Abstract
Alcohol consumption has been established to be a major factor in the development and progress of cancer. Genetic polymorphisms of alcohol-metabolism genes result in differences between individuals in exposure to acetaldehyde, leading to possible carcinogenic effects. Arg47His (rs1229984 G > A) in ADH1B have been frequently studied for its potential effect on carcinogenesis. However, the findings are as yet inconclusive. To gain a more precise estimate of this potential association, we conducted a meta-analysis including 66 studies from 64 articles with 31999 cases and 50964 controls. The pooled results indicated that ADH1B Arg47His polymorphism is significantly associated with the decreased risk of overall cancer (homozygous model, odds ratio (OR) = 0.62, 95% confidence interval (CI) = 0.49–0.77; heterozygous model, OR = 0.71, 95% CI = 0.60–0.84; recessive model, OR = 0.83, 95% CI = 0.76–0.91; dominant model, OR = 0.62, 95% CI = 0.53–0.72; and allele comparison, OR = 0.82, 95% CI = 0.75–0.89). Stratified analysis by cancer type and ethnicity showed that a decreased risk was associated with esophageal cancer and head and neck cancer amongst Asians. In conclusion, our meta-analysis suggested that ADH1B Arg47His polymorphism was significantly associated with decreased overall cancer risk. These findings need further validation in large multicenter investigations.
Keywords: ADH1B, cancer, meta-analysis, polymorphism, risk
Introduction
Cancer is a major public health problem worldwide. According to GLOBOCAN worldwide estimates, an estimated 14.1 million new cancer cases and 8.2 million cancer-related deaths occurred in 2012 [1]. In addition, the incidence of cancer is predicted to reach 25 million worldwide by 2032 [2]. This growing cancer burden is expected as populations expand and age. Meanwhile, certain lifestyles, such as alcohol consumption, are likely to further boost the burden [1–3].
Alcohol consumption is the third-largest risk factor for global health burden [4]. Approximately 3.3 million deaths, almost 5.9% of total deaths worldwide in 2012, were attributable to alcohol consumption [5]. As early as 2002, approximately 3.6% of all cancers and 3.5% of all cancer deaths were reported due to alcohol consumption [3]. It is well established that alcohol is first catalytically oxidized to acetaldehyde, mainly by alcohol dehydrogenases (ADH), and then to harmless acetate by aldehyde dehydrogenases (ALDH) [6,7]. Acetaldehyde may stimulate carcinogenesis by disrupting DNA synthesis and repair, inhibiting DNA methylation, and by interacting with retinoid metabolism [8,9]. Genetic polymorphisms of alcohol-metabolism genes result in differences between individuals in exposure to acetaldehyde, leading to possible carcinogenic effects [10]. Amongst them, Arg47His (rs1229984 G > A) in ADH1B have been frequently studied for its potential effect on the carcinogenesis. Compared with the Arg/Arg individuals, the His/His individuals have a 40-fold higher enzyme activity oxidized alcohol to toxic acetaldehyde [7].
Epidemiologic studies have extensively explored the association between ADH1B Arg47His polymorphism and cancer risk. However, the findings are as yet inconclusive. Several meta-analyses published before 2016 associated this polymorphism only with esophageal, head and neck, gastric, colorectal, and upper aerodigestive tract cancer [11–16]. However, no meta-analyses have ever investigated the association between ADH1B Arg47His polymorphism and overall cancer risk, including other types of cancer. In addition, several more studies with larger sample size were published since 2016 [17–24]. Therefore, we performed an updated meta-analysis including the most recent and relevant studies to clarify the association between ADH1B Arg47His polymorphism and the overall cancer risk, involving 66 studies with 31999 cases and 50964 controls [17–80].
Methods
Identification of relevant studies
A systematic literature search was conducted in the following electronic databases: Medline and Embase database up to 1 July 2018. The following search terms were used: ‘ADH1B or ADH2’ or ‘polymorphism or variant’ or ‘cancer or carcinoma or tumor’. In addition, reviews and references lists of eligible studies were manually searched to identify additional relevant articles.
Inclusion and exclusion criteria
The eligible articles must meet the following criteria. The inclusion criteria were as follows: (i) studies evaluating the association between ADH1B Arg47His polymorphism and overall cancer risk; (ii) case–control studies; (iii) studies with sufficient information to calculate the odds ratio (OR) and its 95% confidence interval (CI). The major exclusion criteria were as follows: (i) no control group; (ii) duplicate publication; (iii) reviews, meta-analyses, conference reports, or editorial articles; (iv) no available data.
Data extraction
Investigators independently extracted the relevant information from all eligible studies according to the inclusion and exclusion criteria listed above. A final consensus was achieved regarding each selected study. The following information was extracted from each study: first author’s surname, publication year, country, ethnicity, cancer type, control source, genotyping method, number of cases and controls with different genotypes, and Hardy–Weinberg equilibrium (HWE) of genotypes in controls.
Statistical analysis
The strength of the association between ADH1B Arg47His polymorphism and overall cancer risk was evaluated by calculating ORs and 95% CIs. The pooled ORs were also estimated using homozygous model (His/His vs. Arg/Arg), heterozygous model (Arg/His vs. Arg/Arg), recessive model [His/His vs. (Arg/His + Arg/Arg)], dominant model [(Arg/His + His/His) vs. Arg/Arg], as well as allele comparison (His vs. Arg). Stratification analyses were further conducted according to ethnicity, cancer type, control source, and HWE. Chi square-based Q-test was applied to assess between-study heterogeneity. If no heterogeneity (P>0.10) was found, the fixed-effect model (Mantel–Haenszel method) was performed [81]. Otherwise, the random-effect model (DerSimonian and Laird method) was used [82]. Sensitivity analysis was carried out to assess the stability of the results, and potential publication bias was assessed with Begg’s funnel plot and Egger’s linear regression test [83]. All the statistical analyses were calculated using STATA software (version 11.0, Stata Corporation, College Station, TX). A P-value less than 0.05 was considered statistically significant.
Results
Study characteristics
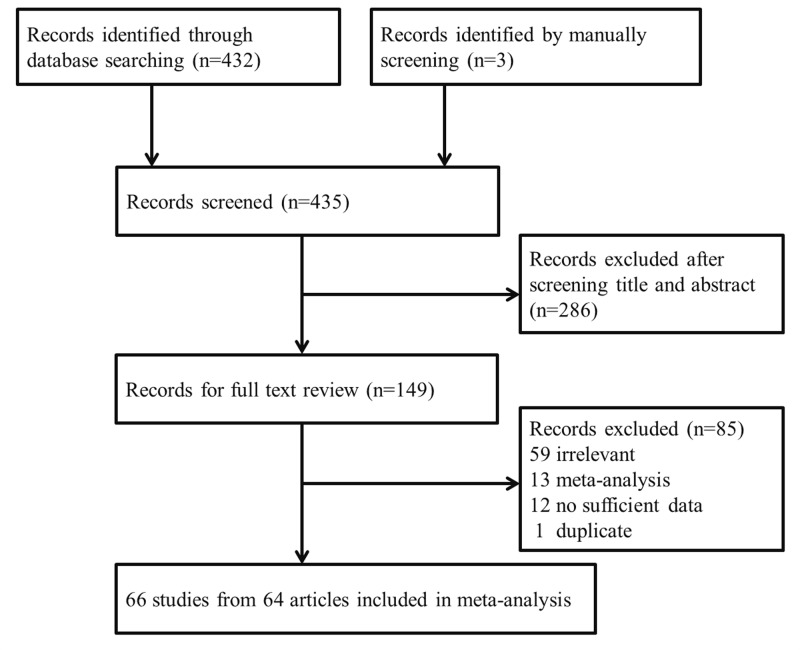
As listed in Figure 1, a total of 432 potential records were initially identified from Medline and Embase using the search terms listed above. After a screening of the titles and abstracts, 146 publications were subjected for further evaluation. Of them, 59 articles were excluded for irrelevant information, 13 for only meta-analysis, 12 for no sufficient data, and 1 was excluded for duplicate study. In addition, three studies were manually identified from reviews and references lists of the eligible studies. Ultimately, 64 articles investigating the association between ADH1B Arg47His polymorphism and cancer risk were included in the final meta-analysis [17–80].
Figure 1. Flow chart of studies included in our meta-analysis.
Overall, 66 studies from 64 articles with 31999 cases and 50964 controls were finally included in our meta-analysis. As shown in Table 1, there were 48 studies conducted amongst Asians, 15 amongst Caucasians, and 3 amongst mixed ethnic group. With respect to cancer type, 23 studies addressed esophageal cancer, 16 head and neck cancer, 10 colorectal cancer, 6 gastric cancer, 4 hepatocellular, 3 upper aerodigestive tract cancer, 2 pancreatic and 1 bladder and breast cancer. Regarding control source, 34 studies were hospital-based and 32 studies were population-based. With respect to HWE, 52 met HWE, 5 departed from HWE, and 9 had not enough information.
Table 1. Main characteristics of included studies in our meta-analysis.
| Author | Year | Country | Ethnicity | Cancer type | Control source | Genotyping method | Number of cases | Number of controls | HWE | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | GA | AA | GG | GA | AA | ||||||||
| Zhong | 2016 | China | Asian | Colorectal | HB | PCR-RFLP | 85 | 125 | 64 | 152 | 172 | 34 | Yes |
| Masaoka | 2016 | Japan | Asian | Bladder | HB | TaqMan | 3 | 38 | 33 | 27 | 265 | 448 | Yes |
| Liu | 2016 | China | Asian | Hepatocellular | HB | Affymetrix | 48 | 262 | 283 | 236 | 1229 | 1748 | Yes |
| Kagemoto | 2016 | Japan | Asian | Esophageal | PB | Multiplex PCR | 31 | 36 | 50 | 60 | 389 | 676 | Yes |
| Chen | 2016 | China | Asian | Gastric | HB | PCR-RFLP | 83 | 117 | 46 | 104 | 125 | 45 | Yes |
| Ji | 2015 | Korea | Asian | Head and neck | HB | TaqMan | 26 | 107 | 127 | 15 | 125 | 190 | Yes |
| Hidaka | 2015 | Japan | Asian | Gastric | PB | TaqMan | 32 | 173 | 252 | 35 | 168 | 254 | Yes |
| Bediaga | 2015 | Spain | Caucasian | Head and neck | PB | TaqMan | 78 | 61 | 61 | 203 | 391 | 39 1 | NA |
| Ye | 2014 | China | Asian | Esophageal | HB | PCR-RFLP | 224 | 400 | 377 | 150 | 578 | 663 | Yes |
| Tsai | 2014 | China | Asian | Head and neck | HB | TaqMan | 47 | 165 | 224 | 25 | 221 | 268 | No |
| Chung | 2014 | China | Asian | UADT | HB | MassARRAY | 68 | 76 | 108 | 25 | 111 | 125 | Yes |
| Yuan | 2013 | China | Asian | Head and neck | PB | TaqMan | 42 | 180 | 170 | 72 | 362 | 455 | Yes |
| Wu | 2013 | China | Asian | Esophageal | PB | TaqMan | 138 | 309 | 355 | 101 | 410 | 510 | Yes |
| Gao | 2013 | China | Asian | Esophageal | PB | TaqMan | 252 | 907 | 939 | 199 | 909 | 1155 | Yes |
| Dura | 2013 | Dutch | Caucasian | Esophageal | PB | TaqMan | 326 | 20 | 0 | 406 | 23 | 0 | Yes |
| Crous-Bou | 2013 | Spain | Caucasian | Colorectal | PB | Illumina | 457 | 324 | 79 | 513 | 360 | 54 | Yes |
| Liang | 2012 | Island | Mixed | Head and neck | PB | TaqMan | 530 | 38 | 5 | 593 | 76 | 15 | No |
| Gu | 2012 | China | Asian | Esophageal | HB | MassArray | 53 | 168 | 158 | 26 | 170 | 182 | Yes |
| Ferrari | 2012 | France | Caucasian | Colorectal | PB | TaqMan | 1129 | 97 | 5 | 1800 | 176 | 6 | Yes |
| Duell | 2012 | Spain | Caucasian | Gastric | PB | Illumina | 317 | 45 | 2 | 1133 | 132 | 6 | Yes |
| Chiang | 2012 | China | Asian | Colorectal | HB | PCR-RFLP | 7 | 34 | 62 | 43 | 205 | 297 | Yes |
| Yin | 2011 | Japan | Asian | Colorectal | PB | PCR-RFLP | 25 | 161 | 268 | 71 | 393 | 588 | Yes |
| Wang | 2011 | China | Asian | Esophageal | HB | PCR-CTPP | 15 | 34 | 33 | 17 | 67 | 78 | Yes |
| McKay | 2011 | France | Caucasian | UADT | PB | Illumina | 6776 | 4161 | 4161 | 7742 | 9071 | 907 1 | NA |
| Marichalar-Mendia | 2011 | Spain | Caucasian | Head and neck | PB | TaqMan | 80 | 71 | 71 | 203 | 391 | 39 1 | NA |
| Ji | 2011 | Korea | Asian | Head and neck | HB | TaqMan | 30 | 87 | 108 | 15 | 112 | 174 | Yes |
| Hakenewerth | 2011 | U.S.A. | Mixed | Head and neck | PB | Illumina | 1192 | 311 | 311 | 1243 | 791 | 791 | NA |
| Wei | 2010 | U.S.A. | Caucasian | Head and neck | HB | PCR-RFLP | 1059 | 51 | 0 | 1075 | 52 | 2 | Yes |
| Tanaka | 2010 | Japan | Asian | Esophageal | HB | Affymetrix | 151 | 5911 | 5911 | 44 | 7761 | 776 1 | NA |
| Soucek | 2010 | Czech | Caucasian | Head and neck | HB | TaqMan | 101 | 21 | 0 | 111 | 10 | 1 | Yes |
| Mohelnikova- Duchonova | 2010 | Czech | Caucasian | Pancreatic | PB | TaqMan | 213 | 22 | 0 | 242 | 22 | 1 | Yes |
| Garcia | 2010 | Brazil | Mixed | Head and neck | HB | PCR-RFLP | 195 | 12 | 0 | 213 | 29 | 2 | Yes |
| Cao | 2010 | China | Asian | Gastric | PB | DHPLC | 40 | 148 | 194 | 29 | 160 | 193 | Yes |
| Yang | 2009 | China | Asian | Colorectal | HB | SNPLex | 39 | 181 | 205 | 62 | 319 | 370 | Yes |
| Oze | 2009 | Japan | Asian | UADT | HB | TaqMan | 71 | 222 | 292 | 53 | 408 | 709 | Yes |
| Kawase | 2009 | Japan | Asian | Breast | HB | TaqMan | 25 | 162 | 265 | 47 | 322 | 539 | Yes |
| Kanda | 2009 | Japan | Asian | Pancreatic | HB | TaqMan | 4 | 55 | 101 | 74 | 551 | 975 | Yes |
| Ding | 2009 | China | Asian | Esophageal | PB | DHPLC | 8 | 75 | 108 | 19 | 96 | 106 | Yes |
| Cui | 2009 | Japan | Asian | Esophageal | PB | Illumina | 194 | 363 | 510 | 151 | 986 | 1626 | Yes |
| Akbari | 2009 | Iran | Asian | Esophageal | PB | MassARRAY | 21 | 232 | 490 | 73 | 471 | 827 | Yes |
| Solomon | 2008 | India | Asian | Head and neck | HB | PCR-RFLP | 13 | 56 | 57 | 8 | 38 | 54 | Yes |
| Lee | 2008 | China | Asian | Esophageal | HB | PCR-RFLP | 117 | 149 | 140 | 46 | 275 | 335 | Yes |
| Guo | 2008 | China | Asian | Esophageal | HB | PCR-RFLP | 17 | 25 | 38 | 24 | 168 | 288 | Yes |
| Gao | 2008 | China | Asian | Colorectal | PB | DHPLC | 15 | 73 | 102 | 20 | 109 | 93 | Yes |
| Ding | 2008 | China | Asian | Hepatocellular | PB | PCR-RFLP | 21 | 132 | 54 | 26 | 97 | 84 | Yes |
| Zhang | 2007 | U.S.A. | Caucasian | Gastric | PB | TaqMan | 261 | 31 | 1 | 352 | 48 | 1 | Yes |
| Yin | 2007 | Japan | Asian | Colorectal | PB | PCR-RFLP | 40 | 294 | 345 | 37 | 289 | 452 | Yes |
| Yang | 2007 | China | Asian | Esophageal | PB | PCR-CTPP | 33 | 80 | 78 | 22 | 76 | 100 | Yes |
| Hiraki | 2007 | Japan | Asian | Head and neck | HB | TaqMan | 26 | 75 | 138 | 31 | 213 | 471 | Yes |
| Asakage | 2007 | Japan | Asian | Head and neck | PB | PCR-RFLP | 31 | 223 | 388 | 19 | 28 | 49 | No |
| Sakamoto | 2006 | Japan | Asian | Hepatocellular | HB | PCR-CTPP | 12 | 73 | 124 | 13 | 103 | 159 | Yes |
| Matsuo | 2006 | Japan | Asian | Colorectal | HB | PCR-CTPP | 19 | 102 | 136 | 36 | 259 | 473 | Yes |
| Hashibe | 2006 | France | Caucasian | Head and neck | HB | TaqMan | 719 | 471 | 471 | 877 | 1081 | 108 1 | NA |
| Hashibe | 2006 | France | Caucasian | Esophageal | HB | TaqMan | 163 | 41 | 41 | 792 | 951 | 95 1 | NA |
| Chen | 2006 | China | Asian | Esophageal | HB | PCR-RFLP | 88 | 117 | 125 | 39 | 240 | 313 | Yes |
| Yang | 2005 | China | Asian | Esophageal | HB | PCR-CTPP | 6 | 85 | 74 | 22 | 168 | 304 | Yes |
| Wu | 2005 | China | Asian | Esophageal | PB | PCR-RFLP | 39 | 49 | 46 | 16 | 191 | 130 | No |
| Landi | 2005 | France | Caucasian | Colorectal | HB | Millipore | 292 | 54 | 2 | 263 | 48 | 3 | Yes |
| Risch | 2003 | Germany | Caucasian | Head and neck | PB | PCR-RFLP | 227 | 18 | 0 | 227 | 24 | 0 | Yes |
| Chao | 2003 | China | Asian | Esophageal | HB | PCR-RFLP | 19 | 41 | 28 | 7 | 43 | 55 | Yes |
| Yokoyama | 2002 | Japan | Asian | Esophageal | PB | PCR-RFLP | 51 | 73 | 110 | 31 | 220 | 383 | Yes |
| Boonyaphiphat | 2002 | Thailand | Asian | Esophageal | HB | APLP | 15 | 86 | 101 | 28 | 139 | 94 | No |
| Yokoyama | 2001 | Japan | Asian | Esophageal | PB | PCR-RFLP | 56 | 561 | 561 | 145 | 3811 | 381 1 | NA |
| Yokoyama | 2001 | Japan | Asian | Gastric | PB | PCR-RFLP | 28 | 101 | 101 | 145 | 3811 | 381 1 | NA |
| Takeshita | 2000 | Japan | Asian | Hepatocellular | PB | PCR-RFLP | 3 | 36 | 63 | 8 | 43 | 74 | Yes |
| Hori | 1997 | Japan | Asian | Esophageal | HB | PCR-RFLP | 20 | 31 | 40 | 5 | 20 | 43 | Yes |
Abbreviations: APLP, amplified product length polymorphism; DHPLC, denaturing high-performance liquid chromatography; HB, hospital-based, NA, not applicable; PB, population-based; PCR-CTPP, PCR with the confronting-two-pair primer; PCR-RFLP, PCR-restriction fragment length polymorphism; UADT, upper aerodigestive tract.
The number of GA + AA.
Meta-analysis results
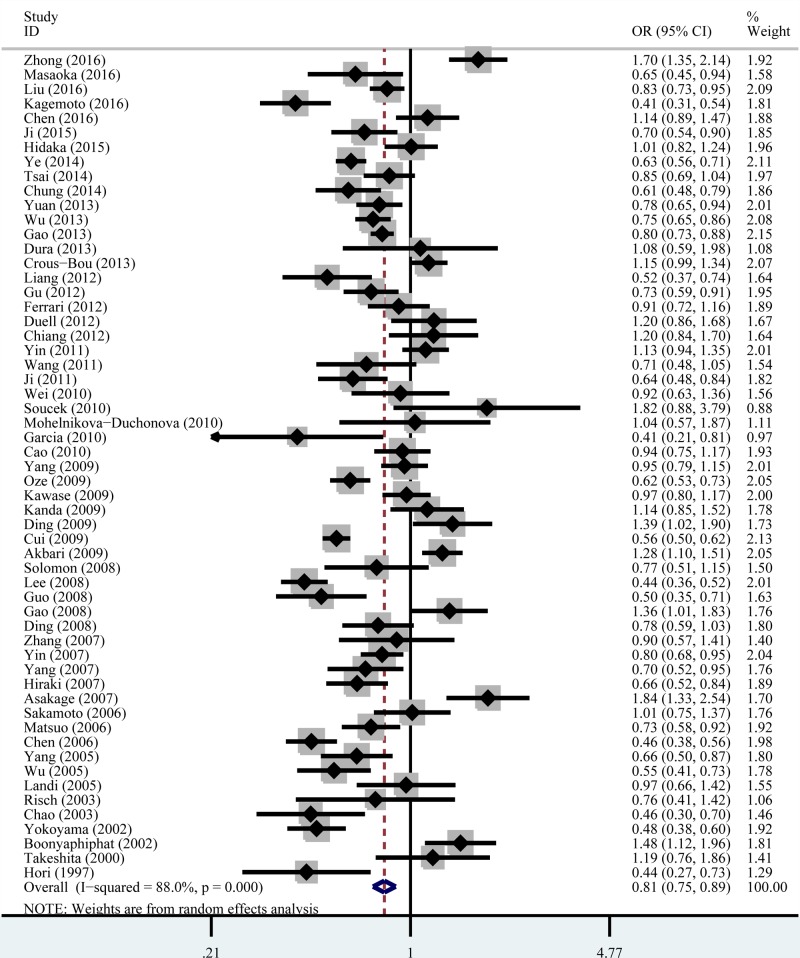
The main results for the association between ADH1B Arg47His polymorphism and cancer risk are shown in Table 2 and Figure 2. We found that ADH1B Arg47His polymorphism significantly associated with the decreased risk of overall cancer under all the five genetic models: homozygous model, OR = 0.62, 95% CI = 0.49–0.77; heterozygous model, OR = 0.71, 95% CI = 0.60–0.84; recessive model, OR = 0.83, 95% CI = 0.76–0.91; dominant model, OR = 0.62, 95% CI = 0.53–0.72; and allele comparison, OR = 0.82, 95% CI = 0.75–0.89.
Table 2. Meta-analysis of the association between the ADH1B Arg47His and cancer risk.
| Variables | Sample size Case/control | Homozygous | Heterozygous | Recessive | Dominant | Allele comparison | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| His/His vs. Arg/Arg | Arg/His vs. Arg/Arg | His/His vs. (Arg/His + Arg/Arg) | (Arg/His + His/His) vs. Arg/Arg | His vs. Arg | |||||||
| OR (95% CI) | Phet | OR (95% CI) | Phet | OR (95% CI) | Phet | OR (95% CI) | Phet | OR (95% CI) | Phet | ||
| Total | 31999/50964 | 0.62 (0.49–0.77) | <0.001 | 0.71 (0.60–0.84) | <0.001 | 0.83 (0.76–0.91) | <0.001 | 0.62 (0.53–0.72) | <0.001 | 0.82 (0.75–0.89) | <0.001 |
| Ethnicity | |||||||||||
| Asian | 17057/31885 | 0.60 (0.48–0.76) | <0.001 | 0.66 (0.53–0.81) | <0.001 | 0.82 (0.75–0.91) | <0.001 | 0.58 (0.47–0.72) | <0.001 | 0.80 (0.72–0.88) | <0.001 |
| Caucasian | 12970/16908 | 1.45 (1.05–2.02) | 0.727 | 1.01 (0.90–1.13) | 0.570 | 1.45 (1.05–2.00) | 0.712 | 0.81 (0.64–1.03) | <0.001 | 1.06 (0.96–1.17) | 0.569 |
| Mixed | 1972/2171 | 0.35 (0.13–0.93) | 0.743 | 0.53 (0.37–0.75) | 0.606 | 0.37 (0.14–0.98) | 0.751 | 0.46 (0.36–0.60) | 0.651 | 0.50 (0.36–0.68) | 0.545 |
| Cancer type | |||||||||||
| Colorectal | 4821/7697 | 1.19 (0.82–1.72) | <0.001 | 0.99 (0.88–1.11) | 0.857 | 1.19 (0.91–1.55) | <0.001 | 1.03 (0.88–1.21) | 0.099 | 1.05 (0.90–1.23) | <0.001 |
| Hepatocellular | 1111/3820 | 0.84 (0.64–1.10) | 0.541 | 1.16 (0.84–1.61) | 0.328 | 0.81 (0.61–1.08) | 0.041 | 0.98 (0.76–1.28) | 0.452 | 0.88 (0.76–1.02) | 0.270 |
| Esophageal | 9117/15930 | 0.39 (0.28–0.55) | <0.001 | 0.47 (0.34–0.64) | <0.001 | 0.72 (0.62–0.83) | <0.001 | 0.41 (0.31–0.54) | <0.001 | 0.67 (0.57–0.78) | <0.001 |
| Gastric | 1770/2930 | 1.02 (0.76–1.36) | 0.637 | 1.03 (0.84–1.27) | 0.356 | 1.02 (0.86–1.22) | 0.973 | 0.77 (0.48–1.23) | <0.001 | 1.03 (0.92–1.16) | 0.629 |
| Head and neck | 6646/7901 | 0.55 (0.31–0.97) | <0.001 | 0.77 (0.52–1.12) | <0.001 | 0.78 (0.66–0.93) | 0.092 | 0.64 (0.47–0.87) | <0.001 | 0.80 (0.66–0.96) | <0.001 |
| UADT | 7613/9173 | 0.31 (0.23–0.42) | 0.921 | 0.33 (0.21–0.53) | 0.161 | 0.70 (0.57–0.86) | 0.260 | 0.39 (0.26–0.58) | 0.010 | 0.62 (0.54–0.71) | 0.924 |
| Pancreatic | 395/1865 | 1.65 (0.62–4.38) | 0.345 | 1.29 (0.76–2.20) | 0.430 | 1.09 (0.78–1.52) | 0.513 | 1.26 (0.75–2.13) | 0.358 | 1.12 (0.86–1.45) | 0.774 |
| Control source | |||||||||||
| HB | 10560/20932 | 0.53 (0.40–0.71) | <0.001 | 0.64 (0.51–0.81) | <0.001 | 0.79 (0.69–0.90) | <0.001 | 0.56 (0.44–0.72) | <0.001 | 0.77 (0.68–0.87) | <0.001 |
| PB | 21439/30032 | 0.75 (0.52–1.07) | <0.001 | 0.79 (0.61–1.02) | <0.001 | 0.89 (0.78–1.02) | <0.001 | 0.68 (0.54–0.85) | <0.001 | 0.87 (0.76–0.99) | <0.001 |
| HWE | |||||||||||
| YES | 20769/37678 | 0.60 (0.48–0.76) | <0.001 | 0.71 (0.60–0.84) | <0.001 | 0.81 (0.74–0.89) | <0.001 | 0.67 (0.56–0.81) | <0.001 | 0.81 (0.74–0.88) | <0.001 |
| NO | 1987/1892 | 0.75 (0.22–2.65) | <0.001 | 0.66 (0.23–1.90) | <0.001 | 1.08 (0.76–1.55) | 0.006 | 0.72 (0.25–2.09) | <0.001 | 0.92 (0.59–1.45) | <0.001 |
Abbreviations: HB, hospital-based; PB, population-based; UADT, upper aerodigestive tract.
Values in bold indicate P<0.05.
Figure 2. Forest plot of the association between ADH1B Arg47His polymorphism and the overall cancer risk under the allele comparison model.
Regarding the stratified analysis by ethnicity, a decreased cancer risk was also detected amongst Asians under all the genetic models: homozygous model, OR = 0.60, 95% CI = 0.48–0.76; heterozygous model, OR = 0.66, 95% CI = 0.53–0.81; recessive model, OR = 0.82, 95% CI = 0.75–0.91; dominant model, OR = 0.58, 95% CI = 0.47–0.72; and allele comparison, OR = 0.80, 95% CI = 0.72–0.88, and amongst mixed ethnic group: homozygous model, OR = 0.35, 95% CI = 0.13–0.93; heterozygous model, OR = 0.53, 95% CI = 0.37–0.75; recessive model, OR = 0.37, 95% CI = 0.14–0.98; dominant model, OR = 0.46, 95% CI = 0.36–0.60; and allele comparison, OR = 0.50, 95% CI = 0.36–0.68. However, an increased risk of cancer was detected amongst Caucasians under homozygous model (OR = 1.45, 95% CI = 1.05–2.02) and recessive model (OR = 1.45, 95% CI = 1.05–2.00).
Regarding the stratified analysis by cancer type, the ADH1B Arg47His polymorphism significantly decreased the risk of esophageal cancer: homozygous model, OR = 0.39, 95% CI = 0.28–0.55; heterozygous model, OR = 0.47, 95% CI = 0.34–0.66; recessive model, OR = 0.72, 95% CI = 0.62–0.83; dominant model, OR = 0.41, 95% CI = 0.31–0.54; and allele comparison, OR = 0.67, 95% CI = 0.57–0.78; upper aerodigestive tract cancer: homozygous model, OR = 0.31, 95% CI = 0.23–0.42; heterozygous model, OR = 0.33, 95% CI = 0.21–0.53; recessive model, OR = 0.70, 95% CI = 0.57–0.86; dominant model, OR = 0.39, 95% CI = 0.26–0.58; and allele comparison, OR = 0.62, 95% CI = 0.54–0.71; and head and neck cancer: homozygous model, OR = 0.55, 95% CI = 0.31–0.97; recessive model, OR = 0.78, 95% CI = 0.66–0.93; dominant model, OR = 0.64, 95% CI = 0.47–0.87; and allele comparison, OR = 0.80, 95% CI = 0.66–0.96.
Regarding the stratified analysis by control source and HWE, a decreased cancer risk was detected in hospital-based studies: homozygous model, OR = 0.53, 95% CI = 0.40–0.71; heterozygous model, OR = 0.64, 95% CI = 0.51–0.81; recessive model, OR = 0.79, 95% CI = 0.69–0.90; dominant model, OR = 0.56, 95% CI = 0.44–0.72; and allele comparison, OR = 0.77, 95% CI = 0.68–0.87; population-based studies: dominant model, OR = 0.68, 95% CI = 0.54–0.85; and allele comparison, OR = 0.87, 95% CI = 0.76–0.99; and also the studies in agreement with HWE: homozygous model, OR = 0.60, 95% CI = 0.48–0.76; heterozygous model, OR = 0.71, 95% CI = 0.60–0.84; recessive model, OR = 0.81, 95% CI = 0.74–0.89; dominant model, OR = 0.67, 95% CI = 0.56–0.81; and allele comparison, OR = 0.81, 95% CI = 0.74–0.88.
Sensitivity analysis and publication bias
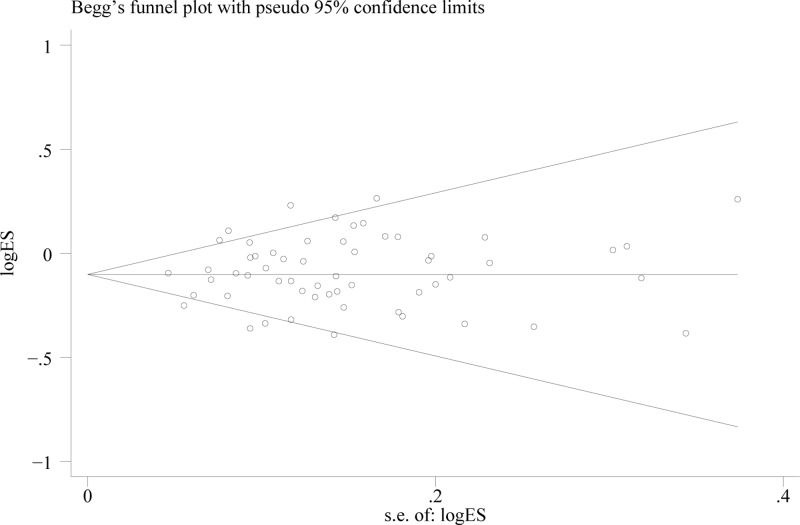
Substantial heterogeneities were found under all the five genetic models (P<0.001). Therefore, the random-effect model was adopted to assess the ORs and 95% CIs. Furthermore, the leave-one-out sensitivity analyses indicated that no single study could change the pooled ORs. The results of the Begg’s funnel plot and Egger’s linear regression test showed no evidence of publication bias (homozygous model, P=0.227; heterozygous model, P=0.697; recessive model, P=0.663; dominant model, P=0.599; and allele comparison P=0.342, see Figure 3).
Figure 3. Funnel plot analysis to detect publication bias for ADH1B Arg47His polymorphism under the allele comparison model.
Discussion
Alcohol consumption has been established to be a major factor in the development and progress of cancer [13]. Alcohol is first catalytically oxidized to acetaldehyde, mainly by ADH, and then to harmless acetate by ALDH [6,7]. Acetaldehyde, a Group I human carcinogen classified by the International Agency for Research on Cancer (IARC), may stimulate carcinogenesis by disrupting DNA synthesis and repair [8,9,84]. Therefore, to reduce the risk of cancer, it is important to modulate exposure levels to acetaldehyde in the liver. ADH1B gene, also known as ADH2, is located on chromosome 4q22 and is the locus responsible for the majority of activities of ADH function [25]. Arg47His (rs1229984 G > A) in ADH1B led to a single amino acid substitution of arginine (Arg) for histidine (His) at codon 47. Compared with the Arg/Arg individuals, the His/His individuals have a 40-fold higher enzyme activity oxidized alcohol to toxic acetaldehyde, thereby inducing tumorigenesis [25,85].
To the best of our knowledge, this is the first meta-analysis investigating the association between ADH1B Arg47His polymorphism and the overall cancer risk. A total of 66 studies from 64 articles with 31999 cases and 50964 controls were included, and the large sample size provided adequate power to detect this association. Overall, ADH1B Arg47His polymorphism was associated with a decreased risk of overall cancer under all the five genetic models. Stratified analysis by ethnicity revealed that ADH1B Arg47His polymorphism reduced cancer risk amongst Asians and mixed ethnicity group but increased risk amongst Caucasians. Stratified analysis by cancer type revealed that ADH1B Arg47His polymorphism reduced risk in esophageal cancer, upper aerodigestive tract cancer, and head and neck cancer, while no effect was found on colorectal, hepatocellular, gastric and pancreatic cancer. In stratified analysis by control source and HWE, a decreased cancer risk was detected in hospital-based studies, population-based studies, and also the studies in agreement with HWE.
There were several meta-analyses focussed on ADH1B Arg47His polymorphism and only one particular type of cancer risk, such as esophageal, head and neck, gastric and colorectal cancer [11–15]. For esophageal cancer, Mao et al. [11] found that the 47His allele was significantly associated with the reduced risk of this cancer when compared with the 47Arg allele. And these findings were replicated in our meta-analysis. For head and neck cancer, the 47His allele was also found to be associated with decreased risk of head and neck cancer amongst Asians only under the dominant model [12]. However, similar results were found under the other three models in our analysis, which may be attributed to a larger sample size including eight more studies. Interestingly, Chen et al. [15] found that ADH1B Arg47His polymorphism was associated with decreased risk of colorectal cancer supported by four studies. However, this decreased risk was not present in the current one including six more studies. It was noteworthy that we found that ADH1B Arg47His polymorphism was associated with decreased cancer risk amongst Asians while increased cancer risk amongst Caucasians. In Caucasian population, the A allele was found to associate with an increased risk of colorectal cancer [32]. The opposite findings may result from the difference of ethnicity with the 47His allele occupied more than 90% amongst Asians but fewer than 20% amongst Caucasians [7]. Furthermore, we re-analyzed the ethnic groups of Asian and Caucasian people. Amongst Asians, a decreased cancer risk was also detected in esophageal cancer and head and neck cancer. While in Caucasians, we did not repeat the results, but an increased cancer risk was detected in colorectal cancer (homozygous model, OR = 1.55, 95% CI = 1.10–2.20 and recessive model, OR = 1.55, 95% CI = 1.11–2.18).
Several limitations in the current meta-analysis should be addressed. First, a number of studies adopted in our meta-analysis had relatively small sample size for each cancer type, like bladder and breast cancer. Second, because of the absence of original data, our analyses were based on unadjusted estimates of ORs without adjustment for other confounding factors. Third, there were substantial heterogeneities in all the five genetic models, hence the random-effect model was adopted and might present unstable results. Overall, due to these limitations, the findings in the current meta-analysis should be interpreted with caution.
In conclusion, our meta-analysis suggested that ADH1B Arg47His polymorphism was significantly associated with the decreased overall cancer risk, especially for esophageal cancer and head and neck cancer amongst Asians.
Abbreviations
- ADH
alcohol dehydrogenase
- ALDH
aldehyde dehydrogenase
- CI
confidence interval
- HWE
Hardy–Weinberg equilibrium
- OR
odds ratio
Funding
This work was supported by the Natural Science Foundation of Hunan Province, China [grant number 2017JJ2156].
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Author contribution
All authors contributed significantly to this work. B.T. designed the research study. B.T. and N.N. performed the research study, analyzed the data, and wrote the paper. Both the authors reviewed the paper.
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J. and Jemal A. (2015) Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.Bender E. (2014) Developing world: global warning. Nature 509, S64–S65 10.1038/509S64a [DOI] [PubMed] [Google Scholar]
- 3.Boffetta P., Hashibe M., La Vecchia C., Zatonski W. and Rehm J. (2006) The burden of cancer attributable to alcohol drinking. Int. J. Cancer 119, 884–887 10.1002/ijc.21903 [DOI] [PubMed] [Google Scholar]
- 4.Sridhar D. (2012) Health policy: regulate alcohol for global health. Nature 482, 302 10.1038/482302a [DOI] [PubMed] [Google Scholar]
- 5.Ratna A. and Mandrekar P. (2017) Alcohol and cancer: mechanisms and therapies. Biomolecules 7, 10.3390/biom7030061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith M. (1986) Genetics of human alcohol and aldehyde dehydrogenases. Adv. Hum. Genet. 15, 249–290 [DOI] [PubMed] [Google Scholar]
- 7.Bosron W.F. and Li T.K. (1986) Genetic polymorphism of human liver alcohol and aldehyde dehydrogenases, and their relationship to alcohol metabolism and alcoholism. Hepatology 6, 502–510 10.1002/hep.1840060330 [DOI] [PubMed] [Google Scholar]
- 8.Brooks P.J. and Zakhari S. (2014) Acetaldehyde and the genome: beyond nuclear DNA adducts and carcinogenesis. Environ. Mol. Mutagen. 55, 77–91 10.1002/em.21824 [DOI] [PubMed] [Google Scholar]
- 9.Seitz H.K. and Stickel F. (2007) Molecular mechanisms of alcohol-mediated carcinogenesis. Nat. Rev. Cancer 7, 599–612 10.1038/nrc2191 [DOI] [PubMed] [Google Scholar]
- 10.Druesne-Pecollo N., Tehard B., Mallet Y., Gerber M., Norat T., Hercberg S.. et al. (2009) Alcohol and genetic polymorphisms: effect on risk of alcohol-related cancer. Lancet Oncol. 10, 173–180 10.1016/S1470-2045(09)70019-1 [DOI] [PubMed] [Google Scholar]
- 11.Mao N., Nie S., Hong B., Li C., Shen X. and Xiong T. (2016) Association between alcohol dehydrogenase-2 gene polymorphism and esophageal cancer risk: a meta-analysis. World J. Surg. Oncol. 14, 191 10.1186/s12957-016-0937-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y., Gu N., Miao L., Yuan H., Wang R. and Jiang H. (2015) Alcohol dehydrogenase-1B Arg47His polymorphism is associated with head and neck cancer risk in Asian: a meta-analysis. Tumour Biol. 36, 1023–1027 10.1007/s13277-014-2727-x [DOI] [PubMed] [Google Scholar]
- 13.Zhang L., Jiang Y., Wu Q., Li Q., Chen D., Xu L.. et al. (2014) Gene-environment interactions on the risk of esophageal cancer among Asian populations with the G48A polymorphism in the alcohol dehydrogenase-2 gene: a meta-analysis. Tumour Biol. 35, 4705–4717 10.1007/s13277-014-1616-7 [DOI] [PubMed] [Google Scholar]
- 14.Wang H.L., Zhou P.Y., Liu P. and Zhang Y. (2014) ALDH2 and ADH1 genetic polymorphisms may contribute to the risk of gastric cancer: a meta-analysis. PLoS ONE 9, e88779 10.1371/journal.pone.0088779 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Chen C., Wang L., Liao Q., Xu L., Huang Y., Zhang C.. et al. (2014) Association between six genetic polymorphisms and colorectal cancer: a meta-analysis. Genet. Test. Mol. Biomarkers 18, 187–195 10.1089/gtmb.2013.0425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo H., Zhang G. and Mai R. (2012) Alcohol dehydrogenase-1B Arg47His polymorphism and upper aerodigestive tract cancer risk: a meta-analysis including 24,252 subjects. Alcohol Clin. Exp. Res. 36, 272–278 10.1111/j.1530-0277.2011.01621.x [DOI] [PubMed] [Google Scholar]
- 17.Zhong Q., Wu R.R. and Zeng Z.M. (2016) Association of ADH1B Arg47His and ALDH2 Glu487Lys polymorphisms with risk of colorectal cancer and their interaction with environmental factors in a Chinese population. Genet. Mol. Res. 15, 1–8 10.4238/gmr.15038682 [DOI] [PubMed] [Google Scholar]
- 18.Masaoka H., Ito H., Soga N., Hosono S., Oze I., Watanabe M.. et al. (2016) Aldehyde dehydrogenase 2 (ALDH2) and alcohol dehydrogenase 1B (ADH1B) polymorphisms exacerbate bladder cancer risk associated with alcohol drinking: gene-environment interaction. Carcinogenesis 37, 583–588 10.1093/carcin/bgw033 [DOI] [PubMed] [Google Scholar]
- 19.Liu J., Yang H.I., Lee M.H., Jen C.L., Hu H.H., Lu S.N.. et al. (2016) Alcohol drinking mediates the association between polymorphisms of ADH1B and ALDH2 and Hepatitis B-related hepatocellular carcinoma. Cancer Epidemiol. Biomarkers Prev. 25, 693–699 [DOI] [PubMed] [Google Scholar]
- 20.Kagemoto K., Urabe Y., Miwata T., Oka S., Ochi H., Kitadai Y.. et al. (2016) ADH1B and ALDH2 are associated with metachronous SCC after endoscopic submucosal dissection of esophageal squamous cell carcinoma. Cancer Med. 5, 1397–1404 10.1002/cam4.705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z.H., Xian J.F. and Luo L.P. (2016) Analysis of ADH1B Arg47His, ALDH2 Glu487Lys, and CYP4502E1 polymorphisms in gastric cancer risk and interaction with environmental factors. Genet. Mol. Res. 15, 1–9 10.4238/gmr15048904 [DOI] [PubMed] [Google Scholar]
- 22.Ji Y.B., Lee S.H., Kim K.R., Park C.W., Song C.M., Park B.L.. et al. (2015) Association between ADH1B and ADH1C polymorphisms and the risk of head and neck squamous cell carcinoma. Tumour Biol. 36, 4387–4396 10.1007/s13277-015-3078-y [DOI] [PubMed] [Google Scholar]
- 23.Hidaka A., Sasazuki S., Matsuo K., Ito H., Sawada N., Shimazu T.. et al. (2015) Genetic polymorphisms of ADH1B, ADH1C and ALDH2, alcohol consumption, and the risk of gastric cancer: the Japan Public Health Center-based prospective study. Carcinogenesis 36, 223–231 10.1093/carcin/bgu244 [DOI] [PubMed] [Google Scholar]
- 24.Bediaga N.G., Marichalar-Mendia X., Rey-Barja N., Setien-Olarra A., Gonzalez-Garcia J.A., de Pancorbo M.M.. et al. (2015) Polymorphisms in alcohol and tobacco metabolism genes in head and neck cancer in the Basque Country. J. Oral Pathol. Med. 44, 769–775 10.1111/jop.12305 [DOI] [PubMed] [Google Scholar]
- 25.Ye B., Ji C.Y., Zhao Y., Li W., Feng J. and Zhang X. (2014) Single nucleotide polymorphism at alcohol dehydrogenase-1B is associated with risk of esophageal squamous cell carcinoma. Cancer Cell Int. 14, 12 10.1186/1475-2867-14-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai S.T., Wong T.Y., Ou C.Y., Fang S.Y., Chen K.C., Hsiao J.R.. et al. (2014) The interplay between alcohol consumption, oral hygiene, ALDH2 and ADH1B in the risk of head and neck cancer. Int. J. Cancer 135, 2424–2436 10.1002/ijc.28885 [DOI] [PubMed] [Google Scholar]
- 27.Chung C.S., Lee Y.C., Liou J.M., Wang C.P., Ko J.Y., Lee J.M.. et al. (2014) Tag single nucleotide polymorphisms of alcohol-metabolizing enzymes modify the risk of upper aerodigestive tract cancers: HapMap database analysis. Dis. Esophagus 27, 493–503 10.1111/j.1442-2050.2012.01437.x [DOI] [PubMed] [Google Scholar]
- 28.Yuan H., Ma H., Lu F., Yuan Z., Wang R., Jiang H.. et al. (2013) Genetic variants at 4q23 and 12q24 are associated with head and neck cancer risk in China. Mol. Carcinog. 52, E2–E9 10.1002/mc.21929 [DOI] [PubMed] [Google Scholar]
- 29.Wu M., Chang S.C., Kampman E., Yang J., Wang X.S., Gu X.P.. et al. (2013) Single nucleotide polymorphisms of ADH1B, ADH1C and ALDH2 genes and esophageal cancer: a population-based case-control study in China. Int. J. Cancer 132, 1868–1877 10.1002/ijc.27803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Y., He Y., Xu J., Xu L., Du J., Zhu C.. et al. (2013) Genetic variants at 4q21, 4q23 and 12q24 are associated with esophageal squamous cell carcinoma risk in a Chinese population. Hum. Genet. 132, 649–656 10.1007/s00439-013-1276-5 [DOI] [PubMed] [Google Scholar]
- 31.Dura P., Berkers T., van Veen E.M., Salomon J., te Morsche R.H., Roelofs H.M.. et al. (2013) Polymorphisms in alcohol-metabolizing enzymes and esophageal carcinoma susceptibility: a Dutch Caucasian case-control study. J. Hum. Genet. 58, 742–748 10.1038/jhg.2013.95 [DOI] [PubMed] [Google Scholar]
- 32.Crous-Bou M., Rennert G., Cuadras D., Salazar R., Cordero D., Saltz Rennert H.. et al. (2013) Polymorphisms in alcohol metabolism genes ADH1B and ALDH2, alcohol consumption and colorectal cancer. PLoS ONE 8, e80158 10.1371/journal.pone.0080158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang C., Marsit C.J., Houseman E.A., Butler R., Nelson H.H., McClean M.D.. et al. (2012) Gene-environment interactions of novel variants associated with head and neck cancer. Head Neck 34, 1111–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu H., Gong D., Ding G., Zhang W., Liu C., Jiang P.. et al. (2012) A variant allele of ADH1B and ALDH2, is associated with the risk of esophageal cancer. Exp. Ther. Med. 4, 135–140 10.3892/etm.2012.547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferrari P., McKay J.D., Jenab M., Brennan P., Canzian F., Vogel U.. et al. (2012) Alcohol dehydrogenase and aldehyde dehydrogenase gene polymorphisms, alcohol intake and the risk of colorectal cancer in the European Prospective Investigation into Cancer and Nutrition study. Eur. J. Clin. Nutr. 66, 1303–1308 10.1038/ejcn.2012.173 [DOI] [PubMed] [Google Scholar]
- 36.Duell E.J., Sala N., Travier N., Munoz X., Boutron-Ruault M.C., Clavel-Chapelon F.. et al. (2012) Genetic variation in alcohol dehydrogenase (ADH1A, ADH1B, ADH1C, ADH7) and aldehyde dehydrogenase (ALDH2), alcohol consumption and gastric cancer risk in the European Prospective Investigation into Cancer and Nutrition (EPIC) cohort. Carcinogenesis 33, 361–367 10.1093/carcin/bgr285 [DOI] [PubMed] [Google Scholar]
- 37.Chiang C.P., Jao S.W., Lee S.P., Chen P.C., Chung C.C., Lee S.L.. et al. (2012) Expression pattern, ethanol-metabolizing activities, and cellular localization of alcohol and aldehyde dehydrogenases in human large bowel: association of the functional polymorphisms of ADH and ALDH genes with hemorrhoids and colorectal cancer. Alcohol 46, 37–49 10.1016/j.alcohol.2011.08.004 [DOI] [PubMed] [Google Scholar]
- 38.Yin G., Hamajima N., Morita M., Tajima O., Tabata S. and Kono S. (2011) Lack of influence of the ADH1B Arg47His genetic polymorphism on risk of colorectal adenoma in middle-aged Japanese men. Asian Pacific J. Cancer Prev. 12, 297–302 [PubMed] [Google Scholar]
- 39.Wang Y., Ji R., Wei X., Gu L., Chen L., Rong Y.. et al. (2011) Esophageal squamous cell carcinoma and ALDH2 and ADH1B polymorphisms in Chinese females. Asian Pacific J. Cancer Prev. 12, 2065–2068 [PubMed] [Google Scholar]
- 40.McKay J.D., Truong T., Gaborieau V., Chabrier A., Chuang S.C., Byrnes G.. et al. (2011) A genome-wide association study of upper aerodigestive tract cancers conducted within the INHANCE consortium. PLoS Genet. 7, e1001333 10.1371/journal.pgen.1001333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marichalar-Mendia X., Acha-Sagredo A., Rodriguez-Tojo M.J., Rey-Barja N., Hernando-Rodriguez M., Aguirregaviria J.I.. et al. (2011) Alcohol-dehydrogenase (ADH1B) Arg48His polymorphism in Basque Country patients with oral and laryngeal cancer: preliminary study. Anticancer Res. 31, 677–680 [PubMed] [Google Scholar]
- 42.Ji Y.B., Tae K., Ahn T.H., Lee S.H., Kim K.R., Park C.W.. et al. (2011) ADH1B and ALDH2 polymorphisms and their associations with increased risk of squamous cell carcinoma of the head and neck in the Korean population. Oral Oncol. 47, 583–587 10.1016/j.oraloncology.2011.04.007 [DOI] [PubMed] [Google Scholar]
- 43.Hakenewerth A.M., Millikan R.C., Rusyn I., Herring A.H., North K.E., Barnholtz-Sloan J.S.. et al. (2011) Joint effects of alcohol consumption and polymorphisms in alcohol and oxidative stress metabolism genes on risk of head and neck cancer. Cancer Epidemiol. Biomarkers Prev. 20, 2438–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei S., Liu Z., Zhao H., Niu J., Wang L.E., El-Naggar A.K.. et al. (2010) A single nucleotide polymorphism in the alcohol dehydrogenase 7 gene (alanine to glycine substitution at amino acid 92) is associated with the risk of squamous cell carcinoma of the head and neck. Cancer 116, 2984–2992 10.1002/cncr.25058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka F., Yamamoto K., Suzuki S., Inoue H., Tsurumaru M., Kajiyama Y.. et al. (2010) Strong interaction between the effects of alcohol consumption and smoking on oesophageal squamous cell carcinoma among individuals with ADH1B and/or ALDH2 risk alleles. Gut 59, 1457–1464 10.1136/gut.2009.205724 [DOI] [PubMed] [Google Scholar]
- 46.Soucek P., Susova S., Mohelnikova-Duchonova B., Gromadzinska J., Moraviec-Sztandera A., Vodicka P.. et al. (2010) Polymorphisms in metabolizing enzymes and the risk of head and neck squamous cell carcinoma in the Slavic population of the central Europe. Neoplasma 57, 415–421 10.4149/neo_2010_05_415 [DOI] [PubMed] [Google Scholar]
- 47.Mohelnikova-Duchonova B., Vrana D., Holcatova I., Ryska M., Smerhovsky Z. and Soucek P. (2010) CYP2A13, ADH1B, and ADH1C gene polymorphisms and pancreatic cancer risk. Pancreas 39, 144–148 10.1097/MPA.0b013e3181bab6c2 [DOI] [PubMed] [Google Scholar]
- 48.Garcia S.M., Curioni O.A., de Carvalho M.B. and Gattas G.J. (2010) Polymorphisms in alcohol metabolizing genes and the risk of head and neck cancer in a Brazilian population. Alcohol Alcohol 45, 6–12 10.1093/alcalc/agp078 [DOI] [PubMed] [Google Scholar]
- 49.Cao H.X., Li S.P., Wu J.Z., Gao C.M., Su P., Liu Y.T.. et al. (2010) Alcohol dehydrogenase-2 and aldehyde dehydrogenase-2 genotypes, alcohol drinking and the risk for stomach cancer in Chinese males. Asian Pacific J. Cancer Prev. 11, 1073–1077 [PubMed] [Google Scholar]
- 50.Yang H., Zhou Y., Zhou Z., Liu J., Yuan X., Matsuo K.. et al. (2009) A novel polymorphism rs1329149 of CYP2E1 and a known polymorphism rs671 of ALDH2 of alcohol metabolizing enzymes are associated with colorectal cancer in a southwestern Chinese population. Cancer Epidemiol. Biomarkers Prev. 18, 2522–2527 [DOI] [PubMed] [Google Scholar]
- 51.Oze I., Matsuo K., Suzuki T., Kawase T., Watanabe M., Hiraki A.. et al. (2009) Impact of multiple alcohol dehydrogenase gene polymorphisms on risk of upper aerodigestive tract cancers in a Japanese population. Cancer Epidemiol. Biomarkers Prev. 18, 3097–3102 [DOI] [PubMed] [Google Scholar]
- 52.Kawase T., Matsuo K., Hiraki A., Suzuki T., Watanabe M., Iwata H.. et al. (2009) Interaction of the effects of alcohol drinking and polymorphisms in alcohol-metabolizing enzymes on the risk of female breast cancer in Japan. J. Epidemiol. 19, 244–250 10.2188/jea.JE20081035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanda J., Matsuo K., Suzuki T., Kawase T., Hiraki A., Watanabe M.. et al. (2009) Impact of alcohol consumption with polymorphisms in alcohol-metabolizing enzymes on pancreatic cancer risk in Japanese. Cancer Sci. 100, 296–302 10.1111/j.1349-7006.2008.01044.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding J.H., Li S.P., Cao H.X., Wu J.Z., Gao C.M., Su P.. et al. (2009) Polymorphisms of alcohol dehydrogenase-2 and aldehyde dehydrogenase-2 and esophageal cancer risk in Southeast Chinese males. World J. Gastroenterol. 15, 2395–2400 10.3748/wjg.15.2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cui R., Kamatani Y., Takahashi A., Usami M., Hosono N., Kawaguchi T.. et al. (2009) Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology 137, 1768–1775 10.1053/j.gastro.2009.07.070 [DOI] [PubMed] [Google Scholar]
- 56.Akbari M.R., Malekzadeh R., Shakeri R., Nasrollahzadeh D., Foumani M., Sun Y.. et al. (2009) Candidate gene association study of esophageal squamous cell carcinoma in a high-risk region in Iran. Cancer Res. 69, 7994–8000 10.1158/0008-5472.CAN-09-1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Solomon P.R., Selvam G.S. and Shanmugam G. (2008) Polymorphism in ADH and MTHFR genes in oral squamous cell carcinoma of Indians. Oral Dis. 14, 633–639 10.1111/j.1601-0825.2007.01437.x [DOI] [PubMed] [Google Scholar]
- 58.Lee C.H., Lee J.M., Wu D.C., Goan Y.G., Chou S.H., Wu I.C.. et al. (2008) Carcinogenetic impact of ADH1B and ALDH2 genes on squamous cell carcinoma risk of the esophagus with regard to the consumption of alcohol, tobacco and betel quid. Int. J. Cancer 122, 1347–1356 10.1002/ijc.23264 [DOI] [PubMed] [Google Scholar]
- 59.Guo Y.M., Wang Q., Liu Y.Z., Chen H.M., Qi Z. and Guo Q.H. (2008) Genetic polymorphisms in cytochrome P4502E1, alcohol and aldehyde dehydrogenases and the risk of esophageal squamous cell carcinoma in Gansu Chinese males. World J. Gastroenterol. 14, 1444–1449 10.3748/wjg.14.1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao C.M., Takezaki T., Wu J.Z., Zhang X.M., Cao H.X., Ding J.H.. et al. (2008) Polymorphisms of alcohol dehydrogenase 2 and aldehyde dehydrogenase 2 and colorectal cancer risk in Chinese males. World J. Gastroenterol. 14, 5078–5083 10.3748/wjg.14.5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ding J., Li S., Wu J., Gao C., Zhou J., Cao H.. et al. (2008) Alcohol dehydrogenase-2 and aldehyde dehydrogenase-2 genotypes, alcohol drinking and the risk of primary hepatocellular carcinoma in a Chinese population. Asian Pacific J. Cancer Prev. 9, 31–35 [PubMed] [Google Scholar]
- 62.Zhang F.F., Hou L., Terry M.B., Lissowska J., Morabia A., Chen J.. et al. (2007) Genetic polymorphisms in alcohol metabolism, alcohol intake and the risk of stomach cancer in Warsaw, Poland. Int. J. Cancer 121, 2060–2064 10.1002/ijc.22973 [DOI] [PubMed] [Google Scholar]
- 63.Yin G., Kono S., Toyomura K., Moore M.A., Nagano J., Mizoue T.. et al. (2007) Alcohol dehydrogenase and aldehyde dehydrogenase polymorphisms and colorectal cancer: the Fukuoka Colorectal Cancer Study. Cancer Sci. 98, 1248–1253 10.1111/j.1349-7006.2007.00519.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang S.J., Wang H.Y., Li X.Q., Du H.Z., Zheng C.J., Chen H.G.. et al. (2007) Genetic polymorphisms of ADH2 and ALDH2 association with esophageal cancer risk in southwest China. World J. Gastroenterol. 13, 5760–5764 10.3748/wjg.v13.i43.5760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hiraki A., Matsuo K., Wakai K., Suzuki T., Hasegawa Y. and Tajima K. (2007) Gene-gene and gene-environment interactions between alcohol drinking habit and polymorphisms in alcohol-metabolizing enzyme genes and the risk of head and neck cancer in Japan. Cancer Sci. 98, 1087–1091 10.1111/j.1349-7006.2007.00505.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Asakage T., Yokoyama A., Haneda T., Yamazaki M., Muto M., Yokoyama T.. et al. (2007) Genetic polymorphisms of alcohol and aldehyde dehydrogenases, and drinking, smoking and diet in Japanese men with oral and pharyngeal squamous cell carcinoma. Carcinogenesis 28, 865–874 10.1093/carcin/bgl206 [DOI] [PubMed] [Google Scholar]
- 67.Sakamoto T., Hara M., Higaki Y., Ichiba M., Horita M., Mizuta T.. et al. (2006) Influence of alcohol consumption and gene polymorphisms of ADH2 and ALDH2 on hepatocellular carcinoma in a Japanese population. Int. J. Cancer 118, 1501–1507 10.1002/ijc.21505 [DOI] [PubMed] [Google Scholar]
- 68.Matsuo K., Wakai K., Hirose K., Ito H., Saito T., Suzuki T.. et al. (2006) A gene-gene interaction between ALDH2 Glu487Lys and ADH2 His47Arg polymorphisms regarding the risk of colorectal cancer in Japan. Carcinogenesis 27, 1018–1023 10.1093/carcin/bgi282 [DOI] [PubMed] [Google Scholar]
- 69.Hashibe M., Boffetta P., Zaridze D., Shangina O., Szeszenia-Dabrowska N., Mates D.. et al. (2006) Evidence for an important role of alcohol- and aldehyde-metabolizing genes in cancers of the upper aerodigestive tract. Cancer Epidemiol. Biomarkers Prev. 15, 696–703 [DOI] [PubMed] [Google Scholar]
- 70.Chen Y.J., Chen C., Wu D.C., Lee C.H., Wu C.I., Lee J.M.. et al. (2006) Interactive effects of lifetime alcohol consumption and alcohol and aldehyde dehydrogenase polymorphisms on esophageal cancer risks. Int. J. Cancer 119, 2827–2831 10.1002/ijc.22199 [DOI] [PubMed] [Google Scholar]
- 71.Yang C.X., Matsuo K., Ito H., Hirose K., Wakai K., Saito T.. et al. (2005) Esophageal cancer risk by ALDH2 and ADH2 polymorphisms and alcohol consumption: exploration of gene-environment and gene-gene interactions. Asian Pacific J. Cancer Prev. 6, 256–262 [PubMed] [Google Scholar]
- 72.Wu C.F., Wu D.C., Hsu H.K., Kao E.L., Lee J.M., Lin C.C.. et al. (2005) Relationship between genetic polymorphisms of alcohol and aldehyde dehydrogenases and esophageal squamous cell carcinoma risk in males. World J. Gastroenterol. 11, 5103–5108 10.3748/wjg.v11.i33.5103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Landi S., Gemignani F., Moreno V., Gioia-Patricola L., Chabrier A., Guino E.. et al. (2005) A comprehensive analysis of phase I and phase II metabolism gene polymorphisms and risk of colorectal cancer. Pharmacogenet. Genomics 15, 535–546 10.1097/01.fpc.0000165904.48994.3d [DOI] [PubMed] [Google Scholar]
- 74.Risch A., Ramroth H., Raedts V., Rajaee-Behbahani N., Schmezer P., Bartsch H.. et al. (2003) Laryngeal cancer risk in Caucasians is associated with alcohol and tobacco consumption but not modified by genetic polymorphisms in class I alcohol dehydrogenases ADH1B and ADH1C, and glutathione-S-transferases GSTM1 and GSTT1. Pharmacogenetics 13, 225–230 10.1097/00008571-200304000-00007 [DOI] [PubMed] [Google Scholar]
- 75.Chao Y.C., Wang S.J., Chu H.C., Chang W.K. and Hsieh T.Y. (2003) Investigation of alcohol metabolizing enzyme genes in Chinese alcoholics with avascular necrosis of hip joint, pancreatitis and cirrhosis of the liver. Alcohol Alcohol 38, 431–436 10.1093/alcalc/agg106 [DOI] [PubMed] [Google Scholar]
- 76.Yokoyama A., Kato H., Yokoyama T., Tsujinaka T., Muto M., Omori T.. et al. (2002) Genetic polymorphisms of alcohol and aldehyde dehydrogenases and glutathione S-transferase M1 and drinking, smoking, and diet in Japanese men with esophageal squamous cell carcinoma. Carcinogenesis 23, 1851–1859 10.1093/carcin/23.11.1851 [DOI] [PubMed] [Google Scholar]
- 77.Boonyaphiphat P., Thongsuksai P., Sriplung H. and Puttawibul P. (2002) Lifestyle habits and genetic susceptibility and the risk of esophageal cancer in the Thai population. Cancer Lett. 186, 193–199 10.1016/S0304-3835(02)00354-3 [DOI] [PubMed] [Google Scholar]
- 78.Yokoyama A., Muramatsu T., Omori T., Yokoyama T., Matsushita S., Higuchi S.. et al. (2001) Alcohol and aldehyde dehydrogenase gene polymorphisms and oropharyngolaryngeal, esophageal and stomach cancers in Japanese alcoholics. Carcinogenesis 22, 433–439 10.1093/carcin/22.3.433 [DOI] [PubMed] [Google Scholar]
- 79.Takeshita T., Yang X., Inoue Y., Sato S. and Morimoto K. (2000) Relationship between alcohol drinking, ADH2 and ALDH2 genotypes, and risk for hepatocellular carcinoma in Japanese. Cancer Lett. 149, 69–76 10.1016/S0304-3835(99)00343-2 [DOI] [PubMed] [Google Scholar]
- 80.Hori H., Kawano T., Endo M. and Yuasa Y. (1997) Genetic polymorphisms of tobacco- and alcohol-related metabolizing enzymes and human esophageal squamous cell carcinoma susceptibility. J. Clin. Gastroenterol. 25, 568–575 10.1097/00004836-199712000-00003 [DOI] [PubMed] [Google Scholar]
- 81.Mantel N. and Haenszel W. (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 22, 719–748 [PubMed] [Google Scholar]
- 82.DerSimonian R. and Laird N. (1986) Meta-analysis in clinical trials. Control. Clin. Trials 7, 177–188 10.1016/0197-2456(86)90046-2 [DOI] [PubMed] [Google Scholar]
- 83.Egger M., Davey Smith G., Schneider M. and Minder C. (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634 10.1136/bmj.315.7109.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boffetta P. and Hashibe M. (2006) Alcohol and cancer. Lancet Oncol. 7, 149–156 10.1016/S1470-2045(06)70577-0 [DOI] [PubMed] [Google Scholar]
- 85.Lee C.H., Wu D.C., Wu I.C., Goan Y.G., Lee J.M., Chou S.H.. et al. (2009) Genetic modulation of ADH1B and ALDH2 polymorphisms with regard to alcohol and tobacco consumption for younger aged esophageal squamous cell carcinoma diagnosis. Int. J. Cancer 125, 1134–1142 10.1002/ijc.24357 [DOI] [PubMed] [Google Scholar]