Abstract
DNA polymerases influence genome stability through their involvement in DNA replication, response to DNA damage, and DNA repair processes. Saccharomyces cerevisiae possess four non-essential DNA polymerases, Pol λ, Pol η, Pol ζ, and Rev1, which have varying roles in genome stability. In order to assess the contribution of the non-essential DNA polymerases in genome stability, we analyzed the pol4Δ rev1Δ rev3Δ rad30Δ quadruple mutant in microhomology mediated repair, due to recent studies linking some of these DNA polymerases to this repair pathway. Our results suggest that the length and quality of microhomology influence both the overall efficiency of repair and the involvement of DNA polymerases. Furthermore, the non-essential DNA polymerases demonstrate overlapping and redundant functions when repairing double-strand breaks using short microhomologies containing mismatches. Then, we examined genome-wide mutation accumulation in the pol4Δ rev1Δ rev3Δ rad30Δ quadruple mutant compared to wild type cells. We found a significant decrease in the overall rate of mutation accumulation in the quadruple mutant cells compared to wildtype, but an increase in frameshift mutations and a shift towards transversion base-substitution with a preference for G:C to T:A or C:G. Thus, the non-essential DNA polymerases have an impact on the nature of the mutational spectrum. The sequence and functional homology shared between human and S. cerevisiae non-essential DNA polymerases suggest these DNA polymerases may have a similar role in human cells.
Keywords: DNA polymerases, Genome stability, Mutation accumulation, DNA repair, yeast
1. Introduction
Replication fidelity, under favorable conditions, ensures the preservation of nucleic acid sequences and continued genomic stability of daughter cells in subsequent generations. However, genetic variation due to mutations impact an organism’s ability to survive and reproduce in changing environmental conditions (1-3). While mutations allow for genetic variation to exist in a population, the process by which mutations accumulate in a genome over generations has been difficult to study. Analysis of mutation rates in the budding yeast Saccharomyces cerevisiae has primarily focused on the use of reporter gene assays, such as CAN1, which is thought to serve as a proxy for mutation accumulation in the entire genome (4-8). The use of reporter genes has several advantages: (1) mutants are easily identified because they use selectable read-out, and (2) the small size of each gene reduces the cost and time required to characterize each mutation. However, mutation rates are not uniform across the entire genome (9). In particular, gene expression patterns and genomic location can influence the mutation rate observed (6,10-12). Thus, conclusions drawn from mutation rates of single gene reporter assays since they may not represent mutation rates at other genomic loci.
Advances in genome sequencing technology, has reduced the cost and time required to sequence and analyze individual samples (13). This allowed the use of whole genome sequencing to determine the spontaneous mutation rates and spectrum in model organisms, with relatively small genomes (14-16). Continued reduction in costs has resulted in the inclusion of genome sequencing and analysis in clinical care, such as sequencing of cells from human patients who have a disease where the genetic cause is unknown or individual tumors (17-19). Taken together, the identification of single-nucleotide spontaneous mutations and genome rearrangements within a genome are now feasible for both research and clinical care. Of particular interest is determining how the rate of spontaneous mutations and overall genome stability is influenced by the loss of candidate genes involved in genome maintenance, due to the effect it would have on the lifetime risk of diseases such as cancer (20,21). Several studies have started to establish the genome-wide mutation spectrum in the absence of genes required for genome stability (22,23), but an examination of the impact DNA polymerases have on the process has yet to be explored.
DNA polymerases play an essential role in genome stability through their contributions to DNA replication, repair, and recombination (24,25). These processes help cells avoid mutations and genome rearrangements, which may be detrimental to cellular viability (26). Mutations within the exonuclease domain of DNA polymerase delta (Pol δ) and epsilon (Pol ε), which are responsible for the vast majority of nuclear DNA replication, are associated with elevated mutagenesis and genome rearrangements (27). In addition to Pol δ and Pol ε, cells possess specialized low-fidelity DNA polymerases which function in specific pathways such as DNA repair and damage tolerance (27). For instance, DNA polymerase eta (Pol η) inserts two adenines opposite a thymine dimer and DNA polymerase zeta (Pol ζ) along with Rev1 extend primer-templates possessing a distortion in the helical structure (28). This process can be error-free or error-prone depending on the type of lesion and which DNA polymerase is recruited, thereby impacting mutagenesis and the development of cancer or disease (28-30). DNA polymerase lambda (Pol λ) is another low-fidelity DNA polymerase, which facilitates the use of microhomologies (MH) to repair double-strand breaks (DSBs) (31-33).
In S. cerevisiae, the repair of DSBs using MHs of 5-25 bp is known as microhomology-mediated repair (MHMR) and relies on proteins used in other DSB repair pathways (32-34). One such protein is Pol δ, which functions to extend MH pairing intermediates of varying lengths and quality (31,35). The involvement of Pol δ in MHMR is at least partially dependent on Pol λ to extend mispaired nucleotides from the 3’-hydroxyl end before taking over extension (31). While the involvement of Pol δ in MHMR is more defined, the precise involvement of the non-essential polymerases in yeast (Pol λ, Pol η, Pol ζ and Rev1) remains unclear. Before its cooperative role with Pol δ was elucidated, Pol λ was originally implicated in promoting MHMR in both S. cerevisiae (32) and Schizosaccharomyces pombe (33). However, studies examining Pol η and Pol ζ in MHMR have shown conflicting results depending on the assay system used (32,35), while the role of Rev1 has not been reported.
In the current study, we report the involvement of Pol λ, Pol η, Pol ζ, and Rev1 in genome stability by assessing their impact on MHMR and whole-genome mutation accumulation in S cerevisiae. Previous reports have shown the functional overlap among different DNA polymerases allows for a complete or partial substitution of function (36,37). Therefore, to reduce the likelihood of functional redundancy when determining the spectrum of mutations (22), we examine mutation accumulation on wild type and quadruple mutant pol4Δ rev1Δ rev3Δ rad30Δ cells.
2. Materials and Methods
2.1. Strain and plasmid construction.
Standard techniques for yeast growth and genetic manipulation were used throughout this study (38). Yeast strains used in the study of MHMR were isogenic with W303-1A and are wild type for the RAD5 gene (39). Yeast strains used in the study of mutation accumulation were derived in the BY4741 background (40). PCR was used to generate gene disruptions using primers specific to the selectable marker of interest and flanked by sequence homologous to the upstream and downstream sequence of the ORF for each gene to be disrupted. Dr. Lorraine Symington kindly provided the pol32::KANMX mutant (41). Creation of the interchromosomal MHMR substrates were described previously (31). Intrachromosomal his3-Δ3’-HOcs-URA3-HOcs-his3Δ5’ MHMR substrates (14-2-2 bp, 14-2-4 bp, 14-2-9 bp, and 20 bp) were generated by PCR and subsequent recombination to introduce the substrate into the HIS3 locus on chromosome XV. Briefly, the first round of PCR was performed (42) using the interchromosomal MHMR his3-Δ3’-HOcs substrates (list strains/plasmids), URA3 from Kluyveromyces lactis (list strains/plasmids), and his3-Δ5’-HOcs (list strains/plasmids) as templates. A second round of PCR was used to fuse the PCR products of each his3Δ3’-HOcs substrate with varying microhomologies to a truncated ura3Δ3’ PCR product and a second PCR fusion reaction using his3Δ5’-HOcs with a truncated ura3Δ5’ PCR product. The resulting his3Δ3’-HOcs-ura3Δ3’ and his3Δ5’-HOcs-ura3Δ5’ fragments were cotransformed into cells and homologous recombination between the overlapping ura3 segments generated the his3Δ3’-HOcs-URA3-his3Δ5’-HOcs substrates. Verification by PCR was used to confirm proper genomic integration.
2.2. Microhomology-mediated repair assay.
Diploid cells of the appropriate genotype were identified and single colonies were used to inoculate 2 mL of YP-Raffinose medium (1 % yeast extract, 2 % peptone, and 2 % raffinose) as described previously (31). Cells containing the intrachromosomal MHMR assay were maintained on synthetic medium lacking uracil (SD-Ura) to maintain the his3Δ3’-HOcs-URA3-his3Δ5’-HOcs substrate before inoculation and induction. The median translocation frequencies and 95% confidence intervals from at least 15 independent cultures was determined.
2.3. Pulse-field gel electrophoresis, gross chromosomal rearrangement and CAN1 mutation rate assays
WT and pol4Δ rev1Δ rev3Δ rad30Δ quadruple mutants possessing both CAN1 and URA3 on chromosome V (43) were identified from dissection plates. These cells from the dissection plates and following 24 serial bottleneck passages (approximately 600 mitoses) were grown in 5 mL of YPD medium for 24 hrs at 30 °C. The next day, an appropriate dilution of cells were plated onto YPD and synthetic medium containing canavanine (SD+canavanine), which were incubated at 30 °C to allow colony formation. The remaining portion of the YPD culture was used to make agarose plugs for pulse-field gel electrophoresis. Chromosomes were separated and visualized using ethidium bromide as previously described (44). Visible colonies from YPD and SD+canavanine plates were counted and colonies appearing on the SD+canavanine were replica plated to SD-Ura medium and allowed to grow at 30 °C overnight. The CAN1 mutation frequency was calculated by counting the number of colonies growing on both SD+canavanine and SD-Ura media over the total number of viable cells plated. The median CAN1 mutation rates and 95% confidence intervals were determined using the CAN1 mutation frequencies from at least 15 independent cultures (45). Cells which grew on SD+canavanine but not SD-Ura were categorized as having gone through a gross chromosomal rearrangement (GCR). GCR rate was calculated by fluctuation analysis from at least 15 independent cultures (46). Comparison of GCR rates and generation of a p-value was done using a contingency table.
2.4. Serial bottleneck passaging and mutome analysis
WT and pol4Δ rev1Δ rev3Δ rad30Δ quadruple mutants identified following dissection were subject to serial bottleneck passaging by first streaking the original colony (~25 generations) from the dissection plate to single cells on a fresh YPD plate, which were grown at 30 °C to form colonies. From this plate (~50 generations) a single colony was selected and used to restreak onto a fresh YPD plate to generate new single colonies. Each colony was estimated to have gone through approximately 25 generations. In total, 24 serial bottleneck passages were performed on each independent line (16 WT and 16 quadruple mutants) for a total of 600 mitoses each. Genomic DNA from 10 quadruple mutants lines having gone through 600 mitoses analyzed by next-generation sequencing as described previously (22). Base substitutions and frameshifts identified through Illumina whole-genome sequencing were verified by PCR and subsequent Sanger sequencing. De novo mutations were identified by bioinformatic analysis using standard pipeline (Suppl. Fig. 1A).
2.5. DNA Polymerase λ Microhomology Extension reactions
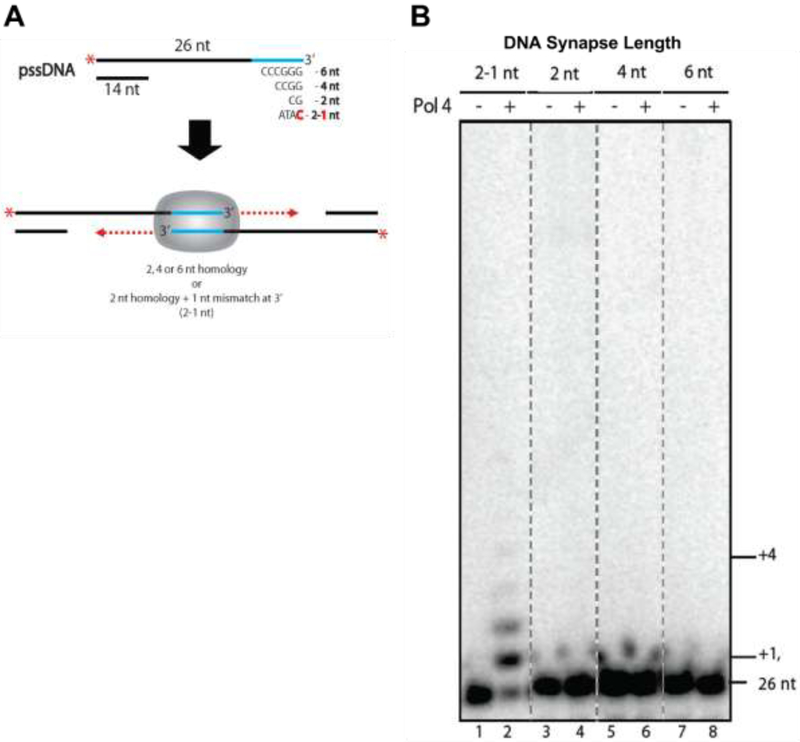
Reactions were carried as described in (47) Briefly, 100 nM 5’-32P radiolabeled pssDNA preincubated for 10’ at 30 °C (25 mM Tris-HCl, pH 8, 10% glycerol, 1 mM DTT, 10 mM MgCl2, and 0.1 mg/ml BSA) was mixed with 100 nM Pol4 for 15 min; this was followed by the addition of 500 μM dNTPs for another 30 min at 30 °C in a total volume of 20 μl. Reactions were terminated by the addition of 4 μl of stop buffer (10 mg/ml proteinase K, 100 mM EDTA, and 1.0% SDS); Radiolabeled DNA was resolved in denaturing 10% polyacrylamide gels. Gels will be dried, imaged with a Molecular Dynamics Phosphor Imager and analyzed using ImageQuant software. All DNA was obtained from Integrated DNA Technologies and purified by 10% denaturing acrylamide gel electrophoresis. Substrates were prepared as described in (47). Shortly, 26 mer oligonucleotides were 32P-5’-radiolabeled with T4 polynucleotide kinase (NEB) and [γ-32P]ATP (PerkinElmer). All pssDNA templates were formed by the annealing of 14 mer oligonucleotide (5’- CTAAGCTCACAGTG-3’) to the following 32P-5’-radiolabeled 26 mer: pssDNA6 (5’ - CACTGTGAGCTTAGGGTTAGCCCGGG –3’); pssDNA4 (5’CACTGTGAGCTTAGGGTTAGAGCCGG-3’); pssDNA2 – (5’-CACTGTGAGCTTAGGGTTAGAGATCG-3’) and pssDNA2-1 – (5’ - CACTGTGAGCTTAGGGTTAGAGATAC-3’). DNA polymerase 4 purification will be described in a manuscript in preparation.
3. Results
To assess the role of the four non-essential nuclear DNA polymerases on mutation accumulation and genome stability, we designed two experimental systems to study MHMR in diploid S. cerevisiae using varying MHs (Fig. 1). We focus on diploid cells, as they represent the natural state of budding yeast, which is a diplontic organism as most eukaryote species (48).
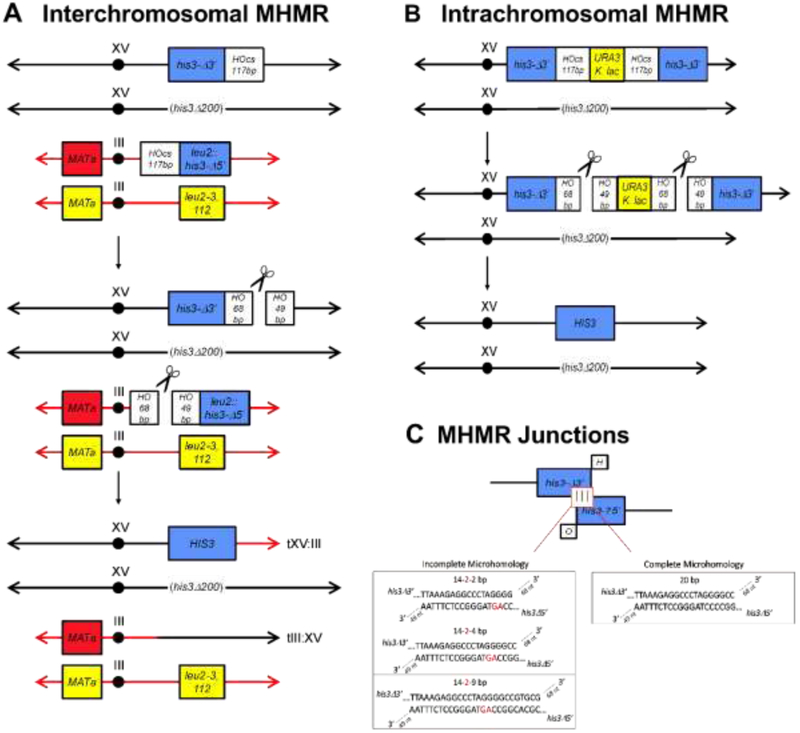
Figure 1. Microhomology mediated repair assays use varying locations, lengths, and continuity of microhomology.
(A and B) Two mutant alleles of his3, his3Δ3’ and his3Δ5’, are located on different chromosomes (A, interchromosomal) or the same chromosome (B, intrachromosomal) and share varying lengths and continuity of microhomolgy (C). The homologous chromosome possesses a complete deletion of the HIS3 ORF, the his3Δ200 allele. HO endonuclease, located at the TRP1 locus and induced in the presence of galactose (44), creates two simultaneous DSBs within the 117 bp HO endonuclease recognition site (HO), located next to each his3 allele. Microhomology mediated repair results in His+ recombinants and in the case of intrachromosomal repair, Ura auxotrophs. (C) Microhomology between his3Δ3’ and his3Δ5’ varies in length (16-25 bp) and is complete or contains a two base pair mismatch (shown in red) located at varying locations along the microhomology. Microhomology pairing intermediates contain the same non-homologous 3’-tails (68 nt and 49 nt) derived from cutting HO.
3.1. DNA polymerases impact on microhomology-mediated repair is dependent on the length, continuity, and chromosomal position of microhomology
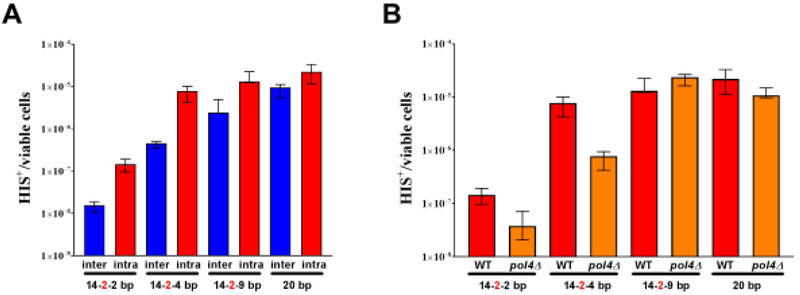
Pol δ , a heterotrimeric protein encoded by POL3, POL31, and POL32 in S. cerevisiae, facilitates the repair of DSBs using MH of varying lengths and quality . The modest but reproducible and significant 2-fold reduction in the absence of Pol λ, encoded by POL4 in S. cerevisiae, suggested a role for Pol λ in MHMR (31). In order to explore the role of Pol λ in MHMR, we determined the repair frequency between two his3 alleles, his3-Δ3’ and his3-Δ5’, located on different chromosomes, interchromosomal MHMR (Fig. 1A), or on the same chromosome, intrachromosomal MHMR (Fig. 1B). Both assay systems generate histidine prototrophs. The his3-Δ3’ and his3-Δ5’ alleles share MH with varying lengths of incomplete homology (14-2-2 bp, 14-2-4 bp or 14-2-9 bp, where the bold-faced number signifies two mismatches), 20 bp of complete homology, or no homology with his3Δ200 (Fig. 1C). The presence of a HO endonuclease recognition sequence allows for DSBs to be generated due to a galactose-inducible HO endonuclease (49). Examination of wild type cells show an increase in MHMR frequency, when the his3-Δ3’ and his3-Δ5’ alleles are located on the same chromosome (Fig. 2A, Table 1). In agreement with previous results examining interchromosomal MHMR (31), an increase in MH length lead to a corresponding increase in MHMR frequency (Figure 2A and Table 1). In particular, the increase was most dramatic between 14-2-2 bp and 14-2-4 bp of MH (Fig. 2A, Table 1). Interestingly, the Pol λ homozygous mutant pol4Δ/pol4Δ showed a significant decrease in MHMR, when the MHs were shorter (14-2-2 bp and 14-2-4 bp) and contained a mismatch (Fig. 2B, Table 1). These results are identical to pol4-D367E/pol4-D367E homozygous mutant we examined, which possesses a single amino acid change eliminating the polymerase catalytic activity (Table 1) (32). This decrease was independent of his3-Δ3’ and his3-Δ5’ allele genomic location (Table 1) and suggests a specialized role for Pol λ in MHMR when the MH is relatively short and contains terminal mismatches. To confirm our results, we tested in vitro whether Pol4 performs DNA synthesis of MMEJ substrates containing variable microhomology lengths (47) (Fig. 3A). Partially single-strand DNA (pssDNA) substrates were incubated with Pol4, in the presence of deoxy-ribonucleotides (dNTPs), for 30 min and terminated by protein degradation by proteinase K to determine the ability of Pol4 to incorporate nucleotides in front of DNA substrates. Confirming the preference for mismatched substrates, Pol4 did not incorporate any nucleotide in front DNA synapses with 2, 4 or 6 nucleotides of microhomology (pssDNA2, pssDNA4 and pssDNA6 respectively), but was efficient in incorporating up to 4 nucleotides, in presence of a 1 nucleotide mismatch at 3’-OH termini (pssDNA2-1) (Fig. 3B).
Figure 2. Genomic location and sequence continuity of shared microhomology impact repair in wild type and pol4Δ mutants.
(A and B) The median frequency of MHMR, ± 95% CI, was determined from a minimum of 15 independent cultures which possess the his3Δ3’ and his3Δ5’ mutant alleles sharing complete (20 bp) or incomplete (14-2-2 bp, 14-2-4 bp, or 14-2-9 bp) microhomologies. (A) MHMR frequencies of wild type cells with the his3Δ3’ and his3Δ5’ mutant alleles located on the same chromosome (intra) or ectopic chromosomes (inter). (B) Impact of pol4Δ single mutants (pol4Δ) on intrachromosomal MHMR. The frequencies of intrachromosomal MHMR in wild type (WT) cells using 14-2-2 bp, 14-2-4 bp, 14-2-9 bp, and 20 bp is represented in panel A and B.
Table 1:
Frequencies of interchomosomal and intrachromosomal MMEJ in wild type and mutant diploid strains
| Interchromosomal | Intrachromosomal | |||||||
|---|---|---|---|---|---|---|---|---|
| Genotypea | 14-2-2 bp | 14-2-4 bp | 14-2-9 bp | 20 bp | 14-2-2 bp | 14-2-4 bp | 14-2-9 bp | 20 bp |
| Wild type | 1.57×10−8 (1)d [1.1-1.9] |
4.51×10−7 (1)d [3.5-5.0] |
2.41×10−6 (1)d [1.7-4.9] |
9.4×10−6 (1)d [5.4-11.2] |
1.45×10−7 (1) [0.96-6.2] |
7.8×10−6 (1) [4.3-9.9] |
1.3×10−5 (1) [1.2-2.3] |
2.2×10−5 (1) [1.1-3.3] |
| pol4Δ/pol4Δ | 4.3×10−10 (↓37) [2.7-26.8] |
2.15×10−7 (↓2.1)d [1.8-2.5] |
2.2×10−6 (↓1.1) [1.4-3.8] |
2.3×10−6 (↓4.1) [1.3-5.7] |
3.8×10−8 (↓3.8) [2.1-7.2] |
7.8×10−7 (↓10) [4.2-9.4] |
2.4×10−5 (↑1.8) [1.6-2.7] |
1.1×10−5 (↓2) [1.0-5.2] |
| pol4-D367E/pol4-D367E | 6.3×10−9 (↓2.5) [3.8-7.9] |
1.0×10−7 (↓4.5) [0.5-2.0] |
3.5×10−6 (↑1.5) [1.1-6.2] |
6.5×10−6 (↓1.5) [4.4-13.8] |
6.5×10−8 (↓2.2) [4.4-8.6] |
1.4×10−6 (↓5.6) [1.2-2.2] |
9.5×10−6 (↓1.4) [5.1-11.8] |
ND |
| pol32Δ/pol32Δ | 3.8×10−9 (↓4.2)d [2.1-6.3] |
5.2×10−8 (↓8.3)d [2.1-6.9] |
5.1×10−8 (↓50)d [3.3-10.5] |
5.5×10−8 (↓167)d [1.4-9.6] |
3.8×10−9 (↓38) [2.1-6.3] |
1.6×10−7 (↓49) [0.9-6.9] |
1.8×10−7 (↓72) [0.85-4.5] |
5.2×10−7 (↓42) [4.2-13] |
| rev1Δ/rev1Δ | 1.0×10−8 (↓1.6) [0.8-1.3] |
1.8×10−7 (↓2.5) [1.1-2.4] |
1.2×10−6 (↓2) [0.6-4.4] |
NDb | 2.5×10−8 (↓5.8) [1.2-3.8] |
1.0×10−6 (↓7.8) [0.6-2.6] |
2.5×10−5 (↑1.9) [1.5-5.1] |
ND |
| rev3Δ/rev3Δ | 1.2×10−8 (↓1.3)d [1.0-3.0] |
2.4×10−7 (↓1.9)d [1.2-2.7] |
1.2×10−6 (↓2)d [0.6-1.6] |
8.6×10−6 (↓1.6)d [3.9-11.9] |
9.7×10−8 (↓1.5) [0.6-15.7] |
1.3×10−6 (↓6) [0.98-2.5] |
9.0×10−6 (↓1.4) [3.2-28.3] |
ND |
| rad30Δ/rad30Δ | 1.1×10−8 (↓1.4) [0.67-1.5] |
2.4×10−7 (↓1.9) [2.1-3.6] |
1.6×10−6 (↓1.5) [0.9-5.1] |
1.2×10−5 (↑1.3) [0.6 −2.7] |
2.9×10−8 (↓5) [2.0-7.5] |
2.5×10−6 (↓3.1) [1.0-2.9] |
1.3×10−5 (1) [0.4-2.8] |
ND |
|
pol4Δ/pol4Δ pol32Δ/pol32Δ |
3.0×10−11 (↓523)c | 1.7×10’−10 (↓2441)d [0.8-2.1] |
8.5×10−8 (↓28) [4.5-12.5] |
8.4×10−8 (↓112) [1.8-15.9] |
2.6×10−10 (↓558) [0.8-7.7] |
2.3×10−9 (↓3391) [1.1-4.6] |
1.0×10−6 (↓13) [0.3-2.5] |
1.5×10−6 (↓14.7) [0.5-4.2] |
|
pol4Δ/pol4Δ rev1Δ/rev1Δ |
2.0×10−9 (↓7.9) [1.1-5.2] |
1.7×10−7 (↓2.7) [1.2-1.9] |
1.9×10−6 (↓1.3) [0.8-3.0] |
2.3×10−5 (↑2.4) [1.1-2.9] |
ND | ND | ND | ND |
|
pol4Δ/pol4Δ rev3Δ/rev3Δ |
ND | 1.4×10−7 (↓3.2)d [0.8-1.9] |
1.5×10−6 (↓1.6) [0.7-2.0] |
ND | ND | ND | ND | ND |
|
pol4Δ/pol4Δ rad30Δ/rad30Δ |
3.4×10−9 (↓4.6) [1.9-5.1] | 1.7×10−7 (↓2.7) [1.4-2.2] |
2.2×10−6 (↓1.1) [1.2-2.8] |
ND | ND | ND | ND | ND |
|
pol4Δ/pol4Δ rev1Δ/rev1Δ rad1Δ/rad1Δ |
ND | 5.7×10−8 (↓7.9) [4.3-10.5] |
1.4×10−7 (↓1.7) [0.8-1.8] |
4.8×10−6 (↓1.9) [2.9-6.4] |
ND | ND | ND | ND |
|
pol4Δ/pol4Δ rev1Δ/rev1Δ rev3Δ/rev3Δ rad30Δ/rad30Δ |
3.3×10−9 (↓4.8) [2.3-5.7] |
2.7×10−8 (↓16.7) [1.5-3.4] |
1.7×10−6 (↓1.4) [1.4-5.2] |
3.8×10−6 (↓2.5) [2.8-7.6] |
ND | ND | ND | ND |
|
pol4Δ/pol4Δ
rev1Δ/rev1Δ rev3Δ/rev3Δ rad30Δ/rad30Δ pol32Δ/pol32Δ |
<2.1×10−10 (↓75)e | <3.4×10−10 (↓1326)e | ND | 2.1×10−7 (↓45) [1.4-2.6] |
ND | ND | ND | ND |
The median translocation frequency was determined for each strain from at least 15 independent cultures. Fold increase (↑) or decrease (↓), rounded to integers, from the median frequency obtained with the wild type strain are indicated in parentheses. The lower and upper 95% confidence intervals for each median were determined using the formulae (n/2 – 1.96√n/2) and (1 + n/2 + 1.96√n/2), respectively, and are indicated in brackets.
All strains possess the his3-Δ5’ substrate at the LEU2 locus on one copy of chromosome III and the his3-Δ3’ substrate at the HIS3 locus (Fig. 1). The his3-Δ5’ and his3-Δ3’ substrates share varying microhomologies of HIS3 coding sequence indicated above each column. Complete genotypes are given in Table S1.
Frequency not determined (ND).
Number represents a single His+ colony from 20 independent cultures.
Frequency was reported previously (31).
Estimate based on assuming a single colony from 15 independent cultures.
Figure3. DNA Polymerase Lambda extends from a mismatched 3’-hydroxy terminus.
(A) Model of MMEJ reaction: pssDNAs were incubated to obtain paired 3’-overhangs of variable microhomology lengths and continuity, followed by the addition of Pol4 to determine the extension activity of Pol4 for each substrate. (B) Denaturing sequencing gel showing MMEJ reactions with each pssDNA (pssDNA2-1: Lanes 1 and 2, pssDNA2: Lanes 3 and 4, pssDNA4: Lanes 5 and 6, pssDNA6: Lanes 7 and 8) in the absence (−) or presense (+) of Pol4.
3.2. Loss of function among multiple DNA polymerases reveal functional redundancies and specialized usage of MHs in MHMR
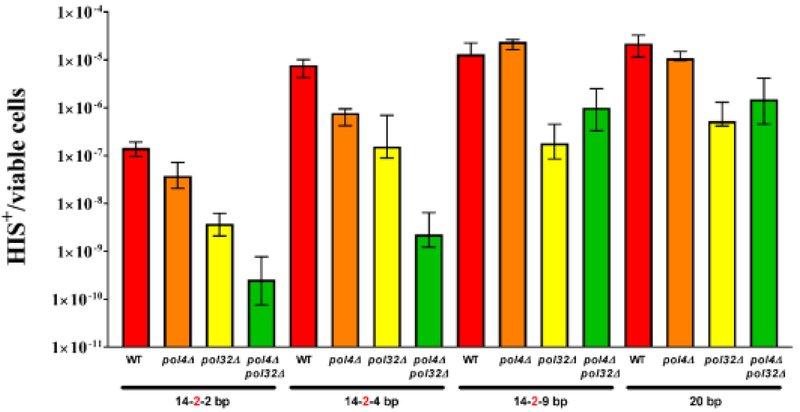
In our previous work, we observed a synergistic decrease in pol4Δ/pol4Δ pol3-ct/pol3-ct and pol4Δ/pol4Δ pol32Δ/pol32Δ homozygous double mutants using 14-2-4 bp of MH located on different chromosomes, suggesting a cooperation between Pol λ and Pol δ (31). We explored this further by determining how the length and quality of MH used for repair influenced MHMR frequency in the pol4Δ pol32Δ double mutant. In agreement with our previous result with MHs of 14-2-4 bp, the pol4Δ pol32Δ double mutants showed a synergistic decrease in MHMR frequency when using 14-2-2 bp of MH (Fig. 4, Table 1). However, the use of 14-2-9 bp or 20 bp of MH for repair in pol4Δ pol32Δ double mutants resulted in MHMR frequencies similar to pol32Δ single mutants (Fig. 4, Table 1). These results suggest the cooperation between Pol λ and Pol δ in MHMR is dependent on the length and quality of MH. This dependence is dictated by the substrate specificity of Pol λ due to the preferential repair of MHs which are short and contain a mismatch (Fig. 4, Table 1).
Figure 4. Cooperation between Pol4 and Pol32 when microhomology is limiting and mismatches are close to the 3’-hydroxyl end.
The median frequency of MHMR, ± 95% CI, was determined for wild type, pol4Δ and pol32Δ single mutants, and pol4Δ pol32Δ double mutant cells. A minimum of 15 independent cultures were examined which possess the his3Δ3’ and his3Δ5’ mutant alleles sharing complete (20 bp) or incomplete (14-2-2 bp, 14-2-4 bp, or 14-2-9 bp) microhomologies located on the same chromosome.
In order to explore the role of other low-fidelity DNA polymerases in MHMR, we explored how the loss of Pol ζ, Pol η, and Rev1 influences the repair process. Pol ζ in S. cerevisiae is a heterodimeric protein encoded by REV3 and REV7, while Rev1 is encoded by REV1 (50). These two DNA polymerases function together to bypass DNA lesions and are responsible for at least 50% of spontaneous mutations due to the ability of Rev1 to provide structural support to Pol ζ, which extends mismatch 3’-hydroxy termini (50,51). The MHMR frequency in rev3Δ/rev3Δ and rev1Δ/rev1Δ homozygous mutants show a modest reduction when 14-2-4 bp of MH is used for repair, regardless of the chromosomal position of his3-Δ3’ and his3-Δ5’ (Fig. 5, Table 1). However, rev3Δ and rev1Δ mutants use MHs of other lengths and quality (14-2-2 bp, 14-2-9 bp, and 20 bp) on the same or different chromosomes with the same efficiency as wild type cells, with the exception of intrachromosomal 14-2-2 bp rev1Δ mutants (Table 1). Finally, the impact of Pol η, which is encoded by RAD30, was examined in rad30Δ/rad30Δ homozygous mutants and showed a modest decrease in the repair of intrachromosomal his3 substrates sharing 14-2-2 bp and 14-2-4 bp of MH, but wild type for all other MHs tested (Fig. 5, Table 1). These results suggest the involvement of low-fidelity DNA polymerases in MHMR is dependent on the MH length, quality, and chromosomal position.
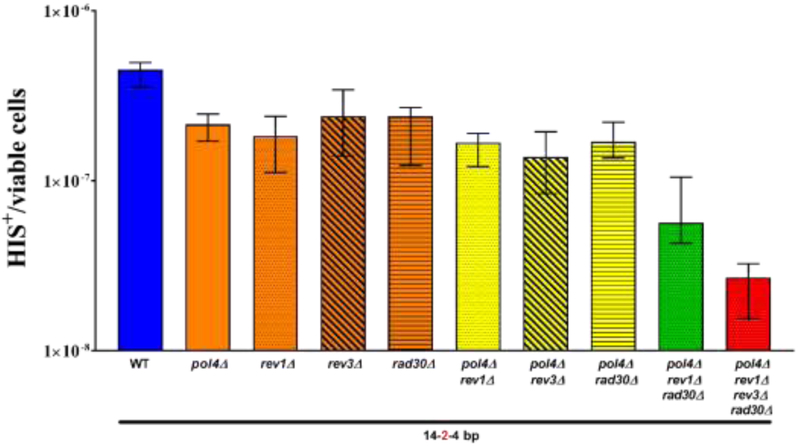
Figure 5. TLS DNA polymerases cooperate and serve redundant functions in microhomology mediated repair.
The median frequency of MHMR, ± 95% CI, was determined for wild type (WT), single mutants of each non-essential DNA polymerase (pol4Δ, rev1Δ, rev3Δ, and rad30Δ), select double (pol4Δ rev1Δ, pol4Δ rev3Δ, and pol4Δ rad30Δ) and triple mutants (pol4Δ rev1Δ rad30Δ), and quadruple mutant (pol4Δ rev1Δ rev3Δ rad30Δ) cells. A minimum of 15 independent cultures, which possess the his3Δ3’ and his3Δ5’ mutant alleles on ectopic chromosomes sharing 14-2-4 bp of microhomolgy were examined.
The involvement of Pol λ, Pol ζ, Pol η, and Rev1 in MHMR prompted an analysis of various mutant combinations to explore the possibility of functional overlap among the low-fidelity DNA polymerases, which might explain the modest reduction in MHMR frequencies of the single pol4Δ, rev3Δ, rev1Δ, and rad30Δ mutants. Since Pol λ had a clear function in MHMR using short MHs with terminal mismatches, we tested rev3Δ, rev1Δ, and rad30Δ mutants in combination with pol4Δ mutants using his3-Δ3’ and his3-Δ5’ substrates on different chromosomes. Each of the homozygous double mutants explored, pol4Δ/pol4Δ rev3Δ/rev3Δ, pol4Δ/pol4Δ rev1Δ/rev1Δ, and pol4Δ/pol4Δ rad30Δ/rad30Δ, were no different than the pol4Δ single mutants (Fig. 5, Table 1). However, a synergistic reduction in MHMR frequency of 8-fold was observed in the homozygous pol4Δ/pol4Δ rev1Δ/rev1Δ rad30Δ/rad30Δ triple mutant and 17-fold for the pol4Δ/pol4Δ rev1Δ/rev1Δ rev3Δ/rev3Δ rad30Δ/rad30Δ quadruple mutant (Fig. 5, Table 1). This suggests functional redundancy between the low-fidelity DNA polymerase. Therefore, the loss of function in one low-fidelity DNA polymerase is potentially compensated by the function of the remaining polymerases. The overall reduction in MHMR frequency of 17-fold observed for the pol4Δ rev1Δ rev3Δ rad30Δ quadruple mutant is not as large as that observed for pol4Δ pol32Δ double mutants (2,441-fold reduction, Table 1). While Pol32 is a subunit of both Pol ζ and Pol δ (52,53), the much stronger effect of the pol4Δ pol32Δ double mutant compared to the pol4Δ rev1Δ rev3Δ rad30Δ quadruple mutant is apparently owed to the significant contribution of Polδ to MHMR. In order to determine the contributions of the low-fidelity DNA polymerases and Pol δ to MHMR, we examined pol4Δ rev1Δ rev3Δ rad30Δ pol32Δ quintuple mutants using 14-2-2 bp, 14-2-4 bp, or 20 bp of MH on different chromosomes for repair. As expected from the extremely low frequency in the pol4 pol32 double mutant, we observed no His+ recombinants when 14-2-2 bp or 14-2-4 bp was used, suggesting a synergistic decrease in MHMR that is similar to our observations in the pol4Δ pol32Δ double mutant (Table 1). However, the frequency of His+ recombinants using 20 bp of MH in the pol4Δ pol32Δ double mutant is sufficiently high to document a potential further decrease. We observed the frequency in the pol4Δ rev1Δ rev3Δ rad30Δ pol32Δ quintuple mutant to not be significantly different from the pol4Δ pol32Δ double mutant (Table 1). Taken together, these data illustrate the complex interactions between the low-fidelity DNA polymerases and Pol δ in MHMR, which is dependent on MH length and continuity.
3.3. Genome stability is influenced by the loss of non-essential DNA polymerase function
Our results above demonstrate that deletions of POL4, REV1, REV3 and RAD30 have the greatest impact on MHMR compared to any other loss-of-function combination tested. In agreement with these results is the impact the individual low-fidelity DNA polymerases Rev1, Rev3, and Rad30 have on specific types of genome rearrangements and mutagenesis (29,54-56). Mutations in these genes reduce forward mutation rates in CAN1 1-2.5 fold and increase the gross chromosomal rearrangement (GCR) rate 1-2 fold. In addition, we determined the CAN1 mutation and GCR rates for pol4Δ single mutants to be 1.56 × 10−7 and 3.52 × 10−9, respectively, and to be not different from wildtype (p < 0.05). We therefore wanted to determine if genome stability is altered in the pol4Δ rev1Δ rev3Δ rad30Δ quadruple mutant by examining mutation rates and genome rearrangements compared to wild type. The CAN1 forward mutation rate was 2-fold lower in the quadruple mutant compared to wild type (Fig. 6A), while genome rearrangements as measured by GCR were similar in both genotypes (Fig. 7A). These results support a role of the low-fidelity DNA polymerases in mutagenesis, but not in the creation of large-scale genomic rearrangements typical of proteins involved in DNA repair (43).
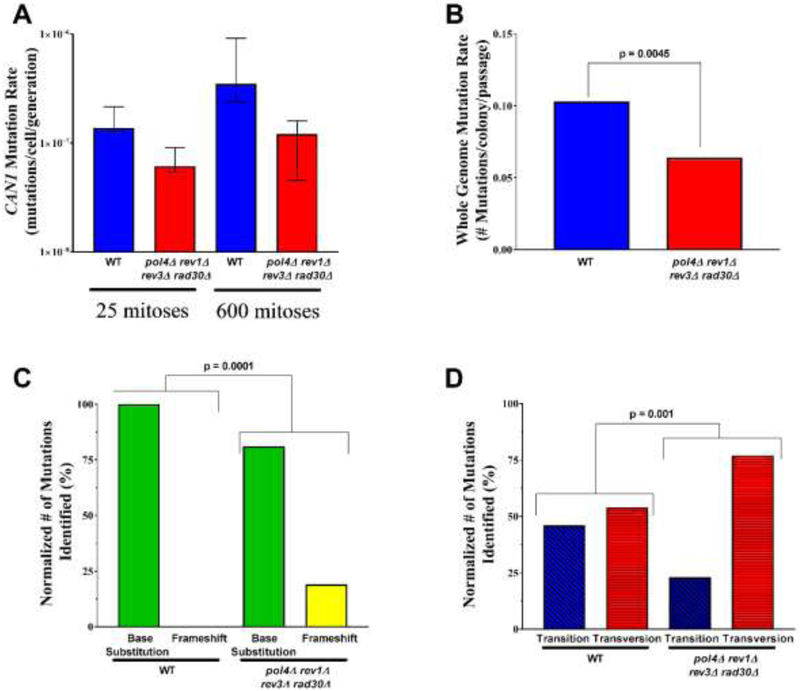
Figure 6. The absence of non-essential DNA polymerases alters the accumulation of mutations and mutational profile over time.
(A) The median CAN1 forward mutation rate and 95% CI from 16 independent cultures, was determined for wild type (WT) and pol4Δ rev1Δ rev3Δ rad30Δ quadruple mutant cells (B) The whole genome mutation rate was determined by taking the number of identified mutations following whole genome sequencing and dividing by the number of parallel cultures analyzed and the number of serial bottleneck passages (4 clones 100 passages for WT, 10 clones 25 passages for quad). A total of 4 wild type (WT) (22) and 10 pol4Δ rev1Δ rev3Δ rad30Δ quadruple mutant cells were used to determine the whole genome mutation rate and mutation rates were compared using a contingency table. (C) The normalized number of mutation classes (base substitutions and frameshifts) between wild type (WT) and pol4Δ rev1Δ rev3Δ rad30Δ quadruple mutant cells was compared using Fisher’s exact test. (D) The normalized number of base substitution types (transitions and transversions) between wild type (WT) and pol4Δ rev1Δ rev3Δ rad30Δ quadruple mutant cells was compared using Fisher’s exact test. WT data from a previous study (22) was used in panels B-D.
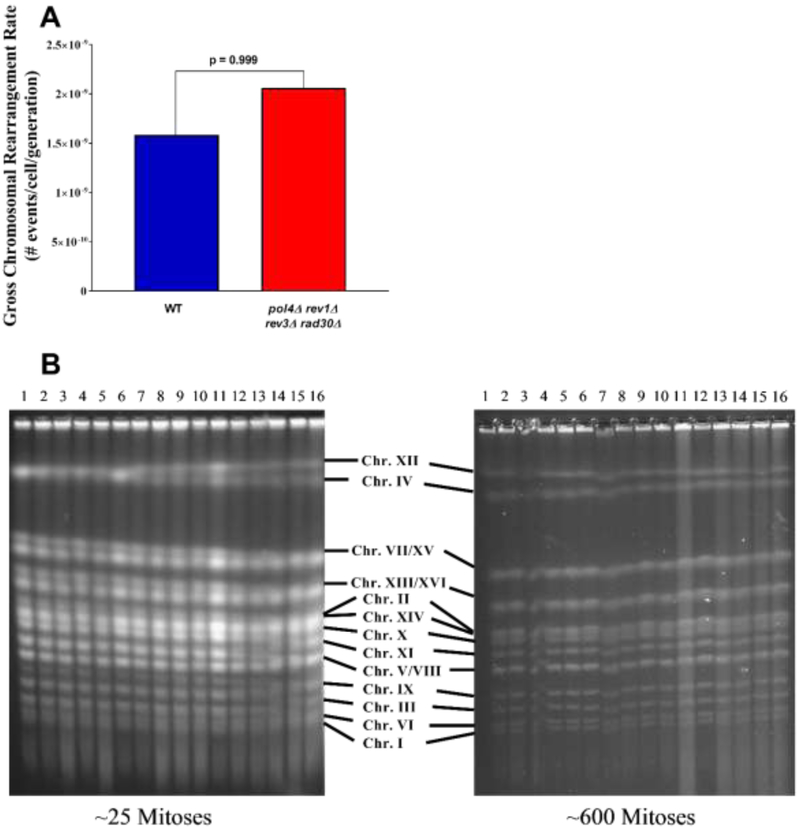
Figure 7. Non-essential DNA polymerases do not contribute to gross-chromosomal rearrangements.
(A) The rate of gross chromosomal rearrangements, measured as the simultaneous loss of CAN1 and hxt13::URA3, was determined for wild type (WT) and pol4Δ rev1Δ rev3Δ rad30Δ quadruple mutant cells after 25 serial bottleneck passages. A total of 16 independent cultures were used to calculate the gross chromosomal rearrangement rate and compared using Fisher’s exact test. (B) Whole chromosomes of pol4Δ rev1Δ rev3Δ rad30Δ quadruple mutant colonies taken from a tetrad dissection plate following sporulation and genotyping, (~25 mitoses) and following 25 serial bottleneck passages (~600 mitoses), were analyzed by pulse-field gel electrophoresis. A total of 16 independent pol4Δ rev1Δ rev3Δ rad30Δ quadruple mutant colonies were analyzed for large-scale genomic rearrangements at ~25 and ~600 mitoses.
The impact of pol4Δ rev1Δ rev3Δ rad30Δ quadruple mutants on mutagenesis prompted us to examine the accumulation of mutations in these cells over time. In order to test this, ten independent quadruple mutant clones were passaged over 25 bottlenecks (~ 600 mitoses) and analyzed by whole genome sequencing to identify de novo spontaneous mutations (Materials and Methods, Suppl. Fig. 1). It revealed a significant decrease in the number of accumulated mutations in the quadruple mutant compared to wild type cells (Fig. 6B). No large-scale genomic rearrangements nor variations in chromosome copy number were identified (data not shown). Consistently, no rearrangements were detected upon chromosome pulsed-field gel electrophoresis (PFGE) in cells before and after serial bottleneck passages (Fig. 7B). Thus, our results support the role of the low fidelity polymerases in the accumulation of de novo point mutations.
To further assess the low-fidelity DNA polymerases involvement in mutation accumulation, we characterized the specific mutations identified by whole genome sequencing in the pol4Δ rev1Δ rev3Δ rad30Δ mutant. In agreement with previous CAN1 mutation analysis in rev3 mutants, a significant shift in the proportion of base-substitution and frameshift mutations was observed in the DNA polymerase quadruple mutants compared to wild type (Fig. 6C, Table 2) (57). The 3 de novo frameshift mutations observed in the quadruple TLS mutant are two +1T and one −1A indels located in poly-T sequence on chromosome 1 and 10, and in a poly-A sequence on chromosome 16, respectively (Suppl. Fig. 1). Thus, as generally observed in WT and for example, mismatches repair mutants, frameshift mutations occur most often within mononucleotide runs (58-60). Examination of base-substitution mutations revealed a shift toward transversions versus transitions in the pol4Δ rev1Δ rev3Δ rad30Δ mutants from the near equal proportion seen in wild type cells (22/41 versus 10/14, respectively) (Fig. 6D). Furthermore, the preference in transversions of G:C base pairs to T:A or C:G base pairs in DNA polymerase quadruple mutants compared to wild type (10/10 versus 12/22) demonstrates a mutation spectrum (Table 2).
Table 2:
Types of Genomic Mutations Identified in Wildtype and Mutant Haploidsa
| Genotypeb | Base Substitutions | Frameshifts | Total Mutations |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Transition | Transversion | ||||||||
| A:T → G:C | G:C → A:T | A:T → T:A | G:C → T:A | A:T → C:G | G:C → C:G | +1 | −1 | ||
| Wild type | 6 | 13 | 6 | 8 | 4 | 4 | 0 | 0 | 41 |
| pol4Δ rev1Δ rev3Δ rad30Δ | 2 | 1 | 0 | 6 | 0 | 4 | 2 | 1 | 16 |
Genomic de novo mutations were identified from four independent strains for WT (22) and ten for pol4Δ rev1Δ rev3Δ rad30Δ using whole-genome Illumina sequencing and analysis.
All strains are derived from BY4741 and 4742 and subjected to 100 passages for WT, and 25 passages for pol4Δ rev1Δ rev3Δ rad30Δ.
4. Discussion
The maintenance of genome stability is necessary for the proper functioning of cells and in the prevention of disease and cancer (61). While many repair pathways in cells can respond to specific types of DNA damage, if DNA replication occurs before repair than the damaged DNA can become a fixed mutation, which can exert its effects slowly over time (62). In addition, decreased fidelity of DNA replication polymerases δ and ε results in a mutator phenotype associated with certain cancers (63). DNA damage can also promote spontaneous mutagenesis through the action of low-fidelity DNA polymerases, which facilitate tolerance to DNA damage during lesion bypass and extension of improperly paired nucleotides (28-30). In this study we examined how the low-fidelity DNA polymerases, Pol η, Pol ζ, Pol λ, and Rev1 contribute to genome stability by testing their involvement in MHMR and mutagenesis.
While previous studies examined low-fidelity DNA polymerases individually in their contribution to MHMR, we examined loss-of-function double, triple, and quadruple mutants to determine if they exhibit any functional redundancy or cooperation during repair using MHs (31-33,35). We observed a clear specialization of function for each DNA polymerase which was dependent on the proximity of a mismatch to the 3’-hydroxyl end, the length of MH, and to a lesser extent the location of MHs used for repair. The most consistent decrease in MHMR frequency was observed in low-fidelity DNA polymerase mutants using 14-2-4 bp of MH for repair. In fact, the pol4Δ rev1Δ rev3Δ rad30Δ quadruple mutant resulted in the most dramatic decrease in MHMR frequency. This suggests functional redundancy between these DNA polymerases in MHMR, such that the loss of function in one can fully or partially compensated for by the function of the remaining DNA polymerases. Results from the pol4Δ rev1Δ rev3Δ rad30Δ pol32Δ quintuple mutant suggest the functional overlap between the low-fidelity DNA polymerases also depends on a fully functional Pol δ and the sequence context of MH used for repair. Since MHs have been observed in several chromosomal repair events, the DNA polymerase engaged may depend on the length and quality of MH used for repair, cell type, and phase of the cell cycle (64-67).
The synergistic effects observed in MHMR from the pol4Δ rev1Δ rev3Δ rad30Δ mutant prompted us to examine the impact genome-wide. Our results demonstrate that the accumulation of spontaneous mutations is dependent, in part, on low-fidelity DNA polymerases. This is most likely due to their specialized functions in lesion bypass, extension and repair (28,29). In fact, it is this specialization of function among the low-fidelity DNA polymerases, which gives them specificity in facilitating MHMR when certain MHs are used. This is supported by mutation spectra analysis of accumulated mutations in the pol4Δ rev1Δ rev3Δ rad30Δ mutant, which show a shift in the types of mutations observed, compared to wild type. The appearance of frameshift mutations and a change in the types of base-substitutions observed in DNA polymerase mutants compared to wild type strongly suggest a direct impact of these low-fidelity DNA polymerases on spontaneous mutation accumulation. Furthermore, it is highly likely that in the presence of a DNA damaging agent, such as UV radiation, the total number and spectra of mutations would also deviate from wild type in pol4Δ rev1Δ rev3Δ rad30Δ mutants (68-70). Finally, in the absence of the low-fidelity DNA polymerases it is highly likely that the replicative DNA polymerases, Pol δ and Pol ε, contribute to the observed mutation spectra which may represent the unique mis-incorporation profiles of these DNA polymerases (71,72)
Our data support a role for the low-fidelity DNA polymerases in maintaining genome stability by facilitating repair and impacting spontaneous mutation accumulation. While mutation and MHMR genome rearrangements are associated with negative cellular consequences related to fitness, it is not always a certainty (20,65). Studies in S. cerevisiae examining fitness-altering mutations during mutation accumulation found up to 13% of such mutations were beneficial (73,74). This suggests a positive evolutionary role for these low-fidelity DNA polymerases and explains why they are relatively well conserved in many eukaryotes (75). Therefore, DNA polymerases that contribute to mutation accumulation may support genetic diversity and minimize other types of mutations, such as frameshifts, which are more likely to have a negative impact on fitness. In a similar manner, MHMR may prevent chromosome loss by allowing for the repair of DSBs, which are not repaired by homologous recombination or non-homologous end-joining. The resulting deletion or translocation, following MHMR, is potentially less detrimental to organismal fitness than chromosome loss. Taken together, the response to DNA damage is a trade-off between survival and overall fitness where the consequences of repair, mutations or genomic rearrangements, are less detrimental than the alternatives in the absence of these repair pathways.
Supplementary Material
Highlights.
Location and quality of microhomology influence DNA polymerase repair efficiency
Non-essential DNA polymerases function in overlapping roles during MHMR
Non-essential DNA polymerases influence mutation accumulation
Non-essential DNA polymerases contribute to specific types of mutations.
Acknowledgements
We thank William Wright and Jie Liu for helpful discussion and comments on the manuscript, and Adam Bailis and Lorraine Symington for providing strains. We thank the Institut Curie, NGS platform for NGS sequencing. DM was partially supported by fellowship 15IB-0109 from the CBCRP and the Training Grant in Oncogenic Signals and Chromosome Biology CA108459. BXHF had partial support from a Presidential Undergraduate Fellowship from the University of California. This work was supported by grant GM58015 the National Institutes of Health to WDH, France-Berkeley Fund (2015-32) to WDH and AN, and Cancer Center Core Support Grant NCI P30CA093373 to UCD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reed DH and Frankham R (2003) Correlation between Fitness and Genetic Diversity. Conservation Biology, 17, 230–237. [Google Scholar]
- 2.Markert JA, Champlin DM, Gutjahr-Gobell R, Grear JS, Kuhn A, McGreevy TJ, Roth A, Bagley MJ and Nacci DE (2010) Population genetic diversity and fitness in multiple environments. BMC Evo Biol, 10, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charlesworth B (2015) Causes of natural variation in fitness: evidence from studies of Drosophila populations. Proc Natl Acad Sci USA, 112, 1662–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Umar A, Schweitzer PA, Levy NS, Gearhart JD and Gearhart PJ (1991) Mutation in a reporter gene depends on proximity to and transcription of immunoglobulin variable transgenes. Proc Natl Acad Sci USA, 88, 4902–4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aquilina G and Bignami M (2001) Mismatch repair in correction of replication errors and processing of DNA damage. J Cell Physiol, 187, 145–154. [DOI] [PubMed] [Google Scholar]
- 6.Hawk JD, Stefanovic L, Boyer JC, Petes TD and Farber RA (2005) Variation in efficiency of DNA mismatch repair at different sites in the yeast genome. Proc Natl Acad Sci USA, 102, 8639–8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tran HT, Keen JD, Kricker M, Resnick MA and Gordenin DA (1997) Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol Cell Biol, 17, 2859–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lang GI and Murray AW (2008) Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics, 178, 67–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith NG, Webster MT and Ellegren H (2002) Deterministic mutation rate variation in the human genome. Genome Res, 12, 1350–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson ST and Petes TD (1992) Instability of simple sequence DNA in Saccharomyces cerevisiae. Mol Cell Biol, 12, 2749–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park C, Qian W and Zhang J (2012) Genomic evidence for elevated mutation rates in highly expressed genes. EMBO Rep, 13, 1123–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishant KT, Singh ND and Alani E (2009) Genomic mutation rates: what high-throughput methods can tell us. Bioessays, 31, 912–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muir P, Li S, Lou S, Wang D, Spakowicz DJ, Salichos L, Zhang J, Weinstock GM, Isaacs F, Rozowsky J et al. (2016) The real cost of sequencing: scaling computation to keep pace with data generation. Genome Biol, 17, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denver DR, Dolan PC, Wilhelm LJ, Sung W, Lucas-LledÃ3 JI, Howe DK, Lewis SC, Okamoto K, Thomas WK, Lynch M et al. (2009) A genome-wide view of Caenorhabditis elegans base-substitution mutation processes. Proc Natl Acad Sci USA, 106, 16310–16314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynch M, Sung W, Morris K, Coffey N, Landry CR, Dopman EB, Dickinson WJ, Okamoto K, Kulkarni S, Hartl DL et al. (2008) A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci USA, 105, 9272–9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keightley PD, Trivedi U, Thomson M, Oliver F, Kumar S and Blaxter ML (2009) Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res, 19, 1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ipe J, Swart M, Burgess KS and Skaar TC (2017) High-Throughput Assays to Assess the Functional Impact of Genetic Variants: A Road Towards Genomic-Driven Medicine. Clin Transl Sci, 10, 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caulfield T, Evans J, McGuire A, McCabe C, Bubela T, Cook-Deegan R, Fishman J, Hogarth S, Miller FA, Ravitsky V et al. (2013) Reflections on the cost of "low-cost" whole genome sequencing: framing the health policy debate. PLoS Biol, 11, e1001699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen KD, Dukhovny D, Siebert U and Green RC (2015) Assessing the Costs and Cost-Effectiveness of Genomic Sequencing. J Pers Med, 5, 470–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson AL and Loeb LA (1998) The Mutation Rate and Cancer. Genetics, 148, 1483–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeggo PA, Pearl LH and Carr AM (2016) DNA repair, genome stability and cancer: a historical perspective. Nat Rev Cancer, 16, 35–42. [DOI] [PubMed] [Google Scholar]
- 22.Serero A, Jubin C, Loeillet S, Legoix-Ne P and Nicolas AG (2014) Mutational landscape of yeast mutator strains. Proc Natl Acad Sci USA, 111, 1897–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lang GI, Parsons L and Gammie AE (2013) Mutation rates, spectra, and genomewide distribution of spontaneous mutations in mismatch repair deficient yeast. G3, 3, 1453–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McVey M, Khodaverdian VY, Meyer D, Cerqueira PG and Heyer WD (2016) Eukaryotic DNA Polymerases in Homologous Recombination. Annu Rev Genet, 50, 393–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng DQ, Zhang K, Wu XC, Mieczkowski PA and Petes TD (2016) Global analysis of genomic instability caused by DNA replication stress in Saccharomyces cerevisiae. Proc Natl Acad Sci USA, 113, E8114–E8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Branzei D and Foiani M (2010) Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol, 11, 208–219. [DOI] [PubMed] [Google Scholar]
- 27.Lange SS, Takata K and Wood RD (2011) DNA polymerases and cancer. Nat Rev Cancer, 11, 96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghosal G and Chen J (2013) DNA damage tolerance: a double-edged sword guarding the genome. Transl Cancer Res, 2, 107–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prakash S, Johnson RE and Prakash L (2005) Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem, 74, 317–353. [DOI] [PubMed] [Google Scholar]
- 30.Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV and Walker GC (2009) Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev, 73, 134–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer D, Fu BX and Heyer WD (2015) DNA polymerases delta and lambda cooperate in repairing double-strand breaks by microhomology-mediated end-joining in Saccharomyces cerevisiae. Proc Natl Acad Sci USA, 112, E6907–6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee K and Lee SE (2007) Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics, 176, 2003–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Decottignies A (2007) Microhomology-mediated end joining in fission yeast is repressed by pku70 and relies on genes involved in homologous recombination. Genetics, 176, 1403–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daley JM and Wilson TE (2005) Rejoining of DNA double-strand breaks as a function of overhang length. Mol Cell Biol, 25, 896–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villarreal DD, Lee K, Deem A, Shim EY, Malkova A and Lee SE (2012) Microhomology directs diverse DNA break repair pathways and chromosomal translocations. PLoS Genet, 8, e1003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jansen JG, Temviriyanukul P, Wit N, Delbos F, Reynaud CA, Jacobs H and de Wind N (2014) Redundancy of mammalian Y family DNA polymerases in cellular responses to genomic DNA lesions induced by ultraviolet light. Nucleic Acids Res, 42, 11071–11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braithwaite EK, Kedar PS, Stumpo DJ, Bertocci B, Freedman JH, Samson LD and Wilson SH (2010) DNA polymerases beta and lambda mediate overlapping and independent roles in base excision repair in mouse embryonic fibroblasts. PLoS One, 5, e12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherman F, Fink GR and Hicks JB (1982) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor NY. [Google Scholar]
- 39.Thomas BJ and Rothstein R (1989) The genetic control of direct-repeat recombination in Saccharomyces: the effect of rad52 and rad1 on mitotic recombination at GAL10, a transcriptionally regulated gene. Genetics, 123, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P and Boeke JD (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast, 14, 115–132. [DOI] [PubMed] [Google Scholar]
- 41.Ho CK, Mazon G, Lam AF and Symington LS (2010) Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol Cell, 40, 988–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erdeniz N, Mortensen UH and Rothstein R (1997) Cloning-free PCR-based allele replacement methods. Genome Res, 7, 1174–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen C and Kolodner RD (1999) Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet, 23, 81–85. [DOI] [PubMed] [Google Scholar]
- 44.Pannunzio NR, Manthey GM and Bailis AM (2008) RAD59 is required for efficient repair of simultaneous double-strand breaks resulting in translocations in Saccharomyces cerevisiae. DNA Repair, 7, 788–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lea DE and Coulson CA (1949) The distribution of the numbers of mutants in bacterial populations. J Genet, 49, 264–285. [DOI] [PubMed] [Google Scholar]
- 46.Luria SE and Delbruck M (1943) Mutations of bacteria from virus sensitivity to virus resistance. Genetics, 28, 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kent T, Chandramouly G, McDevitt SM, Ozdemir AY and Pomerantz RT (2015) Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nat Struct Mol Biol, 22, 230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurtzman CP and Fell JW (1998) The Yeasts - A Taxonomic Study. 4th ed. Amsterdam: Elsevier. [Google Scholar]
- 49.Pannunzio NR, Manthey GM and Bailis AM (2008) RAD59 is required for efficient repair of simultaneous double-strand breaks resulting in translocations in Saccharomyces cerevisiae. DNA Repair (Amst), 7, 788–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prakash S and Prakash L (2002) Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev, 16, 1872–1883. [DOI] [PubMed] [Google Scholar]
- 51.Lawrence CW, Gibbs PE, Murante RS, Wang XD, Li Z, McManus TP, McGregor WG, Nelson JR, Hinkle DC and Maher VM (2000) Roles of DNA polymerase zeta and Rev1 protein in eukaryotic mutagenesis and translesion replication. Cold Spring Harb Symp Quant Biol, 65, 61–69. [DOI] [PubMed] [Google Scholar]
- 52.Johnson RE, Prakash L and Prakash S (2012) Pol31 and Pol32 subunits of yeast DNA polymerase delta are also essential subunits of DNA polymerase zeta. Proc Natl Acad Sci USA, 109, 12455–12460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Makarova AV, Stodola JL and Burgers PM (2012) A four-subunit DNA polymerase zeta complex containing Pol delta accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res, 40, 11618–11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ragu S, Faye G, Iraqui I, Masurel-Heneman A, Kolodner RD and Huang ME (2007) Oxygen metabolism and reactive oxygen species cause chromosomal rearrangements and cell death. Proc Natl Acad Sci USA, 104, 9747–9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Motegi A, Kuntz K, Majeed A, Smith S and Myung K (2006) Regulation of gross chromosomal rearrangements by ubiquitin and SUMO ligases in Saccharomyces cerevisiae. Mol Cell Biol, 26, 1424–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lis ET, O'Neill BM, Gil-Lamaignere C, Chin JK and Romesberg FE (2008) Identification of pathways controlling DNA damage induced mutation in Saccharomyces cerevisiae. DNA Repair (Amst), 7, 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang ME, Rio AG, Galibert MD and Galibert F (2002) Pol32, a subunit of Saccharomyces cerevisiae DNA polymerase delta, suppresses genomic deletions and is involved in the mutagenic bypass pathway. Genetics, 160, 1409–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gragg H, Harfe BD and Jinks-Robertson S (2002) Base composition of mononucleotide runs affects DNA polymerase slippage and removal of frameshift intermediates by mismatch repair in Saccharomyces cerevisiae. Mol Cell Biol, 22, 8756–8762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Greene CN and Jinks-Robertson S (1997) Frameshift intermediates in homopolymer runs are removed efficiently by yeast mismatch repair proteins. Mol Cell Biol, 17, 2844–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walker CJ, Miranda MA, O'Hern MJ, Blachly JS, Moyer CL, Ivanovich J, Kroll KW, Eisfeld AK, Sapp CE, Mutch DG et al. (2016) MonoSeq Variant Caller Reveals Novel Mononucleotide Run Indel Mutations in Tumors with Defective DNA Mismatch Repair. Hum Mutat, 37, 1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoeijmakers JHJ (2001) Genome maintenance mechanisms for preventing cancer. Nature, 411, 366–374. [DOI] [PubMed] [Google Scholar]
- 62.Pray LA (2008) DNA Replication and Causes of Mutation. Nature Education, 1, 214. [Google Scholar]
- 63.Heitzer E and Tomlinson I (2014) Replicative DNA polymerase mutations in cancer. Curr Opin Genet Dev, 24, 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA et al. (2011) Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell, 144, 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McVey M and Lee SE (2008) MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet, 24, 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu P, Carvalho CM, Hastings PJ and Lupski JR (2012) Mechanisms for recurrent and complex human genomic rearrangements. Curr Opin Genet Dev, 22, 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cortizas EM, Zahn A, Hajjar ME, Patenaude AM, Di Noia JM and Verdun RE (2013) Alternative end-joining and classical nonhomologous end-joining pathways repair different types of double-strand breaks during class-switch recombination. J Immunol, 191, 5751–5763. [DOI] [PubMed] [Google Scholar]
- 68.Stary A, Kannouche P, Lehmann AR and Sarasin A (2003) Role of DNA polymerase eta in the UV mutation spectrum in human cells. J Biol Chem, 278, 18767–18775. [DOI] [PubMed] [Google Scholar]
- 69.Lemontt JF (1971) Mutants of yeast defective in mutation induced by ultraviolet light. Genetics, 68, 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lawrence CW, Das G and Christensen RB (1985) REV7, a new gene concerned with UV mutagenesis in yeast. Mol Gen Genet, 200, 80–85. [DOI] [PubMed] [Google Scholar]
- 71.Nick McElhinny SA, Stith CM, Burgers PM and Kunkel TA (2007) Inefficient proofreading and biased error rates during inaccurate DNA synthesis by a mutant derivative of Saccharomyces cerevisiae DNA polymerase delta. J Biol Chem, 282, 2324–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shcherbakova PV, Pavlov YI, Chilkova O, Rogozin IB, Johansson E and Kunkel TA (2003) Unique error signature of the four-subunit yeast DNA polymerase epsilon. J Biol Chem, 278, 43770–43780. [DOI] [PubMed] [Google Scholar]
- 73.Hall DW, Mahmoudizad R, Hurd AW and Joseph SB (2008) Spontaneous mutations in diploid Saccharomyces cerevisiae: another thousand cell generations. Genet Res, 90, 229–241. [DOI] [PubMed] [Google Scholar]
- 74.Joseph SB and Hall DW (2004) Spontaneous mutations in diploid Saccharomyces cerevisiae: more beneficial than expected. Genetics, 168, 1817–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hubscher U, Maga G and Spadari S (2002) Eukaryotic DNA polymerases. Annu Rev Biochem, 71, 133–163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.