Abstract
Background
Historically, the major cause of meningococcal epidemics in the meningitis belt of sub-Saharan Africa has been Neisseria meningitidis serogroup A (NmA), but the incidence has been substantially reduced since the introduction of a serogroup A conjugate vaccine starting in 2010. We performed whole-genome sequencing on isolates collected post-2010 to assess their phylogenetic relationships and inter-country transmission.
Methods
A total of 716 invasive meningococcal isolates collected between 2011 and 2016 from 11 meningitis belt countries were whole-genome sequenced for molecular characterization by the three WHO Collaborating Centers for Meningitis.
Findings
We identified three previously-reported clonal complexes (CC): CC11 (n = 434), CC181 (n = 62) and CC5 (n = 90) primarily associated with NmW, NmX, and NmA, respectively, and an emerging CC10217 (n = 126) associated with NmC. CC11 expanded throughout the meningitis belt independent of the 2000 Hajj outbreak strain, with isolates from Central African countries forming a distinct sub-lineage within this expansion. Two major sub-lineages were identified for CC181 isolates, one mainly expanding in West African countries and the other found in Chad. CC10217 isolates from the large outbreaks in Nigeria and Niger were more closely related than those from the few cases in Mali and Burkina Faso.
Interpretations
Whole-genome based phylogenies revealed geographically distinct strain circulation as well as inter-country transmission events. Our results stress the importance of continued meningococcal molecular surveillance in the region, as well as the development of an affordable vaccine targeting these strains.
Fund
Meningitis Research Foundation; CDC's Office of Advanced Molecular Detection; GAVI, the Vaccine Alliance.
Keywords: Meningitis, Meningitis belt, Invasive meningococcal disease, Phylogenetics, Neisseria meningitidis
Research in context.
Evidence before this study
We searched NCBI's PubMed database for published literature relating to meningococcal strains in the meningitis belt leading up to and during our study's time period, with the goal of obtaining information on important events and outbreaks that occurred in the region. The key words that were used were “meningitis belt”, “Neisseria meningitidis meningitis belt” and “meningitis belt 2011 2016.” We reviewed the resulting articles and the cited articles that were of interest. In 2010, a meningococcal serogroup A conjugate vaccine (MACV) was introduced to the region, and was rolled out over the subsequent years to the various countries of the meningitis belt. Several large scale meningococcal outbreaks and epidemics occurred in the region during our study's time period, specifically a clonal complex (CC) 11 N.meningitidis serogroup W (NmW) epidemic in Burkina Faso in 2012, and large outbreaks of CC10217 NmC in Nigeria (2013–2016) and Niger (2015). These strains, along with CC181 NmX, have become prominent causes of invasive meningococcal disease in the region.
Added value of this study
We used whole-genome sequence based analyses to provide a macro-regional overview of the circulating and emerging invasive meningococcal strains in meningitis belt countries between 2011 and 2016. Using phylogenetic analysis, we provide information regarding the evolutionary relationships of the invasive lineages of the meningitis belt and their inter-country transmission. Notably, we identified geographically distinct strains within CC11, CC5, and CC181 that were circulating and expanding in Central and West African countries during this time period. We also build on past findings describing the primary lineage of CC11 NmW in the meningitis belt, showing the diversity within this lineage, and how it has spread across the region. Our work provides a comprehensive phylogeny for the emerging CC10217, including outbreak and epidemic isolates from Nigeria and Niger as well as invasive isolates collected from Mali and Burkina Faso. For CC181, our results show that at least two strains of this clonal complex were circulating during the time period, each of which appear to be expanding in distinct regions.
Implications of all the available evidence
While epidemics of serogroup A have been largely eliminated from the meningitis belt, other serogroups have risen as major causes of meningitis in the region. Understanding the regional spreading and molecular characteristics of these strains is critical for implementing effective surveillance strategies and for developing vaccines targeting these strains.
Alt-text: Unlabelled Box
1. Introduction
With an estimated population of 400 million, the meningitis belt of sub-Saharan Africa, a region stretching from the coast of Senegal to Ethiopia, holds the highest reported meningitis incidence rate globally [1]. The majority of meningococcal epidemics have historically been caused by N. meningitidis serogroup A (NmA), but the incidence has been substantially reduced since the introduction of a serogroup A conjugate vaccine (MACV, MenAfriVac™) in 2010 [3]. Since then, major causes of meningococcal meningitis in the region have shifted to other serogroups, such as serogroup W (NmW) [4], serogroup X (NmX) [5], and more recently, serogroup C (NmC) [6]. Of note, NmC, prior to recent years, had been very rare and had not caused large outbreaks in the region since 1979 [7].
Between 2011 and 2016, over half (62%; n = 3309) of the confirmed meningococcal meningitis cases reported to the World Health Organization (WHO) in meningitis belt countries were NmW [8]. This included an epidemic in Burkina Faso in 2012 where 2353 confirmed meningitis cases were reported; 1451 (61%) of which were confirmed to be NmW [4]. These NmW cases primarily belonged to clonal complex 11 (CC11), a hyper-invasive lineage typically associated with NmC or NmW and less commonly B or Y [9]. Studies revealed that the epidemic strain shares a common ancestor with the outbreak strain collected in 2000 during the Hajj to Saudi Arabia [10].
While NmW has been the dominant cause of meningococcal meningitis in the region since 2010, NmC has also been a major contributor, accounting for 12.5% (n = 666) of the cases reported to the WHO between 2011 and 2016 [8]. In 2013 and 2014, NmC outbreaks occurred in Nigeria and were caused by a sequence type (ST) that had not been previously detected (ST-10217) [11]. In 2015, ST-10217 was responsible for an epidemic in Niger; phylogenetic analysis of 81 ST-10217 isolates from this epidemic compared with global NmC isolates from eight different clonal complexes showed that the Niger ST-10217 isolates formed a distinct cluster [6].
Along with NmW and NmC, NmX has remained a consistent cause of meningococcal meningitis in the region, contributing 7.6% (n = 406) of the confirmed cases reported to the WHO between 2011 and 2016 [8]. In 2006, NmX was responsible for outbreaks in Niger, comprising 51% of 1139 confirmed meningococcal meningitis cases [12]. Shortly after, sporadic cases and outbreaks of NmX were reported in 2007 in Togo and in 2010 and 2012 in Burkina Faso [5]. All of these reported NmX cases were identified to be ST-181 belonging to clonal complex 181 (CC181).
As continued epidemics and outbreaks of non-A meningococcal serogroups occur in the region, there is a growing need for the use of multivalent conjugate vaccines. In addition to MenAfriVac, several other polysaccharide conjugate vaccines are available, but many are currently not affordable for most African countries [13]. Two protein-based meningococcal vaccines targeting N. meningitidis serogroup B (NmB) are approved for use in the United States and Europe: MenB-FHbp (Trumenba) [14] and MenB-4C (Bexsero) [15]. Although NmB is rarely found in the meningitis belt, previous studies have demonstrated the potential cross-protective effect of these vaccines on diverse strains [16], including those from the meningitis belt [17], raising interest in assessing the potential protection offered on other meningococcal serogroups.
Proper understanding of the major and emerging causes of meningococcal meningitis in the region is critical for guiding surveillance strategies and vaccine development. We used whole-genome sequencing (WGS) to characterize these strains and to determine their phylogenetic relationships, regional spread, and allelic variants of interest.
2. Methods
2.1. Isolate collection
A total of 716 invasive meningococcal isolates from major outbreaks and sporadic cases were collected by the three WHO Collaborating Centers for Meningitis: Centers for Disease Control and Prevention (CDC), Atlanta GA; Norwegian Institute of Public Health (NIPH) Oslo, Norway; Institut Pasteur (IP) Paris, France. All isolates were collected throughout 2011 to 2016 according to WHO laboratory methods [18] from the surveillance programs of the following 11 meningitis belt countries: Benin, Burkina Faso, Cameroon, Central African Republic, Chad, Guinea, Ivory Coast, Mali, Niger, Nigeria, and Togo (Supplement 1). The isolates collected represent a convenient collection obtained from the countries, and were not selected based on a specific criteria. Table 1 contains the number of laboratory-confirmed cases of meningococcal meningitis reported to the WHO from each country along with the number of isolates collected for our study. In several instances, the number of isolates received by collaborating centers exceeded the number of reported cases. A total of 119 historical isolates collected from the years 1984 to 2010 were included to provide evolutionary context for the phylogenetic analyses. These historical isolates were selected based on the following criteria: the isolates belonged to a clonal complex that was being considered in our analysis, were collected on or before 2010 and were collected from a meningitis belt country or one that borders the meningitis belt. Information about these historical isolates is provided in Supplement 2. The serogroup of each isolate was determined by slide agglutination testing or PCR targeting capsule-defining genes [18].
Table 1.
Confirmed meningococcal meningitis cases reported to WHO and isolates collected.
| Countries | 2011 |
2012 |
2013 |
2014 |
2015 |
2016 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Isolates | Cases | Isolates | Cases | Isolates | Cases | Isolates | Cases | Isolates | Cases | Isolates | |
| Mali | 29 | 6 | 94 | 30 | 6 | 0 | 12 | 0 | 23 | 16 | 44 | 0 |
| Burkina Faso | 257 | 41 | 843 | 167 | 180 | 20 | 210 | 4 | 258 | 11 | 176 | 5 |
| Niger | 373 | 17 | 22 | 0 | 11 | 0 | 24 | 0 | 1390 | 102 | 352 | 0 |
| Nigeria | 4 | 0 | 4 | 0 | 10 | 7 | 38 | 5 | 20 | 9 | 22 | 14 |
| Chad | 104 | 47 | 47 | 23 | 3 | 24 | 0 | 1 | 1 | 1 | 1 | 0 |
| Cameroon | 92 | 0 | 19 | 4 | 2 | 0 | 0 | 0 | 6 | 0 | 2 | 0 |
| Benin | 0 | 0 | 6 | 41 | 5 | 0 | 4 | 0 | 4 | 0 | 13 | 0 |
| Togo | 2 | 0 | 9 | 0 | 4 | 0 | 1 | 16 | 36 | 12 | 307 | 42 |
| Ivory coast | 0 | 0 | 89 | 7 | 0 | 0 | 0 | 0 | 2 | 1 | 3 | 0 |
| Guinea | 0 | 4 | 0 | 0 | 15 | 9 | 13 | 0 | 74 | 0 | 13 | 0 |
| CAR | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 56 | 23 |
| Total | 861 | 115 | 1137 | 272 | 236 | 60 | 302 | 26 | 1814 | 159 | 989 | 84 |
The table above contains the confirmed meningococcal meningitis cases and isolates collected for each country per year of the study. The confirmed cases were obtained from the WHO Meningitis Weekly Bulletin Reports for the years 2011–2016.
2.2. Whole genome sequencing and molecular typing
Of the 716 isolates obtained, 14 were sequenced using the Pacific Biosystems (PacBio) RSII instrument and assembled using PacBio's Hierarchical Genome Assembly Process version 3 (HGAP) [19]. These assemblies were then corrected using 250-bp paired-end Illumina read data generated by the Illumina MiSeq. The remaining 702 isolates were sequenced using the Illumina MiSeq. Each WHO Collaborating Center sequenced its own isolates using either 150-bp paired-end or 250-bp paired-end Illumina kits. The genome assemblies for all isolates used in this study have been deposited to PubMLST (https://pubmlst.org/bigsdb?db=pubmlst_neisseria_isolates) [20]. The accession numbers for the 716 isolates used in the study can be found in supplement 1, and the accession numbers for the historical isolates can be found in supplement 2.
All Illumina sequencing reads were trimmed using CutAdapt 1.8.1 [21] to remove identified adapter sequences and bases with quality score of <Q20. The trimmed reads were then assembled using the de novo assembler SPAdes 3.7.0 [22]. The assemblies were then filtered to remove contigs with less than 10% of the assembly-wide average coverage using in-house scripts. These final assemblies were used to identify isolate's ST, CC and vaccine antigen variant profile by comparing against the PubMLST database [20].
2.3. Phylogenetic analysis
The phylogenies for the individual clonal complexes were generated by first creating a SNP alignment using Snippy version 3.1 (https://github.com/tseemann/snippy). The full SNP alignment was used as input for Gubbins [23], which uses RAxML [24] to generate an initial maximum likelihood phylogeny. Gubbins employs a scan statistic to identify and mask regions of recombination on each individual branch, followed by an additional phylogeny building step. The resulting phylogeny was used as input for RAxML once more to correct for ascertainment bias and to obtain bootstrap values. The CC11 phylogeny was produced by taking the full SNP alignment produced by Snippy, identifying and masking recombination sites using Gubbins, and providing the resulting alignment as the input for BEAST 1.8.2 [25]. An ascertainment bias correction step was included by providing the count of the non-core sites for each nucleotide in the reference within the BEAST input XML file. The HKY substitution model was used with relaxed clock rate with an exponential growth tree model. The phylogeny in the first figure containing all 716 isolates used in the study was generated using Parsnp from the Harvest suite of tools [26] with the FAM18 strain of N.meningitidis used as the reference.
2.4. Statistical analysis
All phylogenies, except for CC11, were generated using the maximum likelihood statistical method for estimating unknown parameters. The CC11 phylogeny was generated using BEAST, a Bayesian statistical framework, enabling prior information about the data to be considered in the analysis.
2.5. Ethics statement
This evaluation was determined by human subjects review at the CDC to be public health non-research and review by the Institutional Review Board was not required.
3. Results
3.1. Clonal complex distribution across 11 countries
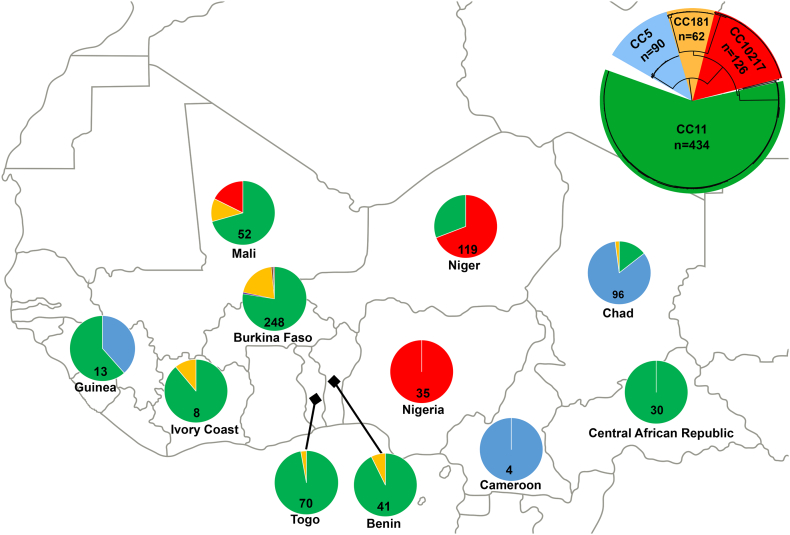
As shown in Fig. 1, CC11 predominated, constituting 60.6% (n = 434) of the isolates in the study. CC11 was also the most widespread CC, as it was found among isolates from 9 of the 11 countries. CC10217 was the next major CC identified, forming 17.5% (n = 126) of the isolate collection; these isolates were primarily collected from Niger, Nigeria, and Mali, as well as one from Burkina Faso. CC5 contributed 12.5% (n = 90) of the total isolates, and isolates from this CC were collected from Guinea, Cameroon and Chad. Finally, CC181 was the last major CC identified in our collection, constituting 8.6% (n = 62) of the isolates and was found in 6 of the 11 countries. CC175 and CC23 were minor CCs represented in our collection by two isolates each; all from Burkina Faso, and were serogroup W and Y, respectively.
Fig. 1.
Geographic clonal complex distribution of study isolates: The clonal complex distribution for all 716 meningococcal isolates collected between 2011 and 2016 are shown. The top right includes a circular phylogeny of these isolates and presents the colour key for each clonal complex, which is as follows: CC11 (green), CC10217 (red), CC181 (dark yellow), CC5 (light blue). Two additional clonal complexes were identified (CC175 and CC23) from Burkina Faso, which are not represented with a colour in the figure. Within each country, a pie chart represents the proportions of each clonal complex identified, as well as the total number of isolates included from that country. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Phylogenetic relationships and regional spread
3.2.1. CC11
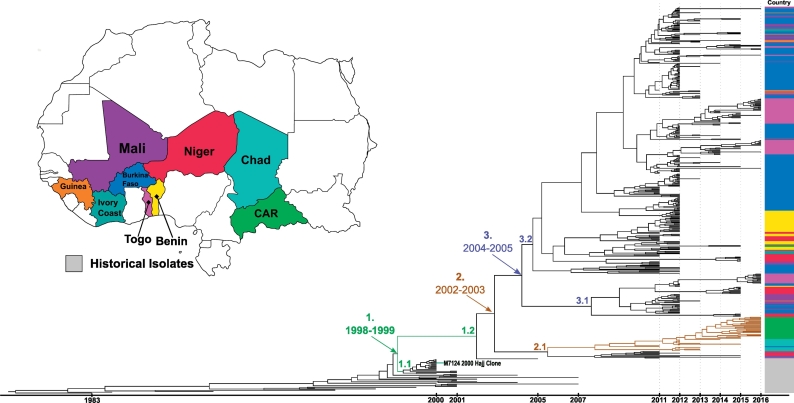
All CC11 isolates in our collection were serogroup W. A timed phylogeny including historical isolates (Fig. 2) was generated for CC11 to provide greater resolution for determining expansion of the CC across the meningitis belt. Three clades are marked 1–3, indicating the estimated year range for the divergence from the common ancestor. Clade 1 (1998–1999) marks the common ancestor which diverged into sub-clades 1.1 and 1.2. Sub-clade 1.1 consisted primarily of the 2000–2001 Hajj-related outbreak isolates, including M7124, the 2000 Hajj outbreak clone, while sub-clade 1.2 appeared to have continued expanding throughout the meningitis belt. Prior to clade 1, an ancestral lineage containing isolates ranging from years 2000–2007 was present that is no longer seen in the region. Clade 2 (2002−2003) marks the common ancestor which diverged into two main clades, sub-clade 2.1 and clade 3. Within sub-clade 2.1, there is a sub-lineage that primarily contained isolates from the two Central African countries, Chad and Central African Republic. Clade 3 primarily consisted of the dominant strains which have been circulating throughout West Africa, and it contained all of the 2016 West African isolates. This clade diverged into sub-clades 3.1 and 3.2. Sub-clade 3.1 contains ten isolates from the Burkina Faso 2012 epidemic, along with isolates from nearby West African countries such as Mali, Benin and Guinea. Sub-clade 3.2 contains substantial diversity, with various clusters found throughout the clade, including a large cluster of 2012 Benin and 2011–2012 Niger isolates. Throughout this sub-clade, there are numerous clusters containing isolates from the Burkina Faso 2012 epidemic. These clusters often contain isolates from neighboring countries such as Togo, Guinea and Mali, suggesting inter-country transmission during that time period. In some cases, these epidemic isolates form outgroups to sub-clades that continue expanding within the respective country; this can be seen with the two predominantly Togo sub-clades in sub-clade 3.2, and another in sub-clade 3.1.
Fig. 2.
CC11 molecular clock timed phylogeny: A molecular clock based phylogeny was created for CC11 consisting of 434 study isolates and 40 historical (2007 or earlier) African isolates. The map represents the colour key for each country included in the phylogeny, with gray representing the historical isolates. The scale bar at the bottom indicates which year the isolates were collected from. Three clades of interest are labeled one to three, indicating the estimated date range for the origin of the common ancestor. The brown branches in sub-clade 2.1 represent a sub-lineage is primarily expanding in two Central African countries, Chad and Central African Republic. The 2000 Hajj outbreak clone (M7124) is also noted in the figure. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
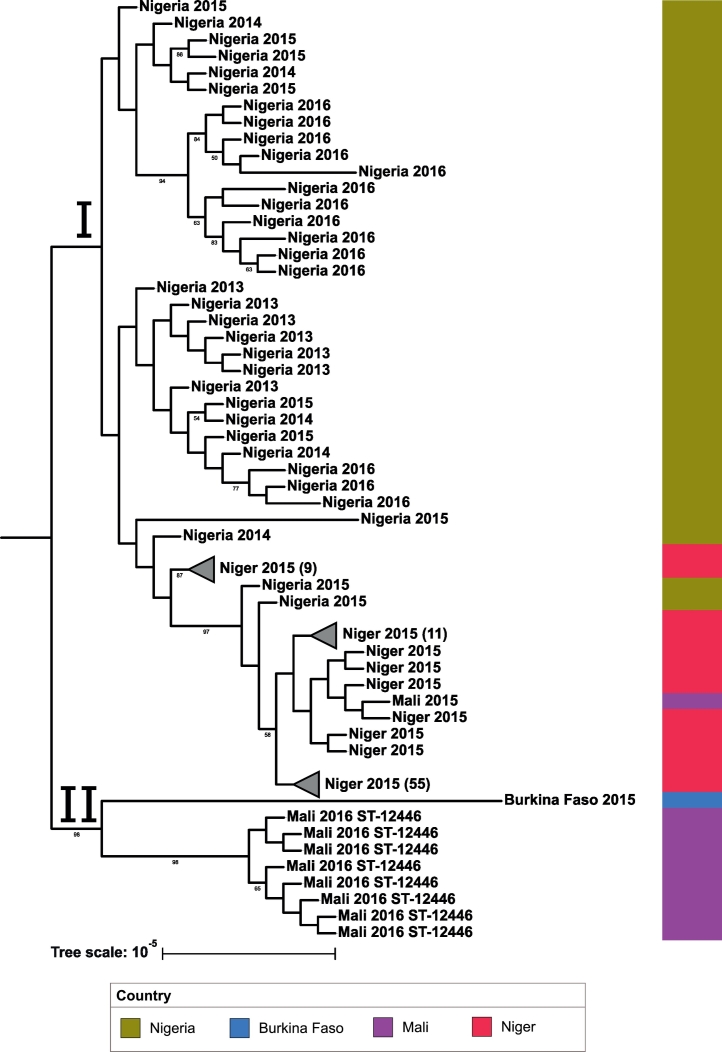
3.2.2. CC10217
All CC10217 isolates were serogroup C. As this is an emerging CC, there were no historical genomes available for comparison. The phylogeny is shown in Fig. 3. All CC10217 isolates from Niger, Nigeria, and Burkina Faso were ST-10217; the 2015 Mali isolate also belonged to this ST. The 2016 Mali isolates were ST-12446, differing from ST-10217 at the adk locus. The resulting phylogeny indicates two major clades for this clonal complex. Clade I contains all the Niger and Nigeria isolates and the 2015 Mali isolate, while clade II contains the 2016 Mali isolates and the one 2015 Burkina Faso isolate. The 2015 Niger epidemic isolates are found in one clade divided into two distinct subclades, each of which is well supported with bootstrap values above 85. This Niger 2015 epidemic subclade contains two Nigeria 2015 isolates, as well as a Nigeria 2014 isolate as an outgroup to the clade. The 2015 Mali isolate clusters with Niger 2015 isolates found within this subclade. The Nigeria isolates form several distinct clusters throughout clade I. While the Burkina Faso isolate was ST-10217, it appears divergent from same-ST isolates from Niger and Nigeria and shared a clade with the Mali isolates. Even though they share a clade, the isolates from Mali and Burkina Faso appear to be distinct from each other and the rest of the CC10217 collection, possibly indicating different strains from those seen in Niger and Nigeria.
Fig. 3.
CC10217 phylogenetic tree: A maximum likelihood phylogenetic tree was created for CC10217 consisting of 126 study isolates. The tree scale is in units of average substitutions per site along that length of the branch. Each tip is labeled with the isolate's country and year that the isolate was received. All isolates are ST-10217 unless labeled otherwise. Bootstrap values are visible for values greater than 50. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
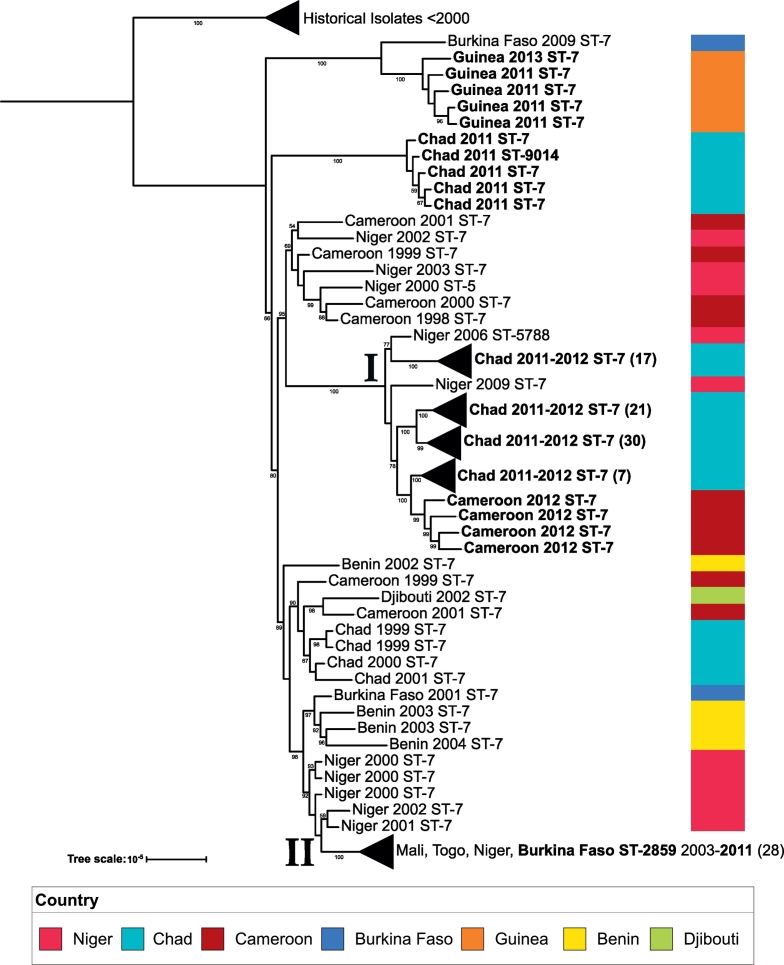
3.2.3. CC5
All CC5 isolates were serogroup A, the serogroup target for the MACV. While the MACV was introduced in the region in December 2010, only Burkina Faso, and parts of Niger and Mali, were initially vaccinated, followed by vaccination campaigns in other countries of the meningitis belt in later years [27]. Isolates were collected from 2011 to 2013 from cases caused by CC5 NmA in Chad, Guinea and Cameroon. In addition, one isolate was collected from an unvaccinated individual in Burkina Faso in 2011. The CC5 phylogeny is shown in Fig. 4. Clade I contains most of the Chad 2011–2012 and 2012 isolates from Cameroon. There was also a cluster of Chad 2011 isolates which formed an outgroup to the collection. A geographical trend was also observed in the phylogeny, as most isolates from West African countries such as Benin, Burkina Faso, Mali, and Togo are found in clade II; additionally, the 2011 Burkina Faso isolate from the unvaccinated individual fell within this clade. Isolates obtained from Guinea form an outgroup from the rest of the collection, and share a clade with a historical Burkina Faso isolate from 2009, indicating a strain circulating within Guinea that was different to the one circulating throughout most of West Africa.
Fig. 4.
CC5 phylogenetic tree: A maximum likelihood phylogenetic tree was created for CC5 consisting of 90 study isolates (boldface) and 66 historical African isolates (2010 and earlier). The tree scale is in units of average substitutions per site along that length of the branch. Each tip is labeled with the isolate's country, year that the isolate was received and sequence type of that isolate. Bootstrap values are visible for values greater than 50. Two clades of interest are labeled (I and II) respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2.4. CC181
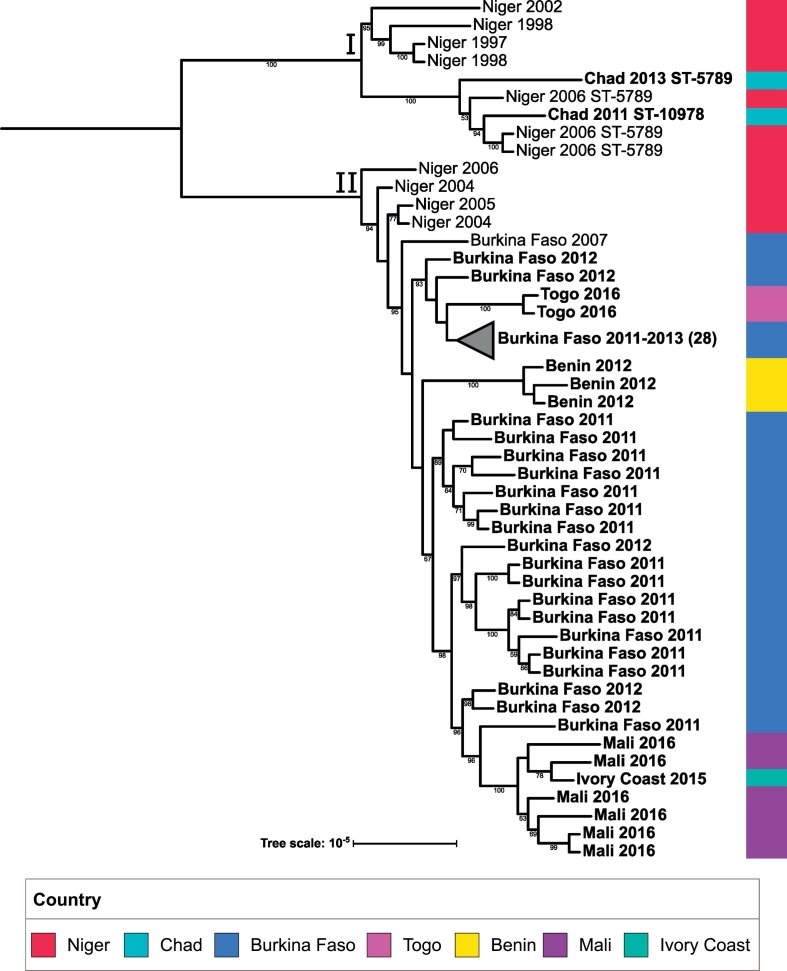
All CC181 isolates were serogroup X. A phylogeny was constructed (Fig. 5) which included all CC181 isolates from the study, as well as several historical CC181 isolates from African countries. All isolates from the study collection are shown in bold. The isolates included in this phylogeny produced two major clades. Clade I contains historical isolates from Niger, as well as more recent isolates from Chad, while clade II contains isolates exclusively from West African countries; clade II additionally contains an outgroup consisting of historical Niger isolates. This suggests a geographical split of two expanding strains, with the clade I strain primarily expanding in Chad, and the clade II strain primarily expanding in West African countries. Within clade II, several subclades contain isolates from multiple bordering countries. This can be clearly seen in the subclade containing Mali isolates, which also contains an isolate from the Ivory Coast. Isolates collected from Togo share a subclade with Burkina Faso isolates, while Benin isolates form a distinct outgroup from the rest of the West African isolate collection. Finally, isolates collected from Burkina Faso appear to have more diversity than the rest of the countries in clade II, as they appear to form several distinct groups throughout clade II.
Fig. 5.
CC181 phylogenetic tree: A maximum likelihood phylogenetic tree was created for CC181 consisting of 62 study isolates (boldface) and 11 historical African isolates (2007 and earlier). The tree scale is in units of average substitutions per site along that length of the branch. Each tip is labeled with the isolate's country, year that the isolate was received. All isolates are ST-181 unless labeled otherwise. Bootstrap values are visible for values greater than 50. Two clades of interest are labeled (I and II) respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Vaccine antigen variants
We also looked at the peptide types of the Bexsero and Trumenba vaccine protein targets present in our isolates. The NhbA, PorA, NadA and FHbp peptide types for each of the major CCs are shown in Table 2. FHbp, NhbA and PorA were found in all isolates of the major CCs. FHbp showed the highest amount of variation, with more than one peptide type being detected in CC11 and CC181 and one in CC10217 and CC5. Only one peptide type of NhbA and PorA was detected in each CC. NadA was detected in CC11 (NadA2/3.6) and CC5 (NadA2/3.8), but not in CC10217; only one CC181 isolate contained NadA2/3.6.
Table 2.
Vaccine antigen compositions for each CC.
| NhbA peptide | PorA type | NadA peptide | FHbp peptide | |
|---|---|---|---|---|
| CC11 | 96 | P1.5,2 | 2/3.6 | 94% 1.9; 4% 3.11, 2% other⁎ |
| CC10217 | 798 | P1.21–15,16 | Not found | 100% 2.27 |
| CC5 | 126 | P1.20,9 | 2/3.8 | 100% 1.5 |
| CC181 | 359 | P1.5–1,10–1 | Not found for 61, one 2/3.6 | 94% 1.74, 3% 1.391, 3% 3.976 |
The table above contains the antigen composition for the indicated peptide for each major clonal complex in the study. The counts for each CC were as follows: 434 CC11, 126 CC10217, 90 CC5, and 62 CC181. The vaccine antigen targets for MenB-FHbp (Trumenba) consist of FHbp peptides 1.55 and 3.45. The vaccine antigen targets for MenB-4C (Bexsero) consist of FHbp peptide 1.1, NadA peptide 2/3.8, NhbA peptide 2, and PorA type VR2–4.
The 2% other FHbp variants for CC11 consist of the following: 1 1.613, 1 1.841, 5 1.843.
4. Discussion
We provide a genomic insight into the changing epidemiological landscape of meningococcal strains in the meningitis belt in the period 2011–2016. This follows the introduction of MACV to the region in 2010 and the subsequent vaccination campaigns throughout several countries in the meningitis belt. Prior to the introduction of MACV, during the years 2004–2010, NmA CC5 isolates dominated with 74.8% of the isolates collected, while NmW CC11 isolates represented 8.8% and NmX CC181 isolates accounted for 6.2% [28]. Post-MACV introduction, NmW CC11 isolates represented 60% of the collection, while NmA CC5 isolates only constituted only 12.5% of the collected isolates and were collected in the earlier years of the study (2011−2013) from regions or individuals that had not yet received vaccination. Furthermore, NmX CC181 remained fairly constant throughout the two time-periods, accounting for 8.6% of the isolates collected in 2011–2016. Finally, the emerging NmC CC10217 isolates constituted 17.5% of our isolate collection, and were collected only in the latter years (2013–2016) of the study.
CC11 was predominant and was collected from nine of 11 countries included in the study. Retchless et al. [10] described four major clades (I,II,III,IV) containing NmW CC11 isolates, with isolates from Burkina Faso 2011–2012 and Mali 2012 falling into subclade IVa within clade IV. Our results corroborate their findings of the primary lineage (subclade IVa) expanding throughout the meningitis belt sharing a common ancestor with the 2000 Hajj-related outbreak strain. Our time-based molecular clock phylogeny suggests that this common ancestor emerged between 1998 and 1999, and diverged into two clades: clade 1.1 contained the Hajj-related outbreak strain and did not appear to continue expanding in the region, while clade 1.2 continued to expand throughout the belt. We also identified a sub-lineage (sub-clade 2.1) which has primarily been expanding throughout Central African countries such as the Central African Republic and Chad. Clade 3 dates back to a common ancestor estimated to have been introduced into the region between 2004 and 2005, which appears to be the source for the main strains circulating throughout West Africa. This clade shows the diversity present in West Africa during the Burkina Faso 2012 epidemic, with evidence of inter-country spreading and introduction events of these strains across the countries of West Africa.
Along with CC11, CC10217 has emerged as a major contributor to meningitis in the region. Kretz et al. [6] hypothesized that comparisons between ST-10217 invasive isolates and a nongroupable, ST-9367 (6/7 MLST alleles shared with ST-10217) carriage isolate from Burkina Faso 2012 may illuminate the transition of this strain from carriage to invasive. Brynildsrud et al. [29] performed this comparison and identified the acquisition of virulence factors in the invasive strain, providing evidence for the origins of this epidemic prone strain. In our results, the isolates associated with the Niger 2015 epidemic were found in one clade which formed two distinct subclades in the phylogeny, and also contained two isolates from Nigeria 2015. As cases of meningococcal meningitis caused by CC10217 were first reported during outbreaks in Nigeria 2013, the presence of Nigeria isolates in the clade containing Niger 2015 isolates strongly suggests Nigeria as the transmission source of the Niger 2015 epidemic strain. This finding is consistent with the geographical range of this ST described by Brynildsrud et al. [29]. Additionally, the Nigeria isolates formed several distinct clades, potentially indicating that several strains from this sequence type have diverged in the country. Furthermore, our results show that the Niger and Nigeria strains were more closely related to each other than those from Mali or Burkina Faso. While the Mali isolates harbor a novel ST (ST-12446), the Burkina Faso isolate shares the same ST as the rest of the collection (ST-10217) while still being divergent from the rest of the collection, possibly indicating a new strain. The divergence of ST-10217 in Nigeria, along with the emergence of ST-12446 in Mali, conveys the risk of the continued genetic evolution of this virulent clonal complex.
While NmA CC5 isolates were included in our study, these strains have not caused major outbreaks in the region since the introduction of the MACV. Our results suggest that there were several strains circulating within Chad during the 2011–2013 time period. Furthermore, a sub-lineage of CC5 NmA derived from ST-7 and comprised of ST-2859 has been expanding throughout most West African countries between the early 2000s and 2011. These results are consistent with the study by Lamelas et al. [30], wherein the researchers concluded that ST-2859 strains most likely emerged in Africa from ST-7 meningococci a few years prior to 2003.
Agnememel et al. [31] showed the presence of two main sub-lineages for CC181 African isolates, one of which contained historical (1990s) and more recent (since 2006) isolates, while the other contained only more recent isolates (since 2006). Our work expands on this finding by showing the spread of these sub-lineages throughout the region. Within our collection of CC181 isolates, we identified two major clades, one of which contained 2011–2013 Chad isolates while the other contained 2011–2015 West African isolates from Burkina Faso, Mali, Ivory Coast, Togo and Benin. As clade I only consisted of historical Niger and recent Chad isolates, there is not enough data to suggest that this strain is circulating throughout Central Africa, as it may also be a strain primarily circulating in Chad. Additional sampling of CC181 isolates from neighboring countries will provide greater resolution on this topic. Regardless, our results suggest at least two major strains of CC181 expanding in certain geographical regions of the meningitis belt. Finally, the presence of historical Niger isolates in both clades I and II suggests that Niger may have been a transmission source for the introduction of these strains to the region.
Our vaccine antigen composition results suggest that the protein-based vaccines Trumenba and Bexsero may offer protection against some of these strains. While none of the FHbp variants identified matched the Bexsero variant 1.1 target, we identified several strains with FHbp variants within family 1, and studies examining the antigenicity of other subvariant targets within this family have previously shown promising results, specifically against NmX CC181 isolates [17]. Furthermore, serum bactericidal assays on diverse FHbp sequences raised against Trumenba sera have suggested a broad range of protection against various strains [16].
5. Conclusion
Previously-circulating strains of NmX CC181 and NmW CC11, as well as emerging strains of NmC from CC10217, have risen as major causes of meningococcal meningitis in the meningitis belt. While our results corroborate the effectiveness of MACV, they also stress the importance of continued meningococcal molecular surveillance in the region. Understanding the strains involved in clonal expansion throughout the region is critical for guiding these surveillance strategies and informing implementation of future vaccines. Our results convey the importance of developing affordable multivalent conjugate vaccines targeting these emerging strains.
Acknowledgements and funding
We would like to acknowledge the Meningitis Research Foundation, GAVI, and the CDC's Office of Advanced Molecular detection for supporting this work. We would also like to acknowledge the clinical and public health labs of the following countries for providing isolates that were used in this study: Benin, Burkina Faso, Cameroon, Central African Republic, Chad, Guinea, Ivory Coast, Mali, Niger, Nigeria, and Togo. We also acknowledge Adam C. Retchless and Sandeep J. Joseph for their technical discussions. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention. The funder of the study had no role in study design, data collection, data analysis, data interpretation, writing of the report or the decision to submit the paper for publication. The corresponding author had full access to the study data and had final responsibility for the decision to submit to publication.
Author contributions
NT, DAC, MKT, OBB, EH and XW all contributed to the study design, data generation and data interpretation. ND, AED were involved in the data generation. RO, SO, KG, BMNL, SD were involved in collecting and sharing isolates that were used in the study. NT prepared the manuscript draft with contributions from DAC, MKT, OBB, LMF and XW. XW supervised the study. All authors critically reviewed the manuscript and gave approval for publication.
Declaration of interests
Dr. Caugant reports grants from Meningitis Research Foundation during the conduct of the study; all other authors have nothing to disclose.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.02.054.
Appendix A. Supplementary data
Supplementary material 1: Study isolates metadata.
Supplementary material 2: Historical isolate metadata.
References
- 1.Meningococcal disease in countries of the African meningitis belt 2012 - emerging needs and future perspectives. Releve epidemiologique hebdomadaire. 2013;88(12):129–136. [PubMed] [Google Scholar]
- 3.Novak R.T., Kambou J.L., Diomandé F.V. Serogroup A meningococcal conjugate vaccination in Burkina Faso: analysis of national surveillance data. Lancet Infect Dis. 2012;12(10):757–764. doi: 10.1016/S1473-3099(12)70168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jessica R.M., Isaïe M., Daouda K. Neisseria meningitidis Serogroup W, Burkina Faso, 2012. Emerg Infect Dis J. 2014;20(3):402. doi: 10.3201/eid2003.131407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delrieu I., Yaro S., Tamekloé T.A.S. Emergence of epidemic Neisseria meningitidis serogroup X meningitis in Togo and Burkina Faso. PLoS ONE. 2011;6(5) doi: 10.1371/journal.pone.0019513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kretz C.B., Retchless A.C., Sidikou F. Whole-genome characterization of epidemic Neisseria meningitidis serogroup C and resurgence of serogroup W, Niger, 2015. Emerg Infect Dis. 2016;22(10):1762. doi: 10.3201/eid2210.160468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broome C.V., Rugh M.A., Yada A.A. Epidemic group C meningococcal meningitis in upper Volta, 1979. Bull World Health Organ. 1983;61(2):325–330. [PMC free article] [PubMed] [Google Scholar]
- 8.Africa WHOROf . 2011-2016. Weekly feedback bulletin on cerebrospinal meningitis. Meningitis Weekly Bulletin. [Google Scholar]
- 9.Lucidarme J., Hill D.M.C., Bratcher H.B. Genomic resolution of an aggressive, widespread, diverse and expanding meningococcal serogroup B, C and W lineage. J Inf Secur. 2015;71(5):544–552. doi: 10.1016/j.jinf.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Retchless A.C., Hu F., Ouédraogo A.-S. The establishment and diversification of epidemic-associated serogroup w meningococcus in the african meningitis belt, 1994 to 2012. mSphere. 2016;1(6) doi: 10.1128/mSphere.00201-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Funk A., Uadiale K., Kamau C., Caugant D.A., Ango U., Greig J. Sequential outbreaks due to a new strain of Neisseria meningitidis serogroup C in northern Nigeria, 2013-14. PLoS curr. 2014;6 doi: 10.1371/currents.outbreaks.b50c2aaf1032b3ccade0fca0b63ee518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boisier P., Nicolas P., Djibo S. Meningococcal meningitis: unprecedented incidence of serogroup X—related cases in 2006 in Niger. Clin Infect Dis. 2007;44(5):657–663. doi: 10.1086/511646. [DOI] [PubMed] [Google Scholar]
- 13.LaForce F.M., Konde K., Viviani S., Préziosi M.-P. The meningitis vaccine project. Vaccine. 2007;25:A97–A100. doi: 10.1016/j.vaccine.2007.04.049. [DOI] [PubMed] [Google Scholar]
- 14.Pharmaceuticals W. 2014. Trumenba: prescribing information. [Google Scholar]
- 15.Diagnostics NVa . 2015. Bexsero: highlights of prescribing information. [Google Scholar]
- 16.Jiang H.-Q., Hoiseth S.K., Harris S.L. Broad vaccine coverage predicted for a bivalent recombinant factor H binding protein based vaccine to prevent serogroup B meningococcal disease. Vaccine. 2010;28(37):6086–6093. doi: 10.1016/j.vaccine.2010.06.083. [DOI] [PubMed] [Google Scholar]
- 17.Hong E., Giuliani M.M., Deghmane A.-E. Could the multicomponent meningococcal serogroup B vaccine (4CMenB) control Neisseria meningitidis capsular group X outbreaks in Africa? Vaccine. 2013;31(7):1113–1116. doi: 10.1016/j.vaccine.2012.12.022. [DOI] [PubMed] [Google Scholar]
- 18.Organization WH . WHO manual. laboratory methods for the diagnosis of meningitis caused by neisseria meningitidis, Streptococcus pneumoniae and haemophilus influenzae WHO manual. 2011. Laboratory methods for the diagnosis of meningitis caused by Neisseria meningitidis, Streptococcus pneumoniae and Haemophilus influenzae. [(Ed. 2)] [Google Scholar]
- 19.Chin C.-S., Alexander D.H., Marks P. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10(6):563. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 20.Jolley K.A., Maiden M.C. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC bioinforma. 2010;11(1):595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–12. [Google Scholar]
- 22.Bankevich A., Nurk S., Antipov D. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Croucher N.J., Page A.J., Connor T.R. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43(3) doi: 10.1093/nar/gku1196. [e15-e] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drummond A.J., Suchard M.A., Xie D., Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Treangen T.J., Ondov B.D., Koren S., Phillippy A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15(11):524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Djingarey M.H., Diomandé F.V.K., Barry R. Introduction and rollout of a new group a meningococcal conjugate vaccine (psa-tt) in african meningitis belt countries, 2010–2014. Clin Infect Dis. 2015;61(suppl_5):S434–S441. doi: 10.1093/cid/civ551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caugant D.A., Kristiansen P.A., Wang X. Molecular characterization of invasive meningococcal isolates from countries in the African Meningitis Belt before introduction of a serogroup a conjugate vaccine. PLoS ONE. 2012;7(9) doi: 10.1371/journal.pone.0046019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brynildsrud O.B., Eldholm V., Bohlin J., Uadiale K., Obaro S., Caugant D.A. Acquisition of virulence genes by a carrier strain gave rise to the ongoing epidemics of meningococcal disease in West Africa. Proc Natl Acad Sci. 2018;115(21):5510–5515. doi: 10.1073/pnas.1802298115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamelas A., Harris S.R., Röltgen K. Emergence of a new epidemic Neisseria meningitidis serogroup a clone in the African meningitis belt: high-resolution picture of genomic changes that mediate immune evasion. MBio. 2014;5(5) doi: 10.1128/mBio.01974-14. [e01974–14] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agnememel A., Hong E., Giorgini D., Nuñez-Samudio V., Deghmane A.E., Taha M.K. Neisseria meningitidis serogroup X in sub-Saharan Africa. Emerg Infect Dis. 2016;22(4):698–702. doi: 10.3201/eid2204.150653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1: Study isolates metadata.
Supplementary material 2: Historical isolate metadata.