42) |
36.92 (36.10, 37.64) |
40.01 (39.11, 40.80) |
43.74 (42.53, 45.46) |
|
| Age | 39.34 ± 9.65 | 38.46 ± 9.47 | 38.84 ± 9.76 | 38.23 ± 9.22 | 0.12 |
| Male, n (%) | 256 (54.46%) | 256 (54.46%) | 248 (52.65%) | 233 (49.57%) | 0.11 |
| BMI (mean ± SD, kg/m2) | 23.13 ± 3.13 | 23.23 ± 3.02 | 23.43 ± 3.26 | 23.61 ± 3.28 | 0.01 |
| Baseline | |||||
| SBP (mean ± SD, mm Hg) | 114.51 ± 14.04 | 116.28 ± 14.17 | 118.47 ± 13.72 | 118.58 ± 14.68 | 9.9×10−7 |
| DBP (mean ± SD, mm Hg) | 72.41 ± 10.26 | 73.41 ± 10.34 | 74.90 ± 10.11 | 74.27 ± 10.41 | 1.0×10−3 |
| MAP (mean ± SD, mm Hg) | 86.44 ± 10.82 | 87.70 ± 10.98 | 89.42 ± 10.39 | 89.04 ± 11.15 | 2.5×10−5 |
BMI=Body mass index; DBP=Diastolic blood pressure; GRS= Genetic risk score; IQR=Inter-quartile range; MAP=Mean arterial pressure; SBP=Systolic blood pressure; SD=Standard deviation.
Results from 24-hour urinary excretions of sodium and potassium showed excellent compliance to the feeding study (Table S2, please see http://hyper.ahajournals.org). Consistent with these findings, SBP, DBP and MAP decreased significantly when switching from baseline to the low-sodium intervention and from the high sodium to the potassium intervention, while BP values increased significantly when switching from the low-sodium to the high-sodium intervention (all P<0.0001; Figure S1, please see http://hyper.ahajournals.org).
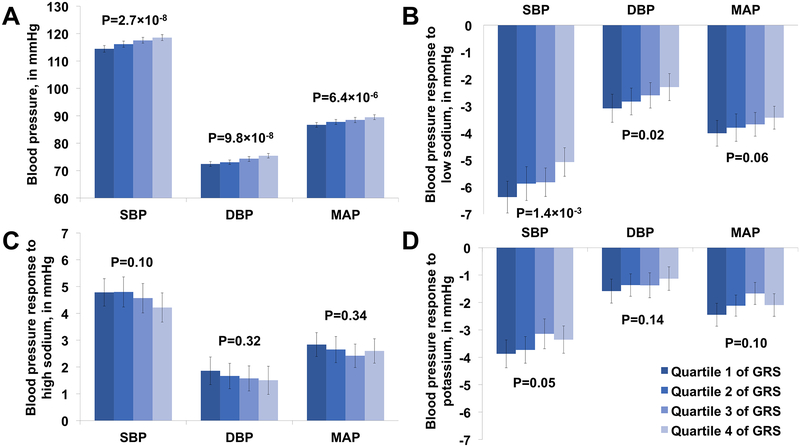
Figure 1a–d depicts adjusted mean baseline BP and BP responses according to quartile of SBP, DBP, and MAP GRS. As expected SBP, DBP, and MAP all increased per quartile of GRS (P=2.7×10−8, 9.8×10−8, and 6.4×10−6, respectively), indicating that participants who had higher BP GRS tended to have higher baseline BP (Figure 1a). In contrast, GRS was inversely associated with the magnitude of BP responses to the low-sodium intervention. Those with higher GRS had significantly smaller decreases in SBP and DBP and marginally smaller decreases in MAP in response to the low-sodium intervention (P=1.4×10−3, 0.02, and 0.06, respectively; Figure 1b). Those with higher GRS also had smaller decreases in SBP in response to the potassium intervention (P=0.05) (Figure 1d). SBP responses to the high-sodium intervention and DBP and MAP responses to the high-sodium (Figure 1c) and potassium (Figure 1d) interventions did not significantly differ according to GRS quartile. Results of sensitivity analyses using a caGRS, not restricted to loci identified in East Asians, were similar to those of the primary analyses (Table S3 and Figure S2, please see http://hyper.ahajournals.org).
Figure 1. Blood pressure and blood pressure responses to interventions.
a. Baseline blood pressure, in mmHg, per quartile of each GRS.
b. Blood pressure response to a low-sodium intervention, in mmHg, per quartile of each GRS.
c. Blood pressure response to a high-sodium intervention, in mmHg, per quartile of each GRS.
d. Blood pressure response to a potassium intervention while maintaining a high-sodium diet, in mmHg, per quartile of each GRS.
DBP=Diastolic blood pressure; GRS=Genetic risk score; MAP=Mean arterial pressure; SBP=Systolic blood pressure
Beta coefficients, standard errors and P values corresponding to each standard deviation increase in GRS for baseline BP and BP responses to the interventions are shown in Table 2. As expected, significant positive associations between SBP, DBP and MAP GRSs and respective baseline SBP, DBP, and MAP levels were identified after adjusting for age, gender, BMI, and field center (P=3.4×10−9, 1.9×10−7, and 8.0×10−6, respectively). Conversely, GRSs for SBP, DBP, and MAP associated with smaller respective SBP (P=3.0×10−4), DBP (P=0.05), and MAP (P=0.05) decreases in response to low-sodium diet after adjusting for age, gender, BMI, field center and baseline BP. Similarly, SBP GRS associated with smaller increases in SBP in response to high-sodium (P=0.04) and high-sodium plus potassium supplementation (P=0.04), after multivariable adjustment. No associations between DBP and MAP GRSs and corresponding DBP and MAP responses to the high-sodium and high-sodium plus potassium supplementation interventions were identified. Furthermore, addition of a quadratic term to models showing significant linear relations did not identify any non-linear trends (data not shown), and addition of interaction terms for age, gender, and BMI did not identify any significant interactions (data not shown).
Table 2.
Baseline blood pressure and blood pressure response to dietary interventions per standard deviation increase in GRS.
| SBP | DBP | MAP | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Study Phase | β* | SE | P-value | β* | SE | P-value | β* | SE | P-value |
| Baseline† | 1.64 | 0.28 | 3.4×10−9 | 1.11 | 0.21 | 1.9×10−7 | 1.00 | 0.22 | 8.0×10−6 |
| Low Sodium‡ | 0.50 | 0.14 | 3.0×10−4 | 0.24 | 0.12 | 0.05 | 0.22 | 0.11 | 0.05 |
| High Sodium‡ | −0.28 | 0.14 | 0.04 | −0.08 | 0.13 | 0.55 | −0.12 | 0.12 | 0.31 |
| Potassium‡ | 0.27 | 0.13 | 0.04 | 0.13 | 0.10 | 0.19 | 0.10 | 0.10 | 0.31 |
BP=Blood pressure; DBP=Diastolic blood pressure; MAP=Mean arterial pressure; SBP=Systolic blood pressure; SE=Standard error.
Results of a linear mixed model indicating change in BP, in mmHg, per standard deviation increase in BP GRS.
Adjusted for age, gender, and baseline body mass index.
Adjusted for age, gender, baseline body mass index, and baseline blood pressure.
Post-hoc analyses exploring the role of usual dietary sodium and potassium intake in the association between BP GRS and BP responses to dietary sodium and potassium were conducted. In univariate analyses, increasing BP GRS marginally positively correlated with habitual dietary sodium and potassium intake (P=0.06 and 0.06, respectively) (Table S4, please see http://hyper.ahajournals.org). However, further adjustment for baseline sodium and potassium had no influence on the association between BP GRS and BP responses to the dietary sodium and potassium interventions (Table S5 and Figure S3, please see http://hyper.ahajournals.org).
Additional analyses examining prGRSs, restricted to loci near salt sensitivity genes, and BP responses to dietary sodium and potassium are presented in Figure S4 and Tables S6 and S7 (please see http://hyper.ahajournals.org). Consistent with the full GRS, those in higher prGRS quartiles had significantly smaller decreases in SBP, DBP and MAP in response to the low-sodium intervention (P=5.2×10−3, 0.02, and 0.02, respectively; Figure S4b please see http://hyper.ahajournals.org). Similarly, each standard deviation increase in prGRSs for SBP, DBP, and MAP associated with smaller respective SBP (P=1.1×10−3), DBP (P=0.02), and MAP (P=1.4×10−3) decreases in response to low-sodium diet after adjusting for age, gender, BMI, field center and baseline BP (Table S7 please see http://hyper.ahajournals.org). Similar results were obtained when prGRSs comprised of loci identified in any population were utilized (data not shown).
Discussion
In the first study to explore the relationship between BP GRS and BP responses to dietary sodium and potassium, we identified a significant inverse relation between BP GRS and salt- and potassium-sensitivity of BP. In other words, as the number of BP risk alleles increased BP responses to dietary sodium and potassium intake tended to decrease. These findings were consistent across BP responses to the low-sodium intervention and SBP responses to high dietary sodium and potassium intake. Furthermore, results were similar when examining BP GRS according to quartile or as a continuous variable. In contrast, BP GRS was significantly and positively associated with baseline BP levels. While the direction of association between BP GRS and baseline BP was consistent with our a priori expectation, its inverse relations with the BP response measures were in contrast to our original hypotheses. These data may provide novel implications regarding the relationship between BP responses to dietary sodium and potassium intakes and BP regulation.
As presumed BP endophenotypes, we hypothesized that those with higher BP GRS would display increased salt- and potassium-sensitivity compared to those with lower BP GRS. Our finding of an inverse relation between BP GRS and magnitude of BP responses to the dietary interventions was unexpected. Previous studies have suggested that high dietary sodium intake may impair endothelial dependent vasodilation (36, 37). Based on these findings, we speculated that habitual high sodium intake could blunt endothelium-dependent vasodilation and subsequent BP responses to acute salt-depletion and salt-loading. In our study, usual dietary sodium intake, defined as baseline urinary sodium excretion, did indeed marginally correlate with BP GRS in post-hoc univariate analyses. However, additional adjustment for usual sodium intake had no influence on the observed associations between BP GRS and salt- and potassium-sensitivity. Thus, our preliminary findings do not support usual sodium intake as a driver of the inverse relation between BP GRS and salt- and potassium-sensitivity, suggesting that other possible explanations for the observed relationship should be considered. Given recent evidence from GWAS meta-analyses refuting a single dominant effect of renal sodium handling on BP (8), we also speculated that the observed association of BP GRS with salt-resistance may reflect relatively larger influences of BP variants in pathways unrelated to salt- and potassium-sensitivity when compared to those linked to BP through the BP response endophenotypes. However, analyses restricted to loci near known ‘salt- and potassium-sensitivity genes’ were consistent with those of our primary analysis, and, therefore, do not support this alternate explanation.
Numerous studies have identified salt-sensitivity as a risk factor for hypertension (18, 19, 38). Interestingly, our findings suggest that the relationship between GRS and elevated BP could also be mediated by salt-resistance. While there is a paucity of research exploring whether salt- and potassium-resistance may associate with altered BP regulation and hypertension phenotypes in humans, animal evidence supports this possibility (39, 40). Atrial natriuretic peptide, which increases in response to high sodium, has been shown to reduce BP by increasing renal excretion of salt and water, promoting vasodilation, and increasing vascular permeability (41–43). Interestingly, mice lacking the guanylyl cyclase-A receptor for atrial natriuretic peptide had chronically elevated BP that remained unchanged in response to minimal or high-salt diet (39). Thus, our findings add to the sparse but accumulating literature suggesting a salt-resistant model of hypertension. Future studies that prospectively explore the association between salt-resistance and hypertension development in humans are needed to fully articulate this relationship.
As expected, BP GRS was significantly and positively associated with baseline BP. Our finding of SBP and DBP increases of 1.64 and 1.11 mmHg, respectively, per standard deviation increase in BP GRS were nearly identical to the corresponding increases of 1.65 and 1.06 mmHg increases in SBP and DBP, respectively, reported previously by Ehret and colleagues (6). These data strongly suggest that the observed inverse association between BP GRS and salt- and potassium-sensitivity cannot be attributed to errors in designation and coding of risk alleles. Furthermore, although the associations between BP GRS and baseline BP values were similar using East Asian specific and all ancestry BP GRS, effect sizes per standard deviation increase in GRS were roughly 0.26, 0.13, and 0.10 mmHg larger for SBP, DBP, and MAP, respectively, using the all ancestry BP GRS. These findings add to the accumulating evidence that variants identified in specific ancestry groups may be relevant to populations with distinct LD structures.
This study has several strengths. To our knowledge, this is the first study to examine the relationship between BP GRS and salt- and potassium-sensitivity of BP. The recruitment of all Han Chinese participants who were also similar with respect to lifestyle risk factors such as diet and physical activity should have minimized environmental variation and increased power to detect BP GRS-phenotype associations. The response rates to the dietary interventions were high, and compliance, as assessed by urinary excretion of sodium and potassium during each intervention period, was excellent. Measurement error was reduced and power enhanced by the large number of BP measurements that were collected for each participant. Finally, stringent quality control procedures were used during measurement of BP and the other study covariables, conduct of the dietary interventions, genotyping, and marker data cleaning. This study also has several limitations. We hypothesized that habitual sodium intake could have mediated the inverse associations of BP GRS with salt- and potassium-sensitivity. To explore this hypothesis, we assessed whether the observed association was attenuated when additionally adjusting for baseline urinary sodium excretion values. Since baseline urinary sodium excretion may not accurately represent habitual sodium intake, in part due to infradian rhythmic sodium retention and excretion patterns (44, 45), these findings should be considered preliminary. However, the utilization of urinary sodium excretion measured over three-day periods should be a more accurate proxy of sodium intake than traditional 24-hour urinary sodium measures. Numerous longitudinal measures of urinary sodium excretion, each measured over multiple days, may be needed to better represent habitual dietary sodium intake and address this research question. In addition, to test whether our findings were consistent we employed a prGRS, which included only variants near genes with known influence on salt- and potassium-sensitivity or renal electrolyte handling. However, given the strong LD of variants across regions that frequently include many genes, it is difficult to discern whether all variants in this analysis truly exert their influence through the presumed nearby sodium- or potassium-related gene.
Supplementary Material
Perspectives.
In the first study to explore the association between BP GRS and BP responses to dietary sodium and potassium intake, we observed an unexpected, inverse relation of BP GRS with salt- and potassium-sensitivity of BP. As expected, BP GRS was positively and significantly correlated with baseline BP. Based on these findings, we cautiously speculate that salt- and potassium-resistance could act as mediators between genetic background and elevated BP, representing novel markers of future hypertension risk. However, prospective studies are warranted to clearly delineate whether individuals resistant to the typical BP altering effects of dietary sodium and potassium may have an increased risk of hypertension and related CVD events.
Novelty and significance.
- What Is New?
- Blood pressure salt sensitivity was examined using a genetic risk score.
- What Is Relevant?
- Salt- and potassium-resistance may mediate the relationship between genetic risk for hypertension and elevated blood pressure.
- Summary
- A blood pressure genetic risk score was used to examine blood pressure responses to dietary sodium and potassium interventions.
- When controlling for baseline blood pressure, participants with lower genetic risk scores were more salt sensitive and participants with higher genetic risk scores were less salt sensitive.
Acknowledgements
The Genetic Epidemiology Network of Salt Sensitivity (GenSalt) is supported by a cooperative agreement project grant (U01HL072507, R01HL087263, and R01HL090682) from the National Heart, Lung and Blood Institute, National Institutes of Health, Bethesda, MD.
Source(s) of funding
The Genetic Epidemiology Network of Salt Sensitivity (GenSalt) is supported by a cooperative agreement project grant (U01HL072507, R01HL087263, and R01HL090682) from the National Heart, Lung and Blood Institute, National Institutes of Health, Bethesda, MD. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of manuscript.
Footnotes
Conflicts of interest and disclosures: none
References
- 1.Lim SS, Vos T, Flaxman AD, et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2224–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mills KT, Bundy JD, Kelly TN, Reed JE, Kearney PM, Reynolds K, Chen J, He J. Global Disparities of Hypertension Prevalence and Control: A Systematic Analysis of Population-Based Studies From 90 Countries. Circulation. 2016;134:441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He J, Gu D, Chen J, Wu X, Kelly TN, Huang JF, Chen JC, Chen CS, Bazzano LA, Reynolds K, Whelton PK, Klag MJ. Premature deaths attributable to blood pressure in China: a prospective cohort study. Lancet. 2009;374:1765–1772. [DOI] [PubMed] [Google Scholar]
- 4.Kelly TN, He J. Genomic epidemiology of blood pressure salt sensitivity. J Hypertens. 2012;30:861–873. [DOI] [PubMed] [Google Scholar]
- 5.Biino G, Parati G, Concas MP, Adamo M, Angius A, Vaccargiu S, Pirastu M. Environmental and genetic contribution to hypertension prevalence: data from an epidemiological survey on Sardinian genetic isolates. PLoS One. 2013;8:e59612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehret GB, Munroe PB, Rice KM, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato N, Loh M, Takeuchi F, et al. Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat Genet. 2015;47:1282–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehret GB, Ferreira T, Chasman DI, et al. The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nat Genet. 2016;48:1171–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newton-Cheh C, Johnson T, Gateva V, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy D, Ehret GB, Rice K, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wain LV, Verwoert GC, O’Reilly PF, et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet. 2011;43:1005–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu C, Kraja AT, Smith JA, et al. Meta-analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci. Nat Genet. 2016;48:1162–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aaron KJ, Sanders PW. Role of dietary salt and potassium intake in cardiovascular health and disease: a review of the evidence. Mayo Clin Proc. 2013;88:987–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mente A, O’Donnell MJ, Rangarajan S, et al. Association of urinary sodium and potassium excretion with blood pressure. N Engl J Med. 2014;371:601–611. [DOI] [PubMed] [Google Scholar]
- 15.Li C, Yang X, He J, Hixson JE, Gu D, Rao DC, Shimmin LC, Huang J, Gu CC, Chen J, Li J, Kelly TN. A gene-based analysis of variants in the serum/glucocorticoid regulated kinase (SGK) genes with blood pressure responses to sodium intake: the GenSalt Study. PLoS One. 2014;9:e98432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelly TN, Hixson JE, Rao DC, et al. Genome-wide linkage and positional candidate gene study of blood pressure response to dietary potassium intervention: the genetic epidemiology network of salt sensitivity study. Circ Cardiovasc Genet. 2010;3:539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Svetkey LP, McKeown SP, Wilson AF. Heritability of salt sensitivity in black Americans. Hypertension. 1996;28:854–858. [DOI] [PubMed] [Google Scholar]
- 18.Barba G, Galletti F, Cappuccio FP, Siani A, Venezia A, Versiero M, Della Valle E, Sorrentino P, Tarantino G, Farinaro E, Strazzullo P. Incidence of hypertension in individuals with different blood pressure salt-sensitivity: results of a 15-year follow-up study. J Hypertens. 2007;25:1465–1471. [DOI] [PubMed] [Google Scholar]
- 19.Weinberger MH, Fineberg NS. Sodium and volume sensitivity of blood pressure. Age and pressure change over time. Hypertension. 1991;18:67–71. [DOI] [PubMed] [Google Scholar]
- 20.Group GCR. GenSalt: rationale, design, methods and baseline characteristics of study participants. J Hum Hypertens. 2007;21:639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He J, Kelly TN, Zhao Q, et al. Genome-wide association study identifies 8 novel loci associated with blood pressure responses to interventions in Han Chinese. Circ Cardiovasc Genet. 2013;6:598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perloff D, Grim C, Flack J, Frohlich ED, Hill M, McDonald M, Morgenstern BZ. Human blood pressure determination by sphygmomanometry. Circulation. 1993;88:2460–2470. [DOI] [PubMed] [Google Scholar]
- 23.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuchsberger C, Abecasis GR, Hinds DA. minimac2: faster genotype imputation. Bioinformatics. 2015;31:782–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacArthur J, Bowler E, Cerezo M, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017;45:D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ganesh SK, Chasman DI, Larson MG, et al. Effects of long-term averaging of quantitative blood pressure traits on the detection of genetic associations. Am J Hum Genet. 2014;95:49–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato N, Takeuchi F, Tabara Y, et al. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet. 2011;43:531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelly TN, Takeuchi F, Tabara Y, et al. Genome-wide association study meta-analysis reveals transethnic replication of mean arterial and pulse pressure loci. Hypertension. 2013;62:853–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franceschini N, Fox E, Zhang Z, et al. Genome-wide association analysis of blood-pressure traits in African-ancestry individuals reveals common associated genes in African and non-African populations. Am J Hum Genet. 2013;93:545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu X, Wang L, Lin X, et al. Genome-wide association study in Chinese identifies novel loci for blood pressure and hypertension. Hum Mol Genet. 2015;24:865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganesh SK, Tragante V, Guo W, et al. Loci influencing blood pressure identified using a cardiovascular gene-centric array. Hum Mol Genet. 2013;22:1663–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson T, Gaunt TR, Newhouse SJ, et al. Blood pressure loci identified with a gene-centric array. Am J Hum Genet. 2011;89:688–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tragante V, Barnes MR, Ganesh SK, et al. Gene-centric meta-analysis in 87,736 individuals of European ancestry identifies multiple blood-pressure-related loci. Am J Hum Genet. 2014;94:349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DuPont JJ, Hill MA, Bender SB, Jaisser F, Jaffe IZ. Aldosterone and vascular mineralocorticoid receptors: regulators of ion channels beyond the kidney. Hypertension. 2014;63:632–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jablonski KL, Racine ML, Geolfos CJ, Gates PE, Chonchol M, McQueen MB, Seals DR. Dietary sodium restriction reverses vascular endothelial dysfunction in middle-aged/older adults with moderately elevated systolic blood pressure. J Am Coll Cardiol. 2013;61:335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamelas PM, Mente A, Diaz R, Orlandini A, Avezum A, Oliveira G, Lanas F, Seron P, Lopez-Jaramillo P, Camacho-Lopez P, O Donnell MJ, Rangarajan S, Teo K, Yusuf S. Association of Urinary Sodium Excretion With Blood Pressure and Cardiovascular Clinical Events in 17,033 Latin Americans. Am J Hypertens. 2016;29:796–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez MJ, Wong SK, Kishimoto I, Dubois S, Mach V, Friesen J, Garbers DL, Beuve A. Salt-resistant hypertension in mice lacking the guanylyl cyclase-A receptor for atrial natriuretic peptide. Nature. 1995;378:65–68. [DOI] [PubMed] [Google Scholar]
- 40.Rieg T, Bundey RA, Chen Y, Deschenes G, Junger W, Insel PA, Vallon V. Mice lacking P2Y2 receptors have salt-resistant hypertension and facilitated renal Na+ and water reabsorption. FASEB J. 2007;21:3717–3726. [DOI] [PubMed] [Google Scholar]
- 41.Curry FR. Atrial natriuretic peptide: an essential physiological regulator of transvascular fluid, protein transport, and plasma volume. J Clin Invest. 2005;115:1458–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campese VM, Tawadrous M, Bigazzi R, Bianchi S, Mann AS, Oparil S, Raij L. Salt intake and plasma atrial natriuretic peptide and nitric oxide in hypertension. Hypertension. 1996;28:335–340. [DOI] [PubMed] [Google Scholar]
- 43.Lang RE, Unger T, Ganten D. Atrial natriuretic peptide: a new factor in blood pressure control. J Hypertens. 1987;5:255–271. [DOI] [PubMed] [Google Scholar]
- 44.Lerchl K, Rakova N, Dahlmann A, et al. Agreement between 24-hour salt ingestion and sodium excretion in a controlled environment. Hypertension. 2015;66:850–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rakova N, Jüttner K, Dahlmann A, et al. Long-term space flight simulation reveals infradian rhythmicity in human Na(+) balance. Cell Metab. 2013;17:125–131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.