Abstract
Background:
Alcohol use disorder (AUD) is a wide-spread, heritable brain disease, but few studies have linked genetic variants or epigenetic factors to brain structures related to AUD in humans, due to many factors including the high-dimensional nature of imaging and genomic data.
Methods:
To provide potential insights into the links among epigenetic regulation, brain structure and AUD, we have performed an integrative analysis of brain structural imaging and blood DNA methylome data from 52 AUD and 58 healthy control subjects collected in the Nathan Kline Institute-Rockland Sample (NKI-RS).
Results:
We first found that AUD subjects had significantly larger insular surface area than healthy controls in both left and right hemispheres. We then found that 7,827 DNA methylation probes on the HumanMethylation450K Beadchip had significant correlations with the right insular surface area (FDR<0.05). Furthermore, we showed that 44 of the insular surface area-correlated methylation probes were also strongly correlated with AUD status (FDR<0.05). These AUD-correlated probes are annotated to 36 protein coding genes, with 16 genes (44%) having been reported by others to be related to AUD or alcohol response, including TAS2R16 and PER2. The remaining 20 genes, in particular ARHGAP22, might represent novel genes involved in AUD or responsive to alcohol.
Conclusion:
We have identified 36 insular surface area- and AUD-correlated protein-coding genes that are either known to be AUD- or alcohol-related or not yet reported by prior studies. Therefore, our study suggests that the brain imaging-guided epigenetic analysis has a potential of identifying possible epigenetic mechanisms involved in AUD.
Keywords: Alcohol use disorder, alcohol response, insular cortex, surface area, DNA methylation
INTRODUCTION
Alcohol use disorder (AUD) has been considered a brain disease, and extensive brain imaging studies have revealed brain structural and functional alterations related to AUD or alcohol response (Litten et al., 2015). Moreover, AUD has been found to be 40–70% heritable, and numerous genetic, epigenetic and genomic studies have identified many genes that are linked to or associate with AUD or are responsive to alcohol uptake (Reilly et al., 2017, Warden and Mayfield, 2017, Palmisano and Pandey, 2017). Clearly, in order to develop effective prevention or treatment strategies, it is essential to identify the genetic variants that link to brain features causing or impacted by AUD.
The insular cortex has been suggested to be involved in or impacted by AUD and other addictive behaviors, given its critical role in multiple brain processes including cognitive functioning, perception, motor control, self-consciousness, and emotional regulation (Naqvi and Bechara, 2009). For example, subjects with alcohol dependence or cocaine addiction have reduced insular cortical thickness (Makris et al., 2008, Durazzo et al., 2011) or reduced posterior insula activity during risky choice tasks (Stewart et al., 2014). However, the opposite relationships between insular structural features or activity and AUD or other addictive behaviors have also been reported. Bilateral insula damage was found to associate with loss of addictive craving (Naqvi et al., 2007), indicating a requirement for the insular activity in addiction. Similarly, it has been reported that the insula activity increased in response to drug-related cues (Schacht et al., 2013), and cue-induced insula activity was positively associated with severity of alcohol addiction (Claus et al., 2011). In our recent study using the Nathan Kline Institute-Rockland Sample (NKI-RS) and the Human Connectome Project (HCP) sample, we did not find a group difference between AUD and healthy controls (HC) in the insular cortical thickness, although AUD impacted the correlation of insular cortex thickness and neuroticism (Zhao et al., 2017). To interpret these conflicting reports, in particular activation of the insula, an interoceptive tuning mechanism has been proposed (Droutman et al., 2015). That is, the insular function likely helps optimize one’s choice of stimulus by internally adjusting the reward value of that stimulus so that the stimulus best satisfies individual’s internal and external environmental needs. Overall, these studies have supported an important role for the insular cortex in alcohol and other drug addictions, and thus the insula has been the subject of our current study.
Many genetic and genomic studies have led to the identification of genes which are linked to or associated with AUD or exhibit differential expression or epigenetic modifications in response to alcohol treatment [reviewed in (Reilly et al., 2017, Warden and Mayfield, 2017, Palmisano and Pandey, 2017, Zhang and Gelernter, 2017, Tulisiak et al., 2017)]. Based on the Ethanol-Related Gene Resource (ERGR) database (Guo et al., 2009), in human alone 3,311 alcohol-related genes have been found. As epigenetic regulation is critical for regulation of gene expression, emerging epigenetic or epigenomic studies have been performed to reveal epigenetic variants, in particular DNA methylation, related to AUD or alcohol response (Schuebel et al., 2016, Reilly et al., 2017, Warden and Mayfield, 2017, Palmisano and Pandey, 2017). Because of the convenience of collecting blood samples combined with the problem of access into the brain tissues of living subjects, blood has been frequently used in DNA methylation studies, leading to the identification of hundreds to thousands of differentially methylated probes (Philibert et al., 2012, Zhang et al., 2013a, Zhang et al., 2013b, Zhao et al., 2013, Ruggeri et al., 2015, Xu et al., 2017, Liu et al., 2018). Despite these encouraging progress, the major genes responsible for the AUD pathogenesis have not been identified yet, which is a common problem, dubbed “missing heritability”, for many other psychiatric diseases (Zhao and Castellanos, 2016). Furthermore, it is even more challenging to identify the genes that link to brain structure and function and eventually to behaviors caused by alcohol drinking or AUD, with limited progress so far (Ruggeri et al., 2015). Several factors have been known to account for this situation, including the heterogeneity of AUD, the lack of large samples collected systematically across different levels of analysis, and the need to develop methods for integrated analysis of large-scale, high-dimensional brain imaging and genetic datasets.
In this study, we integrate imaging endophenotypes with genome-wide methylome analysis to boost statistical power using a community cohort dataset, NKI-RS, which contains brain imaging data and blood DNA sample. Our results showed that integrated imaging epigenetic analysis will help identify significant AUD-associated genes that would not be possibly detected by associating directly methylation probes with AUD in studies with small sample size. This is presumably due to the stronger link between brain structure and AUD than that between methylation probes and AUD status. In our prior work we used the NKI-RS dataset to reveal that AUD diagnostic history impacts the correlation of personality traits and the brain cortical thicknesses (Zhao et al., 2017). As the insular cortical thickness was not correlated with AUD, we therefore tested whether other measurements of the insular cortex might show correlations with AUD. We first found that AUD subjects had significantly larger insular surface area in both left and right hemispheres than HC. We then identified the DNA methylation probes that were correlated with the right insular surface area, while no DNA methylation probes were found to associate with the left insular surface area after multiple testing adjustment. Finally we found that 44 of those right insular surface area-correlated probes turned out to correlate with AUD status.
MATERIALS AND METHODS
Participants
A sample of 122 participants with both brain image and blood DNA samples available were selected from NKI-RS (Nooner et al., 2012). Specifically, there were 61 subjects with lifetime AUD status as of March 2015. All AUD subjects except one had past AUD diagnosis with an average of 26.1 (±16.1) years of regular alcohol consumption defined as drinking at least once per month. Five of AUD subjects reported no alcohol use within the past 12 months. More than half of AUD subjects (52%) had one or more comorbid conditions. Among those with co-morbid conditions, 12 (44.4%) AUDs co-morbid with cannabis use disorders (CUD), 3 (11.1%) comorbid with cocaine use disorders (COD), and 5 (18.5%) comorbid with both CUD and COD. Those 61 AUDs were group matched with 61 healthy controls (HC) on age, gender, and race (i.e., White vs. others). Healthy controls were defined as those who had no diagnosis or condition on Axis I disorders. None of the study subjects had a medical history of convulsion, seizures, epilepsy, schizophrenia, serious head injury, or psychotic illness. After DNA methylation profiling, brain image pre-processing and behavioral data checking, we have complete data on a total of 110 subjects including 52 AUDs and 58 HCs.
Structural brain image preprocessing
T1-weighted Structural MRI image pre-processing using the recon-all pipeline from FreeSurfer version 5.3.0 and data quality checking were described elsewhere (Zhao et al., 2017). Briefly, mean cortical surface area of 68 gyral regions based on the Desikan-Killiany atlas (Desikan et al., 2006) were extracted for each participant. Quality control of cortical measurements followed the steps outlined in the ENIGMA protocol (http://enigma.ini.usc.edu/protocols/imaging-protocols/). In this project, we were only interested in insular surface area at both hemispheres.
DNA methylation profiling and initial data preprocessing
The blood draw was performed on the first day of the visit by the NKI-RS study team (http://fcon_1000.projects.nitrc.org/indi/enhanced/sched.html). DNA from peripheral blood cells was prepared and maintained in the Biologic Core (Rutgers University’s RUCDR Infinite Biologics) and ordered through the NIMH Genetics Repository (https://www.nimhgenetics.org). Illumina Infinium HumanMethylation450K BeadChip array, covering approximately 480 K CpG sites, was used to profile the blood CpG-based DNA methylome. DNA purification, labeling, hybridization, scanning and image data processing were performed in the Microarray Core Facility at University of Texas Southwestern Medical Center.
The data pre-processing and quality checking were done using R package minfi (Aryee et al., 2014). Briefly, probes with a detection P value > 0.01 in over 5% of the samples were removed from analysis. Subset-quantile Within Array Normalization (SWAN) normalization (Maksimovic et al., 2012) was used to remove technical variability. The predicted sex was compared with the self-reported sex. We visually inspected the median of the meth and unmeth signals for each array. The M-value, defined as the log2 ratio of the intensities of methylated probe versus unmethylated probe, was used in our analysis. M-value was reported to be more statistically valid than beta value, defined as meth/(meth+unmeth), for differential analysis of methylation data (Du et al., 2010).
Imaging-guided methylation data analysis
A two-step imaging-guided approach was used to identify methylation probes correlated with AUD. Specifically, the first step is to identify a set of methylation probes significantly associated with residual insular surface area, where residual surface area was the residuals obtained by regressing surface area on ICV only to compensate individual variability in head size. By restricting the probes identified in the first step, the second step is to find out which of them are associated with AUD. In step 1, we built a linear regression model where the response is M-value and the predictors are AUD diagnosis and residual insular surface, controlling for age, sex, race, years of alcohol consumption, cannabis and tobacco use status, Hollingshead socioeconomic status (SES) score, symptomatic distress total score from trauma symptom checklist – 40 (TSC40) (Elliott and Briere, 1992), and laterality index from Edinburgh Handedness Questionnaire (handedness) (http://www.brainmapping.org/shared/Edinburgh.php). To remove unwanted variation (RUV) due to hidden batch effects (Gagnon- Bartsch et al., 2013), cell type heterogeneity, and other confounders common in epigenome-wide association studies (Liang and Cookson, 2014), we used the RUVfit() function in R package missMethyl (Phipson et al., 2016) to detect unwanted factors using control genes and included them in the model as covariates. Empirical Bayes (eBayes) p-values (Smyth, 2004) were obtained to estimate the associations between methylation probes and insular surface area and the associations between methylation probes and AUD for all probes. In step 1, we identify probes significantly associated with insular area controlling for the false discovery rate (FDR) at 0.05. In step 2, we calculated the FDR adjusted q-value for AUD only on the probes significantly related to the insula surface area. All analyses were done using functions in R package missMethyl (Phipson et al., 2016).
Bioinformatic and protein-protein interaction network analyses
DNA methylation probes were annotated to the human genome following the Illumina protocol. Protein-coding genes whose methylation level was correlated with the insular surface area were selected for Gene Ontology (GO) analysis with a focus on biological processes, using the PANTHER web tool (Mi et al., 2013). Protein pathway analysis was also performed in PANTHER. To find out which of AUD-correlated genes identified in our study are related to AUD in other studies, a list of 3,311 human genes from the Ethanol-Related Gene Resource (ERGR) database (Guo et al., 2009) was extracted and compared with our study’s AUD-correlated genes. In addition, a total of 771 genes related to AUD or alcoholism collected by the Public Health Genomics Knowledge Base (v4.0), which was based on published literature and updated on September 6, 2018, were also included for analysis. For protein-protein interaction network analysis, those insular surface area-correlated protein coding genes were mapped to human protein-protein interactions collected and updated by the BioGRID database (Chatr-Aryamontri et al., 2017) on Aug. 25, 2018. Protein-protein interaction networks were visualized by using Cytoscape.
RESULTS
Correlation of the insular surface area with AUD diagnosis status
The HC and AUD subjects do not differ significantly on age, sex, race, handedness, and social economic status (Table 1). The average Hollingshead SES scores ranged from 43 (HC) to 44.6 (AUD), indicating a predominantly upper-middle class SES for our study sample. The study samples were mainly right-handed as evidenced by the mean handedness scores of 65.5 and 66.2 for HC and AUD, respectively. There was a significant difference in tobacco and cannabis use status between HC and AUD subjects. Approximately 57.7% and 55.8% AUD subjects were regular smokers and cannabis users, respectively, compared to only 37.9% and 24.1% in HC subjects. In this sample, 17.3% of AUD subjects had one comorbid condition (with psychiatric or substance use disorder), and 36.5% had two or more comorbid conditions. There was no significant group difference in brain total intracranial volume.
Table 1.
Summary of the AUD subjects and health controls used in this study.
| Variables | HC | AUD |
|---|---|---|
| Number of subjects | 58 | 52 |
| Age1 | 45.8 (16.7) | 45.8 (16.4) |
| Gender: Female | 33 (56.9) | 29 (55.8) |
| Race: White | 37 (63.8) | 40 (76.9) |
| SES1 | 43 (14.2) | 44.6 (10.1) |
| Handedness1 | 65.5 (50.4) | 66.2 (40.4) |
| TSC401* | 15.74 (11.48) | 23.23 (13.52) |
| Brain ICV1 (cm3) | 1471.9 (152.2) | 1491.8 (140.0) |
| Regular smoker* | 22 (37.9) | 30 (57.7) |
| Regular cannabis user* | 14 (24.1) | 29 (55.8) |
| Number of comorbid conditions | ||
| 0 | 24 (46.2) | |
| 1 | 9 (17.3) | |
| 2 | 8 (15.4) | |
| 3 | 6 (11.5) | |
| >=4 | 5 (9.6) | |
Notes: HC, healthy controls; AUD, alcohol use disorders; SD, standard deviation; ICV, intracranial volume.
indicates significant at 0.05 level.
report Mean (SD). Otherwise, we report Frequency (%).
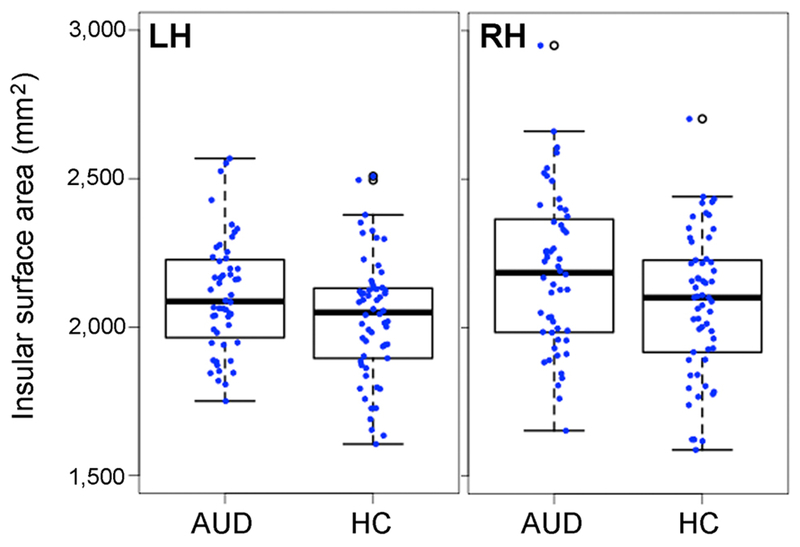
We hypothesized that the insular surface area could correlate with AUD diagnostic status and thus we used linear regression analysis to test the group difference in left and right insular surface areas. Controlling for age, sex, race, cannabis and tobacco use status, SES, TSC40, and handedness, AUD subjects had significantly larger residual insular surface areas than HC controls in both right (alpha=109 mm2, p-value < 0.01) and left (alpha=60.0 mm2, p-value = 0.03) hemispheres (Fig. 1).
Fig. 1.
Box plots showing increased left (LH) and right (RH) insular surface area in AUD group. AUD, alcohol use disorders; HC, healthy control; LH, left hemisphere; RH, right hemisphere.
Analysis of DNA methylation profiles correlated with right insular surface area
We then used the Empirical Bayes (eBayes) statistical approach to identify the probes that exhibit a significant correlation of their DNA methylation level (M-value) with the insular surface area of left and right insular cortex. A total of 7,827 DNA CpG methylation probes were significantly correlated with the right insular cortical surface area (FDR <0.05), but none correlated with the left insular surface area after the multiple testing adjustment. The top 1,000 probes are listed in Suppl. Table S1, and the top 25 probes in Table 2. Among these 25 probes, eight were negatively correlated with the right insular surface area, while the majority (68%) of the probes showed positive correlations. Furthermore, all but one probe (cg07149055 on chromosome 6) were mapped to known protein-coding genes. Among these, 14 probes have DNA methylation probes corresponding to regulatory regions including transcription start sites (TSS) and untranslated regions (UTR) of the protein coding genes, while the others are located to either gene body or specifically 1st exon. One probe, cg13201342, represents two genes, ARNT and CTSK, and thus these 24 probes represent a total of 25 protein-coding genes.
Table 2.
List of top 25 insular surface area-correlated methylation probes ranked by FDR.
| Ilumina Probe ID | Chr. | Gene Symbol | Genomic Location | Alpha | FDR | Effect Size |
|---|---|---|---|---|---|---|
| cg04566037 | 17 | KRTAP3–1 | TSS1500 | 0.00052 | 0.0021 | 0.58 |
| cg05945401 | 6 | BAT1 | Body | 0.00053 | 0.0021 | 0.58 |
| cg24026230 | 5 | NR3C1 | 5’UTR;TSS1500 | −0.00037 | 0.0021 | −0.58 |
| cg02210887 | 4 | GUCY1A3 | TSS1500 | 0.00059 | 0.0047 | 0.55 |
| cg04564935 | 17 | MED13 | TSS1500 | −0.00037 | 0.0047 | −0.56 |
| cg23677229 | 14 | GPHN | TSS1500 | 0.00045 | 0.0047 | 0.55 |
| cg24450157 | 6 | DHX16 | 1stExon;Body | −0.00039 | 0.0047 | −0.56 |
| cg27253887 | 6 | PPIL4 | TSS200 | −0.00057 | 0.0047 | −0.54 |
| cg27585345 | 1 | LDLRAD2 | TSS1500 | 0.00038 | 0.0047 | 0.55 |
| cg23719342 | 7 | TAS2R16 | TSS1500 | 0.00040 | 0.0068 | 0.54 |
| cg01513516 | 2 | TMEM163 | Body | 0.00047 | 0.0075 | 0.53 |
| cg03249947 | 2 | FBLN7 | 5’UTR;1stExon | −0.00043 | 0.0075 | −0.53 |
| cg03393445 | 8 | SH2D4A | Body | 0.00048 | 0.0075 | 0.53 |
| cg05907034 | 2 | GPR113 | TSS1500;Body | 0.00040 | 0.0075 | 0.53 |
| cg07149055 | 6 | 0.00038 | 0.0075 | 0.53 | ||
| cg25829666 | 3 | THRB | TSS1500 | −0.00057 | 0.0075 | −0.53 |
| cg26205859 | 7 | PION | Body | 0.00057 | 0.0075 | 0.53 |
| cg03044249 | 7 | WBSCR17 | TSS1500 | −0.00061 | 0.0075 | −0.52 |
| cg07107055 | 6 | ARID1B | Body | 0.00042 | 0.0075 | 0.53 |
| cg16456625 | 8 | MAK16 | TSS1500 | 0.00039 | 0.0075 | 0.53 |
| cg22505907 | 3 | TCTEX1D2 | Body | 0.00044 | 0.0075 | 0.53 |
| cg03770410 | 20 | ZNF831 | 1stExon | 0.00029 | 0.0076 | 0.54 |
| cg08758387 | 7 | AGAP3 | Body | 0.00038 | 0.0076 | 0.52 |
| cg10061361 | 4 | TNIP3 | Body | −0.00073 | 0.0076 | −0.51 |
| cg13201342 | 1 | ARNT; CTSK | 3’UTR;TSS1500 | 0.00043 | 0.0076 | 0.52 |
Notes: Alpha, regression coefficient; Chr., Chromosome number; UTR, untranslated region; TSS, transcription start site; FDR, false discovery rate; Effect size, the effect size for AUD. Empty cells indicate unknown annotation for the genomic regions represented by the probes. Note that four Probe ID’s each have two genomic locations for the same probe and one Probe ID has two genes with their corresponding genomic locations separated by semicolon.
To provide the biological insights for the insular surface area-correlated protein coding genes, a list of Top 300 hits was further analyzed. All together these 300 probes represent a total of 247 protein coding genes (Suppl. Table S2). Among these, six protein-coding genes (ARHGAP22, BAHCC1, MCC, NR3C1, PARP1, and RBMS3) have two probes, and one gene (PTPRF) has three probes. GO analysis of these 247 genes found that only 13 biological processes were overrepresented, which belong to two major categories, metabolic regulation and organism development (Suppl. Table S2). Pathway analysis indicated that 55 pathways are represented by these proteins (Suppl. Table S2). These include angiogenesis, blood coagulation, and insulin-related pathways, which is consistent with the fact that the blood samples were used in our study. Interestingly, several pathways which are known to be involved in AUD or alcohol response, such as adrenergic receptor signaling, glutamate receptor group, GABA-B receptor II, and Rho GTPase-regulated cytoskeleton signaling, are also represented by some of the insular-correlated genes. We then mapped the 247 proteins into the human protein-protein interaction database collected in BioGRID and found that a total of 81 proteins exhibited at least one binary interactions (Suppl. Fig. S1). Among these, 56 proteins formed a protein-protein interaction network, with BAG3, RPA2 and PARP1 forming the hubs. The co-methylation pattern for those genes encoding proteins that display physical interactions indicates a possible epigenetic co-regulatory system for those interacting protein genes.
Identification of DNA methylation profiles correlated with AUD
Among 7,827 methylation probes significantly correlated with the right insular surface area, a total of 44 were also correlated with AUD with FDR < 0.05. All but four probes have negative correlations (Table 3). The majority of the 44 probes could be mapped to genomic regions corresponding to 36 protein coding genes, with three of these probes each representing two genes. In addition, two probes (cg23719342, and cg03770410, representing TAS2R16 and ZNF831 respectively) were on the list of the Top 25 right insular surface area-related probes (Table 1).
Table 3.
List of top AUD-correlated methylation probes ranked by FDR (<=0.05). The Alpha estimation and FDR for right insular surface area and AUD are separately listed. The methylation levels, as expressed by M-value, are also listed for HC and AUD respectively.
| Ilumina Probe ID | Chr. | Gene Symbol | Genomic Location | AUD | Insular Area | M-value | Effect Size | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Alpha | FDR | Alpha | FDR | HC | AUD | |||||
| cg18116574 | 3 | −0.218 | 0.016 | 0.00034 | 0.032 | 3.99 | 3.84 | −1.34 | ||
| cg19632594 | 5 | FBXL7 | TSS1500 | 0.171 | 0.029 | −0.00028 | 0.044 | −2.85 | −2.73 | 1.20 |
| cg03192874 | 2 | ITGAV | Body | −0.137 | 0.032 | 0.00026 | 0.025 | 3.50 | 3.39 | −1.17 |
| cg03411791 | 6 | −0.171 | 0.032 | 0.00031 | 0.031 | 3.21 | 3.07 | −1.16 | ||
| cg25023257 | 1 | MYBPH | TSS1500 | −0.133 | 0.032 | 0.00025 | 0.032 | 3.22 | 3.09 | −1.16 |
| cg21352837 | 2 | CYS1 | Body | −0.181 | 0.032 | 0.00033 | 0.033 | 3.79 | 3.64 | −1.15 |
| cg04301102 | 5 | SEPT8 | TSS200; 5’UTR | 0.173 | 0.034 | −0.00042 | 0.011 | −4.91 | −4.75 | 1.11 |
| cg07619656 | 7 | −0.147 | 0.034 | 0.00034 | 0.013 | 3.32 | 3.20 | −1.11 | ||
| cg04418999 | 7 | MDH2 | Body | −0.161 | 0.034 | 0.00029 | 0.039 | 3.04 | 2.89 | −1.10 |
| cg08081390 | 16 | FUS | Body | −0.161 | 0.037 | 0.00032 | 0.028 | 2.93 | 2.81 | −1.09 |
| cg09534070 | 2 | −0.167 | 0.039 | 0.00036 | 0.020 | 3.08 | 2.92 | −1.07 | ||
| cg02705758 | 5 | TRIO | Body | −0.127 | 0.039 | 0.00026 | 0.029 | 2.80 | 2.72 | −1.10 |
| cg00311667 | 2 | THAP4 | Body | −0.125 | 0.039 | 0.00023 | 0.038 | 3.48 | 3.38 | −1.10 |
| cg13636462 | 3 | COL6A6 | Body | −0.157 | 0.040 | 0.00028 | 0.047 | 3.35 | 3.20 | −1.07 |
| cg01966522 | 3 | UROC1 | 1stExon | −0.130 | 0.042 | 0.00024 | 0.037 | 3.15 | 3.04 | −1.08 |
| cg00579717 | 5 | RANBP17 | Body | 0.140 | 0.044 | −0.00032 | 0.020 | −3.61 | −3.51 | 1.04 |
| cg04014997 | 3 | ZDHHC3 | 5’UTR | −0.148 | 0.044 | 0.00027 | 0.044 | 3.16 | 3.06 | −1.05 |
| cg19683608 | 10 | C10orf108; DIP2C | TSS1500; Body | −0.130 | 0.044 | 0.00024 | 0.047 | 2.42 | 2.30 | −1.07 |
| cg23719342 | 7 | TAS2R16 | TSS1500 | −0.130 | 0.044 | 0.00040 | 0.007 | 1.92 | 1.80 | −1.05 |
| cg13240962 | 1 | USH2A | Body | −0.165 | 0.045 | 0.00038 | 0.020 | 1.96 | 1.84 | −1.03 |
| cg08163494 | 17 | CSH1 | TSS1500 | −0.128 | 0.045 | 0.00029 | 0.020 | 2.65 | 2.55 | −1.04 |
| cg07841132 | 20 | COL9A3 | Body | −0.131 | 0.045 | 0.00029 | 0.024 | 2.72 | 2.62 | −1.02 |
| cg26926973 | 19 | −0.152 | 0.045 | 0.00031 | 0.034 | 4.02 | 3.91 | −1.01 | ||
| cg19823452 | 19 | USE1 | Body | −0.110 | 0.045 | 0.00022 | 0.034 | 2.44 | 2.35 | −1.03 |
| cg24864707 | 11 | −0.143 | 0.045 | 0.00027 | 0.048 | 3.27 | 3.17 | −1.01 | ||
| cg00069391 | 7 | SDK1 | Body | −0.143 | 0.045 | 0.00027 | 0.049 | 2.69 | 2.57 | −1.02 |
| cg22912497 | 19 | RYR1 | Body | −0.131 | 0.045 | 0.00032 | 0.014 | 3.72 | 3.62 | −1.01 |
| cg13467459 | 1 | −0.125 | 0.046 | 0.00027 | 0.030 | 3.24 | 3.16 | −1.01 | ||
| cg05606089 | 5 | AP3B1 | Body | −0.160 | 0.046 | 0.00032 | 0.037 | 3.32 | 3.19 | −1.00 |
| cg16775752 | 10 | ARHGAP22 | Body | −0.175 | 0.047 | 0.00033 | 0.048 | 4.36 | 4.20 | −0.99 |
| cg05348366 | 1 | −0.152 | 0.047 | 0.00033 | 0.031 | 3.42 | 3.31 | −0.99 | ||
| cg03770410 | 20 | ZNF831 | 1stExon | −0.096 | 0.049 | 0.00029 | 0.008 | 1.97 | 1.88 | −1.01 |
| cg15342873 | 18 | −0.141 | 0.049 | 0.00040 | 0.009 | 3.36 | 3.29 | −0.98 | ||
| cg12421075 | 10 | ZRANB1 | TSS200 | −0.122 | 0.049 | 0.00027 | 0.030 | 2.69 | 2.59 | −0.99 |
| cg04093645 | 5 | FBN2 | Body | −0.136 | 0.049 | 0.00030 | 0.031 | 3.13 | 3.03 | −0.98 |
| cg21615663 | 16 | IQCK; C16orf88 | 5’UTR; 5’UTR | −0.115 | 0.049 | 0.00023 | 0.041 | 3.48 | 3.40 | −1.00 |
| cg16559259 | 10 | −0.137 | 0.049 | 0.00027 | 0.043 | 3.12 | 3.00 | −0.98 | ||
| cg13788000 | 9 | −0.151 | 0.049 | 0.00031 | 0.046 | 2.93 | 2.83 | −0.98 | ||
| cg03896542 | 16 | GNAO1 | Body; 3’UTR | −0.099 | 0.049 | 0.00025 | 0.014 | 1.94 | 1.86 | −1.00 |
| cg26048388 | 10 | ATP5C1; KIN | TSS1500; Body | 0.106 | 0.050 | −0.00022 | 0.046 | −3.70 | −3.62 | 0.99 |
| cg06980460 | 11 | FOLH1 | TSS1500 | −0.136 | 0.050 | 0.00027 | 0.047 | 3.06 | 2.94 | −0.97 |
| cg01791906 | 1 | RNF220 | Body | −0.111 | 0.050 | 0.00028 | 0.018 | 2.77 | 2.68 | −0.98 |
| cg26373541 | 4 | MRFAP1 | 3’UTR | −0.123 | 0.050 | 0.00028 | 0.026 | 2.51 | 2.40 | −0.97 |
| cg11903188 | 2 | PER2 | 5’UTR | −0.100 | 0.050 | 0.00021 | 0.046 | 3.18 | 3.08 | −0.99 |
Notes: Alpha, regression coefficient; Chr., Chromosome No.; UTR, untranslated region; TSS, transcription start site; FDR, false discovery rate; HC, healthy controls; AUD, alcohol use disorders; Effect size, the effect size for AUD. Empty cells indicate unknown annotation for the genomic regions represented by the probes. Note that two Probe ID’s each have two genomic locations for the same probe and three Probe ID’s have two genes for the same probe, with their corresponding genomic locations separated by semicolon.
Bioinformatic analysis of the protein coding genes showing AUD-correlated DNA methylation
To provide initial clues to the possible involvement in AUD of those 36 protein coding genes identified above, we searched various databases and recent publications to assess which of them have been reported to be related to AUD or alcohol response. First, we compared our list with 3,311 human genes collected in the ERGR database (Guo et al., 2009). ERGR is a comprehensive alcohol-related gene resource, including similar number of genes in animal models that have been shown to be responsible for or related to AUD based on the genetic linkage, association study and microarray expression analyses. We found that 14 out of 36 genes were present in the ERGR database, including C10orf108, DIP2C, FBXL7, CYS1, SEPT8, MDH2, FUS, TRIO, TAS2R16, USH2A, SDK1, RYR1, PER2, and KIN (Suppl. Table S3). Because ERGR was not updated, we then compared our list with a total of 771 AUD-related genes that have been identified in literature through the Public Health Genomics Knowledge Base. This search resulted in an additional alcohol-responsive gene, FBN2 (Joslyn et al., 2010). In addition, USE1 was also reported by a study to be affected by AUD (Zhang et al., 2014). In total, 16 out of 36 (i.e. 44%) AUD-correlated protein coding genes identified in our study have been reported to be AUD-related or alcohol responsive. Pathway analysis result showed that adrenergic receptor signaling, glutamate receptor group II, GABA-B receptor II, and opioid signaling are among the pathways represented by those genes, consistent with their roles in AUD or drug addiction in general.
To gain further information on the possible role of those 36 proteins, we constructed a protein-protein interaction network. Using a first-degree interaction in network construction, we found that 25 out of 36 AUD-correlated proteins interact with 656 other proteins (Suppl. Table S3). Several of the AUD-correlated proteins identified here are the major hubs in this network, including FUS, MDH2, TRIO, PER2, and SEPT8 (Suppl. Figure S2). FUS is the largest hub in this network (having 335 interacting proteins), followed by MDH2 which interacts with 50 proteins. In addition, we found that this network contains a total of 192 AUD-related or alcohol-responsive proteins (nodes coded in yellow, see Suppl. Figure S2), which represent 28% of total proteins in the network. Among these, FUS, PER2 and MDH2 are of particular interest, as 92 out of 335 FUS-interacting proteins, 15 out of 50 MDH2-interacting proteins, and 9 out of 18 PER2-interacting proteins are also alcohol-related. Together, this network analysis indicates that 25 of the AUD-related proteins identified in our study interact with a sizable number of proteins that are known to be related to AUD or alcohol response.
DISCUSSION
Use of the brain imaging-guided methylation analysis approach to reveal the blood epigenetic profiles linked to the variations in brain structure and AUD.
It has been fully recognized that finding or establishing the gene-brain-behavior link, which is critical for dissecting pathogenic mechanisms of many mental disorders including addiction, remains a daunting challenge (Zhao and Castellanos, 2016, Bogdan et al., 2017). One major obstacle is analysis of the large-scale and complex data generated by advanced omics and various brain imaging technologies. This is because the data generated by omics (such as DNA sequencing, RNA sequencing and methylome chip) and brain imaging (such as MRI) is very high-dimensional, while the human subjects are limited in sample size. To circumvent this problem, various statistical or Big Data approaches and use of endophenotypes or intermediate brain phenotypes in aiding genetic or epigenetic studies have been proposed (Zhao and Castellanos, 2016, Bogdan et al., 2017).
In AUD research, it has been reported that use of brain imaging-based electrophysiological endophenotypes in genetic screen has resulted in the identification of alcohol-related genes (Dick et al., 2006). In our work, the insular cortical surface area was used to guide DNA methylation analysis. To the best of our knowledge, we haven’t found any published studies that reported a difference in the insular cortical surface area between AUD and HC subjects. However, a recent addiction study (Kaag et al., 2014) did find that cocaine users had a larger insular surface area than HC and that this surface area was related to trait impulsivity. Given our similar observation in AUD subjects (who also had a larger surface area than HC), the insular cortical surface area might have a potential role in alcohol, cocaine and other drug addiction although its exact neurobiological property in relation to AUD remains to be further studied. We have found that using the insular surface area-guided methylation analysis greatly increased power in detecting AUD related probes. Without using the brain-imaging guided strategy, no AUD-correlated DNA methylation probe could be identified, showing that the integrated imaging epigenetic analysis can boost statistical power. In contrast, when we integrated brain structure into epigenetic profiling, we were able to narrow down from approximately 480 thousand probes to less than 8 thousand that are correlated with the right insular surface area (which we found has a significant difference between AUD and HC subjects). Built on this result, a total of 44 methylation probes, which represent 36 protein-coding genes annotated so far, were found to correlate with AUD. We believe that this integrated approach is effective in revealing the AUD-related information hidden from the high-dimensional and complex brain imaging and methylome data. Although those AUD-correlated genes need to be functionally confirmed for their involvement in AUD or response to alcohol, the proportion of the genes that are collected in ERGR can be used to roughly assess the approach effectiveness. Out of 19,194 protein-coding genes in the human genome, 3,311 were collected in ERGR, i.e. 17% of the human genome. Notably, 39% (14 out of 36) of the AUD-correlated genes identified here are present in ERGR. This indicates a dramatic enrichment of alcohol-related gene identification in the final stage.
We recognize that, without further studies, it is virtually impossible to determine whether the genes that are most strongly correlated with the right insular surface area have any biological relevance to the insular function. This is also attributed by the fact that very few brain postmortem omic studies have closely examined the insular cortex. Despite the difficulty in interpreting the gene-to-insula link, the finding that 81 out of 247 proteins encoded by those genes showing correlations of their methylation level with the right insular surface area are present in the protein-protein interaction network (Suppl. Fig. S1) suggests a pattern of co-methylation of those interacting genes. This could be an indication that a common epigenetic regulatory system is possibly related to insular functioning.
In addition, it should also be noted that the majority of the most significantly insular-correlated genes were not strongly correlated with AUD status in our study. For example, only two of the Top 25 insular surface area-related genes, TAS2R16 and ZNF831, were significantly correlated with AUD. Moreover, we have found that some of the insular-correlated genes are known to be related to AUD or alcohol response, and yet they were not correlated with AUD status in our sample. For example, RPA2 and PARP1, the two hubs in the protein-protein interaction network (Suppl. Fig. S1), are present in ERGR, and thus they may also be involved in AUD or response to alcohol. Interestingly, these two proteins interact with several other proteins related to AUD or alcohol response, such as NR3C1, TUBB and FUBP3 (Suppl. Fig. S1). Furthermore, analysis of the Top 300 insular-related genes showed that 51 out of 247 protein-coding genes are in the ERGR database (Suppl. Table S2), but the majority of them were not correlated with AUD in our study. Although this may raise a concern on the limitation of using an insular feature to identify the AUD-related genes, it is understandable that not all brain functions of the insular cortex mediate alcohol response or are impacted by alcohol. Moreover, heterogeneity of AUD indicates that AUD likely involves multiple sets of genes, and thus some of those alcohol-related genes reported by prior studies cannot be found in our study with a relatively small, specific sample.
Possibility of revealing novel AUD-related genes
With the identification of the 36 insular- and AUD-correlated protein coding genes reported here, two major questions remain to be answered: how to interpret their roles in AUD or alcohol response, and whether they represent some novel alcohol-related genes that have not been identified before. First, analysis of the protein-protein interaction network involving 25 out of the 36 proteins (Suppl. Fig. S2) reveals that these insular-correlated, differentially methylated genes interact with other alcohol-related genes that are not correlated with the insular surface area and are not regulated at the DNA methylation level. Perhaps, multiple levels of regulation exist for those alcohol-related, interacting genes, for example through transcription or translation. By interacting with other alcohol-related genes, many of these 36 AUD-correlated genes may together play an important role in AUD pathogenesis or in response to alcohol uptake.
Second, it is encouraging that among those 36 genes, 44% has been reported to be alcohol-related based on ERGR and most recent alcohol-related literature, and importantly, two of them (TAS2R16 and PER2) have been functionally linked to AUD, alcohol consumption or response to alcohol. TAS2R16, which is one of Top 25 differentially methylated genes related to the insular surface area, belongs to a family of seven-transmembrane taste receptors within the G protein-coupled receptor superfamily. A single-nucleotide polymorphism in the TAS2R16 coding region shows significant associations with alcohol dependence or consumption in particular for African American (Hinrichs et al., 2006, Wang et al., 2007). Given that another member TAS2R38 also associates with alcohol consumption (Duffy et al., 2004, Wang et al., 2007) and that these two receptors are involved in controlling the sensitivity to bitter compounds, it indicates a very intriguing link between oral sensation of bitterness and alcohol drinking behaviors or AUD (Duffy et al., 2004, Carrai et al., 2017). Thus, the sensory components of alcohol may affect the onset of alcohol drinking, and genetic variants in the taste-sensing receptors may lead to differential sensitivity to alcohol taste, consequently affecting the choice of alcoholic beverages and individual’s drinking behaviors (Carrai et al., 2017). The second most interesting gene is PER2 encoding a transcription coactivator critical for circadian regulation. While its role in biological clock and sleep disorder is well expected, its involvement in alcohol initially was a surprise. It was first indicated from the animal model studies showing alteration of circadian pacemaker system, such as circadian pattern of Per2 transcription, in the mice or rats selectively bred for ethanol preference or in response to increased ethanol consumption (Chen et al., 2004). Subsequently, Per2 mutant mice were found to increase ethanol consumption, providing convincing genetic evidence regarding the role of Per2 in alcohol drinking (Spanagel et al., 2005). In humans, it was identified from a 4-cM dense whole-genome linkage study involving 484 Irish families by its linkage to the empirically derived alcohol withdrawal symptoms factor score (Kuo et al., 2006). Variations in PER2 were also shown to associate with increased alcohol consumption in other human populations (Spanagel et al., 2005, Comasco et al., 2010, Blomeyer et al., 2013), although one human study did not find its significant association with alcohol consumption or other measurements of alcohol drinking behavior (Kovanen et al., 2010). However, in that same study (Kovanen et al., 2010), several other clock-related genes have been found to associate with AUD or alcohol drinking behaviors. Overall, these human genetic studies pinpointed an important role for PER2 in alcohol drinking. How PER2 and other clock-related genes are regulated by alcohol or impact alcohol response or drinking behavior has received increasing attentions in particular with the use of mouse models (Partonen, 2015, Davis et al., 2018). One possible mechanism is that Per2 in mouse may control glutamate transport and accumulation in the brain (Spanagel et al., 2005). In addition, Per2 likely acts to control the ethanol seeking behavior via regulating both ethanol metabolism and reward response (Gamsby et al., 2013). Another mouse study shows that Per2 may be critically involved in regulating beta-endorphin neuronal function in responses to both acute and chronic ethanol challenges (Agapito et al., 2010). The impact of ethanol exposure in altering the circadian expression of Per2 and a beta-endorphin precursor gene in the mouse hypothalamus has been reported (Chen et al., 2004). Given that the insular and the hypothalamus are important brain structures in the reward system, it is possible that differentially methylated PER2 in the insular cortex controls the reinforcement mechanism in alcohol seeking behavior.
Besides TAS2R16 and PER2, another potentially interesting gene is FUS, which encodes an RNA binding protein involved in regulation of gene expression, maintenance of genomic integrity and mRNA/microRNA processing in neuron cells (Masuda et al., 2016). FUS is one of the alcohol-response genes in the ERGR database (Guo et al., 2009), and in Drosophila it is also differentially regulated following alcohol exposure (Morozova et al., 2006). In our study, FUS is the largest hub in the AUD-related protein-protein interaction network analyzed here (Suppl. Fig. 2), and 92 out of 335 FUS-interaction proteins are also related to alcohol. Thus, if this key regulator of gene expression can be shown to be functionally involved in AUD or alcohol response, it will provide key insights into transcriptional regulation in response to alcohol consumption. In addition, the TRIO gene also collected in ERGR (Guo et al., 2009) is of potential significance. It encodes Rho guanine nucleotide exchange factor. Although Rho GTPase, a key signaling switch in numerous cellular events, has not been demonstrated to function in human alcohol response, work in animal models has demonstrated that Rho GTPase is critical for mediating alcohol response (Guasch et al., 2003, Rothenfluh et al., 2006, Romero et al., 2010, Selva and Egea, 2011). In humans, TRIO has been shown to be related to intellectual disability and synapse function (Ba et al., 2016), and thus it might play an important role in the brain in mediating ethanol response. In addition, other genes have also been reported to associate with or link to alcohol-related behaviors or symptoms, such as USH2A (Johnson et al., 2006, Wang et al., 2011), SDK1 (Hill et al., 2004), FBN2 (Joslyn et al., 2010), and C10orf108 (Hill et al., 2004). Several other genes show differential responses to alcohol drinking or in AUD subjects by altering either transcription, such as USE1 (Zhang et al., 2014), SEPT8 (Lewohl et al., 2000, McClintick et al., 2013), FBXL7 (Iwamoto et al., 2004), and KIN (Saba et al., 2006), or by impacting DNA methylation level such as DIP2C (Ruggeri et al., 2015). Therefore, identification of those known AUD-related genes supports the effectiveness of our approach and has a potential of providing epigenetic regulatory insights into AUD pathogenesis involving the insula.
Third, while it needs to be further investigated whether the remaining 20 protein-coding genes represent novel alcohol-related genes, most of them are expressed in the brain tissues in addition to blood and some of them have been reported to function in brain or might be indirectly implicated in alcohol response. The protein products for these genes and their assigned pathways are listed in Suppl. Table S3, but two groups of genes have potential significance and thus are discussed here in detail. One is related to GTP-binding proteins or GTPases and their signaling. GNAO1, which encodes heterotrimeric GTP-binding protein G(o) subunit alpha, has been shown to control motor function in humans (Danti et al., 2017), assigned to the GABA-B receptor II signaling and metabotropic glutamate receptor group II pathways (Suppl. Table S3), and proposed to be a genetic contributor to variation in physical dependence on opioids in mice (Kest et al., 2009). ARHGAP22 is a Rho GTPase activating protein regulating Rho activity, and RANBP17 is a Ran GTPase binding protein controlling nucleocytoplasmic transport. In a post-hoc analysis, we used Comb-p algorithm (Pedersen et al., 2012) to identify whether there are any differentially methylated regions (DMRs) in close proximity to the CpG probes for those 36 insular surface area-and AUD-correlated protein-coding genes. The genomic autocorrelation was set to 200 bp, and the p-value threshold for a DMR with a minimum of 2 CpG sites was set at p < 0.05. Interestingly, ARHGAP22 is the only gene which had eight CpG sites that together showed differential methylation between AUD and HC subjects (adjusted p-value <0.05). Although these GTP-binding proteins or GTPases and their regulators have not been shown to function in the brain yet, a similar Rho activating protein called RhoGAP18B in Drosophila has been demonstrated to mediate the sedating effects of ethanol by regulating Rho1 and Rac GTPase activity (Rothenfluh et al., 2006). Thus, together with the possible involvement of TRIO in alcohol use discussed above, our results indicate a potentially important role for heterotrimeric GTP-binding protein and monomeric GTPase signaling in AUD or alcohol response. The other group of genes is related to cytoskeleton organization and cell surface modification, including two collagens (COL6A6, and COL9A3), FBN2 (fibrillin-2), ITGAV (integrin subunit alpha V), and MYBPH (myosin binding protein H). SEPT8 (a septin), reported by others to be alcohol-response as discussed above, also regulates cytoskeleton organization. Furthermore, Rho GTPase signaling network provides a major regulatory mechanism in cytoskeleton organization. Taken together, although the remaining 20 insular- and AUD-correlated genes have not been reported to be related to alcohol before, they might represent novel genes involved in AUD or alcohol response. This intriguing possibility might be the case in particular for those genes demonstrated or assigned to function in GTPase signaling, cytoskeleton organization and extracellular matrix modifications. These genes are co-methylated and thus they may act together to exert the coordinated response to alcohol and ultimately contribute to the AUD development.
Limitations and conclusion
Several limitations in our study should be discussed here. First, the results were derived from a community-based NKI-RS dataset without replication. Our study only included a relatively small sample size (110 subjects in total) and thus our findings need to be replicated. Second, we acknowledge the importance of replicating the DNA methylation findings. However, due to limited DNA samples, the 44 CpG probes identified in this study have not been validated via pyrosequencing or bisulfite sequencing. Third, the DNA methylation level is less likely correlated with expression for all the genes identified. This is because gene expression is a complex regulatory process, involving not just DNA methylation. Other epigenetic modifications, such as acetylation or methylation of histone, are also critical for the robust control of gene expression in response to ethanol exposure. Fourth, linking DNA methylation from blood to brain needs to be cautious. Given the tissue or cell type difference, it is expected that some proportion of genes may not match the methylation levels in blood and brain. However, due to the brain access constraint, it is virtually impossible to verify this using the brain DNA samples from subjects in a community cohort. Despite this concern, strong correlations in DNA methylation between blood and brain tissues tested have also been reported (Horvath et al., 2012), and thus we speculate that a certain proportion of the genes identified here might exhibit similar methylation level in the insula. Indeed, we found that at least three of those 36 genes identified here were also reported by two prior studies using postmortem brain tissues (Hagerty et al., 2016, Wang et al., 2016). These include TRIO as one of the Rho GTPase signaling genes identified in the prefrontal cortex (Wang et al., 2016), and SDK1 and ARHGAP22 in the precuneus of AUD subjects (Hagerty et al., 2016). Similar to our finding, ARHGAP22 was hypomethylated in AUD subjects (Hagerty et al., 2016). Furthermore, this differential methylation also occurred in buccal cells. As discussed above, we found that ARHGAP22 has eight CpG sites that together are also hypomethylated in blood of AUD subjects. Therefore, given the demonstrated importance of Rho signaling in alcohol drinking response in non-human animal models (Guasch et al., 2003, Rothenfluh et al., 2006, Romero et al., 2010, Selva and Egea, 2011), this Rho GTPase signaling-related gene may be a potential candidate for follow-up studies in the future. Fifth, our result is correlational instead of causal-effective. DNA methylation may provide a differential epigenetic signal to regulate brain response to alcohol and other drugs of abuse, and yet the causal-effect relationship between DNA methylation and AUD needs to be further studied (Schuebel et al., 2016). Thus, although 44% of the insular surface area- and AUD-correlated genes identified in our study have been implicated by prior studies to function in AUD or ethanol response, these and other genes reported here need to be validated by large-scale population genetic studies in humans or functionally tested in well-designed reverse genetic studies in animal models.
In conclusion, we have used the blood DNA samples to identify 36 protein-coding genes whose DNA methylation level is correlated with insular surface area and AUD status. This was achieved by using an integrated brain imaging-based epigenetic screen. First, we have found that the insular surface areas in both left and right hemispheres are correlated with AUD, showing larger areas in AUD than HC subjects. We then identified a total of 44 DNA methylation probes, among approximately 8,000 probes that are correlated with the right insular surface area. Although it remains a challenge to link the DNA methylation level in blood samples to that in the brain or even mRNA expression levels in the brain, we have found that 44% of those 36 protein coding genes have been implicated in AUD or response to alcohol by prior studies, in particular TAS2R16 and PER2 which have been functionally linked to AUD or alcohol consumption. This indicates that a significant proportion of those insular surface area- and AUD-correlated genes identified in our study, in particular ARHGAP22, may potentially be involved in alcohol response or related to AUD, with some of them possibly being novel alcohol-related genes. While future replication and functional validation studies are needed to determine the physiological relevance of those insular surface area- and AUD-correlated genes, our work suggests that integrated analysis of brain imaging and epigenetic data with the help of the Big Data approach has a potential of revealing hidden information that provides the basis for future mechanistic studies of AUD pathogenesis.
Supplementary Material
Acknowledgments
We are grateful to Drs. F. Xavier Castellanos and Michael P. Milham for help with obtaining the brain imaging data and blood DNA samples. We also appreciate Dr. F. Xavier Castellanos for his suggestion on the project and Dr. Quan-Zhen Li for performing methylation analysis in the Genomics and Microarray Core Facility at University of Texas Southwestern Medical Center.
Support sources: This study was supported from an NIH-NIAAA grant (R21AA023800 to Y.Z.) and in part by a neuroscience fellowship from the Leon Levy Foundation (to Y.Z.).
Role of funding source
This study was supported from an NIH-NIAAA grant (R21AA023800 to Y.Z.) and in part by a neuroscience fellowship from the Leon Levy Foundation (to Y.Z.). None of the funding sources had any role in study design, data collection and analysis, result interpretation, manuscript writing and submission.
Footnotes
Conflict of interest
The authors declared no conflict of interest related to this research.
References cited
- Agapito M, Mian N, Boyadjieva NI, Sarkar DK (2010) Period 2 gene deletion abolishes beta-endorphin neuronal response to ethanol. Alcoholism, clinical and experimental research 34:1613–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA (2014) Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30:1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ba W, Yan Y, Reijnders MR, Schuurs-Hoeijmakers JH, Feenstra I, Bongers EM, Bosch DG, De Leeuw N, Pfundt R, Gilissen C, De Vries PF, Veltman JA, Hoischen A, Mefford HC, Eichler EE, Vissers LE, Nadif Kasri N, De Vries BB (2016) TRIO loss of function is associated with mild intellectual disability and affects dendritic branching and synapse function. Human molecular genetics 25:892–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomeyer D, Buchmann AF, Lascorz J, Zimmermann US, Esser G, Desrivieres S, Schmidt MH, Banaschewski T, Schumann G, Laucht M (2013) Association of PER2 genotype and stressful life events with alcohol drinking in young adults. PloS one 8:e59136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan R, Salmeron BJ, Carey CE, Agrawal A, Calhoun VD, Garavan H, Hariri AR, Heinz A, Hill MN, Holmes A, Kalin NH, Goldman D (2017) Imaging Genetics and Genomics in Psychiatry: A Critical Review of Progress and Potential. Biological psychiatry 82:165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrai M, Campa D, Vodicka P, Flamini R, Martelli I, Slyskova J, Jiraskova K, Rejhova A, Vodenkova S, Canzian F, Bertelli A, Dalla Vedova A, Bavaresco L, Vodickova L, Barale R (2017) Association between taste receptor (TAS) genes and the perception of wine characteristics. Scientific reports 7:9239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-Aryamontri A, Oughtred R, Boucher L, Rust J, Chang C, Kolas NK, O’Donnell L, Oster S, Theesfeld C, Sellam A, Stark C, Breitkreutz BJ, Dolinski K, Tyers M (2017) The BioGRID interaction database: 2017 update. Nucleic acids research 45:D369–D379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CP, Kuhn P, Advis JP, Sarkar DK (2004) Chronic ethanol consumption impairs the circadian rhythm of pro-opiomelanocortin and period genes mRNA expression in the hypothalamus of the male rat. Journal of neurochemistry 88:1547–1554. [DOI] [PubMed] [Google Scholar]
- Claus ED, Ewing SW, Filbey FM, Sabbineni A, Hutchison KE (2011) Identifying neurobiological phenotypes associated with alcohol use disorder severity. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology 36:2086–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comasco E, Nordquist N, Gokturk C, Aslund C, Hallman J, Oreland L, Nilsson KW (2010) The clock gene PER2 and sleep problems: association with alcohol consumption among Swedish adolescents. Upsala journal of medical sciences 115:41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danti FR, Galosi S, Romani M, Montomoli M, Carss KJ, Raymond FL, Parrini E, Bianchini C, McShane T, Dale RC, Mohammad SS, Shah U, Mahant N, Ng J, McTague A, Samanta R, Vadlamani G, Valente EM, Leuzzi V, Kurian MA, Guerrini R (2017) GNAO1 encephalopathy: Broadening the phenotype and evaluating treatment and outcome. Neurology. Genetics 3:e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BTt Voigt RM, Shaikh M Forsyth CB, Keshavarzian A (2018) Circadian Mechanisms in Alcohol Use Disorder and Tissue Injury. Alcoholism, clinical and experimental research 42:668–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ (2006) An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage 31:968–980. [DOI] [PubMed] [Google Scholar]
- Dick DM, Jones K, Saccone N, Hinrichs A, Wang JC, Goate A, Bierut L, Almasy L, Schuckit M, Hesselbrock V, Tischfield J, Foroud T, Edenberg H, Porjesz B, Begleiter H (2006) Endophenotypes successfully lead to gene identification: results from the collaborative study on the genetics of alcoholism. Behavior genetics 36:112–126. [DOI] [PubMed] [Google Scholar]
- Droutman V, Read SJ, Bechara A (2015) Revisiting the role of the insula in addiction. Trends in cognitive sciences 19:414–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, Lin SM (2010) Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC bioinformatics 11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy VB, Davidson AC, Kidd JR, Kidd KK, Speed WC, Pakstis AJ, Reed DR, Snyder DJ, Bartoshuk LM (2004) Bitter receptor gene (TAS2R38), 6-n-propylthiouracil (PROP) bitterness and alcohol intake. Alcoholism, clinical and experimental research 28:1629–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durazzo TC, Tosun D, Buckley S, Gazdzinski S, Mon A, Fryer SL, Meyerhoff DJ (2011) Cortical thickness, surface area, and volume of the brain reward system in alcohol dependence: relationships to relapse and extended abstinence. Alcoholism, clinical and experimental research 35:1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott DM, Briere J (1992) Sexual abuse trauma among professional women: validating the Trauma Symptom Checklist-40 (TSC-40). Child abuse & neglect 16:391–398. [DOI] [PubMed] [Google Scholar]
- Gagnon- Bartsch J, Laurent Jacob L, Speed TP. Removing unwanted variation from high dimensional data with negative controls. online]. http://statistics.berkeley.edu/tech-reports/820.
- Gamsby JJ, Templeton EL, Bonvini LA, Wang W, Loros JJ, Dunlap JC, Green AI, Gulick D (2013) The circadian Per1 and Per2 genes influence alcohol intake, reinforcement, and blood alcohol levels. Behavioural brain research 249:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasch RM, Tomas M, Minambres R, Valles S, Renau-Piqueras J, Guerri C (2003) RhoA and lysophosphatidic acid are involved in the actin cytoskeleton reorganization of astrocytes exposed to ethanol. Journal of neuroscience research 72:487–502. [DOI] [PubMed] [Google Scholar]
- Guo AY, Webb BT, Miles MF, Zimmerman MP, Kendler KS, Zhao Z (2009) ERGR: An ethanol-related gene resource. Nucleic acids research 37:D840–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerty SL, Bidwell LC, Harlaar N, Hutchison KE (2016) An Exploratory Association Study of Alcohol Use Disorder and DNA Methylation. Alcoholism, clinical and experimental research 40:1633–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SY, Shen S, Zezza N, Hoffman EK, Perlin M, Allan W (2004) A genome wide search for alcoholism susceptibility genes. American journal of medical genetics. Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics 128B:102–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs AL, Wang JC, Bufe B, Kwon JM, Budde J, Allen R, Bertelsen S, Evans W, Dick D, Rice J, Foroud T, Nurnberger J, Tischfield JA, Kuperman S, Crowe R, Hesselbrock V, Schuckit M, Almasy L, Porjesz B, Edenberg HJ, Begleiter H, Meyerhof W, Bierut LJ, Goate AM (2006) Functional variant in a bitter-taste receptor (hTAS2R16) influences risk of alcohol dependence. American journal of human genetics 78:103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Zhang Y, Langfelder P, Kahn RS, Boks MP, van Eijk K, van den Berg LH, Ophoff RA (2012) Aging effects on DNA methylation modules in human brain and blood tissue. Genome biology 13:R97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Yamamoto M, Ozawa H, Saito T, Kato T (2004) Decreased expression of NEFH and PCP4/PEP19 in the prefrontal cortex of alcoholics. Neuroscience research 49:379–385. [DOI] [PubMed] [Google Scholar]
- Johnson C, Drgon T, Liu QR, Walther D, Edenberg H, Rice J, Foroud T, Uhl GR (2006) Pooled association genome scanning for alcohol dependence using 104,268 SNPs: validation and use to identify alcoholism vulnerability loci in unrelated individuals from the collaborative study on the genetics of alcoholism. American journal of medical genetics. Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics 141B:844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joslyn G, Ravindranathan A, Brush G, Schuckit M, White RL (2010) Human variation in alcohol response is influenced by variation in neuronal signaling genes. Alcoholism, clinical and experimental research 34:800–812. [DOI] [PubMed] [Google Scholar]
- Kaag AM, Crunelle CL, van Wingen G, Homberg J, van den Brink W, Reneman L (2014) Relationship between trait impulsivity and cortical volume, thickness and surface area in male cocaine users and non-drug using controls. Drug and alcohol dependence 144:210–217. [DOI] [PubMed] [Google Scholar]
- Kest B, Smith SB, Schorscher-Petcu A, Austin JS, Ritchie J, Klein G, Rossi GC, Fortin A, Mogil JS (2009) Gnao1 (G alphaO protein) is a likely genetic contributor to variation in physical dependence on opioids in mice. Neuroscience 162:1255–1264. [DOI] [PubMed] [Google Scholar]
- Kovanen L, Saarikoski ST, Haukka J, Pirkola S, Aromaa A, Lonnqvist J, Partonen T (2010) Circadian clock gene polymorphisms in alcohol use disorders and alcohol consumption. Alcohol and alcoholism 45:303–311. [DOI] [PubMed] [Google Scholar]
- Kuo PH, Neale MC, Riley BP, Webb BT, Sullivan PF, Vittum J, Patterson DG, Thiselton DL, van den Oord EJ, Walsh D, Kendler KS, Prescott CA (2006) Identification of susceptibility loci for alcohol-related traits in the Irish Affected Sib Pair Study of Alcohol Dependence. Alcoholism, clinical and experimental research 30:1807–1816. [DOI] [PubMed] [Google Scholar]
- Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA (2000) Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcoholism, clinical and experimental research 24:1873–1882. [PubMed] [Google Scholar]
- Liang L, Cookson WO (2014) Grasping nettles: cellular heterogeneity and other confounders in epigenome-wide association studies. Human molecular genetics 23:R83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litten RZ, Ryan ML, Falk DE, Reilly M, Fertig JB, Koob GF (2015) Heterogeneity of alcohol use disorder: understanding mechanisms to advance personalized treatment. Alcoholism, clinical and experimental research 39:579–584. [DOI] [PubMed] [Google Scholar]
- Liu C, Marioni RE, Hedman AK, Pfeiffer L, Tsai PC, Reynolds LM, Just AC, Duan Q, Boer CG, Tanaka T, Elks CE, Aslibekyan S, Brody JA, Kuhnel B, Herder C, Almli LM, Zhi D, Wang Y, Huan T, Yao C, Mendelson MM, Joehanes R, Liang L, Love SA, Guan W, Shah S, McRae AF, Kretschmer A, Prokisch H, Strauch K, Peters A, Visscher PM, Wray NR, Guo X, Wiggins KL, Smith AK, Binder EB, Ressler KJ, Irvin MR, Absher DM, Hernandez D, Ferrucci L, Bandinelli S, Lohman K, Ding J, Trevisi L, Gustafsson S, Sandling JH, Stolk L, Uitterlinden AG, Yet I, Castillo-Fernandez JE, Spector TD, Schwartz JD, Vokonas P, Lind L, Li Y, Fornage M, Arnett DK, Wareham NJ, Sotoodehnia N, Ong KK, van Meurs JBJ, Conneely KN, Baccarelli AA, Deary IJ, Bell JT, North KE, Liu Y, Waldenberger M, London SJ, Ingelsson E, Levy D (2018) A DNA methylation biomarker of alcohol consumption. Molecular psychiatry 23:422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makris N, Gasic GP, Kennedy DN, Hodge SM, Kaiser JR, Lee MJ, Kim BW, Blood AJ, Evins AE, Seidman LJ, Iosifescu DV, Lee S, Baxter C, Perlis RH, Smoller JW, Fava M, Breiter HC (2008) Cortical thickness abnormalities in cocaine addiction--a reflection of both drug use and a pre-existing disposition to drug abuse? Neuron 60:174–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimovic J, Gordon L, Oshlack A (2012) SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome biology 13:R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda A, Takeda J, Ohno K (2016) FUS-mediated regulation of alternative RNA processing in neurons: insights from global transcriptome analysis. Wiley interdisciplinary reviews. RNA 7:330–340. [DOI] [PubMed] [Google Scholar]
- McClintick JN, Xuei X, Tischfield JA, Goate A, Foroud T, Wetherill L, Ehringer MA, Edenberg HJ (2013) Stress-response pathways are altered in the hippocampus of chronic alcoholics. Alcohol 47:505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Thomas PD (2013) PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic acids research 41:D377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova TV, Anholt RR, Mackay TF (2006) Transcriptional response to alcohol exposure in Drosophila melanogaster. Genome biology 7:R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi NH, Bechara A (2009) The hidden island of addiction: the insula. Trends in neurosciences 32:56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi NH, Rudrauf D, Damasio H, Bechara A (2007) Damage to the insula disrupts addiction to cigarette smoking. Science 315:531–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nooner KB, Colcombe SJ, Tobe RH, Mennes M, Benedict MM, Moreno AL, Panek LJ, Brown S, Zavitz ST, Li Q, Sikka S, Gutman D, Bangaru S, Schlachter RT, Kamiel SM, Anwar AR, Hinz CM, Kaplan MS, Rachlin AB, Adelsberg S, Cheung B, Khanuja R, Yan C, Craddock CC, Calhoun V, Courtney W, King M, Wood D, Cox CL, Kelly AM, Di Martino A, Petkova E, Reiss PT, Duan N, Thomsen D, Biswal B, Coffey B, Hoptman MJ, Javitt DC, Pomara N, Sidtis JJ, Koplewicz HS, Castellanos FX, Leventhal BL, Milham MP (2012) The NKI-Rockland Sample: A Model for Accelerating the Pace of Discovery Science in Psychiatry. Frontiers in neuroscience 6:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmisano M, Pandey SC (2017) Epigenetic mechanisms of alcoholism and stress-related disorders. Alcohol 60:7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partonen T (2015) Clock genes in human alcohol abuse and comorbid conditions. Alcohol 49:359–365. [DOI] [PubMed] [Google Scholar]
- Pedersen BS, Schwartz DA, Yang IV, Kechris KJ (2012) Comb-p: software for combining, analyzing, grouping and correcting spatially correlated P-values. Bioinformatics 28:2986–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Plume JM, Gibbons FX, Brody GH, Beach SR (2012) The impact of recent alcohol use on genome wide DNA methylation signatures. Frontiers in genetics 3:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B, Maksimovic J, Oshlack A (2016) missMethyl: an R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinformatics 32:286–288. [DOI] [PubMed] [Google Scholar]
- Reilly MT, Noronha A, Goldman D, Koob GF (2017) Genetic studies of alcohol dependence in the context of the addiction cycle. Neuropharmacology 122:3–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero AM, Esteban-Pretel G, Marin MP, Ponsoda X, Ballestin R, Canales JJ, Renau-Piqueras J (2010) Chronic ethanol exposure alters the levels, assembly, and cellular organization of the actin cytoskeleton and microtubules in hippocampal neurons in primary culture. Toxicological sciences : an official journal of the Society of Toxicology 118:602–612. [DOI] [PubMed] [Google Scholar]
- Rothenfluh A, Threlkeld RJ, Bainton RJ, Tsai LT, Lasek AW, Heberlein U (2006) Distinct behavioral responses to ethanol are regulated by alternate RhoGAP18B isoforms. Cell 127:199–211. [DOI] [PubMed] [Google Scholar]
- Ruggeri B, Nymberg C, Vuoksimaa E, Lourdusamy A, Wong CP, Carvalho FM, Jia T, Cattrell A, Macare C, Banaschewski T, Barker GJ, Bokde AL, Bromberg U, Buchel C, Conrod PJ, Fauth-Buhler M, Flor H, Frouin V, Gallinat J, Garavan H, Gowland P, Heinz A, Ittermann B, Martinot JL, Nees F, Pausova Z, Paus T, Rietschel M, Robbins T, Smolka MN, Spanagel R, Bakalkin G, Mill J, Sommer WH, Rose RJ, Yan J, Aliev F, Dick D, Kaprio J, Desrivieres S, Schumann G, Consortium I (2015) Association of Protein Phosphatase PPM1G With Alcohol Use Disorder and Brain Activity During Behavioral Control in a Genome-Wide Methylation Analysis. The American journal of psychiatry 172:543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba L, Bhave SV, Grahame N, Bice P, Lapadat R, Belknap J, Hoffman PL, Tabakoff B (2006) Candidate genes and their regulatory elements: alcohol preference and tolerance. Mammalian genome : official journal of the International Mammalian Genome Society 17:669–688. [DOI] [PubMed] [Google Scholar]
- Schacht JP, Anton RF, Myrick H (2013) Functional neuroimaging studies of alcohol cue reactivity: a quantitative meta-analysis and systematic review. Addiction biology 18:121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuebel K, Gitik M, Domschke K, Goldman D (2016) Making Sense of Epigenetics. The international journal of neuropsychopharmacology/official scientific journal of the Collegium Internationale Neuropsychopharmacologicum 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selva J, Egea G (2011) Ethanol increases p190RhoGAP activity, leading to actin cytoskeleton rearrangements. Journal of neurochemistry 119:1306–1316. [DOI] [PubMed] [Google Scholar]
- Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology 3:Article3. [DOI] [PubMed] [Google Scholar]
- Spanagel R, Pendyala G, Abarca C, Zghoul T, Sanchis-Segura C, Magnone MC, Lascorz J, Depner M, Holzberg D, Soyka M, Schreiber S, Matsuda F, Lathrop M, Schumann G, Albrecht U (2005) The clock gene Per2 influences the glutamatergic system and modulates alcohol consumption. Nature medicine 11:35–42. [DOI] [PubMed] [Google Scholar]
- Stewart JL, Connolly CG, May AC, Tapert SF, Wittmann M, Paulus MP (2014) Striatum and insula dysfunction during reinforcement learning differentiates abstinent and relapsed methamphetamine-dependent individuals. Addiction 109:460–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulisiak CT, Harris RA, Ponomarev I (2017) DNA modifications in models of alcohol use disorders. Alcohol 60:19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Xu H, Zhao H, Gelernter J, Zhang H (2016) DNA co-methylation modules in postmortem prefrontal cortex tissues of European Australians with alcohol use disorders. Scientific reports 6:19430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Hinrichs AL, Bertelsen S, Stock H, Budde JP, Dick DM, Bucholz KK, Rice J, Saccone N, Edenberg HJ, Hesselbrock V, Kuperman S, Schuckit MA, Bierut LJ, Goate AM (2007) Functional variants in TAS2R38 and TAS2R16 influence alcohol consumption in high-risk families of African-American origin. Alcoholism, clinical and experimental research 31:209–215. [DOI] [PubMed] [Google Scholar]
- Wang KS, Liu X, Aragam N, Jian X, Mullersman JE, Liu Y, Pan Y (2011) Family-based association analysis of alcohol dependence in the COGA sample and replication in the Australian twin-family study. J Neural Transm (Vienna) 118:1293–1299. [DOI] [PubMed] [Google Scholar]
- Warden AS, Mayfield RD (2017) Gene expression profiling in the human alcoholic brain. Neuropharmacology 122:161–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Wang F, Kranzler HR, Gelernter J, Zhang H (2017) Alcohol and nicotine codependence-associated DNA methylation changes in promoter regions of addiction-related genes. Scientific reports 7:41816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gelernter J (2017) Review: DNA methylation and alcohol use disorders: Progress and challenges. The American journal on addictions 26:502–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Herman AI, Kranzler HR, Anton RF, Zhao H, Zheng W, Gelernter J (2013a) Array-based profiling of DNA methylation changes associated with alcohol dependence. Alcoholism, clinical and experimental research 37 Suppl 1:E108–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang F, Xu H, Liu Y, Liu J, Zhao H, Gelernter J (2014) Differentially co-expressed genes in postmortem prefrontal cortex of individuals with alcohol use disorders: influence on alcohol metabolism-related pathways. Human genetics 133:1383–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Miao Q, Wang C, Zhao R, Li W, Haile CN, Hao W, Zhang XY (2013b) Genome-wide DNA methylation analysis in alcohol dependence. Addiction biology 18:392–403. [DOI] [PubMed] [Google Scholar]
- Zhao R, Zhang R, Li W, Liao Y, Tang J, Miao Q, Hao W (2013) Genome-wide DNA methylation patterns in discordant sib pairs with alcohol dependence. Asia-Pacific psychiatry : official journal of the Pacific Rim College of Psychiatrists 5:39–50. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Castellanos FX (2016) Annual Research Review: Discovery science strategies in studies of the pathophysiology of child and adolescent psychiatric disorders - promises and limitations. Journal of child psychology and psychiatry, and allied disciplines 57:421–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zheng ZL, Castellanos FX (2017) Analysis of alcohol use disorders from the Nathan Kline Institute-Rockland Sample: Correlation of brain cortical thickness with neuroticism. Drug and alcohol dependence 170:66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.