Abstract
Expression of the non-classical human leukocyte antigen-G (HLA-G) promotes cancer progression in various malignancies including epithelial ovarian cancer (EOC). As single nucleotide polymorphisms (SNPs) in the HLA-G 3′ untranslated region (UTR) regulate HLA-G expression, we investigated HLA-G 3′UTR haplotypes arranged by SNPs in healthy controls (n = 75) and primary EOC patients (n = 79) and determined soluble HLA-G (sHLA-G) levels. Results were related to the clinical status and outcome. Although haplotype frequencies were similar in patients and controls, (i) sHLA-G levels were increased in EOC independent of the haplotype, (ii) homozygosity for UTR-1 or UTR-2 genotypes were significantly associated with metastases formation and presence of circulating tumor cells before therapy, whereas (iii) the UTR-5 and UTR-7 haplotypes were significantly associated with a beneficial clinical outcome regarding negative nodal status, early FIGO staging, and improved overall survival. Lastly, (iv) the ambivalent impact on clinical EOC aspects could be deduced to specific SNPs in the HLA-G 3′UTR: +3187G, +3196G and +3035T alleles. Our results give evidence that even if the genetic background of the HLA-G 3′UTR is identical between patients and controls, certain SNPs have the potential to contribute to diametrical clinical status/outcome in EOC.
Introduction
The human leukocyte antigen G (HLA-G), a non-classical human leukocyte antigen, belongs to the immune checkpoint molecules that regulate/control immune effector responses. The HLA-G molecule is associated with anti-inflammatory and immune-modulatory properties through interaction with inhibitory receptors such as immunoglobulin-like transcript (ILT)2, ILT4 and killer-cell immunoglobulin-like receptor (KIR)2DL4 expressed on different immune-competent cells1. HLA-G inhibits B cells, T cells, and natural killer cells, and induces regulatory T cells thereby mediating escape from the host immune surveillance2. In contrast to classical HLA molecules, HLA-G displays limited allelic variations, but exists in seven isoforms due to alternative splicing3. These isoforms can be expressed at the cell surface (HLA-G1, -G2, -G3 and -G4)4 or as secreted molecules (HLA-G5, -G6, and -G7)5,6. Soluble forms can be released by healthy cells e.g. trophoblasts, adult and embryonic stem cells, and monocytes7 or by malignant cells including breast cancer8,9, melanoma7, renal cancer10, and ovarian cancer cells9,11.
Physiologically, HLA-G cell surface expression is restricted to the maternal-fetal interface in placental mammals, where it mediates immune tolerance, and to immune privileged adult tissues12. In contrast, neo-ectopic or aberrant expression of HLA-G and its soluble forms has been associated with a vast variety of pathological situations13. In the context of malignancies, HLA-G has been implicated in cancer invasiveness and metastatic progression such as epithelial ovarian cancer (EOC)2,14,15, the most lethal of gynecologic malignancies16. EOC is classified into four surgical stages according to the International Federation of Gynecology and Obstetrics (FIGO); this classification considers the extent of ovary affection, the extent of spreading outside the ovaries and outside the pelvis16. The microenvironment of EOC provides a driving force for cancer cell invasion and metastasis17. Although most EOC initially respond to primary treatment, tumor recurrence is frequently not limited to drug-resistant tumors, resulting in an impaired overall patient survival (OS)18. Dissemination of single tumor cells (DTCs) into the blood-stream plays a major role in drug resistance and thereby disease recurrence and metastases formation in EOC19. Likewise, the presence of circulating tumor cells (CTCs) negatively correlates with OS in EOC patients20,21.
HLA-G is located on chromosome 6p21.3 and is composed of eight exons and seven introns, and so far 58 HLA-G allelic variations have been identified22. Importantly, the regulation of HLA-G expression and of its soluble forms encompasses post-transcriptional processes among which alternative splicing, altered mRNA stability, microRNA-mediated protein expression and impaired protein transport to the cell surface2. Especially the polymorphic 3′untranslated region (UTR) shared by the HLA-G1 to HLA-G6 transcripts (~370 bp) plays a pivotal role in HLA-G expression by interfering with transcription, splicing, mRNA stability and translation23. Here, sixteen single nucleotide polymorphisms (SNP; +3001C/T, +3003C/T, +3010C/G, +3027C/A, +3032C/G, +3035C/T, +3052C/T, +3092G/T, +3111A/G, +3121C/T, +3142C/G, +3177G/T, +3183A/G, +3187A/G, +3196C/G, and +3227A/G) and a 14 bp insertion/deletion (INS/DEL) located at position +2961 have been identified in the 3′UTR potentially modifying the affinity of gene targeted sequences for post-transcriptional factors24. These polymorphisms arrange as haplotypes, named UTRs (Supplementary Info 1). UTR-1 to UTR-8 and UTR-18 are the most frequent ones22,25. Six already identified microRNA (miR), miR-148a, miR-148b, miR-152, miR-133a, miR-628-5p, and miR-548q, have been reported to bind to certain SNPs in the 3′UTR in a sequence-specific manner, leading to downregulation of HLA-G expression22. Particularly, UTR-7, encompassing the three most studied variations (14 bp INS, +3142G, and +3187G), has been reported to correlate with lower plasma levels of sHLA-G26. Moreover, many types of cancer exhibit an aberrant miR expression profile27,28, which modulates the tumor microenvironment via non-cell-autonomous mechanisms28 and is involved in tumor initiation, progression, metastasis formation and therapy resistance29. We here propose that, in analogy with other clinical disorders30,31, variable HLA-G 3′UTR sites in combination with the cellular microenvironment may be critical in influencing HLA-G expression.
In this retrospective study we (i) defined the HLA-G 3′UTR haplotypes from primary patients with serous ovarian carcinoma, (ii) determined the association of the genetic background with the presence of CTCs, and (iii) analyzed the impact of the genetic background on clinical status and disease outcome of EOC.
Results
HLA-G 3′UTR haplotypes in EOC patients and healthy controls
To define the 3′UTR haplotypes in EOC patients (n = 79) and healthy donors (HD; n = 75), we sequenced and analyzed 15 previously described polymorphic variations in the HLA-G 3′UTR. As expected, a high linkage disequilibrium (LD) was observed among the SNPs showing a minor allele frequency (MAF) >1%. In addition, a nearly perfect LD (D′ = 0.97, r2 = 0.92) was observed between +3010C/G and +3142G/C alleles in healthy women (Supplementary Data 1). Overall, 9 out of 16 different haplotypes were identified with a frequency >1% and were considered in this study (Supplementary Data 2). In line with previous data26,32, UTR-1 (29% and 28%) and UTR-2 (29% and 27%) displayed the most abundant haplotypes in EOC patients and healthy women, respectively. Interestingly, one undesignated UTR haplotype (UTR-undes.) differing from UTR-2 only at position +3142 by a C instead of a G (Supplementary Info 1), was exclusively identified in the patients’ group with a frequency of 3%. Statistically significant differences between the two study groups were not observed for the genotype or haplotype distribution (Supplementary Data 3).
sHLA-G in EOC patients and healthy controls
Mean sHLA-G levels were almost two-fold elevated in EOC patients compared to HD (n = 30; p < 0.0001), independent of the patients’ UTR haplotype (Fig. 1a,b). In EOC patients, median sHLA-G levels increased with ascending FIGO stages without reaching significance (Fig. 1c). Among EOC patients, the nodal status, metastasis formation, presence of CTC and DTC prior to therapy and OS and PFS was not associated with sHLA-G levels (data not shown).
Figure 1.
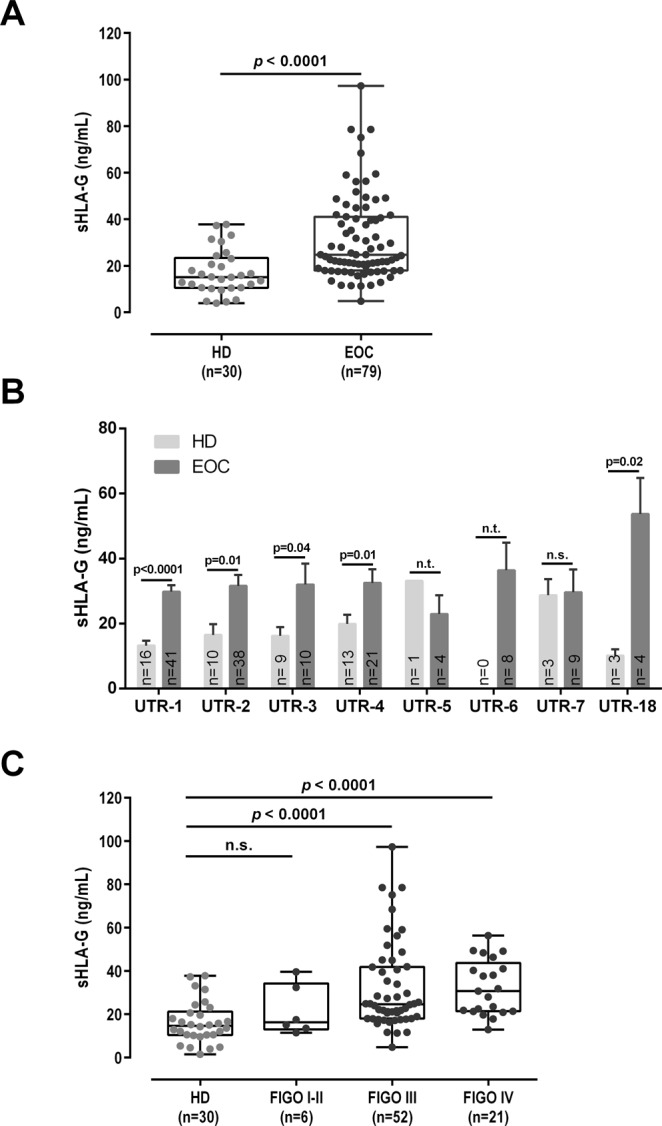
Comparison of sHLA-G levels of EOC patients and healthy donors. (A) sHLA-G is significantly (p < 0.0001) elevated in EOC compared to healthy donors (HD). (B) Elevated sHLA-G levels in EOC are independent of a specific HLA-G 3′UTR haplotype. Bars indicate mean value ± SEM. (C) sHLA-G levels increase with ascending FIGO stage in EOC patients without reaching significance. sHLA-G levels are given in ng/ml. Statistic was performed by Mann-Whitney test (A,C), or Kruskal-Wallis with Dunn’s test for multiple comparison (B). n.s. not significant, n.t. not tested due to low numbers.
HLA-G 3′UTR haplotypes UTR-1 or UTR-2 are associated with metastases and presence of CTCs in EOC patients
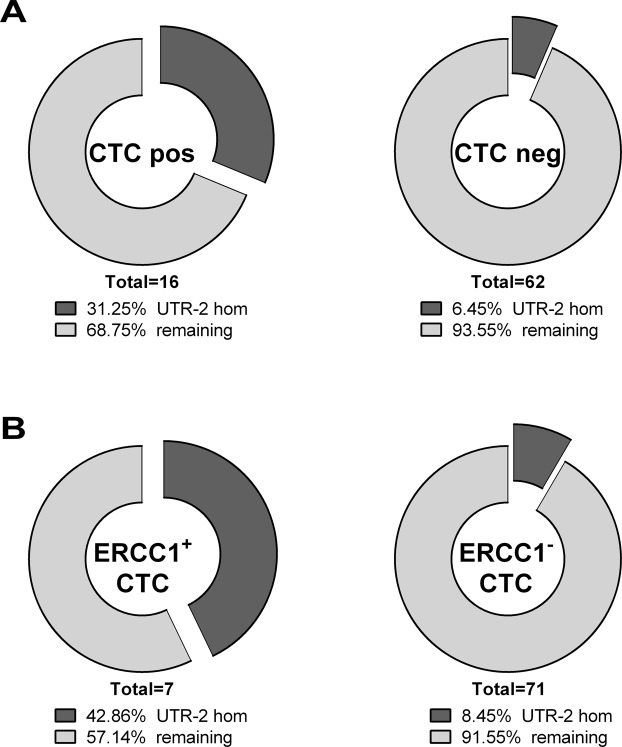
UTR haplotype analysis showed that allelic homozygosity for UTR-1 was significantly associated with the presence of metastases (p = 0.0044, Odds Ratio (OR): 17.81, 95% Confidence Interval (CI): 1.94–163.7), whereas presence of UTR-5 was positively associated with a negative nodal status (p = 0.0392, OR: 0.066, 95% CI: 0.00–1.361) and early FIGO staging (p = 0.001, OR: 0.014, 95% CI: 0.00–0.176; Table 1). Homozygous UTR-2 genotype was significantly associated with the presence of CTCs before therapy (p = 0.0151, OR: 6.59, 95% CI: 1.52–28.51; Table 2 and Fig. 2). Interestingly, concerning CTC specificity, the latter one is significantly associated with ERCC1+ CTCs (p = 0.0312, OR: 8.00, 95% CI: 1.44–44.47). None of the UTR haplotypes or genotypes were associated with CTCs after therapy (data not shown).
Table 1.
HLA-G 3′UTR haplotypes are associated with the clinical status of EOC patients.
| Metastasis formation | M1b | M0b | p a | OR (95% CI) |
|
|---|---|---|---|---|---|
| UTR-1 | UTR-1/UTR-1 | 5 | 1 | 0.0044 | 17.81 (1.94–163.7) |
| Remaining | 16 | 57 | |||
| Nodal status | pN 1 b | pN 0 b | p a |
OR
(95% CI) |
|
| UTR-5 | positive | 0 | 3 | 0.0392 | 0.066 (0.00–1.361) |
| negative | 33 | 15 | |||
| FIGO | III-IV b | I-II b | p a |
OR
(95% CI) |
|
| UTR-5 | positive | 1 | 3 | 0.001 | 0.014 (0.00–0.176) |
| negative | 72 | 3 |
CI – confidence interval; M0 – no metastasis formation; M1 – metastasis formation; pN0 – no nodal infestation; pN1 – nodal infestation; OR – odds ratio.
ap-values were calculated by GraphPad Prism using Fisher’s exact test, alpha <0.05.
bNumbers reflect cases.
Table 2.
Homozygous UTR-2 of the HLA-G 3′UTR is associated with the presence of CTCs, in particular ERCC1 positive CTCs, before therapy.
| UTR-2 homb | remainingb | p a | OR (95% CI) |
|
|---|---|---|---|---|
| CTC pos | 5 | 11 | 0.0151 | 6.591 (1.52–28.51) |
| CTC neg | 4 | 58 | ||
| MUC+ CTC | 3 | 7 | n.s. | 4.429 (0.90–21.75) |
| MUC− CTC | 6 | 62 | ||
| ERCC1+ CTC | 3 | 4 | 0.0312 | 8.000 (1.44–44.47) |
| ERCC1− CTC | 6 | 65 | ||
| EPCAM+ CTC | 1 | 4 | n.s. | 2.031 (0.20–20.50) |
| EPCAM− CTC | 8 | 65 | ||
| HER2+ CTC | 0 | 1 | n.s. | 2.404 (0.09–63.40) |
| HER2− CTC | 9 | 68 |
CI – confidence interval; CTC – circulating tumor cells; EPCAM – epithelial cell adhesion molecule; ERCC1 – Excision Repair cross-complementing group 1; HER-2 – human epidermal growth factor 2; hom – homozygous; MUC – Mucin; n.s. – not significant; OR – odds ratio.
ap-values were calculated by GraphPad Prism using Fisher’s exact test, alpha <0.05.
bNumbers reflect cases.
Figure 2.
Association of HLA-G UTR-2 haplotype with presence of CTCs, especially with ERCC1+ CTCs before therapy. (A) Distribution of homozygous UTR-2 carriers compared to the remaining haplotypes in association with CTCs. CTCs before therapy could not be determined for one patient. (B) Distribution of homozygous UTR-2 carriers compared to the remaining haplotypes in association with ERCC1+ CTCs. ERCC1 gene expression analysis could not be determined for one patient.
HLA G 3′UTR SNP genotypes +3187G/G or +3196G/G are associated with metastases and presence of CTCs and DTCs in EOC patients
Allelic variation at position +3187 distinguished UTR-1 from the others by carrying a G instead of an A (Supplementary Info 1). Interestingly, the +3187GG genotype was significantly associated with metastasis formation (p = 0.0044, OR: 17.81, 95% CI: 1.94–163.7; Table 3). Concerning UTR-2 genotype, homozygous +3196G variant, being exclusively present in UTR-2 and in UTR-undes. in our study, was strongly associated with the presence of both, CTCs and DTCs before therapy (p = 0.0075, OR: 6.84, 95% CI: 1.75–26.76, and p = 0.0156, OR: 5.84, 95% CI: 1.40–24.27, respectively; Table 3). None of the SNP variants were associated with CTCs after therapy (Supplementary Data 4). These observations suggest an association of specific HLA-G 3′UTR-1 and 2 genotypes with oncological phenotypes. Association between the remaining SNPs and the clinical parameters were not found (Supplementary Data 4).
Table 3.
Single nucleotide polymorphisms in the UTR-1 and UTR-2 haplotypes of the HLA-G 3′UTR are associated with aggravated clinical status in EOC patients.
| M1b | M0b | p a | OR (95% CI) |
||
|---|---|---|---|---|---|
| +3187 | GG | 5 | 1 | 0.0044 | 17.81 (1.94–163.7) |
| AA or AG | 16 | 57 | |||
| CTC pos b | CTC neg b | p a |
OR
(95% CI) |
||
| +3196 | GG | 6 | 5 | 0.0075 | 6.840 (1.75–26.76) |
| CC or CG | 10 | 57 | |||
| DTC pos b | DTC neg b | p a |
OR
(95% CI) |
||
| +3196 | GG | 8 | 3 | 0.0156 | 5.841 (1.40–24.27) |
| CC or CG | 21 | 46 |
CI – confidence interval; CTC – circulating tumor cells; DTC – disseminated tumor cells; M0 – no metastasis formation; M1 – metastasis formation; neg – negative; OR – odds ratio; pos – positive; ap-values were calculated by GraphPad Prism using Fisher’s exact test, alpha <0.05.
bNumbers reflect cases.
HLA-G 3′UTR haplotypes UTR-5 or UTR-7 are associated with early FIGO staging and improved clinical outcome in EOC patients
Regarding disease classification, UTR-5 seemed to be a prognostic beneficial factor in EOC, as it was significantly associated with early FIGO: Three out of six (50%) EOC patients classified as FIGO I-II carried UTR-5, whereas only one out of 73 patients (7,7%) with advanced FIGO (III-IV) stages carried UTR-5 (p = 0.001, OR: 0.01, 95% CI: 0.00–0.18; Table 1). Similarly, all patients with nodal infestation were UTR-5 negative (n = 33), whereas in three out of 18 patients without nodal infestation UTR-5 was present (p = 0.0392, OR: 0.06, 95% CI: 0.00–1.36; Table 1). Assessment of OS by combined Kaplan-Meier analysis and Log-rank testing revealed that exclusively patients carrying UTR-7 (n = 9) had a significantly prolonged OS with an undefined median survival time compared to the UTR-7 negative patients (n = 62) with a median survival time of 41 ± 7 months (p = 0.041, HR: 0.17, 95% CI: 0.14–0.95; Fig. 3). None of the UTR haplotypes were associated with PFS.
Figure 3.
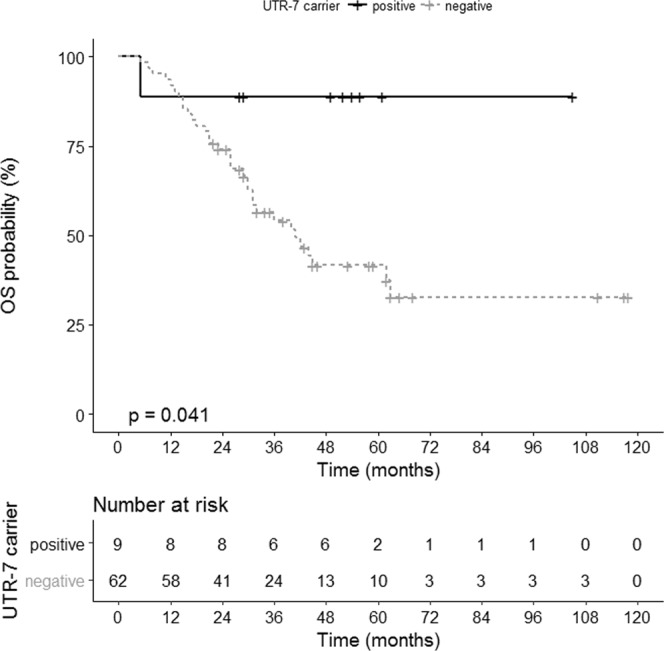
Association of UTR-7 haplotype of HLA-G with an improved OS in EOC patients. Kaplan-Meier plot of the OS of EOC patients carrying UTR-7 (n = 9) and patients not carrying UTR-7 (n = 62).
HLA-G 3′UTR SNP variant +3035T is associated with improved clinical outcome in EOC patients
Both, the UTR-5 and the UTR-7 haplotype exclusively harbor a T instead of a C at position +3035 of the HLA-G 3′UTR (Supplementary Info 1). Taken this SNP variant into consideration, the +3035T carriers (n = 12) revealed a significantly improved PFS (p = 0.029, HR: 0.30, 95% CI: 0.19–0.90; Fig. 4) and OS (p = 0.028, HR: 0.23, 95% CI: 0.17–0.90; Fig. 5) compared to non-T carriers with an undefined median survival time compared to 40 ± 6 and 18 ± 3 month, respectively. These findings suggest that UTR-5 and UTR-7 genotypes are beneficial prognostic factors in EOC, as they appear to significantly associate with early FIGO and enhanced OS, respectively. The +3035T in both UTR-5 and -7 independently associates with PFS and OS in EOC.
Figure 4.

Association of HLA-G 3′UTR SNP variant +3035T with an improved PFS in EOC patients. Kaplan-Meier plot of the PFS of EOC patients bearing the +3035T variant (n = 10) and patients not carrying +3035T (n = 46).
Figure 5.

Association of HLA-G 3′UTR SNP variant +3035T with an improved OS in EOC patients. Kaplan-Meier plot of the OS of EOC patients bearing the +3035T variant (n = 12) and patients not carrying +3035T (n = 59).
Discussion
HLA-G expression is considered as an important immune escape mechanism of cancer cells33. Its expression is regulated at transcriptional, co- and post-transcriptional levels. Regulation at the co- and post-transcriptional level occurs through alternative splicing and/or binding of certain miR within the HLA-G 3′UTR, respectively27. As HLA-G and sHLA-G are reported to be highly expressed in ovarian cancer9,15,34,35, we here examine a possible association between specific HLA-G 3′UTR haplotypes and clinical status of EOC, and whether specific SNPs within the HLA-G 3′UTR region may have prognostic value with regards to EOC treatment and survival. We demonstrate for the first time (i) that haplotype frequencies of the HLA-G 3′UTR between healthy controls and EOC patients are comparable although sHLA-G levels of patients were significantly increased compared to HD independent of their HLA-G 3′UTR haplotype, (ii) that homozygosity for UTR-1 or UTR-2 genotypes are significantly associated with metastases formation and presence of CTCs before therapy, (iii) that the UTR-5 and UTR-7 haplotypes are associated with a beneficial clinical outcome, with regards to negative nodal status, early FIGO staging and improved OS, and lastly (iv) that certain allelic variants, among which +3187G, +3196G and +3035T in the HLA-G 3′UTR have ambivalent impact on clinical aspects of EOC.
Our comparative haplotype analysis between healthy controls and EOC patients revealed that the overall frequencies of both cohorts are very similar to the ones found in worldwide population studies25,26,32. A study of haplotype distribution between papillary thyroid cancer and healthy controls demonstrated significant differences23. As we did not observe such differences in HLA-G 3′UTR distribution, this may point to distinct contribution of HLA-G genotypes to the onset of specific forms of cancer or oncological phenotypes. Although UTR-1 and UTR-2 frequencies encompass more than 50% of all haplotypes, the presence of homozygous status of UTR-1 or UTR-2 is rare. Of note, we found that the frequency of homozygous UTR-1 or UTR-2 was nearly doubled in EOC compared to healthy controls. Although this difference did not reach significance given the current population size, this finding could imply that these genotypes may favor susceptibility to EOC. Indeed, among EOC patients, these genotypes were associated with adverse cancer status: Homozygous UTR-1 genotype is associates with metastases, which may relate to different immune-suppressive microenvironment of advanced CTC compared to the primary tumors. Interestingly, in EOC a cluster of specific miRs has been functionally connected to metastatic lesions36. So far, UTR-2 has not been associated with metastasis by other studies; however a relationship with an increased risk of neurotoxicity after chemotherapy treatment in colorectal cancer was observed37. We20,38 and others39 have already described CTCs as a negative prognostic marker regarding OS as well as chemotherapy resistance in EOC19. Of note, in the present study the detection of CTCs before therapy was associated with a homozygous UTR-2 genotype. In particular, homozygous UTR-2 status in EOC patients appears to be related to the presence of ERCC1 positive CTC subpopulation. In this context, high ERCC1 expression has recently been linked to a worse PFS21 and OS among EOC patients, especially in late stage and poor differentiation serous ovarian patients21,40. Recently, we showed that auxiliary assessment of ERCC1-transcripts expanded the phenotypic spectrum of CTC-detection and we defined a sub-fraction of CTCs41. To the best of our knowledge the current study is the first to demonstrate an association between the presence of CTCs and HLA-G 3′UTR polymorphisms in EOC. Nevertheless, further studies will have to evaluate whether CTCs express HLA-G allowing their survival in the periphery.
In sharp contrast to the UTR-1/2 haplotypes, our study revealed that carriers of the HLA-G 3′UTR haplotypes UTR-5 and UTR-7 have a clinical benefit with respect to long-term OS and negative nodal status. Both haplotypes are associated with a lower HLA-G production compared to UTR-126 thereby preventing HLA-G-mediated immune escape. In vitro studies combining site-directed mutagenesis of HLA-G 3′UTRs with reporter assays demonstrated that 3′UTR polymorphisms have the potential to affect mRNA stability or degradation via RNA-binding proteins and miRs: again, low HLA-G production was previously associated with UTR-5 and UTR-7, whereas UTR-1 was related to high production22. Strikingly, both, UTR-5 and UTR-7, are the only identified haplotypes carrying the rare +3035T variant, which shows to be positively associated with a prolonged OS as well as PFS and being different from all other haplotypes. miR-1224-5p has been reported to bind with high affinity to +3027C/+3035T42,43, which is characteristic for the UTR-5 haplotype. Additionally, miR-187 binds with moderate affinity to the +3027C/A/+3035T haplotype42, targeting both, the HLA-G 3′UTR haplotypes UTR-5 and UTR-7. Our finding that the rare +3035T variant which only occurs in UTR-5 and -7 haplotypes and the +3027A variant which is exclusive for the UTR-7 haplotype, positively associate with increased PFS and OS, is in support of a potential role for sequence-dependent macromolecular control of HLA-G mRNA biology. In line with this notion, +3027A polymorphism was shown to be an independent prognostic factor in pediatric Hodgkin’s lymphoma44.
Hitherto, the most studied SNP of the HLA-G 3′UTR are a 14 bp INS/DEL and +3142G/C. Although not consistent, the majority of studies have associated the14bp DEL as well as +3142C with high HLA-G production potentially promoting an immune-suppressive environment in various malignancies23,45–47. Importantly, the sHLA-G levels of the EOC patients in our study were significantly increased independent of the HLA-G 3′UTR haplotype implying that the tumor burden is the main source for the systemic release of sHLA-G molecules. This concept is supported by the fact that increasing sHLA-G levels were found to be associated with ascending FIGO stage preferentially mirroring the extent of cancer. The +3187G as well as +3196G variants in UTR-1 and UTR-2, respectively, were associated with aggravated clinical outcome considering presence of CTC, DTC and metastasis formation. Homozygous +3187G carriers have already been linked to a worse prognosis in both DFS and OS in colorectal cancer48. However, to deduce this association to the biologic function, it is mandatory to analyze the expression of HLA-G isoforms, the corresponding miRs, and long non-coding RNAs affecting the binding affinity to the HLA-G 3′UTR49 in CTC, DTC and metastases. Although no miR binding sites have been defined yet for +3187G/A and +3196C/G SNPs in the Brazilian population42, it is conceivable that in the context of EOC, these SNPs represent binding sites for as yet unknown HLA-G-targeting miRs. Here, a novel in silico analysis of miR targeting the HLA-G 3′UTR would be of great interest. Alternatively, such nucleotide variants may affect chemical epigenetic modification of mRNAs50. Of note, these two SNPs are in close proximity to the recognized AU-rich motif sequence, which influences alternative pre-mRNA processing, HLA-G mRNA stabilization and, translation (initiation, efficiency) progress and alternative pre-mRNA processing42,51. Only two patients (3%) expressed UTR-undes., both of whom had a homozygous genotype. Hitherto, the 14 bp INS has always been associated with the presence of +3142G and +3187A, which correlates to low HLA-G levels. UTR-undes., however, encompasses the 14 bp INS in combination with +3142C and +3187A. Interestingly, +3142G is suggested to increase the binding of miR-148a, miR-148b, and miR-152, suggesting that the allelic variation +3142C found in our cohort of patients might be disadvantageous by impairing binding of regulatory miRs regulatory control in the HLA-G 3′UTR. This is also in line with recent studies, in which the +3142CC genotype was shown to be associated with increased HLA-G levels and susceptibility to cancer23,52. Further, it already has been shown that some miRs bind to non-polymorphic sequences of the HLA-G 3′UTR in a stable and specific manner, while others bind to polymorphic sequences. Such findings indicate that HLA-G co- and post-transcriptional regulation of mRNA might be depending on both, binding of miR and additional RNA-binding protein factors (RBPs) and is subject to both genetic (variants), as well as epigenetic and microenvironmental control to certain variants present and the miR microenvironment43. In this context, a recent study of renal cell carcinoma analyzed HLA-G transcript, protein and miR expression pattern, infiltrating immune cells, and clinical outcome: A strong post-transcriptional gene regulation of HLA-G by miR-152, -148A, -148B and -133A has been observed53. Immunohistochemical staining revealed an inverse expression of miR-148A and -133A with the HLA-G protein in situ and in vitro indicating a direct interaction with HLA-G regulatory miRs and the HLA-G 3′UTR. Stable miR overexpression caused a downregulation of HLA-G protein enhancing cytotoxic function of immune cells. The association of HLA-G expression with infiltrating cells in HLA-G positive tumors revealed higher numbers of CD3+ and CD8+ T cells but not NK and CD4+ T cells. However, the latter was related to a better disease-specific survival. Concerning ovarian cancer, so far only one study placed emphasis on the analysis of the HLA-G expression in primary tumor lesions and metastatic tissue. In this study it was shown that HLA-G was more frequently expressed in metastatic cells than in primary tumor lesions and the expression of HLA-G inversely associated with the frequency of tumor infiltrating immune cells54. Besides other parameters both, the HLA-G expression and the low infiltration of immune cells contributed to a worse prognosis and overall survival54. Unfortunately, no HLA-G 3′UTR typing was performed in this study to link the increased HLA-G expression in metastatic lesions to a certain HLA-G 3′UTR SNP.
Conclusion
We define multiple different HLA-G 3′UTR haplotypes relating to diametrical clinical status and/or disease outcome in EOC patients. We propose that even if the genetic background of the HLA-G 3′UTR is identical between physiologic and pathologic conditions, presence of certain SNPs within important regulatory sequences in the HLA-G 3′UTR have the potential to contribute to an adverse or beneficial clinical status in EOC. It is worthy to mention that the association of individual SNPs potentially reveals the impact of specific haplotypes on EOC outcome (i.e., at least for +3187G/G and +3196G/G being exclusive for UTR-1 and UTR-2, respectively) due to high LD in HLA-G 3′UTR. In addition, the impact of such genetic variants is likely dependent on the interaction with the tumor microenvironment and tumor phenotype phenotype and tumor heterogeneity, which is partly characterized by the mutational burden. Of note, certain mutations have been associated with a worse clinical outcome in high-grade serous ovarian cancer, e.g. CHEK255. As certain HLA-G 3′UTR SNPs are associated with a worse clinical outcome, here, HLA-G 3′UTR typing or HLA-G expression in tumor lesions may improve the prediction of clinical outcome and thus, the disease management. The present data highlight the complexity of the genetic background of HLA-G affecting the clinical course of EOC. Future studies should be aimed at defining the interactome (ncRNAs as well as protein factors) of HLA-G 3′UTRs. A comprehensive study combining the aspects of HLA-G 3′UTR polymorphisms, HLA-G transcript, protein and miR expression pattern, infiltrating immune cells in primary tumor tissue and metastatic lesions with the clinical outcome of patients would greatly contribute to the understanding of HLA-G mediated tumor immune escape. Improved understanding of HLA-G regulation will contribute to the development of strategies modulating its expression and to optimizing the design of (immune-) therapeutic strategies for the treatment of tumor patients. Enlarging the cohorts of women will strengthen the associations found in the present study.
Methods
Patients’ characteristics
A total of 79 patients diagnosed between 2001 and 2014 at the Department of Gynecology and Obstetrics, University Hospital Essen, with the histologically confirmed primary diagnosis of EOC were analyzed. All patients received the standard treatment consisting of cyto-reductive surgery and adjuvant platinum-based chemotherapy. Patients with EOC stage FIGOIIIB or higher (18 out of 79 patients) received adjuvant bevacizumab treatment. Patients who did not undergo adjuvant platinum-based chemotherapy as well as patients with secondary malignant diseases were excluded from the analysis. Clinical characteristics of the patients are documented in Table 4. 75 healthy female donors (HD) served as control panel for genotyping. Written informed consent was obtained by all participants and the study was approved by the Local Ethics Committees (Essen 05-2870 and 17-7859) and was performed according to the declaration of Helsinki.
Table 4.
Patient characteristics at time of primary diagnosis.
| Total | n = 79 (%) | |
|---|---|---|
| Age | Median: 61 (27–88) | |
| FIGO stage | I-II | 6 (8%) |
| III | 52 (66%) | |
| IV | 21 (26%) | |
| Nodal status | N0 | 18 (23%) |
| N1 | 33 (42%) | |
| Unknown | 28 (35%) | |
| Metastases formation | M0 | 58 (73%) |
| M1 | 21 (27%) | |
| Tumor grading | I-II | 30 (38%) |
| III | 49 (62%) | |
| CTC posa | Before therapy | 16/78 (21%) |
| After therapy | 10/28 (36%) | |
| DTCs posb | Before therapy | 29/78 (37%) |
| Recurrence | No relapse | 23 (29%) |
| Relapse | 42 (53%) | |
| Unknown | 14 (18%) | |
| Platinum-based chemotherapy | No resistance | 51 (65%) |
| Resistance | 13 (16%) | |
| Unknown | 15 (19%) | |
| Overall Survival | 0 | 40 (51%) |
| 1 | 36 (45%) | |
| Unknown | 3 (4%) |
CTC – circulating tumor cell; DTCs – disseminated tumor cells; FIGO – Federation of Gynecology and Obstetrics; M0 – no metastasis formation; M1 – metastasis formation; pN0 – no nodal infestation; pN1 – nodal infestation; aCTCs before therapy could not be determined for one patients, and CTCs after therapy could not be determined for 51 patients; bDTCs before therapy could not be determined for one patient.
Sampling of blood
Two times 5 ml ethylenediaminetetraacetic (EDTA) blood were collected for isolation of CTCs before the application of therapeutic substances with an S-Monovette (Sarstedt AG & Co.) and stored at 4 °C until further examination. The samples were processed within 4 hours after blood collection.
Selection, detection and evaluation of CTCs and DTCs
Enrichment of CTCs and subsequent expression analysis were performed according to Adnatest OvarianCancer (Qiagen, Hilden, Germany). The test has been described in detail21,41. Briefly, CTCs were immunomagnetically selected by using the AdnaTest Ovarian Cancer Select targeting epithelial cell adhesion molecule EpCAM, MUC-1 and Mucin-16 (also known as CA-125). Subsequently, RNA was isolated and gene expression analysis was performed by reverse-transcription (RT) and multiplex RT-PCR detecting EpCAM, MUC-1, and CA-125 (AdnaTest Ovarian Cancer Detect). ERCC1-transcripts were investigated in a separate approach by singleplex RT-PCR (AdnaTest Ovarian Cancer Detect). β-actin served as an internal control. DTCs were analyzed as described before56. In short, DTCs were analyzed by immunocytochemistry using the pan-cytokeratin antibody A45-B/B3.
Quantification of soluble HLA-G components
Soluble HLA-G was quantified as previously described57. Plasma samples were used in a dilution of 1:2 in PBS and purified HLA-G5 served as standard reagent. The sHLA-G levels were determined by four-parameter curve fitting. ELISA detection limit of sHLA-G was 0.25 ng/ml.
HLA-G 3′UTR analysis
After genomic DNA extraction using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions, HLA-G 3′UTR typing was performed by polymerase chain reaction (PCR) as previously described58. The PCR products were directly sequenced using the reverse primer GmiRNA in an ABI 3730 XL DNA sequencer (Applied Biosystems, Foster City, CA, USA) with polymer POP-7 performed in LGC Genomics (LGC Genomics, Berlin, Germany). Sequencing reactions were performed by using BigDye™ Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). HLA-G polymorphism was assessed by interpretation of chromatogram peaks using the FinchTV software version 1.4.0 (available on http://www.geospiza.com/Products/finchtv.shtml). In total, our approach allowed us to evaluate 15 genetic variants (14 bp INS/DEL, +3001C/T, +3003C/T, +3010C/G, +3027A/C, +3032C/G, +3035C/T, +3052C/T, +3092G/T, +3111A/G, +3121C/T, +3142C/G, +3187A/G, +3196C/G, +3227A/G) encompassing the 3′UTR of the HLA-G gene. Linkage disequilibrium (LD) was evaluated by the Haploview software59 through inspections of D′ and r² coefficients in EOC, healthy controls and pooled sample. Haplotype phasing from all individuals were assessed by PHASE 2.1 software using default parameters60. The consistency of the results was checked through 10 independent runs using different seed values. Haplotypes of each subject were inferred with a probability ranging from 0.948 to 1.0 for all subjects. Assessment of haplotype frequencies across different runs showed highly consistent results. HLA-G 3′UTR haplotypes were determined according to previous studies25,32.
To verify differences regarding haplotype frequencies among the patients cohort (n = 79) and controls (n = 75), a residuals analysis was performed (Supplementary Data 2).
Statistical analysis
Statistical analyses were performed by using SPSS 22.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism V6.0 software (GraphPad Software, San Diego, CA, USA). Allele and genotype frequencies of polymorphic sites were calculated by using two-sided Chi-square test. Contribution of allelic variants to clinical parameters was evaluated by Fisher’s exact test as indicated in the table legend. Overall survival (OS) and progression-free survival (PFS) analysis was assessed by the method of Kaplan-Meier and compared using log-rank test implemented in the R package survminer (version 0.4.0; https://CRAN.R-project.or/package=survminer).
Ethics approval and consent to participate
All aspects of this study were approved by the local ethics committee of the University Hospital Essen. Written informed consent was obtained from all enrolled patients, and all relevant investigations were performed according to the principles of the Declaration of Helsinki.
Supplementary information
Supplementary Dataset 1, Supplementary Dataset 2, Supplementary Dataset 3, Supplementary Dataset 4
Acknowledgements
We gratefully thank the patients and healthy controls for participation in the study and for kindly providing the samples. We highly value the technical support by the medical and laboratory team of the Department of Gynecology and Obstetrics and the colleagues of the Institute for Transfusion Medicine (both University Hospital Essen), especially the contribution of Sabine Schramm. Rafael Tomoya Michita was supported by the “Coordenação de Aperfeicoamento de Pessoal de Nível Superior (CAPES) Foundation, Ministry of Education of Brazil, Brasília-DF Brazil (99999.000124/2016-08)”, the “Conselho Nacional de Desenvolvimento Científico e Tecnológico” (CNPq) Foundation, Ministry of Science and Technology of Brazil, Brasília-DF Brazil (142475/2015-7), and by the “Deutscher Akademischer Austauschdienst” (DAAD) Scholarship of the German Federal Ministry of Education and Research. We acknowledge support by the Open Access Publication Fund of the Univerity of Duisburg-Essen.
Author Contributions
E.S. and V.R. conceived and designed research, performed the experiments, interpreted data, performed statistical analysis, wrote the initial draft, and read and approved the final article. R.T.M. performed the experiments, interpreted data, wrote the initial draft, and read and approved the final article. P.B. conceived and designed research, collected and provided clinical data, interpreted data, performed statistical analysis, wrote the initial draft, and read and approved the final article. S.K.B. and R.K. collected and provided clinical data, interpreted data and read and approved the final article. H.R., J.W.V. and P.H. interpreted data, wrote the initial draft, and read and approved the final article.
Data Availability
All data in our study are available upon request.
Competing Interests
S. Kasimir-Bauer is a consultant for Qiagen. Vera Rebmann is a consultant for Bristol-Myers Squibb. All other authors declare no potential conflicts of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41900-z.
References
- 1.Rebmann V, da Silva Nardi F, Wagner B, Horn PA. HLA-G as a tolerogenic molecule in transplantation and pregnancy. J Immunol Res. 2014;2014:297073. doi: 10.1155/2014/297073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amiot L, Ferrone S, Grosse-Wilde H, Seliger B. Biology of HLA-G in cancer: a candidate molecule for therapeutic intervention? Cell Mol Life Sci. 2011;68:417–431. doi: 10.1007/s00018-010-0583-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paul P, et al. Identification of HLA-G 7 as a new splice variant of the HLA-G mRNA and expression of soluble HLA-G5, -G6, and -G7 transcripts in human transfected cells. Hum Immunol. 2000;61:1138–1149. doi: 10.1016/S0198-8859(00)00197-X. [DOI] [PubMed] [Google Scholar]
- 4.Paul P, et al. HLA-G expression in melanoma: a way for tumor cells to escape from immunosurveillance. Proc Natl Acad Sci USA. 1998;95:4510–4515. doi: 10.1073/pnas.95.8.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carosella ED, Paul P, Moreau P, Rouas-Freiss N. HLA-G and HLA-E: fundamental and pathophysiological aspects. Immunol Today. 2000;21:532–534. doi: 10.1016/S0167-5699(00)01707-2. [DOI] [PubMed] [Google Scholar]
- 6.Konig L, et al. The prognostic impact of soluble and vesicular HLA-G and its relationship to circulating tumor cells in neoadjuvant treated breast cancer patients. Hum Immunol. 2016;77:791–799. doi: 10.1016/j.humimm.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Rebmann V, Busemann A, Lindemann M, Grosse-Wilde H. Detection of HLA-G5 secreting cells. Hum Immunol. 2003;64:1017–1024. doi: 10.1016/j.humimm.2003.08.354. [DOI] [PubMed] [Google Scholar]
- 8.Konig L, et al. Elevated levels of extracellular vesicles are associated with therapy failure and disease progression in breast cancer patients undergoing neoadjuvant chemotherapy. Oncoimmunology. 2017;7:e1376153. doi: 10.1080/2162402X.2017.1376153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rebmann V, Regel J, Stolke D, Grosse-Wilde H. Secretion of sHLA-G molecules in malignancies. Semin Cancer Biol. 2003;13:371–377. doi: 10.1016/S1044-579X(03)00028-2. [DOI] [PubMed] [Google Scholar]
- 10.Grange C, et al. Role of HLA-G and extracellular vesicles in renal cancer stem cell-induced inhibition of dendritic cell differentiation. BMC Cancer. 2015;15:1009. doi: 10.1186/s12885-015-2025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singer G, et al. HLA-G is a potential tumor marker in malignant ascites. Clin Cancer Res. 2003;9:4460–4464. [PubMed] [Google Scholar]
- 12.Carosella ED, HoWangYin KY, Favier B, LeMaoult J. HLA-G-dependent suppressor cells: Diverse by nature, function, and significance. Hum Immunol. 2008;69:700–707. doi: 10.1016/j.humimm.2008.08.280. [DOI] [PubMed] [Google Scholar]
- 13.Rebmann V, et al. The Potential of HLA-G-Bearing Extracellular Vesicles as a Future Element in HLA-G Immune Biology. Front Immunol. 2016;7:173. doi: 10.3389/fimmu.2016.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung YW, et al. Correlation of human leukocyte antigen-G (HLA-G) expression and disease progression in epithelial ovarian cancer. Reprod Sci. 2009;16:1103–1111. doi: 10.1177/1933719109342131. [DOI] [PubMed] [Google Scholar]
- 15.Babay, W. et al. Clinicopathologic significance of HLA-G and HLA-E molecules in Tunisian patients with ovarian carcinoma. Hum Immunol, 10.1016/j.humimm.2018.02.012 (2018). [DOI] [PubMed]
- 16.Jelovac D, Armstrong DK. Recent progress in the diagnosis and treatment of ovarian cancer. CA Cancer J Clin. 2011;61:183–203. doi: 10.3322/caac.20113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao L, et al. The RNA binding protein SORBS2 suppresses metastatic colonization of ovarian cancer by stabilizing tumor-suppressive immunomodulatory transcripts. Genome Biol. 2018;19:35. doi: 10.1186/s13059-018-1412-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Housman G, et al. Drug resistance in cancer: an overview. Cancers (Basel) 2014;6:1769–1792. doi: 10.3390/cancers6031769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giannopoulou L, Kasimir-Bauer S, Lianidou ES. Liquid biopsy in ovarian cancer: recent advances on circulating tumor cells and circulating tumor DNA. Clin Chem Lab Med. 2018;56:186–197. doi: 10.1515/cclm-2017-0019. [DOI] [PubMed] [Google Scholar]
- 20.Chebouti I, et al. EMT-like circulating tumor cells in ovarian cancer patients are enriched by platinum-based chemotherapy. Oncotarget. 2017;8:48820–48831. doi: 10.18632/oncotarget.16179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhlmann JD, et al. ERCC1-positive circulating tumor cells in the blood of ovarian cancer patients as a predictive biomarker for platinum resistance. Clin Chem. 2014;60:1282–1289. doi: 10.1373/clinchem.2014.224808. [DOI] [PubMed] [Google Scholar]
- 22.Poras I, et al. Haplotypes of the HLA-G 3′ Untranslated Region Respond to Endogenous Factors of HLA-G+ and HLA-G- Cell Lines Differentially. Plos One. 2017;12:e0169032. doi: 10.1371/journal.pone.0169032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Figueiredo-Feitosa NL, et al. HLA-G 3′ untranslated region polymorphic sites associated with increased HLA-G production are more frequent in patients exhibiting differentiated thyroid tumours. Clin Endocrinol (Oxf) 2017;86:597–605. doi: 10.1111/cen.13289. [DOI] [PubMed] [Google Scholar]
- 24.Silva ID, et al. HLA-G 3′UTR polymorphisms in high grade and invasive cervico-vaginal cancer. Hum Immunol. 2013;74:452–458. doi: 10.1016/j.humimm.2012.11.025. [DOI] [PubMed] [Google Scholar]
- 25.Sabbagh A, et al. Worldwide genetic variation at the 3′ untranslated region of the HLA-G gene: balancing selection influencing genetic diversity. Genes Immun. 2014;15:95–106. doi: 10.1038/gene.2013.67. [DOI] [PubMed] [Google Scholar]
- 26.Martelli-Palomino G, et al. Polymorphic sites at the 3′ untranslated region of the HLA-G gene are associated with differential hla-g soluble levels in the Brazilian and French population. Plos One. 2013;8:e71742. doi: 10.1371/journal.pone.0071742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castelli EC, et al. Insights into HLA-G Genetics Provided by Worldwide Haplotype Diversity. Front Immunol. 2014;5:476. doi: 10.3389/fimmu.2014.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki HI, Katsura A, Matsuyama H, Miyazono K. MicroRNA regulons in tumor microenvironment. Oncogene. 2015;34:3085–3094. doi: 10.1038/onc.2014.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seliger B. Role of microRNAs on HLA-G expression in human tumors. Hum Immunol. 2016;77:760–763. doi: 10.1016/j.humimm.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 30.Rajasekaran A, et al. The impact of HLA-G 3′UTR variants and sHLA-G on risk and clinical correlates of schizophrenia. Hum Immunol. 2016;77:1166–1171. doi: 10.1016/j.humimm.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 31.Michita RT, et al. A tug-of-war between tolerance and rejection - New evidence for 3′UTR HLA-G haplotypes influence in recurrent pregnancy loss. Hum Immunol. 2016;77:892–897. doi: 10.1016/j.humimm.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 32.Castelli EC, et al. The genetic structure of 3′ untranslated region of the HLA-G gene: polymorphisms and haplotypes. Genes Immun. 2010;11:134–141. doi: 10.1038/gene.2009.74. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Yu S, Han Y, Wang Y, Sun Y. Human leukocyte antigen-G expression and polymorphisms promote cancer development and guide cancer diagnosis/treatment. Oncol Lett. 2018;15:699–709. doi: 10.3892/ol.2017.7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jasinski-Bergner S, et al. Identification of novel microRNAs regulating HLA-G expression and investigating their clinical relevance in renal cell carcinoma. Oncotarget. 2016;7:26866–26878. doi: 10.18632/oncotarget.8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rutten MJ, et al. HLA-G expression is an independent predictor for improved survival in high grade ovarian carcinomas. J Immunol Res. 2014;2014:274584. doi: 10.1155/2014/274584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braga EA, Fridman MV, Kushlinskii NE. Molecular Mechanisms of Ovarian Carcinoma Metastasis: Key Genes and Regulatory MicroRNAs. Biochemistry (Mosc) 2017;82:529–541. doi: 10.1134/S0006297917050017. [DOI] [PubMed] [Google Scholar]
- 37.Garziera, M. et al. HLA-G 3′UTR Polymorphisms Predict Drug-Induced G3-4 Toxicity Related to Folinic Acid/5-Fluorouracil/Oxaliplatin (FOLFOX4) Chemotherapy in Non-Metastatic Colorectal Cancer. Int J Mol Sci18, 10.3390/ijms18071366 (2017). [DOI] [PMC free article] [PubMed]
- 38.Aktas B, Kasimir-Bauer S, Heubner M, Kimmig R, Wimberger P. Molecular profiling and prognostic relevance of circulating tumor cells in the blood of ovarian cancer patients at primary diagnosis and after platinum-based chemotherapy. Int J Gynecol Cancer. 2011;21:822–830. doi: 10.1097/IGC.0b013e318216cb91. [DOI] [PubMed] [Google Scholar]
- 39.Fan T, Zhao Q, Chen JJ, Chen WT, Pearl ML. Clinical significance of circulating tumor cells detected by an invasion assay in peripheral blood of patients with ovarian cancer. Gynecol Oncol. 2009;112:185–191. doi: 10.1016/j.ygyno.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao M, et al. Prognostic values of excision repair cross-complementing genes mRNA expression in ovarian cancer patients. Life Sci. 2018;194:34–39. doi: 10.1016/j.lfs.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 41.Chebouti I, et al. ERCC1-expressing circulating tumor cells as a potential diagnostic tool for monitoring response to platinum-based chemotherapy and for predicting post-therapeutic outcome of ovarian cancer. Oncotarget. 2017;8:24303–24313. doi: 10.18632/oncotarget.13286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castelli EC, et al. In silico analysis of microRNAS targeting the HLA-G 3′ untranslated region alleles and haplotypes. Hum Immunol. 2009;70:1020–1025. doi: 10.1016/j.humimm.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 43.Porto IO, et al. MicroRNAs targeting the immunomodulatory HLA-G gene: a new survey searching for microRNAs with potential to regulate HLA-G. Mol Immunol. 2015;65:230–241. doi: 10.1016/j.molimm.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 44.De RV, et al. HLA-G +3027 polymorphism is associated with tumor relapse in pediatric Hodgkin’s lymphoma. Oncotarget. 2017;8:105957–105970. doi: 10.18632/oncotarget.22515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garziera M, et al. Association of the HLA-G 3′UTR polymorphisms with colorectal cancer in Italy: a first insight. Int J Immunogenet. 2016;43:32–39. doi: 10.1111/iji.12243. [DOI] [PubMed] [Google Scholar]
- 46.Carosella ED, Moreau P, Lemaoult J, Rouas-Freiss N. HLA-G: from biology to clinical benefits. Trends Immunol. 2008;29:125–132. doi: 10.1016/j.it.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 47.Li T, et al. Genetic polymorphism in HLA-G 3′UTR 14-bp ins/del and risk of cancer: a meta-analysis of case-control study. Mol Genet Genomics. 2015;290:1235–1245. doi: 10.1007/s00438-014-0985-3. [DOI] [PubMed] [Google Scholar]
- 48.Garziera M, et al. HLA-G 3′UTR Polymorphisms Impact the Prognosis of Stage II-III CRC Patients in Fluoropyrimidine-Based Treatment. Plos One. 2015;10:e0144000. doi: 10.1371/journal.pone.0144000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song B, et al. Long non-coding RNA HOTAIR promotes HLA-G expression via inhibiting miR-152 in gastric cancer cells. Biochem Biophys Res Commun. 2015;464:807–813. doi: 10.1016/j.bbrc.2015.07.040. [DOI] [PubMed] [Google Scholar]
- 50.Polakova K, Bandzuchova E, Tirpakova J, Kuba D, Russ G. Modulation of HLA-G expression. Neoplasma. 2007;54:455–462. [PubMed] [Google Scholar]
- 51.Matoulkova E, Michalova E, Vojtesek B, Hrstka R. The role of the 3′ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol. 2012;9:563–576. doi: 10.4161/rna.20231. [DOI] [PubMed] [Google Scholar]
- 52.Yang YC, et al. Human leucocyte antigen-G polymorphisms are associated with cervical squamous cell carcinoma risk in Taiwanese women. Eur J Cancer. 2014;50:469–474. doi: 10.1016/j.ejca.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 53.Jasinski-Bergner S, et al. Clinical relevance of miR-mediated HLA-G regulation and the associated immune cell infiltration in renal cell carcinoma. Oncoimmunology. 2015;4:e1008805. doi: 10.1080/2162402X.2015.1008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andersson E, et al. Non-classical HLA-class I expression in serous ovarian carcinoma: Correlation with the HLA-genotype, tumor infiltrating immune cells and prognosis. Oncoimmunology. 2016;5:e1052213. doi: 10.1080/2162402X.2015.1052213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ow GS, Ivshina AV, Fuentes G, Kuznetsov VA. Identification of two poorly prognosed ovarian carcinoma subtypes associated with CHEK2 germ-line mutation and non-CHEK2 somatic mutation gene signatures. Cell Cycle. 2014;13:2262–2280. doi: 10.4161/cc.29271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kasimir-Bauer S, et al. Does primary neoadjuvant systemic therapy eradicate minimal residual disease? Analysis of disseminated and circulating tumor cells before and after therapy. Breast Cancer Res. 2016;18:20. doi: 10.1186/s13058-016-0679-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rebmann V, Lemaoult J, Rouas-Freiss N, Carosella ED, Grosse-Wilde H. Report of the Wet Workshop for Quantification of Soluble HLA-G in Essen, 2004. Hum Immunol. 2005;66:853–863. doi: 10.1016/j.humimm.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 58.Zambra FM, et al. Immunogenetics of prostate cancer and benign hyperplasia–the potential use of an HLA-G variant as a tag SNP for prostate cancer risk. HLA. 2016;87:79–88. doi: 10.1111/tan.12741. [DOI] [PubMed] [Google Scholar]
- 59.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 60.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Dataset 1, Supplementary Dataset 2, Supplementary Dataset 3, Supplementary Dataset 4
Data Availability Statement
All data in our study are available upon request.