Abstract
Recent technological advances led to the discovery of hundreds to thousands of peptides and small proteins (microproteins) encoded by small open reading frames (smORFs). Characterization of new microproteins demonstrate their role in fundamental biological processes and highlight the value in discovering and characterizing more microproteins. The elucidation of microprotein-protein interactions (MPIs) is useful for determining the biochemical and cellular roles of microproteins. In this study, we characterize the protein-interaction partners of mitochondrial elongation factor 1 microprotein (MIEF1-MP) using a proximity labeling strategy that relies on APEX2. MIEF1-MP localizes to the mitochondrial matrix where it interacts with the mitochondrial ribosome (mitoribosome). Functional studies demonstrate that MIEF1-MP regulates mitochondrial translation via its binding to the mitoribosome. Loss of MIEF1-MP decreases the mitochondrial translation rate, while elevated level of MIEF1-MP increases the translation rate. The identification of MIEF1-MP reveals a new gene involved in this process.
Keywords: MIEF1, microprotein, MIEF1-MP, mitochondrial translation, APEX2, microprotein- protein interaction
Graphical Abstract
INTRODUCTION
Protein-coding small open reading frames (smORFs) are a group of understudied genes with fundamental roles in biology.1, 2 Most smORFs are non-annotated because gene-finding algorithms require a minimum length requirement for gene assignment.3 About half of all annotated smORFs are upstream of longer open reading frames (ORFs)4 where they function as cis-regulators of protein translation—in most cases, the presence of an upstream ORF (uORF) reduces translation of the downstream ORF.4 However, the discovery of hundreds to thousands of previously non-annotated smORFs and microproteins, which has come about due to advances in computational,5–8 genomic,9–13 and proteomic7, 14–16 technologies, revealed many smORFs that are not uORFs. Evolutionary conservation indicated that some of these smORFs would be translated into functional microproteins and recent work in the field has validated this thinking.17–21
Functional studies in flies and mammals, for instance, have demonstrated that at least some of the microproteins play fundamental biological roles in development, metabolism and muscle function.22–24 Knockout of tarsal-less/polished rice (Tal/Pri) gene in flies led to pronounced developmental phenotypes.25 Tal/Pri is a polycistronic mRNA with four smORFs that encode three 11-amino acid and one 32-amino acid microproteins that are required for proper Drosophila embryogenesis.23, 26 Subsequent studies of the Tal/Pri microproteins revealed that these microproteins bind to the Ubr3 ubiquitin ligase in flies and promotes the limited proteolysis and thus activation of the Shaven baby (Svb) transcription factor driving epidermal differentiation.17, 18
There are also biologically active microproteins in mammals.19–21, 27, 28 For example, myoregulin (MLN) is a conserved microprotein that is encoded by a skeletal muscle-specific transcript that interacts with the SERCA cation channel to impede Ca2+ uptake into sarcoplasmic reticulum (SR) and controls muscle function. Deletion of MLN from mice enhances Ca2+ handling in skeleton muscles and improves exercise performance in vivo, highlighting the role of MLN as a regulator of skeleton muscle physiology.19 CYREN is a mammalian microprotein that interacts with the Ku70/80 heterodimer to inhibit error-prone DNA repair during the S and G2 phases of the cell cycle.20, 21 These examples highlight the importance of studying microproteins to understand critical biological processes.
Here, we focus on the molecular and biochemical characterization of a uORF located in the 5’ UTR of mitochondrial dynamics protein MID51 (MIEF1) gene. MIEF1 regulates mitochondrial fission,29 and the initial characterization of the MIEF1 uORF which encodes the MIEF1 microprotein (MIEF1-MP) revealed a mitochondrial microprotein that is also involved in mitochondrial fission.30 MIEF1-MP is 70-amino acid long and has been detected by proteomic analysis in several cell lines including HEK293, HeLa, and THP1 cells as well as in vivo in the intestinal tissue.7, 31 Publicly available ribosome profiling data also confirmed the translation of the MIEF1 uORF in HEK293, HeLa, PC3, and THP1 cells.32 MIEF1-MP is evolutionarily conserved in vertebrates and contains an LYR motif. The LYR motif is found in 11 mammalian proteins and six of these interact with the mitochondrial respiration machinery.33–37 Others are involved in Fe-S Cluster Biogenesis (LYRM4),38 insulin signaling (LYRM1),39 complex I phylogenetic profile COPP (LYRM5),37 or have unknown functions.40 Previous work indicates that MIEF1-MP promotes mitochondrial fission using the LYR domain, as LYR mutants were inactive.30 One caveat in these functional studies is that they relied on MIEF1-MP overexpression, and in this study, we observe that overexpression results in detection of non-functional MIEF1-MP-protein interactions.
We utilized a biochemical approach to identify the MIEF1-MP protein interactors using an in vivo proximity method that relies on the engineered APEX2 (APEX) protein.41, 42 Using this approach, we found that MIEF1-MP interacts with the mitochondrial ribosome (mitoribosome). The mitochondrial genome encodes 38 genes: 14 protein coding, 22 for mitochondrial tRNA, and 2 for mitochondrial rRNA. The mitoribosome translates several mitochondrially encoded mRNAs to produce the 13 subunits of respiratory complexes I, III, IV, and V.43 We measured the levels of newly synthesized mitochondrial-encoded proteins and found that MIEF1-MP is a regulator of mitochondrial translation. Loss of MIEF1-MP decreases the rate of global mitochondrial translation while elevated levels of MIEF1-MP increase the rate of newly synthesized mitochondrial proteins. These results reveal MIEF1-MP as a new gene involved in regulating mitochondrial translation.
MATERIALS AND METHODS
Immunofluorescence and Confocal Imaging
HeLa cells were seeded onto a coverslip (Fisher Scientific, #12-541-B) in a six-well plate, which was pretreated with 50 μg/mL poly-L-lysine (Sigma, #P1399). The next day, cells were transfected with 1 μg of C-terminal FLAG tagged MIEF1 microprotein (MIEF1-MP-FLAG) or C-terminal APEX-MYC tagged MIEF1-MP (MIEF1-MP-APEX-MYC) plasmid using Lipofectamine 2000. Forty-eight hours post-transfection, cells were fixed with 4% paraformaldehyde (Polysciences, Inc., #18814) and permeabilized with 0.1% saponin (Alfa Aesar, #A18820). After being incubated with 4% BSA in phosphate buffered saline (PBS) for 1 hour at room temperature, cells were stained with primary antibodies (mouse anti-FLAG and rabbit anti-TOM20 or rabbit anti-MYC and mouse anti-TOM20) at a 1:1000 dilution overnight at 4 °C. Cells were washed three times with PBS, followed by incubation with secondary antibodies (goat anti-mouse Alexa Fluor 488 and goat anti-rabbit Alexa Fluor 546 or goat anti-rabbit Alexa Fluor 488 and goat anti-mouse Alexa Fluor 546, 1:500 in PBS) for 1 hour at room temperature. Nuclei were counterstained with Hoechst 33258 (Sigma, # 94403; 1:2000 in PBS). After three PBS washes, the coverslip was mounted with Prolong Gold Antifade Mountant (Life Technologies, # P36930) and analyzed by confocal imaging using a Zeiss LSM 710 laser scanning confocal microscope with a 63× oil immersion objective. Images were analyzed with FIJI software.
Proteinase K Protection Assay for Sub-Mitochondrial Localization
HEK293T cells were seeded in 10 cm cell culture plates and next day the cells were transfected with MIEF1-FLAG plasmid using Lipofectamine 2000. The following day, cells were harvested, suspended in isolation buffer (225 mM Mannitol 75 mM Sucrose 50 mM NaHEPES pH 7.4 supplemented with Roche complete protease inhibitor cocktail tablet), dounce homogenized, and spun to isolate different cellular fractions44. The isolated mitochondrial pellet was washed three times and then suspended in isolation buffer, 2 mM HEPES or 0.3% Triton X-100, followed by addition of Proteinase K to select samples. After 30 minutes incubation on ice, 1 mM phenylmethanesulfonylfluoride (PMSF) was added to quench the protease and trichloroacetic acid (TCA, 12% final concentration) was added to precipitate the samples, which were then washed with acetone. The pellets were dissolved in 2X SDS loading dye, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS– PAGE) and analyzed by Western blotting using the indicated antibodies.
Co-Immunoprecipitation
FLAG-tagged microprotein constructs or the empty pcDNA3.1 (+) vector were transfected into a 10-cm dish of HEK293T cells using Lipofectamine 2000 according to the manufacturer’s protocol. Twenty-four hours post-transfection, cells were harvested and lysed in IP lysis buffer (ThermoFisher, # 87787) supplemented with a Roche complete protease inhibitor cocktail tablet and 1 mM PMSF. Cells were lysed on ice for 10 min followed by centrifugation at 20,000g for 20 min at 4 °C to remove cell debris. Cell lysates were added to prewashed mouse IgG agarose beads (Sigma, # A0919) and rotated at 4 °C for 1 hour. The supernatants were collected and added to prewashed anti-FLAG M2 Affinity Gel (Sigma, # A2220). The suspensions were rotated at 4 °C for 2 hours and washed three times with TBST. Bound proteins were eluted with 3×FLAG peptide (Sigma, # F4799) at 4 °C for 1 hour.
Biotin-Phenol Labeling in Live Cells and Streptavidin Pull Down
Biotin-phenol labeling in live cells was performed according to previously published protocols.41, 45 Briefly, constructs harboring the MIEF1-MP–APEX fusion proteins or APEX (control sample) was transfected into HEK293T cells using Lipofectamine 2000. Twenty-four hours post-transfection, cell culture medium was changed to fresh growth medium containing 500 μM biotin-tyramide (CDX-B0270, Adipogen). After incubation at 37 °C for 30 min, H2O2 was added to each plate at a final concentration of 1 mM, and the plates were gently agitated for 1 min. Cells were then washed three times with a quenching solution (5 mM Trolox, 10 mM sodium azide, and 10 mM sodium ascorbate in PBS), and pelleted by centrifugation at 1000g for 5 min. Cell pellets were lysed on ice for 20 min in RIPA buffer (Thermo catalog no. 89901) supplemented with a Roche complete protease inhibitor cocktail tablet and 1 mM PMSF followed by centrifugation at 20,000g for 20 min at 4 °C to remove cell debris. Cell lysates were added to prewashed streptavidin agarose resin (ThermoFisher, # 20359), rotated at 4 °C overnight, and then washed three times with TBST and 0.5% (v/v) sodium dodecyl sulfate (SDS) at room temperature. Bound proteins were eluted with 2×SDS by boiling at 95 °C and analyzed by proteomics and Western blotting.
Proteomic Analysis of Co-Immunoprecipitation and Biotin-Phenol Labeling
For proteomics, eluted samples were precipitated with trichloroacetic acid (TCA, MP Biomedicals, # 196057) overnight at 4 °C or using Methanol-Chloroform. Dried pellets were dissolved in 8 M urea, reduced with 5 mM tris(2-carboxyethyl) phosphine hydrochloride (TCEP, ThermoFisher, #20491), and alkylated with 10 mM iodoacetamide (Sigma, # I1149). Proteins were then digested overnight at 37 °C with trypsin (Promega, # V5111). The reaction was quenched with formic acid at a final concentration of 5% (v/v). Digested samples were analyzed on a Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer.
Metabolic Labeling of Mitochondrial Translation Products Using AHA
HEK293T cells were transfected with MIEF1-MP-FLAG (1 µg per well) for 24 hours or MIEF1 siRNA (100 pmol) for 72 hours in triplicates with respective controls pcDNA vector and negative siRNA. Similarly, for the rescue experiment, 293T cells were treated with MIEF1 siRNA (100 pmol) for 48 hours followed by pcDNA vector or MIEF1-MP-FLAG (1 µg per well) for 24 hours in triplicates. The pulse labeling of mitochondrial translation products was then performed as per a reported protocol with modifications46. Specifically, cells at 80% confluence were incubated with 100 μM AHA in HBS buffer co ntaining 100 μg/ml of cytosolic translation inhibitor emetine for 3 hours. After AHA labeling, the cells were lysed, normalized and a “click” reaction was performed by incubating the cell lysates with IRDye800® conjugated alkyne (LI-COR, #929–60002) for 3 hours at room temperature. The proteins were TCA precipitated, washed by ice-cold acetone and air-dried before being subjected SDS-PAGE separation using the Bolt 4–12% Bis-Tris Protein Gel (Invitrogen). The gel was visualized, and the images analyzed using LI-COR Odyssey CLx.
RESULTS AND DISCUSSION
MIEF1 microprotein translates from the smORF upstream of the MIEF1 gene
MIEF1 is an outer mitochondrial membrane protein that regulates mitochondrial membrane dynamics. MIEF1 interacts with Drp1 and Mff in a trimeric complex to regulate Mff-induced Drp1 accumulation. Overexpression of MIEF1 sequesters Drp1 to the outer mitochondrial membrane and promotes mitochondrial fusion and elongation. MIEF1 knockdown resulted in mitochondrial fission and fragmentation.47, 48
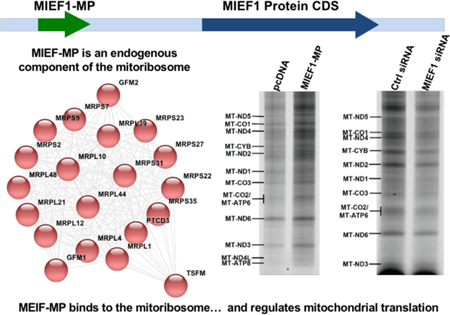
Recently, a protein-coding smORF was identified in the 5’ UTR region of MIEF1 gene that produces the MIEF1 microprotein (MIEF1-MP). Currently, there are no antibodies for MIEF1-MP so we needed to assure ourselves that it is being expressed in the HEK293T cells that we were using. Ribosome profiling (Ribo-Seq) is a ribosome footprinting method that identifies translated regions of the transcriptome. Analysis of publicly available HEK293T Ribo-Seq data indicates translation of MIEF1-smORF (Figure 1A) and our proteomics data validated this result through detection of a tryptic peptide, “YTDRDFYFASIR”, from MIEF1-MP (Figure 1B). MIEF1-MP is a 70-amino acid long microprotein that is conserved between humans and zebrafish (Figure 1C). Previous work indicated a role for MIEF1-MP in mitochondrial fission.30 Here, we sought to obtain a molecular understanding of MIEF1-MP biochemistry and use this information to explain or predict MIEF1-MP function.
Figure 1.
MIEF1-MP expression and conservation. (A) MIEF1-MP is encoded by an upstream smORF (uORF) in the MIEF1 mRNA. RNA-Seq (magenta) and Ribo-Seq (green) data from HEK293T cells indicates translation of the MIEF1-MP smORF and the MIEF1 (ORF) . (B) A tryptic peptide from the MIEF1 microprotein detected by shotgun mass spectrometry validates that MIEF1-MP is a stable member of the proteome. (C) Sequence alignment of the MIEF1-MP demonstrates that this microprotein is conserved in numerous species at the protein level.
MIEF1 microprotein localizes to the mitochondrial matrix
Mitochondria are essential organelles found in mammalian cells and eukaryotes. Mitochondria are cellular powerhouses that produce more than 60% of the ATP in a cell through mitochondrial oxidative phosphorylation (OXPHOS) pathway. OXPHOS requires five multiprotein complexes in the inner mitochondrial membrane comprised of proteins encoded by the nuclear and mitochondrial genomes. In addition to energy production, mitochondria have additional roles, such as mediating programmed cell death (apoptosis).49, 50 Our goal in this study is to understand the mitochondrial pathways regulated by MIEF1-MP.
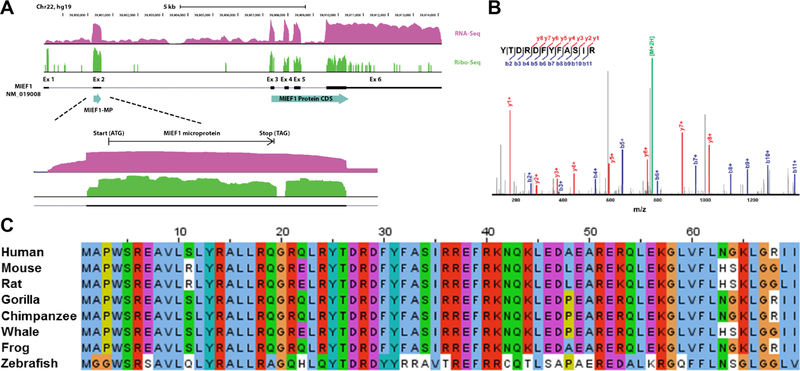
Sequence analysis of MIEF1-MP with TargetP51 predicts mitochondrial localization and MitoFates52 also predicts a TOM20 recognition motif (amino acids 12–16 and 66–70). Transient expression of MIEF1-MP-FLAG resulted in mitochondrial localization (Figure 2A), demonstrating that the FLAG epitope does not disrupt MIEF1-MP localization.30 The localization of MIEF1-MP-FLAG to the mitochondria is consistent with previous reports that utilized higher resolution microscopy to localize GFP-tagged MIEF1-MP to mitochondrial networks.30
Figure 2.
MIEF1-MP is a mitochondrial matrix microprotein. (A) MIEF1-MP-FLAG colocalizes with the mitochondrial marker TOM20 in Hela cells verifying its mitochondrial localization (Hoechst (blue) nuclear stain, TOM20 (red), MIEF1-MP (green), yellow/orange overlap). (B) Proteinase K digestion of MIEF1-MP-FLAG had a similar pattern to HSP60, a mitochondrial matrix marker, confirming its sub-cellular localization to the mitochondrial matrix in HEK293T cells.
The localization of MIEF1-MP within the mitochondria is unknown. Determining the mitochondrial location of MIEF1-MP would help understand its function. We used a proteinase K protection assay, which tests protein stability under conditions that selectively permeabilize the mitochondria to determine where the protein resides between the outer- or inner-mitochondrial membranes or the matrix. Mitochondria were isolated from MIEF1-MP-FLAG expressing 293T cells by differential centrifugation for the subcellular localization experiments.44 The isolated mitochondria were treated under three conditions that differentially regulate proteinase K access to sub-compartments of the mitochondria. Addition of proteinase K to mitochondria in isolation buffer degrades the mitochondrial outer membrane marker TOM20, but leaves TIM50, an inner mitochondrial membrane maker and HSP60, a mitochondrial matrix marker, unperturbed (Figure 2B, lane 2). Pre-treatment of mitochondria with 2 mM HEPES results in osmotic swelling and selective rupture of the outer mitochondrial membrane resulting in the degradation of TIM50 (Figure 2B, lane 4), and solubilizing mitochondria with Triton X-100 is necessary to proteolyze HSP60 (Figure 2B, lane 6). MIEF1-MP-FLAG degradation in the proteinase K assay had an identical pattern to that of HSP60, indicating that MIEF1-MP is a mitochondrial matrix microprotein.
MIEF1 microprotein-protein interactions
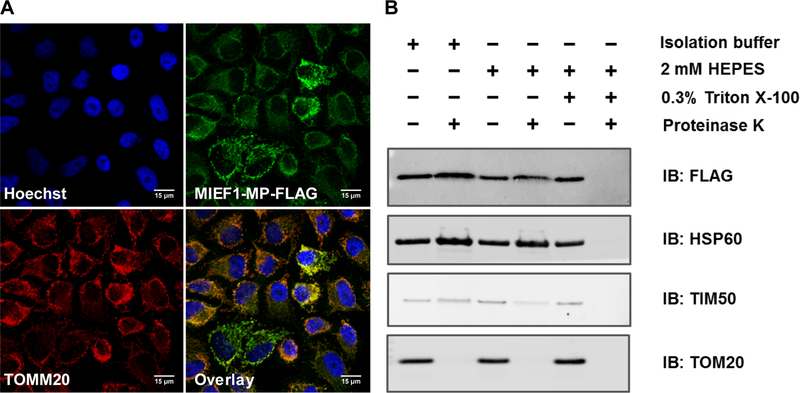
MIEF1-MP contains a Complex1_LYR domain40 that is defined by a conserved tripeptide L-Y-R (leucine/tyrosine/arginine) sequence close to its N-terminus and a downstream F (phenylalanine) residue (Figure 3A). The human genome contains 11 LYR-containing proteins, and 8 of these are mitochondrial proteins.40 Most LYR proteins are subunits (LYRM3 and LYRM6) or assembly factors (LYRM7, LYRM8, ACN9, and FMC1) of the oxidative phosphorylation (OXPHOS) core complexes in mitochondria. Human LYRMs are linked with diseases, such as insulin-resistance (LYRM1),39 muscular hypotonia (LYRM3),53 deficiency of multiple OXPHOS complexes (LYRM4),38 apoptosis in HIV-1 infection (LYRM6),54 encephalopathy and lactic acidosis (LYRM7/MZM1L),34 infantile leukoencephalopathy (LYRM8/succinate dehydrogenase assembly factor (SDHAF) 1)36 and alcohol dependence (acetate non-utilizing protein (ACN) 9/SDHAF3).35
Figure 3.
MIEF1-MP contains an LYR motif and interacts with ACPM/ NDUFAB1 when overexpressed. (A) A LOGO plot of MIEF1-MP with known LYR motif containing proteins highlights the conserved tripeptide L-Y-R (leucine/tyrosine/arginine) and downstream F (phenylalanine) (LYR and F) motif. (B) Immunoprecipitation (IP) of the MIEF1-MP-FLAG enriches ACPM/NDUFAB1 while mutating the LYR and F. to alanines abrogates this binding. (C) Proximity labeling with MIEF1-MP-APEX-MYC demonstrates ACPM/NDUFAB1 enrichment in living cells.
Several of the known LYRMs (LYRM3, LYRM4, LYRM6, and FMC1) interact with ACPM/NDUFAB1 which is part of complex I, 33, 40, 55, 56 and we sought to determine if MIEF1-MP associates with ACPM/NDUFAB1 via its LYR domain. We carried out an immunoprecipitation (IP) of cell lysates overexpressing MIEF1-MP-FLAG or an empty pcDNA vector (mock) using an anti-FLAG antibody and then analyzed the immunoprecipitates by proteomics. We found ACPM/NDUFAB1 enriched in the MIEF1-MP-FLAG overexpressing sample over the mock sample (Figure 3B). We mutated the L-Y-R (leucine/tyrosine/arginine) and downstream F (phenylalanine) residues of MIEF1-MP to alanines and repeated the immunoprecipitation-mass spectrometry experiment and found that the MIEF1-MP: ACPM/NDUFAB1 interaction requires the LYR motif (Figure 3B). This experiment reassured us that MIEF1-MP could partake in predicted protein-protein interactions, but we are always cautious not to overinterpret lysate experiments with structurally unknown microproteins because we might be observing spurious, non-endogenous interactions. We carried out a proximity labeling experiment using MIEF1-MP-APEX fusion protein to obtain cellularly relevant interaction since proximity labeling identifies protein complexes in the context of a living cell.41, 45, 57 In this approach, the APEX fused protein of interest is expressed in cells, followed by treatment with hydrogen peroxide (H2O2) in the presence of biotin-phenol. H2O2 fuels the catalytic oxidation of biotin-phenol by APEX to generate a highly reactive biotin-phenoxy radical. The lifetime of the radical is <1 ms that restricts the labeling to 20 nm radius. This results in the covalent labeling of proteins proximal to the APEX fusion protein with biotin, which are then enriched and analyzed to identify protein interactors in vivo. Before performing the proximity labeling experiment, we validated that MIEF1-MP-APEX-MYC localizes to the mitochondria by immunofluorescence (Figure S1). The proximity labeling experiments indicate that ACPM/NDUFAB1 interacts with the MIEF1-MP-APEX-MYC fusion protein but not MIEF1mut-MP-APEX-MYC (LYR and F are all mutated to alanines), consistent with the FLAG immunoprecipitation experiments (Figure 3C). Together the data confirm that overexpressed MIEF1-MP interacts with the ACPM/NDUFAB1 protein via its LYR motif.
ACPM/NDUFAB1 has a role in lipid biosynthesis and respiratory complex 1 activity,58 and this led us to hypothesize that MIEF1-MP might influence these pathways in the mitochondria. However, we observed no effects of MIEF1-MP using a mitochondrial complex 1 activity assay (Figure S2A) or lipid biosynthesis (Figure S2B) suggesting that MIEF1-MP: ACPM/NDUFAB1 interaction is not functionally relevant. One problem might come from the fact that we observed these protein-protein interactions in cells overexpressing MIEF1-MP, which might promote biologically irrelevant interactions with higher concentrations of MIEF1-MP. Even the functional characterization of MIEF1-MP relied on overexpression since knockdown of MIEF1 mRNA would result in a knockdown of MIEF1-MP and MIEF1. This led us to the realization that we needed a better strategy to find and validate microprotein interactions, especially when we have to overexpress the microprotein.
Bioinformatic Reciprocal Immunoprecipitations to Discover Relevant MIEF1-MP Interactions
We searched the proteomics data from MIEF1-MP-APEX-MYC experiments to determine whether we had missed other potentially significant interactors. Our informatics workflow consisted of analyzing the data using Significance Analysis of INTeractome Express (SAINTexpress),59 which uses a predicted distribution of spectral counts for a real protein-protein interaction to filter out false positives, and Contaminant Repository for Affinity Purification (CRAPome),60 which removes proteins that are repeatedly, and non-specifically enriched in protein-interaction studies. Application of SAINT and CRAPome to the MIEF1-MP-APEX-MYC dataset resulted in 153 enriched proteins with an average spectral count of greater than or equal to 5. Though our list of potential interactors is greatly reduced, validating 153 protein interactions would be an experimental challenge.
Reciprocal immunoprecipitations are the gold standard for validating protein-protein interactions. To do this for 153 proteins would require antibodies for each of the proteins and for human MIEF1-MP, which we do not have and are not commercially available. To solve this problem, we turned to a bioinformatic approach. First, we used the bioinformatics tool STRING61 (Figure 4A) to identify which of the 153 proteins are known to interact with each other. The idea here is that the identification of multiple proteins from known complexes increases confidence that MIEF1-MP is involved in a legitimate protein-protein interaction. We removed enriched proteins with less than or equal to 2 interactors (singletons or proteins that are not part of complexes that contained more than 3 proteins) and identified five MIEF1-MP bound protein clusters (Figure 4A, color-coded clusters). The five clusters include complexes linked to the mitochondrial ribosome, isocitrate/succinate/malate dehydrogenase, pyruvate dehydrogenase, hydroxyacyl CoA dehydrogenase and electron transport chain.
Figure 4.
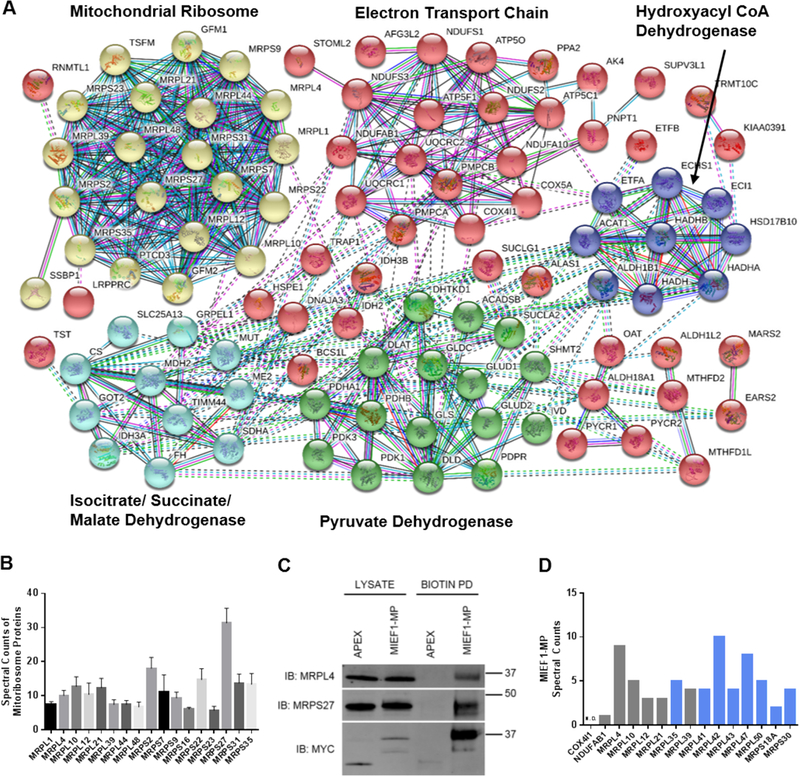
MIEF1-MP interacts with the mitoribosome. (A) The interaction network of the proteins enriched in the MIEF1-MP-APEX-MYC experiment after STRING analysis reveals MIEF1-MP enriched proteins that interact with each other. (B) Spectral counts for different members of the mitoribosome after the MIEF1-MP-APEX-MYC experiment. (C) Western blot validation of MIEF1-MP protein-protein interaction with some members of the mitoribosome (MRPL4 and MRPL27) by proximity labeling. The experiment used an MIEF1-MP-APEX-MYC expression construct for the proximity labeling and expression of MIEF1-MP-APEX-MYC was confirmed by western blot using anti-MYC antibodies. (D) An “in silico” reverse IP searching for MIEF1-MP peptides in mitoribosome protein data from the Bioplex database. Bar graph shows the number of spectral counts for MIEF1-MP peptides in the pulldown data for each of the mitoribosome proteins on the x-axis from the Bioplex database. In addition to the mitoribosome proteins enriched in the proximity labelling experiment (grey) additional mitoribosome proteins that were able to enrich MIEF1-MP were identified (blue).
This analysis reduced the number of likely MIEF1-MP interactors to 100 proteins, but we are still limited by a lack of suitable antibodies to validate these interactions. Instead, we felt that we could obtain the information we needed by searching publicly available proteome-wide, protein-protein interaction datasets. We decided to search the Bioplex protein interaction network database,62 generated from affinity-purification mass spectrometry data for more than 2500 human proteins in HEK293T cells. Lentiviral library of FLAG-HA-tagged ORFs were constructed and infected in HEK293T cells followed by immuno-purification, trypsin digestion, and analysis by LC-MS. Specific interactors for each bait protein was identified and assembled into the Bioplex human interactome network. The resulting database contains more 23,744 interactions among 7,668 proteins. This unbiased mapping of interactions also aids in the characterization of unknown or poorly studied proteins by suggesting possible cellular localization, biological process, and molecular function.62 Our strategy is to use this database to determine whether any MIEF1-MP-binding proteins enriched MIEF1-MP when they were used as bait proteins, providing us with reciprocal IP data without the need for any antibodies.
We downloaded the proteomics data for MIEF1-MP binding partners, which were the baits in this experiment, from the Bioplex database, if available. When the Bioplex data was collected and analyzed, MIEF1-MP was not annotated in the human RefSeq or UniProt databases and, therefore, would not appear as a member of any protein complex during the original analysis of the Bioplex data. We searched the downloaded Bioplex data against the human UniProt database with MIEF1-MP included. The analysis of ACPM/NDUFAB1 detected only a single spectral count from MIEF1-MP and while the mitoribosome proteins MRPL4, MRPL10, MRPL12, MRPL21, and MRPL39 had many spectral counts for MIEF1-MP (Table 1). Multiple mitoribosome proteins had high spectral counts (Figure 4B) in MIEF1-MP-APEX-MYC experiment. Overall, the mitoribosome proteins were the only cluster with substantial evidence of MIEF1-MP binding indicating that under endogenous conditions (i.e., no overexpression of MIEF1-MP) MIEF1-MP interacts primarily with the mitoribosome. We validated the MRPL4 and MRPS27 interaction by Western blot (Figure 4C and Figure S3). Lastly, we analyzed additional mitoribosome proteins in the Bioplex database, though these were not enriched in our MIEF1-MP-APEX-MYC experiment and discovered eight additional mitoribosome proteins that enriched MIEF1-MP (Figure 4D and Table S1). Also, we find that other mitochondrial proteins (COX4I1, Figure 4D) do not enrich MIEF1-MP demonstrating that MIEF1-MP selectively binds to mitoribosome. Our proteomics data support the results from an cryo-EM structure of the human mitoribosome isolated from HEK293S cells that identified MIEF1-MP as binding partner of the mitoribosome32 (referred to as L0R8F8 in this publication). Furthermore, MIEF1-MP was hypothesized to be a mitoribosome assembly factor because of its localization in the mitoribosome structure32, and this proposed model is in agreement with our translation results where the loss of MIEF1-MP inhibits mitoribosome translation while overexpression of MIEF1-MP accelerates translation (See below).
Table 1.
Spectral counts of MIEF1-MP for different baits in the Bioplex database (colors correspond to clusters in Figure 4A).
| Protein Name | MIEF1-MP Spectral Count |
|---|---|
| MRPL4 | 9 |
| MRPL10 | 5 |
| MRPL12 | 3 |
| MRPL21 | 3 |
| MRPL39 | 4 |
| MRPS2 | 0 |
| MRPS7 | 0 |
| MRPS27 | 0 |
| MRPS31 | 0 |
| PTCD3 | 0 |
| FH | 0 |
| GOT2 | 0 |
| IDH3A | 0 |
| SDHA | 0 |
| TIMM44 | 0 |
| ALDH1B1 | 0 |
| ETFA | 0 |
| ECHS1 | 0 |
| HADHB | 0 |
| HSD17B10 | 0 |
| ACADSB | 0 |
| DLD | 0 |
| GLUD2 | 0 |
| IVD | 0 |
| PDHB | 0 |
| PDK1 | 0 |
| PDK3 | 0 |
| SHMT2 | 0 |
| SUCLA2 | 0 |
| ALDH1L2 | 0 |
| IDH3B | 0 |
| ETFB | 0 |
| PYCR1 | 0 |
| PYCR2 | 0 |
| ATP5C1 | 0 |
| ATP5F1 | 0 |
| COX4I1 | 0 |
| COX5A | 0 |
| NDUFAB1 | 1 |
| NDUFS1 | 0 |
| NDUFS3 | 0 |
| NDUFA10 | 0 |
Effect on the Rate of Mitochondrial Translation
The mitoribosome is responsible for the translation of mitochondrial RNAs, and the data indicated a role for MIEF1-MP in this process (Figure S4). Although nuclear DNA encodes most mitochondrial proteins, several vital proteins are encoded by mitochondrial DNA (mtDNA) and synthesized by an independent mitochondrial translation system43. Mitochondrially encoded mRNA produce 13 subunits of respiratory complexes; NADH dehydrogenase subunits of complex I (MT-ND1, MT-ND2, MT-ND3, MT-ND4L, MT-ND4, MT-ND5, MT-ND6), cytochrome b of complex III (MT-CYB), cytochrome c oxidase subunits of complex IV (MT-CO1, MT-CO2, MT-CO3), and ATP synthase subunits of complex V (MT-ATP6, MT-ATP8). To determine whether MIEF1-MP modulates mitochondrial translation, we began by measuring the steady-state levels of the mtDNA encoded proteins by quantitative proteomics in the MIEF1-MP over-expressing cells. We observed a 6-fold increase in MIEF1-MP levels, but no change in the concentrations of mtDNA encoded proteins (Figure S5), indicating that MIEF1-MP does not affect steady state levels of mitochondrial encoded proteins. Next, we used a mitochondrial translational assay that measures newly synthesized proteins to determine whether MIEF1-MP can influence the translation rate of the mitochondrial proteome, which has been observed for other proteins in this complex.63, 64
The newly synthesized mtDNA-encoded proteins were quantified using the BioOrthogonal Non-Canonical Amino Acid Tagging (BONCAT) approach.46 BONCAT introduces a chemical handle into newly synthesized proteins that can be used to enrich and detect the protein(s). BONCAT relies on the introduction of the non-canonical amino acid azidohomoalanine (AHA) in place of methionine into newly synthesized proteins. Labeling with AHA places an azide functional group onto new proteins, and these proteins are subsequently enriched, detected, and quantified by clicking on a fluorophore or biotin.46 We overexpressed or knocked down MIEF1-MP in HEK293T cells and then incubated these cells with azidohomoalanine (AHA) to allow protein synthesis and AHA incorporation. These experiments were carried out in the presence of emetine, a cytosolic translation inhibitor so that we could solely focus on the products of mitochondrial translation. After incubation, the cells were lysed, and click chemistry was used to append an alkyne-bearing dye to newly synthesized mitochondrial proteins.
SDS-PAGE separation of the labeled mitochondrial translation products revealed the role of MIEF1-MP in mitochondrial translation. Comparison of MIEF1-MP overexpressing to control samples revealed increased band densities for most proteins particularly MT-ND4, MT-CYB, MT-CO2/ MT-ATP6 and MT-ND3 indicating that MIEF1-MP promotes mitochondrial translation (Figure 5A and S5A), and we validated overexpression of MIEF1-MP in these samples (Figure S6). By contrast, knockdown of MIEF1-MP led to lower band densities for most proteins including MT-CO1, MT-ND4, MT-CYB, MT-ND2, MT-CO3, MT-CO2/ MT-ATP6, MT-ND6 and MT-ND3, indicating decreased mitochondrial translation (Figure 5B and S5B). Quantitative analysis of the mtDNA-encoded protein band densities revealed a robust increase (54%) in protein levels upon MIEF1-MP overexpression, and significant decreases (32%) in cells lacking MIEF1-MP (Figure 5C). Knockdown of MIEF1-MP also results in the knockdown of MIEF1 and we needed to validate that these changes were due to MIEF1-MP. We expressed a siRNA resistant MIEF1-MP in cells treated with siRNA against the MIEF1-MP transcript and found that expression of MIEF1-MP rescued the loss of protein translation (Figure 6) supporting a role for MIEF1-MP and not MIEF1 in the mitochondrial translation. These numbers compare favorably to other mitoribosome components such as MRPL14 and DAP3 that modulate nascent mitochondrial translation by ~40– 50%.63, 64 MIEF1-MP is a biologically active component of mitochondrial translation pathway through its interaction with the mitoribosome.
Figure 5.
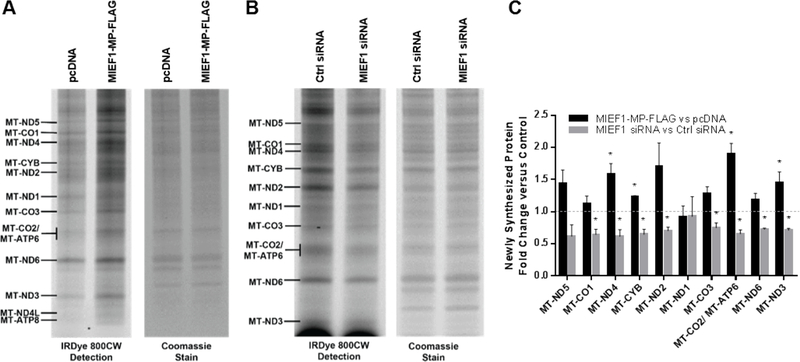
MIEF1-MP modulates the mitochondrial translation rate. (A) Using the BONCAT method, newly synthesized mtDNA encoded proteins are labelled with IRDye® 800CW dye and visualized on a gel. MIEF1-MP-FLAG overexpression in HEK293T cells increases the amount of newly synthesized mitochondrial proteins versus the control (pcDNA vector). (B) MIEF1 siRNA-treated HEK293T cells led to decreased levels of translation of newly synthesized proteins compared to control siRNA treated cells. (C) Quantitative analysis showed robust change for the newly synthesized mtDNA encoded protein levels on MIEF1 microprotein overexpression and knockdown. Comparison of the MIEF1-MP perturbed sample to the control was used to determine statistical significance. For example, MIEF1-MP overexpression leads to a statistically significant increase in MT-ND4 versus the pcDNA control, and MIEF1-MP siRNA leads to a statistically significant decrease in MT-ND4 translation versus the siRNA control (*, p-value <0.05 using t-test).
Figure 6. Rescuing the effect of MIEF1 siRNA treatment on mitochondrial translation rate.
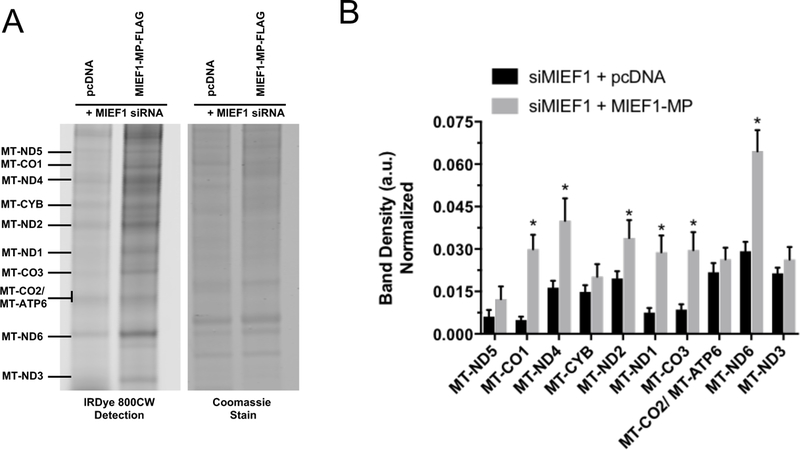
Using the BONCAT method, newly synthesized mtDNA encoded proteins are labelled with IRDye® 800CW dye and visualized on a gel. A) In the MIEF1 siRNA treated sample expressing MIEF1-MP-FLAG increases the amount of newly synthesized mitochondrial proteins versus the control (pcDNA vector). B) Quantitative analysis showed robust change for the newly synthesized mtDNA encoded protein levels on MIEF1 microprotein knockdown and re-expression. Comparison of the MIEF1 siRNA treated with MIEF1-MP re-expressed sample to the control was used to determine statistical significance (*, p-value <0.05 using t-test).
CONCLUSION
Here, we characterized a 70-amino acid microprotein encoded upstream of MIEF1 gene, MIEF1-MP. We demonstrated that MIEF1-MP is a mitochondrial microprotein and interacts with mitoribosome. In addition, exogenous expression of a C-terminal FLAG tagged MIEF1-MP (MIEF-MP-FLAG) showed the same intracellular localization and association to the mitoribosome, suggesting that the FLAG tag does not interfere with MIEF1-MP functions. Previous success with using protein-protein interactions to discover micropeptide functions prompted a similar approach to be used in this study. MIEF1-MP enriched many proteins but functional follow up of the MIEF1-MP and ACPM/NDUFAB1 interaction suggested that this interaction is not functionally relevant. We reasoned that MIEF1-MP overexpression led to the appearance of non-functional interactions, such as ACPM/NDUFAB1, which required a better approach for validating MIEF1-MP interaction partners. We settled on a two-prong computational approach that looked for MIEF1-MP protein interaction partners that are already known to interact, reducing the total number of proteins, and an “in silico” reciprocal IP that utilized Bioplex data to identify proteins that could enrich MIEF1-MP from HEK293T cells. This analysis revealed proteins that are part of the mitoribosome as the most relevant MIEF1-MP interaction partners, which we validated experimentally by IP using MRPL4 and MRPS27 antibodies (Figure 4C). These results are also consistent with an earlier cryo-EM structure of the mitoribosome that detected additional electron density on the mitoribosome that belongs to MIEF1-MP.32 We tested the hypothesis that MIEF1-MP might be a functional component of the mitoribosome and obtained data from overexpression and knockdown studies that indicates that MIEF1-MP can regulate mitochondrial translation, which supports the hypothesis that MIEF1-MP is a mitoribosome assembly factor.32
Mitochondrial protein synthesis is an essential process in mammals responsible for encoding key components of the oxidative phosphorylation (OXPHOS) complexes. Complete molecular details of this deceptively simple process still remain unclear. Problems in mitochondrial protein synthesis are leading cause of OXPHOS dysfunction resulting in metabolic and developmental disorders. Heart and brain are particularly sensitive to defects in the mtDNA-encoded protein synthesis during late embryonic or early postnatal development, due to massive mitochondrial biogenesis at that stage. Impaired mitochondrial translation causes severe pediatric cardiomyopathy and brain disease with OXPHOS abnormalities.65 Furthermore, MIEF1-MP is part of a growing class of recently discovered mitochondrial microproteins. The most interesting being mitoregulin66/MOXI67, which has been shown to regulate beta oxidation67, mitochondrial assembly66, and other mitochondrial functions. Similarly, MIEF1-MP is reported to control mitochondrial fission30 and here we link it to ribosome biosynthesis. These findings could be important as they may reveal a new connection between disparate cellular pathways. For example, we reason that mitochondrial fission by MIEF1 the longer ORF on the MIEF1 gene may require MIEF1-MP promoting translation of mitochondrial proteins. The identification of MIEF1-MP reveals a new gene involved in regulating mitochondrial translation, thus, highlighting the need for further investigation and the value in characterizing microproteins.
Supplementary Material
ACKNOWLEDGMENT
We thank James Moresco for technical support.
Funding Sources
A.R. was funded by Timken Sturgis Foundation Award and Salk Women in Science Fellowship, Q.C. was supported by the George E. Hewitt Foundation for medical research, D.T. was supported by Pioneer Fellowship and T.F.M. was funded by NIH Fellowship GM123685. This work was supported by the Mass Spectrometry Core of the Salk Institute with funding from NIH-NCI CCSG: P30 014195 and the Helmsley Center for Genomic Medicine. This study was supported by the National Institutes of Health with a National Cancer Institute Cancer Center Support Grant P30 (CA014195 MASS core, A.S.), R01 (GM102491, A.S.), (P41 GM103533, J.R.Y.) and (R01 MH067880, J.R.Y.). Additional support was provided by the Leona M. and Harry B. Helmsley Charitable Trust grant (2012-PG-MED002 to A.S.), and Dr. Frederick Paulsen Chair/Ferring Pharmaceuticals (A.S.).
Footnotes
Supporting information
The following Supporting Information files are available free of charge.
Additional experimental details and supplementary figures (PDF)
Proteomics data and analysis (XLSX)
REFERENCES
- [1].Cabrera-Quio LE, Herberg S, and Pauli A (2016) Decoding sORF translation – from small proteins to gene regulation, RNA Biology 13, 1051–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Saghatelian A, and Couso JP (2015) Discovery and characterization of smORF-encoded bioactive polypeptides, Nat Chem Biol 11, 909–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Burge CB, and Karlin S (1998) Finding the genes in genomic DNA, Current opinion in structural biology 8, 346–354. [DOI] [PubMed] [Google Scholar]
- [4].Calvo SE, Pagliarini DJ, and Mootha VK (2009) Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans, Proceedings of the National Academy of Sciences of the United States of America 106, 7507–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Frith MC, Forrest AR, Nourbakhsh E, Pang KC, Kai C, Kawai J, Carninci P, Hayashizaki Y, Bailey TL, and Grimmond SM (2006) The Abundance of Short Proteins in the Mammalian Proteome, PLoS Genetics 2, e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ladoukakis E, Pereira V, Magny EG, Eyre-Walker A, and Couso JP (2011) Hundreds of putatively functional small open reading frames in Drosophila, Genome Biology 12, R118–R118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Vanderperre B, Lucier JF, Bissonnette C, Motard J, Tremblay G, Vanderperre S, Wisztorski M, Salzet M, Boisvert FM, and Roucou X (2013) Direct detection of alternative open reading frames translation products in human significantly expands the proteome, PLoS One 8, e70698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mackowiak SD, Zauber H, Bielow C, Thiel D, Kutz K, Calviello L, Mastrobuoni G, Rajewsky N, Kempa S, Selbach M, and Obermayer B (2015) Extensive identification and analysis of conserved small ORFs in animals, Genome Biology 16, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Smith Jenna E., Alvarez-Dominguez Juan R., Kline N Huynh Nathan J., Geisler S, Hu W, Coller J, and Baker Kristian E. (2014) Translation of Small Open Reading Frames within Unannotated RNA Transcripts in Saccharomyces cerevisiae, Cell Reports 7, 1858–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Aspden JL, Eyre-Walker YC, Phillips RJ, Amin U, Mumtaz MAS, Brocard M, and Couso J-P (2014) Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq, eLife 3, e03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, and Giraldez AJ (2014) Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation, The EMBO Journal 33, 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Crappé J, Van Criekinge W, Trooskens G, Hayakawa E, Luyten W, Baggerman G, and Menschaert G (2013) Combining in silico prediction and ribosome profiling in a genome-wide search for novel putatively coding sORFs, BMC Genomics 14, 648–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ingolia NT, Lareau LF, and Weissman JS (2011) Ribosome Profiling of Mouse Embryonic Stem Cells Reveals the Complexity of Mammalian Proteomes, Cell 147, 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Slavoff SA, Mitchell AJ, Schwaid AG, Cabili MN, Ma J, Levin JZ, Karger AD, Budnik BA, Rinn JL, and Saghatelian A (2013) Peptidomic discovery of short open reading frame-encoded peptides in human cells, Nat Chem Biol 9, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ma J, Ward CC, Jungreis I, Slavoff SA, Schwaid AG, Neveu J, Budnik BA, Kellis M, and Saghatelian A (2014) Discovery of human sORF-encoded polypeptides (SEPs) in cell lines and tissue, J. Proteome Res 13, 1757–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ma J, Diedrich JK, Jungreis I, Donaldson C, Vaughan J, Kellis M, Yates JR 3rd, and Saghatelian A (2016) Improved Identification and Analysis of Small Open Reading Frame Encoded Polypeptides, Anal Chem 88, 3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zanet J, Benrabah E, Li T, Pélissier-Monier A, Chanut-Delalande H, Ronsin B, Bellen HJ, Payre F, and Plaza S (2015) Pri sORF peptides induce selective proteasome-mediated protein processing, Science 349, 1356–1358. [DOI] [PubMed] [Google Scholar]
- [18].Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F, and Kageyama Y (2010) Small Peptides Switch the Transcriptional Activity of Shavenbaby During Drosophila Embryogenesis, Science 329, 336–339. [DOI] [PubMed] [Google Scholar]
- [19].Anderson DM, Anderson KM, Chang C-L, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, and Olson EN (2015) A Micropeptide Encoded by a Putative Long Non-coding RNA Regulates Muscle Performance, Cell 160, 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Slavoff SA, Heo J, Budnik BA, Hanakahi LA, and Saghatelian A (2014) A human short open reading frame (sORF)-encoded polypeptide that stimulates DNA end joining, J Biol Chem 289, 10950–10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Arnoult N, Correia A, Ma J, Merlo A, Garcia-Gomez S, Maric M, Tognetti M, Benner CW, Boulton SJ, Saghatelian A, and Karlseder J (2017) Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN, Nature 549, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lee C, Zeng J, Drew Brian G., Sallam T, Martin-Montalvo A, Wan J, Kim S-J, Mehta H, Hevener Andrea L., de Cabo R, and Cohen P (2015) The Mitochondrial-Derived Peptide MOTS-c Promotes Metabolic Homeostasis and Reduces Obesity and Insulin Resistance, Cell Metabolism 21, 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S, and Kageyama Y (2007) Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA, Nature Cell Biology 9, 660. [DOI] [PubMed] [Google Scholar]
- [24].Magny EG, Pueyo JI, Pearl FMG, Cespedes MA, Niven JE, Bishop SA, and Couso JP (2013) Conserved Regulation of Cardiac Calcium Uptake by Peptides Encoded in Small Open Reading Frames, Science 341, 1116–1120. [DOI] [PubMed] [Google Scholar]
- [25].Tupy JL, Bailey AM, Dailey G, Evans-Holm M, Siebel CW, Misra S, Celniker SE, and Rubin GM (2005) Identification of putative noncoding polyadenylated transcripts in Drosophila melanogaster, Proceedings of the National Academy of Sciences of the United States of America 102, 5495–5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Galindo MI, Pueyo JI, Fouix S, Bishop SA, and Couso JP (2007) Peptides Encoded by Short ORFs Control Development and Define a New Eukaryotic Gene Family, PLoS Biology 5, e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang Q, Vashisht AA, O’Rourke J, Corbel SY, Moran R, Romero A, Miraglia L, Zhang J, Durrant E, Schmedt C, Sampath SC, and Sampath SC (2017) The microprotein Minion controls cell fusion and muscle formation, Nature Communications 8, 15664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].D’Lima NG, Ma J, Winkler L, Chu Q, Loh KH, Corpuz EO, Budnik BA, Lykke-Andersen J, Saghatelian A, and Slavoff SA (2017) A human microprotein that interacts with the mRNA decapping complex, Nat Chem Biol 13, 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Losón OC, Song Z, Chen H, and Chan DC (2013) Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission, Molecular biology of the cell 24, 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Samandi S, Roy AV, Delcourt V, Lucier J-F, Gagnon J, Beaudoin MC, Vanderperre B, Breton M-A, Motard J, Jacques J-F, Brunelle M, Gagnon-Arsenault I, Fournier I, Ouangraoua A, Hunting DJ, Cohen AA, Landry CR, Scott MS, and Roucou X (2017) Deep transcriptome annotation enables the discovery and functional characterization of cryptic small proteins, eLife 6, e27860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kim M-S, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, Thomas JK, Muthusamy B, Leal-Rojas P, Kumar P, Sahasrabuddhe NA, Balakrishnan L, Advani J, George B, Renuse S, Selvan LDN, Patil AH, Nanjappa V, Radhakrishnan A, Prasad S, Subbannayya T, Raju R, Kumar M, Sreenivasamurthy SK, Marimuthu A, Sathe GJ, Chavan S, Datta KK, Subbannayya Y, Sahu A, Yelamanchi SD, Jayaram S, Rajagopalan P, Sharma J, Murthy KR, Syed N, Goel R, Khan AA, Ahmad S, Dey G, Mudgal K, Chatterjee A, Huang T-C, Zhong J, Wu X, Shaw PG, Freed D, Zahari MS, Mukherjee KK, Shankar S, Mahadevan A, Lam H, Mitchell CJ, Shankar SK, Satishchandra P, Schroeder JT, Sirdeshmukh R, Maitra A, Leach SD, Drake CG, Halushka MK, Prasad TSK, Hruban RH, Kerr CL, Bader GD, Iacobuzio-Donahue CA, Gowda H, and Pandey A (2014) A draft map of the human proteome, Nature 509, 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Brown A, Rathore S, Kimanius D, Aibara S, Bai X. c., Rorbach J, Amunts A, and Ramakrishnan V (2017) Structures of the human mitochondrial ribosome in native states of assembly, Nature Structural and Molecular Biology 24, 866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Angerer H, Radermacher M, Mańkowska M, Steger M, Zwicker K, Heide H, Wittig I, Brandt U, and Zickermann V (2014) The LYR protein subunit NB4M/NDUFA6 of mitochondrial complex I anchors an acyl carrier protein and is essential for catalytic activity, Proceedings of the National Academy of Sciences of the United States of America 111, 5207–5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Invernizzi F, Tigano M, Dallabona C, Donnini C, Ferrero I, Cremonte M, Ghezzi D, Lamperti C, and Zeviani M (2013) A Homozygous Mutation in LYRM7/MZM1L Associated with Early Onset Encephalopathy, Lactic Acidosis, and Severe Reduction of Mitochondrial Complex III Activity, Human Mutation 34, 1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dennis RA, and McCammon MT (1999) Acn9 is a novel protein of gluconeogenesis that is located in the mitochondrial intermembrane space, European Journal of Biochemistry 261, 236–243. [DOI] [PubMed] [Google Scholar]
- [36].Ghezzi D, Goffrini P, Uziel G, Horvath R, Klopstock T, Lochmüller H, D’Adamo P, Gasparini P, Strom TM, Prokisch H, Invernizzi F, Ferrero I, and Zeviani M (2009) SDHAF1, encoding a LYR complex-II specific assembly factor, is mutated in SDH-defective infantile leukoencephalopathy, Nature Genetics 41, 654. [DOI] [PubMed] [Google Scholar]
- [37].Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong S-E, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, and Mootha VK (2008) A mitochondrial protein compendium elucidates complex I disease biology, Cell 134, 112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lim SC, Friemel M, Marum JE, Tucker EJ, Bruno DL, Riley LG, Christodoulou J, Kirk EP, Boneh A, DeGennaro CM, Springer M, Mootha VK, Rouault TA, Leimkühler S, Thorburn DR, and Compton AG (2013) Mutations in LYRM4, encoding iron–sulfur cluster biogenesis factor ISD11, cause deficiency of multiple respiratory chain complexes, Human Molecular Genetics 22, 4460–4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhu G-Z, Zhang M, Kou C-Z, Ni Y-H, Ji C-B, Cao X-G, and Guo X-R (2012) Effects of Lyrm1 knockdown on mitochondrial function in 3 T3-L1 murine adipocytes, Journal of Bioenergetics and Biomembranes 44, 225–232. [DOI] [PubMed] [Google Scholar]
- [40].Angerer H (2015) Eukaryotic LYR Proteins Interact with Mitochondrial Protein Complexes, Biology 4, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chu Q, Rathore A, Diedrich JK, Donaldson CJ, Yates JR, and Saghatelian A (2017) Identification of Microprotein–Protein Interactions via APEX Tagging, Biochemistry 56, 3299–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lam SS, Martell JD, Kamer KJ, Deerinck TJ, Ellisman MH, Mootha VK, and Ting AY (2015) Directed evolution of APEX2 for electron microscopy and proximity labeling, Nat Meth 12, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Mai N, Chrzanowska-Lightowlers ZMA, and Lightowlers RN (2017) The process of mammalian mitochondrial protein synthesis, Cell and Tissue Research 367, 5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wieckowski MR, Giorgi C, Lebiedzinska M, Duszynski J, and Pinton P (2009) Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells, Nature Protocols 4, 1582. [DOI] [PubMed] [Google Scholar]
- [45].Hung V, Udeshi ND, Lam SS, Loh KH, Cox KJ, Pedram K, Carr SA, and Ting AY (2016) Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2, Nat. Protocols 11, 456–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Dieterich DC, Lee JJ, Link AJ, Graumann J, Tirrell DA, and Schuman EM (2007) Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging, Nature Protocols 2, 532. [DOI] [PubMed] [Google Scholar]
- [47].Zhao J, Liu T, Jin S, Wang X, Qu M, Uhlén P, Tomilin N, Shupliakov O, Lendahl U, and Nistér M (2011) Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission, The EMBO Journal 30, 2762–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Yu R, Liu T, Jin S-B, Ning C, Lendahl U, Nistér M, and Zhao J (2017) MIEF1/2 function as adaptors to recruit Drp1 to mitochondria and regulate the association of Drp1 with Mff, Scientific Reports 7, 880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bhola PD, and Letai A (2016) Mitochondria – judges and executioners of cell death sentences, Molecular cell 61, 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Green DR, and Reed JC (1998) Mitochondria and Apoptosis, Science 281, 1309–1312. [DOI] [PubMed] [Google Scholar]
- [51].Emanuelsson O, Brunak S, Von Heijne G, and Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools, Nature protocols 2, 953. [DOI] [PubMed] [Google Scholar]
- [52].Fukasawa Y, Tsuji J, Fu S-C, Tomii K, Horton P, and Imai K (2015) MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites, Molecular & Cellular Proteomics, mcp. M114. 043083. [DOI] [PMC free article] [PubMed]
- [53].Haack TB, Madignier F, Herzer M, Lamantea E, Danhauser K, Invernizzi F, Koch J, Freitag M, Drost R, Hillier I, Haberberger B, Mayr JA, Ahting U, Tiranti V, Rötig A, Iuso A, Horvath R, Tesarova M, Baric I, Uziel G, Rolinski B, Sperl W, Meitinger T, Zeviani M, Freisinger P, and Prokisch H (2012) Mutation screening of 75 candidate genes in 152 complex I deficiency cases identifies pathogenic variants in 16 genes including NDUFB9, Journal of Medical Genetics 49, 83. [DOI] [PubMed] [Google Scholar]
- [54].Ladha JS, Tripathy MK, and Mitra D (2005) Mitochondrial complex I activity is impaired during HIV-1-induced T-cell apoptosis, Cell Death And Differentiation 12, 1417. [DOI] [PubMed] [Google Scholar]
- [55].Vinothkumar KR, Zhu J, and Hirst J (2014) Architecture of mammalian respiratory complex I, Nature 515, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zickermann V, Wirth C, Nasiri H, Siegmund K, Schwalbe H, Hunte C, and Brandt U (2015) Mechanistic insight from the crystal structure of mitochondrial complex I, Science 347, 44–49. [DOI] [PubMed] [Google Scholar]
- [57].Han S, Udeshi ND, Deerinck TJ, Svinkina T, Ellisman MH, Carr SA, and Ting AY (2017) Proximity Biotinylation as a Method for Mapping Proteins Associated with mtDNA in Living Cells, Cell chemical biology 24, 404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Feng D, Witkowski A, and Smith S (2009) Down-regulation of Mitochondrial Acyl Carrier Protein in Mammalian Cells Compromises Protein Lipoylation and Respiratory Complex I and Results in Cell Death, The Journal of Biological Chemistry 284, 11436–11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Teo G, Liu G, Zhang J, Nesvizhskii AI, Gingras A-C, and Choi H (2014) SAINTexpress: improvements and additional features in Significance Analysis of Interactome software, Journal of proteomics 100, 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Mellacheruvu D, Wright Z, Couzens AL, Lambert J-P, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, Halim VA, Bagshaw RD, Hubner NC, al-Hakim A, Bouchard A, Faubert D, Fermin D, Dunham WH, Goudreault M, Lin Z-Y, Badillo BG, Pawson T, Durocher D, Coulombe B, Aebersold R, Superti-Furga G, Colinge J, Heck AJR, Choi H, Gstaiger M, Mohammed S, Cristea IM, Bennett KL, Washburn MP, Raught B, Ewing RM, Gingras A-C, and Nesvizhskii AI (2013) The CRAPome: a contaminant repository for affinity purification-mass spectrometry data, Nat Meth 10, 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, and Jensen LJ (2013) STRING v9.1: protein-protein interaction networks, with increased coverage and integration, Nucleic Acids Research 41, D808–D815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Huttlin Edward L., Ting L, Bruckner Raphael J., Gebreab F, Gygi Melanie P., Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, Dong R, Guarani V, Vaites Laura P., Ordureau A, Rad R, Erickson Brian K., Wühr M, Chick J, Zhai B, Kolippakkam D, Mintseris J, Obar Robert A., Harris T, Artavanis-Tsakonas S, Sowa Mathew E., De Camilli P, Paulo Joao A., Harper JW, and Gygi Steven P. (2015) The BioPlex Network: A Systematic Exploration of the Human Interactome, Cell 162, 425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Fung S, Nishimura T, Sasarman F, and Shoubridge EA (2013) The conserved interaction of C7orf30 with MRPL14 promotes biogenesis of the mitochondrial large ribosomal subunit and mitochondrial translation, Molecular Biology of the Cell 24, 184–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Xiao L, Xian H, Lee KY, Xiao B, Wang H, Yu F, Shen H-M, and Liou Y-C (2015) Death-associated Protein 3 Regulates Mitochondrial-encoded Protein Synthesis and Mitochondrial Dynamics, The Journal of Biological Chemistry 290, 24961–24974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kamps R, Szklarczyk R, Theunissen TE, Hellebrekers DMEI, Sallevelt SCEH, Boesten IB, de Koning B, van den Bosch BJ, Salomons GS, Simas-Mendes M, Verdijk R, Schoonderwoerd K, de Coo IFM, Vanoevelen JM, and Smeets HJM (2018) Genetic defects in mtDNA-encoded protein translation cause pediatric, mitochondrial cardiomyopathy with early-onset brain disease, European Journal of Human Genetics 26, 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Stein CS, Jadiya P, Zhang X, McLendon JM, Abouassaly GM, Witmer NH, Anderson EJ, Elrod JW, and Boudreau RL (2018) Mitoregulin: A lncRNA-Encoded Microprotein that Supports Mitochondrial Supercomplexes and Respiratory Efficiency, Cell reports 23, 3710–3720. e3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Makarewich CA, Baskin KK, Munir AZ, Bezprozvannaya S, Sharma G, Khemtong C, Shah AM, McAnally JR, Malloy CR, and Szweda LI (2018) MOXI Is a Mitochondrial Micropeptide That Enhances Fatty Acid β-Oxidation, Cell reports 23, 3701–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


