Abstract
Patulin (PAT), a mycotoxin mainly produced by various species of fungi, is frequently detected in moldy fruit- and vegetable-based products, which pose a health risk to the consumer. Over the past decades, a few studies reported that PAT content could be significantly decreased by microbial fermentation process. However, the physical adsorption mechanism between PAT and yeast during fermentation is still unclear. In this paper, we focused on the physical adsorption of PAT by Saccharomyces cerevisiae CCTCC 93161 during fermentation in aqueous solutions. Firstly, morphology of differently treated yeast cells were analyzed by scanning electron microscope, then the interactions between PAT and yeast cells were investigated by infrared absorption spectra of differently treated S. Cerevisiae cells before and after the adsorption of PAT. The results showed that the efficiency of PAT removal raised significantly with the increase of fermentation temperature and time, whereas it decreased significantly with the increase of initial PAT concentration in the fermentation system. The proteins and polysaccharides in the cell walls of yeast interacted with PAT and accounted for the physical adsorption. The current work would possibly provide some new insights on PAT control for fermented foods.
Keywords: Patulin, Saccharomyces cerevisiae, Fermentation, Physical adsorption
Introduction
Patulin (PAT) is a toxic metabolite produced by various species of fungi, most commonly by Penicillium, Aspergillus and Byssochlamys, and it represents a significant hazard to the food and feed chain (Richard 2007; Neri et al. 2010; Oteiza et al. 2017). It can be found in a number of fruits, most commonly found in rotting apples, in general the amount of PAT in apple products is viewed as a measure of the quality of the apples used in production [Commission regulation (EC) No 1881/2006]. PAT is a lactone that is heat-stable, so it is difficult to be destroyed by pasteurization or thermal denaturation. Food processing can limit PAT levels in the food chain by physical removal and decontamination by chemical or enzymatic transformation of PAT into less toxic products, yet do not eliminate PAT completely. The known strategies for detoxification of PAT in apple juice include physical, biological and chemical methods. In the past few years, biological absorption method is of growing importance to control PAT level in related foods, and is also thought of as one promising approach (Ge et al. 2017). Currently, attempts have been made to remove PAT with high efficiency from contaminated foods by inactivated microorganisms (Yue et al. 2011; Guo et al. 2012; Wang et al. 2015). Besides, live microorganisms can also absorb either by attaching PAT to their cell wall components or by active internalization and accumulation (Harwig et al. 1973; Tannous et al. 2017). A later study showed that 3 strains of Saccharomyces cerevisiae reduced PAT levels during fermentive growth but not aerobic growth. This reduction resulted in the production of 2 major products: E-ascladiol, PAT’s immediate biosynthetic precursor, and its isomer Z-ascladiol (Moss and Long 2002). However, the physical adsorption mechanism of PAT during fermentation by living microorganism cells remains unclear, which became the bottleneck of the further studies on the biodegradation of PAT, since the physical adsorption was considered as the initial step of the biodegradation process. Therefore, the aim of this work was to study the physical adsorption of PAT by yeast cells during fermentation in aqueous solution in order to establish a foundation of the further studies on the mechanisms of PAT biodegradation. Additionally, the effects of various conditions, such as fermentation time, inoculation amount, initial PAT concentration and fermentation temperature, on PAT removal efficiency by S. cerevisiae cells were also investigated.
Materials and methods
Chemicals and yeast strain
Patulin standard (HPLC grade) was purchased from Aladdin Chemistry (Shanghai, China). Immediately prior to use, weighed aliquots of the PAT were dissolved in water that had been acidified with acetic acid (pH 4) according to the method by Li et al. (2018). Snail enzyme, yeast extracts, peptone, and glucose were purchased from Beijing Dingguo Biotechnology (Beijing, China). Methanol, water, and phosphoric acid were of HPLC grade and all other reagents were of analytical grade.
Saccharomyces cerevisiae CCTCC 93161 used in this study were obtained from the Center for Type Culture Collection (Wuhan, China), were maintained viable in 20% glycerol at − 80 °C.
Preparation of yeast cells
Cells of Saccharomyces cerevisiae CCTCC 93161 were scraped from a YEPD plate (10.0 g of yeast extracts, 20.0 g of peptone, 20.0 g of glucose, 20.0 g of agar and 1.0 L of distilled water), and incubated at 30 °C on a rotary shaker at 150 r/min for 12 h according to the method by Li, et al. (2018). Then, the culture (cell density: 1.5 × 106/mL) was centrifuged for 15 min at 5000× g; and the cell pellets were collected, and washed for 3 times with normal saline for the following experiments.
Effects of different fermented conditions on PAT-removing-capacity by yeast cells
Effect of fermentation time
S. cerevisiae (10 g/L) was inoculated into 5 mL of 20% gluconate solution with the PAT initial concentration of 500 μg/L. The fermentation broth was placed in a constant temperature shaker, cultured at 30 °C (150 r/min) for different times, and then centrifuged at 6000 r/min (4 °C) for 15 min.
Effect of PAT initial concentration
S. cerevisiae (10 g/L) was inoculated into 5 mL of 20% gluconate solution with different PAT initial concentrations, and then cultured at 30 °C for 48 h with a shaking rate of 150 r/min.
Effect of fermentation temperature
S. cerevisiae (10 g/L) was inoculated into 5 mL of 20% gluconate solution with PAT initial concentration of 500 μg/L, and then cultured for 48 h with a shaking rate of 150 r/min at different fermentation temperatures.
Physical adsorption of PAT by yeast cells
Morphology of yeast cells
To determine the effects of cell walls on PAT removal, the yeast cells treated with different methods were used to observe the morphology and explore the physical adsorption mechanism of PAT by S. cerevisiae CCTCC 93161. The yeast cells were sampled at their logarithmic growth phase and centrifuged at 5000x g and 4 °C for 5 min; then, the pellets were washed for 3 times with sterilized water to remove the attached matrix (this yeast cells is labelled as Intact Cells). Another portion of intact cells were collected and resuspended in normal saline. Such samples were autoclaved at 121 °C for 20 min (this yeast cells is labelled as Inactivated Cells). A third portion of intact cells were prepared and resuspended in citrate–phosphate buffer (pH 5.4). The solution was incubated with snail enzyme (0.025 g/g wet cell weight) at 37 °C and 150 r/min for 6 h (this yeast cells is labelled as Protoplasts).These different treatment methods made the yeast cells with different cellular wall morphologies. The intact and inactivated cells had whole cell walls, while the protoplasts represented cell wall deficiency and damage.
After the above treatments, 1 mL of each solutions was sampled and centrifuged at 5000× g and 4 °C for 15 min. The pellets were washed with sterilized water for 3 times, and the morphologies of the yeast cells with different treatments were determined with a scanning electron microscope (SEM) (8010, Techcomp Ltd. China) according to the methods of Ge et al. (2017).
Fourier transform infrared spectroscopy (FT-IR)
To elucidate physical adsorption mechanism, FTIR analysis with a NEXUS 470 FTIR spectrophotometer (Nicolet, Madison, USA) equipped with a KBr beam splitter, was used to identify chemical groups of unknown composition and intensity of absorption spectra associated with the molecular composition of the chemical group. The above three types of cells (1.0 g for each) were separately added into 100 mL of acidified normal saline (pH 4.0) with the PAT concentration of 500 µg/L. All the solutions were incubated at 30 °C and 150 r/min for 48 h and then centrifuged to get cell pellets. The cell pellets were freeze-dried for FTIR analyses according to the methods of Luo et al. (2015) and Ge et al.(2017).
HPLC analysis for PAT removal efficient by yeast cells
HPLC analyses were performed as previously described by Li, et al. (2018). The column used was a 250 mm × 4.6 mm i.d., 5 μm, Symmetry C18 (Waters, Massachusetts, USA). The mobile phase was acetonitrile–water (10:90, V/V) with a flow rate of 1 mL/min, an injection volume of 20 μL, and a total run of 20 min. The efficiency of PAT removed by S. cerevisiae cells (Y) was calculated using the Eq. (1).
| 1 |
where and are the initial and final concentration of PAT (µg/L), respectively.
Results and discussion
Effects of different fermented conditions on PAT-removing-capacity by yeast cells
Effects of different fermented conditions on PAT-removing-capacity by yeast cells are shown in Fig. 1. From Fig. 1a, it was found that 53.97% of PAT was removed after 6 h of fermentation, while the PAT removal rate reached 85.88% when the systems were fermented for 24 h. Moreover, it was found that about 22.13% of PAT in the samples was removed from the even beginning of fermentation (0 h), indicating that physical adsorption occurred at the initial stage of biodegradation. After the yeast cells were added into the systems, their cellular surfaces interacted and absorbed PAT immediately, which was consistent with the findings of Du and Guo (2016). After 24 h, the biological degradation also came to play an important role in PAT removal, resulting in further increases in PAT removal. These results indicated that more PAT would be removed from the fermentation systems by both physical adsorption and biological degradation as the fermentation time was elongated.
Fig. 1.
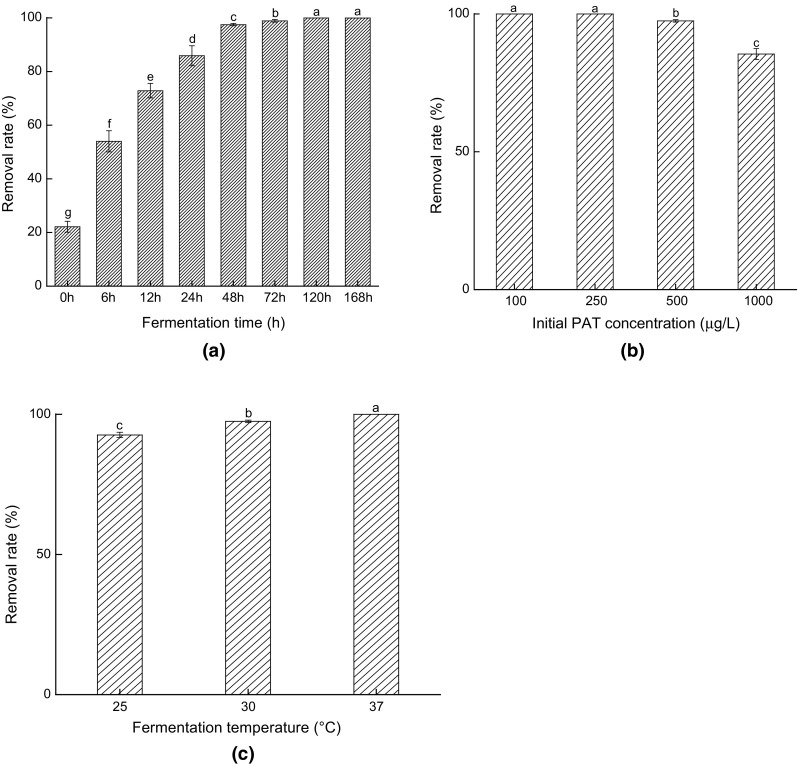
Effects of fermentation time (a), initial PAT concentration (b) and fermentation temperature (c) on PAT-removing-capacity by S. cerevisiae CCTCC 93161 (Bars with different letters are significantly different. p < 0.05)
It was found from Fig. 1b that the high concentration of PAT (> 500 μg/L) evidently declined the removal rate of such mycotoxin, and the yeast cells biodegraded all the PAT in the systems when the PAT concentration was relatively low (< 250 μg/L) after 48 h of fermentation (< 250 μg/L). Taking the results in Fig. 1a into account, it could be concluded that the physical adsorption on the cell walls and the biodegradation inside the cells were continuous procedures and these two stages would limit the removal speed and efficiency of PAT.
It was found from Fig. 1c that PAT removal rate significantly increased with the increase of temperature from 25 to 37 °C (p < 0.05). This phenomenon was primarily caused by more drastic motions of the molecules in the system under higher temperatures which increased the chances of contact of PAT with yeast cells, and thus enhanced PAT adsorption rate. On the other hand, the metabolic activity of yeast cells would also be increased with temperature (Kannamba et al. 2010; Liu et al. 2018).
SEM of different S. cerevisiae cells and their removal efficiencies of PAT
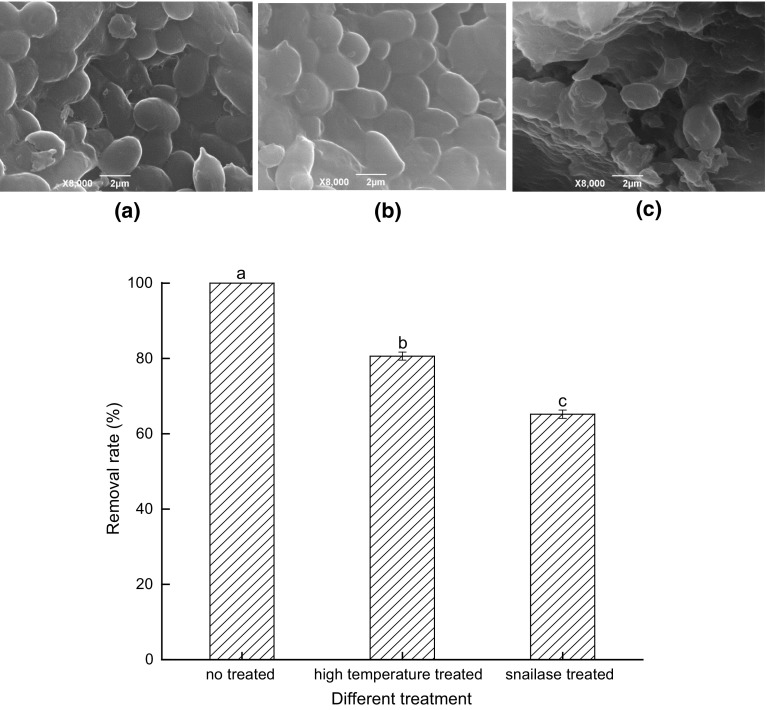
The SEM images of Intact Cells, Inactivated Cells and Protoplasts of S. cerevisiae CCTCC 93161 and their removal efficiencies of PAT are demonstrated in Fig. 2. It can be seen that there is no obvious difference between the shapes of Intact Cells and Inactivated Cells in Fig. 2a, b. The Inactivated Cells that were treated with high-temperature autoclaving, maintained the same plump ellipses as active cells. Compared with the Protoplasts in Fig. 2c, the cell walls of Inactivated Cells (Fig. 2b) were intact after high-temperature autoclaving, while regional indentations were observed on the cellular surfaces of Protoplasts, indicating that the cell walls were successfully removed and thus that the cells were soft and elastic.
Fig. 2.
SEM images of S.cerevisiae CCTCC 93161 cells with different treatments at 8000 × magnification and their removal efficiencies of PAT, Intact Cells (a), Inactivated Cells (b) and Protoplasts (c). Bars with different letters are significantly different at p < 0.05 according to Duncan’s multiple range test
It can be seen from Fig. 2 that there were significant differences in the PAT removal rate between the three types of S. cerevisiae yeast cells. Intact Cells removed all the PAT in the system after 48 h, while the removal rates of Inactivated Cells and Protoplasts were significantly lower (p < 0.05), leading to 19.40% and 34.83% of the PAT remaining in the system, respectively. The Protoplasts exhibited a removal ability for PAT accounting for over 65% of that of the Intact Cells. These results indicated that PAT removal was introduced by both the cellular walls and protoplasts of yeast cells. Thus, from this study it was concluded that the yeast cells during fermentation processes removed PAT through both of the physical adsorption and biological degradation. Some studies have reported that some microorganisms such as yeasts and lactic acid bacteria remove PAT through physical adsorption by functional groups on the cellular walls (Luo et al. 2015; Ge et al. 2017) and biodegradation by enzymes inside the cells (Ricelli et al. 2007), which agreed with the current findings from S. cerevisiae CCTCC 93161.
FTIR analyses of differently treated S. cerevisiae cells
The infrared absorption spectra of differently treated S. cerevisiae cells before and after the adsorption of PAT are summarized in Table 1. As shown in Table 1, the strong and broad absorption peak at 3415.37 cm−1 was attributed to the stretching vibrations of O–H and N–H bonds. The absorption peak at 2925.53 cm−1 resulted from the stretching vibrations of C–H bonds in lipids and proteins. Those peaks at 1654.65 cm−1, 1542.80 cm−1 and 1241.95 cm−1 were due to the stretching vibrations of C=O bonds in the protein amide I bands, the bending vibrations of N–H bonds in the protein amide II band and the C–N stretching vibrations in the protein amide III band, respectively. The peak at 1049.10 cm−1 was from the stretching vibrations of C=O bonds in the polysaccharides, and the peak at 528.41 cm−1 was the characteristic absorption peak of S. cerevisiae. Moreover, it was found that the IR spectra of Intact Cells were shifted upon PAT adsorption. The peak for O–H/N–H shifted to 3403.80 cm−1, indicating that O–H/N–H had a function in PAT adsorption. The peaks at 1654.65 cm−1 and 1542.80 cm−1 were shifted to 1656.58 cm−1 and 1546.66 cm−1, respectively, suggesting that the functional groups on the proteins participated in the PAT adsorption. The peak at 1049.10 cm−1 also decreased to 1047.18 cm−1, which suggested that the C=O in polysaccharides also contributed to PAT adsorption. With respect to Inactivated Cells after PAT absorption, the peaks at 3411.51 cm−1, 1658.51 cm−1, and 1537.01 cm−1 were shifted to 3396.09 cm−1, 1654.65 cm−1, and 1540.87 cm−1, respectively, indicating that the O–H/N–H and C=O bonds in amide I band and the N–H bonds in amide II participated in PAT adsorption. Compared with the Intact Cells, it is noted that all the peaks except for those of the C–H bond and the C–N stretching vibrations in the amide III band were shifted regardless of PAT adsorption, which likely resulted from denaturation of proteins and exposure of more functional groups after high-temperature treatments. Compared with Intact Cells, it was observed that there were also significant changes in the infrared spectra of Protoplasts of S. cerevisiae before PAT adsorption. The C=O bonds in the amide I band and the polysaccharides changed from 1654.65 cm−1 and 1049.10 cm−1 to 1658.51 cm−1 and 1047.18 cm−1, respectively, which was likely due to removal of the cell walls. After PAT adsorption, the C–H and C=O bonds of the amide I band remained unchanged, while the other peaks shifted towards smaller wavenumbers. The results from FTIR clearly demonstrated that the O–H/N–H bonds of proteins and polysaccharides in yeast cell walls participated in the physical adsorption of PAT. Similar observations were reported in previous studies (Guo et al. 2012, Yuan et al. 2014).
Table 1.
FTIR bands observed in different S. cerevisiae CCTCC 93161 cells before and after PAT adsorption
| Functional groups | Wave numbers (cm−1) | ||
|---|---|---|---|
| Non-treated cells | High-temperature-treated cells | Snailase-treated cells | |
| O–H/N–H stretching | 3415.37 (3403.80) | 3411.51 (3396.09) | 3392.23 (3380.66) |
| C–H stretching | 2925.53 (2925.53) | 2925.53 (2925.53) | 2925.53 (2925.53) |
| C=O stretching in amide I | 1654.65 (1656.58) | 1658.51 (1654.65) | 1658.51 (1658.51) |
| N–H stretching in amide II | 1542.80 (1546.66) | 1537.01 (1540.87) | 1542.80 (1537.01) |
| C–N stretching in amide III | 1241.95 (1241.95) | 1241.95 (1241.95) | 1241.95 (1240.03) |
| C–O stretching in polysaccharides | 1049.10 (1047.18) | 1047.18 (1047.18) | 1047.18 (1047.18) |
| C–Br stretching in alkyl halide | 528.41 (530.34) | 578.55 (576.62) | – (–) |
() The infrared characteristic peaks of yeast cells after PAT adsorption
Conclusion
This paper focused on the physical adsorption of PAT by S. cerevisiae CCTCC 93161 during fermentation. In order to elucidate the effects of fermentation conditions on PAT-removing-capacity of yeast cells during fermentation, a series of experiments were performed. The results showed that the efficiency of PAT removal raised significantly with the increase of fermentation temperature and time, whereas it decreased significantly with the increase of initial PAT concentration in fermentation system. It was also found that yeast cells removed PAT by physical adsorption through interactions the between O–H/N–H bonds of proteins and polysaccharides in cell walls and PAT.
Acknowledgements
This research was partially supported by the National Natural Science Foundation of China (No. 31671850) and Natural Science Foundation of Hubei Province of China (No. 2017CFB599).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Du J, Guo C. Adsorption of patulin from apple juice by waste beer yeast. Food Sci. 2016;37(5):56–61. [Google Scholar]
- Ge N, Xu J, Li F, Peng B, Pan S. Immobilization of inactivated microbial cells on magnetic Fe3O4@CTS nanoparticles for constructing a new biosorbent for removal of patulin in fruit juice. Food Control. 2017;82:83–90. doi: 10.1016/j.foodcont.2017.06.027. [DOI] [Google Scholar]
- Guo C, Yue T, Hatab S, Yuan Y. Ability of inactivated yeast powder to adsorb patulin from apple juice. J Food Protect. 2012;75:585–590. doi: 10.4315/0362-028X.JFP-11-323. [DOI] [PubMed] [Google Scholar]
- Harwig J, Scott PM, Kennedy BPC, Chen YK. Disappearance of patulin from apple juice fermented by Saccharomyces spp. Can I Food Sci Tech J. 1973;6(1):45–46. doi: 10.1016/S0315-5463(73)73965-1. [DOI] [Google Scholar]
- Kannamba B, Reddy KL, AppaRao BV. Removal of Cu(II) from aqueous solutions using chemically modified chitosan. J Hazard Mater. 2010;175:939–948. doi: 10.1016/j.jhazmat.2009.10.098. [DOI] [PubMed] [Google Scholar]
- Li M, Chen W, Zhang Z, Zhang Z, Peng B. Fermentative degradation of patulin by saccharomyces cerevisiae in aqueous solution. LWT Food Sci Tech. 2018;97:427–433. doi: 10.1016/j.lwt.2018.07.040. [DOI] [Google Scholar]
- Liu B, Peng X, Meng X. Effective biodegradation of mycotoxin patulin by porcine pancreatic lipase. Front Microbiol. 2018;9:615. doi: 10.3389/fmicb.2018.00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Wang J, Liu B, Wang Z, Yuan Y, Yue T. Effect of yeast cell morphology, cell wall physical structure and chemical composition on patulin adsorption. PLoS one. 2015;10(8):e0136045. doi: 10.1371/journal.pone.0136045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss MO, Long MT. Fate of patulin in the presence of the yeast Saccharomyces cerevisiae. Food Addit Contam A. 2002;19:387–399. doi: 10.1080/02652030110091163. [DOI] [PubMed] [Google Scholar]
- Neri F, Donati I, Veronesi F, Mazzoni D, Mari M. Evaluation of Penicillium expansum isolates for aggressiveness growth and patulin accumulation in usual and less common fruit hosts. Food Microbiol. 2010;143:109–117. doi: 10.1016/j.ijfoodmicro.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Oteiza JM, Khaneghah AM, Campagnollo FB, Granato D, Mahmoudi MR, Sant’Ana AS. Influence of production on the presence of patulin and ochratoxin A in fruit juices and wines of Argentina. LWT Food Sci Tech. 2017;80:200–207. doi: 10.1016/j.lwt.2017.02.025. [DOI] [Google Scholar]
- Ricelli A, Baruzzi F, Solfrizzo M, Morea M, Fanizzi FP. Biotransformation of patulin by Glucono bacteroxydans. Appl Environ Microbiol. 2007;73:785–792. doi: 10.1128/AEM.02032-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard JL. Some major mycotoxins and their mycotoxicoses—an overview. Int J Food Microbiol. 2007;119(1–2):3–10. doi: 10.1016/j.ijfoodmicro.2007.07.019. [DOI] [PubMed] [Google Scholar]
- Tannous J, Snini SP, El KR, Canlet C, Pinton P, Lippi Y. Patulin transformation products and last intermediates in its biosynthetic pathway, E- and Z ascladiol, are not toxic to human cells. Arch Toxicol. 2017;9:2455–2467. doi: 10.1007/s00204-016-1900-y. [DOI] [PubMed] [Google Scholar]
- Wang L, Yue T, Yuan Y, Wang Z, Ye M, Cai R. A new insight into the adsorption mechanism of patulin by the heat-inactive lactic acid bacteria cells. Food Control. 2015;50:104–110. doi: 10.1016/j.foodcont.2014.08.041. [DOI] [Google Scholar]
- Yuan Y, Wang X, Hatab S, Wang Z, Wang Y, Luo Y. Patulin reduction in apple juice by inactivated Alicyclobacillus spp. Lett Appl Microbiol. 2014;59:604–609. doi: 10.1111/lam.12315. [DOI] [PubMed] [Google Scholar]
- Yue T, Dong Q, Guo C, Worobo RW. Reducing patulin contamination in apple juice by using inactive yeast. J Food Prot. 2011;74:149–153. doi: 10.4315/0362-028X.JFP-10-326. [DOI] [PubMed] [Google Scholar]