Abstract
RNA editing is an important form of regulating gene expression and activity. APOBEC1 cytosine deaminase was initially characterized as pairing with a cofactor, A1CF, to form an active RNA editing complex that specifically targets APOB RNA in regulating lipid metabolism. Recent studies revealed that APOBEC1 may be involved in editing other potential RNA targets in a tissue specific manner, and another protein, RBM47, appears to instead be the main cofactor of APOBEC1 for editing APOB RNA. In this report, by expressing APOBEC1 with either A1CF or RBM47 from human or mouse in an HEK293T cell line with no intrinsic APOBEC1/A1CF/RBM47 expression, we have compared direct RNA editing activity on several known cellular target RNAs. By using a sensitive cell-based fluorescence assay that enables comparative quantification of RNA editing through subcellular localization changes of eGFP, the two APOBEC1 cofactors, A1CF and RBM47, showed clear differences for editing activity on APOB and several other tested RNAs, and clear differences were observed when mouse vs. human genes were tested. In addition, we have determined the minimal domain requirement of RBM47 needed for activity. These results provide useful functional characterization of RBM47 and direct biochemical evidence for the differential editing selectivity on a number of RNA targets.
Introduction
RNA editing by deamination of cytosine and adenosine is a form of regulation and diversification of genes involved in important biological functions and diseases in humans [1–5]. In mammals, cytosine-to-uracil RNA editing was first observed on mRNA transcripts of Apob, where deamination at cytosine 6666 of the transcript results in an early stop codon and expression of a truncated version of the translated protein [6], altering its role in lipid and cholesterol metabolism [7, 8]. The active component essential for catalyzing the conversion of cytosine to uracil on RNA is APOBEC-1 (abbreviated here as APO1) [9–12], a member of the AID/APOBEC cytosine deaminase family now known to play many different roles in cells [13–15]. APO1 catalytic activity was initially found to only be possible in the presence of an RNA-binding cofactor, APOBEC1 complementation factor (A1CF) [16–20]. Together these two proteins, APO1 and A1CF, were thought to be the minimum components needed to reconstitute activity of the RNA “editosome”.
More recently, RNA-binding protein 47 (RBM47) was found to confer APOB RNA editing activity in the absence of A1CF [21], and an A1cf gene knockout did not result in changes to in vivo levels of Apob mRNA editing in mice [21, 22]. These findings suggest that RBM47 may instead be the major complementation factor for APOB RNA editing in vivo [23], leaving the actual biological function of A1CF still uncertain. Additionally, transcriptome-sequencing studies of extracted intestinal or liver cells of mice revealed that many other mRNA transcripts appear to be edited by APO1, generally within 3’ UTRs [23–25], implying that APO1 and its cofactors may play a broader biological function beyond regulating APOB expression and cholesterol metabolism [20–22, 24, 26, 27]. These developments raise the intriguing question regarding the exact role of A1CF and RBM47 in targeting cellular RNA targets for cytosine deamination.
It was our goal to better characterize the domains of RBM47 needed for complementation of APO1 editing activity, and to compare the relative editing activity of RBM47 and A1CF in complex with APO1 on both APOB and several other described RNA targets. During the preparation of this manuscript, a new report demonstrated the tissue-specific regulation of APO1 activity by A1CF and RBM47 on multiple endogenous RNA targets through the use of knockout mouse models [27]. We report here a methodology that allows for direct assessment of RNA editing activity in cells using a novel fluorescence-based assay. Using this method, it was possible to probe the specificity of APO1 activity on selectively targeted regions of known RNA substrates in the presence of either of the two reported cofactors, in order to better understand what guides the selection and activity of this RNA editing behavior and provide more context to what has recently been observed in vivo.
RESULTS
Design of a reporter system for RNA editing in cells
Investigation of RNA deamination is difficult when compared to similar enzymatic editing activity on other polynucleotides, as there are few systems that can effectively provide a readout for when RNA editing is occurring. A few have been proposed for adenosine deamination [28–30], and some have been adapted for cytidine deamination [6, 31, 32], although these can sometimes be time consuming, show low sensitivity, or are difficult to quantify precisely. Refining on several of these methods led to our development of a system where RNA editing directly induces a shift in subcellular localization of an engineered eGFP [33] construct. This type of relative fluorescence intensity comparison was preferred as it should not be affected by varying levels of fluorophore expression or strength of excitation. When visualized by a fluorescence system such as a scanning confocal microscope, the nuclear region of a cell can be easily detected with a DNA-binding stain, allowing for accurate calculation of the ratio of nuclear to cytosolic eGFP intensity for an individual cell [34–36].
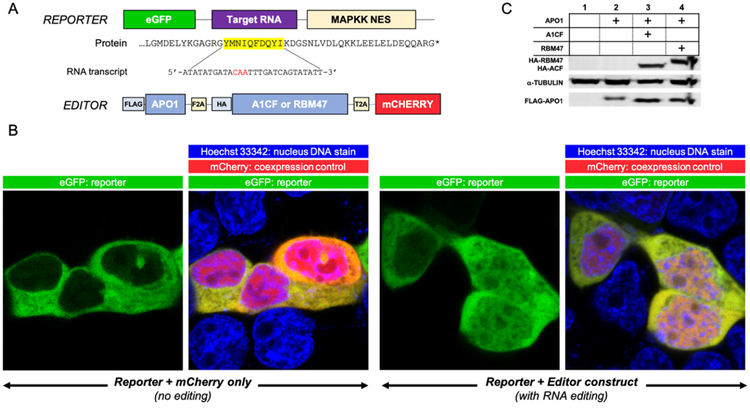
The proposed method utilizes separate reporter and editor constructs as the core components (Figure 1A), with the goal of maximizing flexibility in experiment design. This allows for easily switching out different editing enzymes, cofactors or alternative RNA substrates in the two constructs and allows for high-throughput experimentation while maintaining reliability. In the editor construct, human APO1, A1CF or RBM47, and mCherry [37] are all translated within a single open reading frame but are cleaved into individual proteins by self-cutting 2A peptides [38, 39] to ensure a consistent ratio of APO1 to its cofactor (Figure 1C). The presence of mCherry fluorescence acts as further confirmation of expression of APO1 and its cofactor. HEK293T cells are used for co-transfection of the individual editor and reporter vectors for reconstitution of editosome activity, as no endogenous APO1, A1CF, or RBM47 was detected in this cell line (Sup. Figure 1).
Figure 1. Design of an assay system for RNA editing through C-to-U deamination by the APO1 editosome.
(A) Cartoon depiction of the reporter and editor constructs used in this study. The editor construct expresses FLAG-APO1, a cofactor (HA-A1CF or HA-RBM47), and mCherry as individual, cleaved proteins from a single open reading frame via self-cleaving 2A peptides. mCherry was always included as a control for successful co-transfection of the two separate plasmids per any observed cell. For the reporter construct, eGFP is fused to a MAPKK nuclear export sequence (NES) at the C-terminus, with a peptide encoded by a target RNA substrate transcript sequence inserted between eGFP and NES. Shown here is the minimal 27-base APOB target RNA transcript sequence known to be edited by APO1. Editing of the target RNA transcript (deamination of the target C) of the corresponding mRNA by the co-expressed editor APO1/cofactor proteins results in an early stop codon before the NES, leading to nuclear retention of the NES-less eGFP. (B) Initial test results of the assay system, showing the live-cell eGFP fluorescence images for either an unedited APOB reporter construct (left) or a condition where editing has occurred (right). mCherry fluorescence represents co-expression of the editor plasmid in a particular cell to ensure eGFP localization is occurring in the presence of the editing machinery. Nuclear periphery is demarcated by staining with Hoechst 33342 to enable proper selection of the nuclear vs. cytoplasmic regions when fluorescence intensity values are similar. RNA editing shows a clear shift in the nuclear localization of eGFP fluorescence, which can then be quantified through ImageJ or comparable software. (C) Characteristic western blot from three independent co-transfections of the eGFP reporter with either mCherry alone (1), FLAG-APO1 (2), or FLAG-APO1 with either HA-A1CF (3) or HA-RBM47 (4), showing the successful expression of all proteins after self-cleavage in cells. α-tubulin is the internal control for protein load. Full uncropped western is available in Sup. Figure 2.
APOB mRNA editing has previously been shown to be dependent on localized secondary structure formed by a minimum of 27 highly conserved nucleotides (nt) surrounding the edited cytosine that is recognized by A1CF, as shown via a previous NMR study [40]. It is important to note that the length of RNA taken from the original target may have a large impact on the overall activity due to potential extra interactions between A1CF or APO1 with distal regions of the RNA sequence. However, this NMR study suggests that local secondary structure may play a large role in substrate recognition, and so for the purposes of this system this 27 nt APOB RNA was used for the initial editing assay to limit any such effects and simulate only this localized substrate specificity.
The reporter construct used here thus contains this minimal portion of the target APOB RNA sequence [40, 41] inserted between eGFP and the MAPKK nuclear export signal (NES), and expression of this construct alone shows eGFP fluorescence primarily within the cytosol (Figure 1B), as expected by the action of the expressed NES fused to the C-terminus of eGFP. Upon co-expression of the APO1-A1CF editor construct together with the reporter construct, the reporter mRNA is edited to create an early stop codon prior to the NES, resulting in a shift in GFP fluorescence localization to the nucleus as shown in Figure 1B. The mean localization ratio changes compared to the negative and positive controls were found to be highly significant (P < 0.0001, Sup. Figure 3), indicating editing can quantifiably shift localization of the reporter.
Stronger editing of APOB RNA by APO1/RBM47 than by APO1/A1CF
With this assay system working at hand, we decided to investigate whether there may be a difference in APOB RNA editing by the APO1 editosome when paired with A1CF versus RBM47. The same APOB reporter was co-expressed with the editor construct containing either mCherry alone as a negative control, APO1, APO1+A1CF, or APO1+RBM47 (Figure 2A).
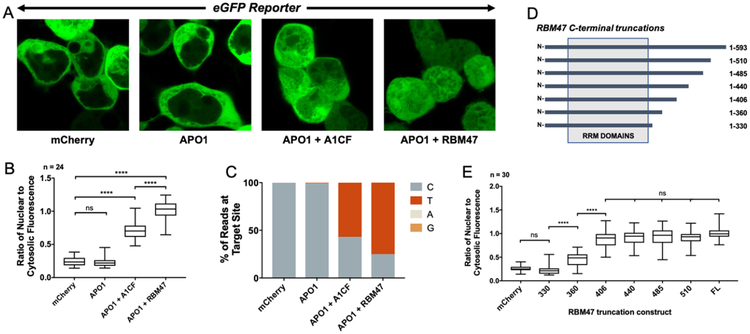
Figure 2. Characterization of the editing activity on APOB RNA by the APO1 editosome paired with either A1CF or RBM47.
(A) Live-cell fluorescence images showing that RNA editing can be visualized directly by a clear shift in subcellular localization; RNA editing only occurred when APO1 is co-expressed with A1CF or RBM47 cofactors. Shown are representative eGFP fluorescence images of the APOB reporter co-transfected with either mCherry alone, APO1 alone, APO1 + A1CF, or APO1 + RBM47. The Hoechst 33342 and mCherry signals have been excluded for clarity. (B) Quantification of the fluorescent localization changes (expressed as changes in the ratio of nuclear eGFP fluorescence divided by cytosolic), with indication of statistical significance with the co-expression of APO1 with either A1CF or RBM47. Displayed is a box-and-whisker plot comparing the mean values of the ratio of nuclear to cytosolic fluorescence for the same transfections described in 2A; analysis is via ANOVA with Tukey post-test for significance, with reported significance levels shown from multiplicity-adjusted P-values (n.s. = not significant when P > 0.05 and **** = P < 0.0001, n = number of cells used in analysis). This experiment was repeated an additional two times and the same trend was observed in all three trials as shown in Sup. Figure 4, implying the result is not due to variation in transfection efficiency. (C): The NGS result of the reporter construct showed that an increase in C-to-T transition mutations within the APOB RNA is highly correlated with the fluorescence localization changes. Paired-end Illumina amplicon sequencing (>50,000 reads) was done on Trizol-extracted mRNA that was reverse-transcribed and amplified around the reporter substrate region. The results showed a dramatic increase in the number of reads containing T at the target site – but not anywhere else – in the presence of either A1CF or RBM47 when compared to APO1 alone or mCherry as a negative control. See Sup. Figure 4 for an example alignment showing such transitions. (D) Characterizing functional domain requirements of RBM47 by a series of C-terminal truncation constructs. The RRM domains have been labeled as annotated in literature. The residue number at which the C-terminal deletion is made is indicated to the left of each construct. (E) The editing assay result of the various RBM47 deletion constructs, showing that last 187 residues of the C-terminus of RBM47 are not necessary for APO1 complementation activity. Box and whisker plots from ANOVA for comparison with significance calculated via a Tukey post-test analysis. Constructs longer than 406 residues showed no significance in calculated multiplicity-adjusted P-values (P > 0.05) when compared to wild-type, and constructs between 406, 360, and 330 residues all showed a significant difference in editing amount, with P-values < 0.0001.
As expected, co-expression with mCherry or APO1 alone showed no change in eGFP localization, i.e. no RNA editing (Figure 2A, 2B). To our surprise, there were significantly different levels of RNA editing by APO1 in the presence of A1CF or RBM47, and these differences were reproducible across three independent trials (Sup. Figure 4). APO1 paired with RBM47 displayed the highest degree of editing on APOB RNA, with a mean ratio of nuclear to cytosolic intensity of 0.997 ± 0.131 across the three trials. A1CF by comparison showed a mean ratio of 0.726 ± 0.177. It is notable that RBM47 was so clearly favored over A1CF in terms of editing activity of APOB RNA, as this aligns well with previous reports suggesting that A1CF may not be the favored cofactor for Apob RNA editing under normal physiological conditions [21, 22]. It is also very consistent with the most recent report that revealed involvement of both RBM47 and A1CF on Apob RNA editing in mouse hepatic cells [27], with RBM47 having a greater impact than A1CF on this editing activity. This consistency with in vivo methods provides excellent validation of the methodology.
To further verify these results and confirm the changes in the re-localization of eGFP were due to bona fide transcript editing of the APOB RNA within the cells, reporter mRNA from one of these experiments was extracted and the resulting cDNA was sent out for Illumina Amplicon next-gen sequencing (Genewiz). As expected, co-expression with mCherry alone or APO1 without any cofactor showed effectively no degree of C to T transitions. However, there was a large increase in the number of C to T transitions within the APOB target region when APO1 was present with either A1CF or RBM47 (Figure 2C), consistent with the change of re-localized eGFP fluorescence results. A1CF showed a transition from cytosine to uracil on 56% of the observed sequences, whereas RBM47 showed conversion on 75%. This editing was highly specific and was only observed at the recognized APOB target site within the approximately 225-nucleotide sequenced region (Sup. Figure 5). Notably, this observed trend in editing percentage correlates very well to the original fluorescence localization ratios: after normalization of the means to the negative and positive control values, the ratio due to A1CF-paired editing was found to be about 55% of the positive control, while RBM47 normalizes to 87%. These data indicate that the observed fluorescence equilibrium may be a good representation of actual RNA editing percentages for each experiment.
In order to evaluate the effect that different lengths of substrates may have on the efficiency of RNA editing, APOB RNA insertions of length 27 nt, 48 nt, and 102 nt were compared with this assay to observe any effects the increased lengths may have on editing efficiency. The original construct containing 27 nt of APOB and the 48 nt substrate both had identical trends for editing efficiency, with A1CF (mean localization ratio of 0.693 ± 0.143) being less efficient than RBM47 (1.01 ± 0.205) as shown in Sup. Figure 6A. When the length of inserted APOB RNA was increased to 102 nt, it was found that APO1+A1CF and APO1+RBM47 showed an equivalent fluorescence shift (1.11 ± 0.190 and 1.07 ± 0.114 respectively) (Sup. Figure 6A). Despite the apparently enhanced levels of RNA editing for A1CF, the Amplicon NGS result of these RNA extracts revealed that both A1CF and RBM47 had an increased degree of C-to-T transitions, with RBM47 still having the greater percentage of overall editing (Sup. Figure 6B). Thus, it appears that the 102 nt substrate somehow increased the overall efficiency of editing for both cofactors, leading to a saturation in fluorescence signal that was too strong for discerning differences via the assay system with the parameters used. This system could be further optimized to fine-tune the fluorescent signals for the proper comparison of longer RNA substrate editing. However, for the purposes of this study, the original minimal APOB sequence of 27 nt was utilized for additional experiments.
Identification of RBM47 domains required for APO1/RBM47 RNA editing activity
Before the discovery of RBM47, several studies sought to characterize the APO1/A1CF interaction and determine the minimal functional domains needed for RNA editing activity [18, 42, 43]. These reports showed that residues 1–320 of A1CF were weakly able to bind APO1 and showed partial editing activity, while residues 1–391 had almost full activity. Using the assay system described herein, we similarly characterized the domains of RBM47 required for RNA editing by testing a series of C-terminal deletions of the full length RBM47 in the editor construct (Figure 2D). The result of this analysis is shown in Figure 2E. No significant difference was observed between mCherry (negative control) and the shortest deletion RBM47–330. Construct RBM47–360 showed significant RNA editing compared to the negative control, and all constructs with 406 residues or longer had activity comparable to that of the full-length protein of 593 amino acids. Notably, a sequence alignment indicates that RBM47 residues 1–406 are roughly analogous to A1CF 1–391 (Sup. Figure 7).
Editing is observed for additional RNA targets by either APO1/A1CF or APO1/RBM47
We next attempted to adapt this system for testing differences in how A1CF and RBM47 can affect APO1 editing of different RNAs using some of the candidate substrates as reported earlier [22, 24, 25]. The NMR structure of the targeted RNA sequence of APOB reveals a stem-loop structure containing the edited cytosine in the loop and a ten-residue mooring sequence in the stem [40], and RNA secondary structure prediction of a sampling of these observed editing sites suggests that some of them may have a similar local structure (Sup. Figure 8) [44, 45]. In an attempt to discover any trends in substrate targeting conferred when APO1 is paired with either cofactor, we replaced the APOB RNA substrate region in the reporter construct with 48-nucleotide segments encoding twelve additional RNA targets. Ten of these were adapted from candidate mice genes [24, 25], whereas another came from an exonic region of the human neurofibromatosis-1 tumor suppressor gene (NF1), for which editing by APO1 has previously been implicated in peripheral nerve-sheath tumors formed during type-I neurofibromatosis [46, 47]. In addition, a designer RNA that was engineered to have no stable predicted secondary structure (flatRNA) was included with the intention of testing if the APO1 editosome can edit linear RNA in this experimental setting. A full description of all tested sequences is shown in Sup. Figure 9.
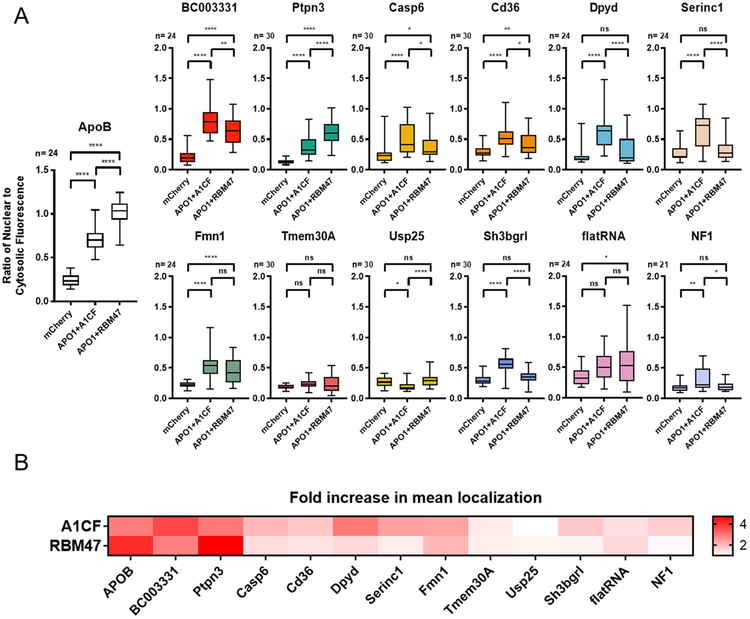
The results of each independent editing assay of these RNA substrates are summarized in Figure 3, with complete data available in Sup. Figure 10. Broadly speaking most of the tested reporter constructs showed some level of editing in the presence of APO1 with either A1CF or RBM47. Similar to what was observed for APOB, Ptpn3 RNA showed higher editing activity in the presence of RBM47 than with A1CF; this was the only other tested substrate that displayed this trend. Conversely, seven of the remaining nine known RNA targets of APO1 showed higher editing by A1CF than by RBM47. The results showed little editing for the reporters adapted from the Tmem30A and Usp25 genes, suggesting that the presence of the canonical mooring sequence alone is not sufficient to be efficiently edited by the APO1 editosome. Interestingly, the NF1 RNA sequence also showed some levels of editing with A1CF but not with RBM47 as a cofactor (P = 0.0014, 0.9615 respectively), although only weak editing activity was observed in this case. The intentionally linear substrate (flatRNA) showed possible editing in the presence of RBM47 (P = 0.0372) but no statistical significance was observed for levels with A1CF (P = 0.1015). Overall, these data suggest that the RNA editing machinery is very selective for specific substrates and is very tightly regulated, even in the context of this overexpression-based system.
Figure 3. Comparison of the editing activity on a set of candidate RNA substrates by the human APO1 editosome paired with either A1CF or RBM47.
(A) Quantification of editing on different RNA targets by measuring the fluorescent localization changes (expressed as changes of the nuclear to cytosolic fluorescence ratio). Different substrates are favored by either A1CF or RBM47, with most of the alternative RNA substrates favoring A1CF, but APOB and Ptpn3 both favoring RBM47. Results are displayed as box-and-whisker plots with the results of transfection with APO1 alone removed for clarity, as no substrate showed any significant difference from mCherry under this condition. Significance was calculated by one-way ANOVA and Tukey post-test with multiplicity-adjusted P-values represented as ns = not significant when P > 0.05, * = P < 0.05, ** = P < 0.01, *** = P < 0.001, **** = P < 0.0001. Full charts with the excluded APO1-only values are available in Sup. Figure 10. (B) Temperature-bar chart summarizing the editing activity of different tested RNA targets by the APO1 editosome paired with either A1CF or RBM47. Data is normalized by dividing the mean value for each test by its comparison mCherry negative control value to get a “x-fold increase” ratio. A majority of the substrates are edited by A1CF, with few edited by RBM47; only two substrates were preferentially edited by the latter cofactor. Statistical analyses of this particular comparison were not completed and it is only intended to help display general trends.
Comparison of RNA editing by mouse APO1/A1CF and APO1/RBM47
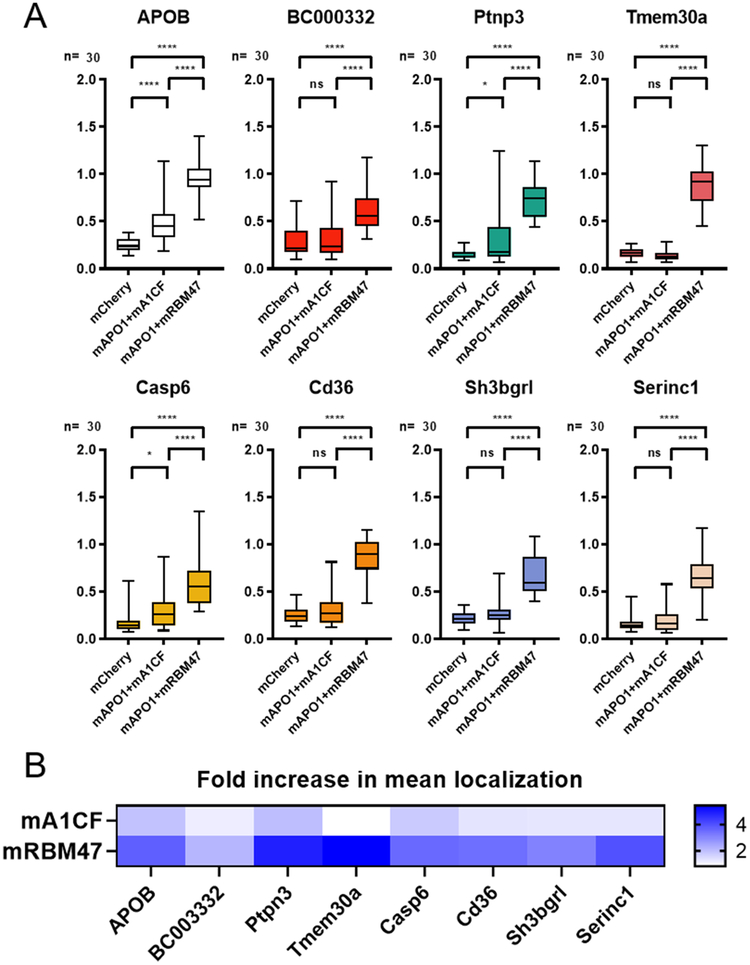
All of the comparisons described above were made using protein sequences from the human APO1, A1CF, and RBM47 genes. Given many of the tested RNA substrates were adapted from mouse mRNA transcripts, it was important to instead see the relative editing efficiency of these RNA substrates using mouse APO1, A1CF, and RBM47. Two new editor constructs were made to pair mouse APO1 (mAPO1) with either mouse A1CF (mA1CF) or RBM47 (mRBM47), and their relative editing efficiency is shown in Figure 4. Remarkably, there are very apparent differences when comparing the overall trend of these mouse proteins to the human equivalents: of the tested sequences, only APOB, Ptpn3, and Casp6 had a statistically significant difference in RNA editing levels in the presence of mA1CF compared to the negative control; the rest showed no significant editing at all. However, all eight tested substrates showed very strong editing in the presence of mRBM47. The remarkable difference observed between human and mouse APO1/A1CF and APO1/RBM47 in this assay system (see Sup. Figure 11 for a side-by-side comparison) suggests that there are many subtleties in how APO1 has evolved to target RNA substrates in different species, opening the door to a number of potential future investigations in the field.
Figure 4. Comparison of the editing activity on candidate RNA substrates with mouse-derived APO1, A1CF and RBM47.
(A) Quantification of fluorescence localization ratio for several of the same RNA reporter constructs, this time in the presence of either mouse APO1 with mouse A1CF, mouse APO1 with mouse RBM47, or mCherry alone as a negative control. Results are displayed as box-and-whisker plots with significance values calculated by one-way ANOVA with Tukey post-test. Multiplicity-adjusted P-values are represented as ns = not significant when P > 0.05, * = P < 0.05, ** = P < 0.01, *** = P < 0.001, **** = P < 0.0001. For the most part, mouse APO1 + RBM47 displayed efficient editing for these substrates. (B) Temperature-bar chart displaying the ratio of the mean localization ratio of A1CF or RBM47 to the negative control for each substrate, in order to give an approximate fold-increase value for comparison of RNA editing. Statistical analyses of these comparisons were not completed and are only for the purpose of making qualitative comparisons.
DISCUSSION
Despite an increasing number of studies looking into the function and implications of APO1 and other RNA editing proteins (such as Adenosine Deaminases Acting on RNA, or ADAR), the techniques for investigating the biochemistry of this class of enzymes remain relatively tedious. We have described here a methodology for making quantitative comparisons of editing by the minimal APO1 RNA editosome (i.e. the APO1 deaminase paired with one of the two known necessary cofactors). Using this method, we have successfully characterized the required domains of RBM47 needed for its complementation activity of APO1 in editing APOB RNA; among the series of deletion constructs tested for RBM47, at least 360 residues of the N-terminus are necessary for partial editing complementation, and including up to residue 406 from the N-terminus is sufficient to recreate nearly wild-type levels of editing activity. Notably, a sequence alignment between A1CF and RBM47 indicates that RBM47 residues 1–406 are roughly analogous to A1CF 1–391, which has previously been shown to be the minimum functional A1CF construct that can convey near-native complementation activity. The domain C-terminal to residue 406 of RBM47 has no obvious homology with A1CF, and the relevant biological functions of these regions have yet to be determined. It may be possible that they are involved with further regulation and tissue-specification of editing activity.
It is clear that human A1CF can mediate APO1 editing activity on APOB RNA in this cell-based assay system, but its effectiveness on activity is about 73% of RBM47; this trend is consistent with the relative impact that knockout of mouse A1cf and Rbm47 has on Apob editing activity in vivo as was recently reported [27]. This same trend was observed when testing the mouse homologs of these proteins with this cell-based assay system, although mA1CF was observed to be slightly weaker for aiding APOB editing activity: mA1CF is only about 52% as effective at inducing editing as mRBM47. There were also stark differences when looking at the effect of human vs. mouse genes on editing alternative substrates. Human APO1 with RBM47 only preferentially favored editing of the APOB and Ptpn3 targets, while a majority of the tested substrates saw more efficient editing with A1CF, and with the exception of APOB none of the tested substrates saw very strong editing overall. By comparison, the mouse homologs efficiently edited all of the mouse-derived targets when mRBM47 was present, and very little activity was generally observed with mA1CF. Given that A1CF and RBM47 are both highly conserved between the two species (93.6 and 94.7 percent identity, respectively), it is possible that the less conserved APO1 (70.7% identity) may play a larger role in substrate selectivity than previously expected. Further studies will need to be done to clarify additional mechanisms of specificity.
For the most part, this data correlates well with what has been suggested by mouse in vivo studies on substrate specificity of the two cofactors [27], and many of the substrates shown to be favored by RBM47 in that past work were similarly favored here. However, a few key differences stand out. First, the human-derived editor constructs showed very different trends from what was observed for several of these substrates in vivo, but these differences may be due to the aforementioned subtleties of substrate specificity between human and mice orthologs. This is particularly interesting given that for the most part, A1CF is generally favored within the human system instead, even though there are often no analogous regions in the 3’UTRs of the equivalent human genes.
Another key difference was that editing of endogenous Sh3bgrl mRNA was preferentially affected by the mouse A1cf knockout in vivo [27], whereas the Sh3bgrl substrate showed no statistically significant levels of editing in the presence of mouse mAPO1 and mA1CF with this assay. This may be due to differences in the system: it is possible the human-derived HEK293T cells failed to make a key post-translational modification the mouse proteins needed, or perhaps some other regulatory cofactor is missing. An in vivo enzymatic regulatory system is often highly complex, and this fluorescence assay also does not take into account the interplay of having both cofactors present at the same time, as would be the case in a living organism. Alternatively, this simulated substrate may have simply failed to recreate a key structural feature that mA1CF needs for targeting, despite seeing editing by mRBM47. Given that the length of the substrate chosen does have an effect on the relative strength of APOB editing mediated by A1CF and RBM47, it is possible that further optimization may be necessary for clarification of this discrepancy.
The assay system described here is very useful as a detailed mechanistic study of the enzyme complexes and substrate specificity in a cellular environment. This cell-based editing system eliminates the need for purification of the individual enzymatic components as soluble proteins, and removes some of the high cost and difficulties of working with RNA in vitro. There is no need to purchase or purify the substrates individually, and it no longer relies on an error-prone reverse transcriptase to detect and display editing as is often done when using the common method of poisoned primer extension [6]. This assay can detect subtle differences in editing thanks to a strong sensitivity in detecting small changes to the RNA transcript equilibrium, and because observation is directly on living cells, it is also relatively low cost and high throughput. This assay design, in principle, allows for adaptation to any desired RNA substrate to be tested. Although this assay system was optimized for RNA editing by APO1 editosome, it is also potentially adaptable towards other RNA deaminases, such as the ADAR family [48, 49]. Although there are still many ways the work described herein could be optimized and improved, it is our hope and anticipation that this strategy can aid further comprehensive understanding of the biochemical and biological functions of APO1 and other RNA-editing enzymes in the future.
Materials and Methods
Cloning
All primers and final DNA sequences are available as a part of Supplemental File 1. All relevant constructs were made using In-Fusion cloning and PrimeStar MAX (Takara) high fidelity PCR as applicable. Mutagenic PCR was done using primers (ordered from IDT DNA) designed to amplify the entire vector as per the manufacturer’s recommended design. Synthetic double-stranded DNA geneblocks were all ordered as GeneStrings from ThermoFisher. Human codon-optimized geneblocks of FLAG-tagged APOBEC1 (NCBI sequence ID # AAA64230.1) and HA-tagged A1CF (NCBI # NP_620310.1) including the F2A and T2A self-cleaving peptides, were ordered as three fragments of 750 to 1000 bases from Invitrogen for insertion into a pcDNA3.1(+) vector. Possibly due to toxicity of either A1CF or APOBEC1 (or the combination of both), it was extremely difficult to clone this construct in E coli thanks to the leaky CMV promoter and so an additional geneblock of FLAG-APO1 with an inserted intronic region from the type 2 adenovirus RNAse gene was ordered; this method was used in the past by our lab to avoid toxicity in E. coli due to a frame shift induced by the intron[50]. This editor construct was then modified, first by replacing A1CF with an RBM47 geneblock (NCBI # NP_001092104.1) to create the alternative cofactor clone, then by removing A1CF completely (keeping APO1, the T2A peptide, and mCherry) and both APO1 and A1CF (leaving only mCherry). These four constructs were the basis for most experiments. Additional RBM47 truncations were generated using primers designed for deletions of the desired areas. The mouse gene equivalent editor constructs for APO1 (NCBI # NM_031159.3), A1CF (NCBI # NM_001081074.2) and RBM47 (NCBI # NM_139065.3) were ordered as two additional fragments, also human-codon optimized, and inserted into the same starting vector.
Reporter constructs were all built on an in-house pCIneo (Promega) foundation with inserted eGFP, where the substrate region and MAPKK nuclear export sequence are located C-terminal to the eGFP domain. The original design contained both a nuclear export sequence upstream of the substrate and a nuclear localization sequence downstream of the substrate, with the expectation that the NLS would be able to override the NES; this was found to not work so well, so was later simplified to only using the nuclear export sequence downstream of the substrate. Accordingly, a single geneblock containing three different strengths of nuclear export sequences (MAPKK, REV, and IκBα), the APOB substrate, and an SV40 NLS was ordered, and the MAPKK sequence was ultimately rearranged with an additional set of primers to remove the NLS and place the NES after the substrate. Further substrates replacing APOB with 48-base insertions from other targets, including the longer versions of APOB, were generated with a single round of PCR using long-oligo (50–60 base) sets. All clones were amplified in E. coli overnight cultures in LB media, miniprepped using a ThermoFisher miniprep kit, and confirmed by Sanger sequencing (Genewiz).
Transfection
These experiments all utilized HEK293T cells passaged in DMEM media with 10% FBS. Transfection growth was done on 8-well glass slides (CellVis) previously coated with 0.1 mg/mL poly-D-lysine (Sigma). After reaching approximately 90% confluency, cells were resuspended in DMEM via trypsin-EDTA digestion and counted using a haemocytometer. The cells were then diluted to an approximate concentration of 250,000 to 300,000 cells/mL – a relatively low value, in order to ensure a clean monolayer for visualization. 250 μL of cells were added to each well. After an initial adherence and growth period of 20 hours, the cells were transfected with X-tremeGENE 9 transfection reagent (Sigma): 50 μL master mixes were made by combining 1 μL of a reporter construct at 50 ng/uL and 5 μL of an editor construct at 100 ng/μL with 44 μL of OPTI-mem reduced serum media (Thermo Fisher), adding 1.5 μL of reagent, and allowing it to sit at room temperature for 30 minutes. 15 μL of each mix was then added dropwise to a particular well, and expression was allowed to occur for 48 hours at 37C, 5% CO2.
Fluorescence confocal microscopy
These experiments made use of live cell microscopy on a Zeiss LSM-700 inverted confocal microscope. It was found that ideal visualization for analysis was through a 40× water-immersion objective to maximize the number of easily countable cells per image. Prior to visualization, the DMEM media was aspirated and cells were washed once with 250 μL of PBS; they were then allowed to incubate with a 5 μg/mL solution of Hoechst 33342 nuclear stain diluted in PBS for 15 minutes. This stain was then aspirated and cells were rinsed with PBS two more times and then stored in imaging buffer (140 mM NaCl, 2.5 mM KCl, 1.8mM CaCl2, 1.0 mM MgCl2, 20 mM Hepes 7.4, 5 mM glucose). All imaging was done at a higher laser intensity (generally around 15–20%) and lower gain (approximately 500–600 units) in order to maximize the observed signal-to-noise. The excitation wavelengths used for Hoechst 33342, eGFP, and mCherry were 405, 488 and 555 nm respectively; emission band-pass filters were set to 400–480 nm, 490–555, and 555–700 nm respectively. Images were captured as multichannel16-bit grayscale intensity images 1012 × 1012 pixels across, using 2-pass line averaging for smoothing and a pixel dwell time of 0.80 μsec. For each well, approximately 3–5 images were captured, allowing for measurement of around 20–30 cells.
RNA extraction for Amplicon NGS
After image capture, the imaging buffer was aspirated off and 250 μL of Trizol (Thermo Fisher) was directly added to the wells, with gentle pipetting to lyse. The manufacturer’s recommended protocol for Trizol was followed from then on, with 50 μL of chloroform added and about 40 μL of aqueous phase extracted. After isopropanol precipitation the total RNA was reverse transcribed with ProtoscriptII (NEB) for one hour at 42C. This reaction made use of a specific primer containing an added sequencing primer region that targeting the region just downstream of the substrate target within the reporter transcript. The resulting cDNA was then amplified with a forward-facing library tag primer for the targeted region of approximately 200 bases total. This PCR product was cleaned up using a spin column PCR cleanup kit (Thermo) and submitted for Amplicon next-gen sequencing by Genewiz. The raw sequencing reads were filtered and the primers trimmed off, then aligned to the reference sequence in UGENE [51] (see Sup. Figure 5). The coverage data was exported in order to find the number of A, T, C or G bases observed at each aligned position, allowing for a comparison of the % of total bases for each, effectively showing the amount of editing occurring for each experimental condition. The edited site (and only major transition observed) appeared at position 87 for this sequence.
Analysis of fluorescence images
All image analysis was done using the LSM Toolbox plugin built into the FIJI distribution of ImageJ2 [52, 53]. The output LSM image file was opened in single-channel color mode and the Hoechst stain channel was used to assess the location of the nucleus for each cell. The mCherry channel was only used as a confirmation that a cell has both the eGFP reporter and mCherry-containing editor construct and was not quantified. Freehand selection was used to outline the nuclear and the cytoplasmic regions respectively within the eGFP channel, and the average intensity of each region was recorded. At least 21 cells, but ideally 30, were calculated this way; very little variation was observed within this sample size. The ratio of average nuclear to cytosolic intensity was calculated for each cell and these values were assessed in Graphpad Prism 7.0d via a one-way ANOVA analysis. The data was assumed to have a Gaussian distribution and a Tukey post-test was done to compare the means within experiments. The calculated P-values from the individual significance tests are accordingly adjusted for the inherent multiplicities and conclusions were drawn based on the observed levels of significance, with all data displayed as box-and-whisker plots showing the median and upper/lower quartiles within the box and whiskers extended to the minimum and maximum for each sample.
Supplementary Material
Acknowledgements
We thank Aida Bareghamyan of the Molecular and Computational Biology department, and the Translational Imaging Center at the University of Southern California for providing the fluorescent microscope for our study. This work is supported by the NIH grant R01GM087986 to X.S.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests
The authors report no conflict of interest with regards to this research.
References:
- [1].Gagnidze K, Rayon-Estrada V, Harroch S, Bulloch K, Papavasiliou FN. A New Chapter in Genetic Medicine: RNA Editing and its Role in Disease Pathogenesis. Trends in Molecular Medicine. 2018;24:294–303. [DOI] [PubMed] [Google Scholar]
- [2].Mingardi J, Musazzi L, De Petro G, Barbon A. miRNA Editing: New Insights into the Fast Control of Gene Expression in Health and Disease. Molecular Neurobiology. 2018;55:7717–27. [DOI] [PubMed] [Google Scholar]
- [3].Qi L, Chan THM, Tenen DG, Chen L. RNA Editome imbalance in hepatocellular carcinoma. Cancer Research. 2014;74:1301–6. [DOI] [PubMed] [Google Scholar]
- [4].Thomas JM, Beal PA. How do ADARs bind RNA? New protein-RNA structures illuminate substrate recognition by the RNA editing ADARs. BioEssays. 2017;39:1600187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Vu LT, Tsukahara T. C-to-U editing and site-directed RNA editing for the correction of genetic mutations. BioScience Trends. 2017;11:243–53. [DOI] [PubMed] [Google Scholar]
- [6].Driscoll D, Wynne J, Wallis S, Scott J. An In Vitro System for the Editing of Apolipoprotein B mRNA. Cell. 1989;58:519–25. [DOI] [PubMed] [Google Scholar]
- [7].Chen SH, Habib G, Yang CY, Gu ZW, Lee BR, Weng SA, et al. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science. 1987;238:363–6. [DOI] [PubMed] [Google Scholar]
- [8].Powell LM, Wallis SC, Pease RJ, Edwards YH, Knott TJ, Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987;50:831–40. [DOI] [PubMed] [Google Scholar]
- [9].Hirano KI, Young SG, Farese RV, Ng J, Sande E, Warburton C, et al. Targeted disruption of the mouse apobec-1 gene abolishes apolipoprotein B mRNA editing and eliminates apolipoprotein B48. Journal of Biological Chemistry. 1996;271:9887–90. [DOI] [PubMed] [Google Scholar]
- [10].Nakamuta M, Chang BHJ, Zsigmond E, Kobayashi K, Lei H, Ishida BY, et al. Complete phenotypic characterization of apobec-1 knockout mice with a wild-type genetic background and a human apolipoprotein B transgenic background, and restoration of apolipoprotein B mRNA editing by somatic gene transfer of apobec-1. Journal of Biological Chemistry. 1996;271:25981–8. [DOI] [PubMed] [Google Scholar]
- [11].Teng B, Burant C, Davidson N. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260:1816–9. [DOI] [PubMed] [Google Scholar]
- [12].Teng BB, Ochsner S, Zhang Q, Soman KV, Lau PP, Chan L. Mutational analysis of apolipoprotein B mRNA editing enzyme (APOBEC1). structure-function relationships of RNA editing and dimerization. J Lipid Res. 1999;40:623–35. [PubMed] [Google Scholar]
- [13].Salter JD, Bennett RP, Smith HC. The APOBEC Protein Family: United by Structure, Divergent in Function. Trends Biochem Sci. 2016;41:578–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat Rev Immunol. 2004;4:868–77. [DOI] [PubMed] [Google Scholar]
- [15].Swanton C, McGranahan N, Starrett GJ, Harris RS. APOBEC Enzymes: Mutagenic Fuel for Cancer Evolution and Heterogeneity. Cancer Discov. 2015;5:704–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Anant S, Davidson NO. Identification and regulation of protein components of the apolipoprotein B mRNA editing enzyme. A complex event. Trends Cardiovasc Med. 2002;12:311–7. [DOI] [PubMed] [Google Scholar]
- [17].Lellek H, Kirsten R, Diehl I, Apostel F, Buck F, Greeve J. Purification and molecular cloning of a novel essential component of the apolipoprotein B mRNA editing enzyme-complex. Journal of Biological Chemistry. 2000;275:19848–56. [DOI] [PubMed] [Google Scholar]
- [18].Mehta A, Driscoll DM. Identification of domains in apobec-1 complementation factor required for rna binding and apolipoprotein-B mRNA editing. Rna. 2002;8:69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sowden M, Hamm JK, Spinelli S, Smith1 HC, Smith HC. Determinants involved in regulating the proportion of edited apolipoprotein B RNAs. Rna. 1996;2:274–88. [PMC free article] [PubMed] [Google Scholar]
- [20].Chester A, Somasekaram A, Tzimina M, Jarmuz A, Gisbourne J, O’Keefe R, et al. The apolipoprotein B mRNA editing complex performs a multifunctional cycle and suppresses nonsense-mediated decay. EMBO J. 2003;22:3971–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Fossat N, Tourle K, Radziewic T, Barratt K, Liebhold D, Studdert JB, et al. C to U RNA editing mediated by APOBEC1 requires RNA-binding protein RBM47. EMBO Rep. 2014;15:903–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Snyder EM, McCarty C, Mehalow A, Svenson K, Murray S, Korstanje R, et al. APOBEC1 complementation factor (A1CF) is not required for C-to-U RNA editing in vivo. RNA (New York, NY). 2017:rna.058818.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rayon-Estrada V, Harjanto D, Hamilton CE, Berchiche YA, Gantman EC, Sakmar TP, et al. Epitranscriptomic profiling across cell types reveals associations between APOBEC1-mediated RNA editing, gene expression outcomes, and cellular function. Proc Natl Acad Sci U S A. 2017;114:13296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Blanc V, Park E, Schaefer S, Miller M, Lin Y, Kennedy S, et al. Genome-wide identification and functional analysis of Apobec-1-mediated C-to-U RNA editing in mouse small intestine and liver. Genome Biology. 2014;15:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rosenberg BR, Hamilton CE, Mwangi MM, Dewell S, Papavasiliou FN. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3′ UTRs. Nature Structural and Molecular Biology. 2011;18:230–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cole DC, Chung Y, Gagnidze K, Hajdarovic KH, Rayon-Estrada V, Harjanto D, et al. Loss of APOBEC1 RNA-editing function in microglia exacerbates age-related CNS pathophysiology. Proc Natl Acad Sci U S A. 2017;114:13272–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Blanc V, Xie Y, Kennedy S, Riordan JD, Rubin DC, Madison BB, et al. APOBEC1 complementation factor (A1CF) and RBM47 interact in tissue-specific regulation of C to U RNA editing in mouse intestine and liver. Rna. 2018:rna.068395.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gommans WM, McCane J, Nacarelli GS, Maas S. A mammalian reporter system for fast and quantitative detection of intracellular A-to-I RNA editing levels. Anal Biochem. 2010;399:230–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Reenan RA. Molecular determinants and guided evolution of species-specific RNA editing. Nature. 2005;434:409–13. [DOI] [PubMed] [Google Scholar]
- [30].Wang Y, Park S, Beal PA. Selective Recognition of RNA Substrates by ADAR Deaminase Domains. Biochemistry. 2018;57:1640–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kankowski S, Förstera B, Winkelmann A, Knauff P, Wanker EE, You XA, et al. A Novel RNA Editing Sensor Tool and a Specific Agonist Determine Neuronal Protein Expression of RNA-Edited Glycine Receptors and Identify a Genomic APOBEC1 Dimorphism as a New Genetic Risk Factor of Epilepsy. Frontiers in Molecular Neuroscience. 2018;10:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Severi F, Conticello SG. Flow-cytometric visualization of C>U mRNA editing reveals the dynamics of the process in live cells. RNA Biology. 2015;12:389–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kilgard R, Heim AB, Tsien RY. Improved green fluorescence. Nature 1995. p. 663–4. [DOI] [PubMed] [Google Scholar]
- [34].Di Ventura B, Kuhlman B. Go in! Go out! Inducible control of nuclear localization. Current Opinion in Chemical Biology. 2016;34:62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Henderson BR, Eleftheriou A. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Experimental Cell Research. 2000;256:213–24. [DOI] [PubMed] [Google Scholar]
- [36].Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: Definition, function, and interaction with importin α. Journal of Biological Chemistry. 2007;282:5101–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature Biotechnology. 2004;22:1567–72. [DOI] [PubMed] [Google Scholar]
- [38].Liu Z, Chen O, Wall JBJ, Zheng M, Zhou Y, Wang L, et al. Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Scientific Reports. 2017;7:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ryan MD, King AMQ, Thomas GP. Cleavage of foot-and-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. Journal of General Virology. 1991;72:2727–32. [DOI] [PubMed] [Google Scholar]
- [40].Maris C, Masse J, Chester A, Navaratnam N, Allain FH-T. NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor. RNA (New York, NY). 2005;11:173–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Smith HC. Analysis of Protein Complexes Assembled on Apolipoprotein B mRNA for Mooring Sequence-Dependent RNA Editing. Methods. 1998;15:27–39. [DOI] [PubMed] [Google Scholar]
- [42].Blanc V, Henderson JO, Kennedy S, Davidson NO. Mutagenesis of Apobec-1 Complementation Factor Reveals Distinct Domains That Modulate RNA Binding, Protein-Protein Interaction with Apobec-1, and Complementation of C to U RNA-editing Activity. Journal of Biological Chemistry. 2001;276:46386–93. [DOI] [PubMed] [Google Scholar]
- [43].Galloway CA, Kumar A, Krucinska J, Smith HC. APOBEC-1 complementation factor (ACF) forms RNA-dependent multimers. Biochemical and Biophysical Research Communications. 2010;398:38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proceedings of the National Academy of Sciences. 2004;101:7287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Reuter JS, Mathews DHD, Eddy S, Mello C, Conte D, Chow J, et al. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mukhopadhyay D, Anant S, Lee RM, Kennedy S, Viskochil D, Davidson NO. C→U Editing of Neurofibromatosis 1 mRNA Occurs in Tumors That Express Both the Type II Transcript and apobec-1, the Catalytic Subunit of the Apolipoprotein B mRNA–Editing Enzyme. The American Journal of Human Genetics. 2002;70:38–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Skuse GR, Cappione AJ, Sowden M, Metheny LJ, Smith HC. The neurofibromatosis type I messenger RNA undergoes base-modification RNA editing. Nucleic Acids Research. 1996;24:478–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Goodman RA, Macbeth MR, Beal PA. ADAR Proteins: Structure and Catalytic Mechanism. Current Topics in Microbiology and Immunology 2011. p. 1–33. [DOI] [PubMed] [Google Scholar]
- [49].Functions Nishikura K. and Regulation of RNA Editing by ADAR Deaminases. Annual Review of Biochemistry. 2010;79:321–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Xiao X, Yang H, Arutiunian V, Fang Y, Besse G, Morimoto C, et al. Structural determinants of APOBEC3B non-catalytic domain for molecular assembly and catalytic regulation. Nucleic Acids Research. 2017;45:7494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Okonechnikov K, Golosova O, Fursov M, Varlamov A, Vaskin Y, Efremov I, et al. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–7. [DOI] [PubMed] [Google Scholar]
- [52].Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 2017;18:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: An open-source platform for biological-image analysis. Nature Methods. 2012;9:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.