ABSTRACT
The Zetaproteobacteria are a class of bacteria typically associated with marine Fe(II)-oxidizing environments. First discovered in the hydrothermal vents at Loihi Seamount, Hawaii, they have become model organisms for marine microbial Fe(II) oxidation. In addition to deep sea and shallow hydrothermal vents, Zetaproteobacteria are found in coastal sediments, other marine subsurface environments, steel corrosion biofilms and saline terrestrial springs. Isolates from a range of environments all grow by autotrophic Fe(II) oxidation. Their success lies partly in their microaerophily, which enables them to compete with abiotic Fe(II) oxidation at Fe(II)-rich oxic/anoxic transition zones. To determine the known diversity of the Zetaproteobacteria, we have used 16S rRNA gene sequences to define 59 operational taxonomic units (OTUs), at 97% similarity. While some Zetaproteobacteria taxa appear to be cosmopolitan, others are enriched by specific habitats. OTU networks show that certain Zetaproteobacteria co-exist, sharing compatible niches. These niches may correspond with adaptations to O2, H2 and nitrate availability, based on genomic analyses of metabolic potential. Also, a putative Fe(II) oxidation gene has been found in diverse Zetaproteobacteria taxa, suggesting that the Zetaproteobacteria evolved as Fe(II) oxidation specialists. In all, studies suggest that Zetaproteobacteria are widespread, and therefore may have a broad influence on marine and saline terrestrial Fe cycling.
Keywords: Zetaproteobacteria, Fe(II) oxidation, microbial ecology, hydrothermal vents, marine Fe cycling, phylogenetic analysis
Review and synthesis of the discovery, ecology, and genetic potential of the most prolific marine Fe-oxidizing clade, the Zetaproteobacteria.
INTRODUCTION
Fe in marine environments is a study in contrasts. It is often a limiting nutrient in the open ocean, while the basaltic ocean crust and many sediments have abundant Fe. This stark difference is due to the redox chemistry of Fe, which is present as Fe(II) in basalt and anoxic groundwater, but rapidly oxidizes to Fe(III) in oxic ocean water, precipitating as Fe(III) minerals. This oxidation was assumed to be dominated by rapid abiotic oxidation at circumneutral pH, but the discovery of the Fe(II)-oxidizing Zetaproteobacteria in marine environments gave proof that the process can be driven by microbes. First proposed as a class in 2007 (Emerson et al. 2007), Zetaproteobacteria have since been widely observed in deep sea and coastal environments. All isolates are obligate autotrophs and can couple Fe(II) oxidation to oxygen respiration, producing highly reactive Fe(III) oxyhydroxides that can adsorb or coprecipitate nutrients and metals (e.g. Laufer et al. 2017). However, despite the biogeochemical importance of microbial Fe(II) oxidation, we are just beginning to learn about Fe(II)-oxidizer distribution and how they function and influence marine ecosystems. With a recent surge of culturing and sequencing, there is now a substantial set of data from which we can glean broader insights into microbial Fe(II) oxidation in marine and other saline habitats.
The goal of this paper is to review our current knowledge of marine Fe(II)-oxidizers through the lens of this increasingly well-established class of Fe(II)-oxidizing bacteria (FeOB). We begin by describing the discovery of this novel class at an Fe(II)-rich hydrothermal system at Loihi Seamount (also spelled Lō’ihi Seamount) in Hawaii. We lay out the evidence for microbially-driven Fe(II) oxidation in this marine system, including new kinetics results from experiments with the Loihi isolate and model Zetaproteobacteria, Mariprofundus ferrooxydans PV-1. Work at Loihi has inspired numerous studies of Zetaproteobacteria isolates, biominerals and environmental distribution. In addition to reviewing these, we present a comprehensive reanalysis of Zetaproteobacteria diversity and distribution, enabled by the newly developed ZetaHunter classification program (McAllister, Moore and Chan 2018), to gain insights into Zetaproteobacteria niches (sets of conditions favorable for growth). We then use current genomic evidence to evaluate whether all members of this class have the potential to oxidize Fe(II) and further describe Zetaproteobacteria niches based on inferred metabolic potential. Finally, we discuss our perspectives on open questions in Zetaproteobacteria evolution, ecology and impacts on geochemical cycling. This article was submitted to an online preprint archive (McAllister et al. 2018).
Zetaproteobacteria: a novel class of marine Fe(II)-oxidizing bacteria
The discovery of Zetaproteobacteria is a story that began decades before the class was proposed. The unusual morphology of biogenic Fe(III) (oxyhydr)oxides have long been used to recognize microbial Fe(II) oxidation in terrestrial environments. The twisted stalks of Gallionella and hollow sheaths containing cells of Leptothrix were described in terrestrial Fe(II)-rich environments as early as the mid-1800s (Ehrenberg 1838; Kützing 1843). However, Winogradsky (1888) was the first to confirm that Leptothrix required Fe(II) for growth, thus linking microbial activity with iron mineral deposition in terrestrial environments (Harder 1919). In the 1980s, similar structures were found in Fe(II)-rich marine environments, including the Red Seamount of the East Pacific Rise, the Explorer Ridge and Loihi Seamount (Fig. S1, Supporting Information) (Alt et al. 1987; Alt 1988; Juniper and Fouquet 1988; Karl et al. 1988; Karl, Brittain and Tilbrook 1989). This led to the assumption that these structures were made by Gallionella and Leptothrix, though these organisms were not detected in subsequent studies of marine Fe(II)-oxidizing microbial mats (Fe mats) based on small subunit ribosomal RNA (SSU rRNA, frequently referred to as 16S rRNA) marker gene surveys (e.g. Moyer, Dobbs and Karl 1995; Davis et al. 2009; Rassa et al. 2009). Instead, Moyer et al. (1995) discovered the first sequence of the novel Zetaproteobacteria class, though it was not recognized at the time because there were no isolates or other closely related sequences. The first isolates, Mariprofundus ferrooxydans strains PV-1 and JV-1, were obtained from samples collected at Loihi Seamount near Hawaii in 1996–98 (Emerson and Moyer 2002; Emerson et al. 2007). Additional surveys from Fe(II)-rich environments provided related 16S rRNA gene sequences (Eder et al. 2001; Dhillon et al. 2003; Davis et al. 2009; Kato et al. 2009b), which helped establish the Zetaproteobacteria as a monophyletic group within the Proteobacteria (Emerson et al. 2007). The association of the Zetaproteobacteria and Fe(II)-rich marine environments has been strengthened since these initial observations, with continued discovery of Zetaproteobacteria within Fe(II)-rich saline environments.
Zetaproteobacteria isolates: model systems for microbial Fe(II) oxidation
The difficulty of culturing FeOB has been one of the main challenges in demonstrating marine microbial Fe(II) oxidation. The first Zetaproteobacteria isolates were obtained using liquid and agarose-stabilized gradient tubes and plates designed to provide both Fe(II) and O2 in opposing gradients (Emerson and Moyer 2002; Emerson and Floyd 2005). With this setup, Fe(II) is gradually released by dissolution of solid reduced Fe minerals (e.g. Fe(0), FeS, or FeCO3) at the bottom of the tube or plate while O2 diffuses from the headspace above. These culturing techniques make it difficult to control O2 and Fe(II) concentrations. To date, Zetaproteobacteria have not been culturable on solid media, so isolation requires serial dilution to extinction, with transfers every ∼2–3 days due to increasing autocatalytic Fe(II) oxidation over time (Lueder et al. 2018). In all, these challenges likely account for why so few Zetaproteobacteria have been isolated.
Despite these hurdles, Zetaproteobacteria representatives from two genera and eight OTUs have been successfully isolated (Table 1). These include seven isolates from microbial mats at Fe(II)-rich hydrothermal vents, and eight from coastal environments. Mariprofundus ferrooxydans PV-1 is the type strain of the most frequently isolated genus, and is an obligate neutrophilic autotrophic Fe(II)-oxidizer. All but two other isolates are similarly obligate Fe(II)-oxidizers. These two, Ghiorsea bivora TAG-1 and SV-108, are facultative Fe(II)-oxidizers that are also capable of growth by H2 oxidation (Mori et al. 2017). Except for this instance, isolates vary primarily in their physiological preferences (Table 2), which are related to characteristics of their source environments.
Table 1.
Isolates of the Zetaproteobacteria and their assigned ZOTUs, with representation in the environment, biomineral and metabolic properties, and references.
| Isolate | ZOTU | ZOTU Envir. abund.a | Isolation source | Primary biomineral morphology | Fe(II) oxidation | H2 oxidation | References |
|---|---|---|---|---|---|---|---|
| Mariprofundus ferrooxydans PV-1 | ZOTU11 | 1.58% | Loihi hydrothermal vents | stalk | X | Emerson and Moyer 2002 | |
| Mariprofundus ferrooxydans JV-1 | ZOTU11 | Loihi hydrothermal vents | stalk | X | Emerson and Moyer 2002 | ||
| Mariprofundus ferrooxydans M34 | ZOTU11 | Loihi hydrothermal vents | stalk | X | McAllister et al. 2011 | ||
| Mariprofundus ferrooxydans SC-2 | ZOTU11 | Big Fisherman's Cove pyrrhotite colonization | stalk | X | Barco et al. 2017 | ||
| Mariprofundus ferrinatatus CP-8 | ZOTU37 | 0.09% | Chesapeake Bay stratified water column | dreads only | X | Chiu et al. 2017 | |
| Mariprofundus aestuarium CP-5 | ZOTU18 | 1.49% | Chesapeake Bay stratified water column | dreads only | X | Chiu et al. 2017 | |
| Mariprofundus micogutta ET2 | ZOTU18 | Bayonnaise hydrothermal vents | thin filaments | X | Makita et al. 2017 | ||
| Mariprofundus sp. DIS-1 | ZOTU18 | West Boothbay Harbor mild steel incubation | stalk | X | Mumford et al. 2016 | ||
| Mariprofundus sp. GSB-2 | ZOTU23 | 0.26% | Great Salt Bay salt marsh Fe mat | stalk | X | McBeth et al. 2011 | |
| Mariprofundus sp. EKF-M39 | ZOTU36 | 0.35% | Loihi hydrothermal vents | stalk | X | Field et al. 2015 | |
| Ghiorsea bivora TAG-1 | ZOTU9 | 8.42% | MAR hydrothermal vents | none | X | X | Mori et al. 2017 |
| Ghiorsea bivora SV-108 | ZOTU9 | Mariana hydrothermal vents | none | X | X | Mori et al. 2017 | |
| Zetaproteobacteria sp. CSS-1 | ZOTU14 | 4.04% | coastal sediment bloodworm microcosm | stalk | X | Beam et al. 2018 | |
| Zetaproteobacteria sp. S1OctC | ZOTU3 | 3.51% | Norsminde Fjord estuary sediments | stalk | X | Laufer et al. 2017 | |
| Zetaproteobacteria sp. S2.5 | ZOTU3 | Kalø Vig beach sediments | stalk | X | Laufer et al. 2017 |
Estimates of ZOTU environmental abundance based on 16S rRNA gene surveys (SILVA release 128), including in the estimate counts for instances where a single published sequence represents multiple clones. ZOTU environmental abundance estimates are given once for each ZOTU.
Table 2.
Growth preferences of Zetaproteobacteria isolates, including optimal growth salinity, temperature, pH and oxygen concentrations.
| Growth salinity (ppt) | Growth temperature (°C) | Growth pH | Growth [O2] | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate | ZOTU | Doubling time (h) | Isolation | Range | Opt. | Isolation | Range | Opt. | Isolation | Range | Opt. | Headspace(% O2) | Range (µM) | References |
| Mariprofundus ferrooxydans PV-1 | ZOTU11 | 12 | 35 (ASW) | 3.5-35a | 28-31.5a | 12-24 | 10-30 | 30 | 6.4-6.5 | 5.5-7.2 | 6.2-6.5 | 1 | – | Emerson and Moyer 2002; Emerson et al. 2007 |
| Mariprofundus ferrooxydans JV-1 | ZOTU11 | 12 | 35 (ASW) | – | – | 12-24 | 10-30 | 30 | 6.4-6.5 | 5.5-7.2 | 6.2-6.5 | 1 | – | Emerson and Moyer 2002; Emerson et al. 2007 |
| Mariprofundus ferrooxydans M34 | ZOTU11 | – | 35 (ASW) | – | – | RT | – | – | – | – | – | 5-15 | – | McAllister et al. 2011 |
| Mariprofundus ferrooxydans SC-2 | ZOTU11 | – | 35 (ASW) | – | – | RT | – | – | 6.2-6.5 | – | – | 3.4 | – | Barco et al. 2017 |
| Mariprofundus ferrinatatus CP-8 | ZOTU37 | 27 | 18 (SEM)b | 7-31.5 | 14-17.5 | RT | 15-35 | 25-30 | 6.2 | 5.5-8.3 | 6.9-7.2 | 1 | 0.07-1.7 | Chiu et al. 2017 |
| Mariprofundus aestuarium CP-5 | ZOTU18 | 19.5 | 18 (SEM)b | 7-31.5 | 14-17.5 | RT | 10-30 | 20-25 | 6.2 | 5.5-8.3 | 6.9-7.2 | 1 | 0.31-2.0 | Chiu et al. 2017 |
| Mariprofundus micogutta ET2 | ZOTU18 | 24 | 35 (ASW) | 10-40 | 27.5 | 25 | 15-30 | 25 | 6.2-6.5 | 5.8-7.0 | 6.4 | 1-3 | – | Makita et al. 2017 |
| Mariprofundus sp. DIS-1 | ZOTU18 | –c | 35 (ASW) | – | – | RT | – | – | 6.1-6.4 | max 8 | – | 5-15 | max ∼220 | Mumford et al. 2016 |
| Mariprofundus sp. GSB-2 | ZOTU23 | 13d | 35 (ASW) | 1.75-35 | – | RT | – | 25 | 6.1-6.4 | max 7.25 | – | 5-15 | – | McBeth et al. 2011 |
| Mariprofundus sp. EKF-M39 | ZOTU36 | – | 35 (ASW) | – | – | – | – | – | 6.1-6.4 | – | – | 0e | – | Field et al. 2015 |
| Ghiorsea bivora TAG-1 | ZOTU9 | 21.8f | 35 (ASW) | – | – | – | 5-30 | 20 | 6.5 | 5.5-7.5 | 6.5-7.0 | 0.4-15 | – | Mori et al. 2017 |
| Ghiorsea bivora SV-108 | ZOTU9 | 20.0f | 35 (ASW) | – | – | – | 5-30 | 20 | 6.5 | 6.0-7.5 | 6.5-7.0 | 0.4-15 | – | Mori et al. 2017 |
| Zetaproteobacteria sp. CSS-1 | ZOTU14 | – | 35 (ASW) | – | – | RT | – | – | – | – | – | 5-10 | opt. 60g | Beam et al. 2018 |
| Zetaproteobacteria sp. S1OctC | ZOTU3 | – | 23 (ASW) | 6.9-23 | – | – | – | – | 7.1 | – | – | 6-10 | – | Laufer et al. 2017 |
| Zetaproteobacteria sp. S2.5 | ZOTU3 | – | 23 (ASW) | 6.9-23 | – | – | – | – | 7.1 | – | – | 6-10 | – | Laufer et al. 2017 |
RT = room temperature; ASW = Artificial Seawater Medium; – = No data available.
Data from Chiu et al. (2017).
SEM = Simulated estuary medium (50:50 MWMM:ASW); MWMM = Modified Wolfe's Mineral Medium.
Cited as comparable with other FeOB.
13 h doubling time in standard gradient tubes; 7–8 h doubling time on metal coupons.
Trace O2 was likely introduced into the culture with aerobic vitamin/mineral solutions and mat innoculum.
Doubling time on Fe(II) shown; Doubling time on H2 was 14.1 h and 16.3 h for TAG-1 and SV-108, respectively.
Reported as approximate optimum.
Zetaproteobacteria isolates are generally microaerophiles originating from oxic-anoxic transition zones, where O2 concentrations are low, i.e. micromolar to submicromolar. Abiotic Fe(II) oxidation is slow at these low O2 concentrations (Stumm and Lee 1961; Millero, Sotolongo and Izaguirre 1987), which allows the Zetaproteobacteria to compete. In terrestrial freshwater circumneutral environments, kinetics experiments near 25°C suggest that biotic Fe(II) oxidation is a significant component of total Fe(II) oxidation below 50 µM and can outcompete abiotic Fe(II) oxidation at 15 µM O2 (Druschel et al. 2008). However, there are no kinetics data from marine FeOB. To understand the conditions where marine biotic Fe(II) oxidation is competitive, we measured Fe(II) oxidation kinetics using M. ferrooxydans PV-1 as a model (see Supplemental Methods). With this experiment, we have shown that PV-1 outcompetes abiotic oxidation below 49 µM O2, and accounts for up to 99% of the Fe(II) oxidation at 10 µM O2 (Fig. 1; Table 3). In cultures of M. aestuarium CP-5 and M. ferrinatatus CP-8, oxygen concentrations ranged from 0.07–2.0 µM O2 within the cell growth band (Chiu et al. 2017). This range of O2 growth conditions is well below the level at which almost all Fe(II) oxidation was biotic for PV-1, suggesting that many Zetaproteobacteria are well adapted to compete and thrive under micromolar and submicromolar O2 concentrations. Such low O2 concentrations are common within the oxic-anoxic transition zones where the Zetaproteobacteria are found (e.g. Chan et al. 2016; Field et al. 2016).
Figure 1.
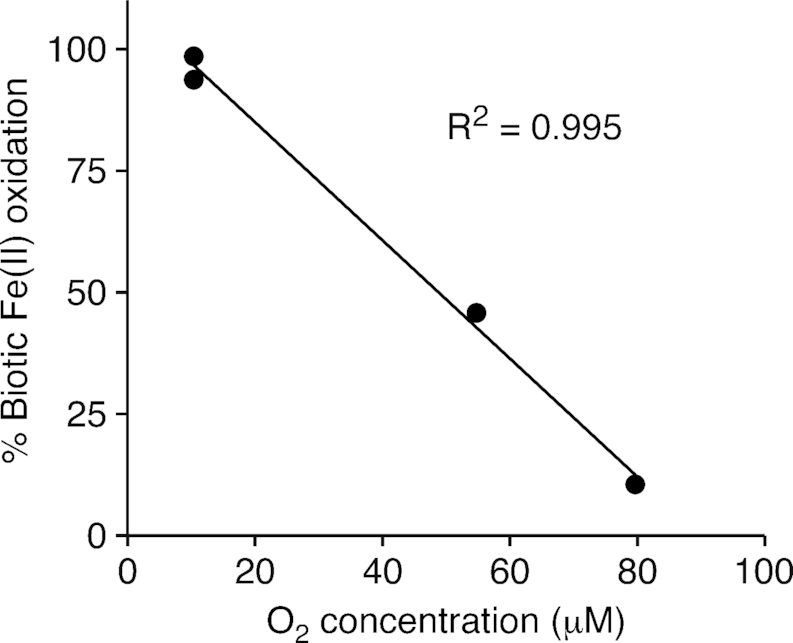
Biotic Fe(II) oxidation rate as a percentage of the total Fe(II) oxidation rate (biotic plus abiotic) at varying oxygen concentrations, using the model Zetaproteobacteria isolate, M. ferrooxydans PV-1. Range of 6.5 – 6.7 pH for experimental conditions. Further details in Supplemental Methods.
Table 3.
Biotic and abiotic Fe(II) oxidation rates of Mariprofundus ferrooxydans PV-1 under a range of O2 concentrations.
| Fe(II) oxidation rate | |||||
|---|---|---|---|---|---|
| O2 conc. | biotic | biotic | abiotic | ||
| µM | µM Fe(II) hr−1 | µM Fe(II) cell−1 hr−1 | µM Fe(II) hr−1 | % Biotic Fe(II) oxidation | pH |
| 10.4 | 21.05 | 5.06E-04 | 0.32 | 98.5 | 6.63 |
| 10.4 | 24.69 | 5.53E-04 | 1.66 | 93.7 | 6.70 |
| 54.8 | 33.32 | 1.00E-03 | 39.43 | 45.8 | 6.67 |
| 79.7 | 5.57 | 1.25E-04 | 47.31 | 10.5 | 6.50 |
Iron biomineral morphologies: form follows function
M. ferrooxydans PV-1 has been a model system for biomineralization by an obligate Fe(II)-oxidizer. PV-1 cells form a twisted stalk (Fig. 2A–C), so similar to the one formed by the terrestrial Fe(II)-oxidizer Gallionella ferruginea that it could be mistaken for a Gallionella stalk (Emerson and Moyer 2002). The stalk consists of individual filaments made of nanoparticulate Fe(III) oxyhydroxides and acidic polysaccharides, controlling Fe mineral growth near the cell surface (Chan et al. 2011). Stalk growth was measured to be 2.2 µm length h−1, or nearly 5x the width of a PV-1 cell per hour (Chan et al. 2011). The combination of this directed mineralization and a near-neutral cell surface charge explains how the cell remains remarkably free of encrustation (Saini and Chan 2012). These encrustation avoidance mechanisms are important for any Fe(II)-oxidizing microbe to avoid cell death by Fe mineral growth inside and outside the cell.
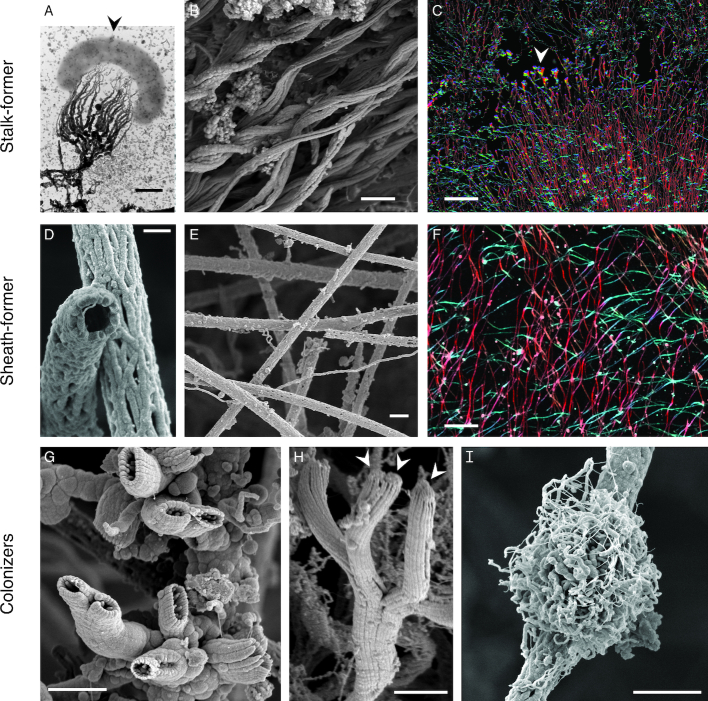
Figure 2.
Morphologies of FeOOH biominerals known or suspected to be formed by the Fe(II)-oxidizing Zetaproteobacteria. (A–C) Twisted stalks from individual cell to intact Loihi Fe microbial mats. (D–F) Sheaths from individual tubes to intact Loihi mats. (G–I) Fe biominerals attached to stalks and sheaths in Loihi mats. (A) A single M. ferrooxydans PV-1 cell (arrow) and its fibrillar stalk. (B) Intact curd-type mats (Fig. 4A) are composed of parallel stalks. (C) Intact mat showing directional stalks that are interrupted (arrow), before biomineral production resumes. See Chan et al. (2016) for details. (D) Hollow sheaths formed by the Zetaproteobacteria. (E) Intact veil-type mats (Fig. 4B) are composed of sheaths. (F) Zoomed out intact mat with multiple sets of sheaths oriented in different directions. (C,F) Color corresponds with filament direction/orientation. (G–H) Short, Y-shaped tubular biominerals formed by Zetaproteobacteria. (H) Arrows show attached cells. (I) Siderocapsa-like nest-type biominerals; it is not known if they are formed by the Zetaproteobacteria. Scale bars: 0.5 µm (A,D), 2 µm (B,E,G–I), 100 µm (C,F). Images reproduced with permission: (A) from Chan et al. (2011), (F, H) from Chan et al. (2016). (B–I) from the samples described in Chan et al. (2016). (D, I) imaged on JEOL-7200 field emission SEM.
While most Zetaproteobacteria isolates form a stalk, some make other biomineral morphologies (Table 1; Fig. 3). Mariprofundus ferrinatatus CP-8 and M. aestuarium CP-5 form shorter filaments that resemble the dreadlock hairstyle (Fig. 3C; Chiu et al. 2017). Dreads were originally observed in terrestrial FeOB Gallionellaceae Ferriphaselus spp., which makes both stalk and dreads (Fig. 3B; Kato et al. 2015b). In both Ferriphaselus and the CP strains, the dreads are shed from cells. This suggests that dreads and similar structures are used specifically to avoid encrustation, whereas the stalk has other functions. PV-1 cells use the stalk as a holdfast to anchor the cell to surfaces (Krepski et al. 2013). As the cell oxidizes Fe(II) and produces new stalk, the cell moves forward, leaving stalk behind. Since the stalk is rigid and anchored, this is a means of motility. Experiments in controlled Fe(II) and O2 gradients showed that PV-1 cells use their stalks to position themselves at an optimum position within that gradient, often forming filaments oriented toward higher O2 (Krepski et al. 2013).
Figure 3.
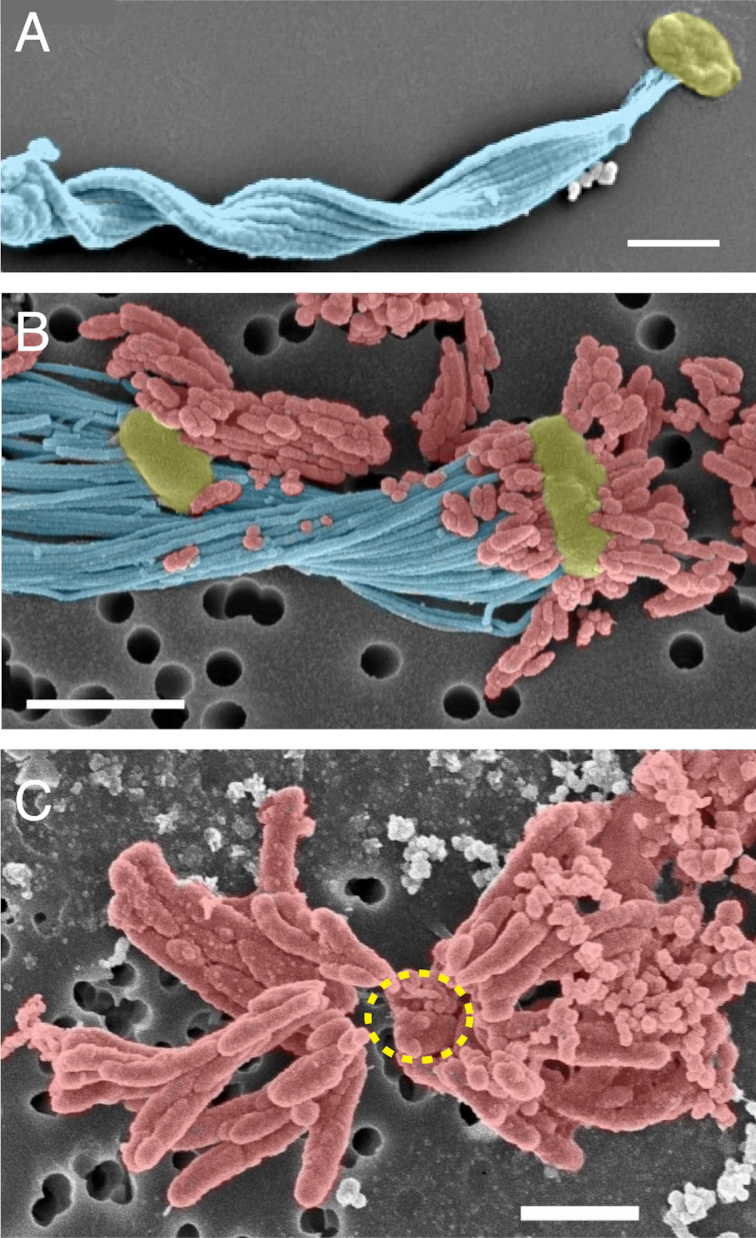
Colorized SEM images of stalk and dread biominerals of selected marine and freshwater FeOB. (A) Twisted stalk (blue) formed by a M. ferrooxydans PV-1 cell (yellow). Most Zetaproteobacteria isolates form stalks. (C) In contrast, M. aestuarium CP-5 and M. ferrinatatus CP-8 produce only short dreads (red), which are easily shed from the cell (inferred cell position indicated by dashed yellow line). (B) Stalk (blue) and dreads (red) of the freshwater Betaproteobacteria FeOB Ferriphaselus sp. R-1 resembled the structures formed by marine FeOB. Scale bars = 1 µm. Images reproduced with permission: (A) from Chan et al. (2016), (B) from Kato et al. (2015b), (C) from Chiu et al. (2017).
In the environment, such oriented filaments are common. At Loihi Seamount, curd-type mats (cohesive Fe mats with a bumpy surface reminiscent of cheese curds) often form directly above a vent orifice (Fig. 4A) (Chan et al. 2016). Micrographs of intact curd mats showed centimeters-long, highly directional twisted stalks forming the mat architecture (Fig. 2A-C). These stalks record the synchronous movements of a community of cells all growing and twisting in the same direction, as well as shifts in directionality in response to changes in the environment (Chan et al. 2016). The mechanism by which these cells actively control their directionality through stalk production is currently unknown.
Figure 4.
Photographs of Zetaproteobacteria habitats. (A–D) Marine hydrothermal vent mats, where Zetaproteobacteria have been found in highest abundance. (A) Curd-type and (B) veil-type Fe mats, from Loihi Seamount. (C) Mn-crusted Fe mat from the Ula Nui site, Loihi. Fe mat visible under broken surface (bottom right). (D) Fe mats on the Golden Horn Chimney, at the Urashima vent site, Mariana Trough. (E) Transition from reduced to Fe(III) (oxyhydr)oxide-stained marine sediments (dashed line) in 26 m below seafloor core from the hydrothermal circulation cell of Iheya North vent field, Okinawa Trough. See Takai et al. (2012) for details. (F) Terrestrial saline CO2-rich spring at Crystal Geyser, UT, USA. (G) Fe(III) (oxyhydr)oxide-coated worm burrows from the beach at Cape Shores, DE, USA. (H) Mild steel corrosion biofilm formed by isolate M. sp. GSB-2. Original photography reproduced with permission: (H) by Joyce M. McBeth, (F) by Chris T. Brown.
Beyond stalks, Loihi Seamount also hosts sheath-rich veil-type mats, which form millimeters-thick Fe mat draped over rock or older Fe mat in diffuse venting environments (Fig. 4B). These mats are created by organisms that form hollow Fe(III) (oxyhydr)oxide sheaths (Fig. 2D–F), similar to those produced by the terrestrial Betaproteobacteria Leptothrix. In the marine environment, however, these sheaths are formed by Zetaproteobacteria (Fleming et al. 2013), informally called zetathrix. From studies based on the terrestrial Leptothrix, sheaths function similarly to stalks, with tens of cells producing a single sheath and leaving it behind as the cells move forward (Chan et al. 2016). In Loihi Seamount intact veil mats, sheaths also leave a record of highly directional growth, despite oxygen profiles of these mats showing a shallow O2 gradient with O2 present throughout the mat (Chan et al. 2016). In both curd- and veil-type mats, Zetaproteobacteria work together to form a highly porous and fluffy mat almost completely composed of biomineral filaments formed by cells, making these Fe mats different from other commonly studied mats or biofilms, which feature cells embedded in an exopolysaccharide matrix. The biomineral filaments forming the structure of the mat also frequently have Fe biominerals attached to them, suggesting FeOB also colonize the mat interior (Fig. S1, Supporting Information). Short branching hollow tubes are formed by the Zetaproteobacteria (Fig. 2G,H) (Emerson et al. 2017) informally called “y-guys.” However, the organisms forming other Fe biominerals have yet to be identified, including nest-like structures reminiscent of freshwater Siderocapsa-like organisms (Fig. 2I) (Emerson, Fleming and McBeth 2010). The range of biomineral morphologies is related to differing biomineral functions, which likely correspond to different geochemical/physical niches within the Fe mat habitat.
Habitats of the Zetaproteobacteria
The Zetaproteobacteria are found in a variety of Fe(II)-rich habitats globally. The detection or observation of Zetaproteobacteria in these habitats is based almost exclusively on the distribution of the 16S rRNA gene. This gene is by far the most frequently used in microbial surveys, making it the best means of comparing Zetaproteobacteria ecology across studies. Isolation, direct observation using fluorescent probes, and metagenomic reconstruction have also been used to identify the Zetaproteobacteria, though in only a few instances, as noted.
Loihi Seamount hydrothermal vents: a Zetaproteobacteria observatory
Most of what we know about Zetaproteobacteria is based on work at the Loihi Seamount, a Hawaiian submarine volcano, from long-term studies including the Iron Microbial Observatory (FeMO). Loihi Seamount is an ideal habitat for Zetaproteobacteria, with hydrothermal fluids rich in CO2 (up to 303 mmol/kg) and Fe(II) (up to 934 µM), and low in sulfide (<50 µM in vent fluids; undetectable in Fe mats) (Karl et al. 1988; Sedwick, McMurtry and Macdougall 1992; Glazer and Rouxel 2009). Background seawater oxygen concentrations are ∼50 µM at the summit of Loihi Seamount, due to its location within the oxygen minimum zone (Glazer and Rouxel 2009). At the base of Loihi Seamount, the Ula Nui site has higher ambient O2 concentrations (145 µM), but lower venting temperatures (1.7°C average compared to ∼42°C average at the summit) (Edwards et al. 2011). Low temperatures and low ambient O2 concentrations favor biotic Fe(II) oxidation by reducing the abiotic rate at and below the mat surface (Millero, Sotolongo and Izaguirre 1987; Emerson et al. 2015). Thus, the conditions at Loihi Seamount have favored the growth of Fe microbial mats ranging from centimeters to meters thick and up to 15 km2 (Fig. 4A–C) (Edwards et al. 2011; Chan et al. 2016). The extensive Fe mats at Loihi Seamount may reflect years- to decades-long stable Fe mat production by the Zetaproteobacteria, based on productivity estimates (Chan et al. 2016; Emerson et al. 2017).
Loihi Seamount studies have provided the cornerstones of Zetaproteobacteria ecology. Since the discovery of Zetaproteobacteria in the 1990s (Moyer, Dobbs and Karl 1995; Emerson and Moyer 2002), five research expeditions from 2006–2013 have focused on Zetaproteobacteria succession, niche and species diversity, and genetic potential. Colonization experiments over 4–10 days showed that Zetaproteobacteria prefer low- to mid-temperature (from 22–60°C, average 40°C) Loihi hydrothermal vents (Rassa et al. 2009). This preference was reflected in longer term observations following the 1996 Loihi eruption, which showed Zetaproteobacteria increasing in abundance as high temperature vents cooled to pre-eruption temperatures and transitioned from sulfide-rich to Fe(II)-rich fluids (Davis et al. 2005; Moyer et al. 2007; Glazer and Rouxel 2009; Emerson and Moyer 2010). The bulk of the omic information on Zetaproteobacteria originates from Loihi Seamount, with the first isolate genome, single cell genomes, metagenome and proteome all from Loihi sources (Singer et al. 2011, 2013; Barco et al. 2015; Field et al. 2015).
Other hydrothermally influenced habitats
Beyond Loihi, Zetaproteobacteria are hosted by many other hydrothermal systems. Extensive Fe mats form around vents at seamounts and island arc systems (Fig. 4D) (Kato et al. 2009a; Emerson and Moyer 2010; Makita et al. 2016; Bortoluzzi et al. 2017; Hager et al. 2017). However, Fe mats have also been found at spreading ridge systems, within diffuse flow at the periphery of high-temperature chimneys and vents (Dekov et al. 2010; Breier et al. 2012; Scott et al. 2015; Vander Roost, Thorseth and Dahle 2017). Most hydrothermal Fe mats consist of biomineral morphologies similar to those at the Loihi Seamount (twisted stalks, sheaths, y-guys, etc.) (Breier et al. 2012; Scott et al. 2015). However, mat textures and lithification can vary as a function of geochemistry (e.g. Mn, Si concentration) and rates of hydrothermal discharge (Li et al. 2012; Johannessen et al. 2017).
Zetaproteobacteria are also found in the marine subsurface. There, oxygenated seawater can mix with anoxic Fe(II)-rich fluids, providing a favorable environment for Fe(II) oxidation. Zetaproteobacteria have been observed by both 16S rRNA gene surveys and metagenomic reconstruction up to 332 meters below the sea floor, within both hydrothermal recharge and cold oxic circulation cells (Fig. 4E) (Yanagawa et al. 2013; Meyer et al. 2016; Tully et al. 2018). In many near surface sediments, shallow mixing introduces O2 into an Fe(II)-rich environment, leading to abundant Zetaproteobacteria populations (Davis et al. 2009; Kato et al. 2009b; Handley et al. 2010; Gonnella et al. 2016). As hydrothermal systems age and cool, basalts and the minerals within inactive sulfide mounds can also serve as Fe(II) sources for Zetaproteobacteria (Sylvan, Toner and Edwards 2012; Kato et al. 2015a; Henri et al. 2016; Barco et al. 2017). These studies show that the Zetaproteobacteria are abundant members of shallow and deep marine subsurface environments, one of the largest underexplored habitats in the oceans.
Coastal and terrestrial habitats
Zetaproteobacteria have only recently been discovered in coastal and terrestrial environments. Colonization experiments showed that Zetaproteobacteria biofilms grow on Fe(II) released from mild and carbon steel that is commonly used in ships and docks, suggesting that these FeOB contribute to corrosion (Fig. 4H) (Dang et al. 2011; McBeth et al. 2011; Lee et al. 2013; McBeth and Emerson 2016; Mumford, Adaktylou and Emerson 2016). [For a review on the role of FeOB in biocorrosion, see Emerson 2019.] Fe(II) can also come from natural sources in coastal environments, originating from mineral weathering and Fe(III) reduction and transported in anoxic groundwater. Fe redox cycling at the oxic-anoxic transition zone of stratified estuaries can support the growth of Zetaproteobacteria, as evidenced by the isolation of M. ferrinatatus CP-8 and M. aestuarium CP-5 (Field et al. 2016; Chiu et al. 2017). In near shore sediments, Fe(II)-rich groundwater can support microbial communities with Zetaproteobacteria at the sediment surface (Rubin-Blum et al. 2014; Laufer et al. 2016; Hassenrück et al. 2016; Otte et al. 2018). Also in these sediments, bioturbation from plant roots and animal burrows provides conduits of O2 to this Fe(II)-rich groundwater. Biotic and abiotic Fe(II) oxidation in these environments leads to the formation of Fe(III) (oxyhydr)oxides, which coat sands, salt grass and mangrove roots and burrows (Fig. 4G) (Taketani et al. 2010; McBeth et al. 2011; McAllister et al. 2015; Beam et al. 2018). Beam et al. (2018) found the abundance of Zetaproteobacteria within Fe(III) oxide-coated worm burrows to be an order of magnitude higher than surrounding bulk sediment, suggesting that Zetaproteobacteria growth and biotic Fe(II) oxidation can be favored in these bioturbated sediments. The Fe(III) (oxyhydr)oxides produced in these environments can sequester toxins that adsorb to the mineral surface (Charette, Sholkovitz and Hansel 2005). Thus, FeOB activity could affect coastal water quality.
Zetaproteobacteria have generally been considered marine FeOB, detected at salinities up to 112 ppt in hypersaline brines (Eder et al. 2001; Guan et al. 2015). Their occurrence in coastal environments provides the opportunity to delineate their minimum salinity requirements. McBeth et al. (2013) surveyed Fe mats along the tidal Sheepscot River, Maine, as it entered the estuary, finding that Zetaproteobacteria only appeared in environments with 5 ppt salinity or higher. This explains why Zetaproteobacteria are not commonly found nor expected to be found in most terrestrial environments.
Thus, it was interesting and novel to find abundant populations of Zetaproteobacteria in CO2-rich terrestrial springs. Surveys of the 16S rRNA genes from carbonic springs at Tierra Amarilla Spring, New Mexico (∼9 ppt salinity) revealed a microbial population up to one third Zetaproteobacteria (Colman et al. 2014). Similarly, 16S rRNA gene and metagenomic work at the CO2-rich Crystal Geyser, Utah, (∼11-14 ppt salinity; Fig. 4F) found the Zetaproteobacteria to be both abundant and consistently present over a year of observation (Emerson et al. 2016; Probst et al. 2017, 2018). These springs represent the first habitat with abundant populations of both Zetaproteobacteria and Betaproteobacteria FeOB (Gallionellaceae), whose abundance is likely driven by cycles of freshwater and saline subsurface groundwater mixing (Probst et al. 2018). The work at Crystal Geyser has produced full-length 16S rRNA gene sequences and the only terrestrial Zetaproteobacteria genomes (Emerson et al. 2016; Probst et al. 2017, 2018). Our analysis of 16S rRNA gene phylogenetic placement and genomic clustering (by average nucleotide identity) suggests that Zetaproteobacteria populations in terrestrial subsurface environments are primarily novel and deeply branching Zetaproteobacteria (see further discussion of habitat selection and niche below).
Common habitat characteristics
In all, Zetaproteobacteria have been found in habitats sharing the following characteristics: 1) brackish to hypersaline water, 2) a supply of Fe(II) and 3) predominantly micro-oxic conditions. These conditions are widespread and found in diverse habitats, likely supplying multiple niches for the diversification and evolution of the Zetaproteobacteria.
Zetaproteobacteria diversity
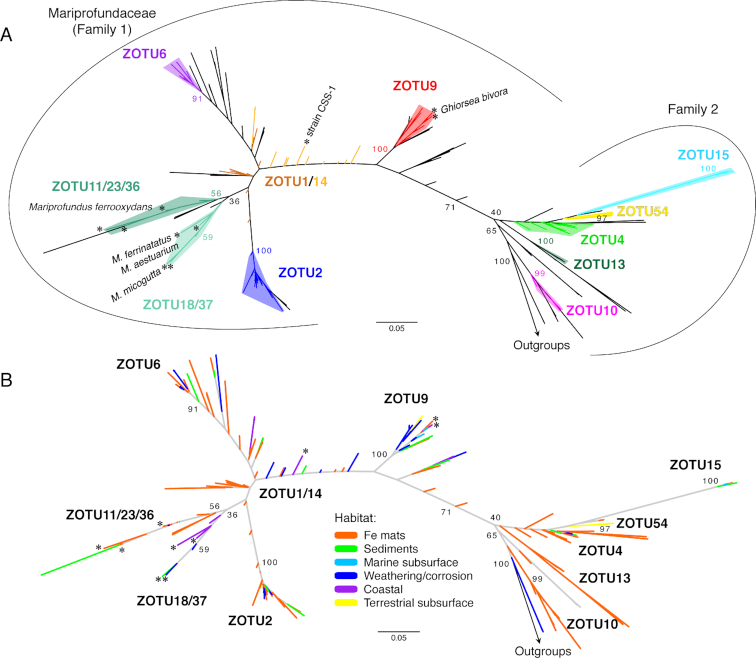
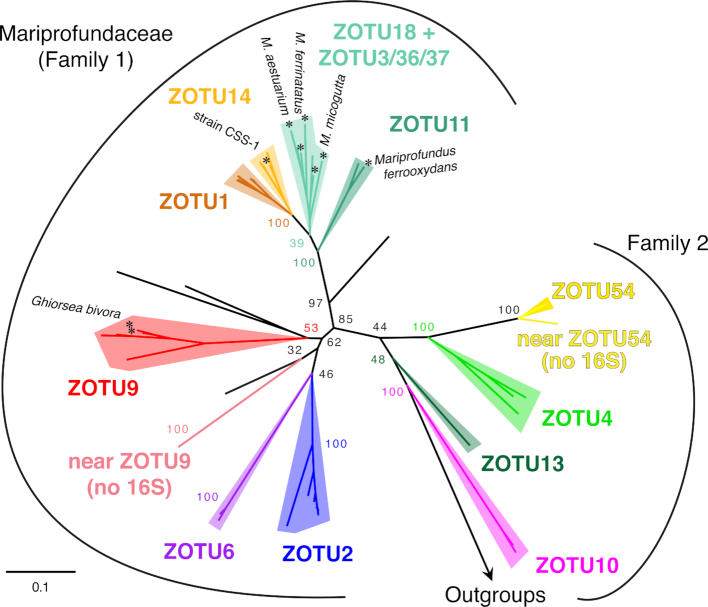
Zetaproteobacteria diversity has been defined using 16S rRNA gene Zetaproteobacteria operational taxonomic units (ZOTU; 97% similarity), based on sequences from isolates and environmental samples (Table 1; Dataset S1, Supporting Information). Since their initial description, the Zetaproteobacteria class has remained a robust taxonomic group within the Proteobacteria (Hug et al. 2016; Parks et al. 2018). A systematic analysis of 227 Zetaproteobacteria full-length 16S rRNA gene sequences yielded 59 ZOTUs (McAllister, Moore and Chan 2018), an increase from 28 ZOTUs in 2011 (McAllister et al. 2011). The majority of these ZOTUs are contained within two families of the Zetaproteobacteria, based on sequence similarity (Fig. 5A, Fig. 6). Fig. 5 shows key ZOTUs, which are frequently sampled and abundant in the environment and are primarily distinct monophyletic taxonomic groups by 16S rRNA gene (see detail in Fig. S2, Supporting Information). These 15 ZOTUs represent 83% of sequences found in the environment. ZOTUs 1 and 14 are the one exception to monophyly by the 16S rRNA gene, yet do form distinct monophyletic groups in a concatenated tree of 12 ribosomal proteins (Fig. 6; see Supplemental Methods). ZOTUs that include isolates represent only 20% of environmental sequences (Table 1), showing that the Zetaproteobacteria are largely uncultivated.
Figure 5.
Maximum likelihood phylogenetic tree showing Zetaproteobacteria 16S rRNA gene diversity, (A) colored by ZOTU and (B) colored by habitat type where the sequences were sampled. A total of 59 ZOTUs have been classified, though only the most frequently sampled are shown in the figure above. ZOTUs 1 and 14 are poorly resolved phylogenetically by the 16S rRNA gene. Published isolates of the Zetaproteobacteria are starred and labeled. Phylogenetic trees were colored automatically using Iroki (Moore et al. 2018). A rectangular version of this tree is represented in Fig. S2 (Supporting Information), which also includes the habitat information for each sequence.
Figure 6.
Maximum likelihood phylogenetic tree showing a more robust phylogenetic placement of ZOTUs based on the concatenated alignments of 12 ribosomal proteins. Data from isolate, SAG and MAG genomes (see Supplemental Methods). In this tree, ZOTUs 1 and 14 are monophyletic entities. Taxa near ZOTU9 and near ZOTU54 are only represented by genomes; these clades have only been found in the terrestrial CO2-rich spring waters of Crystal Geyser, UT.
In order to compare Zetaproteobacteria diversity across studies, we used ZetaHunter, a classification pipeline designed to rapidly and reproducibly classify ZOTUs (McAllister, Moore and Chan 2018). We classified publicly available Zetaproteobacteria full- and partial-length 16S rRNA gene sequences from SILVA (Glöckner et al. 2017) and Integrated Microbial Genomes (Chen et al. 2017), and included data from NCBI SRA (Leinonen, Sugawara and Shumway 2011) as organized by the Integrated Microbial Next Generation Sequencing platform (Lagkouvardos et al. 2016) (total of 1.2 million sequences from 93 studies; summary of samples in Dataset S1; see Supplemental Methods). This work provided the basis for the habitat and diversity analysis below, while also allowing us to correct previous ZOTU assignments (Table S1, Supporting Information).
Connecting Zetaproteobacteria diversity, habitat and niche
Zetaproteobacteria diversity is likely primarily driven by the variety of niches they inhabit. A niche is the set of conditions favorable for growth, which are further influenced, or partitioned, by inter- and intra-species population dynamics in the environment (Holt 2009). A challenge in microbial ecology is to tease apart the niche of an organism through sampling at the appropriate spatial and temporal resolution. For most Zetaproteobacteria environments, we lack the highly resolved chemical and spatial information to describe niches. However, we can look for patterns of associations between different Zetaproteobacteria and their habitats to understand where and the extent to which Zetaproteobacteria niches may overlap.
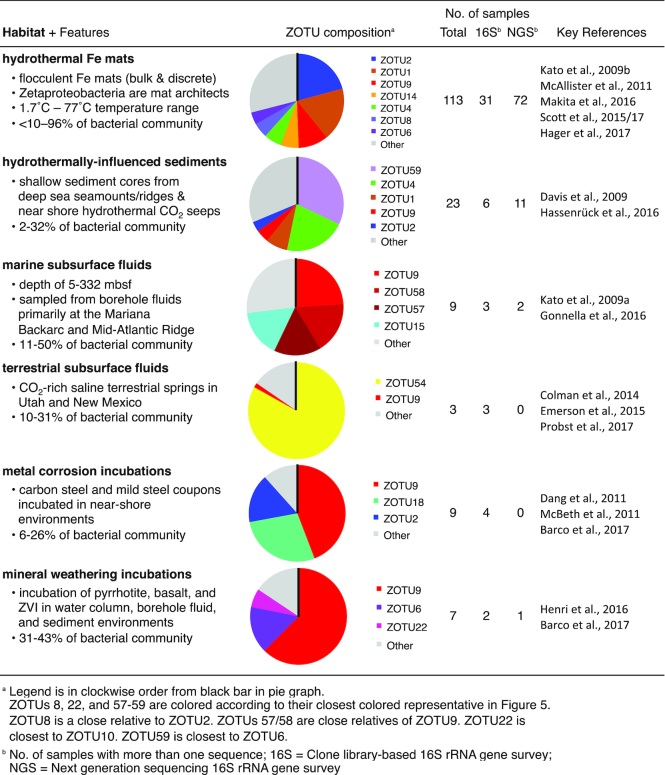
Each habitat displayed distinct and abundant ZOTUs, indicating that habitats can host a specific set of niches that support these ZOTUs (Table 4; Dataset S1, Supporting Information). In particular, dominant ZOTUs differ between habitat types, suggesting that each habitat has a set of dominant niches that favor the growth of those particular ZOTUs. ZOTU54 is a striking example of a dominant ZOTU clearly successful within the terrestrial subsurface fluid environment. ZOTU54 is a deep-branching ZOTU that is primarily limited to this environment (Colman et al. 2014; Emerson et al. 2016; Probst et al. 2017, 2018). The distribution of ZOTU54 suggests that it is endemic or adapted to thrive within terrestrial carbonic Fe(II)-rich springs. However, while it is frequently found at high abundance in terrestrial springs, ZOTU54 is also found in other habitats at very low abundance, including hydrothermal Fe mats (Dataset S1, Supporting Information). In fact, many ZOTUs span habitats (Fig. 5B; Dataset S1, Supporting Information), suggesting that similar niches supporting these ZOTUs can exist in multiple habitats.
Table 4.
Summary of the habitats where the Zetaproteobacteria are found in high abundance using data from Dataset S1 (Supporting Information).
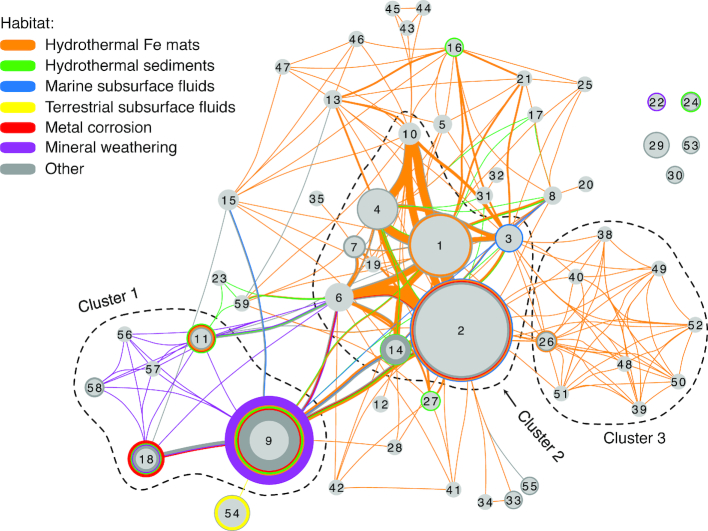
Next, we looked for patterns in ZOTU associations with each other, mapping connections in a ZOTU network (Fig. 7). This network shows which ZOTUs are found in isolation and which co-occur, with connections drawn between ZOTUs if they are found in the same sample. Multiple observations of the same connection result in a thicker line, showing the strength of those connections. Further, the network layout is based on the frequency of co-occurrence, so when two ZOTUs commonly co-occur, they are closer together. Most ZOTUs co-occur with others (Fig. 7), and these connections are not random. Some ZOTUs co-occur more frequently, forming clusters of interconnected ZOTU nodes (Clusters 1–3, Fig. 7). The most abundantly sampled ZOTUs form a central cluster (Cluster 2), sharing a common set of niches most frequently sampled in the hydrothermal Fe mat environment. Cluster 3 centers around ZOTUs found together in hydrothermal Fe mat samples from the Mariana Arc, but which are not common in other environments. Cluster 1 is dominated by ZOTUs from samples associated with metal corrosion and mineral weathering, which suggests that these habitats host niches distinct from those in hydrothermal Fe mat habitats. Overall, these clusters highlight ZOTUs with niches that frequently overlap, suggesting those niches, and thus the growth requirements of these ZOTUs, are compatible.
Figure 7.
Zetaproteobacteria OTU network showing the association of known ZOTUs within the specified habitats. Lines connect ZOTUs that are found in the same sample, with thickness representing the frequency of that association in multiple samples. Colored circles surrounding ZOTU nodes show samples where only a single ZOTU was found. ZOTU nodes are sized according to their environmental abundance. Placement of ZOTUs in the network was determined automatically, based on the frequency of co-occurrence (Cytoscape's edge-weighted, spring embedded layout). Dotted lines denote ZOTU clusters common to the following habitats: Cluster 1, metal corrosion and mineral weathering; Cluster 2, ubiquitous Fe mats; Cluster 3, Mariana Trough Fe mats. Isolate sequences are not shown. Dataset based on SILVA release 128.
A combination of ZOTU environmental distribution, habitat characteristics and isolate physiology can help us better understand a particular ZOTU's niche. Here, we use this approach to describe ZOTU9, as an example. This ZOTU is a key player in marine subsurface fluids, metal corrosion and mineral weathering habitats (see Cluster 1, Fig. 7; Table 4). In mineral weathering habitats, ZOTU9 is frequently the only ZOTU (visualized as a colored circle around the ZOTU node; Fig. 7). For example, ZOTU9 was the only Zetaproteobacteria detected within a basaltic glass weathering enrichment, making up 39% of the bacterial community by 16S rRNA gene sequencing estimates (Henri et al. 2016). These habitat associations suggest that growth of ZOTU9 is favored when metal corrosion or mineral weathering is a source for Fe(II). This association likely relates to these sources producing H2 as well as Fe(II). Metals composed of zero valent Fe produce H2 as a byproduct of anaerobic corrosion (Matheson and Tratnyek 1994). Fe minerals can be a source of H2 because they catalyze hydrolysis and induce radiolysis of water (Bach and Edwards 2003; Dzaugis et al. 2016). The production of H2 is known to benefit some members of ZOTU9, including the Fe(II)- and H2-oxidizing isolates Ghiorsea bivora TAG-1 and SV-108 (Mori et al. 2017). The genetic machinery required for H2 oxidation has also been found in two ZOTU9 single amplified genomes (Field et al. 2015; Scott et al. 2015), which suggests that H2 oxidation may be a feature of other members of ZOTU9, though not necessarily all (since ZOTUs are based on 97% 16S rRNA gene similarity). From these observations, we conclude that both Fe(II) and H2 may play a central role in the niche of ZOTU9 and the habitats where it can be found. By combining isolate physiology with habitat distribution patterns, we can identify key features of a ZOTU's niche.
Spatial and taxonomic resolution in Zetaproteobacteria ecology
Hydrothermal Fe mats have opposing gradients of Fe(II) and O2 and a complex internal structure (e.g. Fig. S1, Supporting Information) (Glazer and Rouxel 2009; Chan et al. 2016). This heterogeneity leads to multiple niches at small spatial scales, suggesting that high-resolution sampling could help us better understand ZOTU niches in this habitat. Initial bulk techniques for sampling collected liters of mat material, and a single sample could contain all major ZOTUs (McAllister et al. 2011). Therefore, new collection devices were engineered to sample small volumes (50–75 mL) at centimeter spatial resolution (Breier et al. 2012). From these discretely sampled Fe mats, we increased the resolution of our ZOTU network (Fig. S3, Supporting Information). Of the 29 ZOTUs found within Fe mat habitats, 17 showed a preference for a specific Fe mat type. However, the high co-occurrence of abundant ZOTUs within a single sample remained, even when considering more highly resolved sampling (Fig. S3, Supporting Information). This result suggests that these ZOTUs share compatible niches at the centimeter scale in Fe mat habitats.
Taxonomic resolution can also affect our understanding of Zetaproteobacteria ecology through the lumping or splitting of ecologically-distinct groups. The ZOTU classification may in certain cases be too coarse, representing multiple related populations that have different niches. For example, Scott et al. (2017) found multiple oligotypes (ecological units defined by informative sequence variability) within each ZOTU. While multiple oligotypes do not necessarily suggest each has a distinct niche, for ZOTU6, only one oligotype differed in abundance over a transect approaching the hydrothermal vent orifice. This abundance change suggested that a subpopulation of ZOTU6 prefers higher flow conditions, warmer temperatures and/or the differing geochemistry found near the vent (Scott, Glazer and Emerson 2017). Results like this warrant a more resolved Zetaproteobacteria taxonomy, which could be aided by whole genome comparisons.
Using genomics to understand metabolic potential and niche
Here, we use Zetaproteobacteria genomes to understand metabolic potential and niche, though interpretations are subject to genome completeness and representation of Zetaproteobacteria diversity. Almost all Zetaproteobacteria isolates have high-quality genomes, greater than 99% complete. However, these isolates represent a small portion of Zetaproteobacteria diversity, requiring genomes from single cells and metagenomes (SAGs and MAGs) to better understand their overall metabolic potential. In most cases, these genomes are much less complete, ranging from <10% to 83% completeness (average 46%) for the SAGs (Field et al. 2015) and from <10% to 100% completeness (average 75%) for the MAGs (Fullerton et al. 2017; Probst et al. 2017). Thus, gene presence is more informative than absence in the SAGs and MAGs.
Carbon fixation
All Zetaproteobacteria isolates are obligate autotrophs, using the Calvin-Benson-Bassham (CBB) cycle to fix carbon. Similarly, all ZOTUs sampled to date have the ribulose-1,5-bisphosphate carboxylase oxygenase gene (RuBisCO; key enzyme in the CBB cycle), suggesting carbon fixation by this pathway is a shared capability across the class. The isolates of Mariprofundus ferrooxydans, strains PV-1, JV-1 and M34, all encode the genes for both Form I (O2-insensitive) and Form II (O2-sensitive) RuBisCO (Singer et al. 2011; Fullerton, Hager and Moyer 2015). Similarly, both forms are encoded by Mariprofundussp. DIS-1, which was specifically isolated to be more aerotolerant (Mumford, Adaktylou and Emerson 2016). However, all other isolates and most Zetaproteobacteria SAG and MAG genomes only encode Form II RuBisCO (Field et al. 2015; Fullerton et al. 2017; Probst et al. 2017), suggesting most Zetaproteobacteria are specifically adapted to lower O2 concentrations.
Energy metabolism: are all Zetaproteobacteria Fe(II)-oxidizers?
The Zetaproteobacteria are often associated with high Fe(II) environments, and all isolates of the Zetaproteobacteria are capable of Fe(II) oxidation. These observations have led to the current assumption that all Zetaproteobacteria are capable of Fe(II) oxidation. To test this assumption, we first have to understand the mechanism of Fe(II) oxidation in the marine environment.
Initial genome analysis of PV-1 led to the proposal of the alternative complex III (ACIII) as part of an iron oxidase complex (Singer et al. 2011). Follow-up studies later changed this model, suggesting ACIII was involved in reverse electron transport (Singer et al. 2013; Barco et al. 2015; Kato et al. 2015b). However, Field et al. (2015) and Chiu et al. (2017) isolated Zetaproteobacteria isolates that lacked ACIII but were still capable of Fe(II) oxidation. Furthermore, only 2 of 23 Zetaproteobacteria SAGs have the ACIII gene, and these 23 SAGs represent the majority of Zetaproteobacteria diversity (Field et al. 2015). Combined, this evidence showed that ACIII is not a critical component of the Fe(II) oxidation pathway.
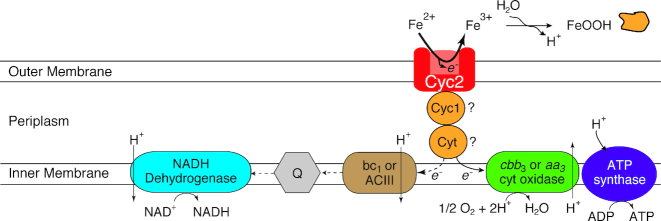
The putative Fe(II) oxidase, Cyc2, and another cytochrome Cyc1 were first identified in Zetaproteobacteria by Barco et al. (2015) through a proteome analysis of PV-1. They were initially identified through comparison of the proteome with the closely related M. ferrooxydans M34 genome. Their presence in the proteome suggested that the cyc1 and cyc2 genes were missing from the PV-1 draft genome due to gaps in the assembly, which was confirmed by resequencing (Barco et al. 2015). The Cyc2 protein from PV-1 is a homolog of the biochemically-characterized Cyc2 Fe(II) oxidase from Acidithiobacillus ferrooxidans (22% amino acid identity) (Castelle et al. 2008). Based on this, the Fe(II) oxidation pathway model for the Zetaproteobacteria was revised (Fig. 8). Cyc2 homologs have been found in other Zetaproteobacteria and other neutrophilic FeOB, strengthening the proposed pathway (He et al. 2017; Chan et al. 2018). In fact, every single ZOTU that has a genomic representative, including ZOTUs without isolates, has a homolog of this putative Fe(II) oxidation gene, consistent with the notion that all Zetaproteobacteria are Fe(II)-oxidizers (Field et al. 2015; Fullerton et al. 2017).
Figure 8.
Model for Fe(II) oxidation in the Zetaproteobacteria modified from Barco et al. (2015). An electron from Fe(II) is passed from Cyc2 to a periplasmic electron carrier (Cyc1 and/or other c-type cytochrome) before being passed to the terminal oxidase (cbb3- or aa3-type cytochrome c oxidases), generating a proton motive force. For reverse electron transport, the electron from the periplasmic carrier is passed to the bc1 complex or alternative complex III (ACIII) before being passed to the quinone pool (Q) where it is used to regenerate NADH.
Genomic clues to niche based on O2 and nitrogen
Genomic evidence suggests that adaptation to differing O2 conditions plays a role in ZOTU niches. Three terminal oxidases potentially used in the putative Fe oxidation pathway have been found within Zetaproteobacteria genomes: cbb3- and aa3-type cytochrome c oxidases and the cytochrome bd-I ubiquinol oxidase. These have different affinities for oxygen, which would influence the niche of each ZOTU; Km’s of 230–300 nM (cbb3, bd-I) to 4.3 µM (aa3) are reported (Bekker et al. 2009; Arai et al. 2014). The cbb3-type terminal oxidase gene is found in most of the Zetaproteobacteria, sometimes in multiple copies, suggesting a predominant preference for very low O2 concentrations (submicromolar) (Field et al. 2015). However, the complete genomes of M. aestuarium and M. ferrinatatus contain only the higher-O2 adapted aa3-type terminal oxidase gene, which helps explain their adaptation to frequently higher O2 concentrations of their tidally-mixed water column habitat (Chiu et al. 2017). Many Zetaproteobacteria genomes have multiple terminal oxidases, suggesting they are adapted to fluctuating oxygen conditions (Field et al. 2015; Fullerton et al. 2017). ZOTU10 and the isolate Mariprofundus sp. DIS-1 may have a higher tolerance for such conditions with increased numbers of genes for O2 radical protection (Field et al. 2015; Mumford, Adaktylou and Emerson 2016).
The genetic potential for nitrogen species transformations differentiates marine and terrestrial Zetaproteobacteria. In the marine environment, most ZOTUs have the potential for assimilatory nitrate reduction to ammonium (nasA, nirBD) (Field et al. 2015; Fullerton et al. 2017). In terrestrial Fe(II)-rich springs such as Crystal Geyser, Zetaproteobacteria genomes lack these genes, but many possess nitrogen fixation genes (e.g. nifH) (Emerson et al. 2016; Probst et al. 2017). In contrast, only three marine isolates (Mariprofundus strains DIS-1, EKF-M39 and M34) and one MAG outside of Mariprofundus possesses nif genes (Field et al. 2015; Mumford, Adaktylou and Emerson 2016; Fullerton et al. 2017), and, as yet, it has not been experimentally shown that these isolates fix N2. Supporting these genomic observations, the nifH gene is rarely detected in marine Fe mats (Jesser et al. 2015). From these patterns, the differences between these Zetaproteobacteria likely correspond with differences in nitrate and ammonium availability in these habitats; nitrate is below detection at Crystal Geyser compared to concentrations up to 32 µM within Loihi Fe mats (Emerson et al. 2016; Sylvan et al. 2017). Nitrogen transformations and O2 tolerance likely play a role in many Zetaproteobacteria niches, though physiological experiments are required for verification. Regardless, there are likely other conditions driving niche diversity yet to be discovered.
Outstanding questions and opportunities
Over the last two decades, Zetaproteobacteria have been established as a diverse, taxonomically-robust class, which thrive in a wide range of Fe(II)-rich habitats. Environmental studies, isolate experiments and genomic analyses have given insight into how they use biomineralization and metabolic strategies to succeed. Building on this work, we are poised to address a number of intriguing questions.
How did the Zetaproteobacteria come to specialize in Fe(II) oxidation?
Thus far, genomic evidence suggests that all Zetaproteobacteria are Fe(II)-oxidizers. If this is true, the Zetaproteobacteria would be an interesting model system in which to explore the selection and evolution of a particular metabolic specialty. The answer to this question likely rests on the complex challenges of Fe(II) oxidation at circumneutral pH. Zetaproteobacteria must position themselves at specific environmental interfaces to gain energy from Fe(II) oxidation. Meanwhile, they must compete with or tolerate abiotic reactions of Fe(II) with O2 and nitrogen compounds, which can form O2 radicals and toxic nitric and nitrous oxides (Winterbourn 1995; Jones et al. 2015). They produce intricate biomineral structures, which allow them to avoid encrustation, control motility and construct mats. Thus, microbial Fe(II) oxidation appears to be a complex physiological trait, which is much more likely to be inherited vertically through descent rather than transmitted horizontally (Martiny, Treseder and Pusch 2013). Since Fe(II) oxidation is a complex trait, this capability was likely acquired by the Zetaproteobacteria prior to their divergence. It is unclear where the Fe(II) oxidation trait originated, but as we determine its genetic basis, phylogenetic comparisons of these genes will allow us to understand FeOB evolutionary relationships.
What are the drivers of Zetaproteobacteria diversification?
The Zetaproteobacteria have diversified into at least 59 operational taxonomic units, which we can now track using ZetaHunter (McAllister, Moore and Chan 2018). Given the increasing number of available genomes, the next logical step is to develop a systematic taxonomy based on both 16S rRNA gene and phylogenomics analysis. Ultimately, diversification is driven by the range in environmental niches. We will improve our understanding as we continue to study environmental distribution, physiology of new isolates, and genomes, especially as we focus our explorations beyond the well-studied hydrothermal vents. We may be able to define niches better via discrete sampling, though there are practical lower limits to sample size and spatial resolution. Although intact samples are challenging to obtain, the effort is worthwhile in order to use imaging-based techniques (e.g. FISH), coupled to high spatial-resolution geochemistry and activity measurements (e.g. elemental mapping, SIP) to discern millimeter- and micron-scale associations. We are just beginning to discover the variety of adaptations across genomes. As genome analyses progress, patterns of functional genes and phylogeny will elucidate the drivers of Zetaproteobacteria diversification. In turn, genomic clues can help us culture novel organisms, which will be key to demonstrating particular biogeochemical roles. The integrated results of these studies will show how these organisms have evolved to occupy particular niches, and how they could work together to influence the geochemistry of Fe(II)-rich habitats.
How do Zetaproteobacteria affect geochemical cycling, and how can we track these effects?
Now that we know the basics of Zetaproteobacteria metabolisms and potential geochemical effects, we can move toward detecting this influence in the environment and determining the controls on those effects. The key will be developing ways to track Zetaproteobacteria activity, and relating this to quantitative effects. There is no clear biotic Fe isotopic signature that can be used to assess the activity of microbially mediated Fe(II) oxidation (Anbar 2004). An alternative is to track activity via gene expression. Traditionally, this would be done via a marker gene for Fe(II) oxidation. The cyc2 gene may work if its expression proves to be specific to Fe(II) oxidation. However, now with (meta)transcriptomic approaches, we can use multiple genes (e.g. the whole Fe(II) oxidation pathway, linked with C fixation and other pathways). With the Zetaproteobacteria, this will be an iterative exercise, as we are still determining/validating the genes involved in Fe(II) oxidation and other metabolisms. This will be most straightforward in Zetaproteobacteria-dominated hydrothermal Fe mat environments, but work in other environments will improve our understanding of the range of their effects on geochemical cycling. As Zetaproteobacteria are widespread in diverse environments, continued work will most likely reveal their broad influence on Fe cycling in marine and saline terrestrial environments.
Supplementary Material
ACKNOWLEDGEMENTS
SMM and CSC drafted the manuscript. All authors contributed to editing. AG, GWL and CSC designed and implemented M. ferrooxydans PV-1 kinetics experiments. SMM and RMM developed ZOTU assignments and networks. SEM images presented in this manuscript were obtained through the work of Deborah Powell at the University of Delaware BioImaging Center. We would like to thank Jarrod Scott and Erin Field for their help on structuring and improving this manuscript.
FUNDING
This work was supported by the National Science Foundation [OCE-1155290 and EAR-1151682 to C.S.C. and OCE-1558738 and MGG OCE-1558712 to G.W.L.], the National Aeronautics and Space Administration [Exobiology NNX12AG20G to C.S.C, D.E., and G.W.L and NNX15AM11G to D.E.], the Office of Naval Research [N00014-17-1-2640 to C.S.C and N00014-17-1-2641 to D.E.], the USDA National Institute of Food and Agriculture [Agriculture and Food Research Initiative grant number 2012-68003-30155 to R.M.M.] and the University of Delaware [Doctoral Dissertation Fellowship to S.M.M.]. Computational infrastructure support by the University of Delaware CBCB Core Facility funded by Delaware INBRE (grant number NIH P20 GM103446) and the Delaware Biotechnology Institute.
Conflicts of interest. None declared.
REFERENCES
- Alt JC. Hydrothermal oxide and nontronite deposits on seamounts in the Eastern Pacific. Mar Geol. 1988;81:227–39. [Google Scholar]
- Alt JC, Lonsdale P, Haymon R et al.. Hydrothermal sulfide and oxide deposits on seamounts near 21°N, East Pacific Rise. Geol Soc Am Bull. 1987;98:157–68. [Google Scholar]
- Anbar AD. Iron stable isotopes: beyond biosignatures. Earth Planet Sci Lett. 2004;217:223–36. [Google Scholar]
- Arai H, Kawakami T, Osamura T et al.. Enzymatic characterization and in vivo function of five terminal oxidases in Pseudomonas aeruginosa. J Bacteriol. 2014;196:4206–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach W, Edwards KJ. Iron and sulfide oxidation within basalt ocean crust: Implications for chemolithoautotrophic microbial biomass production. Geochim Cosmochim Acta. 2003;67:3871–87. [Google Scholar]
- Barco RA, Emerson D, Sylvan JB et al.. New insight into microbial iron oxidation as revealed by the proteomic profile of an obligate iron-oxidizing chemolithoautotroph. Appl Environ Microbiol. 2015;81:5927–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barco RA, Hoffman CL, Ramírez GA et al.. In-situ incubation of iron-sulfur mineral reveals a diverse chemolithoautotrophic community and a new biogeochemical role for Thiomicrospira. Environ Microbiol. 2017;19:1322–37. [DOI] [PubMed] [Google Scholar]
- Beam JP, Scott JJ, McAllister SM et al.. Biological rejuvenation of iron oxides in bioturbated marine sediments. ISME J. 2018;12:1389–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker M, de Vries S, Ter Beek A et al.. Respiration of Escherichia coli can be fully uncoupled via the nonelectrogenic terminal cytochrome bd-II oxidase. J Bacteriol. 2009;191:5510–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortoluzzi G, Romeo T, La Cono V et al.. Ferrous iron- and ammonium-rich diffuse vents support habitat-specific communities in a shallow hydrothermal field off the Basiluzzo Islet (Aeolian Volcanic Archipelago). Geobiology. 2017;15:664–77. [DOI] [PubMed] [Google Scholar]
- Breier JA, Gomez-Ibanez D, Reddington E et al.. A precision multi-sampler for deep-sea hydrothermal microbial mat studies. Deep Sea Res Part I Oceanogr Res Pap. 2012;70:83–90. [Google Scholar]
- Castelle C, Guiral M, Malarte G et al.. A new iron-oxidizing/O2-reducing supercomplex spanning both inner and outer membranes, isolated from the extreme acidophile Acidithiobacillus ferrooxidans. J Biol Chem. 2008;283:25803–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS, Fakra SC, Emerson D et al.. Lithotrophic iron-oxidizing bacteria produce organic stalks to control mineral growth: implications for biosignature formation. ISME J. 2011;5:717–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS, McAllister SM, Garber A et al.. Fe oxidation by a fused cytochrome-porin common to diverse Fe-oxidizing bacteria. bioRxiv. 2018:doi:10.1101/228056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS, McAllister SM, Leavitt AH et al.. The architecture of iron microbial mats reflects the adaptation of chemolithotrophic iron oxidation in freshwater and marine environments. Front Microbiol. 2016;7:796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charette MA, Sholkovitz ER, Hansel CM. Trace element cycling in a subterranean estuary: Part 1. Geochemistry of the permeable sediments. Geochim Cosmochim Acta. 2005;69:2095–109. [Google Scholar]
- Chen I-MA, Markowitz VM, Chu K et al.. IMG/M: Integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 2017;45:D507–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu BK, Kato S, McAllister SM et al.. Novel pelagic iron-oxidizing Zetaproteobacteria from the Chesapeake Bay oxic-anoxic transition zone. Front Microbiol. 2017;8:1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman DR, Garcia JR, Crossey LJ et al.. An analysis of geothermal and carbonic springs in the western United States sustained by deep fluid inputs. Geobiology. 2014;12:83–98. [DOI] [PubMed] [Google Scholar]
- Dang H, Chen R, Wang L et al.. Molecular characterization of putative biocorroding microbiota with a novel niche detection of Epsilon- and Zetaproteobacteria in Pacific Ocean coastal seawaters. Environ Microbiol. 2011;13:3059–74. [DOI] [PubMed] [Google Scholar]
- Davis RE, Moyer CL, McAllister SM et al.. Spatial and temporal variability of microbial communities from pre- and post-eruption microbial mats collected from Loihi Seamount, Hawaii. AGU Fall Meet Abstr. 2005:PS01015. [Google Scholar]
- Davis RE, Stakes DS, Wheat CG et al.. Bacterial variability within an iron-silica-manganese-rich hydrothermal mound located off-axis at the Cleft segment, Juan de Fuca Ridge. Geomicrobiol J. 2009;26:570–80. [Google Scholar]
- Dekov VM, Petersen S, Garbe-Schönberg C-D et al.. Fe–Si-oxyhydroxide deposits at a slow-spreading centre with thickened oceanic crust: the Lilliput hydrothermal field (9°33′S, Mid-Atlantic Ridge). Chem Geol. 2010;278:186–200. [Google Scholar]
- Dhillon A, Teske A, Dillon J et al.. Molecular characterization of sulfate-reducing bacteria in the Guaymas Basin. Appl Environ Microbiol. 2003;69:2765–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druschel GK, Emerson D, Sutka R et al.. Low-oxygen and chemical kinetic constraints on the geochemical niche of neutrophilic iron(II) oxidizing microorganisms. Geochim Cosmochim Acta. 2008;72:3358–70. [Google Scholar]
- Dzaugis ME, Spivack AJ, Dunlea AG et al.. Radiolytic hydrogen production in the subseafloor basaltic aquifer. Front Microbiol. 2016;7:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder W, Jahnke LL, Schmidt M et al.. Microbial diversity of the brine-seawater interface of the Kebrit Deep, Red Sea, studied via 16S rRNA gene sequences and cultivation methods. Appl Environ Microbiol. 2001;67:3077–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KJ, Glazer BT, Rouxel OJ et al.. Ultra-diffuse hydrothermal venting supports Fe-oxidizing bacteria and massive umber deposition at 5000 m off Hawaii. ISME J. 2011;5:1748–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenberg CG. Die infusionsthierchen Als vollkommene organismen. Leipzig: Leopold Voss, 1838. [Google Scholar]
- Emerson D. The role of iron-oxidizing bacteria in biocorrosion: a review. Biofouling, 2019; doi: 10.1080/08927014.2018.1526281 [DOI] [PubMed] [Google Scholar]
- Emerson D, Fleming EJ, McBeth JM. Iron-oxidizing bacteria: an environmental and genomic perspective. Annu Rev Microbiol. 2010;64:561–83. [DOI] [PubMed] [Google Scholar]
- Emerson D, Floyd MM. Enrichment and isolation of iron-oxidizing bacteria at neutral pH. Methods Enzymol. 2005;397:112–23. [DOI] [PubMed] [Google Scholar]
- Emerson D, Moyer CL. Neutrophilic Fe-oxidizing bacteria are abundant at the Loihi Seamount hydrothermal vents and play a major role in Fe oxide deposition. Appl Environ Microbiol. 2002;68:3085–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson D, Moyer CL. Microbiology of seamounts: common patterns observed in community structure. Oceanography. 2010;23:148–63. [Google Scholar]
- Emerson D, Rentz JA, Lilburn TG et al.. A novel lineage of proteobacteria involved in formation of marine Fe-oxidizing microbial mat communities. PLoS One. 2007;2:e667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson D, Scott JJ, Benes J et al.. Microbial iron oxidation in the Arctic tundra and its implications for biogeochemical cycling. Appl Environ Microbiol. 2015;81:8066–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson D, Scott JJ, Leavitt A. et al In situ estimates of iron-oxidation and accretion rates for iron-oxidizing bacterial mats at Lō’ihi Seamount. Deep Res Part I Oceanogr Res Pap. 2017;126:31–9. [Google Scholar]
- Emerson JB, Thomas BC, Alvarez W et al.. Metagenomic analysis of a high carbon dioxide subsurface microbial community populated by chemolithoautotrophs and bacteria and archaea from candidate phyla. Environ Microbiol. 2016;18:1686–703. [DOI] [PubMed] [Google Scholar]
- Field EK, Kato S, Findlay AJ et al.. Planktonic marine iron oxidizers drive iron mineralization under low-oxygen conditions. Geobiology. 2016;14:499–508. [DOI] [PubMed] [Google Scholar]
- Field EK, Sczyrba A, Lyman AE et al.. Genomic insights into the uncultivated marine Zetaproteobacteria at Loihi Seamount. ISME J. 2015;9:857–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming EJ, Davis RE, McAllister SM et al.. Hidden in plain sight: discovery of sheath-forming, iron-oxidizing Zetaproteobacteria at Loihi Seamount, Hawaii, USA. FEMS Microbiol Ecol. 2013;85:116–27. [DOI] [PubMed] [Google Scholar]
- Fullerton H, Hager KW, McAllister SM et al.. Hidden diversity revealed by genome-resolved metagenomics of iron-oxidizing microbial mats from Lō’ihi Seamount, Hawai'i. ISME J. 2017;11:1900–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton H, Hager KW, Moyer CL. Draft genome sequence of Mariprofundus ferrooxydans strain JV-1, isolated from Loihi Seamount, Hawaii. Genome Announc. 2015;3:e01118–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer BT, Rouxel OJ. Redox speciation and distribution within diverse iron-dominated microbial habitats at Loihi Seamount. Geomicrobiol J. 2009;26:606–22. [Google Scholar]
- Glöckner FO, Yilmaz P, Quast C et al.. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J Biotechnol. 2017;261:169–76. [DOI] [PubMed] [Google Scholar]
- Gonnella G, Böhnke S, Indenbirken D et al.. Endemic hydrothermal vent species identified in the open ocean seed bank. Nat Microbiol. 2016;1:16086. [DOI] [PubMed] [Google Scholar]
- Guan Y, Hikmawan T, Ngugi D et al.. Diversity of methanogens and sulfate-reducing bacteria in the interfaces of five deep-sea anoxic brines of the Red Sea. Res Microbiol. 2015;166:688–99. [DOI] [PubMed] [Google Scholar]
- Hager KW, Fullerton H, Butterfield DA et al.. Community structure of lithotrophically-driven hydrothermal microbial mats from the Mariana Arc and Back-Arc. Front Microbiol. 2017;8:1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley KM, Boothman C, Mills RA et al.. Functional diversity of bacteria in a ferruginous hydrothermal sediment. ISME J. 2010;4:1193–205. [DOI] [PubMed] [Google Scholar]
- Harder EC. Iron-deposting bacteria and their geologic relations. US Geol Surv Prof Pap; 1131919:101. [Google Scholar]
- Hassenrück C, Fink A, Lichtschlag A et al.. Quantification of the effects of ocean acidification on sediment microbial communities in the environment: The importance of ecosystem approaches. FEMS Microbiol Ecol. 2016;92:fiw02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Barco RA, Emerson D et al.. Comparative genomic analysis of neutrophilic iron(II) oxidizer genomes for candidate genes in extracellular electron transfer. Front Microbiol. 2017;8:1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henri PA, Rommevaux-Jestin C, Lesongeur F et al.. Structural iron (II) of basaltic glass as an energy source for Zetaproteobacteria in an abyssal plain environment, off the Mid Atlantic Ridge. Front Microbiol. 2016;6:1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt RD. Bringing the Hutchinsonian niche into the 21st century: ecological and evolutionary perspectives. Proc Natl Acad Sci. 2009;106:19659–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug LA, Baker BJ, Anantharaman K et al.. A new view of the tree of life. Nat Microbiol. 2016;1:16048. [DOI] [PubMed] [Google Scholar]
- Jesser KJ, Fullerton H, Hager KW et al.. Quantitative PCR analysis of functional genes in iron-rich microbial mats at an active hydrothermal vent system (Lō’ihi Seamount, Hawai'i). Appl Environ Microbiol. 2015;81:2976–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen KC, Vander Roost J, Dahle H et al.. Environmental controls on biomineralization and Fe-mound formation in a low-temperature hydrothermal system at the Jan Mayen Vent Fields. Geochim Cosmochim Acta. 2017;202:101–23. [Google Scholar]
- Jones LC, Peters B, Lezama Pacheco JS et al.. Stable isotopes and iron oxide mineral products as markers of chemodenitrification. Environ Sci Technol. 2015;49:3444–52. [DOI] [PubMed] [Google Scholar]
- Juniper SK, Fouquet Y. Filamentous iron-silica deposits from modern and ancient hydrothermal sites. Can Mineral. 1988;26:859–69. [Google Scholar]
- Karl DM, Brittain AM, Tilbrook BD. Hydrothermal and microbial processes at Loihi Seamount, a mid-plate hot-spot volcano. Deep Sea Res Part A, Oceanogr Res Pap. 1989;36:1655–73. [Google Scholar]
- Karl DM, McMurtry GM, Malahoff A et al.. Loihi Seamount, Hawaii: a mid-plate volcano with a distinctive hydrothermal system. Nature. 1988;335:532–5. [Google Scholar]
- Kato S, Ikehata K, Shibuya T et al.. Potential for biogeochemical cycling of sulfur, iron and carbon within massive sulfide deposits below the seafloor. Environ Microbiol. 2015a;17:1817–35. [DOI] [PubMed] [Google Scholar]
- Kato S, Kobayashi C, Kakegawa T et al.. Microbial communities in iron-silica-rich microbial mats at deep-sea hydrothermal fields of the Southern Mariana Trough. Environ Microbiol. 2009a;11:2094–111. [DOI] [PubMed] [Google Scholar]
- Kato S, Ohkuma M, Powell DH et al.. Comparative genomic insights into ecophysiology of neutrophilic, microaerophilic iron oxidizing bacteria. Front Microbiol. 2015b;6:1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Yanagawa K, Sunamura M et al.. Abundance of Zetaproteobacteria within crustal fluids in back-arc hydrothermal fields of the Southern Mariana Trough. Environ Microbiol. 2009b;11:3210–22. [DOI] [PubMed] [Google Scholar]
- Krepski ST, Emerson D, Hredzak-Showalter PL et al.. Morphology of biogenic iron oxides records microbial physiology and environmental conditions: toward interpreting iron microfossils. Geobiology. 2013;11:457–71. [DOI] [PubMed] [Google Scholar]
- Kützing FT. Phycologia generalis oder anatomie, physiologie und systemkunde der tange. Leipzig: F. A. Brockhaus, 1843. [Google Scholar]
- Lagkouvardos I, Joseph D, Kapfhammer M et al.. IMNGS: A comprehensive open resource of processed 16S rRNA microbial profiles for ecology and diversity studies. Sci Rep. 2016;6:33721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer K, Nordhoff M, Halama M et al.. Microaerophilic Fe(II)-oxidizing Zetaproteobacteria isolated from low-Fe marine coastal sediments: Physiology and characterization of their twisted stalks. Appl Environ Microbiol. 2017;83:e03118–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer K, Nordhoff M, Schmidt C et al.. Co-existence of microaerophilic, nitrate-reducing, and phototrophic Fe(II)-oxidizers and Fe(III)-reducers in coastal marine sediment. Appl Environ Microbiol. 2016;82:1433–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, McBeth JM, Ray RI et al.. Iron cycling at corroding carbon steel surfaces. Biofouling. 2013;29:1243–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen R, Sugawara H, Shumway M. The sequence read archive. Nucleic Acids Res. 2011;39:D19–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhou H, Peng X et al.. Microbial diversity and biomineralization in low-temperature hydrothermal iron-silica-rich precipitates of the Lau Basin hydrothermal field. FEMS Microbiol Ecol. 2012;81:205–16. [DOI] [PubMed] [Google Scholar]
- Lueder U, Druschel G, Emerson D et al.. Quantitative analysis of O2 and Fe2+ profiles in gradient tubes for cultivation of microaerophilic Iron(II)-oxidizing bacteria. FEMS Microbiol Ecol. 2018;94:fix177. [DOI] [PubMed] [Google Scholar]
- Makita H, Kikuchi S, Mitsunobu S et al.. Comparative analysis of microbial communities in iron-dominated flocculent mats in deep-sea hydrothermal environments. Appl Environ Microbiol. 2016;82:5741–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makita H, Tanaka E, Mitsunobu S. et al Mariprofundus micogutta sp. nov., a novel iron-oxidizing zetaproteobacterium isolated from a deep-sea hydrothermal field at the Bayonnaise knoll of the Izu-Ogasawara arc, and a description of Mariprofundales ord. nov. and Zetaproteobacteria classis. Arch Microbiol. 2017;199:335–46. [DOI] [PubMed] [Google Scholar]
- Martiny AC, Treseder K, Pusch G. Phylogenetic conservatism of functional traits in microorganisms. ISME J. 2013;7:830–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheson LJ, Tratnyek PG. Reductive dehalogenation of chlorinated methanes by iron metal. Environ Sci Technol. 1994;28:2045–53. [DOI] [PubMed] [Google Scholar]
- McAllister SM, Barnett JM, Heiss JW et al.. Dynamic hydrologic and biogeochemical processes drive microbially enhanced iron and sulfur cycling within the intertidal mixing zone of a beach aquifer. Limnol Oceanogr. 2015;60:329–45. [Google Scholar]
- McAllister SM, Davis RE, McBeth JM et al.. Biodiversity and emerging biogeography of the neutrophilic iron-oxidizing Zetaproteobacteria. Appl Environ Microbiol. 2011;77:5445–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister SM, Moore RM, Chan CS. ZetaHunter, a reproducible taxonomic classification tool for tracking the ecology of the Zetaproteobacteria and other poorly resolved taxa. Microbiol Resour Announc. 2018;7:e00932–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister SM, Moore RM, Gartman A et al.. Marine Fe-oxidizing Zetaproteobacteria: Historical, ecological, and genomic perspectives. bioRxiv. 2018:doi:10.1101/416842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBeth JM, Emerson D. In situ microbial community succession on mild steel in estuarine and marine environments: Exploring the role of iron-oxidizing bacteria. Front Microbiol. 2016;7:767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBeth JM, Fleming EJ, Emerson D. The transition from freshwater to marine iron-oxidizing bacterial lineages along a salinity gradient on the Sheepscot River, Maine, USA. Environ Microbiol Rep. 2013;5:453–63. [DOI] [PubMed] [Google Scholar]
- McBeth JM, Little BJ, Ray RI et al.. Neutrophilic iron-oxidizing “Zetaproteobacteria” and mild steel corrosion in nearshore marine environments. Appl Environ Microbiol. 2011;77:1405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JL, Jaekel U, Tully BJ et al.. A distinct and active bacterial community in cold oxygenated fluids circulating beneath the western flank of the Mid-Atlantic ridge. Sci Rep. 2016;6: 22541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millero FJ, Sotolongo S, Izaguirre M. The oxidation kinetics of Fe(II) in seawater. Geochim Cosmochim Acta. 1987;51:793–801. [Google Scholar]
- Moore RM, Harrison AO, McAllister SM et al.. Iroki: automatic customization and visualization of phylogenetic trees. bioRxiv. 2018:doi:10.1101/106138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori JF, Scott JJ, Hager KW et al.. Physiological and ecological implications of an iron- or hydrogen-oxidizing member of the Zetaproteobacteria, Ghiorsea bivora, gen. nov., sp. nov. ISME J. 2017;11:2624–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer CL, Davis RE, Curtis AC et al.. Spatial and temporal variability in microbial communities from pre- and post-eruption microbial mats collected from Loihi Seamount, Hawaii: an update. American Geophysical Union Abstr. 2007;B23G–02., adsabs.harvard.edu/abs/2007AGUFM.B23G..02M. [Google Scholar]
- Moyer CL, Dobbs FC, Karl DM. Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1995;61:1555–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumford AC, Adaktylou IJ, Emerson D. Peeking under the iron curtain: development of a microcosm for imaging the colonization of steel surfaces by Mariprofundus sp. strain DIS-1, an oxygen-tolerant Fe-oxidizing bacterium. Appl Environ Microbiol. 2016;82:6799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otte JM, Harter J, Laufer K et al.. The distribution of active iron-cycling bacteria in marine and freshwater sediments is decoupled from geochemical gradients. Environ Microbiol. 2018;20:2483–99. [DOI] [PubMed] [Google Scholar]
- Parks DH, Chuvochina M, Waite DW et al.. A proposal for a standardized bacterial taxonomy based on genome phylogeny. Nat Biotechnol. 2018;36:996–1004. [DOI] [PubMed] [Google Scholar]
- Probst AJ, Castelle CJ, Singh A et al.. Genomic resolution of a cold subsurface aquifer community provides metabolic insights for novel microbes adapted to high CO2 concentrations. Environ Microbiol. 2017;19:459–74. [DOI] [PubMed] [Google Scholar]
- Probst AJ, Ladd B, Jarett JK et al.. Differential depth distribution of microbial function and putative symbionts through sediment-hosted aquifers in the deep terrestrial subsurface. Nat Microbiol. 2018;3:328–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassa AC, McAllister SM, Safran SA. et al Zeta-Proteobacteria dominate the colonization and formation of microbial mats in low-temperature hydrothermal vents at Loihi Seamount, Hawaii. Geomicrobiol J. 2009;26:623–38. [Google Scholar]
- Vander Roost J, Thorseth IH, Dahle H. Microbial analysis of Zetaproteobacteria and co-colonizers of iron mats in the Troll Wall Vent Field, Arctic Mid-Ocean Ridge. PLoS One. 2017;12:e0185008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin-Blum M, Antler G, Tsadok R et al.. First evidence for the presence of iron oxidizing Zetaproteobacteria at the Levantine continental margins. PLoS One. 2014;9:e91456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini G, Chan CS. Near-neutral surface charge and hydrophilicity prevent mineral encrustation of Fe-oxidizing micro-organisms. Geobiology. 2012;11:191–200. [DOI] [PubMed] [Google Scholar]
- Scott JJ, Breier JA, Luther GW III et al.. Microbial iron mats at the Mid-Atlantic Ridge and evidence that Zetaproteobacteria may be restricted to iron-oxidizing marine systems. PLoS One. 2015;10:e0119284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JJ, Glazer BT, Emerson D. Bringing microbial diversity into focus: high-resolution analysis of iron mats from the Lō’ihi Seamount. Environ Microbiol. 2017;19:301–16. [DOI] [PubMed] [Google Scholar]
- Sedwick P, McMurtry GM, Macdougall J. Chemistry of hydrothermal solutions from Pele's vents, Loihi Seamount, Hawaii. Geochim Cosmochim Acta. 1992;56:3643–67. [Google Scholar]
- Singer E, Emerson D, Webb EA. et al Mariprofundus ferrooxydans, PV-1 the first genome of a marine Fe(II) oxidizing Zetaproteobacterium. PLoS One. 2011;6:e25386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer E, Heidelberg JF, Dhillon A et al.. Metagenomic insights into the dominant Fe(II) oxidizing Zetaproteobacteria from an iron mat at Lō’ihi, Hawai'i. Front Microbiol. 2013;4: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumm W, Lee GF. Oxygenation of ferrous iron. Ind Eng Chem. 1961;53:143–146. [Google Scholar]
- Sylvan JB, Toner BM, Edwards KJ. Life and death of deep-sea vents: Bacterial diversity and ecosystem succession on inactive hydrothermal sulfides. MBio. 2012;3:e00279–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvan JB, Wankel SD, LaRowe DE et al.. Evidence for microbial mediation of subseafloor nitrogen redox processes at Loihi Seamount, Hawaii. Geochim Cosmochim Acta. 2017;198: 131–50. [Google Scholar]
- Takai K, Mottl MJ, Nielsen SHH et al.. IODP expedition 331: Strong and expansive subseafloor hydrothermal activities in the Okinawa trough. Sci Drill. 2012;13:19–27. [Google Scholar]
- Taketani RG, Franco NO, Rosado AS et al.. Microbial community response to a simulated hydrocarbon spill in mangrove sediments. J Microbiol. 2010;48:7–15. [DOI] [PubMed] [Google Scholar]
- Tully BJ, Wheat CG, Glazer BT et al.. A dynamic microbial community with high functional redundancy inhabits the cold, oxic subseafloor aquifer. ISME J. 2018;12:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winogradsky S. Ueber Eisenbakterien. Bot Zeitung. 1888;46:261–270. [Google Scholar]
- Winterbourn CC. Toxicity of iron and hydrogen peroxide: the Fenton reaction. Toxicol Lett. 1995;82/83:969–74. [DOI] [PubMed] [Google Scholar]
- Yanagawa K, Nunoura T, McAllister SM et al.. The first microbiological contamination assessment by deep-sea drilling and coring by the D/V Chikyu at the Iheya North hydrothermal field in the Mid-Okinawa Trough (IODP Expedition 331). Front Microbiol. 2013;4:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.