Abstract
Background
Innate lymphoid cells (ILCs) are a newly discovered family of immune cells that have similar cytokine-secreting profiles as T helper cell subsets. Although ILCs are critical for host defense against infections and tissue homeostasis, their roles in tumor development are not well established.
Methods
We studied the function of ILC3 cells in the liver for the development of hepatocellular carcinoma (HCC) in murine HCC models using flow cytometry, adoptive transfer, and in vitro functional assays.
Findings
We found that ILC3 lacking the natural cytotoxicity-triggering receptor (NCR−ILC3) promoted the development of HCC in response to interleukin 23 (IL-23). IL-23 serum level is elevated in HCC patients and its high expression is associated with poor clinical outcomes. We found that IL-23 could promote tumor development in murine HCC tumor models. IL-23 promoted the expansion of NCR−ILC3 and its differentiation from group 1 ILCs (ILC1s). Furthermore, NCR−ILC3 initiated IL-17 production upon IL-23 stimulation and directly inhibited CD8+ T cell immunity by promoting lymphocyte apoptosis and limiting their proliferation.
Interpretation
Together, our findings suggest that NCR−ILC3 initiates the IL-17-rich immunosuppressive tumor microenvironment and promotes the development of HCC, thus may serve as a promising target for future cancer immunotherapy.
Fund
This work was supported by grants from National Natural Science Foundation of China (81471586, 81571556), the Priority Academic Program Development of Jiangsu Higher Education Institutions, the collaborative Innovation Center of Hematology, start-up grant from National University of Singapore, the Cancer Prevention and Research Institute of Texas CPRIT (RR180017), and the National Cancer Institute's Cancer Center Support (Core) Grant CA016672 (to The University of Texas MD Anderson Cancer Center).
Keywords: IL-23, IL-17, ILC, Tumor microenvironment, HCC
Research in context.
Evidence before this study
Development of hepatocellular carcinoma (HCC) is driven by chronic inflammation. We demonstrated previously that interleukin (IL)-17, a major inflammatory cytokine, could promote HCC development by suppressing anti-tumor immune response. Other studies also showed that both Th17 and Tc17 were enriched in the HCC tumor microenvironment. However, how IL-17 production is initiated in the tumor microenvironment is not clear. IL-23 has been shown to promote IL-17 production by many cell subsets, including ILC3. ILC subsets mainly reside in tissues and represent an early source of cytokines in response to infections or tissue damage. The role of ILC3 in tumor development remains elusive, and it may depend on the tissue microenvironment as well as the type of tumors. Thus, whether ILC3 could respond to inflammatory signals, such as IL-23, produce IL-17 to regulate antitumor adaptive immune response, and contribute to the development of HCC is not known.
Added value of this study
We assessed the development of HCC, the adaptive antitumor immune response, and the kinetics of the development of IL-17-producing cells in an IL-23-rich tumor microenvironment. We found that IL-23 inhibited antitumor adaptive immune response and promoted HCC tumor growth by enhancing IL-17 production. Interestingly, NCR−ILC3 cells were the first responders to IL-23, producing IL-17 during the early phase of tumor development. Their proliferation in vivo was facilitated by IL-23, and adoptive transfer of NCR−ILC3 cells promoted HCC growth. We have confirmed the presence of ILC3 in the liver, and demonstrated, for the first time, that NCR−ILC3 cells, being the initial IL-17-producing cells in response to IL-23 in the liver, could contribute to the immunosuppressive tumor microenvironment.
Implications of all the available evidence
NCR−ILC3 cells are the tissue-resident population in the liver responding to inflammatory cytokines in the early stage of tumor development. The production of IL-17 by ILC3 could set the stage for the differentiation of other IL-17-producing cells, and thus generate an IL-17-rich immunosuppressive tumor microenvironment. Therefore, NCR−ILC3 cells, as well as inflammatory cytokine IL-23, could be targeted for early prevention or therapeutic modulation of HCC.
Alt-text: Unlabelled Box
1. Introduction
Development of hepatocellular carcinoma (HCC) is one example of inflammation-based carcinogenic processes, and chronic inflammatory liver diseases and liver cirrhosis are the main risk factors for the development of HCC [1,2]. We showed previously that interleukin (IL)-17, a major inflammatory cytokine, could promote HCC development by recruiting and promoting the suppressive function of myeloid-derived suppressor cells (MDSCs) [3]. Moreover, Kuang et al. reported that both Th17 [4] and Tc17 [5] were enriched in the HCC tumor microenvironment. While Stat3 has been implicated in the initiation of IL-17 production by Th17 cells [6], the exact kinetics of IL-17 initiation in the tumor microenvironment is not clear.
IL-23 is an inflammatory cytokine characterized as a member of the IL-12 family in 2000 [7]. IL-23 shares the p40 subunit with IL-12 and displays pro-inflammatory functions [8]. Although IL-23 is upregulated in many tumors, its role in tumor development is controversial. IL-23 could reduce tumor growth in mammary cancer [9], colon adenocarcinoma [10], and B16F1 melanoma [11], whereas others found that elevated IL-23 in various human cancers was associated with adverse prognosis and more aggressive diseases [[12], [13], [14], [15], [16]]. A recent study identified IL-23 secreted by MDSCs as a driver of the resistance to androgen-deprivation therapy, a condition known as castration-resistant prostate cancer [17]. The ability of IL-23 to promote IL-17 secretion by Th17 cells was initially characterized in 2003 [18]. Many recent studies have further demonstrated that IL-23 can promote IL-17 and/or IL-22 production by CD8+ T cells [19], γδT cells [20] and group 3 innate lymphoid cells (ILC3) [21]. However, the role of IL-23 in HCC development and whether it depends on the induction of IL-17 and/or IL-22 are not known.
ILC3 cells are defined by their ability to produce IL-17 and/or IL-22 and can be further subclassified as lymphoid tissue-inducer (LTi) cells, natural cytotoxicity triggering receptor-positive (NCR)+ ILC3 cells, and NCR−ILC3 cells [22]. ILC subsets mainly reside in tissues and represent an early source of cytokines in response to infections or tissue damage. A tumor-promoting role has recently been proposed for ILC3-derived IL-22 in colorectal cancer [23]. Another group demonstrated that IL-23 is sufficient to induce de novo gut tumorigenesis by activating ILCs [24]. On the other hand, ILC3 has recently been shown to localize at the edge of tumor-associated tertiary lymphoid structures, which can be used to predict favorable clinical outcome [25]. Eisenring et al. also found that ILC3 displayed anti-tumorigenic potential in a model of malignant melanoma [26], and another study showed that splenic ILC3 suppressed tumor growth in an IL-12-dependent manner [27]. Thus, the role of ILC3 in tumor development remains elusive, and it may depend on the tissue microenvironment as well as the type of tumors. The properties of tissue-resident ILC3 have been reported for intestine, skin, and lung [[28], [29], [30]]. A recent study with Rag1−/− mice detected the presence of ILC3 in the liver [27]. However, whether ILC3 could respond to inflammatory signals, regulate antitumor adaptive immune response, and contribute to the development of HCC is not known.
In the current study, we assessed the development of HCC, the adaptive antitumor immune response, and the kinetics of the development of IL-17-producing cells in an IL-23-rich tumor microenvironment. We found that IL-23 inhibited antitumor adaptive immune response and promoted HCC tumor growth by enhancing IL-17 production. Interestingly, NCR−ILC3 cells were the first responders to IL-23, producing IL-17 during the early phase of tumor development. Their proliferation in vivo was facilitated by IL-23, and adoptive transfer of NCR−ILC3 cells promoted HCC growth. We have confirmed the presence of ILC3 in the liver, and demonstrated, for the first time, that IL-23 could promote HCC development through enhancing IL-17 production, and NCR−ILC3 cells, being the initial IL-17-producing cells in response to IL-23 in the liver, could be critical in establishing the IL-17-rich immunosuppressive tumor microenvironment.
2. Materials and methods
2.1. Mice
C57BL/6 (CD45.2) mice were purchased from the Shanghai Laboratory Animal Center (Shanghai, China). CD45.1 C57BL/6 mice were purchased from Beijing Charles River Laboratory Animal Technology Co., Ltd. (Beijing, China). C57BL/6 IL-17A−/− mice were provided by Dr. Chen Dong (Tsinghua University, Beijing, China) and C57BL/6 RORgtgfp/+ mice were provided by Dr. Ju Qiu (Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China). All mice were housed in specific-pathogen-free facilities and in accordance with the National Animal Care and Use Committee. All animal experiments were approved by the Institutional Laboratory Animal Care and Use Committee of Soochow University (Suzhou, China).
2.2. Cell lines
The murine HCC cell line hepa1-6 and human embryonic kidney line 293T cells were obtained from the American Type Culture Collection (Manassas, VA) and cultured in DMEM with 10% fetal bovine serum (Gibco, Gaithersburg, MD). Human 293T cell line has recently been authenticated by ATCC cell line authentication service (Manassas, VA). The murine HCC cell line hepa1-6 has recently been authenticated by mouse cell line authentication service provided by Applied Biological Materials (Richmond, BC, Canada). An IL-23-expressing lentivirus generated from 293 T cells was used to infect hepa1-6 cells to generate the hepa1-6-IL-23 cell line, and an empty vector was used as a vector control. The YFP-positive monoclonal cells were sorted by a FACSAria III flow cytometer (BD Biosciences, San Jose, CA) into 96-well plates and then screened for stable clones.
2.3. Plasmid construction
IL-23-p19 and IL-12-p40 were amplified from cDNA generated from hepa1-6 cells (IL-23-p19 F: GGATCCATGCTGGATTGCAGAGCAG, R: AGCTGTTGGCACTAAGGGC and IL-12-p40 F: ATGTGGGAGCTGGAGAAAGAC, R: GGATCGGACCCTGCAGG) and linked with a flexible region (gssss)3 to form the IL-23 construct, which was then inserted into lentiviral plasmid (pRRL-venus was provided by Dr. Yun Zhao, Soochow University, Suzhou, China) and minicircle plasmid (pMC.EF1; SBI, Palo Alto, CA). A lentiviral construct was used to generate IL-23 stable-expressing cell line. Minicircle plasmids were delivered hydrodynamically (i.e., 80 μg/2 ml plasmids in PBS were injected via tail vein within 8–10 s) to produce expression of IL-23 in vivo.
2.4. Murine HCC models
Three murine HCC models were used in the study. The orthotopic HCC model was established as described previously with minor modifications [3]. In brief, mice were anesthetized and 1 × 106 hepa1-6 cells in 25 μL PBS were orthotopically implanted in the liver. Two weeks later, the mice were sacrificed and tumor size was measured with fine digital calipers, and tumor volume was calculated as 0.5 × width2 × length. To generate the orthotopic HCC model by hydrodynamic injection, 1 × 106 hepa1-6 cells in 2 ml PBS were injected via tail vein within 8 to 10 s. Mice were sacrificed 3 weeks later, and tumor nodules were counted. The DEN-induced spontaneous HCC model was generated by injecting 14-day-old C57BL/6 mice with 25 mg/kg DEN (Sigma-Aldrich, St Louis, MO) followed by hydrodynamic injection of either minicircle plasmids expressing IL-23 or vector alone every other month from the age of 8 weeks. Eight months later, mice were sacrificed and the livers were removed. Tumor size was measured and the number of tumor nodules was counted.
2.5. Flow cytometry
For cell-surface staining, cells were blocked with CD16/32 FcR-block (BioLegend, San Diego, CA) for 15 min, and stained with fluorescent dye-conjugated mAb for 30 min at 4 °C. For intracellular cytokine staining, cells were stimulated for 4–6 h with PMA (50 ng/ml) and ionomycin (500 ng/ml) in the presence of brefeldin A (10 μg/ml; BD Biosciences, San Jose, CA). Cells were stained for surface markers for 30 min, then fixed with 4% paraformaldehyde, permeabilized with 1% saponin (Sigma-Aldrich, St Louis, MO), and stained for cytokines for 30 min at 4 °C. In Figs. 5 and 7a–c and h, intracellular staining was performed without stimulation by PMA and ionomycin. For staining of RORγt, T-bet, or Foxp3, fixation/permeabilization buffer (eBioscience, San Diego, CA) was used. The antibodies used for FACS staining were FITC-anti-mouse-NK1.1 (PK136), PE-anti-mouse-RORγt (Q31-378), PE/CF594-anti-mouse-CD3 (145-2C11), PE/CF594-anti-mouse-Ly-6A/E (D7), PerCP/Cy5.5-anti-mouse-CD4 (RM4–5), PerCP/Cy5.5-anti-mouse-NK1.1 (PK136), PE/CY7-anti-mouse-CD19 (1D3), Alexa Fluor 647-anti-mouse-RORγt (Q31–378), APC/H7-anti-mouse-CD4 (GK1.5), and ACP/Cy7-anti-mouse-CD11b (M1/70) were purchased from BD Bioscience (San Jose, CA). The FITC-anti-mouse-CD8 (53–6.7), FITC-anti-mouse-CD4 (GK1.5), PE-anti-mouse-IL-22 (Poly5.64), PE-anti-mouse-CD127 (SB/199), PE-anti-mouse-IL-17A (TC11-18H10.1), PE-anti-mouse-IFN-γ (XMG1.2), PE-anti-mouse-I-Ab (AF6–120.1), PE-anti-mouse-H-2Kb (AF6–88.5), PE-Rat-IgG2b, kappa Isotype (RTK4530), PerCP/Cy5.5-anti-mouse-TCRγδ (GL3), PerCP/Cy5.5-anti-mouse-IL-17A (TC11-18H10.1), PerCP/Cy5.5-anti-mouse-CD45.1 (A20), APC-anti-mouse-CD8 (53–6.7), APC-anti-mouse-CD117 (ACK2), Alexa fluor 700-anti-mouse-CD45 (30-F11), APC-anti-mouse-IFN-γ (XMG1.2), PE/Cy7-anti-mouse-TNF-α (MP6-XT22), and PE/Cy7-anti-mouse-T-bet (4B10), APC/Cy7-anti-mouse-CD127 (A7R34) were purchased from BioLegend (San Diego, CA).
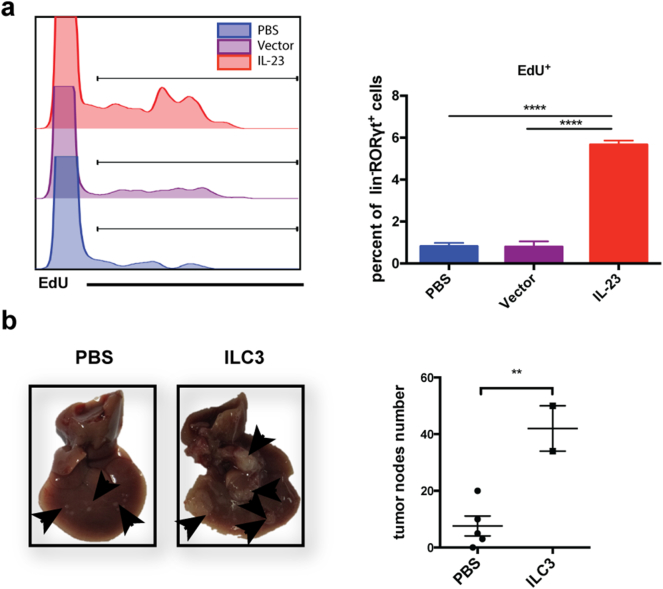
Fig. 5.
ILC3 cells in the liver proliferate upon IL-23 stimulation and promote HCC development. (a) 1 × 106 hepa1–6-IL-23, hepa1–6-vector cells in 2 ml PBS or 2 ml PBS alone were injected into C57BL/6 mice hydrodynamically (n = 5 mice/group). 4 days later, 5 mg/kg EdU was intraperitoneally injected into the recipients. After 4 h, flow cytometry analysis was performed to detect the percentage of EdU+ ILC3 cells in the liver. (b) 1 × 106 hepa1–6-IL-23 cells in 2 ml PBS were injected into C57BL/6 RORgtgfp/+ mice hydrodynamically. 5 days later, ILC3 cells were sorted from tumor-bearing C57BL/6 RORgtgfp/+ mice. 5 × 105 hepa1–6-IL-23 cells with or without isolated ILC3 cells (25,000 cells/mouse) were injected into C57BL/6 mice (n = 2–5 mice/group) hydrodynamically. Tumor nodules in the liver were counted on day 21 after injection. Data are shown as mean ± SEM. One-way ANOVA followed by Tukey's multiple comparisons test (a) or Two-tail unpaired student's t-test (b) was used for statistical analysis. *p < .05, **p < .01, ***p < .001. Data shown are the representative of two independent experiments.
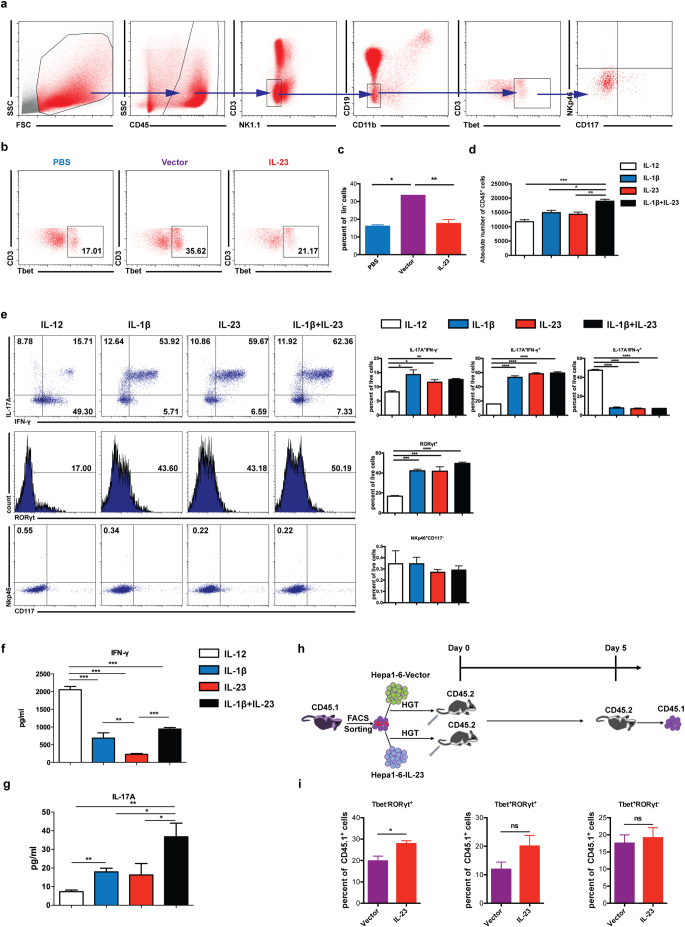
Fig. 7.
IL-23 promotes the differentiation of ILC1 to ILC3 in the tumor microenvironment. (a) 1 × 106 hepa1–6-vector cells in 2 ml PBS were injected into C57BL/6 mice hydrodynamically. 4 days later, ILC1 cells [CD45+lin(CD3,CD19,CD11b,NK1.1)−CD117−NKp46−T-bet+] were detected as shown in the flow plots. (b–c) 1 × 106 hepa1–6-IL-23, hepa1–6-vector cells in 2 ml PBS or 2 ml PBS were injected into C57BL/6 mice hydrodynamically (n = 3 mice/group). The percent of ILC1 cells in CD45+lin− population in the liver was analyzed on day 4 post injection. (d) CD127+ILC1 cells were sorted from hepa1–6 tumor-bearing mice and cultured in the presence of IL-12, IL-1β, IL-23, or IL-1β + IL-23 with IL-2 for 5 days. Cellular yield was shown at the end of the culture. (e) Flow cytometry analysis of the expression of IL-17, IFN-γ, RORγt, NKp46, and CD117 with or without various treatments was shown as dot plots or histograms (left panel) or as the percent of total live cells (right panel), respectively. (f) IFN-γ and (g) IL-17 production in the culture supernatants was measured by ELISA. (h) Experimental sketch of ILC1 adoptive transfer experiments. ILC1 cells were sorted from CD45.1+C57BL/6 tumor-bearing mice and adoptively transferred into CD45.2+C57BL/6 mice implanted with hepa1–6-IL-23 or hepa1–6-vector cells. 5 days later, the phenotypes of CD45.1+ cells in the liver were analyzed. (i) The percent of T-bet−RORγt+, T-bet+RORγt+, T-bet+RORγt− cells in CD45.1+ cells. Data are shown as mean ± SEM. One-way ANOVA followed by Tukey's multiple comparisons test (c,d,e,f,g) or Two-tail unpaired student's t-test (i) was used for statistical analysis. *p < .05, **p < .01, ***p < .001. Data shown are the representative of two independent experiments.
2.6. Co-culture experiments with ILCs and T cells
C57BL/6 RORgtgfp/+ mice were injected with hepa1-6-IL-23 hydrodynamically. Four days later, intrahepatic leukocytes were harvested and CD45+Lin (CD3−CD19 –NK1.1−CD11b−)− CD127+GFP+ cells were sorted as ILC3 cells by a FACSAria III flow cytometer (BD Biosciences, San Jose, CA). In addition, naïve CD4+ or CD8+ T cells were separated by using mouse naïve CD4 or CD8 isolation cocktail kits, respectively (Stem cell, Vancouver, BC). Naïve CD4 or CD8 cells in the suspension were then isolated and co-cultured with sorted ILC3s in a 96-well at a ratio of 30:1 in the presence of 50 U/ml IL-2 (Beijing Four Rings Bio-Pharmaceutical, Beijing, China). Three days later, apoptosis and proliferation of CD4+ or CD8+ T cells were analyzed by the assays described below. In some experiments, naïve CD8 cells in the suspension were isolated and co-cultured with sorted ILC3 in a 96-well at different ratio of 15:1, 30:1, 60:1 or 120:1. Three days later, apoptosis and proliferation of CD8+ T cells were analyzed by the assays described below. In some experiments, anti-mouse-IL-17A antibody (Sungene Biotech, Tianjin, China) was added to the co-culture at in different concentrations (2.5μg/ml or 5μg/ml). Three days later, the concentration of IL-17A was detected by the ELISA and CD8+ T cell apoptosis was analyzed. In some experiments, naïve CD8+ T cells were co-cultured with sorted ILC3s in a 24-well at a ratio of 30:1 or separated by transwell chambers (Corning, New York, USA) at the same ratio. Three days later, lower compartment (CD8+ T cells) was harvested and CD8+ T cell apoptosis was analyzed.
2.7. In vitro apoptosis and proliferation assays
Apoptosis was measured by using a PE Annexin V apoptosis detection kit (BD Biosciences, San Jose, CA) according to the manufacturer's instructions. Proliferation was assessed by Ki67 staining. In brief, cells were harvested and stained for the surface markers for 30 min. After washes, cells were resuspended in the fixation/permeabilization buffer (eBioscience, San Jose, CA) for 30 min at 4 °C in the dark, and then incubated with anti-Ki67 antibody at room temperature for 30 min in the dark. Ki67-positive proliferating cells were detected by a FACSCanto II cytometer (BD Biosciences, San Jose, CA).
2.8. In vitro differentiation of ILC1s to ILC3s
Mice were injected with hepa1-6-vector hydrodynamically. Four days later, splenocytes were harvested and ILC1 cell were sorted according to the expressions of CD45+Lin (CD3− CD19 –CD11b –NK1.1−CD127+CD117−NKp46−). Cells were cultured in medium at 105 per well in a 96-well plate with 50 U IL-2 (Beijing Four Rings Bio-Pharmaceutical, Beijing, China), with 20 ng/ml IL-12, 20 ng/ml IL-1β, or 20 ng/ml IL-23 (all from R&D Systems, Minneapolis, MN), or with 20 ng/ml IL-1β plus 20 ng/ml IL-23. Five days later, cell phenotypes, cytokine expression, and RORγt expression were analyzed by a Gallios cytometer (Beckman Coulter, Brea, CA).
2.9. Adoptive transfer of ILC1s
ILC1 cells were sorted as described above from CD45.1 C57/BL6 mice, and transferred via tail vein into CD45.2 C57/BL6 mice bearing hepa1-6-IL-23 tumors or control tumors (3 × 105/300 μL PBS). Five days later, the expression of RORγt and T-bet in intrahepatic leukocytes was analyzed by a Gallios cytometer (Beckman Coulter, Brea, CA).
2.10. In vivo proliferation assay
Mice were injected with hepa1-6-IL-23 or hepa1-6-vector or PBS hydrodynamically. Four days later, mice were injected intraperitoneally with 5 mg/kg EdU (Invitrogen, Waltham, MA). Four hours later, EdU+ intrahepatic leukocytes were determined by flow cytometry according to the manufacturer's instructions. Cells were then washed twice and analyzed by a FACSCanto II cytometer (BD Biosciences, San Jose, CA).
2.11. Cytometric bead array and enzyme-linked immunosorbent assays
Blood was collected from tumor-bearing mice and serum was separated by centrifugation at 850g for 30 min at room temperature. Supernatant was transferred into a new tube and kept frozen at −80 °C. Serum cytokine levels were detected by using a BD Cytometric Bead Array mouse soluble-protein master buffer kit with a mouse soluble-protein flex set (BD Pharmingen, San Jose, CA) on a FACSCanto II cytometer (BD Biosciences, San Jose, CA). Data were analyzed with BD FCAP Array software (BD Biosciences, San Jose, CA). The flex set includes mouse IL-23, IL-6, IL-9, IL-12 and IL-17A.
Cell culture supernatant was separated by centrifugation at 335g for 5 min at 4 °C, after which the supernatant was transferred into new tubes and kept frozen at −80 °C. Levels of IL-17A or IFN-γ were measured with ELISA kits according to the manufacturer's instructions (BioLegend, San Diego, CA).
3. Analysis of Oncomine data
The expression level of IL-23 and the survival curve related to IL-23 expression level in HCC patients were analyzed within the Oncomine database (www.oncomine.org). This analysis was based on the datasets, including Roessler Liver 2, Chen Liver, Wurmbach Liver, Mas Liver, TCGA Liver and Guichard Liver datasets. Differences were considered to be statistically significant when p < .05.
3.1. Statistical analysis
Data were analyzed with GraphPad Prism 6 software. Student t-tests (nonparametic) were used to compare two groups. Multi-group comparisons were analyzed by one-way analysis of variance (nonparametic). All data were expressed as mean ± SEM. Differences were considered to be statistically significant when p < .05. The significance levels are marked as *p < .05; **p < .01; ***p < .001; and ****p < .0001.
4. Results
4.1. IL-23 promotes the development of murine HCC and induces IL-17 production by both CD4+ and CD8+ T cells
IL-23 expression has been found to be increased in tumor sites from HCC patients [31]. IL-23 gene expression has also been suggested to be involved in HCC carcinogenesis [32,33]. Using the oncomine database, we also found that IL-23 expression was significantly elevated in HCC patients (Supplementary Fig. S1a). Meanwhile, the high IL-23 expression group had lower overall survival rate compared with the low IL-23 expression group (Supplementary Fig. S1b). Therefore, IL-23 is an important inflammatory cytokine that may regulate anti-tumor immune response in HCC patients. However, experimental evidence is lacking supporting the role of IL-23 in HCC and its immune regulatory function is not known.
IL-23-specific subunit p19 knockout mice have been generated, but the study using these mice may be complicated by the increased IL-12 levels since the other subunit p40 can be shared by IL-12. Therefore, to investigate the role of IL-23 without affecting endogenous IL-12 expression in the development of HCC, we established a stable IL-23-expressing murine HCC cell line, hepa1-6-IL-23, which produced higher level of IL-23 than the hepa1-6-vector control cells (Supplementary Fig. S2a). Proliferation of the hepa1-6-IL-23 cell line was also slightly increased compared with the control cells (Supplementary Fig. S2b), with no significant difference in apoptosis (Supplementary Fig. S2c). In an orthotopic HCC model established by hydrodynamic injection of these two cell lines, the IL-23 serum levels were significantly higher in hepa1-6-IL-23 tumor-bearing mice than those in the control group (Supplementary Fig. S2d).
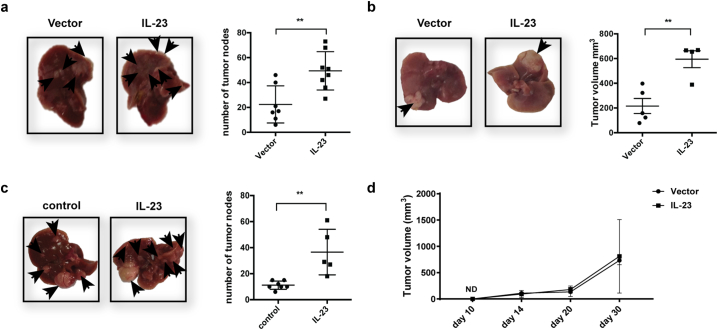
The numbers of tumor nodules in the liver at 3 weeks after tumor-cell injection were greater in the hepa1-6-IL-23 mice than in mice injected with hepa1-6-vector cells (Fig. 1a). We also created another orthotopic HCC model by surgical injection of hepa1-6-IL-23 or hepa1-6-vector cells intrahepatically (Fig. 1b). Measurement of tumor size 2 weeks later showed that overexpression of IL-23 led to increased tumor volumes (Fig. 1b). To confirm this result in a more physiologically relevant spontaneous tumor model, we used diethylmitrosamine (DEN) to induce HCC [34,35] and hydrodynamically injected minicircle-IL-23 every other month to express IL-23 in the liver during the development of HCC (Fig. 1c). Eight months later, mice given minicircle-IL-23 had more tumor nodules in the liver than did those given control vectors (Fig. 1c). However, when control and IL-23-expressing tumors were implanted in the livers of NOD-SCID mice, the tumor-promoting effect of IL-23 disappeared (Fig. 1d). Collectively, these results show that IL-23 promoted HCC development in an immune-dependent manner in murine models.
Fig. 1.
IL-23 promoted the development of murine HCC. (a) 1 × 106 hepa1-6-IL-23 or hepa1-6-vector cells in 2 ml PBS were injected into C57BL/6 mice (n = 7–8 mice/group) hydrodynamically via tail veins. The tumor nodules in the liver from tumor-bearing mice were counted on day 21. The representative of tumor morphology (left panel) is shown and the numbers of tumor nodules are shown as mean ± SEM (right panel). (b) 1 × 106 hepa1-6-IL-23 or hepa1-6-vector cells in 25ul PBS were intrahepatically injected into C57BL/6 mice (n = 4–5 mice/group). The tumor volumes were measured on day 14. The representative of tumor morphology (left panel) is shown and tumor size is shown as mean ± SEM (right panel). (c) 14-day-old C57BL/6 mice (5–7 mice/group) were injected with 25 mg/kg DEN intraperitoneally. Two months later, mice were injected hydrodynamically with 80 mg/2 ml minicircle vector or minicircle-IL-23 plasmids every other month for 8 months. The number of tumor nodules in the liver was counted. The representative of tumor morphology (left panel) is shown and the numbers of tumor nodules are shown as means±SEM (right panel). (d) 1 × 106 hepa1-6-IL-23 or hepa1-6-vector cells in 25ul PBS were intrahepatically injected into NOD-SCID mice (n = 4–5 mice/group). The tumor volumes were measured on days 10, 14, 20, and 30. Tumor size is shown as mean ± SEM, Data were pooled from two independent experiments. Each symbol represents an individual mouse. Two-tail unpaired student's t-test was used for statistical analysis. **p < .01. Data are representative of at least three independent experiments for a–c.
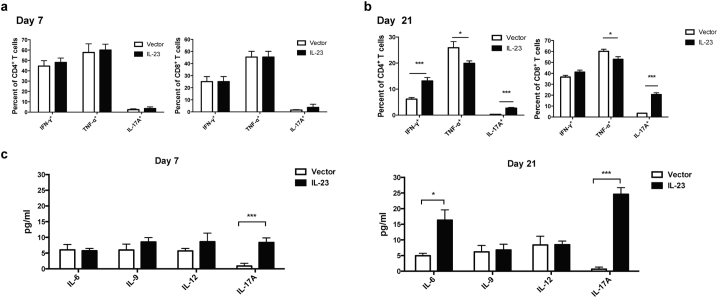
Because T cell–mediated adaptive immune responses are important in both chronic liver inflammation and HCC development, we examined cytokine production by both CD4+ and CD8+ T cells on day 7 and day 21 after tumor implantation (Fig. 2). On day 7, IL-23 expression had no effect on cytokine productions by both CD4+ and CD8+ T cells (Fig. 2a; Supplementary Fig. S3a). On day 21, IL-23 expression led to increased IL-17-producing CD4+ and CD8+ T cells in both spleen and liver (Fig. 2b; Supplementary Fig. S3b). Although the percentage of IFN-γ-producing CD4+ T cells was increased in the liver, the percentages of IFN-γ+ CD8+ T cells in both spleen and liver were not affected by IL-23 expression. Moreover, TNF-α production by CD4+ T cells in the liver and CD8+ T cells in both spleen and liver was decreased by IL-23 expression. We further found that IL-23 did not directly affect the expression of TNF-α or IFN-γ by either CD4+ or CD8+ T cells (Supplementary Fig. S4). As expected, increased levels of IL-17 were detected in the serum on both day 7 and day 21 in the IL-23 group (Fig. 2c). Serum IL-6 was also increased on day 21 in the IL-23-expressing group, but serum levels of IL-12, which shares a common subunit, p40, with IL-23, were not affected by IL-23 expression (Fig. 2c). IL-23 has been shown to inhibit IL-9 production in a melanoma tumor model [36]. However, we did not observe any changes in IL-9 level with IL-23 overexpression in HCC models (Fig. 2c). In summary, IL-23 enhanced IL-17 production by both CD4+ and CD8+ T cells but inhibited TNF-α expression by both CD4+ and CD8+ T cells indirectly in vivo, which could contribute to reduced antitumor immunity.
Fig. 2.
IL-23 enhanced IL-17 production from CD4+and CD8+T cells. 1 × 106 hepa1–6-IL-23 or hepa1–6-vector cells in 2 ml PBS were injected into C57BL/6 mice (n = 5 mice/group) hydrodynamically via tail veins. On day 7 and day 21, T cells were analyzed for cytokine production. (a–b) Frequencies of IFN-γ+, TNF-α+, or IL-17A+ cells among CD3+CD4+ or CD3+CD8+ cells obtained from the livers of tumor-bearing mice on day 7 (a) or day 21 (b) were determined by flow cytometry. (c) IL-6, IL-9, IL-12, and IL-17A production was measured by a cytometric bead array assay in serum of tumor-bearing mice on day 7 (left panel) and day 21 (right panel). Data are shown as mean ± SEM. Two-tail unpaired student's t-test was used for statistical analysis. *p < .05, **p < .01, ***p < .001. Data shown are the representative of three independent experiments.
4.2. Enhanced development of HCC induced by IL-23 depends on IL-17 production
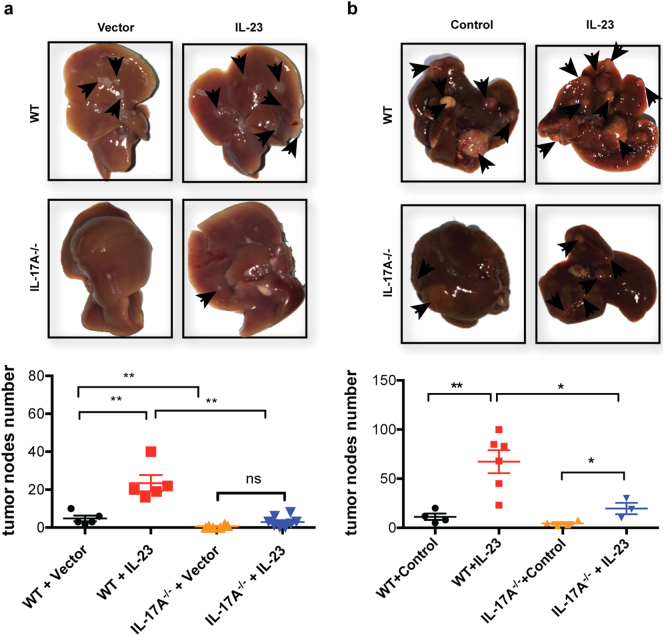
We previously showed that IL-17 could promote HCC development [3]. Since IL-23 mainly enhanced IL-17 production, we examined whether the tumor-promoting function of IL-23 was dependent on IL-17. WT or IL-17-deficient mice were implanted with hepa-1-6-IL-23 or hepa1-6-vector tumors by hydrodynamic injection (Fig. 3) and IL-17 production in those mice was determined by intracellular staining (Supplementary Fig. S5). Consistent with the results shown in Fig. 1, IL-23 significantly promoted tumor growth in WT mice but the tumor-promoting effect of IL-23 was diminished in IL-17-deficient mice (Fig. 3a). We confirmed this result by repeating the experiment with a DEN-induced spontaneous HCC model (Fig. 3b) in which plasmids expressing IL-23 or vector control were hydrodynamically injected through the tail vein every other month during tumor development. Similarly, IL-23 promoted HCC development in WT mice, but had little effect on tumor growth in IL-17-deficient mice. Thus, IL-23 promoted HCC development in an IL-17-dependent manner.
Fig. 3.
The enhanced HCC development induced by IL-23 is IL-17-dependent. (a) 1 × 106 hepa1–6-IL-23 or hepa1–6-vector cells in 2 ml PBS were injected into WT or IL-17-deficient C57BL/6 mice (n = 4–6 mice/group) hydrodynamically via tail veins. The tumor nodules in the liver from tumor-bearing WT or IL-17-deficient mice were counted on day 21. The representative of tumor morphology (top) and the numbers of tumor nodules (bottom) are shown. (b) WT or IL-17-deficient C57BL/6 mice (3–6 mice/group) were injected intraperitoneally with 25 mg/kg DEN on day 14 after birth. After 2 months, mice were injected hydrodynamically with 80 mg minicircle vector or minicircle-IL-23 plasmids in 2 ml PBS every other month for 4 months. The number of tumor nodules in the livers was counted at the end of the eighth month. The representative of tumor morphology (top) and the numbers of tumor nodules (bottom) are shown. The data shown are mean ± SEM. Statistical analysis was performed using one-way ANOVA followed by Tukey's multiple comparisons test. *p < .05, **p < .01, ***p < .001. Data are the representative of three independent experiments.
4.3. IL-17-producing NCR−ILC3 cells are the initial responders to IL-23 in the tumor microenvironment to promote HCC development
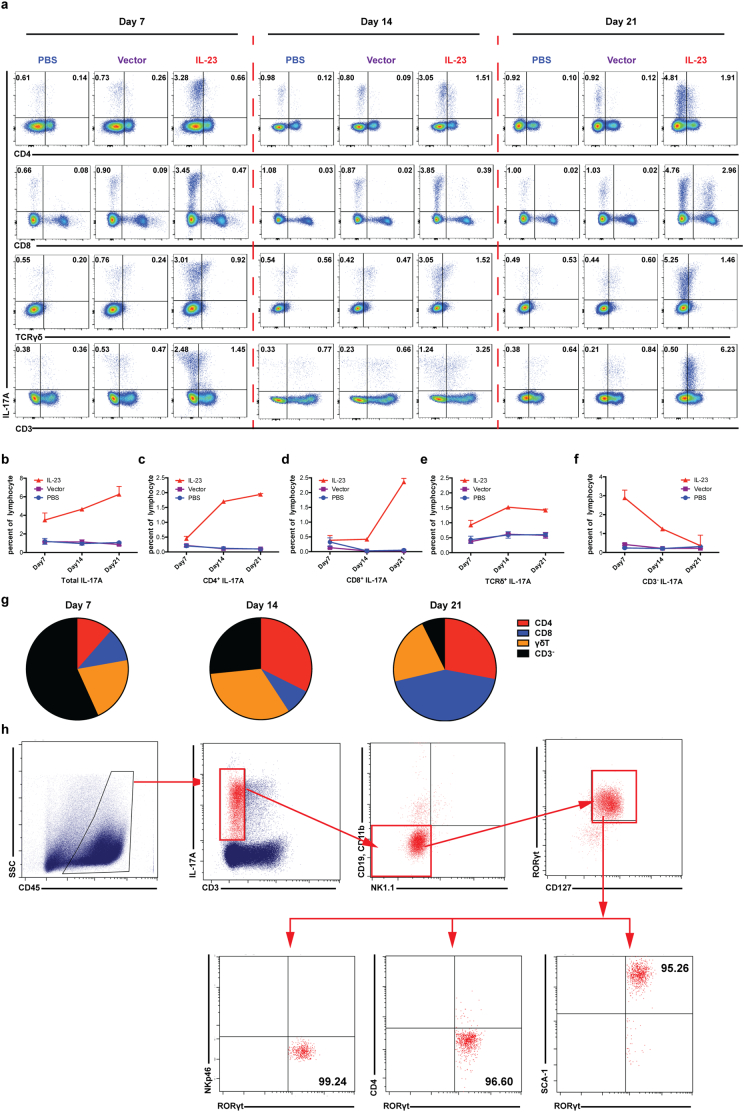
The ability of IL-23 to induce IL-17-production in Th17, Tc17, and γδT17 cells has been established. However, the kinetics of IL-17-producing cells in response to IL-23 in the tumor microenvironment is not clear. The total number of CD45+ cells infiltrating into the tumor was not affect by IL-23 (Supplementary Fig. S6a). By analyzing IL-17-producing cells on day 7, 14 and 21 during HCC development (Fig. 4), we found that the total number of IL-17-producing cells was increased in the presence of IL-23 throughout the 3-week period (Fig. 4a and 4b). IL-23 increased the percentages of Th17, Tc17, and γδT17 cells, with Th17 and Tc17 peaking on day 21 and γδT17 peaking on day 14 (Figs. 4c–4e). Most strikingly, we found that most IL-17-producing cells on day 7 were CD3− cells (Fig. 4f and 4g), the numbers of which peaked on day 7 and gradually dropped to levels near that of the controls by day 21. Similar results were observed in the orthotopic surgical model (Supplementary Fig. 6c and 6d). Other than Th17, Tc17, and γδT17 cells, other reported sources of IL-17 have included iNKT cells [37], NK cells [38], LTi cells [39], neutrophils, and mast cells [40]. To further characterize the IL-17-expressing CD3− cells, we examined their surface expression of CD3, CD19, CD11b, NK1.1, CD127, NKp46, CD4, and Sca-1 as well as the transcription factor RORγt in IL-23-expressing tumor-bearing mice (Fig. 4h, Supplementary Fig. S6b). We found that these IL-17-producing CD3− cells were CD19−CD11b−NK1.1−NKp46−CD127+CD4−Sca-1+ and RORγt+, meaning that they were NCR−ILC3 cells. Therefore, the initial responders to IL-23 to produce IL-17 in the HCC tumor microenvironment were NCR−ILC3 cells.
Fig. 4.
IL-17-producing NCR−ILC3 cells are the initial responders to IL-23 in the tumor microenvironment 1 × 106 hepa1–6-IL-23, hepa1–6-vector cells in 2 ml PBS or PBS alone were injected into C57BL/6 mice hydrodynamically (n = 10 mice/group). Flow cytometry was performed to analyze IL-17-procuding cells every week for 3 weeks. (a) The representative flow cytometry plots for IL-17 expression by CD4+ T, CD8+ T, γδT and CD3− cells isolated from the liver of tumor-bearing or control mice on days 7, 14, and 21 post injection. (b–f) Percentages of IL-17-producing cells at various time points post injection. (g) Cellular components of IL-17-producing cells in the liver of hepa1–6-IL-23 tumor-bearing mice at different time points of HCC development. (h) Phenotyping of IL-17-producing CD3− cells after PMA/Ionomycin (50 ng/ml/500 ng/ml) stimulation. Data are shown as mean ± SEM. Data shown are the representative of three independent experiments.
Because IL-23 has also been shown to promote IL-22 production in NCR−ILC3 cells [41], we also examined IL-22 production in NCR−ILC3s responding to IL-23 (Supplementary Fig. S7). Although IL-23 significantly promoted IL-17 production by NCR−ILC3, the percentages of IL-22 single-positive and IL-22/IL-17 double-positive cells were extremely low even in the presence of IL-23. Thus, NCR−ILC3s, as the initial responders to IL-23 during HCC development, produce mainly IL-17.
To further characterize the expansion of NCR−ILC3s in the presence of IL-23 in the tumor microenvironment, we assessed proliferation in vivo via 5-ethynyl-2′-deoxyuridine (EdU) labeling (Fig. 5a). The results showed that IL-23 significantly promoted the expansion of NCR−ILC3s in the liver. Furthermore, when we adoptively transferred NCR−ILC3 cells in the orthotopic HCC model on day 0, tumor growth was greatly promoted compared with a phosphate-buffered saline (PBS) control (Fig. 5b). In summary, NCR−ILC3s can be expanded by IL-23 in the tumor microenvironment to promote HCC development.
4.4. NCR−ILC3 can directly regulate T-cell responses
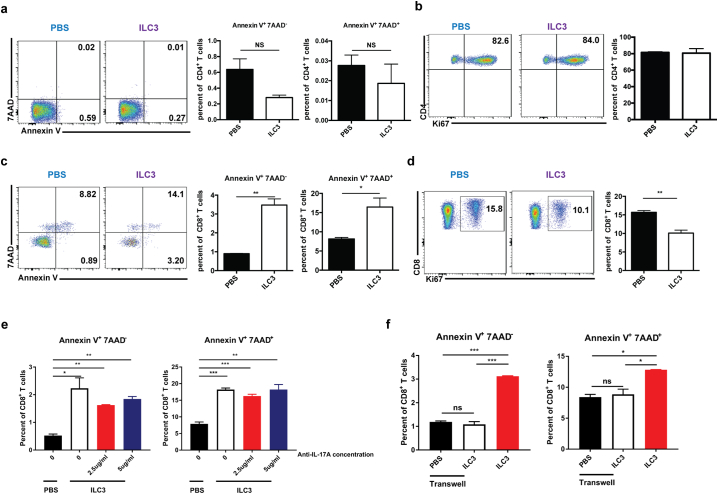
IL-17 has been shown to have immunosuppressive effects in HCC development through the recruitment and enhancement of MDSCs [3]. In addition to producing IL-17, ILC3s have also been shown to express MHC II [42] and MHCII+ILC3s can directly induce commensal bacteria-specific T-cell apoptosis [43]. Thus, we performed co-culture experiments to determine whether NCR−ILC3s could directly affect T-cell apoptosis and proliferation (Fig. 6). The results showed that the presence of NCR−ILC3s did not affect the apoptosis and proliferation of CD4+ T cells (Fig. 6a and 6b). To our surprise, the presence of NCR−ILC3s significantly increased apoptosis of CD8+ T cells (Fig. 6c) and inhibited their proliferation (Fig. 6d). We further found in other co-culture experiments that NCR−ILC3s did not directly affect γδT-cell proliferation, apoptosis, or cytokine production (Supplementary Fig. S8). When anti-IL-17 blocking antibodies were added at different concentrations (Supplementary Fig. S9a), the effect of ILC3 cells inducing CD8+ T cell apoptosis was not affect (Fig. 6e). However, when the co-cultures were separated in the transwell system, this effect diminished (Fig. 6f). Therefore, cell-cell interaction is required for the direct regulatory effect of ILC3 on CD8+ T cells. We further detected both MHC I and MHC II expression on ILC3 (Supplementary Fig. S9b), and this apoptosis-inducing effect is dose-dependent when we performed the co-culture experiments at different ILC3:CD8+ T cell ratios (Supplementary Fig. S9c–e). Collectively, these findings suggest that NCR−ILC3s can directly regulate CD8+ T-cell responses through cell-cell interaction.
Fig. 6.
NCR−ILC3 has the capacity to directly regulate T cell response. ILC3 cells were sorted from hepa1–6-IL-23 tumor-bearing C57BL/6 RORgtgfp/+ mice and co-cultured with anti-CD3/anti-CD28 activated naïve CD4+ T cells (a–b) or naïve CD8+ T cells (c–f). (a) Flow cytometry analysis of the percent of apoptotic CD4+ T cells co-cultured with or without ILC3 for 3 days. (b) Flow cytometry analysis of the proliferation of CD4+ T cells (measured by Ki67 staining) co-cultured with or without ILC3 for 3 days. (c) Flow cytometry analysis of the percent of apoptotic CD8+ T cells co-cultured with or without ILC3 for 3 days. (d) Flow cytometry analysis of the proliferation of CD8+ T cells (measured by Ki67 staining) co-cultured with or without ILC3 for 3 days. (e) Flow cytometry analysis of the percent of apoptotic CD8+ T cells co-cultured with or without ILC3 in addition with different concentrations of anti-IL-17A antibody for 3 days. (f) Flow cytometry analysis of the percent of apoptotic CD8+ T cells co-cultured with or without ILC3 or separately in transwells for 3 days. Data are shown as mean ± SEM. Two-tail unpaired student's t-test was used for statistical analysis. *p < .05, **p < .01, ***p < .001. Data shown are the representative of three independent experiments.
4.5. IL-23 promotes differentiation of ILC1 to ILC3 in the tumor microenvironment
Recently, Bernink et al. showed that IL-23 could drive differentiation of human ILC1s into the ILC3 subset [44]. Thus, we examined whether the expansion of NCR−ILC3s in response to IL-23 could be partially differentiated from ILC1s. First, we examined the percentages of ILC1s (i.e., CD3−CD19−CD11b−NK1.1−NKp46−CD117− and T-bet+) in the presence of IL-23 in our murine HCC model (Fig. 7a) and found that the percentages of ILC1 cells were increased in the livers of tumor-bearing mice compared with the PBS control (Fig. 7b and 7c). However, the percentages of ILC1s were significantly decreased in the presence of IL-23 in the tumor microenvironment.
To further determine whether ILC1s could differentiate to ILC3s in the presence of IL-23 in vitro, we sorted ILC1 cells and cultured them in the presence of different cytokines (Fig. 7d and 7e). The cellular yield was similar except a slight increase in the culture with IL-1β plus IL-23. Culture with IL-12 maintained the ILC1 phenotype and enhanced IFN-γ production. However, when IL-1β, IL-23 or IL-1β plus IL-23 were added to the culture, most ILC1 cells differentiated to ILC3 phenotypes of expressing IL-17 and RORγt. The ILC1s and differentiated ILC3s were also double-negative for both NKp46 and CD117 and thus were of NCR− phenotype. Moreover, the increased production of IL-17 and reduced production of IFN-γ were also confirmed in differentiated ILC3s by enzyme-linked immunosorbent assay (ELISA) of the cell culture supernatant, suggesting that IL-23 could promote ILC1 differentiation to ILC3 in vitro (Fig. 7f and 7g).
To further explore whether IL-23 could drive ILC1 differentiation to ILC3 in vivo, we sorted ILC1s from CD45.1 mice and transferred them into CD45.2 mice via tail vein together with implantation of hepa1-6-vector or hepa1-6-IL-23 tumors (Fig. 7h). Five days after transfer, the CD45.1+ cells from the liver were examined for expression of T-bet and RORγt. As shown in Fig. 7i, about 20% of the transferred cells became RORγt single-positive and 10% became RORγt and T-bet double-positive in the tumor-bearing mice. Furthermore, in the presence of IL-23, the percentage of RORγt single-positive cells was significantly increased to about 30%. Thus, IL-23 is able to induce the differentiation of ILC1s to ILC3s in the tumor microenvironment in vivo.
5. Discussion
IL-23 can promote the differentiation of IL-17-producing cells (e.g., Th17, Tc17, γδT17) which are important for tumor development in many tumor models [[3], [4], [5],16,45,46]. Antibody blocking IL-23 signal pathway can enhance the efficacy of anti-tumor therapy [17,47]. We previously demonstrated that γδT17 cells could promote HCC development by recruiting MDSCs and enhancing their functions. Interestingly, IL-17 produced by γδT17 cells facilitates IL-23 production by myeloid cells to further enhance IL-17 production. Therefore, the IL-17-rich immunosuppressive microenvironment could be established by this positive feedback loop. However, it was not clear which cells are the initial responders to the inflammatory cue to start building this immunosuppressive environment. For the first time, we identified NCR−ILC3 cells as the tissue-resident population to respond to inflammatory cytokines in the early stages of tumor development. The production of IL-17 by ILC3 could set the stage for the differentiation of other IL-17-producing cells, and thus generate an IL-17-rich immunosuppressive tumor microenvironment.
NCR−ILC3s are a subset of ILC3s that were first identified in intestinal tissues in 2010 [41]. Initially named cILC, this subset has been shown to sustain colon cancer development [23]. However, the presence of this population in the liver and its role in HCC development have been controversial. ILC3s were initially found in the liver in 2014 [48]. In a subsequent study, Robinette et al. showed that ILC3s are present mostly in the small intestine but are rare in the liver [49]. The discrepancy of these two studies could reflect the different methods used to identify ILC3s. The first study used PMA/ionomycin stimulation and analyzed the percentage of ILC3s in CD3− cells [48], whereas the second study involved analysis of ILC3s by staining RORγt expression without any stimulation [49]. In the current study, by overexpressing IL-23, we could clearly detect the expansion of the IL-17-producing ILC3 cells in the liver, which were identified by RORγt expression without PMA/ionomycin stimulation. Therefore, we confirmed the presence of ILC3s in the liver. Further analysis showed that the expanded ILC3s in response to IL-23 was NCR−ILC3 cells. As a newly identified population, the role of ILC3s in tumor development has been controversial. Although many studies have suggested an antitumor function [[25], [26], [27]], ILC3s have also been shown to contribute to colon cancer development [23,24] and promote lymph node metastasis of breast cancer [50]. In the current study, we found that ILC3s could promote HCC development by producing IL-17 in response to IL-23. Therefore, the tissue microenvironment could be a key factor determining the function of ILC3s.
IL-17 production could be the most important aspect of the immunosuppressive mechanism of ILC3. We showed previously that IL-17 could promote HCC development by recruiting and promoting the suppressive function of MDSCs [3]. In addition to this indirect immunosuppressive function through IL-17 production and MDSCs, we explored here the direct effect of ILC3 cells on T-cell proliferation and apoptosis in in vitro co-culture experiments. Although ILC3 cells did not directly affect CD4+ T-cell proliferation and apoptosis, they inhibited CD8+ T-cell proliferation, and promoted their apoptosis (Fig. 6). This provides the first evidence that ILC3s could directly affect CD8+ T-cell homeostasis and response. MHC II has been implicated in the role of ILC3s regulating CD4+ T-cell function [42]. We also detected both MHC I and MHC II expressions on the isolated ILCs from tumor-bearing mice. The high MHC I expression could be the reason why ILC3s have more potent effects on CD8+ T-cells than on CD4+ T-cells. Nevertheless, the mechanism of their direct effect on T cells needs further investigation.
The plasticity of the ILC subsets has been shown previously in other models. In one study, ILC3s could develop into ILC1 under the influence of IL-12 [51]; in another, ILC1s could become ILC3s in the presence of IL-1β and IL-23 in human intestinal lamina propria [44]. Therefore, IL-23 in the tumor microenvironment could not only promote the conversion of ILC1s to ILC3s, but also enhance the proliferation of ILC3s. Through these two mechanisms, IL-23 could expand the ILC3 population in the liver within a short period before the rise of IL-17-producing T cells.
In summary, we demonstrated here that NCR−ILC3s were the initial responders to IL-23 to promote HCC development in murine HCC models. Their presence and function are yet to be determined in other HCC models involving different carcinogenesis conditions, as well as in HCC patients. Nevertheless, their role in establishing the IL-17-rich immunosuppressive tumor microenvironment suggests that they could be targeted for early prevention or therapeutic modulation of HCC.
Acknowledgments
Acknowledgements
We thank Dr. Yiqiang Wang, Dr. Yun Zhao and Dr. Jianhong Chu of Soochow University for helpful discussions.
The authors also thank C. Wogan of MD Anderson Cancer Center's Division of Radiation Oncology for editorial assistance.
Funding sources
This work was supported by grants from National Natural Science Foundation of China (81471586, 81571556), the Priority Academic Program Development of Jiangsu Higher Education Institutions, the Collaborative Innovation Center of Hematology, a start-up grant from the National University of Singapore, the Cancer Prevention and Research Institute of Texas CPRIT (PR180017), and the National Cancer Institute‘s Cancer Center Support (Core) Grant CA016672 (to The University of Texas MD Anderson Cancer Center). All funding sources have no role in study design, in the collection, analysis, and interpretation of data, in the writing of the manuscript, and in the decision to submit the paper for publication.
Declaration of interests
The authors declare no competing financial interests.
Author contributions
YL and HL designed the experiments; YL, YS, DL, LL, YM, ZJ, HG, YZ, BH, YZ, LZ, and HYT performed the experiments; YL, YS, WJ, DW, YH, and HL analyzed data and interpreted results; YL, YS, YH and HL wrote the manuscript, JQ and CD provided critical experimental materials, all authors commented on the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.02.050.
Contributor Information
Depei Wu, Email: wudepei@suda.edu.cn.
Yuhui Huang, Email: huangyh@suda.edu.cn.
Haiyan Liu, Email: micliuh@nus.edu.sg.
Appendix A. Supplementary data
Supplementary material
References
- 1.Amann T., Bataille F., Spruss T. Activated hepatic stellate cells promote tumorigenicity of hepatocellular carcinoma. Cancer Sci. 2009;100(4):646–653. doi: 10.1111/j.1349-7006.2009.01087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruix J., Boix L., Sala M., Llovet J.M. Focus on hepatocellular carcinoma. Cancer Cell. 2004;5(3):215–219. doi: 10.1016/s1535-6108(04)00058-3. [DOI] [PubMed] [Google Scholar]
- 3.Ma S., Cheng Q., Cai Y. IL-17A produced by gammadelta T cells promotes tumor growth in hepatocellular carcinoma. Cancer Res. 2014;74(7):1969–1982. doi: 10.1158/0008-5472.CAN-13-2534. [DOI] [PubMed] [Google Scholar]
- 4.Kuang D.M., Peng C., Zhao Q., Wu Y., Chen M.S., Zheng L. Activated monocytes in peritumoral stroma of hepatocellular carcinoma promote expansion of memory T helper 17 cells. Hepatology. 2010;51(1):154–164. doi: 10.1002/hep.23291. [DOI] [PubMed] [Google Scholar]
- 5.Kuang D.M., Peng C., Zhao Q. Tumor-activated monocytes promote expansion of IL-17-producing CD8+ T cells in hepatocellular carcinoma patients. J Immunol. 2010;185(3):1544–1549. doi: 10.4049/jimmunol.0904094. [DOI] [PubMed] [Google Scholar]
- 6.Wu S., Rhee K.J., Albesiano E. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15(9):1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oppmann B., Lesley R., Blom B. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13(5):715–725. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 8.Teng M.W., Bowman E.P., McElwee J.J. IL-12 and IL-23 cytokines: from discovery to targeted therapies for immune-mediated inflammatory diseases. Nat Med. 2015;21(7):719–729. doi: 10.1038/nm.3895. [DOI] [PubMed] [Google Scholar]
- 9.Liu L., Shan B., Feng Y. Antitumor effects and immunoregulation mechanisms of IL-23 gene in mouse mammary cancer mediated by retrovirus. Cell Immunol. 2009;258(2):181–187. doi: 10.1016/j.cellimm.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Shan B.E., Hao J.S., Li Q.X., Tagawa M. Antitumor activity and immune enhancement of murine interleukin-23 expressed in murine colon carcinoma cells. Cell Mol Immunol. 2006;3(1):47–52. [PubMed] [Google Scholar]
- 11.Lo C.H., Lee S.C., Wu P.Y. Antitumor and antimetastatic activity of IL-23. J Immunol. 2003;171(2):600–607. doi: 10.4049/jimmunol.171.2.600. [DOI] [PubMed] [Google Scholar]
- 12.Hu W.H., Chen H.H., Yen S.L., Huang H.Y., Hsiao C.C., Chuang J.H. Increased expression of interleukin-23 associated with progression of colorectal cancer. J Surg Oncol. 2017;115(2):208–212. doi: 10.1002/jso.24505. [DOI] [PubMed] [Google Scholar]
- 13.Li J., Lau G., Chen L. Interleukin 23 promotes hepatocellular carcinoma metastasis via NF-kappa B induced matrix metalloproteinase 9 expression. PLoS One. 2012;7(9):e46264. doi: 10.1371/journal.pone.0046264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grivennikov S.I., Wang K., Mucida D. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012;491(7423):254–258. doi: 10.1038/nature11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schetter A.J., Nguyen G.H., Bowman E.D. Association of inflammation-related and microRNA gene expression with cancer-specific mortality of colon adenocarcinoma. Clin Cancer Res. 2009;15(18):5878–5887. doi: 10.1158/1078-0432.CCR-09-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langowski J.L., Zhang X., Wu L. IL-23 promotes tumour incidence and growth. Nature. 2006;442(7101):461–465. doi: 10.1038/nature04808. [DOI] [PubMed] [Google Scholar]
- 17.Calcinotto A., Spataro C., Zagato E. IL-23 secreted by myeloid cells drives castration-resistant prostate cancer. Nature. 2018;559(7714):363–369. doi: 10.1038/s41586-018-0266-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aggarwal S., Ghilardi N., Xie M.H., de Sauvage F.J., Gurney A.L. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J Biol Chem. 2003;278(3):1910–1914. doi: 10.1074/jbc.M207577200. [DOI] [PubMed] [Google Scholar]
- 19.He D., Wu L., Kim H.K., Li H., Elmets C.A., Xu H. CD8+ IL-17-producing T cells are important in effector functions for the elicitation of contact hypersensitivity responses. J Immunol. 2006;177(10):6852–6858. doi: 10.4049/jimmunol.177.10.6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lockhart E., Green A.M., Flynn J.L. IL-17 production is dominated by gammadelta T cells rather than CD4 T cells during mycobacterium tuberculosis infection. J Immunol. 2006;177(7):4662–4669. doi: 10.4049/jimmunol.177.7.4662. [DOI] [PubMed] [Google Scholar]
- 21.Takatori H., Kanno Y., Watford W.T. Lymphoid tissue inducer-like cells are an innate source of IL-17 and IL-22. J Exp Med. 2009;206(1):35–41. doi: 10.1084/jem.20072713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spits H., Artis D., Colonna M. Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13(2):145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 23.Kirchberger S., Royston D.J., Boulard O. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J Exp Med. 2013;210(5):917–931. doi: 10.1084/jem.20122308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan I.H., Jain R., Tessmer M.S. Interleukin-23 is sufficient to induce rapid de novo gut tumorigenesis, independent of carcinogens, through activation of innate lymphoid cells. Mucosal Immunol. 2014;7(4):842–856. doi: 10.1038/mi.2013.101. [DOI] [PubMed] [Google Scholar]
- 25.Dieu-Nosjean M.C., Antoine M., Danel C. Long-term survival for patients with non-small-cell lung cancer with intratumoral lymphoid structures. J Clin Oncol Off J Am Soc Clin Oncol. 2008;26(27):4410–4417. doi: 10.1200/JCO.2007.15.0284. [DOI] [PubMed] [Google Scholar]
- 26.Eisenring M., vom Berg J., Kristiansen G., Saller E., Becher B. IL-12 initiates tumor rejection via lymphoid tissue-inducer cells bearing the natural cytotoxicity receptor NKp46. Nat Immunol. 2010;11(11):1030–1038. doi: 10.1038/ni.1947. [DOI] [PubMed] [Google Scholar]
- 27.Nussbaum K., Burkhard S.H., Ohs I. Tissue microenvironment dictates the fate and tumor-suppressive function of type 3 ILCs. J Exp Med. 2017;214(8):2331–2347. doi: 10.1084/jem.20162031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carrega P., Loiacono F., Di Carlo E. NCR(+)ILC3 concentrate in human lung cancer and associate with intratumoral lymphoid structures. Nat Commun. 2015;6:8280. doi: 10.1038/ncomms9280. [DOI] [PubMed] [Google Scholar]
- 29.Villanova F., Flutter B., Tosi I. Characterization of innate lymphoid cells in human skin and blood demonstrates increase of NKp44+ ILC3 in psoriasis. J Invest Dermatol. 2014;134(4):984–991. doi: 10.1038/jid.2013.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Satoh-Takayama N., Vosshenrich C.A., Lesjean-Pottier S. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29(6):958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Jiang R., Tan Z., Deng L. Interleukin-22 promotes human hepatocellular carcinoma by activation of STAT3. Hepatology. 2011;54(3):900–909. doi: 10.1002/hep.24486. [DOI] [PubMed] [Google Scholar]
- 32.Mas V.R., Maluf D.G., Archer K.J. Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma. Mol Med. 2009;15(3–4):85–94. doi: 10.2119/molmed.2008.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guichard C., Amaddeo G., Imbeaud S. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44(6):694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tolba R., Kraus T., Liedtke C., Schwarz M., Weiskirchen R. Diethylnitrosamine (DEN)-induced carcinogenic liver injury in mice. Lab Anim. 2015;49(1 Suppl):59–69. doi: 10.1177/0023677215570086. [DOI] [PubMed] [Google Scholar]
- 35.Vesselinovitch S.D., Mihailovich N. Kinetics of diethylnitrosamine hepatocarcinogenesis in the infant mouse. Cancer Res. 1983;43(9):4253–4259. [PubMed] [Google Scholar]
- 36.Purwar R., Schlapbach C., Xiao S. Robust tumor immunity to melanoma mediated by interleukin-9-producing T cells. Nat Med. 2012;18(8):1248–1253. doi: 10.1038/nm.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monteiro M., Almeida C.F., Agua-Doce A., Graca L. Induced IL-17-producing invariant NKT cells require activation in presence of TGF-beta and IL-1beta. J Immunol. 2013;190(2):805–811. doi: 10.4049/jimmunol.1201010. [DOI] [PubMed] [Google Scholar]
- 38.Passos S.T., Silver J.S., O'Hara A.C., Sehy D., Stumhofer J.S., Hunter C.A. IL-6 promotes NK cell production of IL-17 during toxoplasmosis. J Immunol. 2010;184(4):1776–1783. doi: 10.4049/jimmunol.0901843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cupedo T., Crellin N.K., Papazian N. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nat Immunol. 2009;10(1):66–74. doi: 10.1038/ni.1668. [DOI] [PubMed] [Google Scholar]
- 40.Lin A.M., Rubin C.J., Khandpur R. Mast cells and neutrophils release IL-17 through extracellular trap formation in psoriasis. J Immunol. 2011;187(1):490–500. doi: 10.4049/jimmunol.1100123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buonocore S., Ahern P.P., Uhlig H.H. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464(7293):1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinette M.L., Colonna M. Innate lymphoid cells and the MHC. Hla. 2016;87(1):5–11. doi: 10.1111/tan.12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hepworth M.R., Fung T.C., Masur S.H. Immune tolerance. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria-specific CD4(+) T cells. Science. 2015;348(6238):1031–1035. doi: 10.1126/science.aaa4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernink J.H., Krabbendam L., Germar K. Interleukin-12 and -23 control plasticity of CD127(+) group 1 and group 3 innate lymphoid cells in the intestinal Lamina Propria. Immunity. 2015;43(1):146–160. doi: 10.1016/j.immuni.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 45.Zhang B., Rong G., Wei H. The prevalence of Th17 cells in patients with gastric cancer. Biochem Biophys Res Commun. 2008;374(3):533–537. doi: 10.1016/j.bbrc.2008.07.060. [DOI] [PubMed] [Google Scholar]
- 46.Wu P., Wu D., Ni C. γδT17 cells promote the accumulation and expansion of myeloid-derived suppressor cells in human colorectal cancer. Immunity. 2014;40(5):785–800. doi: 10.1016/j.immuni.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.von Scheidt B., Leung P.S., Yong M.C. Combined anti-CD40 and anti-IL-23 monoclonal antibody therapy effectively suppresses tumor growth and metastases. Cancer Res. 2014;74(9):2412–2421. doi: 10.1158/0008-5472.CAN-13-1646. [DOI] [PubMed] [Google Scholar]
- 48.Jie Z., Liang Y., Hou L. Intrahepatic innate lymphoid cells secrete IL-17A and IL-17F that are crucial for T cell priming in viral infection. J Immunol. 2014;192(7):3289–3300. doi: 10.4049/jimmunol.1303281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinette M.L., Fuchs A., Cortez V.S. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol. 2015;16(3):306–317. doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Irshad S., Flores-Borja F., Lawler K. RORgammat+ innate lymphoid cells promote lymph node metastasis of breast cancers. Cancer Res. 2017;77(5):1083–1096. doi: 10.1158/0008-5472.CAN-16-0598. [DOI] [PubMed] [Google Scholar]
- 51.Bernink J.H., Peters C.P., Munneke M. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol. 2013;14(3):221–229. doi: 10.1038/ni.2534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material