Abstract
The accumulation of lipids within drusen, the epidemiologic link of a high fat diet, and the identification of polymorphisms in genes involved in lipid metabolism that are associated with disease risk, have prompted interest in the role of lipid abnormalities in AMD. Despite intensive investigation, our understanding of how lipid abnormalities contribute to AMD development remains unclear. Lipid metabolism is tightly regulated, and its dysregulation can trigger excess lipid accumulation within the RPE and Bruch’s membrane. The high oxidative stress environment of the macula can promote lipid oxidation, impairing their original function as well as producing oxidation-specific epitopes (OSE), which unless neutralized, can induce unwanted inflammation that additionally contributes to AMD progression. Considering the multiple layers of lipid metabolism and inflammation, and the ability to simultaneously target multiple pathways, microRNA (miRNAs) have emerged as important regulators of many age-related diseases including atherosclerosis and Alzheimer’s disease. These diseases have similar etiologic characteristics such as lipid-rich deposits, oxidative stress, and inflammation with AMD, which suggests that miRNAs might influence lipid metabolism in AMD. In this review, we discuss the contribution of lipids to AMD pathobiology and introduce how miRNAs might affect lipid metabolism during lesion development. Establishing how miRNAs contribute to lipid accumulation in AMD will help to define the role of lipids in AMD, and open new treatment avenues for this enigmatic disease.
Keywords: Age-related Macular Degeneration, Retinal pigmented epithelium, Lipids, Oxidative stress, inflammation, microRNA
1. Introduction
Age-related Macular Degeneration (AMD) is the leading cause of blindness among the elderly in western societies (Resnikoff et al., 2004). Patients with early and intermediate non-neovascular or dry AMD with 20/20 visual acuity can have reduced light sensitivity or contrast sensitivity as their primary visual complaint while in advanced disease, visual acuity loss is the main complaint (Chen et al., 1992; Eisner et al., 1992; Midena et al., 1997; Owsley et al., 2016). For dry AMD, antioxidant micronutrients can slow the progression to advanced disease in patients with intermediate disease (Age-Related Eye Disease Study 2 Research, 2013; Age-Related Eye Disease Study Research, 2001). However, no treatment for any stage of dry AMD can restore vision in large part, because its pathophysiologic mechanisms remain unclear, which prevents the development of therapy that targets critical pathways at the appropriate disease stage (Xu et al., 2018). Epidemiologically, a high fat diet is an established risk factor for developing AMD (Clemons et al., 2005; Heiba et al., 1994; Vingerling et al., 1995). A number of genetic variants that have function related to lipid biology are associated with risk for advanced AMD (Edwards et al., 2005; Fritsche et al., 2016; Grassmann et al., 2017; Hageman et al., 2005; Haines et al., 2005; Klein et al., 2005). Lipid-rich deposits and their oxidation products including lipoproteins within drusen, have been identified in AMD tissue, and preclinical studies have shown that lipid dysregulation can contribute to AMD lesion development in animal models that simulate phenotypic features of AMD (Crabb et al., 2002; Dithmar et al., 2000; Fujihara et al., 2009; Malek et al., 2005; Spaide et al., 1999; Toomey et al., 2015). Interestingly, some of these lipid related pathologic characteristics are shared with other age-related diseases such as atherosclerosis (AS) and Alzheimer’s disease (AD) (Xu et al., 2018).
Recent studies indicate that epigenetic mechanisms, such as microRNA (miRNA) regulation of gene expression, are relevant to AMD pathobiology (Berber et al., 2017). miRNAs are single-stranded non-coding RNAs (sncRNA) of 18–24 nucleotides that play a pivotal role in regulating target gene expression (Hutvagner and Zamore, 2002). Recently, Pogue and Lukiw identified 7 miRNAs that are found with increased abundance in both Alzheimer’s Disease brains and AMD maculas (Pogue and Lukiw, 2018). These miRNAs can potentially downregulate 9460 target mRNAs, or about 3.5% of the genome, that are involved in oxidative stress and inflammation. Importantly, four of these miRNAs, including miR-27a, miR-34a, miR-146a and miR-155, play a role in lipid metabolism in Alzheimer’s Disease and atherosclerosis. These works highlight the potentially powerful and comprehensive impact that selected miRNAs can have on disease progression. Recent studies have reported miRNAs in multiple samples of AMD patients and in animal models of AMD, suggesting a role for miRNAs in AMD pathobiology (Askou et al., 2018; Ertekin et al., 2014; Grassmann et al., 2014; Menard et al., 2016; Ren et al., 2017; Szemraj et al., 2015). However, how miRNAs may play a role in lipid regulation and inflammation in the eye, especially in the RPE, and whether they are involved in AMD pathobiology, is largely unknown. In this review, we will discuss the potential contribution of oxidized lipids and inflammation to AMD pathology and summarize the miRNAs that are known to regulate lipid metabolism in other disease models that could contribute to AMD progression.
2. Lipid Accumulation in AMD
Drusen, or focal accumulations of heterogeneous material including lipids, lipoproteins, and oxidized lipids within Bruch’s membrane (BrM), are a hallmark AMD lesion. The accumulation of these lipids are considered one important, early event in AMD pathobiology (Curcio et al., 2001; Curcio et al., 2005a). In addition, a number of genetic variants in genes regulating lipid metabolism and the transfer of lipids among lipoproteins are associated with risk for advanced AMD (Edwards et al., 2005; Fritsche et al., 2016; Grassmann et al., 2017; Hageman et al., 2005; Haines et al., 2005; Klein et al., 2005). These include variations in hepatic lipase (LIPC), lipoprotein lipase (LPL), cholesterol ester transferase (CETP), ABC binding cassettes A1 (ABCA1) and apolipoprotein E (APOE) (Chen et al., 2010; Conley et al., 2005; Neale et al., 2010), suggesting that dysregulated lipid metabolism may influence the course of AMD pathobiology.
Lipid metabolism is governed by apolipoproteins (apo), which are structural proteins that are integral to the metabolism of triglycerides (TG) and cholesterol (Schaefer et al., 1978). Systemic lipoproteins, which are classified into 5 different types by density, transport exogenous (dietary) and endogenous (de novo) lipids between tissues (Miller, 1979). Unlike those present in the systemic circulation, Bruch’s membrane lipoproteins (BrM-LP) represent a distinct class of lipoproteins that contain apo B100, E and AI and are similar in size to very low density lipoproteins (VLDL), but are rich in esterified cholesterol like low density lipoproteins (LDLs) (Li et al., 2006; Wang et al., 2010). Retinal pigment epithelial (RPE) cells express components that are necessary for lipoprotein ingestion, cholesterol synthesis, lipoprotein synthesis, and secretion, as well as reverse cholesterol transport (RCT) (Curcio et al., 2011; Li et al., 2006; Li et al., 2005; Malek et al., 2003; Mullins et al., 2000; Tserentsoodol et al., 2006; Zheng et al., 2012). In addition, the RPE produces the receptors for low density lipoproteins, such as LDL-R, and high density lipoproteins (HDL), including scavenger receptor BI, SR-BI and SR-BII, which are localized on the basolateral side (Duncan et al., 2009; Tserentsoodol et al., 2006), enabling lipoprotein uptake. Thus, the RPE appears to be centrally involved in lipid metabolism within the ocular system.
The ambiguous association of serum cholesterol with AMD (Curcio et al., 2011) has led to the theory that an ocular production of lipoproteins is the main source of lipoproteins found in drusen (Dashti et al., 2006). While it is possible that BrM-LP are modified plasma lipoproteins, much evidence indicates that lipoproteins are secreted by the RPE in part, as a response to lipid overload (Curcio et al., 2011; Li et al., 2006; Li et al., 2005; Malek et al., 2003; Mullins et al., 2000; Tserentsoodol et al., 2006; Wu et al., 2010). The phagocytosis and degradation of photoreceptor outer segments (POS) are a major lipid source that the RPE processes (Ershov and Bazan, 2000). The ingestion of systemically delivered lipoproteins can add to the RPE’s lipid burden, particularly in individuals who consume a high fat diet. As with cardiac myocytes, the RPE can eliminate lipids by secreting apoB100 containing lipoproteins to avoid lipo-apoptosis (Fujihara et al., 2014; Nielsen et al., 2002; Nielsen et al., 1998; Yokoyama et al., 2004). Thus, BrM-LPs appear to be secreted from the RPE for transit through Bruch’s membrane to the choroid for elimination (Pikuleva and Curcio, 2014).
The RPE also has an active reverse cholesterol transport system. Excess cholesterol is removed by HDLs after they are loaded with cellular cholesterol by ABC transporters, ultimately returning excess cholesterol to the liver. Ishida et al. demonstrated that HDLs, lipid free apoA-I, and apoA-I containing vesicles, stimulate efflux of radiolabeled lipids of POS origin from the basal surface of RPE cells (Ishida et al., 2006). Recent work by Biswas et al. showed that the mitochondrial cholesterol trafficking protein, translocator protein (TSPO), is highly expressed by human and mouse RPE cells and significantly increases cholesterol efflux from RPE cells to ApoE, ApoAI and HDLs (Biswas et al., 2017). With aging, the RPE has decreased TSPO expression and impaired cholesterol efflux, which could increase the burden of eliminating lipids by the BrM-LPs that are secreted by the RPE.
Subretinal drusenoid deposits (SDD), or reticular pseudodrusen, are associated with AMD progression. Like drusen, SDD contain a mixture of cholesterol and apoE (Ebrahimi et al., 2013; Oak et al., 2014; Rudolf et al., 2008; Sarks et al., 2011). This composition suggests that reverse cholesterol transport is important at the apical RPE for transporting cholesterol from the RPE to photoreceptors. SDDs also contain inflammatory proteins including the complement C5b-9 regulator CD59 that is secreted by the RPE, as well as complement factor H (CFH) and vitronectin (Ebrahimi et al., 2013; Oak et al., 2014; Rudolf et al., 2008). These findings suggest that inflammation, including complement activation, contributes to SDD formation. With SDD accumulation in the subretinal space, the normal transport of nutrients including oxygen from the choriocapillaris to photoreceptors is compromised, potentially contributing to outer retinal atrophy.
Interestingly, SDD accumulation is localized to the rod-rich perifoveal macula while drusen are located in the cone-rich fovea (Curcio et al., 2013). Pikuleva and Curcio proposed a two lesion, two compartment model based on the differential cholesterol content between rod and cone outer segments to explain the distribution of drusen and SDD, respectively (Pikuleva and Curcio, 2014). In the cone-rich fovea where drusen develop, the RPE processes cholesterol derived from cholesterol-rich cone OS that have been phagocytosed, the delivery of lipoproteins from the plasma, and endogenously synthesized cholesterol. The excess cholesterol in the RPE is secreted in VLDL-like lipoproteins basolaterally into Bruch’s membrane where drusen form. On the other hand, rod outer segments have a decreasing cholesterol gradient such that distal outer segments are cholesterol poor relative to cones. Since HDLs cycle cholesterol between the RPE and photoreceptors, it has been proposed that HDLs accumulate the released cholesterol by rod outer segments in the subretinal space due to impaired lipid transport by dysfunctional RPE.
3. Effects of lipid oxidation on AMD Pathobiology
The accumulation of lipoproteins in Bruch’s membrane is not sufficient for drusen formation. Our lab showed that genetically modified mice predominantly producing apoB100 instead of apoB48 in mice to simulate humans, accumulate apoB100 lipoproteins in Bruch’s membrane by 2 months, but with aging, do not develop basal deposits or drusen (Fujihara et al., 2014). The accumulation of inflammatory debris within drusen indicates that inflammation is an additional requirement for drusen formation. The well-studied “response-to-retention” hypothesis of atherosclerosis suggests that following their retention in the inner arterial wall, lipoproteins become oxidized to activate innate immunity including complement, which unleashes pro-inflammatory events that contribute to atherosclerotic lesion formation (Tabas et al., 2007). Thus, oxidized lipids are a primary choice as one trigger for AMD pathobiology due to the high oxidative stress environment of the macula, the reduced antioxidant capability of the RPE with aging, and their ability to induce a pro-inflammatory response (Handa et al., 2017).
With aging, Bruch’s membrane undergoes structural changes that can impair the transit of lipoproteins. For example, our lab found that advanced glycation end product (AGE) formation in Bruch’s membrane induces the expression of lipoprotein lipase, which by a non-enzymatic process, retains lipoproteins in Bruch’s membrane (Cano et al., 2011). Indeed, BrM-LPs first accumulate as linear tracks in the inner collagenous layer adjacent to the elastic layer, and then toward the RPE, eventually coalescing to form a lipid wall, which contributes to drusen formation (Curcio et al., 2001; Curcio et al., 2005b). The macula is a high oxidative stress microenvironment due to the high blood flow needed to meet the high metabolism of vision, the generation of reactive oxygen species (ROS) from the daily phagocytosis of photoreceptor outer segments, photo-oxidative stress from sunlight exposure, and for those who choose to smoke, the additional ingestion of chemical oxidants (Ham et al., 1978; Rangasamy et al., 2004; Smith and Hansch, 2000; Winkler et al., 1999). Thus, lipoproteins retained within Bruch’s membrane are susceptible to oxidation that can initiate a cascade of pro-inflammatory events with similarity to atherosclerosis (Handa, 2007; Handa et al., 1999; Hewitt et al., 1989; Kliffen et al., 1996; Lakkaraju et al., 2007; Newsome et al., 1987; Tabas, 1999; Zarbin, 2004). Furthermore, Thompson et al. showed that small, hollow hydroxyapatite (HAP) spherules with cholesterol-containing cores are present in sub-RPE deposits, provide a scaffold on which proteins, including CFH, vitronectin, and amyloid beta (Aβ), adhere and accumulate (Thompson et al., 2015). Pilgrim et al. confirmed the production of HAP and recapitulated sub-RPE deposit formation in primary porcine and human RPE cell cultures (Pilgrim et al., 2017) to further suggest that sub-RPE deposit formation is initiated, and regulated by the RPE.
Many forms of oxidized lipids have been detected in AMD including carboxyethylpyrrole, oxidized phospholipids, 4-hydroxynonenal, malondialdehyde, isolevuglandins, and 7-ketocholesterol (Charvet et al., 2013; Crabb et al., 2002; Moreira et al., 2009; Shen et al., 2007; Spaide et al., 1999; Suzuki et al., 2007; Weismann et al., 2011). The oxidized lipids or lipoproteins can directly impair cellular function. Due to their high reactivity, the degradation products of oxidized lipids can modify self-molecules and thereby generate structural neo-epitopes that are recognized by immune system receptors (Palinski et al., 1989; Palinski et al., 1990). These neo-epitopes have been termed “oxidation-specific epitopes” (OSEs), and represent a common set of epitopes present on various oxidatively modified proteins and lipids (Palinski et al., 1994). OSEs serve as markers of oxidatively modified endogenous structures, and allow the immune system through pattern recognition receptors to mediate their clearance in order to maintain homeostasis (Chou et al., 2008). However, either an increase in OSE generation or impaired innate immune neutralization can result in the accumulation of OSEs, which can trigger a pro-inflammatory immune response that includes complement activation, pro-inflammatory cytokine expression, and macrophage/microglia recruitment which if severe enough, can induce tissue dysfunction such as RPE degeneration during AMD development. The possible lipid-related pathogenic mechanisms in AMD are summarized in Figure 1.
Figure 1.
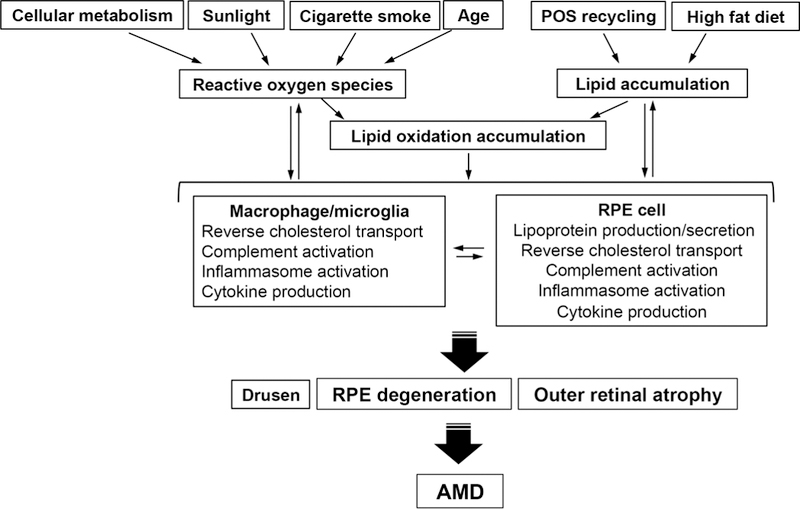
Lipid-related pathogenic mechanisms in AMD. Lipids accumulate in the sub-RPE and sub-retinal space. Various oxidative sources including aging, cigarette smoking, cellular metabolism, and sunlight cause the oxidation of lipid molecules, which induces inflammation in part, through complement and inflammasome activation, and cytokine production in the RPE and/or microglia/macrophages. In addition, a high fat diet and the phagocytosis of photoreceptor outer segments (POS) by the RPE are major sources of lipid accumulation. Monocytes/macrophages/microglia are recruited and further differentiated by various cytokines, to induce inflammation and modify lipid transport metabolism. These processes worsen lipid oxidation and promote the accumulation of inflammatory molecules during drusenogenesis. The combination of direct injury from oxidized lipids and dysregulated, chronic inflammation can contribute to RPE degeneration that can mediate to photoreceptor death (outer retinal atrophy) resulting vision loss and AMD.
4. miRNA Biogenesis and a Possible Role in AMD
miRNAs have been investigated for their role in eye development and retina/RPE cell function (Sundermeier and Palczewski, 2016), or as biomarkers and therapeutic targets for AMD (Askou et al., 2018; Ertekin et al., 2014; Grassmann et al., 2014; Menard et al., 2016; Ren et al., 2017; Szemraj et al., 2015). miRNAs are evolutionarily conserved, small (~22 nucleotides), single-stranded non-coding RNAs that were originally discovered in C. elegans in 1984 as genes responsible for defects in neurological development (Ambros and Horvitz, 1984; Ferguson et al., 1987). Almost a decade later, miRNAs are now known to play an important role in regulating gene expression by blocking mRNA translation and promoting mRNA degradation. miRNAs are transcribed by RNA polymerase II into a precursor form called primary-miRNA (Pri-miRNA), which is bound by DiGeorge Syndrome Critical Region 8 (DGCR8) and DROSHA. After DROSHA cleaves the pri-miRNA into pre-miRNAs, pre-miRNAs are exported to the cytoplasm where they are cleaved into mature miRs by DICER. After being incorporated into the RNA-induced silencing complex (RISC) and binding to the 3’ untranslated region of the target mRNA, the mature miRNA interacts with its target mRNA to ultimately fine-tune protein synthesis, and regulate normal tissue development and homeostasis (Lee et al., 1993; Wightman et al., 1993). Given that an miRNA can bind to multiple target 3’UTRs and multiple miRNAs can target the same gene, it is estimated that more than 60% of protein-coding genes are directly regulated by miRNAs (Friedman et al., 2009). Abnormal expression or activity of miRNAs by i) changes in miRNA biogenesis machinery, ii) up/down regulation of miRNA expression either by transcriptional or epigenetic modification, or iii) mutations in either an miRNA or the target gene, could cause an imbalance in cellular homeostasis that leads to disease.
For example, the maturation of specific miRNAs is necessary for proper lipid metabolism in the liver (de novo) and intestine (dietary). Deletion of DICER in the liver reduces liver-specific miR-122, miR-148a, miR-192 and miR-194, resulting in the accumulation of lipid droplets that contain cholesterol esters, triglycerides, free cholesterol, and free fatty acids (Sekine et al., 2009). The lipid droplet accumulation is due in part, to impaired lipoprotein packaging and secretion from reduced miR-122 and miR-148a. Using mice with an inducible deletion of DICER in the liver, Sahasrabuddhe et al. found that miRNAs including miR-124, are decreased, and that the majority of the upregulated proteins involved in lipid metabolism are known PPARα targets and involved in peroxisomal β-oxidation of fatty acids (Sahasrabuddhe et al., 2014). Huang et al. found that systemic reduction of DICER in adult mice have reduced levels of miR-215, miR-192, and miR-194 with a marked decrease in microsomal triglyceride transfer protein (MTP), long-chain fatty acyl-CoA ligase 5, fatty acid finding protein, and very-long-chain fatty acyl-CoA dehydrogenase, among others that coincided with Oil Red O stained lipid accumulations in the small intestines (Huang et al., 2012).
Sundermeier et al. highlight how miRNA biogenesis can influence lipid metabolism in the RPE (Sundermeier et al., 2017). Conditional knock out of DGCR8, a cofactor for the miRNA-processing nuclease DROSHA, caused the accumulation of phagosomes and lipid droplets in the RPE, coinciding with shortened photoreceptor outer segments. These lipid accumulations in the RPE are similar to those seen in the liver or intestines of DICER deletion mice. These data suggest that DGCR8 deficiency in the RPE causes either a defect in digesting phagocytosed material or lipid metabolism. Sundermeier et al. also showed that both miRNA-processing enzymes DGCR8 and DICER are essential for RPE function and survival (Sundermeier et al., 2017). In RPE-specific conditional knockout mice, either DGCR8 or DICER1 inactivation, which disrupts two independent steps of miRNA biogenesis, led to strikingly similar defects in RPE survival and function. The deletion of DGCR8 in RPE cells dramatically reduced RPE cell-enriched miRNAs including miR-204 and miR-211. These two miRNAs play a crucial role in the differentiation and function not only of the RPE, but also of the retina, lens, and ciliary body (Adijanto et al., 2012; Conte et al., 2010). These results suggest that the loss of certain mature miRNAs may impair lipid metabolism, contributing to RPE cell death, subsequent loss of visual function, and thus, play a role in AMD pathobiology. However, the role of the miRNA biogenesis machinery is incomplete because Kaneko et al. demonstrated that a decrease in DICER1 induced the accumulation of toxic alu RNAs, which activated the NLRP3 inflammasome to induce RPE cell death and geographic atrophy, a mechanism that was independent of impaired miRNA maturation (Kaneko et al., 2011).
5. Overview of miRNAs in Lipid Metabolism
miRNAs regulate all phases of lipoprotein metabolism (Elmen et al., 2008; Esau et al., 2006; Marquart et al., 2010; Rayner et al., 2010). miRNAs repress the expression of genes involved in lipoprotein packaging (ApoB) and secretion (MTP), uptake (LDLR and SR-B1), and cholesterol efflux (ABCA1), and can influence hyperlipidemia and the development of atherosclerosis, as reviewed in (Aryal et al., 2017; Feinberg and Moore, 2016; Zhou et al., 2016). miRNAs also influence lipid metabolism in Alzheimer’s disease, as reviewed in (Goedeke et al., 2014; Yoon et al., 2016). In Alzheimer’s disease, miRNAs regulate cholesterol efflux (ABCA1) and ApoE lipidation that are involved in Aβ accumulation. Given that lipid abnormalities are also implicated in AMD pathobiology, miRNAs that regulate lipid metabolism could play an important role in AMD. The possible lipid-regulating miRNAs in AMD are summarized in Figure 2.
Figure 2.

Scheme of lipid metabolism and miRNAs with the potential to regulate lipid pathways within the RPE during AMD progression. The RPE cell in the cholesterol poor rod (green) dominated perifoveal macula on the left is ingesting systemically delivered lipoproteins via receptors like LDLR and SR-BI for LDL and HDL uptake, or oxidized LDLs through CD36 on its basolateral side. In addition, the apical RPE is removing cholesterol for photoreceptors by reverse cholesterol transport. The miRNAs that can regulate these processes are listed. Dysregulation of this process can result in subretinal drusenoid deposit (SDD) formation.
5.1. miRNAs Involved in Cholesterol and TG synthesis
The sterol regulatory element–binding proteins (SREBPs) are central transcription factors that target genes involved in cellular cholesterol synthesis, efflux, uptake and fatty acid oxidation (Magana and Osborne, 1996; Shimomura et al., 1997a; Shimomura et al., 1997b; Tontonoz et al., 1993). The SREBPs work in concert with multiple miRNAs. For example, miR-33a and miR-33b (miR-33a/b), by their location within the introns of the SREBF2 and SREBF1 genes, respectively, are co-transcribed with their host genes. While the SREBPs promote lipid synthesis and uptake, miR-33a and miR-33b help prevent the loss of cellular lipids by targeting genes involved in cholesterol trafficking and efflux, such as Niemann Pick C (NPC1), ATP-Binding Casette A1 (ABCA1), and ATP-Binding Casette G1 (ABCG1), and fatty acid β-oxidation, including carnitine palmitoyltransferase 1A (CPT1A), carnitine O-octanoyltransferase (CROT), and hydroxyacyl-CoA dehydrogenase/3-ketoacy-CoA thiolase/enoyl-CoA hydratase β subunit (HADHB) (Davalos et al., 2011; Gerin et al., 2010; Marquart et al., 2010; Najafi-Shoushtari et al., 2010; Rayner et al., 2010). Recently, miR-185 was shown to regulate cholesterol homeostasis in coordination with genes encoded by the SREBPs. In particular, miR-185 is transcriptionally activated by SREBP1c and negatively regulates SREBP2 expression, thereby inhibiting de novo cholesterol biosynthesis and LDL uptake (Yang et al., 2014). In addition to miR-33 and miR-185, the locus comprising miR-96, miR-182, and miR-183 (miR-96/182/183) act in a feedback loop to regulate cholesterol metabolism (Goedeke et al., 2015).
Vickers et al. also identified miR-27b and miR-223 as key post-transcriptional regulatory hubs that control cholesterol and lipoprotein metabolism gene networks (Vickers et al., 2014; Vickers et al., 2013). Using an unbiased in silico target site search, miR-27b was predicted to target 151 lipid metabolism genes. Indeed, microarray/real-time qPCR based gene expression in human hepatocytes confirmed robust miR-27b-mediated regulation of lipid metabolism genes including peroxisome proliferator-activated receptor gamma (PPARG), Angiopoietin-like 3 (ANGPTL3), N-deacetylase/N-sulfotransferase 1 (NDST1), and glycerol-3-phosphate acyltransferase 1, mitochondrial (GPAM). ANGPTL3 is expressed by the liver and secreted into the circulation (Clark et al., 2003), and suppresses the activity of lipoprotein lipase (Shimizugawa et al., 2002) and endothelial lipase (Shimamura et al., 2007), which regulate TG and HDL-cholesterol, respectively. GPAM catalyzes the first committed step in de novo TG synthesis and is implicated in regulating cholesterol synthesis (Gonzalez-Baro et al., 2007). miR-27a/b can also regulate macrophage cholesterol homeostasis by targeting cholesterol esterification such as acetyl-CoA acetyltransferase 1, and efflux, including ABCA1(Zhang et al., 2014), and is upregulated in the liver of high-fat/high-cholesterol fed APOE deficient mice, inducing severe hypercholesterolemia and advanced atherosclerosis (Vickers et al., 2013). In addition to regulating HDL cholesterol uptake by directly repressing the HDL receptor SR-BI, miR-223 inhibits cholesterol biosynthesis by repressing HMG-CoA synthase 1 and methylsterol monooxygenase 1(Vickers et al., 2014). Collectively, these studies elucidate the importance of these miRNAs in regulating the circuitry of cholesterol metabolism, particularly with SREBPs-mediated transcriptional regulation.
miR-27b and miR-223 are elevated in the plasma of patients with AMD relative to control patients (Ertekin et al., 2014). Furthermore, miR-223 is increased in the RPE/choroid of mice that develop RPE degeneration after intravitreal injection of Aβ1–40 (Huang et al., 2017; Yoshida et al., 2005). These studies suggest that miR-27b and miR-223 influence cholesterol and TG synthesis in the RPE.
5.2. miRNAs Regulate Lipoprotein Packaging and Secretion.
The liver is centrally involved in both the production and clearance of lipoproteins. Not surprisingly, many hepatic-enriched miRNAs have been found to regulate lipoprotein metabolism (Feinberg and Moore, 2016). For example, miR-30c, miR-34a, and miR-122 modulate hepatic production of apoB-containing lipoproteins (Zhou et al., 2016). miR-30c, an evolutionarily conserved miRNA, regulates the assembly and secretion of apoB100-containing lipoproteins by modulating MTP activity, which regulates apoB100 lipidation (Soh et al., 2013). Overexpression of miR-30c in mice potently reduced hepatic MTP expression and activity, plasma cholesterol levels, and the production of triglyceride-rich VLDL when compared to controls. Hepatic overexpression of miR-30c also reduced de novo fatty acid synthesis by targeting lysophosphatidylglycerol acyltransferase 1. In APOE knockout mice, hepatic overexpression of miR-30c mitigated hyperlipidemia and atherosclerosis while maintaining liver health without inducing steatosis, unlike conventional pharmacological MTP inhibitors. These studies demonstrate that miR-30c safely reduces hepatic lipoprotein production and lipid synthesis to lower plasma lipids. As the RPE produces both MTP and ApoB, miR-30c is an attractive target for reducing lipoprotein production and their release into BrM to retard or halt lipid wall formation.
Mice made obese after being given a high-fat diet have a 10-fold increase in miR-34a over controls (Xu et al., 2015). Xu et al. also showed that miR-34a inhibits VLDL production by targeting transcription factor HNF4α to regulate apoB and MTP expression. In fact, miR-34a and its target HNF4α also have an inverse relationship in non-alcoholic fatty liver disease (Choi et al., 2013; Xu et al., 2015). Increased hepatic expression of miR-34a attenuated atherosclerotic lesion size by >50% in apoE or LDLR deficient mice by reducing VLDL secretion to lower plasma VLDL and LDL cholesterol. However, the retention of lipids by impaired VLDL secretion caused liver steatosis. In contrast, miR-34a deficient mice have increased plasma TG and cholesterol, and decreased hepatic TGs. These data demonstrate that miR-34a regulates lipid metabolism by controlling hepatic lipoprotein synthesis and secretion. miR-34a can also inhibit a number of cholesterol biogenesis genes including sterol regulatory element-binding protein 1c, acetyl-CoA carboxylase 1 and 2, and HMG-CoA reductase. These results suggest that the miR-34a-HNFα pathway is activated under metabolic stress and regulates lipoprotein metabolism.
Importantly, miR-34a is significantly upregulated in both the whole retina and the macular region of AMD patients compared to controls (Bhattacharjee et al., 2016). Furthermore, miR-34a is expressed by ARPE19 cells (Hou et al., 2013) and miR-34a is significantly increased with aging in the RPE and retina of mice (Smit-McBride et al., 2014). Given its regulatory role in lipoprotein metabolism, it is possible that miR-34a overexpression in the RPE limits apoB100 lipoprotein secretion, resulting in lipid accumulation in the RPE, which as in liver, could induce cellular dysfunction or lipo-apoptosis. Further elucidation of the effects of miR-34a on RPE lipid metabolism will enable us to understand its role when the RPE is under high lipid metabolic stress, and determine the extent that it is a treatment target.
Unlike miR30c and miR-34a, miR-122 deficiency reduces hepatic MTP expression, VLDL production, and plasma lipids, an effect that is ameliorated by overexpressing MTP in the liver (Tsai et al., 2012). How miR-122 regulates MTP expression is unknown as MTP is not a predicted target (Xu et al., 2015). Since inhibiting miR-122 is expected to upregulate its direct targets, the decreased gene expression raises the possibility that miR-122 targets a transcriptional inhibitor. In fact, treatment with an antisense oligonucleotide against miR-122 decreased the expression of many key genes that regulate lipid metabolism. miR-122 inhibition in normal mice reduced plasma cholesterol, increased hepatic fatty-acid oxidation, and decreased hepatic fatty-acid and cholesterol synthesis (Esau et al., 2006). Similar effects were shown in non-human primates treated with LNA-mediated miR-122 silencing (Elmen et al., 2008). These results identified miR-122 as a crucial regulator of cholesterol and fatty acid synthesis, and thus, lipoprotein homeostasis.
Recent studies have found that miR-122 plays a role in retinal development in dogs (Genini et al., 2014) and is detected in the aqueous humor of human patients, which raises the possibility that this miRNA regulates lipid metabolism genes in the eye (Drewry et al., 2018). Importantly, the miR-122 gene variant rs41292412: C>T (Chr18:56118358) in the stem loop regions of pre-miR-122 is associated with AMD risk with reduced expression of the mature miR-122 when tested in vitro (Ghanbari et al., 2017). These data suggest that decreased miR-122 by its genetic variant, reduces MTP expression during AMD. Further clarification of the miR-122 pathway that mediates MTP expression as well as its impact on RPE cell lipid metabolism is warranted.
5.3. miRNAs are Involved in Lipoprotein Uptake
miRNAs target the LDL receptor (LDLR) to regulate plasma LDL levels by promoting the clearance of circulating LDLs. Two GWAS studies have identified polymorphisms in miR-148a with atherosclerosis risk, and further investigation found that miR-148a is a negative regulator of LDLR since inhibiting miR-148a increased LDL cholesterol by inducing hepatic LDLR expression in mice (Goedeke et al., 2015; Wagschal et al., 2015). Both studies showed that inhibiting miR-148a increased hepatic LDLR expression and decreased plasma LDL cholesterol. In addition, polymorphisms in the miR-148a promoter are associated with altered LDL cholesterol in people, further suggesting its role in dyslipidemia (Wagschal et al., 2015). Wagschal et al. also identified miR-128–1, miR-130b, and miR-301b that were predicted to target LDLR. Introduction of these miRNAs as well as miR-148a precursor oligonucleotides into human hepatoma cells decreased LDLR. In high fat diet fed C57BL/6J and ApoE KO mice, overexpression and antisense targeting of miR-128–1 or miR-148a altered hepatic LDLR and ABCA1 expression, respectively, and modulated circulating lipoprotein-cholesterol and TG. miR-148a-3p is increased in the vitreous and subretinal fluid of eyes with rhegmatogenous retinal detachment where free floating RPE cells are present (Takayama et al., 2016; Tsunekawa et al., 2017). As in the liver, it is therefore possible that miR-148a could play a role LDL uptake in the RPE.
Macrophages have an important role in LDL and oxLDL uptake, a critical early step in foam cell formation during atherogenesis, a process that is regulated by miRNAs. For example, miR-27a/b can regulate macrophage lipoprotein uptake by targeting LDLR and CD36 gene expression (Zhang et al., 2014). Likewise, miR-125a and miR-146a decrease lipid uptake in oxLDL–stimulated macrophages (Chen et al., 2009; Yang et al., 2011). miR-155 also appears to regulate lipoprotein uptake by macrophages since it is increased in the plasma and macrophages of ApoE KO mice, and is induced in macrophages by oxLDL. Importantly, inhibiting miR-155 by systemically delivered antagomiR-155 decreased lipid-loading in macrophages and reduced atherosclerosis plaques in ApoE deficient mice (Tian et al., 2014).
Of the above miRNAs that can regulate lipoprotein uptake, miR-27a, miR-146a and miR-155 are upregulated in both Alzheimer’s Disease brains and importantly, in AMD retinas (Pogue and Lukiw, 2018). Among them, miR-146a and miR-155 are also of particular interest because they can downregulate the expression of complement factor H (CFH), and both have been found to be increased in the plasma, vitreous, and retina of AMD patients (Lukiw et al., 2012). In addition, SanGiovanni et al. identified polymorphisms in the 3’UTR of CFH, which could interfere with the miR-146a/miR-155 binding site that is associated with AMD (SanGiovanni et al., 2017). These data suggest that the individual miRNA profile and its target gene polymorphism can interfere with miRNA-mRNA binding. Aside from its regulatory role in the alternative complement pathway, the ability of CFH to block the uptake of malondialdehyde-modified proteins and minimize inflammation raises the possibility that miRNAs modulating CFH could play an important role in regulating oxidized lipid uptake in the RPE or macrophages in AMD (Weismann et al., 2011). Interestingly, the 402H CFH variant, which is associated with increased AMD risk, has a reduced ability to bind malondialdehyde, suggesting that malondialdehyde induced inflammation is an alternative mechanism to impaired complement regulation during AMD lesion development. Thus, careful investigation is needed to fully attribute a phenotype to a specific miRNA.
5.4. Reverse Cholesterol Transport
Multiple miRNAs can regulate reverse cholesterol transport. miR-128 and miR-148a directly reduce the expression of ABCA1, which promotes cellular cholesterol efflux to APOA1 during HDL biogenesis in order to remove excess cholesterol from hepatocytes, macrophages, and RPE cells (Feinberg and Moore, 2016; Goedeke et al., 2015). Likewise, miR-223 indirectly promotes ABCA1 expression by regulating SP3 to enhance cellular cholesterol efflux (Vickers et al., 2014). In atherosclerosis, many miRNAs have been identified that promote foam cell formation by reducing ABCA1 to inhibit macrophage cholesterol efflux, including miR-26, miR-33, miR-106, miR-128–1, miR-130b, miR-144, miR-148a, miR-301b, miR-302a, and miR-758 (de Aguiar Vallim et al., 2013; Feinberg and Moore, 2016; Marquart et al., 2010). This high degree of miRNA targeting of ABCA1 points toward a need for careful fine-tuning of macrophage cholesterol efflux to maintain cholesterol homeostasis. Interestingly, miR-33, by targeting ABCA1, inhibits apoE lipidation to reduce cholesterol efflux and induces Aβ in the brain, which suggests that this miRNA is involved in Alzheimer’s disease progression (Kim et al., 2015). Their role in the RPE or ocular macrophages is unknown.
5.5. miRNAs Regulate HDL Uptake
Circulating HDL cholesterol is regulated by hepatic clearance via the scavenger receptor SR-BI, which is targeted by miR-96, miR-125a, miR-185, miR-223, and miR-455–5p (Hu et al., 2012; Wang et al., 2013). In addition, HDLs can carry miRNAs such as miR-92a, miR-126, and miR-223, which can be internalized by macrophages and endothelial cells through SR-B1 to repress their target gene expression (Feinberg and Moore, 2016). Genetic ablation of miR-223, which is also known to control monocyte differentiation through NF-κB signaling, can increase HDL cholesterol, and total hepatic and plasma cholesterol in mice (Johnnidis et al., 2008; Li et al., 2010). Importantly, miR-223 is upregulated in the RPE/choroid after intravitreal injection of Aβ1–40, which induces RPE degeneration (Huang et al., 2017; Yoshida et al., 2005). In this model, it is possible that miR-223 impaired HDL uptake into the RPE. The dual role on both cholesterol uptake and monocyte differentiation raises the intriguing possibility of its involvement in AMD pathobiology.
6. Conclusions
Excess lipid accumulation within the RPE can trigger either the reverse cholesterol efflux pathway or the secretion of apoB containing lipoproteins that can eventually form the “lipid wall” in Bruch’s membrane. With aging related changes to Bruch’s membrane, chronic oxidative stress can oxidize these lipid accumulations, inducing inflammation, which cumulatively, can have a pivotal impact on AMD pathobiology. Given the evidence for lipoprotein accumulation in Bruch’s membrane, their oxidation, and subsequent inflammation during drusenogenesis, investigations that attempt to lower lipoprotein uptake into the RPE or macrophages, regulate lipoprotein secretion from these cells, reduce their retention in Bruch’s membrane, and/or reduce their oxidation may yield effective treatment for AMD. miRNAs have a significant role in mediating lipid metabolism in atherosclerosis and to some extent, Alzheimer’s Disease. Gaining an understanding of the tissue specific lipid metabolism miRNA profile in the eye, and how these miRNAs control/impact lipid biogenesis pathways in the RPE will deepen our understanding of how lipid metabolism contributes to AMD pathobiology. Furthermore, circulating miRNAs that are carried by nanosized exosomes or microvesicles in the blood or vitreous have been identified in age-related diseases, and can potentially be valuable diagnostic or prognostic biomarkers for age-related diseases including AMD (Grassmann et al., 2014).
To date, pharmacologic intervention of these pathways, which have enjoyed success by reducing cardiovascular disease by 20–40%, such as reducing lipoproteins with the HMG-CoA reductase inhibitor Statins, or anti-sense to apoB (Mipomersen), which was approved by the FDA in 2013 for use in patients with familial hypercholesterolemia (Thomas et al., 2013; Thomas and Ginsberg, 2010). A clinical trial targeting lipid metabolism has not been conducted for AMD. One exception is the pilot study by Vavvas et al. Over a 12 month period, high-dose atorvastatin resulted in regression of drusen deposits in 10 of 23 patients (Vavvas et al., 2016). Given the potential to correct lipid metabolism abnormalities, future studies should consider testing high dose statins and these other targets as treatment for AMD. Lipid metabolism and inflammation are complex interweaving and interactive networks. By simultaneously targeting multiple pathways, miRNAs offer a unique treatment advantage to comprehensively control dysregulated pathways such as the complexity of lipid metabolism. However, a full understanding of how lipids contribute to AMD pathobiology, both in terms of the magnitude and timing of their contribution, and how miRNAs influence lipid metabolism during the development of AMD would likely enable the identification of novel miRNAs or miRNA inhibitors that might someday restore lipid metabolic homeostasis in the macula.
The RPE in the cholesterol rich cone (blue) dominated fovea on the right is synthesizing cholesterol and fatty acids and packaging them into apoB lipoproteins for secretion into Bruch’s membrane for release into the choriocapillaris. Once in Bruch’s membrane, the lipids and lipoproteins bind to hydroxyapatite (HAP), and become oxidized, which induces inflammation and drusen formation. The miRNAs that can regulate these lipid pathways are listed. miRNAs that are expressed by RPE cells or related to AMD, are in bold.
Highlights.
Lipids accumulate in the macula in AMD
Oxidative stress oxidizes lipids
Oxidized lipids can damage tissue and induce innate immunity
MicroRNAs are important regulators of lipid metabolism
This review discusses how microRNAs might regulate lipid metabolism in AMD
Acknowledgments
Drs. Jun, Cano, and Handa receive grant support from Bayer Pharmaceutical, Inc. on an unrelated topic.
Funding: EY027691 (JTH), Macular Degeneration Foundation (JTH), Research to Prevent Blindness (Wilmer Eye Institute). Dr. Handa is the Robert Bond Welch Professor.
Abbreviations:
- AMD
Age-related Macular Degeneration
- RPE
Retinal pigmented epithelium
- LDL
Low-density lipoprotein
- ox-LDL
Oxidized LDL
- miR
microRNA
Footnotes
The other authors have no conflicts.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adijanto J, Castorino JJ, Wang ZX, Maminishkis A, Grunwald GB, Philp NJ, 2012. Microphthalmia-associated transcription factor (MITF) promotes differentiation of human retinal pigment epithelium (RPE) by regulating microRNAs-204/211 expression. J Biol Chem 287, 20491–20503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Age-Related Eye Disease Study 2 Research, G., 2013. Lutein + zeaxanthin and omega-3 fatty acids for age-related macular degeneration: the Age-Related Eye Disease Study 2 (AREDS2) randomized clinical trial. JAMA 309, 2005–2015. [DOI] [PubMed] [Google Scholar]
- Age-Related Eye Disease Study Research, G., 2001. A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report no. 8. Arch Ophthalmol 119, 1417–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Horvitz HR, 1984. Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226, 409–416. [DOI] [PubMed] [Google Scholar]
- Aryal B, Singh AK, Rotllan N, Price N, Fernandez-Hernando C, 2017. MicroRNAs and lipid metabolism. Curr Opin Lipidol 28, 273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askou AL, Alsing S, Holmgaard A, Bek T, Corydon TJ, 2018. Dissecting microRNA dysregulation in age-related macular degeneration: new targets for eye gene therapy. Acta Ophthalmol 96, 9–23. [DOI] [PubMed] [Google Scholar]
- Berber P, Grassmann F, Kiel C, Weber BH, 2017. An Eye on Age-Related Macular Degeneration: The Role of MicroRNAs in Disease Pathology. Mol Diagn Ther 21, 31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S, Zhao Y, Dua P, Rogaev EI, Lukiw WJ, 2016. microRNA-34a-Mediated Down-Regulation of the Microglial-Enriched Triggering Receptor and Phagocytosis-Sensor TREM2 in Age-Related Macular Degeneration. PLoS One 11, e0150211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas L, Zhou X, Dhillon B, Graham A, Shu X, 2017. Retinal pigment epithelium cholesterol efflux mediated by the 18 kDa translocator protein, TSPO, a potential target for treating age-related macular degeneration. Hum Mol Genet 26, 4327–4339. [DOI] [PubMed] [Google Scholar]
- Cano M, Fijalkowski N, Kondo N, Dike S, Handa J, 2011. Advanced glycation endproduct changes to Bruch’s membrane promotes lipoprotein retention by lipoprotein lipase. Am J Pathol 179, 850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvet CD, Saadane A, Wang M, Salomon RG, Brunengraber H, Turko IV, Pikuleva IA, 2013. Pretreatment with pyridoxamine mitigates isolevuglandin-associated retinal effects in mice exposed to bright light. J Biol Chem 288, 29267–29280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Fitzke FW, Pauleikhoff D, Bird AC, 1992. Functional loss in age-related Bruch’s membrane change with choroidal perfusion defect. Invest Ophthalmol Vis Sci 33, 334–340. [PubMed] [Google Scholar]
- Chen T, Huang Z, Wang L, Wang Y, Wu F, Meng S, Wang C, 2009. MicroRNA-125a-5p partly regulates the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages. Cardiovasc Res 83, 131–139. [DOI] [PubMed] [Google Scholar]
- Chen W, Stambolian D, Edwards AO, Branham KE, Othman M, Jakobsdottir J, Tosakulwong N, Pericak-Vance MA, Campochiaro PA, Klein ML, Tan PL, Conley YP, Kanda A, Kopplin L, Li Y, Augustaitis KJ, Karoukis AJ, Scott WK, Agarwal A, Kovach JL, Schwartz SG, Postel EA, Brooks M, Baratz KH, Brown WL, Group, C.o.A.-R.M.D.P.T.R., Brucker AJ, Orlin A, Brown G, Ho A, Regillo C, Donoso L, Tian L, Kaderli B, Hadley D, Hagstrom SA, Peachey NS, Klein R, Klein BE, Gotoh N, Yamashiro K, Ferris Iii F, Fagerness JA, Reynolds R, Farrer LA, Kim IK, Miller JW, Corton M, Carracedo A, Sanchez-Salorio M, Pugh EW, Doheny KF, Brion M, Deangelis MM, Weeks DE, Zack DJ, Chew EY, Heckenlively JR, Yoshimura N, Iyengar SK, Francis PJ, Katsanis N, Seddon JM, Haines JL, Gorin MB, Abecasis GR, Swaroop A, 2010. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc Natl Acad Sci U S A 107, 7401–7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SE, Fu T, Seok S, Kim DH, Yu E, Lee KW, Kang Y, Li X, Kemper B, Kemper JK, 2013. Elevated microRNA-34a in obesity reduces NAD+ levels and SIRT1 activity by directly targeting NAMPT. Aging Cell 12, 1062–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou MY, Hartvigsen K, Hansen LF, Fogelstrand L, Shaw PX, Boullier A, Binder CJ, Witztum JL, 2008. Oxidation-specific epitopes are important targets of innate immunity. J Intern Med 263, 479–488. [DOI] [PubMed] [Google Scholar]
- Clark HF, Gurney AL, Abaya E, Baker K, Baldwin D, Brush J, Chen J, Chow B, Chui C, Crowley C, Currell B, Deuel B, Dowd P, Eaton D, Foster J, Grimaldi C, Gu Q, Hass PE, Heldens S, Huang A, Kim HS, Klimowski L, Jin Y, Johnson S, Lee J, Lewis L, Liao D, Mark M, Robbie E, Sanchez C, Schoenfeld J, Seshagiri S, Simmons L, Singh J, Smith V, Stinson J, Vagts A, Vandlen R, Watanabe C, Wieand D, Woods K, Xie MH, Yansura D, Yi S, Yu G, Yuan J, Zhang M, Zhang Z, Goddard A, Wood WI, Godowski P, Gray A, 2003. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: a bioinformatics assessment. Genome Res 13, 2265–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons TE, Milton RC, Klein R, Seddon JM, Ferris FL 3rd, Age-Related Eye Disease Study Research, G., 2005. Risk factors for the incidence of Advanced Age-Related Macular Degeneration in the Age-Related Eye Disease Study (AREDS) AREDS report no. 19. Ophthalmology 112, 533–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley YP, Thalamuthu A, Jakobsdottir J, Weeks DE, Mah T, Ferrell RE, Gorin MB, 2005. Candidate gene analysis suggests a role for fatty acid biosynthesis and regulation of the complement system in the etiology of age-related maculopathy. Hum Mol Genet 14, 1991–2002. [DOI] [PubMed] [Google Scholar]
- Conte I, Carrella S, Avellino R, Karali M, Marco-Ferreres R, Bovolenta P, Banfi S, 2010. miR-204 is required for lens and retinal development via Meis2 targeting. Proc Natl Acad Sci U S A 107, 15491–15496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb JW, Miyagi M, Gu X, Shadrach K, West KA, Sakaguchi H, Kamei M, Hasan A, Yan L, Rayborn ME, Salomon RG, Hollyfield JG, 2002. Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc Natl Acad Sci U S A 99, 14682–14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio CA, Johnson M, Rudolf M, Huang JD, 2011. The oil spill in ageing Bruch membrane. Br J Ophthalmol 95, 1638–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio CA, Messinger JD, Sloan KR, McGwin G, Medeiros NE, Spaide RF, 2013. Subretinal drusenoid deposits in non-neovascular age-related macular degeneration: morphology, prevalence, topography, and biogenesis model. Retina 33, 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio CA, Millican CL, Bailey T, Kruth HS, 2001. Accumulation of cholesterol with age in human Bruch’s membrane. Invest Ophthalmol Vis Sci 42, 265–274. [PubMed] [Google Scholar]
- Curcio CA, Presley JB, Malek G, Medeiros NE, Avery DV, Kruth HS, 2005a. Esterified and unesterified cholesterol in drusen and basal deposits of eyes with age-related maculopathy. Exp Eye Res 81, 731–741. [DOI] [PubMed] [Google Scholar]
- Curcio CA, Presley JB, Millican CL, Medeiros NE, 2005b. Basal deposits and drusen in eyes with age-related maculopathy: evidence for solid lipid particles. Exp Eye Res 80, 761–775. [DOI] [PubMed] [Google Scholar]
- Dashti N, McGwin G, Owsley C, Curcio CA, 2006. Plasma apolipoproteins and risk for age related maculopathy. Br J Ophthalmol 90, 1028–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos A, Goedeke L, Smibert P, Ramirez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC, Esplugues E, Fisher EA, Penalva LO, Moore KJ, Suarez Y, Lai EC, Fernandez-Hernando C, 2011. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A 108, 9232–9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Aguiar Vallim TQ, Tarling EJ, Kim T, Civelek M, Baldan A, Esau C, Edwards PA, 2013. MicroRNA-144 regulates hepatic ATP binding cassette transporter A1 and plasma high-density lipoprotein after activation of the nuclear receptor farnesoid X receptor. Circ Res 112, 1602–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dithmar S, Curcio CA, Le NA, Brown S, Grossniklaus HE, 2000. Ultrastructural changes in Bruch’s membrane of apolipoprotein E-deficient mice. Invest Ophthalmol Vis Sci 41, 2035–2042. [PubMed] [Google Scholar]
- Drewry MD, Challa P, Kuchtey JG, Navarro I, Helwa I, Hu Y, Mu H, Daniel Stamer W, Kuchtey RW, Liu Y, 2018. Differentially expressed microRNAs in the aqueous humor of patients with exfoliation glaucoma or primary open-angle glaucoma. Hum Mol Genet 27, 1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan KG, Hosseini K, Bailey KR, Yang H, Lowe RJ, Matthes MT, Kane JP, LaVail MM, Schwartz DM, Duncan JL, 2009. Expression of reverse cholesterol transport proteins ATP-binding cassette A1 (ABCA1) and scavenger receptor BI (SR-BI) in the retina and retinal pigment epithelium. Br J Ophthalmol 93, 1116–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi KB, Fijalkowski N, Cano M, Handa JT, 2013. Decreased membrane complement regulators in the retinal pigmented epithelium contributes to age-related macular degeneration. J Pathol 229, 729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AO, Ritter R 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA, 2005. Complement factor H polymorphism and age-related macular degeneration. Science 308, 421–424. [DOI] [PubMed] [Google Scholar]
- Eisner A, Klein ML, Zilis JD, Watkins MD, 1992. Visual function and the subsequent development of exudative age-related macular degeneration. Invest Ophthalmol Vis Sci 33, 3091–3102. [PubMed] [Google Scholar]
- Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S, 2008. LNA-mediated microRNA silencing in non-human primates. Nature 452, 896–899. [DOI] [PubMed] [Google Scholar]
- Ershov AV, Bazan NG, 2000. Photoreceptor phagocytosis selectively activates PPARgamma expression in retinal pigment epithelial cells. J Neurosci Res 60, 328–337. [DOI] [PubMed] [Google Scholar]
- Ertekin S, Yildirim O, Dinc E, Ayaz L, Fidanci SB, Tamer L, 2014. Evaluation of circulating miRNAs in wet age-related macular degeneration. Mol Vis 20, 1057–1066. [PMC free article] [PubMed] [Google Scholar]
- Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP, 2006. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 3, 87–98. [DOI] [PubMed] [Google Scholar]
- Feinberg MW, Moore KJ, 2016. MicroRNA Regulation of Atherosclerosis. Circ Res 118, 703–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson EL, Sternberg PW, Horvitz HR, 1987. A genetic pathway for the specification of the vulval cell lineages of Caenorhabditis elegans. Nature 326, 259–267. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP, 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19, 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche LG, Igl W, Bailey JN, Grassmann F, Sengupta S, Bragg-Gresham JL, Burdon KP, Hebbring SJ, Wen C, Gorski M, Kim IK, Cho D, Zack D, Souied E, Scholl HP, Bala E, Lee KE, Hunter DJ, Sardell RJ, Mitchell P, Merriam JE, Cipriani V, Hoffman JD, Schick T, Lechanteur YT, Guymer RH, Johnson MP, Jiang Y, Stanton CM, Buitendijk GH, Zhan X, Kwong AM, Boleda A, Brooks M, Gieser L, Ratnapriya R, Branham KE, Foerster JR, Heckenlively JR, Othman MI, Vote BJ, Liang HH, Souzeau E, McAllister IL, Isaacs T, Hall J, Lake S, Mackey DA, Constable IJ, Craig JE, Kitchner TE, Yang Z, Su Z, Luo H, Chen D, Ouyang H, Flagg K, Lin D, Mao G, Ferreyra H, Stark K, von Strachwitz CN, Wolf A, Brandl C, Rudolph G, Olden M, Morrison MA, Morgan DJ, Schu M, Ahn J, Silvestri G, Tsironi EE, Park KH, Farrer LA, Orlin A, Brucker A, Li M, Curcio CA, Mohand-Said S, Sahel JA, Audo I, Benchaboune M, Cree AJ, Rennie CA, Goverdhan SV, Grunin M, Hagbi-Levi S, Campochiaro P, Katsanis N, Holz FG, Blond F, Blanche H, Deleuze JF, Igo RP Jr., , Truitt B, Peachey NS, Meuer SM, Myers CE, Moore EL, Klein R, Hauser MA, Postel EA, Courtenay MD, Schwartz SG, Kovach JL, Scott WK, Liew G, Tan AG, Gopinath B, Merriam JC, Smith RT, Khan JC, Shahid H, Moore AT, McGrath JA, Laux R, Brantley MA Jr., Agarwal A, Ersoy L, Caramoy A, Langmann T, Saksens NT, de Jong EK, Hoyng CB, Cain MS, Richardson AJ, Martin TM, Blangero J, Weeks DE, Dhillon B, van Duijn CM, Doheny KF, Romm J, Klaver CC, Hayward C, Gorin MB, Klein ML, Baird PN, den Hollander AI, Fauser S, Yates JR, Allikmets R, Wang JJ, Schaumberg DA, Klein BE, Hagstrom SA, Chowers I, Lotery AJ, Leveillard T, Zhang K, Brilliant MH, Hewitt AW, Swaroop A, Chew EY, Pericak-Vance MA, DeAngelis M, Stambolian D, Haines JL, Iyengar SK, Weber BH, Abecasis GR, Heid IM, 2016. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet 48, 134–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujihara M, Bartels E, Nielsen LB, Handa JT, 2009. A human apoB100 transgenic mouse expresses human apoB100 in the RPE and develops features of early AMD. Exp Eye Res 88, 1115–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujihara M, Cano M, Handa JT, 2014. Mice that produce ApoB100 lipoproteins in the RPE do not develop drusen yet are still a valuable experimental system. Invest Ophthalmol Vis Sci 55, 7285–7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genini S, Guziewicz KE, Beltran WA, Aguirre GD, 2014. Altered miRNA expression in canine retinas during normal development and in models of retinal degeneration. BMC Genomics 15, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, Leclercq IA, MacDougald OA, Bommer GT, 2010. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem 285, 33652–33661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanbari M, Erkeland SJ, Xu L, Colijn JM, Franco OH, Dehghan A, Klaver CCW, Meester-Smoor MA, 2017. Genetic variants in microRNAs and their binding sites within gene 3’UTRs associate with susceptibility to age-related macular degeneration. Hum Mutat 38, 827–838. [DOI] [PubMed] [Google Scholar]
- Goedeke L, Aranda JF, Fernandez-Hernando C, 2014. microRNA regulation of lipoprotein metabolism. Curr Opin Lipidol 25, 282–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedeke L, Rotllan N, Canfran-Duque A, Aranda JF, Ramirez CM, Araldi E, Lin CS, Anderson NN, Wagschal A, de Cabo R, Horton JD, Lasuncion MA, Naar AM, Suarez Y, Fernandez-Hernando C, 2015. MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nat Med 21, 1280–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Baro MR, Lewin TM, Coleman RA, 2007. Regulation of Triglyceride Metabolism. II. Function of mitochondrial GPAT1 in the regulation of triacylglycerol biosynthesis and insulin action. Am J Physiol Gastrointest Liver Physiol 292, G1195–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassmann F, Heid IM, Weber BH, International AMDGC, 2017. Recombinant Haplotypes Narrow the ARMS2/HTRA1 Association Signal for Age-Related Macular Degeneration. Genetics 205, 919–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassmann F, Schoenberger PG, Brandl C, Schick T, Hasler D, Meister G, Fleckenstein M, Lindner M, Helbig H, Fauser S, Weber BH, 2014. A circulating microrna profile is associated with late-stage neovascular age-related macular degeneration. PLoS One 9, e107461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, Smith RJ, Silvestri G, Russell SR, Klaver CC, Barbazetto I, Chang S, Yannuzzi LA, Barile GR, Merriam JC, Smith RT, Olsh AK, Bergeron J, Zernant J, Merriam JE, Gold B, Dean M, Allikmets R, 2005. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A 102, 7227–7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, Schnetz-Boutaud N, Agarwal A, Postel EA, Pericak-Vance MA, 2005. Complement factor H variant increases the risk of age-related macular degeneration. Science 308, 419–421. [DOI] [PubMed] [Google Scholar]
- Ham WT Jr., Ruffolo JJ Jr., Mueller HA, Clarke AM, Moon ME, 1978. Histologic analysis of photochemical lesions produced in rhesus retina by short-wave-length light. Invest Ophthalmol Vis Sci 17, 1029–1035. [PubMed] [Google Scholar]
- Handa JT, 2007. New molecular histopathologic insights into the pathogenesis of age-related macular degeneration. Int Ophthalmol Clin 47, 15–50. [DOI] [PubMed] [Google Scholar]
- Handa JT, Cano M, Wang L, Datta S, Liu T, 2017. Lipids, oxidized lipids, oxidation-specific epitopes, and Age-related Macular Degeneration. Biochim Biophys Acta 1862, 430–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa JT, Verzijl N, Matsunaga H, Aotaki-Keen A, Lutty GA, te Koppele JM, Miyata T, Hjelmeland LM, 1999. Increase in the advanced glycation end product pentosidine in Bruch’s membrane with age. Invest Ophthalmol Vis Sci 40, 775–779. [PubMed] [Google Scholar]
- Heiba IM, Elston RC, Klein BE, Klein R, 1994. Sibling correlations and segregation analysis of age-related maculopathy: the Beaver Dam Eye Study. Genet Epidemiol 11, 51–67. [DOI] [PubMed] [Google Scholar]
- Hewitt AT, Nakazawa K, Newsome DA, 1989. Analysis of newly synthesized Bruch’s membrane proteoglycans. Invest Ophthalmol Vis Sci 30, 478–486. [PubMed] [Google Scholar]
- Hou Q, Tang J, Wang Z, Wang C, Chen X, Hou L, Dong XD, Tu L, 2013. Inhibitory effect of microRNA-34a on retinal pigment epithelial cell proliferation and migration. Invest Ophthalmol Vis Sci 54, 6481–6488. [DOI] [PubMed] [Google Scholar]
- Hu Z, Shen WJ, Kraemer FB, Azhar S, 2012. MicroRNAs 125a and 455 repress lipoprotein-supported steroidogenesis by targeting scavenger receptor class B type I in steroidogenic cells. Mol Cell Biol 32, 5035–5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Sun J, Wang F, Luo X, Feng J, Gu Q, Liu T, Sun X, 2017. MicroRNA Expression Patterns Involved in Amyloid Beta-Induced Retinal Degeneration. Invest Ophthalmol Vis Sci 58, 1726–1735. [DOI] [PubMed] [Google Scholar]
- Huang TC, Sahasrabuddhe NA, Kim MS, Getnet D, Yang Y, Peterson JM, Ghosh B, Chaerkady R, Leach SD, Marchionni L, Wong GW, Pandey A, 2012. Regulation of lipid metabolism by Dicer revealed through SILAC mice. J Proteome Res 11, 2193–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD, 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297, 2056–2060. [DOI] [PubMed] [Google Scholar]
- Ishida BY, Duncan KG, Bailey KR, Kane JP, Schwartz DM, 2006. High density lipoprotein mediated lipid efflux from retinal pigment epithelial cells in culture. Br J Ophthalmol 90, 616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD, 2008. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 451, 1125–1129. [DOI] [PubMed] [Google Scholar]
- Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, Kariko K, Yoo JW, Lee DK, Hadziahmetovic M, Song Y, Misra S, Chaudhuri G, Buaas FW, Braun RE, Hinton DR, Zhang Q, Grossniklaus HE, Provis JM, Madigan MC, Milam AH, Justice NL, Albuquerque RJ, Blandford AD, Bogdanovich S, Hirano Y, Witta J, Fuchs E, Littman DR, Ambati BK, Rudin CM, Chong MM, Provost P, Kugel JF, Goodrich JA, Dunaief JL, Baffi JZ, Ambati J, 2011. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 471, 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yoon H, Horie T, Burchett JM, Restivo JL, Rotllan N, Ramirez CM, Verghese PB, Ihara M, Hoe HS, Esau C, Fernandez-Hernando C, Holtzman DM, Cirrito JR, Ono K, Kim J, 2015. microRNA-33 Regulates ApoE Lipidation and Amyloid-beta Metabolism in the Brain. J Neurosci 35, 14717–14726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J, 2005. Complement factor H polymorphism in age-related macular degeneration. Science 308, 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliffen M, Mooy CM, Luider TM, Huijmans JG, Kerkvliet S, de Jong PT, 1996. Identification of glycosaminoglycans in age-related macular deposits. Arch Ophthalmol 114, 1009–1014. [DOI] [PubMed] [Google Scholar]
- Lakkaraju A, Finnemann SC, Rodriguez-Boulan E, 2007. The lipofuscin fluorophore A2E perturbs cholesterol metabolism in retinal pigment epithelial cells. Proc Natl Acad Sci U S A 104, 11026–11031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V, 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. [DOI] [PubMed] [Google Scholar]
- Li CM, Clark ME, Chimento MF, Curcio CA, 2006. Apolipoprotein localization in isolated drusen and retinal apolipoprotein gene expression. Invest Ophthalmol Vis Sci 47, 3119–3128. [DOI] [PubMed] [Google Scholar]
- Li CM, Presley JB, Zhang X, Dashti N, Chung BH, Medeiros NE, Guidry C, Curcio CA, 2005. Retina expresses microsomal triglyceride transfer protein: implications for age-related maculopathy. J Lipid Res 46, 628–640. [DOI] [PubMed] [Google Scholar]
- Li T, Morgan MJ, Choksi S, Zhang Y, Kim YS, Liu ZG, 2010. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat Immunol 11, 799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ, Surjyadipta B, Dua P, Alexandrov PN, 2012. Common micro RNAs (miRNAs) target complement factor H (CFH) regulation in Alzheimer’s disease (AD) and in age-related macular degeneration (AMD). Int J Biochem Mol Biol 3, 105–116. [PMC free article] [PubMed] [Google Scholar]
- Magana MM, Osborne TF, 1996. Two tandem binding sites for sterol regulatory element binding proteins are required for sterol regulation of fatty-acid synthase promoter. J Biol Chem 271, 32689–32694. [DOI] [PubMed] [Google Scholar]
- Malek G, Johnson LV, Mace BE, Saloupis P, Schmechel DE, Rickman DW, Toth CA, Sullivan PM, Bowes Rickman C, 2005. Apolipoprotein E allele-dependent pathogenesis: a model for age-related retinal degeneration. Proc Natl Acad Sci U S A 102, 11900–11905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek G, Li CM, Guidry C, Medeiros NE, Curcio CA, 2003. Apolipoprotein B in cholesterol-containing drusen and basal deposits of human eyes with age-related maculopathy. Am J Pathol 162, 413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquart TJ, Allen RM, Ory DS, Baldan A, 2010. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A 107, 12228–12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard C, Rezende FA, Miloudi K, Wilson A, Tetreault N, Hardy P, SanGiovanni JP, De Guire V, Sapieha P, 2016. MicroRNA signatures in vitreous humour and plasma of patients with exudative AMD. Oncotarget 7, 19171–19184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midena E, Degli Angeli C, Blarzino MC, Valenti M, Segato T, 1997. Macular function impairment in eyes with early age-related macular degeneration. Invest Ophthalmol Vis Sci 38, 469–477. [PubMed] [Google Scholar]
- Miller NE, 1979. Plasma lipoproteins, lipid transport, and atherosclerosis: recent developments. J Clin Pathol 32, 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira EF, Larrayoz IM, Lee JW, Rodriguez IR, 2009. 7-Ketocholesterol is present in lipid deposits in the primate retina: potential implication in the induction of VEGF and CNV formation. Invest Ophthalmol Vis Sci 50, 523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RF, Russell SR, Anderson DH, Hageman GS, 2000. Drusen associated with aging and age-related macular degeneration contain proteins common to extracellular deposits associated with atherosclerosis, elastosis, amyloidosis, and dense deposit disease. FASEB J 14, 835–846. [PubMed] [Google Scholar]
- Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM, 2010. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 328, 1566–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Fagerness J, Reynolds R, Sobrin L, Parker M, Raychaudhuri S, Tan PL, Oh EC, Merriam JE, Souied E, Bernstein PS, Li B, Frederick JM, Zhang K, Brantley MA Jr., Lee AY, Zack DJ, Campochiaro B, Campochiaro P, Ripke S, Smith RT, Barile GR, Katsanis N, Allikmets R, Daly MJ, Seddon JM, 2010. Genome-wide association study of advanced age-related macular degeneration identifies a role of the hepatic lipase gene (LIPC). Proc Natl Acad Sci U S A 107, 7395–7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome DA, Hewitt AT, Huh W, Robey PG, Hassell JR, 1987. Detection of specific extracellular matrix molecules in drusen, Bruch’s membrane, and ciliary body. Am J Ophthalmol 104, 373–381. [DOI] [PubMed] [Google Scholar]
- Nielsen LB, Bartels ED, Bollano E, 2002. Overexpression of apolipoprotein B in the heart impedes cardiac triglyceride accumulation and development of cardiac dysfunction in diabetic mice. J Biol Chem 277, 27014–27020. [DOI] [PubMed] [Google Scholar]
- Nielsen LB, Veniant M, Boren J, Raabe M, Wong JS, Tam C, Flynn L, Vanni-Reyes T, Gunn MD, Goldberg IJ, Hamilton RL, Young SG, 1998. Genes for apolipoprotein B and microsomal triglyceride transfer protein are expressed in the heart: evidence that the heart has the capacity to synthesize and secrete lipoproteins. Circulation 98, 13–16. [DOI] [PubMed] [Google Scholar]
- Oak AS, Messinger JD, Curcio CA, 2014. Subretinal drusenoid deposits: further characterization by lipid histochemistry. Retina 34, 825–826. [DOI] [PubMed] [Google Scholar]
- Owsley C, Huisingh C, Clark ME, Jackson GR, McGwin G Jr., 2016. Comparison of Visual Function in Older Eyes in the Earliest Stages of Age-related Macular Degeneration to Those in Normal Macular Health. Curr Eye Res 41, 266–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palinski W, Ord VA, Plump AS, Breslow JL, Steinberg D, Witztum JL, 1994. ApoE-deficient mice are a model of lipoprotein oxidation in atherogenesis. Demonstration of oxidation-specific epitopes in lesions and high titers of autoantibodies to malondialdehyde-lysine in serum. Arterioscler Thromb 14, 605–616. [DOI] [PubMed] [Google Scholar]
- Palinski W, Rosenfeld ME, Yla-Herttuala S, Gurtner GC, Socher SS, Butler SW, Parthasarathy S, Carew TE, Steinberg D, Witztum JL, 1989. Low density lipoprotein undergoes oxidative modification in vivo. Proc Natl Acad Sci U S A 86, 1372–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palinski W, Yla-Herttuala S, Rosenfeld ME, Butler SW, Socher SA, Parthasarathy S, Curtiss LK, Witztum JL, 1990. Antisera and monoclonal antibodies specific for epitopes generated during oxidative modification of low density lipoprotein. Arteriosclerosis 10, 325–335. [DOI] [PubMed] [Google Scholar]
- Pikuleva IA, Curcio CA, 2014. Cholesterol in the retina: the best is yet to come. Prog Retin Eye Res 41, 64–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilgrim MG, Lengyel I, Lanzirotti A, Newville M, Fearn S, Emri E, Knowles JC, Messinger JD, Read RW, Guidry C, Curcio CA, 2017. Subretinal Pigment Epithelial Deposition of Drusen Components Including Hydroxyapatite in a Primary Cell Culture Model. Invest Ophthalmol Vis Sci 58, 708–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue AI, Lukiw WJ, 2018. Up-regulated Pro-inflammatory MicroRNAs (miRNAs) in Alzheimer’s disease (AD) and Age-Related Macular Degeneration (AMD). Cell Mol Neurobiol [DOI] [PMC free article] [PubMed]
- Rangasamy T, Cho CY, Thimmulappa RK, Zhen L, Srisuma SS, Kensler TW, Yamamoto M, Petrache I, Tuder RM, Biswal S, 2004. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J Clin Invest 114, 1248–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C, 2010. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 328, 1570–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C, Liu Q, Wei Q, Cai W, He M, Du Y, Xu D, Wu Y, Yu J, 2017. Circulating miRNAs as Potential Biomarkers of Age-Related Macular Degeneration. Cell Physiol Biochem 41, 1413–1423. [DOI] [PubMed] [Google Scholar]
- Resnikoff S, Pascolini D, Etya’ale D, Kocur I, Pararajasegaram R, Pokharel GP, Mariotti SP, 2004. Global data on visual impairment in the year 2002. Bull World Health Organ 82, 844–851. [PMC free article] [PubMed] [Google Scholar]
- Rudolf M, Malek G, Messinger JD, Clark ME, Wang L, Curcio CA, 2008. Sub-retinal drusenoid deposits in human retina: organization and composition. Exp Eye Res 87, 402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahasrabuddhe NA, Huang TC, Ahmad S, Kim MS, Yang Y, Ghosh B, Leach SD, Gowda H, Somani BL, Chaerkady R, Pandey A, 2014. Regulation of PPAR-alpha pathway by Dicer revealed through proteomic analysis. J Proteomics 108, 306–315. [DOI] [PubMed] [Google Scholar]
- SanGiovanni JP, SanGiovanni PM, Sapieha P, De Guire V, 2017. miRNAs, single nucleotide polymorphisms (SNPs) and age-related macular degeneration (AMD). Clin Chem Lab Med 55, 763–775. [DOI] [PubMed] [Google Scholar]
- Sarks J, Arnold J, Ho IV, Sarks S, Killingsworth M, 2011. Evolution of reticular pseudodrusen. Br J Ophthalmol 95, 979–985. [DOI] [PubMed] [Google Scholar]
- Schaefer EJ, Eisenberg S, Levy RI, 1978. Lipoprotein apoprotein metabolism. J Lipid Res 19, 667–687. [PubMed] [Google Scholar]
- Sekine S, Ogawa R, Ito R, Hiraoka N, McManus MT, Kanai Y, Hebrok M, 2009. Disruption of Dicer1 induces dysregulated fetal gene expression and promotes hepatocarcinogenesis. Gastroenterology 136, 2304–2315 e2301–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JK, Dong A, Hackett SF, Bell WR, Green WR, Campochiaro PA, 2007. Oxidative damage in age-related macular degeneration. Histol Histopathol 22, 1301–1308. [DOI] [PubMed] [Google Scholar]
- Shimamura M, Matsuda M, Yasumo H, Okazaki M, Fujimoto K, Kono K, Shimizugawa T, Ando Y, Koishi R, Kohama T, Sakai N, Kotani K, Komuro R, Ishida T, Hirata K, Yamashita S, Furukawa H, Shimomura I, 2007. Angiopoietin-like protein3 regulates plasma HDL cholesterol through suppression of endothelial lipase. Arterioscler Thromb Vasc Biol 27, 366–372. [DOI] [PubMed] [Google Scholar]
- Shimizugawa T, Ono M, Shimamura M, Yoshida K, Ando Y, Koishi R, Ueda K, Inaba T, Minekura H, Kohama T, Furukawa H, 2002. ANGPTL3 decreases very low density lipoprotein triglyceride clearance by inhibition of lipoprotein lipase. J Biol Chem 277, 33742–33748. [DOI] [PubMed] [Google Scholar]
- Shimomura I, Bashmakov Y, Shimano H, Horton JD, Goldstein JL, Brown MS, 1997a. Cholesterol feeding reduces nuclear forms of sterol regulatory element binding proteins in hamster liver. Proc Natl Acad Sci U S A 94, 12354–12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura I, Shimano H, Horton JD, Goldstein JL, Brown MS, 1997b. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J Clin Invest 99, 838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit-McBride Z, Forward KI, Nguyen AT, Bordbari MH, Oltjen SL, Hjelmeland LM, 2014. Age-dependent increase in miRNA-34a expression in the posterior pole of the mouse eye. Mol Vis 20, 1569–1578. [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, Hansch C, 2000. The relative toxicity of compounds in mainstream cigarette smoke condensate. Food Chem Toxicol 38, 637–646. [DOI] [PubMed] [Google Scholar]
- Soh J, Iqbal J, Queiroz J, Fernandez-Hernando C, Hussain MM, 2013. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat Med 19, 892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaide RF, Ho-Spaide WC, Browne RW, Armstrong D, 1999. Characterization of peroxidized lipids in Bruch’s membrane. Retina 19, 141–147. [DOI] [PubMed] [Google Scholar]
- Sundermeier TR, Palczewski K, 2016. The impact of microRNA gene regulation on the survival and function of mature cell types in the eye. FASEB J 30, 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundermeier TR, Sakami S, Sahu B, Howell SJ, Gao S, Dong Z, Golczak M, Maeda A, Palczewski K, 2017. MicroRNA-processing Enzymes Are Essential for Survival and Function of Mature Retinal Pigmented Epithelial Cells in Mice. J Biol Chem 292, 3366–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Kamei M, Itabe H, Yoneda K, Bando H, Kume N, Tano Y, 2007. Oxidized phospholipids in the macula increase with age and in eyes with age-related macular degeneration. Mol Vis 13, 772–778. [PMC free article] [PubMed] [Google Scholar]
- Szemraj M, Bielecka-Kowalska A, Oszajca K, Krajewska M, Gos R, Jurowski P, Kowalski M, Szemraj J, 2015. Serum MicroRNAs as Potential Biomarkers of AMD. Med Sci Monit 21, 2734–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I, 1999. Nonoxidative modifications of lipoproteins in atherogenesis. Annu Rev Nutr 19, 123–139. [DOI] [PubMed] [Google Scholar]
- Tabas I, Williams KJ, Boren J, 2007. Subendothelial lipoprotein retention as the initiating process in atherosclerosis: update and therapeutic implications. Circulation 116, 1832–1844. [DOI] [PubMed] [Google Scholar]
- Takayama K, Kaneko H, Hwang SJ, Ye F, Higuchi A, Tsunekawa T, Matsuura T, Iwase T, Asami T, Ito Y, Ueno S, Yasuda S, Nonobe N, Terasaki H, 2016. Increased Ocular Levels of MicroRNA-148a in Cases of Retinal Detachment Promote Epithelial-Mesenchymal Transition. Invest Ophthalmol Vis Sci 57, 2699–2705. [DOI] [PubMed] [Google Scholar]
- Thomas GS, Cromwell WC, Ali S, Chin W, Flaim JD, Davidson M, 2013. Mipomersen, an apolipoprotein B synthesis inhibitor, reduces atherogenic lipoproteins in patients with severe hypercholesterolemia at high cardiovascular risk: a randomized, double-blind, placebo-controlled trial. J Am Coll Cardiol 62, 2178–2184. [DOI] [PubMed] [Google Scholar]
- Thomas T, Ginsberg H, 2010. Development of apolipoprotein B antisense molecules as a therapy for hyperlipidemia. Curr Atheroscler Rep 12, 58–65. [DOI] [PubMed] [Google Scholar]
- Thompson RB, Reffatto V, Bundy JG, Kortvely E, Flinn JM, Lanzirotti A, Jones EA, McPhail DS, Fearn S, Boldt K, Ueffing M, Ratu SG, Pauleikhoff L, Bird AC, Lengyel I, 2015. Identification of hydroxyapatite spherules provides new insight into subretinal pigment epithelial deposit formation in the aging eye. Proc Natl Acad Sci U S A 112, 1565–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian FJ, An LN, Wang GK, Zhu JQ, Li Q, Zhang YY, Zeng A, Zou J, Zhu RF, Han XS, Shen N, Yang HT, Zhao XX, Huang S, Qin YW, Jing Q, 2014. Elevated microRNA-155 promotes foam cell formation by targeting HBP1 in atherogenesis. Cardiovasc Res 103, 100–110. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Kim JB, Graves RA, Spiegelman BM, 1993. ADD1: a novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol Cell Biol 13, 4753–4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomey CB, Kelly U, Saban DR, Bowes Rickman C, 2015. Regulation of age-related macular degeneration-like pathology by complement factor H. Proc Natl Acad Sci U S A 112, E3040–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, Chen HC, Lee CH, Tsai TF, Hsu MT, Wu JC, Huang HD, Shiao MS, Hsiao M, Tsou AP, 2012. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest 122, 2884–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tserentsoodol N, Gordiyenko NV, Pascual I, Lee JW, Fliesler SJ, Rodriguez IR, 2006. Intraretinal lipid transport is dependent on high density lipoprotein-like particles and class B scavenger receptors. Mol Vis 12, 1319–1333. [PubMed] [Google Scholar]
- Tsunekawa T, Kaneko H, Takayama K, Hwang SJ, Oishi A, Nagasaka Y, Ye F, Iwase T, Nonobe N, Ueno S, Ito Y, Yasuda S, Matsuura T, Shimizu H, Suzumura A, Kataoka K, Terasaki H, 2017. Correlation between miR-148 Expression in Vitreous and Severity of Rhegmatogenous Retinal Detachment. Biomed Res Int 2017, 3427319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavvas DG, Daniels AB, Kapsala ZG, Goldfarb JW, Ganotakis E, Loewenstein JI, Young LH, Gragoudas ES, Eliott D, Kim IK, Tsilimbaris MK, Miller JW, 2016. Regression of Some High-risk Features of Age-related Macular Degeneration (AMD) in Patients Receiving Intensive Statin Treatment. EBioMedicine 5, 198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers KC, Landstreet SR, Levin MG, Shoucri BM, Toth CL, Taylor RC, Palmisano BT, Tabet F, Cui HL, Rye KA, Sethupathy P, Remaley AT, 2014. MicroRNA-223 coordinates cholesterol homeostasis. Proc Natl Acad Sci U S A 111, 14518–14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers KC, Shoucri BM, Levin MG, Wu H, Pearson DS, Osei-Hwedieh D, Collins FS, Remaley AT, Sethupathy P, 2013. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology 57, 533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vingerling JR, Klaver CC, Hofman A, de Jong PT, 1995. Epidemiology of age-related maculopathy. Epidemiol Rev 17, 347–360. [DOI] [PubMed] [Google Scholar]
- Wagschal A, Najafi-Shoushtari SH, Wang L, Goedeke L, Sinha S, deLemos AS, Black JC, Ramirez CM, Li Y, Tewhey R, Hatoum I, Shah N, Lu Y, Kristo F, Psychogios N, Vrbanac V, Lu YC, Hla T, de Cabo R, Tsang JS, Schadt E, Sabeti PC, Kathiresan S, Cohen DE, Whetstine J, Chung RT, Fernandez-Hernando C, Kaplan LM, Bernards A, Gerszten RE, Naar AM, 2015. Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nat Med 21, 1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Clark ME, Crossman DK, Kojima K, Messinger JD, Mobley JA, Curcio CA, 2010. Abundant lipid and protein components of drusen. PLoS One 5, e10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Jia XJ, Jiang HJ, Du Y, Yang F, Si SY, Hong B, 2013. MicroRNAs 185, 96, and 223 repress selective high-density lipoprotein cholesterol uptake through posttranscriptional inhibition. Mol Cell Biol 33, 1956–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weismann D, Hartvigsen K, Lauer N, Bennett KL, Scholl HP, Charbel Issa P, Cano M, Brandstatter H, Tsimikas S, Skerka C, Superti-Furga G, Handa JT, Zipfel PF, Witztum JL, Binder CJ, 2011. Complement factor H binds malondialdehyde epitopes and protects from oxidative stress. Nature 478, 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G, 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862. [DOI] [PubMed] [Google Scholar]
- Winkler BS, Boulton ME, Gottsch JD, Sternberg P, 1999. Oxidative damage and age-related macular degeneration. Mol Vis 5, 32. [PMC free article] [PubMed] [Google Scholar]
- Wu T, Tian J, Cutler RG, Telljohann RS, Bernlohr DA, Mattson MP, Handa JT, 2010. Knockdown of FABP5 mRNA decreases cellular cholesterol levels and results in decreased apoB100 secretion and triglyceride accumulation in ARPE-19 cells. Lab Invest 90, 906–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Cao S, Rajapakse S, Matsubara JA, 2018. Understanding AMD by analogy: systematic review of lipid-related common pathogenic mechanisms in AMD, AD, AS and GN. Lipids Health Dis 17, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zalzala M, Xu J, Li Y, Yin L, Zhang Y, 2015. A metabolic stress-inducible miR-34a-HNF4alpha pathway regulates lipid and lipoprotein metabolism. Nat Commun 6, 7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, He YS, Wang XQ, Lu L, Chen QJ, Liu J, Sun Z, Shen WF, 2011. MiR-146a inhibits oxidized low-density lipoprotein-induced lipid accumulation and inflammatory response via targeting toll-like receptor 4. FEBS Lett 585, 854–860. [DOI] [PubMed] [Google Scholar]
- Yang M, Liu W, Pellicane C, Sahyoun C, Joseph BK, Gallo-Ebert C, Donigan M, Pandya D, Giordano C, Bata A, Nickels JT Jr., 2014. Identification of miR-185 as a regulator of de novo cholesterol biosynthesis and low density lipoprotein uptake. J Lipid Res 55, 226–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama M, Yagyu H, Hu Y, Seo T, Hirata K, Homma S, Goldberg IJ, 2004. Apolipoprotein B production reduces lipotoxic cardiomyopathy: studies in heart-specific lipoprotein lipase transgenic mouse. J Biol Chem 279, 4204–4211. [DOI] [PubMed] [Google Scholar]
- Yoon H, Flores LF, Kim J, 2016. MicroRNAs in brain cholesterol metabolism and their implications for Alzheimer’s disease. Biochim Biophys Acta 1861, 2139–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Ohno-Matsui K, Ichinose S, Sato T, Iwata N, Saido TC, Hisatomi T, Mochizuki M, Morita I, 2005. The potential role of amyloid beta in the pathogenesis of age-related macular degeneration. J Clin Invest 115, 2793–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarbin MA, 2004. Current concepts in the pathogenesis of age-related macular degeneration. Arch Ophthalmol 122, 598–614. [DOI] [PubMed] [Google Scholar]
- Zhang M, Wu JF, Chen WJ, Tang SL, Mo ZC, Tang YY, Li Y, Wang JL, Liu XY, Peng J, Chen K, He PP, Lv YC, Ouyang XP, Yao F, Tang DP, Cayabyab FS, Zhang DW, Zheng XL, Tian GP, Tang CK, 2014. MicroRNA-27a/b regulates cellular cholesterol efflux, influx and esterification/hydrolysis in THP-1 macrophages. Atherosclerosis 234, 54–64. [DOI] [PubMed] [Google Scholar]
- Zheng W, Reem RE, Omarova S, Huang S, DiPatre PL, Charvet CD, Curcio CA, Pikuleva IA, 2012. Spatial distribution of the pathways of cholesterol homeostasis in human retina. PLoS One 7, e37926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Irani S, Sirwi A, Hussain MM, 2016. MicroRNAs regulating apolipoprotein B-containing lipoprotein production. Biochim Biophys Acta 1861, 2062–2068. [DOI] [PubMed] [Google Scholar]


