Abstract
Purpose:
In this study, we explore the feasibility of a novel framework for MR-based attenuation correction for PET/MR imaging based on deep learning via convolutional neural networks, which enables fully automated and robust estimation of a pseudo CT image based on ultrashort echo time (UTE), fat, and water images obtained by a rapid MR acquisition.
Methods:
MR images for MRAC are acquired using dual echo ramped hybrid encoding (dRHE), where both UTE and out-of-phase echo images are obtained within a short single acquisition (35 sec). Tissue labeling of air, soft tissue, and bone in the UTE image is accomplished via a deep learning network that was pre-trained with T1-weighted MR images. UTE images are used as input to the network, which was trained using labels derived from co-registered CT images. The tissue labels estimated by deep learning are refined by a conditional random field based correction. The soft tissue labels are further separated into fat and water components using the two-point Dixon method. The estimated bone, air, fat, and water images are then assigned appropriate Hounsfield units, resulting in a pseudo CT image for PET attenuation correction. To evaluate the proposed MRAC method, PET/MR imaging of the head was performed on 8 human subjects, where Dice similarity coefficients of the estimated tissue labels and relative PET errors were evaluated through comparison to a registered CT image.
Result:
Dice coefficients for air (within the head), soft tissue, and bone labels were 0.76±0.03, 0.96±0.006, and 0.88±0.01. In PET quantification, the proposed MRAC method produced relative PET errors less than 1% within most brain regions.
Conclusion:
The proposed MRAC method utilizing deep learning with transfer learning and an efficient dRHE acquisition enables reliable PET quantification with accurate and rapid pseudo CT generation.
Keywords: MR-based attenuation correction, deep learning, transfer learning
I. INTRODUCTION
MR-based attenuation correction (MRAC) is essential but challenging in simultaneous PET-MR imaging due to the nonlinear relationship between MR signals and photon attenuation, which makes estimation of attenuation coefficients based on MR images challenging. Atlas-based MRAC methods 1–5 have been introduced and are used in contemporary clinical PET/MR systems for brain imaging, but errors in attenuation correction can be large for subjects with irregular anatomy. Alternatively, methods based on tissue segmentation of MR images have been proposed in literature6–13 to realize more direct, subject-specific attenuation maps. More advanced methods have been recently investigated to achieve clinically feasible brain MRAC14. However, it remains challenging to achieve rapid, accurate, and fully-automated tissue segmentation for MRAC due to low image contrast between bone and air in MR images. To improve air and bone detection in MRAC, ultrashort echo time (UTE) imaging has been proposed, which has been shown to be efficient at differentiating bone and air15–20. However, long image acquisition time (2~5min) imposed by the additional UTE imaging for MRAC can impede PET/MR imaging workflow. Moreover, ambiguities at air-bone interfaces are exacerbated by partial volume effects in the UTE images.
Deep learning has received much attention lately as a promising method in computer vision for object recognition and image segmentation21–23. In medical imaging, deep Convolutional Neural Networks (CNN) have been shown to be especially effective at various tasks24,25 such as segmenting cardiac structures26, brain structures27, brain tumors28, and musculoskeletal tissues29. Similar to CNNs are Convolutional Encoder-Decoder (CED) networks, which consist of a paired encoder and decoder, which are particularly favorable in multiple tissue segmentation studies due to their high efficiency and applicability29–35. The encoder network performs efficient image data compression while estimating robust and spatial invariant image features. The corresponding decoder network takes encoded features then performs feature map up-sampling to recover high-resolution pixel-wise image labeling. The U-Net developed by Ronneberger et.al.30 has a CED structure with direct skip connections between encoder and decoder for improving spatial segmentation accuracy. The SegNet proposed by Badrinarayanan et.al.36 utilizes deep structure of VGG16 network37 for encoder and an efficient up-sampling technique in decoder via index restoration. Recent biomedical studies implementing these CED networks have shown successful and efficient tissue segmentation for high-resolution neuronal structures in electron microscopy30, liver tumor in CT31, cartilage and bone in MR29.
In this study, we explore feasibility of a new framework for rapid and robust MRAC utilizing efficient dual echo RHE (dRHE) acquisition38,39 and a CED29 network powered by transfer learning. In our dRHE acquisition, UTE, fat, and water images are acquired within a very short scan time (35 seconds). The obtained UTE image is used as input to the deep learning network, which is trained to segment the images into 3 tissue labels (air, soft tissue, and bone) using a discretized co-registered CT image as ground truth. For more reliable training, a transfer learning approach is utilized, where the network is pre-trained with T1 weighted MR images (due to their greater abundance in our institutional MR database32) and then re-trained using UTE images. A three-dimensional (3D) fully-connected Conditional Random Field (CRF)40 is applied to the model’s output to refine tissue labels estimated by the deep learning network. The estimated tissue labels – along with fat and water images obtained in dRHE – are used to generate a pseudo CT image. Feasibility of the proposed approach was evaluated in human subjects scanned on 3T simultaneous PET/MR system.
II. MATERIALS AND METHODS
A. Deep learning based MRAC using dual echo RHE
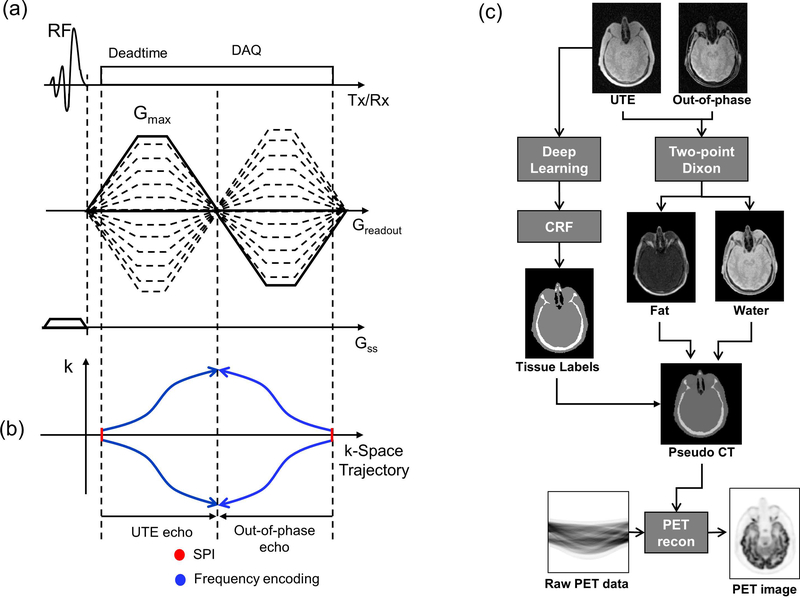
In our dRHE acquisition41, a UTE and an out-of-phase image with high spatial resolution (1 mm3) are obtained within a short single acquisition (scantime = 35 sec), where a 3D UTE image is rapidly acquired after slab selection with Shinnar-La Roux (SLR) RF pulse with minimum phase 42, and an out-of-phase image (TE≈1.1ms in 3T MR) is acquired during fly-back to the center of k-space as shown in Figure 1-a. Both k-spaces in UTE and out-of-phase echo are hybrid-encoded to improve image quality in which central k-space is encoded by single point imaging (SPI), and outer k-space is acquired by frequency encoding39 as illustrated in Figure 1-b.
Fig. 1.
(a) Pulse sequence diagram for rapid dual echo acquisition, (b) k-space trajectories for UTE and out-of-phase echo, and (c) a framework of proposed deep learning-based MRAC. Note that in (a) out-of-phase echo is acquired in fly-back to the center of k-space for rapid acquisition and also hybrid-encoded. The two acquired images (UTE and out-of-phase echo image) are used for two-point Dixon reconstruction to obtain fat and water separated images, while UTE image is processed with deep learning to yield air, soft tissue, and bone labels as shown in (c).
Figure 1-c illustrates the proposed MRAC framework combining dRHE and deep learning. In this method, a pseudo CT image is synthesized by using two 3D images (UTE and out-of-phase echo image) as an input. The UTE and out-of-phase images are processed with two-point Dixon fat-water separation to generate fat and water separated images43. Meanwhile, a UTE image is processed with deep learning network to obtain 3-class tissue labels (air, soft tissue, and bone). The tissue labels estimated by deep learning network is refined using conditional random field (CRF) based correction. The tissue labels from deep learning, water image, and fat image are combined to synthesize a pseudo CT image. The generated pseudo CT image is used for attenuation correction during PET reconstruction.
B. Convolutional encoder-decoder architecture
The essential component of the proposed method is a deep CED network, SegNet, which provides robust tissue labeling and has been successfully used to segment bone and soft tissue in MR images29,32. The specific details of the CED network used in this study are described in our previous study 29, which is also illustrated in Figure 2. The same 13 convolutional layers from the VGG16 network37 is utilized in the encoder. The encoder network is followed by a decoder network with reversed layers of the encoder network to reconstruct pixel-wise tissue labels. The max-pooling in the decoder is substituted by an up-sampling process to consistently resample image features and hence fully recover the original image size. A multi-class soft-max classifier44, the final layer of the decoder network, produces class probabilities for each pixel.
Fig. 2.
Schematic illustration of the deep learning workflow and CED network. The model is iteratively optimized to learn CED network parameters for 3-class tissue labels (air, soft tissue, and bone) in a training phase. (BN: Batch normalization. ReLU: Rectified-linear activation)
C. Network training procedure
The training data consists of MR head images as input and reference tissue labels for air, bone and soft tissue obtained from non-contrast CT data from the same subject. Ground truth reference data are automatically obtained by using CT images as in previous works in literatures8,19,32, where CT images were co-registered to UTE MR images using a rigid Euler transformation followed by non-rigid B-spline transformation using existing image registration tools45. For each training dataset, the reference 3-class tissue labels were created by pixel-intensity-based thresholding (HU > 300 for bone, HU < −300 for air, and otherwise soft tissue) of the co-registered CT.
For training the network, 3D MR and CT volume data were input into the model as a stack of 2D axial images. All 2D input images were normalized using local contrast normalization46 and resampled to 340 × 340 using bilinear interpolation before being used as input to the CED. The encoder and decoder network weights were initialized using pre-trained network weights on a head and neck MR image dataset consisting of 30 high-resolution T1-weighted images (using GE T1-BRAVO sequence) from our previous study32. The network weights were then updated using Stochastic Gradient Descent (SGD)47 with a fixed learning rate of 0.01 and momentum of 0.9. The CED network iteratively estimates output tissue labels and compares them to the reference mask generated from CT data. Reference CT data ensures that network learns the relationship between MR image contrast and reference tissue labels. The network was trained using multi-class cross-entropy loss23 as an objective function where the loss is calculated in a mini-batch of 4 images in each iteration. The network training was performed with 20,000 iterations corresponding to 76 epochs for our training data. The training data was shuffled before each epoch to create randomization in batch training and each image slice in the training data set was used once per epoch. Once the training phase was complete, the CED network structure was fixed and was used for labeling of bone, air, and soft tissue for new MR images, which were subsequently processed into pseudo CT images. Description of all datasets can be found in Table I.
Table I.
Dataset. 30 training datasets were retrospectively acquired from institutional clinical MR/CT database, while 14 datasets were progressively acquired.
| Pre-Training | Training | Evaluation | |
|---|---|---|---|
| MR Scanner | GE 1.5T Signa HDxt GE 1.5T MR450w | GE 3T Signa PET/MR | GE 3T Signa PET/MR |
| MR Type | T1 Weighted | PD Weighted UTE | PD Weighted UTE |
| CT Scanner | GE Optima CT 660 GE Discovery GE CT750HD GE Revolution GSI | GE Discovery 710 GE Discovery VCT | GE Discovery 710 GE Discovery VCT |
| Size | 30 | 6 | 8 |
In this study, the deep learning framework was implemented in a computing environment involving Python, MATLAB, and C/C++. The CED network was modified based on the Caffe implementation with GPU parallel computing support44.
D. Conditional random field
Since the deep leaning tissue labeling is done slice by slice, we added maximum a posteriori (MAP) inference in a conditional random field (CRF) defined over voxels in the 3D brain volume to take into account the 3D contextual relationships between voxels and to maximize the label agreement among similar voxels. The fully connected CRF model takes the output of the deep learning model and builds unary potentials for each voxel and takes the original 3D volume to build the pairwise potentials on all pairs of voxels. The Gibbs energy for the fully connected CRF is
| (1) |
where x is the assigned label for each voxel, and i and j range from 1 to the total number of voxels. ψu(x) is the unary potential which is defined as the negative logarithm of the probability for a particular label from the deep learning prediction, while ψp(xi,xj) is the pairwise potential
| (2) |
pi and pj are the positions of each voxel, and Ii and Ij are the Intensity of each voxel. The first exponential term is the appearance kernel accounting for the assumption that voxels close to each other or having similar contrasts tend to share the same label, and is controlled by θβ and θα. The second exponential term is the smoothness kernel that helps to remove isolated small regions and is controlled by θγ. ω1 and ω2 are the weights. μ(xi,xj) is the compatibility function and is defined as the Potts model, μ(xi,xj) ≠ [xi ≠ xj]. To achieve a highly efficient convergence, an algorithm with a mean approximation to the CRF distribution is chosen. The mean field update of all variables can be conducted with Gaussian filtering in the feature space. As a result, the computational complexity can be reduced from quadratic to linear in the number of variables.40
E. Pseudo CT generation
A pseudo CT map is generated using the tissue labels (air, soft tissue, and bone) estimated by deep learning network, and fat/water separated images. Continuous CT value for soft tissue region is generated based on fat fraction as (1 − ffat)HUwater + ffatHUfat, where ffat is a fat fraction image obtained by two point Dixon, and HUwater and HUfat are Hounsfield unit values (HU) for water and fat, respectively. HU values, −1000, −42, 42, and 1018 were used for air, fat, water, and bone, respectively, which is consistent with values used in other studies5,48–50. The generated CT image is spatially filtered using a Gaussian kernel with standard deviation of 2 pixels (2 mm).
F. Experimental setup
The dataset for pre-training of the network consisted of 30 subject images retrospectively obtained with approval of our institutional IRB. All subjects for pre-training underwent a T1-weighted post contrast 3D MR scan and a non-contrast CT scan on the same day. The MR images were acquired on 1.5T scanners Signa HDxt or MR450w (GE Healthcare, Waukesha, WI) with the following imaging parameters: 0.46–0.52 mm transaxial voxel dimensions, 1.2 mm slice thickness, 450 ms inversion time, 8.9–10.4 ms repetition time, 3.5–3.8 ms echo time, 13 degree flip angle, utilizing an 8-channel receive-only head coil. The CT images were acquired on three scanners (Optima CT 660, Discovery CT750HD, Revolution GSI; GE Healthcare) with the following range of acquisition/reconstruction settings: 0.43–0.46 mm transaxial voxel dimensions, 1.25–2.5 mm slice thicknesses, 120 kVp, automatic exposure control with GE noise index of 2.8–12.4, and 0.53 helical pitch.
An additional 14 subjects were prospectively recruited for UTE based training and evaluation, where patients underwent simultaneous PET/MR imaging on a 3T GE Signa PET/MR system prior to clinical 18F-FDG PET/CT imaging (from either a Discovery 710 or Discovery VCT PET/CT scanner) under an IRB approved protocol. MR imaging consisted of the system-default MRAC scan and the 35-second dRHE acquisition with a 5-minute or 10-minute PET acquisition of the head. The dRHE imaging was performed with the following parameters: Gmax=33 mT/m, slewrate=118 mT/m/ms, desired FOV=350 mm3, voxel size=1 mm3, TR=4.2 ms, TE=52 μs for a UTE image, 1172 μs for an out-of-phase image, readout duration for each echo=0.64 ms, scantime=35 sec, sampling bandwidth=250 Hz, FA=1 degree, number of radial spokes=7442, and number of SPI encoding=925 utilizing a 40-channel HNU coil. For training, 3D UTE images of 6 subjects were used (number of total slices=1050), not including the 30 subjects used for pre-training. The remaining 8 subjects were used for evaluation, where UTE and out-of-phase images were used to yield tissue labels, fat and water images, and the resultant pseudo CT image as shown in Figure 1-c.
To reconstruct the MR images, convolution gridding reconstruction51 using a kernel size of 5 and oversampling ratio of 1.5 was applied, and then phase-arrayed coil images were combined52. Images were reconstructed with matrix size of 301×301×301 and voxel size of 1 mm3. After image reconstruction, fat and water separated images were obtained using a 2-point Dixon reconstruction from within the GE Healthcare Orchestra SDK. Signal bias in UTE images is corrected in the inverse log domain similar to 53. All training and testing were performed on a desktop computer running a 64-bit Linux operating system with an Intel Xeon W3520 quad-core CPU, 12 GB DDR3 RAM, and a Nvidia Quadro K4200 graphic card (1344 CUDA cores, 4GB GDDR5 RAM).
G. Evaluation and data analysis
Prospectively acquired data from 8 subjects invisible to the training phase of the CED network were used to evaluate the MRAC method. To evaluate the accuracy of tissue labeling, Dice coefficients for soft tissue, air, and bone were calculated between estimated labels and ground truth labels generated using co-registered CT image. To evaluate the efficacy of the CRF correction, Dice coefficients were calculated with the tissue labels with and without application of the CRF correction. To evaluate the proposed MRAC method in terms of reconstructed PET error, PET reconstruction (PET Toolbox, GE Healthcare) was performed using the system’s available MRAC methods (soft tissue only [MRAC-1] and atlas-based [MRAC-2]), an MRAC based on the dRHE acquisition using histogram-based image segmentation41 [MRAC-3], the proposed MRAC using deep learning and dRHE acquisition, and with the registered CT [CTAC] as the reference for comparison. Note that MRAC-3 and the proposed MRAC were performed using same dRHE data in evaluation.
PET reconstruction parameters were as follows: 256×256 matrix, 300×300 mm2 field of view, TOF-OSEM reconstruction algorithm, 28 iterations, 4 subsets, SharpIR, and 4 mm post filter. PET images reconstructed using MRAC-1, MRAC-2, MRAC-3, and the proposed MRAC were compared with a reference PET image reconstructed using CTAC, and pixelwise percent errors were calculated within 23 different region-of-interest (ROI) in brain. The ROIs were obtained using the IBASPM parcellation software with a brain atlas54. Repeated-measures one-way ANOVA were used to compare absolute errors within each ROI for MRAC-1, MRAC-2, MRAC-3, and the proposed MRAC. Paired-sample t-test was used for pairwise comparison between the proposed MRAC and MRAC-1, MRAC-2, or MRAC-3. Statistical analysis was performed with statistical significance defined as a p < 0.05. All image reconstructions and data analysis were performed in Matlab (2013a, MathWorks, Natick, USA) using a computer equipped with Opeteron 6134.
III. RESULTS
A. Tissue labeling
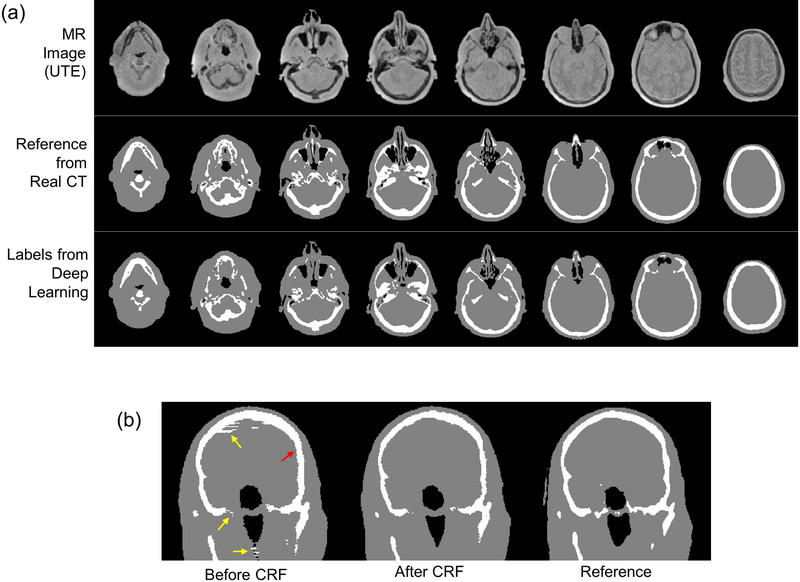
Figure 3-a shows input MR images, tissue labels estimated with the deep learning network, and the reference labels obtained from CT images. As shown, accurate tissue labels were obtained by deep learning where even small bone and air structures were detected. Figure 3-b shows tissue labels before and after CRF correction and the reference labels. CRF improved the tissue labels overall, alleviating discontinuity between slices as indicated by yellow arrows. Over-estimation of bone structure was corrected by the CRF correction as indicated by a red arrow. Dice coefficients across all 8 testing subjects for air (within the head), soft tissue, and bone were 0.75±0.03, 0.95±0.003, and 0.85±0.01 respectively before CRF correction. Dice coefficients across all 8 testing subjects for air, soft tissue, and bone were 0.76±0.03, 0.96±0.006, and 0.88±0.01 respectively after CRF correction, indicating that CRF correction did indeed improve bone and air segmentation.
Fig. 3.
Deep learning based tissue labeling. (a) Input MR image, reference labels, and labels estimated by deep learning, and (b) before and after application of CRF correction and reference labels. Note that CRF correction improves tissue labeling as indicated by yellow and red arrows in (b).
B. Pseudo CT images and PET reconstruction
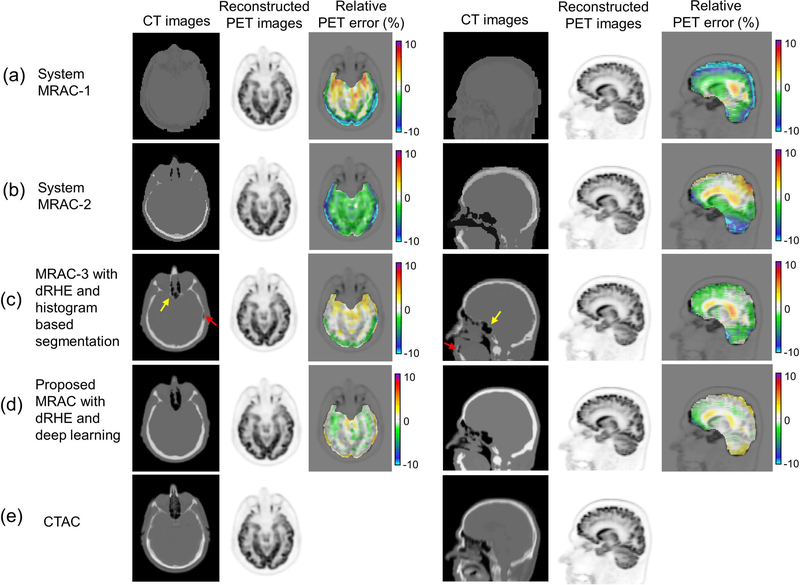
Figure 4 shows axial and sagittal slices of a CT image (real and pseudo), a reconstructed PET image, and pixelwise PET error in (a) MRAC-1, (b) MRAC-2, (c) MRAC-3, and (d) the proposed MRAC relative to (e) CTAC. The proposed MRAC shows the most accurate pseudo CT image among all MRACs, where MRAC-1 and MRAC-2 often produce poor estimates of head anatomy, while MRAC-3 shows errors in tissue boundaries due to partial volume effect (yellow arrow) and can struggle with small regions of air (red arrow). The propose MRAC improved PET quantification with errors less than 1% in most of brain regions. Table II shows mean and standard deviation of errors within 23 ROIs using MRAC-1, 2, 3, and the proposed MRAC. In the most brain regions, the proposed method showed significantly lower error (p < 0.05) compared with MRAC-1, 2, and 3. Differences between the proposed MRAC and system MRACs (MRAC-1 and MRAC-2) were greater than the differences between the proposed MRAC and MRAC-3, since those two methods are based on same dRHE acquisitions.
Fig. 4.
PET reconstruction. An axial slice and a coronal slice in a CT images, a reconstructed PET image, and a relative PET error map in (a) MRAC-1, (b) MRAC-2, (c) MRAC-3, the proposed MRAC with deep learning, and (e) CTAC. As seen in PET error maps the proposed MRAC shows much reduced percent error (< 1%) in most brain regions over other MRACs.
Table II.
PET error (mean ± standard deviation) relative to CT attenuation correction, utilizing MRAC-1, MRAC-2, MRAC-3, and proposed MRAC using deep learning and dRHE in 23 brain regions of 8 subjects. The proposed MRAC shows overall lower mean and standard deviation than MRAC-1, -2, and -3.
| Brain Regions | MRAC-1 error [%] | MRAC-2 error [%] | MRAC-3 error [%] | Proposed MRAC error [%] | p-value with MRAC-1 | p-value with MRAC-2 | p-value with dRHE-MRAC | ANOVA |
|---|---|---|---|---|---|---|---|---|
| Frontal Lobe Left | −7.7 ± 2.0 | −1.4 ± 4.2 | −1.4 ± 1.0 | −0.4 ± 0.9 | 5.1E-5 | 0.02 | 0.04 | 6.9E-7 |
| Frontal Lobe Right | −9.3 ± 1.8 | 0.1 ± 4.7 | −1.5 ± 1.1 | 0.2 ±1.2 | 2.8E-5 | 0.03 | 0.22 | 2.2E-7 |
| Temporal Lobe Left | −2.6 ± 2.8 | −4.8 ± 2.8 | −0.4 ± 1.2 | −0.8 ± 0.8 | 0.05 | 0.01 | 0.86 | 7.9E-4 |
| Temporal Lobe Right | −6.1 ± 2.3 | −4.1 ± 2.9 | −0.9 ± 1.0 | −0.1 ± 0.8 | 1.7E-3 | 7.6E-4 | 0.02 | 3.2E-6 |
| Parietal Lobe Left | −6.7 ± 2.2 | 0.9 ± 2.6 | −0.8 ± 0.9 | 0.6 ± 0.8 | 1.2E-3 | 0.20 | 0.49 | 2.2E-5 |
| Parietal Lobe Right | −7.5 ± 2.3 | 1.8 ± 3.1 | −0.8 ± 0.9 | 0.7 ± 0.8 | 5.5E-4 | 0.14 | 0.55 | 3.3E-5 |
| Occipital Lobe Left | −9.5 ± 2.5 | −3.1 ± 2.2 | −1.8 ± 1.0 | 0.8 ± 1.3 | 7.2E-4 | 0.05 | 0.41 | 1.2E-7 |
| Occipital Lobe Right | −11.3 ± 2.4 | −2.2 ± 2.1 | −1.8 ± 1.0 | 1.1 ± 1.3 | 1.5E-4 | 0.04 | 0.38 | 5.2E-10 |
| Cerebellum Left | −5.0 ± 3.7 | −5.9 ± 2.2 | −3.2 ± 1.8 | 0.2± 0.9 | 0.02 | 4.8E-4 | 0.01 | 4.2E-4 |
| Cerebellum Right | −5.0 ± 3.8 | −5.7 ± 2.4 | −2.9 ± 2.0 | −0.2 ± 0.9 | 0.02 | 9.6E-4 | 0.01 | 7.6E-4 |
| Brainstem | −1.4 ± 2.7 | −3.6 ± 2.6 | −0.7 ± 1.3 | −0.2 ± 0.7 | 0.08 | 4.3E-3 | 0.05 | 1.8E-3 |
| Caudate Nucleus Left | −2.7 ± 1.9 | −1.5 ± 1.8 | −0.4 ± 1.5 | −0.2 ± 0.8 | 0.02 | 0.11 | 0.06 | 0.03 |
| Caudate Nucleus Right | −2.4 ± 1.7 | −1.2 ± 2.3 | −0.5 ± 1.6 | −0.6 ± 0.9 | 0.03 | 0.09 | 0.22 | 0.10 |
| Putamen Left | −2.4 ± 2.1 | −1.8 ± 2.1 | −0.4 ± 1.6 | −0.9 ± 0.6 | 0.07 | 0.06 | 0.19 | 0.08 |
| Putamen Right | −1.9 ± 1.6 | −1.5 ± 2.0 | −0.3 ± 1.4 | −1.0 ± 0.6 | 0.16 | 0.17 | 0.46 | 0.28 |
| Thalamus Left | −2.1 ± 2.0 | −1.1 ± 1.8 | 0.4 ± 1.6 | 0.0 ± 0.6 | 0.03 | 0.09 | 0.05 | 0.08 |
| Thalamus Right | −1.6 ± 2.1 | −0.9 ± 2.1 | 0.1 ± 1.6 | −0.4 ± 0.6 | 0.02 | 0.11 | 0.04 | 0.10 |
| Subthalamic Nucleus Left | −2.3 ± 2.6 | −1.4 ± 2.1 | 0.4 ± 1.7 | −0.2 ± 0.6 | 0.03 | 0.02 | 0.02 | 0.04 |
| Subthalamic Nucleus Right | −0.8 ± 2.0 | −1.2 ± 1.4 | 0.4 ± 1.5 | −0.4 ± 0.3 | 0.10 | 2.3E-3 | 0.01 | 0.07 |
| Globus Pallidus Left | −3.2 ± 2.2 | −2.1 ± 1.9 | −0.5 ± 1.7 | −0.3 ± 0.6 | 0.01 | 3.1E-3 | 0.02 | 2.6E-3 |
| Globus Pallidus Right | −2.0 ± 1.5 | −1.8 ± 1.6 | −0.2 ± 1.6 | −0.6 ± 0.6 | 0.04 | 0.03 | 0.15 | 0.04 |
| Cingulate Region Left | −4.1 ± 1.7 | −1.0 ± 2.3 | −0.9 ± 1.4 | −0.5 ± 0.9 | 2.9E-3 | 0.09 | 0.17 | 4.5E-4 |
| Cingulate Region Right | −3.9 ± 1.5 | −0.8 ± 2.5 | −1.0 ± 1.4 | −0.5 ± 0.9 | 3.4E-3 | 0.11 | 0.16 | 1.1E-3 |
| All | −4.4 ± 3.7 | −1.9 ± 3.1 | −0.8 ± 1.7 | −0.2 ± 1.0 |
IV. DISCUSSION
The proposed framework for MRAC utilizes rapid dRHE acquisition (35 sec) providing high resolution (1 mm3) UTE, fat, and water images in a single scan. Compared to other UTE-based MRACs in literatures that require longer acquisition time (2~5 min)17,19, the proposed method more clinically-feasible and less likely to impede imaging workflow. Moreover, dRHE enables better image quality with reduced chemical shift artifacts of the second kind (intravoxel interference) and blurriness of short T2* species (i.e. bone in MRAC) by performing fastest encoding with minimized readout duration (~0.6ms). This is advantageous over zero echo time (ZTE) based methods where encoding speed is limited by RF pulse bandwidth.
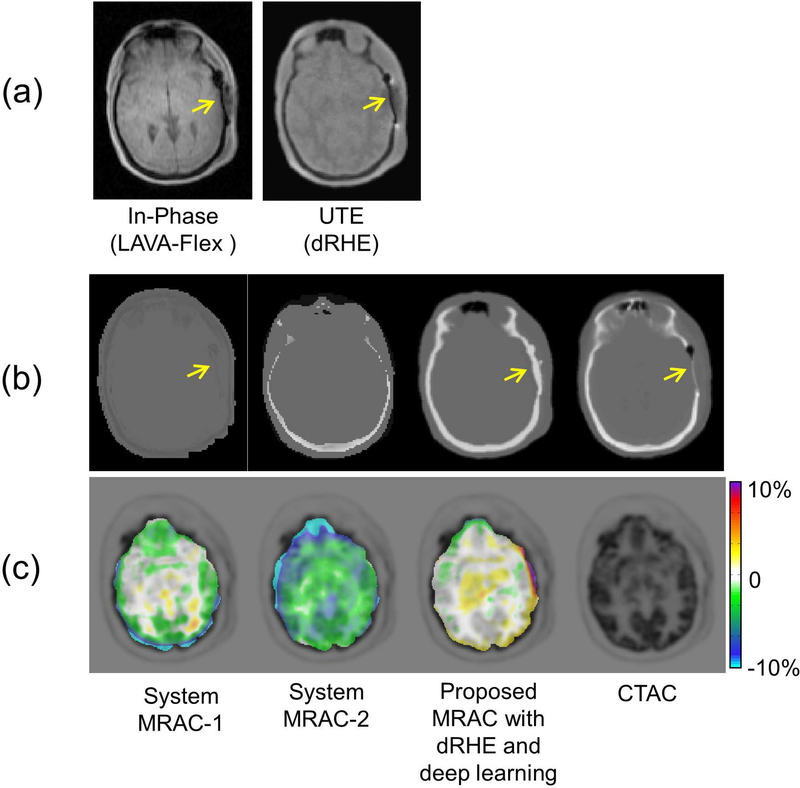
Although the proposed deep learning based method achieves reliable tissue labelling, accuracy of the labeling depends on a quality of the input MR image. Figure 5 shows an example of PET reconstruction performed with a patient who had a metallic implant in the skull. Figure 5-a shows the MR images used for MRAC, which show strong susceptibility artifact near metallic implant (yellow arrows). Figure 5-b shows the generated pseudo CT image by system MRAC-1, MRAC-2, and the proposed MRAC, together with the real CT image. System MRAC-2 incorrectly estimated the anatomy of the head in the pseudo CT image, and both system MRAC-1 and the proposed MRAC were affected by susceptibility artifacts. However, the proposed MRAC achieves lower PET error in this subject (less than 1%) than with the system MRACs. As this example illustrates, the proposed method may not provide accurate tissue labeling in special cases with anatomical abnormality, however, this appears to have small impact on overall PET error in the brain as shown in Figure 5-c. Moreover, training with a larger dataset that includes more cases of anatomical abnormalities will improve the tissue labeling.
Fig. 5.
Reconstructed PET reconstruction image for a patient with a metallic implant in a skull. (a) In-Phase MR images used in system MRACs and the proposed MRAC, (b) generated pseudo CT images and a real CT image, and (c) percent pixel-wise PET error maps. MR images are prone to susceptibility artifact near metallic implant (yellow arrows), which is challenging issue in for most MRAC approaches. Despite the mis-estimation of bone near the metallic implant shown in a pseudo CT image, the proposed method shows improved better PET errors compared than with system MRAC-1 and −2.
Deep learning networks based on CNNs can provide robust and rapid tissue labelling in images. Pre-training of our network with T1 weighted images took approximately 34 hours, while training with the UTE images took 14 hours. Once training was completed, tissue labeling of one UTE image took only 1 min. The additional CRF correction did not increase processing time by much, where processing time was 1~2 min for one subject. We have shown that the CRF correction improved bone and air detection in our model and corrects for discretization artifacts in tissue labels. These discretization artifacts are present because our deep learning model performs tissue labeling with 2D axial slices of UTE images, which can become discontinuous in the superior-inferior direction when combined into 3D labels. The CRF correction noticeably improved the 3D tissue labels, resulting in more continuous tissue labeling between neighboring pixels.
V. CONCLUSIONS
In this study, we have evaluated the performance of a new framework for MRAC based on rapid (35 sec) UTE imaging combined with a deep learning network (1 min) and a CRF correction (1~2 min). We showed that our method can accurately differentiate bone and air in MR images, and improved the accuracy of PET quantification when compared to clinically-used system MRACs.
Supplementary Material
ACKNOWLEDGEMENTS
The work was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health [grant number 1R21EB013770]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
DISCLOSURE OF CONFLICTS OF INTEREST
The authors have no relevant conflicts of interest to disclose.
REFERENCES
- 1.Hofmann M, Steinke F, Scheel V, et al. MRI-based attenuation correction for PET/MRI: a novel approach combining pattern recognition and atlas registration. J Nucl Med. 2008;49(11):1875–1883. doi: 10.2967/jnumed.107.049353. [DOI] [PubMed] [Google Scholar]
- 2.Wollenweber SD, Ambwani S, Delso G, et al. Evaluation of an atlas-based PET head attenuation correction using PET/CT & MR Patient Data. IEEE Trans Nucl Sci. 2013;60(5):3383–3390. doi: 10.1109/TNS.2013.2273417. [DOI] [Google Scholar]
- 3.Schreibmann E, Nye J a, Schuster DM, Martin DR, Votaw J, Fox T. MR-based attenuation correction for hybrid PET-MR brain imaging systems using deformable image registration. Med Phys. 2010;37(5):2101–2109. doi: 10.1118/1.3377774. [DOI] [PubMed] [Google Scholar]
- 4.Hofmann M, Bezrukov I, Mantlik F, et al. MRI-based attenuation correction for whole-body PET/MRI: quantitative evaluation of segmentation- and atlas-based methods. J Nucl Med. 2011;52(9):1392–1399. doi: 10.2967/jnumed.110.078949. [DOI] [PubMed] [Google Scholar]
- 5.Malone IB, Ansorge RE, Williams GB, Nestor PJ, Carpenter TA, Fryer TD. Attenuation correction methods suitable for brain imaging with a PET/MRI scanner: a comparison of tissue atlas and template attenuation map approaches. J Nucl Med. 2011;52(7):1142–1149. doi: 10.2967/jnumed.110.085076. [DOI] [PubMed] [Google Scholar]
- 6.Zaidi H, Montandon M-L, Slosman DO. Magnetic resonance imaging-guided attenuation and scatter corrections in three-dimensional brain positron emission tomography. Med Phys. 2003;30(5):937–948. doi: 10.1118/1.1569270. [DOI] [PubMed] [Google Scholar]
- 7.Wagenknecht G, Kops ER, Tellmann L, Herzog H. Knowledge-based segmentation of attenuation-relevant regions of the head in T1-weighted MR images for attenuation correction in MR/PET systems. IEEE Nucl Sci Symp Conf Rec. 2009:3338–3343. doi: 10.1109/NSSMIC.2009.5401751. [DOI] [Google Scholar]
- 8.Wagenknecht G, Kops ER, Kaffanke J, et al. CT-based evaluation of segmented head regions for attenuation correction in MR-PET systems. IEEE Nucl Sci Symp Conf Rec. 2010:2793–2797. doi: 10.1109/NSSMIC.2010.5874301. [DOI] [Google Scholar]
- 9.Wagenknecht G, Rota Kops E, Mantlik F, et al. Attenuation correction in MR-BrainPET with segmented T1-weighted MR images of the patient’s head - A comparative study with CT. IEEE Nucl Sci Symp Conf Rec. 2012:2261–2266. doi: 10.1109/NSSMIC.2011.6153858. [DOI] [Google Scholar]
- 10.Kops ER, Wagenknecht G, Scheins J, Tellmann L, Herzog H. Attenuation correction in MR-PET scanners with segmented T1-weighted MR images. IEEE Nucl Sci Symp Conf Rec. 2009:2530–2533. doi: 10.1109/NSSMIC.2009.5402034. [DOI] [Google Scholar]
- 11.Hu Z, Ojha N, Renisch S, et al. MR-based attenuation correction for a whole-body sequential PET/MR system. IEEE Nucl Sci Symp Conf Rec. 2009:3508–3512. doi: 10.1109/NSSMIC.2009.5401802. [DOI] [Google Scholar]
- 12.Schulz V, Torres-Espallardo I, Renisch S, et al. Automatic, three-segment, MR-based attenuation correction for whole-body PET/MR data. Eur J Nucl Med Mol Imaging. 2011;38(1):138–152. doi: 10.1007/s00259-010-1603-1. [DOI] [PubMed] [Google Scholar]
- 13.Akbarzadeh A, Ay MR, Ahmadian A, Riahi Alam N, Zaidi H. Impact of using different tissue classes on the accuracy of MR-based attenuation correction in PET-MRI. IEEE Nucl Sci Symp Conf Rec. 2012:2524–2530. doi: 10.1109/NSSMIC.2011.6152682. [DOI] [Google Scholar]
- 14.Ladefoged CN, Law I, Anazodo U, et al. A multi-centre evaluation of eleven clinically feasible brain PET/MRI attenuation correction techniques using a large cohort of patients. Neuroimage. 2017;147(June):346–359. doi: 10.1016/j.neuroimage.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keereman V, Fierens Y, Broux T, De Deene Y, Lonneux M, Vandenberghe S. MRI-based attenuation correction for PET/MRI using ultrashort echo time sequences. J Nucl Med. 2010;51(5):812–818. doi: 10.2967/jnumed.109.065425. [DOI] [PubMed] [Google Scholar]
- 16.Berker Y, Franke J, Salomon a., et al. MRI-Based Attenuation Correction for Hybrid PET/MRI Systems: A 4-Class Tissue Segmentation Technique Using a Combined Ultrashort-Echo-Time/Dixon MRI Sequence. J Nucl Med. 2012;53(5):796–804. doi: 10.2967/jnumed.111.092577. [DOI] [PubMed] [Google Scholar]
- 17.Juttukonda MR, Mersereau BG, Chen Y, et al. MR-based attenuation correction for PET/MRI neurological studies with continuous-valued attenuation coefficients for bone through a conversion from R2* to CT-Hounsfield units. Neuroimage. 2015;112:160–168. doi: 10.1016/j.neuroimage.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santos Ribeiro A, Rota Kops E, Herzog H, Almeida P. Skull segmentation of UTE MR images by probabilistic neural network for attenuation correction in PET/MR. Nucl Instruments Methods Phys Res Sect A Accel Spectrometers, Detect Assoc Equip. 2013;702:114–116. doi: 10.1016/j.nima.2012.09.005. [DOI] [Google Scholar]
- 19.Leynes AP, Yang J, Shanbhag DD, et al. Hybrid ZTE/Dixon MR-based attenuation correction for quantitative uptake estimation of pelvic lesions in PET/MRI. Med Phys. 2017;44(3):902–913. doi: 10.1002/mp.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiesinger F, Sacolick LI, Menini A, et al. Zero TE MR bone imaging in the head. Magn Reson Med. 2015;114(October 2014):107–114. doi: 10.1002/mrm.25545. [DOI] [PubMed] [Google Scholar]
- 21.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 22.Krizhevsky A, Sutskever I, Hinton GE. ImageNet Classification with Deep Convolutional Neural Networks. Adv Neural Inf Process Syst. 2012:1–9. doi: 10.1016/j.protcy.2014.09.007. [DOI] [Google Scholar]
- 23.Long J, Shelhamer E, Darrell T. Fully Convolutional Networks for Semantic Segmentation. ArXiv e-prints. 2014;1411. [DOI] [PubMed]
- 24.Greenspan Bram van HG, Summers RM. Guest Editorial Deep Learning in Medical Imaging: Overview and Future Promise of an Exciting New Technique. IEEE Trans Med Imaging. 35(5):1153–1159. [Google Scholar]
- 25.Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42(1995):60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 26.Avendi MR, Kheradvar A, Jafarkhani H. A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Med Image Anal. 2016;30:108–119. doi: 10.1016/j.media.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 27.Moeskops P, Viergever MA, Mendrik AM, de Vries LS, Benders MJ, Isgum I. Automatic segmentation of MR brain images with a convolutional neural network. IEEE Trans Med Imaging. 2016. doi: 10.1109/TMI.2016.2548501. [DOI] [PubMed] [Google Scholar]
- 28.Pereira S, Pinto A, Alves V, Silva CA. Brain Tumor Segmentation using Convolutional Neural Networks in MRI Images. IEEE Trans Med Imaging. 2016. doi: 10.1109/TMI.2016.2538465. [DOI] [PubMed] [Google Scholar]
- 29.Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, Kijowski R. Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn Reson Med. 2017;25:5662. doi: 10.1002/mrm.26841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. May 2015:234–241. doi: 10.1007/978-3-319-24574-4_28. [DOI] [Google Scholar]
- 31.Christ PF, Ettlinger F, Grün F, et al. Automatic Liver and Tumor Segmentation of CT and MRI Volumes Using Cascaded Fully Convolutional Neural Networks.
- 32.Liu F, Jang H, Kijowski R, Bradshaw T, McMillan AB. Deep Learning MR Imaging–based Attenuation Correction for PET/MR Imaging. Radiology. 2018;286(2):676–684. doi: 10.1148/radiol.2017170700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zha W, Kruger SJ, Johnson KM, et al. Pulmonary ventilation imaging in asthma and cystic fibrosis using oxygen-enhanced 3D radial ultrashort echo time MRI. J Magn Reson Imaging. October 2017:1–11. doi: 10.1002/jmri.25877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao G, Liu F, Oler JA, Meyerand ME, Kalin NH, Birn RM. Bayesian convolutional neural network based MRI brain extraction on nonhuman primates. Neuroimage. 2018;175(October 2017):32–44. doi: 10.1016/j.neuroimage.2018.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Z, Zhao G, Kijowski R LF. Deep Convolutional Neural Network for Segmentation of Knee Joint Anatomy. Magn Reson Med. 2018;In Press. doi: 10.1002/mrm.27229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Badrinarayanan V, Kendall A, Cipolla R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39(12):2481–2495. doi: 10.1109/TPAMI.2016.2644615. [DOI] [PubMed] [Google Scholar]
- 37.Simonyan K, Zisserman A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv Prepr. 2014:1–10. doi: 10.1016/j.infsof.2008.09.005. [DOI]
- 38.Jang H, Liu F, Bradshaw T, McMillan AB. Rapid dual-echo ramped hybrid encoding MR-based attenuation correction (dRHE-MRAC) for PET/MR. Magn Reson Med. 2018;79(6):2912–2922. doi: 10.1002/mrm.26953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jang H, Wiens CN, McMillan AB. Ramped hybrid encoding for improved ultrashort echo time imaging. Magn Reson Med. 2016;76(3):814–825. doi: 10.1002/mrm.25977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krahenbuhl P, Koltun V. Efficient Inference in Fully Connected CRFs with Gaussian Edge Potentials. In: Neural Information Processing Systems.; 2011:109–117. [Google Scholar]
- 41.Jang H, Mcmillan AB. Rapid Dual Echo UTE MR-Based Attenuation Correction. In: The ISMRM 25th Annual Meeting Honolulu, Hawaii, USA; 2017:Abstract 3886. [Google Scholar]
- 42.Pauly J, Le Roux P, Nishimura D, Macovski A. Parameter relations for the Shinnar-Le Roux selective excitation pulse design algorithm (NMR imaging). IEEE Trans Med Imaging. 1991;10(1):53–65. doi: 10.1109/42.75611. [DOI] [PubMed] [Google Scholar]
- 43.Coombs BD, Szumowski J, Coshow W. Two-point Dixon technique for water-fat signal decomposition with B0 inhomogeneity correction. Magn Reson Med. 1997;38(6):884–889. doi: 10.1002/mrm.1910380606. [DOI] [PubMed] [Google Scholar]
- 44.Jia Y, Shelhamer E, Donahue J, et al. Caffe: Convolutional Architecture for Fast Feature Embedding. ArXiv e-prints. 2014;1408.
- 45.Klein S, Staring M, Murphy K, Viergever MA, Pluim JPW. Elastix: A toolbox for intensity-based medical image registration. IEEE Trans Med Imaging. 2010;29(1):196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- 46.Jarret K, Kavukcuoglu K, Ranzato M, LeCun Y. What is the Best Multi-Stage Architecture for Object Recognition. Proc IEEE Int Conf Comput Vis. 2009. doi:citeulike-article-id:13828245. [Google Scholar]
- 47.Bottou L Large-Scale Machine Learning with Stochastic Gradient Descent. 19th Int Conf Comput Stat. 2010:177–186. doi: 10.1007/978-3-7908-2604-3_16LB-Bottou2010. [DOI] [Google Scholar]
- 48.Catana C, van der Kouwe A, Benner T, et al. Toward implementing an MRI-based PET attenuation-correction method for neurologic studies on the MR-PET brain prototype. J Nucl Med. 2010;51(9):1431–1438. doi: 10.2967/jnumed.109.069112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez-Möller A, Souvatzoglou M, Delso G, et al. Tissue classification as a potential approach for attenuation correction in whole-body PET/MRI: evaluation with PET/CT data. J Nucl Med. 2009;50(4):520–526. doi: 10.2967/jnumed.108.054726. [DOI] [PubMed] [Google Scholar]
- 50.Akbarzadeh A, Ay MR, Ahmadian A, Riahi Alam N, Zaidi H. MRI-guided attenuation correction in whole-body PET/MR: Assessment of the effect of bone attenuation. Ann Nucl Med. 2013;27(2):152–162. doi: 10.1007/s12149-012-0667-3. [DOI] [PubMed] [Google Scholar]
- 51.Beatty PJ, Nishimura DG, Pauly JM. Rapid gridding reconstruction with a minimal oversampling ratio. IEEE Trans Med Imaging. 2005;24(6):799–808. doi: 10.1109/TMI.2005.848376. [DOI] [PubMed] [Google Scholar]
- 52.Walsh DO, Gmitro AF, Marcellin MW. Adaptive reconstruction of phased array MR imagery. Magn Reson Med. 2000;43(5):682–690. doi:. [DOI] [PubMed] [Google Scholar]
- 53.Wiesinger F, Sacolick LI, Menini A, et al. Zero TE MR Bone Imaging in the Head. Magn Reson Med. 2015;0(October 2014):n/a–n/a. doi: 10.1002/mrm.25545. [DOI] [PubMed] [Google Scholar]
- 54.Alemán-Gómez Y, Melie-García L, Valdés-Hernandez P. IBASPM: Toolbox for automatic parcellation of brain structures. In: The 12th Annual Meeting of the Organization for Human Brain Mapping Florence, Italy.; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.