Abstract
Lenvatinib is a substrate of cytochrome P450 (CYP) 3A and ATP-binding cassette (ABC) transporters. In this study, we aimed to evaluate how CYP3A4/5 and ABC transporter polymorphisms affected the mean steady-state dose-adjusted plasma trough concentrations (C0) of lenvatinib in a cohort of 40 Japanese patients with thyroid cancer. CYP3A4 20230G > A (*1G), CYP3A5 6986A > G (*3), ABCB1 1236C > T, ABCB1 2677G > T/A, ABCB1 3435C > T, ABCC2 −24C > T, and ABCG2 421C > A genotypes were determined using polymerase chain reaction-restriction fragment length polymorphism. In univariate analysis, there were no significant differences in the mean dose-adjusted C0 values of lenvatinib between the ABCB1, ABCG2, and CYP3A5 genotypes. However, the mean dose-adjusted C0 values of lenvatinib in patients with the CYP3A4*1/*1 genotype and ABCC2 −24T allele were significantly higher than those in patients with the CYP3A4*1G allele and −24C/C genotype, respectively (P = 0.018 and 0.036, respectively). In multivariate analysis, CYP3A4 genotype and total bilirubin were independent factors influencing the dose-adjusted C0 of lenvatinib (P = 0.010 and 0.046, respectively). No significant differences were found in the incidence rates of hypertension, proteinuria, and hand-foot syndrome following treatment with lenvatinib between the genotypes of CYP3A4/5 and ABC transporters. Lenvatinib pharmacokinetics were significantly influenced by the CYP3A4*1G polymorphism. If the target plasma concentration of lenvatinib for efficacy or toxicity is determined, elucidation of the details of the CYP3A4*1G genotype may facilitate decision-making related to the appropriate initial lenvatinib dosage to achieve optimal plasma concentrations.
Introduction
Lenvatinib is an oral tyrosine kinase inhibitor (TKI) that inhibits vascular endothelial growth factor receptor (VEGFR) 1–3, fibroblast growth factor receptor (FGFR) 1–4, platelet-derived growth factor receptor α (PDGFRα), stem cell factor receptor (KIT), and rearranged during transfection (RET)1–4.
Lenvatinib is primarily metabolized by cytochrome P450 (CYP) 3A4 and acts as a substrate for ATP-binding cassette (ABC) transporters, such as P-glycoprotein and breast cancer resistance protein (BCRP), which are encoded by the ABCB1 and ABCG2 genes, respectively5–8. P-glycoprotein and BCRP are expressed in the small intestine, liver, kidney, and blood-brain barrier and are associated with drug disposition, functioning to regulate the absorption and elimination of substrate drugs9. Co-administration of the potent CYP3A, P-glycoprotein, and BCRP inhibitor ketoconazole has been reported to increase the maximum plasma concentration of lenvatinib compared with placebo; however, the elimination half-life of lenvatinib was not altered5. This result suggested that CYP3A, P-glycoprotein, and BCRP in the small intestine contribute to the pharmacokinetics of lenvatinib. Therefore, the pharmacokinetics of lenvatinib could be affected by polymorphisms in the CYP3A4, ABCB1, and ABCG2 genes. Furthermore, CYP3A4 and CYP3A5 generally have similar catalytic specificities, although the activity of CYP3A5 is less than that of CYP3A410,11. Polymorphic CYP3A5 expression in the small intestine and liver is strongly correlated with a single nucleotide polymorphism (SNP), 6986A > G, within intron 3 of CYP3A5, which is designated CYP3A5*312. Therefore, lenvatinib pharmacokinetics may be influenced by CYP3A5 polymorphisms. However, the effects of CYP3A4/5, ABCB1, and ABCG2 polymorphisms on the plasma concentrations of lenvatinib have not been evaluated to date.
In addition, in humans, three ABC subfamilies, B, C, and G, contain efflux transporters for a large number of drugs. Among these ABC subfamilies, ABCB1, ABCG2, and ABCC2 (multidrug resistance-associated protein [MRP] 2) are important for the drug transporters. To date, however, in both in vitro and in vivo studies, the influence of MRP2 on membrane transport of lenvatinib has not been reported. In addition to P-glycoprotein and BCRP, MRP2 may also play a role in lenvatinib pharmacokinetics. In addition, no studies have reported the effects of ABCC2 polymorphisms on lenvatinib plasma concentrations. Therefore, in the present study, we investigated the effects of polymorphisms in CYP3A4 20230G > A, CYP3A5 6986A > G, ABCB1 1236C > T, ABCB1 2677G > T/A, ABCB1 3435C > T, ABCG2 421C > A, and ABCC2 −24C > T on the mean steady-state dose-adjusted plasma trough concentration (C0) of lenvatinib in 40 patients with thyroid cancer.
Methods
Patients and protocols
Forty Japanese patients with thyroid cancer (27 women and 13 men) taking lenvatinib (LENVIMA; Eisai Co., Ltd., Tokyo, Japan) were consecutively enrolled in this study. All patients were treated at Ito Hospital between June 2015 and April 2018. The eligibility criteria were as follows: adequate bone marrow function (neutrophil count of 1500/μL or more, platelet count of 100,000/μL or more, and haemoglobin level of 8.0 g/dL or more), renal function (serum creatinine levels less than 2.0 mg/dL), and hepatic function (total bilirubin level less than 2.0 mg/dL, aspartate transferase and alanine transferase levels less than or equal to 2.5 times the upper limit of the normal range). Table 1 lists the clinical characteristics of patients prior to lenvatinib therapy. Patients were excluded if they had taken drugs that affected CYP3A, P-glycoprotein, or BCRP function. Lenvatinib was administered as medical care for patients with thyroid cancer according to a Drug Interview Form8. The study was approved by the Ethics Committee of Ito Hospital and Akita University School of Medicine and was carried out according to the recommendations outlined in the 1964 Declaration of Helsinki and its later amendments. All patients who participated in this study provided written informed consent prior to enrolment.
Table 1.
Clinical characteristics of patients receiving lenvatinib.
| Characteristics | |
|---|---|
| Total number | 40 |
| Female:Male | 27:13 |
| Age (years) | 63.0 ± 10.9 (33–78) |
| Body weight (kg) | 56.0 ± 11.0 (35–82) |
| Laboratory test values at lenvatinib initiation | |
| Aspartate transaminase (IU/L) | 25.9 ± 8.9 (13–56) |
| Alanine transaminase (IU/L) | 22.3 ± 9.1 (11–45) |
| Serum albumin (g/dL) | 3.9 ± 0.5 (2.7–5.1) |
| Total bilirubin (mg/dL) | 0.6 ± 0.2 (0.4–1.5) |
| Serum creatinine (mg/dL) | 0.76 ± 0.29 (0.41–1.78) |
| ABCB1 1236C > T | C/C: C/T: T/T = 8: 22: 10 |
| ABCB1 2677G > T/A | G/G: G/T: G/A: T/T: T/A: A/A = 9: 15: 4: 4: 5: 3 |
| ABCB1 3435C > T | C/C: C/T: T/T = 14: 21: 5 |
| ABCG2 421C > A | C/C: C/A: A/A = 20: 18: 2 |
| ABCC2 −24C > T | C/C: C/T: T/T = 24: 14: 2 |
| CYP3A4 20230 G > A(*1G) | *1/*1: *1/*1G: *1G/*1G = 24: 15: 1 |
| CYP3A5 6986 A > G(*3) | *1/*1: *1/*3: *3/*3 = 1: 13: 26 |
Data are presented as number or mean ± standard deviation (range).
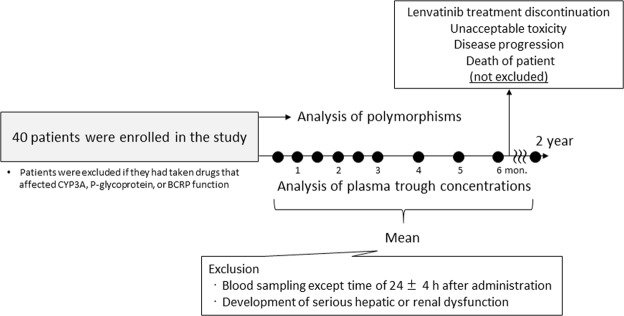
Oral lenvatinib administration was carried out at dosages of 4–24 mg once daily. The daily dose of lenvatinib was reduced as needed based on the grade of each side effect, as determined by the Common Terminology Criteria for Adverse Events version 4.0. Intake of foods or drugs known to alter CYP3A, P-glycoprotein, or BCRP function was not permitted during the treatment period (withdrawal criteria). The plasma concentrations of lenvatinib were monitored 24 ± 4 h after administration, and trough plasma concentrations (C0) were evaluated every 2 weeks for the first 3 months after initiation of lenvatinib therapy and then every 1 month thereafter. Assays of lenvatinib C0 were performed monthly until lenvatinib treatment discontinuation (unacceptable toxicity, disease progression, or death of patient) or for a maximum of 2 years (maximum 27 sampling points). Blood samples except the sampling time of 24 ± 4 h after administration and the steady-state was excluded from analysis. In addition, plasma samples from patients who developed serious hepatic or renal dysfunction after initiation of lenvatinib therapy were excluded from analysis because of the influence on lenvatinib pharmacokinetics (Fig. 1). Plasma was isolated by centrifugation at 1,900 × g for 15 min and was stored at −40 °C until analysis.
Figure 1.
Study profile.
Pharmacokinetics analysis
Plasma concentrations of lenvatinib were measured by high-performance liquid chromatography (HPLC), as has been used previously for quantitative analysis of gefitinib plasma concentrations13. Analysis of the calibration curve for lenvatinib in human plasma showed that the curve was linear for the concentration range from 5 to 1000 ng/mL. Moreover, for this assay, the limit of quantification (LOQ) of lenvatinib was 5 ng/mL. For the concentration range from 5 to 1000 ng/mL, the coefficients of variation (CVs) were lower than 12.6%, and the accuracies of the intra- and interday assays were within 10.6%.
Pharmacogenomics analysis
Extraction of DNA from peripheral blood samples was carried out using a QIAamp Blood Mini Kit (Qiagen, Tokyo, Japan). Genotypes CYP3A4 20230G > A (*1G) (rs2242480), within intron 10 of the human CYP3A4 gene, and CYP3A5 6986A > G (*3) (rs776746) were identified using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP)14. Genotyping procedures identifying the C and T alleles in exon 12 (1236C > T, rs1128503), the G and T/A alleles in exon 21 (2677G > T/A, rs2032582), and the C and T alleles in exon 26 (3435C > T, rs1045642) of the ABCB1 gene were performed using PCR-RFLP as previously reported15–17. PCR-RFLP was used for genotyping of the ABCG2 421C > A (rs2231142) polymorphism, as described by Kobayashi et al.18. Genotyping procedures identifying the C and T alleles in exon 1 (−24C > T, rs717620) of the ABCC2 gene were carried out using PCR-RFLP, as described by Naesens et al.19. All frequencies for the different analysed loci were at Hardy-Weinberg equilibrium.
Statistical analyses
The Kolmogorov-Smirnov test was applied to assess the distribution in each data set. The results used the average of dose-adjusted C0 values of lenvatinib during therapy for one patient. Dose-adjusted C0 values for each genotype were presented as medians (minimum–maximum). To elucidate differences between these groups, we applied Kruskal-Wallis tests or Mann-Whitney U tests. Correlations in continuous variables between groups were determined using Spearman’s rank correlation coefficient test, with results presented as correlation coefficients (r). The effects of factors in univariate analysis were evaluated using stepwise multiple linear regression analysis. For each patient, dummy variables were used to replace the genotypes of CYP3A4/5 and ABC transporters (1 and 0 in 2 groups; 1 and 0, 0 and 0, and 0 and 1 in 3 groups). Chi-square tests were used to examine differences in categorical data. For post-hoc power analysis, the effect size calculated in comparison with the lenvatinib dose-adjusted C0 between CYP3A4 or ABCC2 genotypes showed significant differences. An effect size of greater than 0.5 was considered clinically meaningful20. Power was calculated using G*Power ver. 3.1 software. Results with p values of less than 0.05 were considered significant, and SPSS 20.0 for Windows (SPSS IBM Japan Inc., Tokyo, Japan) was used for all statistical analyses.
Results
The genotype frequencies for the polymorphisms in ABC transporters and CYP3A4/5 in 40 Japanese patients with thyroid cancer are shown in Table 1. The median (range) CV values of the lenvatinib C0 at the steady state for one patient (intrapatient) for the same daily dose during lenvatinib therapy were 20.8% (8.9–54.4%) at 24 mg, 15.3% (5.7–63.8%) at 20 mg, 25.6% (3.4–66.2%) at 14 mg, 24.2% (4.8–72.9%) at 10 mg, 32.8% (16.3–50.9%) at 8 mg, and 23.5% (7.1–39.8%) at 4 mg. Therefore, the mean lenvatinib plasma concentration during lenvatinib therapy was used. The median (range) duration of lenvatinib therapy was 394 (64–735) days.
There were no significant differences in the mean dose-adjusted C0 of lenvatinib between genotypes for the ABCB1, ABCG2, and CYP3A5 polymorphisms, although the dose-adjusted C0 of lenvatinib in patients with the ABCG2 421A allele or CYP3A5*3/*3 tended to be higher than those with other corresponding genotypes (Table 2). However, the mean dose-adjusted C0 of lenvatinib in patients with the CYP3A4*1/*1 genotype was significantly higher than that in patients with the CYP3A4*1G allele (P = 0.018, effect size = 0.863; Table 2). In addition, the mean dose-adjusted C0 of lenvatinib in patients with the ABCC2 −24T allele was significantly higher than that in patients with −24C/C genotypes (P = 0.036, effect size = 0.605; Table 2). There were also significant correlations with total bilirubin at lenvatinib initiation and the mean dose-adjusted C0 of lenvatinib at the steady state (P = 0.037; Table 2). The dose-adjusted C0 of lenvatinib in patients with both the CYP3A4*1/*1 and ABCC2 −24T alleles (median 6.70 ng/mL/mg) was about 1.5-fold higher than that in patients with both the CYP3A4*1G/*1G and ABCC2 −24C/C genotypes (median 4.42 ng/mL/mg; P = 0.007; Fig. 2).
Table 2.
Comparison of dose-adjusted mean trough concentrations of lenvatinib between genotypes of ABC transporters and CYP3A4/5.
| Genotype | n | Dose-adjusted C0 (ng/mL/mg) | P value | |
|---|---|---|---|---|
| Median | Minimum - Maximum | |||
| ABCB1 1236C > T | ||||
| C/C | 8 | 4.37 | (1.67–10.4) | 0.303a |
| C/T | 22 | 5.87 | (1.40–12.6) | |
| T/T | 10 | 5.85 | (3.24–11.0) | |
| ABCB1 2677G > T/A | ||||
| G/G | 9 | 5.08 | (3.09–12.6) | 0.755a |
| G/T + G/A | 19 | 5.67 | (1.40–11.0) | |
| T/T + T/A + A/A | 12 | 5.62 | (2.06–11.0) | |
| ABCB1 3435C > T | ||||
| C/C | 14 | 4.90 | (1.40–12.6) | 0.724a |
| C/T | 21 | 5.52 | (2.06–11.0) | |
| T/T | 5 | 6.02 | (4.24–7.46) | |
| ABCG2 421C > A | ||||
| C/C | 20 | 4.73 | (1.40–12.6) | 0.063b |
| C/A + A/A | 20 | 6.34 | (1.67–10.4) | |
| ABCC2 −24C > T | ||||
| C/C | 24 | 5.03 | (1.40–12.6) | 0.036b |
| C/T + T/T | 16 | 6.57 | (4.06–11.0) | |
| CYP3A4 20230G > A(*1G) | ||||
| *1/*1 | 24 | 6.34 | (1.40–12.6) | 0.018b |
| *1/*1 G + *1 G/*1G | 16 | 4.77 | (1.67–7.52) | |
| CYP3A5 6986 A > G(*3) | ||||
| *1/*1 + *1/*3 | 14 | 4.77 | (1.67–10.4) | 0.063b |
| *3/*3 | 26 | 6.22 | (1.40–12.6) | |
| Sex | ||||
| Female | 27 | 5.67 | (1.67–12.6) | 0.252b |
| Male | 13 | 5.08 | (1.40–10.4) | |
| Correlation coefficient (r) | ||||
| Age (years) | 0.061 | 0.817 | ||
| Body weight (kg) | −0.220 | 0.414 | ||
| Laboratory test values at lenvatinib initiation | ||||
| Aspartate transaminase | 0.273 | 0.098 | ||
| Alanine transaminase | 0.151 | 0.388 | ||
| Serum albumin | 0.189 | 0.249 | ||
| Total bilirubin | 0.336 | 0.037 | ||
| Serum creatinine | −0.151 | 0.357 | ||
aKruskal Wallis, bMann-Whitney.
The dose-adjusted C0 of lenvatinib for each patient was calculated using the average of several C0 values at the steady-state.
Figure 2.
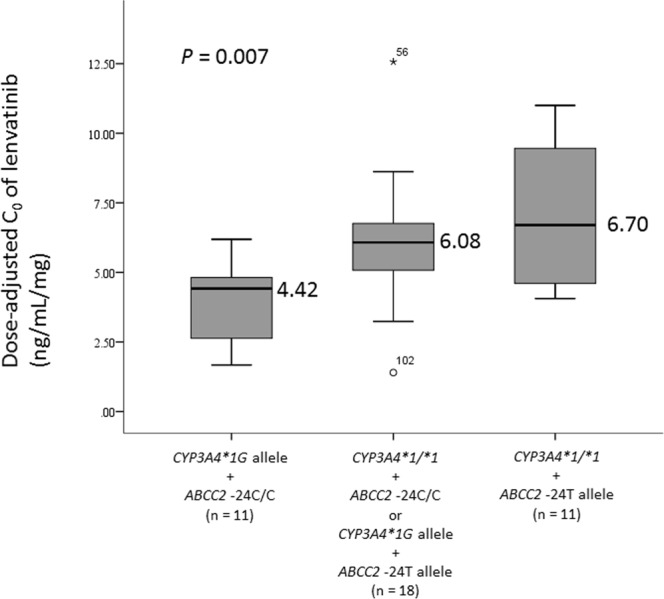
Effects of CYP3A4*1G and ABCC2 −24C > T polymorphisms on the steady-state mean plasma trough concentrations of lenvatinib. Graphical analysis was performed using a box and whiskers plot. The box spanned data between 2 quartiles (IQR), with the median represented as a bold horizontal line. The ends of the whiskers (vertical lines) represent the smallest and largest values that are not outliers. Outliers (circle) are values between 1.5 and 3 IQRs from the end of the box. Values more than three IQRs from the end of the box are defined as extremes (stars).
Stepwise selection multiple linear regression analysis of explanatory variables for the dose-adjusted C0 values of lenvatinib is shown in Table 3. The CYP3A4 genotype (CYP3A4*1/*1) and total bilirubin were independent factors influencing the dose-adjusted C0 of lenvatinib (P = 0.010 and 0.046, respectively); however, the determination coefficient for the dose-adjusted C0 of lenvatinib was 0.226.
Table 3.
Stepwise multiple regression analysis of explanatory variables for the dose-adjusted C0 of lenvatinibTable 3. Stepwise multiple regression analysis of explanatory variables for the dose-adjusted C0 of lenvatinib.
| Explanatory variable | Slope | SE | SRC | P- value | R 2 |
|---|---|---|---|---|---|
| 0.226 | |||||
| CYP3A4 genotype (*1G allele = 1) | −2.05 | 0.75 | −0.397 | 0.010 | |
| Total bilirubin (mg/dL) | 3.67 | 1.77 | 0.300 | 0.046 | |
| Intercept = | 6.39 | 1.66 | |||
C0, predose concentration.
SE, standard error; SRC, standardized regression coefficient.
No significant differences were found in the incidence rates of hypertension, proteinuria, and hand-foot syndrome following treatment with lenvatinib between the genotypes of CYP3A4/5 and ABC transporters (Table 4).
Table 4.
The frequencies of lenvatinib-induced adverse events between genotypes of ABC transporters and CYP3A4/5.
| Adverse events | Hypertension | P value | Proteinuria | P value | Hand-foot syndrome | P value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTCAE grade | 0 | 1 | 2 | 3 | 0 | 1 | 2 | 3 | 0 | 1 | 2 | |||
| ABCB1 1236C>T | ||||||||||||||
| C/C | 0 | 4 | 3 | 1 | 0.140 | 3 | 2 | 3 | 0 | 0.599 | 0 | 6 | 2 | 0.431 |
| C/T | 0 | 3 | 16 | 3 | 11 | 3 | 6 | 2 | 0 | 14 | 8 | |||
| T/T | 0 | 1 | 6 | 3 | 6 | 3 | 1 | 0 | 1 | 7 | 2 | |||
| ABCB1 2677G>T/A | ||||||||||||||
| G/G | 0 | 0 | 8 | 1 | 0.211 | 3 | 2 | 4 | 0 | 0.387 | 0 | 7 | 2 | 0.702 |
| G/T + G/A | 0 | 4 | 10 | 5 | 11 | 4 | 2 | 2 | 1 | 11 | 7 | |||
| T/T + T/A + A/A | 0 | 4 | 7 | 1 | 6 | 2 | 4 | 0 | 0 | 9 | 3 | |||
| ABCB1 3435C>T | ||||||||||||||
| C/C | 0 | 3 | 10 | 1 | 0.798 | 4 | 3 | 7 | 0 | 0.093 | 0 | 13 | 1 | 0.150 |
| C/T | 0 | 4 | 12 | 5 | 13 | 3 | 3 | 2 | 1 | 11 | 9 | |||
| T/T | 0 | 1 | 3 | 1 | 3 | 2 | 0 | 0 | 0 | 3 | 2 | |||
| ABCG2 421C>A | ||||||||||||||
| C/C | 0 | 5 | 11 | 4 | 0.606 | 11 | 4 | 4 | 1 | 0.896 | 0 | 14 | 6 | 0.595 |
| C/A + A/A | 0 | 3 | 14 | 3 | 9 | 4 | 6 | 1 | 1 | 13 | 6 | |||
| ABCC2 −24C>T | ||||||||||||||
| C/C | 0 | 3 | 17 | 4 | 0.305 | 11 | 5 | 7 | 1 | 0.866 | 1 | 15 | 8 | 0.574 |
| C/T + T/T | 0 | 5 | 8 | 3 | 9 | 3 | 3 | 1 | 0 | 12 | 4 | |||
| CYP3A4 20230G>A( *1G ) | ||||||||||||||
| *1/*1 | 0 | 6 | 14 | 4 | 0.625 | 14 | 3 | 6 | 1 | 0.457 | 1 | 14 | 9 | 0.281 |
| *1/*1G + *1G/*1G | 0 | 2 | 11 | 3 | 6 | 5 | 4 | 1 | 0 | 13 | 3 | |||
| CYP3A5 6986A>G(*3) | ||||||||||||||
| *1/*1 + *1/*3 | 0 | 3 | 9 | 2 | 0.923 | 5 | 4 | 4 | 1 | 0.577 | 0 | 12 | 2 | 0.185 |
| *3/*3 | 0 | 5 | 16 | 5 | 15 | 4 | 6 | 1 | 1 | 15 | 10 | |||
Data are presented as patients numbers.
Discussion
To the best of our knowledge, this is the first study to report the effects of CYP3A4/5 and ABC transporter polymorphisms on lenvatinib pharmacokinetics in patients with thyroid cancer. Our findings showed that lenvatinib pharmacokinetics were significantly influenced by the CYP3A4 20230G > A polymorphism; however, the ABCB1, ABCG2, and CYP3A5 polymorphisms had no effect. Notably, univariate analysis showed that the dose-adjusted C0 of lenvatinib was significantly higher in patients having the ABCC2 −24T allele than in patients having the −24C/C genotype (P = 0.036). Accordingly, in patients with both the CYP3A4*1G/*1G and ABCC2 −24C/C genotypes, the dose-adjusted C0 of lenvatinib was significantly lower than that in patients with other genotypes. However, the appearances of hypertension, proteinuria, and hand-foot syndrome following lenvatinib treatment appeared to be unrelated to these genetic polymorphisms.
Eighty-eight percent of cases of the CYP3A4*1G polymorphism 20230G > A have been reported to be linked to CYP3A5*121. Notably, in a previous study of patients harboring the CYP3A4*1G allele, metabolic activity for the substrate of CYP3A4 was increased; that is, patients harboring the CYP3A4*1G allele showed significantly lower dose-adjusted C0 values of tacrolimus than patients harboring the CYP3A4*1/*1 genotype14. As previously demonstrated21, patients with the CYP3A4*1G allele exhibited significantly lower dose-adjusted C0 values for lenvatinib than patients having the CYP3A4*1/*1 genotype. However, the dose-adjusted C0 values of lenvatinib did not differ significantly between the 2 CYP3A5 genotypes. Twelve percent of CYP3A4*1G polymorphisms that are not linked to CYP3A5*1 seem to show increased correlations between CYP3A4*1G polymorphisms and lenvatinib pharmacokinetics.
In an in vitro study, the ABCC2 −24T variant was reported to be associated with decreased transport activity of MRP222. Similarly, in the present study, the dose-adjusted C0 of lenvatinib was also significantly increased in patients having the ABCC2 −24T allele compared with that in patients having the −24C/C genotype. Therefore, ABCC2 polymorphism may also contribute to interindividual differences in lenvatinib pharmacokinetics. However, the SNP in position 421C > A of ABCG2 is the most prevalent allele in Japanese individuals (33%) and exhibits functional importance23. The level and function of ABCG2 protein expressed from the 421A allele are reduced compared with those of the 421C/C protein18. To date, the plasma concentrations of other TKIs, particularly imatinib, in patients with the ABCG2 421A allele have been reported to be higher than those in patients with the 421C/C genotype24,25. Notably, our current findings demonstrated that in patients having the ABCG2 421A allele, the dose-adjusted C0 of lenvatinib tended to be higher than in patients having the 421C/C genotype, although this difference was not significant. Furthermore, each polymorphism in the human ABCB1 gene (1236C > T, 2677G > T/A, and 3435C > T) has been associated with altered expression and function of P-glycoprotein in various human tissues15–17. The dose-adjusted C0 of lenvatinib in patients with the ABCB1 1236 T/T or 3435 T/T genotype also tended to be higher than that in patients with the corresponding 1236C/C or 3435C/C genotype. Accordingly, further studies with larger sample sizes are needed; however, in addition to P-glycoprotein and BCRP, MRP2 may also contribute to the absorption and disposition of lenvatinib. MRP2 is localized at the apical membrane of enterocytes in the intestine and plays a key role in limiting the absorption of substrate drugs incorporated orally. In the liver, MRP2 localizes to the canalicular membrane of hepatocytes and mediates the efflux of substrate drugs into the bile26.
Our current results showed that patients having both the CYP3A4*1/*1 and ABCC2 −24T alleles administered the same daily dose of lenvatinib exhibited lenvatinib C0 values about 1.5-fold higher than those in patients having both the CYP3A4*1G/*1G and ABCC2 −24C/C genotypes. Higher lenvatinib plasma concentrations may be associated with higher rates of some adverse events. In lenvatinib therapy, evaluation of lenvatinib plasma concentrations may be necessary to provide individual treatment through dose adjustment because of improved efficacy or to avoid adverse events. Identification of therapeutic target plasma concentrations indicating the relationships of drug exposure-efficacy or toxicity will then be required. When target plasma concentrations of lenvatinib are elucidated, the present results may be useful clinically. Specifically, elucidation of information regarding the primary CYP3A4 genotype and minor ABCC2 genotype prior to initiation of lenvatinib therapy may facilitate decision-making related to the appropriate daily dosage for obtaining optimal lenvatinib plasma exposure. Further studies are necessary in the future to examine this possibility.
Our study had several limitations. First, the number of patients treated with lenvatinib therapy was only 40, which may have hindered the precise determination of lenvatinib pharmacogenomics. Second, because some polymorphisms were of low frequency, the associations observed may have been accidental. Therefore, further studies with larger patient cohorts are needed to confirm our results. However, we investigated the validity of the sample size from the results obtained from the study and found that the effect sizes in comparison with the lenvatinib dose-adjusted C0 values between CYP3A4 or ABCC2 genotypes were 0.863 and 0.605, respectively, supporting the clinical significance of the results.
Conclusion
Overall, our results demonstrated that the mean steady-state dose-adjusted C0 of lenvatinib was significantly affected by CYP3A4 polymorphism but not ABCB1, ABCG2, or CYP3A5 polymorphisms. In addition to P-glycoprotein and BCRP, MRP2 may also contribute to the absorption and disposition of lenvatinib. If the target plasma concentration of lenvatinib for efficacy or toxicity is determined, elucidation of details of the CYP3A4*1G genotype may facilitate decision-making related to the appropriate initial lenvatinib dosage to achieve optimal plasma concentrations.
Acknowledgements
This work was supported by a grant (no. 17K08408) from the Japan Society for the Promotion of Science, Tokyo, Japan.
Author Contributions
M.N., A.S., K.S. and M.M. participated in the design of the study and reviewed the results. M.N., A.S., K.S. and K.I. were responsible for the patient collection and involved in acquisition of data. T.O. and K.F. carried out genotyping. M.M. analysed plasma concentrations. T.O., M.N. and M.M. were responsible for the statistical analysis. T.O., M.N. and M.M. drafted the manuscript. K.F., A.S., K.S. and K.I. helped to draft the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Matsui J, et al. E7080, a novel inhibitor that targets multiple kinases, has potent antitumor activities against stem cell factor producing human small cell lung cancer H146, based on angiogenesis inhibition. Int J Cancer. 2008;122:664–71. doi: 10.1002/ijc.23131. [DOI] [PubMed] [Google Scholar]
- 2.Okamoto K, et al. Antitumor activities of the targeted multi-tyrosine kinase inhibitor lenvatinib (E7080) against RET gene fusion-driven tumor models. Cancer Lett. 2013;340:97–103. doi: 10.1016/j.canlet.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Yamamoto Y, et al. Lenvatinib, an angiogenesis inhibitor targeting VEGFR/FGFR, shows broad antitumor activity in human tumor xenograft models associated with microvessel density and pericyte coverage. Vasc Cell. 2014;6:18. doi: 10.1186/2045-824X-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tohyama O, et al. Antitumor activity of lenvatinib (e7080): an angiogenesis inhibitor that targets multiple receptor tyrosine kinases in preclinical human thyroid cancer models. J Thyroid Res. 2014;2014:638747. doi: 10.1155/2014/638747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shumaker R, et al. Effects of ketoconazole on the pharmacokinetics of lenvatinib (E7080) in healthy participants. Clin Pharmacol Drug Dev. 2015;4:155–60. doi: 10.1002/cpdd.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta A, et al. Population pharmacokinetic analysis of lenvatinib in healthy subjects and patients with cancer. Br J Clin Pharmacol. 2016;81:1124–33. doi: 10.1111/bcp.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shumaker RC, et al. Effect of rifampicin on the pharmacokinetics of lenvatinib in healthy adults. Clin Drug Investig. 2014;34:651–9. doi: 10.1007/s40261-014-0217-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LENVIMA (Lenvatinib) Drug Interview Form (Package insert), Eisai Co., Ltd., Tokyo, Japan (2015).
- 9.Lin JH. Transporter-mediated drug interactions: clinical implications and in vitro assessment. Expert Opin Drug Metab Toxicol. 2007;3:81–92. doi: 10.1517/17425255.3.1.81. [DOI] [PubMed] [Google Scholar]
- 10.Lin YS, et al. Co-regulation of CYP3A4 and CYP3A5 and contribution to hepatic and intestinal midazolam metabolism. Mol Pharmacol. 2002;62:162–72. doi: 10.1124/mol.62.1.162. [DOI] [PubMed] [Google Scholar]
- 11.Williams JA, et al. Comparative metabolic capabilities of CYP3A4, CYP3A5, and CYP3A7. Drug Metab Dispos. 2002;30:883–891. doi: 10.1124/dmd.30.8.883. [DOI] [PubMed] [Google Scholar]
- 12.Hustert E, et al. The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics. 2001;11:773–9. doi: 10.1097/00008571-200112000-00005. [DOI] [PubMed] [Google Scholar]
- 13.Miura M, et al. A limited sampling strategy for estimation of the area under the plasma concentration-time curve of gefitinib. Ther Drug Monit. 2014;36:24–29. doi: 10.1097/FTD.0b013e31829dabbc. [DOI] [PubMed] [Google Scholar]
- 14.Fukushima-Uesaka H, et al. Haplotypes of CYP3A4 and their close linkage with CYP3A5 haplotypes in a Japanese population. Hum Mutat. 2004;23:100. doi: 10.1002/humu.9210. [DOI] [PubMed] [Google Scholar]
- 15.Wu L, et al. MDR1 gene polymorphisms and risk of recurrence in patients with hepatocellular carcinoma after liver transplantation. J Surg Oncol. 2007;96:62–8. doi: 10.1002/jso.20774. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka H, et al. Acute myelogenous leukemia with PIG-A gene mutation evolved from aplastic anemia-paroxysmal nocturnal hemoglobinuria syndrome. Int J Hematol. 2001;73:206–12. doi: 10.1007/BF02981939. [DOI] [PubMed] [Google Scholar]
- 17.Cascorbi I, et al. Frequency of single nucleotide polymorphisms in the P-glycoprotein drug transporter MDR1 gene in white subjects. Clin Pharmacol Ther. 2001;69:169–74. doi: 10.1067/mcp.2001.114164. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi D, et al. Functional assessment of ABCG2 (BCRP) gene polymorphisms to protein expression in human placenta. Drug Metab Dispos. 2005;33:94–101. doi: 10.1124/dmd.104.001628. [DOI] [PubMed] [Google Scholar]
- 19.Naesens M, et al. Multidrug resistance protein 2 genetic polymorphisms influence mycophenolic acid exposure in renal allograft recipients. Transplantation. 2006;82:1074–84. doi: 10.1097/01.tp.0000235533.29300.e7. [DOI] [PubMed] [Google Scholar]
- 20.Cohen J. A power primer. Psychol Bull. 1992;112:155–9. doi: 10.1037/0033-2909.112.1.155. [DOI] [PubMed] [Google Scholar]
- 21.Miura M, et al. Impact of the CYP3A4*1G polymorphism and its combination with CYP3A5 genotypes on tacrolimus pharmacokinetics in renal transplant patients. Pharmacogenomics. 2011;12:977–984. doi: 10.2217/pgs.11.33. [DOI] [PubMed] [Google Scholar]
- 22.Laechelt S, et al. Impact of ABCC2 haplotypes on transcriptional and posttranscriptional gene regulation and function. Pharmacogenomics J. 2011;11:25–34. doi: 10.1038/tpj.2010.20. [DOI] [PubMed] [Google Scholar]
- 23.Imai Y, et al. C421A polymorphism in the human breast cancer resistance protein gene is associated with low expression of Q141K protein and low-level drug resistance. Mol Cancer Ther. 2002;1:611–6. [PubMed] [Google Scholar]
- 24.Takahashi N, et al. Influence of CYP3A5 and drug transporter polymorphisms on imatinib trough concentration and clinical response among patients with chronic phase chronic myeloid leukemia. J Hum Genet. 2010;55:731–7. doi: 10.1038/jhg.2010.98. [DOI] [PubMed] [Google Scholar]
- 25.Miura M. Therapeutic drug monitoring of imatinib, nilotinib, and dasatinib for patients with chronic myeloid leukemia. Biol Pharm Bull. 2015;38:645–54. doi: 10.1248/bpb.b15-00103. [DOI] [PubMed] [Google Scholar]
- 26.Tocchetti GN, et al. Modulation of expression and activity of intestinal multidrug resistance-associated protein 2 by xenobiotics. Toxicol Appl Pharmacol. 2016;303:45–57. doi: 10.1016/j.taap.2016.05.002. [DOI] [PubMed] [Google Scholar]