Abstract
Background
LncRNAs have been found to be involved in various aspects of biological processes. In this study, we aimed to uncover the molecular mechanisms of lncRNA EPB41L4A-AS1 in regulating glycolysis and glutaminolysis in cancer cells.
Methods
The expression of EPB41L4A-AS1 in cancer patients was analyzed in TCGA and GEO datasets. The level of cellular metabolism was determined by extracellular flux analyzer. The relationship between p53 and EPB41L4A-AS1 was explored by qRT-PCR, luciferase assay and ChIP assay. The interactions between EPB41L4A-AS1 and HDAC2 or NPM1 were determined by RNA immunoprecipitation, RNA pull-down assay and RNA-FISH- immunofluorescence.
Findings
EPB41L4A-AS1 was a p53-regulated gene. Low expression and deletion of lncRNA EPB41L4A-AS1 were found in a variety of human cancers and associated with poor prognosis of cancer patients. Knock down EPB41L4A-AS1 expression triggered Warburg effect, demonstrated as increased aerobic glycolysis and glutaminolysis. EPB41L4A-AS1 interacted and colocalized with HDAC2 and NPM1 in nucleolus. Silencing EPB41L4A-AS1 reduced the interaction between HDAC2 and NPM1, released HDAC2 from nucleolus and increased its distribution in nucleoplasm, enhanced HDAC2 occupation on VHL and VDAC1 promoter regions, and finally accelerated glycolysis and glutaminolysis. Depletion of EPB41L4A-AS1 increased the sensitivity of tumor to glutaminase inhibitor in tumor therapy.
Interpretation
EPB41L4A-AS1 functions as a repressor of the Warburg effect and plays important roles in metabolic reprogramming of cancer.
Keywords: EPB41L4A-AS1, HDAC2, Glycolysis, Glutaminolysis, Cancer metabolism
Research in context.
Evidence before this study
High rate of glycolysis and glutaminolysis are common phenomena in cancers to maintain tumor cell proliferation. Recently, accumulating data show that lncRNAs function as crucial regulators of glycolysis and glutaminolysis. LncRNA EPB41L4A-AS1 can translate a small peptide and its ectopic expression inhibits the colony-formation of tumor cells and tumor growth in soft agar. The clinical data and gene expression information of cancer patients were from TCGA and GEO (GSE44001).
Added value of this study
We indicated lncRNA EPB41L4A-AS1, a p53-regulated gene, was downregulated in cancers through analysis of RNA expression profiles of different types of cancer from TCGA. Clinically, the expression of EPB41L4A-AS1 was associated with poor prognosis. EPB41L4A-AS1 could alter glycolysis and glutaminolysis levels of cancer cells. Furthermore, EPB41L4A-AS1 knockdown plus glutaminase inhibitor suppressed tumor progression both in vitro and in vivo. Mechanistically, we demonstrated that EPB41L4A-AS1 interacted and colocalized with HDAC2 and NPM1 in nucleolus, controlling HDAC2 occupation and H3k27ac modification levels on VHL and VDAC1 promoter regions and regulating their expression, which affected glycolysis and glutaminolysis.
Implications of all the available evidence
EPB41L4A-AS1 regulates glycolysis and glutaminolysis by altering the nucleolar translocation of HDAC2. Our work provides new insights in understanding the mechanisms of lncRNAs regulating glycolysis and glutaminolysis and altering nucleolar translocation of chromatin regulators. Characterization and analysis of EPB41L4A-AS1 revealed possibilities of EPB41L4A-AS1 acts as prognostic predictor or targeted treatment combination with glutaminase inhibitors.
Alt-text: Unlabelled Box
1. Introduction
As a pioneer of cancer metabolism, Otto Warburg and his co-workers found that cancer cells metabolize more glucose by glycolysis, producing lactate as an end product, despite their high oxygen environment. This phenomenon is known as the Warburg effect [1,2], confirmed by recent clinical evidences through FDG-PET imaging [3,4]. Recently, the high rate of glutaminolysis is also reported as a hallmark of cancer metabolism and links to the Warburg effect because of its association with glycolysis [5]. In order to maintain cell proliferation and survival under glucose deprivation, glutamine-derived TCA cycle increases [6]. GLS1 is a glutaminase which converts glutamine to glutamate. GLS1 involves the transcriptional inhibition of TXNIP (thioredoxin interacting protein), resulting in reduced glucose uptake and glycolysis [7]. Warburg effect is considered as the result of a passive metabolic reprogramming to supply the higher energy requirement and macromolecular synthesis in cancer cells [[8], [9], [10], [11], [12]]. Identifying the genes in regulating cancer metabolism and their biological pathways is a major challenge in understanding cancer metabolic reprogramming. Recently, increasing evidences demonstrate that some oncogenes or tumor suppressors are involved in the metabolic reprogramming of cancer cells [8,9]. It has been reported that both MYC and p53 regulate glycolysis and glutamine metabolism. The oncogene c-Myc stimulates glycolysis and alters mitochondrial metabolism through exogenous glutamine metabolism via regulating ASCT2, GLS1 and GLS2 expression [13,14]. The tumor suppressor p53 and its mutations regulate many aspects of metabolism, including glycolysis, glutaminolysis, mitochondrial oxidative phosphorylation, pentose phosphate pathway, fatty acid synthesis and oxidation, to maintain the homeostasis of cellular metabolism [15,16].
It has been reported that lncRNAs, a class of ncRNA longer than 200nt, regulate glycolysis and glutamine metabolism [[17], [18], [19]]. For instance, the androgen-induced prostate-specific lncRNA PCGEM1 (prostate cancer gene expression marker 1) regulates glycolysis and glutamine metabolism pathways via c-Myc [18]. LncRNA CCAT2 (colon cancer associated transcript 2) mediates cellular glucose and glutamine metabolism by regulating the alternative splicing of glutaminase [19]. LincRNA-p21 and PVT1, the downstream genes of p53, also regulate metabolic reprogramming in cancer cells [[20], [21], [22], [23]]. These studies indicate that lncRNAs play critical roles in the link between glycolysis and glutamate metabolism.
Here we report that lncRNA EPB41L4A-AS1 regulates both glycolysis and glutamine metabolism. EPB41L4A-AS1 locates in the 5q22.2 region of the genome, which is closely associated with tumorigenesis because of the occurrence of frequent DNA fragment deletion. However, the role of EPB41L4A-AS1 in cancer is still unknown. In this study, we show that EPB41L4A-AS1 is a p53 and PGC-1α inducible gene and its low expression is frequently related to poor prognosis in many human cancers. EPB41L4A-AS1 knockdown accelerates aerobic glycolysis and glutamine metabolism. Glutamine metabolism dependency induced by EPB41L4A-AS1 silencing enhances the efficacy of glutaminase inhibitor in tumor therapy. Therefore, our study provides new insights for Warburg effect and p53 mediated regulation of tumor metabolism.
2. Materials and methods
2.1. Cell lines and transfection
HeLa (ATCC, CCL-2) and HepG2 (ATCC, HB-8065) cells were cultured in modified Eagle's medium (Thermo Fisher Scientific, Cat.No: 12100046), supplemented with 10% FBS (Biowest, Cat.No: S1810) and 10 U/ml penicillin/streptomycin in 5% CO2-humidified incubator at 37 °C. Plasmids carrying EPB41L4A-AS1 RNAi were purchased from Shanghai Genechem Co., Ltd. (Shanghai, China). To obtain EPB41L4A-AS1 stable knockdown cell line, HeLa cells were transfected with ShEPB41L4A-AS1 plasmid, the empty vector GV248 was used as negative control, followed by 1 μg/ml final concentration of puromycin (Thermo Fisher Scientific, Cat.No: A1113803) selection. For transient transfection, EPB41L4A-AS1 Plasmid or siRNAs were transfected using Lipofectamine 3000 as described by the manufacturer protocol.
2.2. Colony formation assay
Cells with EPB41L4A-AS1 stable knockdown were plated at a concentration of 5 × 103 cell per well in 6-well. GV248 plasmid was used as control and the cells were plated as well. The glutaminase inhibitor compound 968 (SIGMA-ALDRICH, Cat.No: SML1327) was added to the cells at a final concentration of 10 nM every three days after cell adhesion. Ten days later, cells were fixed with 1% formaldehyde and stained with 1% crystal violet.
2.3. Bioinformatics analysis
Clinical data and gene expression information were downloaded from UCSC Xena or GEO (gene expression omnibus) database. Mann-Whitney test was used to compare two groups of clinical patients' genes expression. Kaplan-Meier analysis was performed for survival analysis. GO (Gene Ontology) enrichment was performed to analyze TIGA1 binding proteins. GSEA (Gene Enrichment Analysis) was performed using GSEA 2–2.2.2 software and “h.al.v5.1.symbols.gmt” gene sets. Chromosomal copy number aberrations were analyzed by Progenetix website. Expression correlation between EPB41L4A-AS1 and p53 or VDAC1 in cervical and liver cancers was examined by Spearman correlation test.
2.4. Seahorse assay
XFp Extracellular Flux Analyzer (Seahorse Bioscience) was used to determine ECAR (extracellular acidification rate), OCR (oxygen consumption rate) and mitochondria fuels dependency. Cells were plated in 8-well (8000 cells/well) Seahorse Assay plates and incubated overnight. Sensor cartridge was hydrated at 37 °C in CO2-free incubator overnight. ECAR was determined using ECAR determination kit (Agilent Technologies, Cat.No: 103017). OCR was analyzed using OCR determination kit (Agilent Technologies, Cat.No: 103010–100). Mitochondria fuels dependency was measured using seahorse XFp mito fuel flex test kit (Agilent Technologies, Cat.No: 103270–100). All measurements were normalized with total protein concentration on each well. Each assay was run in triplicate per each condition.
2.5. Measurements of glucose, lactate, glutamate, α-KG and ATP levels
Glucose was determined by mixing culture medium with glucose detection mix, the absorbance was read at 460 nm wavelength (Biosino Bio-Technology and Science Incorporation, Cat.No: 00002010). Lactate levels were measured by Lactate Colorimetric/Fluorometric Assay Kit (BioVision, Cat.No: K607–100) as described by the manufacturer protocol. To determined intercellular glutamate and α-KG (alpha-ketoglutarate) levels, the cells were harvested and diluted in ice-cold RIPA buffer for 20 min. Glutamate level was determined using Glutamic Acid/Glutamate Oxidase Assay Kit (Thermo Fisher Scientific, Cat.No: A12221). ATP level was determined using ATP Determination Kit according to the manufacturer protocol (Thermo Fisher Scientific, Cat.No: A22066).
2.6. Chromatin immunoprecipitation
Cells were fixed using 1% formaldehyde and harvested on ice with ChIP lysis buffer (50 mM Tris-HCl ph 8.0, 5 mM EDTA, 0.1% deoxycholated, 1% Triton X-100, 150 mM NaCl and proteinase inhibitor). Subsequently, cells were sonicated and the supernatant was collected and incubated with dynabeads protein G and primary antibody, IgG was used as negative control. After 2 h, the complex was washed three times and DNA was purified and condensed. Finally, the DNA fraction was analyzed by qRT-PCR.
2.7. Co-immunoprecipitation
HeLa cells were transfected with TIGA1 plasmid and the empty vector pEGFP-C1 for 48 h, the cells were harvested and dissolved in RIPA buffer on ice, then the supernatant was transferred to dynabeads protein G-antibody (anti-GFP, Abcam, ab290) complex. The complex was washed 3 times with 200 μl washing buffer and loading buffer was added to elute the samples. The supernatant was transferred to a new Eppendorf for western blot or mass spectra analysis.
2.8. Real-time quantitative PCR
Total RNA was isolated using RNA iso Plus (Takara, Cat. No: D9108B). Reverse transcription of the isolated total RNA was performed using ReverTra Ace qPCR RT Kits (TOYOBO, Cat.No: FSQ-101). Real-time qRT-PCR was carried out by SYBR® Green Real time PCR Master Mix (TOYOBO, Cat.No: QPK-201). Samples were repeated in triplicate, data were normalized to ACTB and 2-ΔΔCt was used to calculate the fold expression.
2.9. Western blot
Proteins were extracted from cultured cells or animal tissues by cell extract buffer (50 mM Tris-HCl, pH 8.0, 4 M urea and 1% Triton X-100) with protease inhibitor mixture (Roche Diagnostics, Cat.No: 04693132001). Cell lysates were resolved by SDS-PAGE and then analyzed by western blotting. The following antibodies were used: ACTB (Proteintech, #20536-1-AP, 1:5000), PFKL (Cell Signaling Technology, #8175, 1:1000), PFKFB3 (Cell Signaling Technology, #13123, 1:1000), HK2 (Cell Signaling Technology, #2867, 1:1000), pyruvate dehydrogenase (Cell Signaling Technology, #3205, 1:1000), SDHA (Cell Signaling Technology, #11998, 1:1000), VDAC (Cell Signaling Technology, #4661, 1:1000), ATF4 (Cell Signaling Technology, #11815, 1:1000), P-eIF2α (Cell Signaling Technology, #3398, 1:1000), α-tubulin (SIGMA-ALDRICH, T9026, 1:500), SLC38A5 (abcam, ab72717, 1:1000), TIGA1 (Santa Cruz, sc-244,315, 1:200), PGC-1a (Cell Signaling Technology, #2178, 1:1000), P53 (Millipore, 17-613, 1:1000), SN2 (abcam, ab72717, 1:1000), HIF-1a (BD Pharmingen, 565,924, 1:200), H3K27ac (abcam, ab4729,1:1000), Anti-Rabbit IgG (KPL, 074-1506, 1:5000), Anti-mouse IgG (KPL, 074-1806, 1:5000), Anti-Goat IgG (KPL, 14-13-06, 1:5000).
2.10. FISH and immunofluorescence
RNA-FISH was performed with EPB41L4A-AS1 specific probe (Biosearch Technologies). Cells were fixed with 3% paraformaldehyde for 10 min and then incubated with EPB41L4A-AS1 probe overnight at 37 °C. Then the cells were washed and blocked by 3% BSA. Subsequently, incubating with HDAC2 antibody (Abcam, ab219054) for 1 h. Then cells were washed and incubated with rabbit secondary antibodies conjugated with Alexa Fluor® 488. After that, the cells were stained with DAPI and imaged with an Olympus FV1000 confocal microscope.
2.11. Immunohistochemistry
Anti-TIGA1 antibody (Proteintech, 24,698–1-AP, 1:1500) was used for immunohistochemical staining of cervical and liver tumor tissue. Slides were scanned using microscope Olympus BX50 (Olympus, Aartselaar, Belgium). Staining intensity was converted to TIGA1 expression levels using a protocol as previously described [24]. Briefly, two observers (PD and RM) examined the spots independently and blindly to both pathological and clinical data. Cytoplasmic immunopositivity was measured as follows: Positive staining rate was evaluated using scores from 0 to 3: 0, none; 1, 1–30%; 2, 31–60%; 3, >60%). Intensity was also evaluated using scores from 0 to 3: 0, none; 1, weak staining; 2, moderate staining; 3, strong staining). Combination of extent (E) and intensity (I) of staining was obtained. – means score 0; + means score 1–3, ++ means score 4–6, +++ means score 7–9.
2.12. RNA-pulldown assay
Different fractions of EPB41L4A-AS1 were construction with pcDNA3.1 plasmid containing T7 promoter. The plasmids were linearized and then transcription in vitro with MAXIscript™ T7 transcription kit (Thermos fisher, AM1312). After transcription, the RNA fractions were added poly(A) with Poly(A) tailing kit (Thermos fisher, AM1350). The antisense of EPB41L4A-AS1 is the gene of EPB41L4A, therefore the same amount of non-poly(A) RNA fractions were used as negative control. The RNA was precipitation and incubated with cells lysis and Dynabeads Oligo (dT) 25 for 2 h at 4 °C. Washing and harvesting the samples and analysis with western blot.
2.13. RNA immunoprecipitation assay
Cells were harvested and lysed with polysome lysis buffer (10 mM KCl, 5 mM MgCl2, 10 mM HEPES pH 7.0, 0.5% NP-40, 1 mM DTT, 100 U/ml RRI, 20 μl/ml protein inhibitor calculator, 2 mM vanadyl ribonucleotide complex solution). Then incubated with 2 μg HDAC2, HDAC1 (Abcam, ab7028) or p300 (Abcam, ab14984) antibody overnight at 4 °C. Then washed the non-specifically binding antibodies and incubated with Dynabeads protein A for 4 h at 4 °C. Then washed and precipitated the RNA with ethanol and sodium acetate. The RNA fraction was reverse transcription and analyzed with qRT-PCR.
2.14. Animal experiment
Four weeks old athymic nude mice were purchased from the Experimental Animal Center of Guang Zhou University of Chinese Medicine (Guang Zhou, China) and were acclimated to the animal facilities for 1 week before treatment. Mice were randomly divided into two groups, 7 mice for each group. 5 × 105 HeLa cells with EPB41L4A-AS1 stable knockdown or control cells were subcutaneous injected into flank of mice. After 13 days, when tumor growth reached a volume of approximately 75 mm3, compound 968 (100 μg per mice) or vehicle (1% of DMSO in PBS) as treatment control was intraperitoneally injected every 2 days. Mice were sacrificed 4 weeks after the injection of cancer cells, and tumors weight was analyzed. EPB41L4A-AS1 and TIGA1 expression in tumors was determined by qRT-PCR and western blot.
2.15. Statistical analysis
Data plotting were performed by Prism Graph Pad 6.0 and statistical analysis by SPSS. A value of P < 0.05 was considered statistically significant. Unpaired, two-tail Student's t-test, Z test, Log-Rank test or Mann-Whitney test was used to compare the results between two groups.
3. Results
3.1. EPB41L4A-AS1 is down-regulated in some human cancers
EPB41L4A-AS1 is a lncRNA located within the frequently deleted region across 475 cancer sample sets surveyed: 5q22.2 (Fig. 1A). Deletion of EPB41L4A-AS1 gene occurred in a variety of human cancers (Supplementary Fig. 1A). We investigated the clinical significance of EPB41L4A-AS1 in human cancers. The low expression of EPB41L4A-AS1 was associated with poor survival in several cancer types, including cervix, liver, breast, bladder and other cancers (Fig. 1B; Supplementary Fig. 1B). The first exon of EPB41L4A-AS1 gene also translates a peptide with 120 amino acid residues, named TIGA1 (Supplementary Fig. 1C). The immunohistochemical analysis from 125 cervical and 92 liver cancer patients revealed that the protein level of TIGA1 was also down regulated in both cervical and liver cancer tissues, compared with adjacent normal tissues (Fig. 1C and D).
Fig. 1.
EPB41L4A-AS1 expression was downregulated in human cancers. A. Analysis the copy numbers of EPB41L4A-AS1 across all chromosomes from 475 cancer samples by the Progenetix histoplot. B. EPB41L4A-AS1 is significantly downregulated in cervical, liver, breast and bladder cancer compared with normal tissues (upper panels). Kaplan-Meier survival curves analyzing EPB41L4A-AS1 expression in these four types of cancer tissues (lower panels). C-D. Immunohistochemical staining of TIGA1 in cervical cancer (C) and liver cancer D) tissues. Quantitative analysis of TIGA1 intensity in 125 cervical cancer patients (C, right) and 92 liver cancer patients (D, right). –, score 0; +, score 1–3; ++, score 4–6; +++, score 7–9. Data are represented as means ± SD, *P < 0.05; **P < 0.01; ***P < 0.001, Mann-Whitney test.
3.2. The expression of EPB41L4A-AS1 is regulated by p53 and PGC-1α
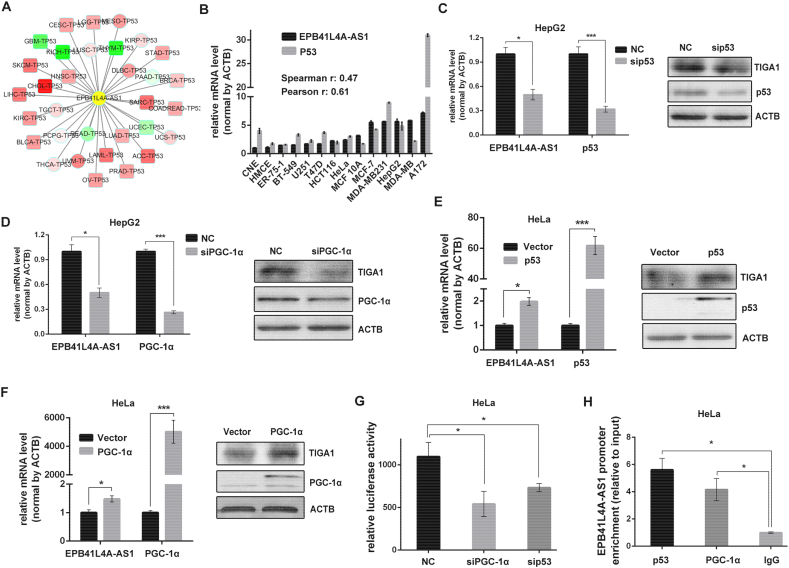
In the gene co-expression network, the expression of EPB41L4A-AS1 and p53 was positively correlated in most types of human cancers, indicating that p53 may regulate EPB41L4A-AS1 expression (Fig. 2A). The result of qPCR from 14 different cell lines demonstrated a positive correlation between EPB41L4A-AS1 and p53 expression (Fig. 2B). It has been reported that TIGA1 is a mitochondrial membrane protein [25], therefore, we wondered if PGC-1α, a transcriptional coactivator of energy metabolism would regulate EPB41L4A-AS1 expression. We knocked down p53 or PGC-1α in HepG2 cells expressing wild-type p53, both siRNAs reduced EPB41L4A-AS1 expression (Fig. 2C and D). Then we overexpressed GFP-p53 or GFP-PGC-1α in HeLa cells with p53 deficiency, overexpression of p53 or PGC-1α increased the level of EPB41L4A-AS1 (Fig. 2E and F). We next analyzed whether p53 and PGC-1α transcriptionally regulated EPB41L4A-AS1 expression. EPB41L4A-AS1 promoter, the 781bp nucleotides of EPB41L4A-AS1 upstream fragment, was cloned into pGL3-enhancer luciferase reporter. When the luciferase reporter was co-transfected with sip53 or siPGC-1α into HepG2 cells, the luciferase activity was markedly reduced (Fig. 2G). ChIP (chromatin immunoprecipitation) assay also revealed that both p53 or PGC-1α could bind to EPB41L4A-AS1 promoter (Fig. 2H). Together, these results suggested that EPB41L4A-AS1 expression was transcriptionally regulated by p53 and PGC-1α.
Fig. 2.
EPB41L4A-AS1 expression was regulated by p53 and PGC-1α.
A. Correlation between p53 mRNA and EPB41L4A-AS1 expression in different types of cancer. The -Spearman correlation coefficient is shown as color intensity, red indicates EPB41L4A-AS1 positive relevant to p53 and green indicates negative correlation. The square frame indicates P < 0.05 and circles indicates P ≧ 0.05. B. Correlation between p53 mRNA and EPB41L4A-AS1 expression in 14 different cancer cell lines by qRT-PCR (n = 3). C-D. EPB41L4A-AS1 and TIGA1 expression in HepG2 cells depleted with p53 or PGC-1α (n = 3). E–F. EPB41L4A-AS1 and TIGA1 expression in HeLa cells transfected with GFP-p53 or GFP-PGC-1α plasmids (n = 3). G. PGL3-enhancer vector containing EPB41L4A-AS1 promoter was co-transfected with NC, sip53 or siPGC-1α into HepG2 cells, relative luciferase activity was determined by bioluminescence (n = 3). H. HeLa cells transfected with GFP-p53 or GFP-PGC-1α for 48 h, p53 or PGC-1α occupation on EPB41L4A-AS1 promoter was evaluated by ChIP-qPCR, IgG was used as negative control (n = 3). Data are represented as means ± SD, *P < 0.05; **P < 0.01, ***P < 0.001, unpaired, two-tailed, Student's t-test. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. EPB41L4A-AS1 regulates glycolysis through VHL/HIF-1α pathway
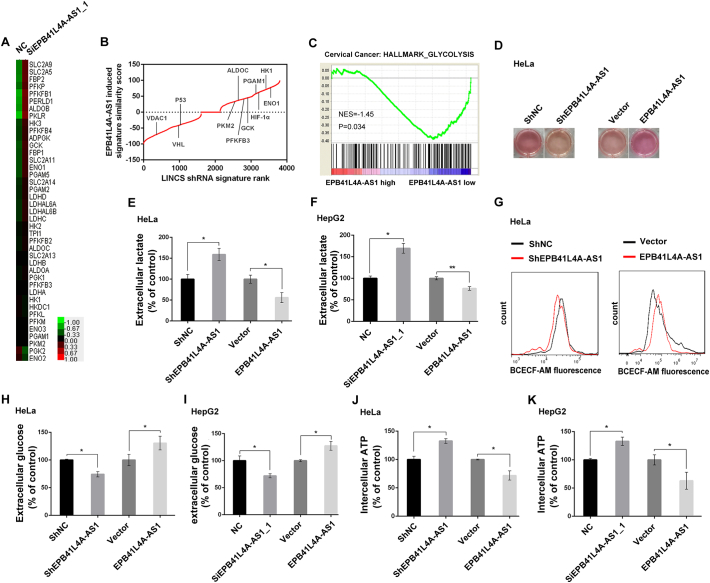
To understand the potential role of EPB41L4A-AS1 in cancer, we performed microarray analysis. Knockdown of EPB41L4A-AS1 resulted in the upregulation of several enzymes related to glycolysis, such as hexokinase and phosphofructokinase (Fig. 3A; Supplementary Fig. 2A; Table S1). The genes with >2-fold change in EPB41L4A-AS1 knockdown cells were defined as EPB41L4A-AS1 regulated signature. We used LINCS (Library of Integrated Network-based Cellular Signatures) database to evaluate the gene signature patterns. Knockdown of glycolysis related genes, including PKM2, PFKFB3, GCK, ENO1, HIF-1α, PGAM1, HK1, and ALDOC resulted in gene signature patterns positively correlated to EPB41L4A-AS1 regulated signature, while knockdown of p53, VDAC1, and VHL resulted in gene signature patterns inversely correlated with that of EPB41L4A-AS1 regulated signature (Fig. 3B). GSEA (gene set enrichment analysis) also confirmed that genes related to glycolysis were markedly enriched in cervical cancer patients with EPB41L4A-AS1 low expression (Fig. 3C), indicating that EPB41L4A-AS1 may negatively regulate glycolysis. Interestingly, we found that 10 glycolytic enzymes were upregulated in most of cervical cancer patients, the patients with alterations of these genes showed a significant poor survival ratio, compared with patients without these alterations (Supplementary Fig. 2B and C).
Fig. 3.
EPB41L4A-AS1 regulates glucose metabolism. A. Heatmap analysis of cDNA microarray of glycolysis related genes expression in EPB41L4A-AS1 knockdown HeLa cells. B. Evaluation of the EPB41L4A-AS1 signature in the LINCS. In the plot shown, genes are ranked according to the similarity score in their induced expression patterns with those of the EPB41L4A-AS1 signature. C. GSEA enrichment score curve shows glycolysis related to low expression of EPB41L4A-AS1 in 300 cervical carcinoma patients (GEO: GSE44001). The green curve indicates the enrichment score (ES). The negative enrichment score in EPB41L4A-AS1 low expression end indicates up-regulation of glycolysis pathway in the samples with EPB41L4A-AS1 low expression. D. HeLa cells with EPB41L4A-AS1 stable knockdown or transient overexpression were cultured for 48 h. The culture medium was more acidified in EPB41L4A-AS1 knockdown cells and less in EPB41L4A-AS1 overexpressing cells, as indicated by the colors. E-F. Extracellular lactate levels were measured in HeLa (E) and HepG2 (F) cells with EPB41L4A-AS1 knockdown or overexpression (n = 3). G. Intercellular pH by flow cytometry using a pH-sensitive dye (BCECF-AM) in HeLa cells after EPB41L4A-AS1 stable knockdown (left) and overexpression (right) for 48 h. H—I. Extracellular glucose levels were measured in HeLa (H) and HepG2 (I) cells with EPB41L4A-AS1 knockdown or overexpression (n = 3). J-K. Intracellular ATP levels were measured in HeLa (J) and HepG2 (K) cells with EPB41L4A-AS1 knockdown or overexpression (n = 3). Data are represented as means ± SD, *P < 0.05; **P < 0.01, unpaired, two-tailed, Student's t-test. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Next, we investigated the effect of EPB41L4A-AS1 on metabolism in cancer cells. Stable knockdown of EPB41L4A-AS1 changed the culture medium color from red to yellow due to medium acidification, while EPB41L4A-AS1 overexpression showed the opposite effect (Fig. 3D; Supplementary Fig. 2D). Consistent with the color change in the culture medium, we observed a 59% increase and a 45% decrease in extracellular lactate production in HeLa cells with EPB41L4A-AS1 stable knockdown or overexpression, respectively (Fig. 3E). Transient transfection of EPB41L4A-AS1 siRNA showed the similar results (Supplementary Fig. 2E and F). The same phenomenon also occurred in HepG2 and L02 cells (Fig. 3F; Supplementary Fig. 2G). Not surprisingly, EPB41L4A-AS1 stable knockdown decreased intracellular pH, while overexpression of EPB41L4A-AS1 showed the opposite effect (Fig. 3G). We also measured extracellular glucose level, EPB41L4A-AS1 stable knockdown cells showed reduction while EPB41L4A-AS1 overexpressing cells showed increase in glucose content (Fig. 3H and I; Supplementary Fig. 2H and I). Intercellular ATP levels also markedly modified by EPB41L4A-AS1 knockdown or overexpression in both HeLa and HepG2 cells (Fig. 3J and K), suggesting that EPB41L4A-AS1 knockdown enhanced glucose uptake and catabolism.
ECAR (extracellular acidification rate) assay showed that EPB41L4A-AS1 knockdown markedly increased the level of glycolysis and glycolytic capacity (Fig. 4A and C; Supplementary Fig. 2J). However, exogenous EPB41L4A-AS1 significantly reduced glycolysis, glycolytic capacity and glycolytic reserve in both HeLa and HepG2 cells (Fig. 4B and D). It has been reported that VHL/HIF-1α pathway is critical for glycolysis by modulating the expression of multiple glycolytic enzymes. Immunoblotting revealed EPB41L4A-AS1 stable knockdown dramatically upregulated the expression of HIF-1α and its targets genes but reduced the levels of VHL protein exogenous EPB41L4A-AS1 showed the opposite effect (Fig. 4E). We also examined the effect of EPB41L4A-AS1 on HIF-1α expression under hypoxic condition. Knockdown of EPB41L4A-AS1 significantly upregulated HIF-1α whereas exogenous EPB41L4A-AS1 decreased the level of HIF-1α (Supplementary Fig. 3A). qRT-PCR demonstrated that EPB41L4A-AS1 shRNA downregulated VHL but not HIF-1α mRNA expression under both normaxia and hypoxia (Fig. 4F; Supplementary Fig. 3B). The metabolic related enzymes HK1, PGK1 and PKM2 were upregulated in EPB41L4A-AS1 stable knockdown cells under hypoxia (Supplementary Fig. 3C). EPB41L4A-AS1 knockdown also enhanced glycolysis in HeLa, HepG2 and L02 cells under hypoxic (Supplementary Fig. 3D–F). These results suggested that EPB41L4A-AS1 knockdown triggered glycolysis through VHL/HIF-1α pathway.
Fig. 4.
EPB41L4A-AS1 regulates glycolysis through VHL/HIF-1α pathway. A-D. Seahorse XFp demonization ECAR in HeLa and HepG2 cells with EPB41L4A-AS1 knockdown (A and C) or overexpression (B and D). Black arrows indicate the time point of cell treatment. The lower panels are the quantification of glycolysis, glycolytic capacity and glycolytic reserve from the upper panels, respectively (n = 3). E. Immunoblotting analysis of HIF-1α, HK2, PFKL, PFKPB3 and ACTB in HeLa cells with EPB41L4A-AS1 stable knockdown or transient overexpression. F. The mRNA expression of VHL and HIF-1α in HeLa cells with EPB41L4A-AS1 stable knockdown (n = 3). Data are represented as means ± SD, *P < 0.05; **P < 0.01, unpaired, two-tailed, Student's t-test.
3.4. EPB41L4A-AS1 modulates glutamine metabolism
It has been reported that cancer cells enhance glutamine metabolism. Analysis of glutamine metabolism related genes showed that their expression was upregulated in some cervical cancer patients (Supplementary Fig. 4A). Next, we determined whether EPB41L4A-AS1 regulated glutamine metabolism. The evaluation of intercellular glutamate and α-KG (alpha-ketoglutarate), the intermediate metabolites of glutamine, revealed that both these intermediates were significantly increased in EPB41L4A-AS1 knockdown cells, while exogenous EPB41L4A-AS1 reduced the levels of intercellular glutamate and α-KG (Fig. 5A–D; Supplementary Fig. 4B–C). Furthermore, mitochondria fuels dependency assay showed that EPB41L4A-AS1 knockdown resulted in a 28% and 51% increase of glutamine metabolism dependency in HeLa and HepG2 cells, respectively. However, overexpression EPB41L4A-AS1 had no effect in glutamate dependency in both cells (Fig. 5E and F).
Fig. 5.
EPB41L4A-AS1 regulates glutamate metabolism. A-D. Intercellular glutamate (A-B) and α-KG (C-D) levels were measured in HeLa and HepG2 cells with EPB41L4A-AS1 knockdown or overexpression (n = 3). E-F. Mitochondria fuels oxidation dependency assay in HeLa (E) and HepG2 (F) cells with EPB41L4A-AS1 knockdown or overexpression for (n = 3). Data are represented as means ± SD, *P < 0.05; **P < 0.01, unpaired, two-tailed, Student's t-test.
Glutamine carbon could integrate into the TCA (tricarboxylic acid cycle) to replenish the mitochondrial respiration [6]. We found that the OCR (oxygen consumption rate) increased significantly in the EPB41L4A-AS1 knockdown cells and reduced in EPB41L4A-AS1 overexpressing cells (Fig. 6A–D; Supplementary Fig. 4D). EPB41L4A-AS1 stable knockdown increased ROS production, whereas exogenous EPB41L4A-AS1 decreased ROS level (Fig. 6E). Increased ROS contributes to cellular stress, which in turn activated of P-eIF2α/ATF4 pathway [26,27]. Immunoblotting revealed that knockdown of EPB41L4A-AS1 upregulated glutamine transporter SN2 and P-eIF2α/ATF4, whereas exogenous EPB41L4A-AS1 reduced their expression. Knockdown of EPB41L4A-AS1 also significantly upregulated the expression of TCA related genes, including SDHA and PDH, whereas exogenous EPB41L4A-AS1 decreased the expression of these genes (Fig. 6F). Knockdown of EPB41L4A-AS1 also increased the expression of glutamine metabolism related genes, including ASCT2, GLS1, GLS1, ME1 and ME2 (Supplementary Fig. 4E). Interestingly, we found that the expression of P-eIF2α and ATF4 was also upregulated in cells transfected with siVDAC1 or treated with erastin, a VDAC targeting compound, indicating that VDAC1 might be involved in glutaminolysis mediated by EPB41L4A-AS1 knockdown (Fig. 6G and H). Both qPCR and immunoblotting demonstrated that EPB41L4A-AS1 knockdown decreased VDAC1 mRNA and protein expression in HeLa cells (Fig. 6I and J; Supplementary Fig. 4F). Correlation analysis between VDAC1 and EPB41L4A-AS1 mRNA expression in 309 cervical cancer patients showed the positive correlation (Fig. 6K). We examined the effect of pH on VDAC expression, both VDAC and H3K27ac were downregulated in cells treated with acidic solution, suggesting that intracellular pH might be a crosslink between glycolysis and glutamine metabolism (Supplementary Fig. 4G).
Fig. 6.
EPB41L4A-AS1 regulates mitochondria respiratory through glutamine metabolism. A-D. Seahorse XFp assays measured OCR in HeLa and HepG2 cells with EPB41L4A-AS1 knockdown (A and C) or overexpression (B and D). Black arrows indicate the time point of cells treatment. The lower panels are quantification of basal respiration, ATP production, maximal respiration, spare respiratory capacity, proton leak and non-mitochondria respiration from the upper panels, respectively (n = 3). E. ROS levels by flow cytometry using mitosox in HeLa cells after EPB41L4A-AS1 stable knockdown (left) or overexpression (right). F. Immunoblotting analysis of SN2, ATF4, phosphor-eIF2α, SDHA, PDH and ACTB in HeLa with EPB41L4A-AS1 stable knockdown or instant overexpression for 48 h. G. Immunoblotting analysis of ATF4, VDAC, and phosphor-eIF2α in HeLa cells transfected with NC or siVDAC1. H. Immunoblotting analysis of ATF4, VDAC, and phosphor-eIF2α in HeLa cells treatment with erastin (50 μM, 24 h). I-J. VDAC protein (I) and mRNA (J) level was determined in HeLa cells with EPB41L4A-AS1 stable knockdown. K. Correlation between EPB41L4A-AS1 and VDAC1 mRNA expression was determined by Spearman coefficient analysis in 309 cervical carcinoma patients in TCGA database. All qRT-PCR were normalized by ACTB. Data are represented as means ± SD, *P < 0.05; **P < 0.01, unpaired, two-tailed, Student's t-test.
3.5. EPB41L4A-AS1 regulates VDAC1 and VHL expression through interaction with HDAC2
To distinguish it is lncRNA or protein derived from EPB41L4A-AS1 gene regulate metabolism, we mutated ATG (start codon of TIGA1 protein) in EPB41L4A-AS1 plasmid. Both VHL and VDAC1 mRNA levels were upregulated in cells transfected with EPB41L4A-AS1 or EPB41L4A-AS1 ATG mutation (Fig. 7A). Cell fractionation analysis revealed that EPB41L4A-AS1 RNA distributed in both nucleus and cytoplasm, but more concentrated in the nucleus (Fig. 7B). Then we determined whether EPB41L4A-AS1 regulated gene expression by altering histone modification. EPB41L4A-AS1 shRNA dramatically reduced H3K27 acetylation and decreased the occupation of H3K27ac on the promoter regions of VHL and VDAC1, whereas exogenous EPB41L4A-AS1 had the opposite effect (Fig. 7C and D). RIP (RNA immunoprecipitation) assay showed that HDAC2 but not HDAC1 or p300 interacted with EPB41L4A-AS1 (Fig. 7E). We also performed RNA-pulldown assay, both full-length RNA of EPB41L4A-AS1, ORF region with or without ATG mutation and short fragment of EPB41L4A-AS1 could associate with HDAC2 and H3K27ac. ORF region with or without ATG mutation seemed to show stronger interaction with HDAC2 (Fig. 7F). Surprisingly, we found that EPB41L4A-AS1 also interacted with NPM1. RNA-FISH (fluorescent in situ hybridization) revealed that EPB41L4A-AS1 colocalized with both HDAC2 and NPM1 in nucleolus (Fig. 7G–I; Supplementary Fig. 5A). It has been reported that nucleolar translocation of HDAC2 is involved in regulation of transcriptome [28]. However, the mechanisms of how HDAC2 shuttles between nucleolus and nucleoplasm is still unknown. Interestingly, we found that knockdown of EPB41L4A-AS1 reduced the interaction between HDAC2 and NPM1, leading to the translocation of HDAC2 from nucleolus to nucleoplasm and discretely distributed in nucleoplasm (Fig. 7I–J; Supplementary Fig. 5B). HDAC2 was significantly enriched in VHL and VDAC1 promoters in EPB41L4A-AS1 knockdown cells, whereas exogenous EPB41L4A-AS1 markedly reduced HDAC2 enrichment (Fig. 7K). The protein level of HDAC2 was not affected by EPB41L4A-AS1, however both VDAC1 and VHL mRNA expression were regulated by HDAC2 siRNA (Fig. 7L; Supplementary Fig. 5C). Knockdown HDAC2 or treatment with TSA (trichostatin A, histone deacetylase inhibitor) restored VDAC1 and VHL expression via increasing H3K27ac level of these genes in EPB41L4A-AS1 knockdown cells, suggesting that EPB41L4A-AS1 regulated VDAC1 and VHL expression through HDAC2 (Fig. 7M–N; Supplementary Fig. 5D). These results demonstrated that EPB41L4A-AS1 interacted with HDAC2, EPB41L4A-AS1 knockdown enhanced the occupation of HDAC2 on VHL and VDAC1 promoters, and finally reducing VHL and VDAC1 expression via histone modification.
Fig. 7.
EPB41L4A-AS1 regulates VHL and VDAC1 expression through interaction with HDAC2. A. The mRNA expression of VHL and VDAC1 in HeLa cells transfected with EPB41L4A-AS1 plasmids with or without ATG mutation for 48 h (n = 3). B. The distribution of EPB41L4A-AS1 RNA in cytosol and nucleus, NEAT1 was used as a positive control (n = 3). C. Immunoblotting analysis of H3K27ac level in HeLa cells with EPB41L4A-AS1 stable knockdown. D. ChIP-qPCR analysis H3K27ac enrichment on VDAC1 and VHL promoters (n = 3). E. RIP-qPCR analysis EPB41L4A-AS1 interaction with HDAC2, HDAC1 or p300, IgG as negative control (n = 3). F. The interaction of different fragments of EPB41L4A-AS1 RNA with HDAC2 and H3K27ac was analyzed by RNA-pulldown assay. G. RIP-qPCR analysis EPB41L4A-AS1 interaction with HDAC2 and NPM1 (n = 3). H. RNA-FISH and immunofluorescence staining of EPB41L4A-AS1 RNA (red) and NPM1 protein (green) in the nucleolus. I. RNA-FISH and immunofluorescence staining of EPB41L4A-AS1 RNA (red) and HDAC2 protein (green) in the nucleus. J. HeLa cells with or without EPB41L4A-AS1 stable knockdown were performed for immunoprecipitation with anti-HDAC2 and anti-NPM1 antibody. Immunoprecipitation was analyzed by western blotting. K. HDAC2 enrichment in VDAC1 and VHL promoters was determined by ChIP-qPCR analysis (n = 3). L. VHL and VDAC1 expression in HeLa cells transfected with siHDAC2 for 48 h (n = 3). M. EPB41L4A-AS1 stable knockdown cells or control cells transfected with NC or siHDAC2 for 48 h, qRT-PCR analyzing the expression of VHL and VDAC1 (n = 3). N. VDAC1 and VHL mRNA levels in EPB41L4A-AS1 stable knockdown cells treatment with TSA for 4 h (n = 3). Data are represented as means ± SD, *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, unpaired, two-tailed, Student's t-test. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Next, we investigated whether the small protein TIGA1 translated from EPB41L4A-AS1 gene regulated cancer cell metabolism. Consistent with previous reports, TIGA1 located in mitochondria (Supplementary Fig. 6A). We investigated potential TIGA1 binding proteins by co-immunoprecipitation and mass spectrum. GO (gene ontology) analysis suggested that TIGA1 interacted with cytoskeletal and mitochondrial proteins (Supplementary Fig. 6B; Table S2). The binding between TIGA1 and α-tubulin was further confirmed by co-immunoprecipitation in HeLa cells transfected with GFP-TIGA1 or GFP-α-tubulin (Supplementary Fig. 6C–D). Immunofluorescence staining also demonstrated that TIGA1 co-localized with α-tubulin in the nucleus end of microtubules (Supplementary Fig. 6E). As an α-tubulin-binding protein located on mitochondrial membrane, TIGA1 might mediate the connection between microtubules and mitochondria. Silencing TIGA1 decreased the co-localization between mitochondria and microtubules and changed the distribution of microtubules (Supplementary Fig. 6F). We also examined the effect of TIGA1 on microtubule stability by examining the level of acetylated tubulin. Microtubule depolymerizing agent nocodazole reduced the level of acetylated tubulin, whereas microtubule polymerizing agent taxol increased the expression of acetylated tubulin. Immunoblotting revealed that TIGA1 knockdown did not change the expression of total α-tubulin, but the level of acetylated α-tubulin was markedly reduced by TIGA1 shRNA (Supplementary Fig. 6G–H), indicating that knockdown of TIGA1 may induce microtubule depolymerization. It has been reported that free α-tubulin can interact with VDAC and trigger VDAC block, which is functionally important in regulating mitochondrial respiration and cellular stress [[29], [30], [31]]. Immunoprecipitation results showed that more α-tubulin interacted with VDAC in TIGA1 knockdown cells (Supplementary Fig. 6I), suggesting that TIGA1 knockdown mediated depolymerization of microtubules releases free tubulin, which in turn increased the amount of VDAC bound tubulin. Cellular stresses activate p38 MAPK and the activation of p38 MAPK leads to HIF-1α accumulation [32]. Therefore, we examined if TIGA1 knockdown activated p38 MAPK and caused HIF-1α accumulation. Knockdown of TIGA1 increased the levels of phosphorylated p38 and HIF-1α (Supplementary Fig. 6J). The phosphorylation of p38 and accumulation of HIF-1α were also increased in cells treated with nocodazole (Supplementary Fig. 6K). Knockdown VDAC by siRNA had similar effect to increase HIF-1α expression (Supplementary Fig. 6L). These results suggested that TIGA1 was an important mediator between microtubules and mitochondria. Depletion of TIGA1 destabilized microtubule and induced the functional block of VDAC and cellular stress, which finally activated p38 MAPK and enhanced HIF-1α accumulation.
3.6. Depletion of EPB41L4A-AS1 increases the therapeutic effect of glutaminase inhibitor
Since depletion of EPB41L4A-AS1 increased glutamine metabolism dependency, we hypothesized that EPB41L4A-AS1 knockdown might increase the antitumor activity of glutaminase inhibitor. The colony formation assay demonstrated that EPB41L4A-AS1 stable knockdown increased the number of colonies compared with the control cells. Glutaminase inhibitor, compound 968, significantly reduced colony formation. Tumor cell proliferation was nearly completely inhibited by combined treatment of glutaminase inhibitor and EPB41L4A-AS1 shRNA, indicating that EPB41L4A-AS1 knockdown cells largely relied on glutamine metabolism to promote cancer cell growth (Fig. 8A).
Fig. 8.
EPB41L4A-AS1 knockdown increases the antitumor effect of glutaminase inhibitor in vivo. A. Colony formation assay in control HeLa cells or EPB41L4A-AS1 stable knockdown cells treated with or without compound 968 (10 nM, 10 days), DMSO as control. B. HeLa cells stably transfected with ShEPB41L4A-AS1 or ShNC were injected subcutaneously into the flank of nude mice. ShEPB41L4A-AS1 promotes tumorigenesis compared with ShNC at day 8, 10, 12, and 14 of injection. C. ShEPB41L4A-AS1 promotes tumor growth in vivo. When the average size of the tumor reached approximately 75 mm3, mice were treated with compound 968 or DMSO every other day by intraperitoneal injections. Tumor volumes were measured and plotted in a graph. D-E. Tumor images (D) and weight (E). F-G. The expression levels of EPB41L4A-AS1 RNA (F) and TIGA1 protein (G) in tumors. H. Model depicting the role of EPB41L4A-AS1 in glycolysis and glutamine metabolism reprogramming. Data are represented as means ± SD, *P < 0.05; **P < 0.01, unpaired, two-tailed, Student's t-test.
Finally, we tested whether EPB41L4A-AS1 regulated tumor growth by modulating cancer cell metabolism in vivo. Control cells and EPB41L4A-AS1 stable knockdown cells were subcutaneously injected into the flanks of nude mice. Time of tumor formation was earlier and tumor volume was also significantly larger in mice injected with EPB41L4A-AS1 stable knockdown cells than the mice injected with the control cells (Fig. 8B and C). Compound 968 effectively inhibited tumor growth and EPB41L4A-AS1 knockdown markedly increased the anti-tumor effect of compound 968 (Fig. 8D–G). EPB41L4A-AS1 knockdown cells largely depended on glutamine metabolism to promote cancer cells growth and proliferation. Depletion of EPB41L4A-AS1 enhanced the therapeutic effect of glutaminase inhibitor. Collectively, our results demonstrated that EPB41L4A-AS1 is a p53 and PGC-1a regulated gene. It regulates cell metabolism in both lncRNA and protein forms. LncRNA EPB41L4A-AS1 regulates glycolysis and glutamine metabolism by modulating the epigenetic regulation of VDAC1 and VHL through HDAC2. (Fig. 8H).
4. Discussion
In this study, we reported that EPB41L4A-AS1, a p53 inducible long non-coding gene, regulates glycolysis and glutamine metabolism. As a key regulator, p53 controls a broad and flexible network with complex functions including cell cycle checkpoint and DNA repair, apoptosis, cellular plasticity, tissue remodeling, pluripotency repression [15,33,34]. The functions regulated by p53 network also include ROS production and metabolism [15]. The roles of p53 in metabolism are complex. It downregulates glycolysis and lipid synthesis but upregulates oxidative phosphorylation and lipid catabolism in both normal and tumor cells [16]. P53 suppresses glycolysis via transcriptionally inhibiting GLUT1 and GLUT4, or transactivating RRAD and TIGAR, the inhibitors of glycolysis [[35], [36], [37], [38]]. P53 also suppresses glucose metabolism by inhibits glucose-6-phosphate dehydrogenase [39]. On the other hand, p53 stimulates oxidative phosphorylation by upregulation of SCO2 (cytochrome c oxidase assembly) and glutaminase GLS2 [40]. In some tumor cells, p53 mutants positively regulate Warburg effect partially through increasing the levels of GLUT1 and GLUT4 and reducing the levels of SCO2 and GLS2 to favor aerobic glycolysis over oxidative phosphorylation [41,42]. As one of the downstream genes of p53, knockdown of EPB41L4A-AS1 not only triggers glycolysis but also stimulates glutamine metabolism. EPB41L4A-AS1 knockdown increases glycolysis via HIF-1α pathway and enhances glutamine metabolism by P-eIF2α/ATF4 pathway.
Knockdown of EPB41L4A-AS1 reduced VDAC1 expression and activated P-eIF2α/ATF4 pathway. As an important cellular metabolite transporter of mitochondria, VDAC mediates the exchange of many metabolites, such as pyruvate, malate, succinate, glutamate citrate and NADH between the cytosol and mitochondria [43,44]. VDAC closure or reduction not only decreases metabolites exchange but also increases intramitochondrial oxidative stress via blocking efflux of O2– from the IMS (intermembrane space) to the cytosol, finally enhances cellular stress and ROS, which can activate P-eIF2α/ATF4 pathway [[45], [46], [47]]. Previous studies suggest that P-eIF2α up-regulation could downregulate global protein synthesis but selectively upregulate ATF4 translation [48]. ATF4 is a regulator of amino acid transporters [13]. It is possible that up-regulation of ATF4 activates expression of amino acid transporters, including ASCT2 and SN2, the glutamine transporters, and finally induces glutaminolysis.
The activation of VHL/HIF-1α pathway mediated by EPB41L4A-AS1 knockdown is critical to the increases of glycolysis. In this investigation, we found that two mechanisms were involved in the accumulation of HIF-1α. On one hand, knockdown of EPB41L4A-AS1 upregulates HIF-1α by reducing the expression of VHL. On the other hand, VDAC reduction or closure induces cellular stress, leading to p38 MAPK activation and HIF-1α accumulation [32,45]. It has been reported that VDAC reduction or closure induced ROS also stabilizes HIF-1α through prolyl hydroxylases (PHDs) inactivation [[49], [50], [51]].
TIGA1 pathway is related to both glycolysis and glutaminolysis. Increasing evidences show that lncRNAs encode biological functional small proteins or peptides. Yabuta reported that EPB41L4A-AS1 gene encodes a small mitochondrion located protein, named TIGA1, and its ectopic expression inhibits the colony-formation of tumor cells and tumor growth in soft agar [25]. Here, we found that TIGA1 interacts with α-tubulin, like VDAC. Knockdown of TIGA1 destabilizes microtubules and increases free α-tubulin and its binding with VDAC1, leading to the partially blocking of VDAC channel. VDAC blocking finally induces HIF-1α accumulation and activates P-eIF2α/ATF4 pathway.
We found that three pathways are involved in EPB41L4A-AS1 mediates glycolysis and glutaminolysis. However, it is hard to say which one is more important. Firstly, both glucose and glutamine are two principal nutrients that support survival and biosynthesis in mammalian cells. Cancer cells meet their requirement for higher energy and macromolecular synthesis though high rate of glycolysis to obtained diverse carbon intermediates and glutaminolysis-derived NADH, FADH2 and NADPH, which provides reducing power for a wide variety of biosynthetic reactions [52]. Furthermore, there is a very complex interaction among three pathways. McBrian et al. reports that intracellular pH can regulate histone acetylation [53]. We found that high glycolysis induces accumulation of intracellular lactate and the low intracellular pH can also inhibit VDAC1 expression, finally, increase glutamine metabolism. On the other hand, the down-regulation of VDAC1 expression mediated by lncRNA EPB41L4A-AS1 knockdown or VDAC1 blocking induced via TIGA1 silencing enhances ROS generation which increases the stability of HIF-1α.
The down-regulation of VHL and VDAC1 by EPB41L4A-AS1 knockdown is mediated by the interaction between EPB41L4A-AS1 and HDAC2. It has been reported that lncRNAs such as Lnc34a and lnc-Smad3 regulate gene expression through recruitment HDAC1 to the promoter region of their target genes [54,55]. However, no reports show that lncRNAs interact with HDAC2. Here, we demonstrated that lncRNA EPB41L4A-AS1 interacted with both HDAC2 and NPM1, and mediated HDAC2 locating in the nucleolus. Knockdown of EPB41L4A-AS1 reduced the interaction between HDAC2 and NPM1, leading to the translocation of HDAC2 between nucleolus and nucleoplasm. Nucleolar translocation of chromatin regulators like HDAC1, HDAC2, SIRT1 have been reported, with chromatin remodeling and interfering with global transcription [28,56]. However, the mechanisms in regulating the chromatin regulators shuttling between nucleoplasm and nucleolus are still unknown. In addition, how chromatin regulators are recruited to the correct chromatin sites is also unclear. Here, we reported that lncRNA EPB41L4A-AS1 interacted and colocalized with HDAC2 in nucleolus. Knockdown of EPB41L4A-AS1 induced the translocation of HDAC2 from nucleolus to nucleoplasm and discretely distributed in nucleoplasm, increasing its interaction with the promoters of VHL, VDAC1 and other undetermined target genes and reducing H3K27ac occupation on the promoter regions, and finally reducing gene transcription. However, further studies are required to demonstrate why EPB41L4A-AS1 specifically interacts with HDAC2 but not HDAC1.
The low expression of EPB41L4A-AS1 in human cancers is associated with poor prognosis, suggesting that EPB41L4A-AS1 may can be used as a potential prognostic predictor in cancers. Furthermore, low expression of EPB41L4A-AS1 increases glutamine dependency in cancers, according to the results of our study. Therefore, the cancer patients with low expression of EPB41L4A-AS1 maybe more dependent on glutamine metabolism, compared with the patients with normal expression of EPB41L4A-AS1. Using EPB41L4A-AS1 as screening indicators should be helpful to select the patients who are better response to the treatment of glutaminase inhibitor and enhance the efficacy of the antitumor medicine. In the future, the combination of EPB41L4A-AS1 siRNA and glutaminase inhibitors may be as a new and powerful therapeutic strategy for tumor treatment of the patient with normal expression of EPB41L4A-AS1.
In conclusion, our results highlighted a key role of EPB41L4A-AS1 in cancer metabolic reprogramming. Since EPB41L4A-AS1 knockdown cancer cells largely depended on glutamine metabolism, the simultaneous application of EPB41L4A-AS1 shRNA and glutaminase inhibitor might be a powerful strategy in cancer therapy and repressor of Warburg effect.
Funding
This works was supported by the National Natural Science Foundation of China (31571400), basic research fund of Shenzhen [JCYJ20170405103953336] and special project of Suzhou-Tsinghua innovation leading action (2016SZ3012).
Conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
Meijian Liao and Weijie Liao designed and conducted the experiments, analyzed the data, and revised the manuscript. Yuanchang Zhu, Weidong Xie, Shikuang Zhang, Xiduan Wei, Bing Sun and Deheng Chen conducted the experiments. Bing Li conducted the bioinformation analysis. Liu Cao, Naihan Xu and Burton B Yang revised the manuscript. Yaou Zhang designed the experiments, supervised the project, and wrote the manuscript.
Supplementary data
Supplementary Figure S1-S6
Table S1: Genes alteration in EBP41L4A-AS1 knockdown
Table S2: TIGA1-interating proteins by MS
Contributor Information
Liu Cao, Email: caoliu@mail.cmu.edu.cn.
Burton B. Yang, Email: byang@sri.utoronto.ca.
Yaou Zhang, Email: zhangyo@sz.tsinghua.edu.cn.
References
- 1.Warburg O., Wind F., Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8(6):519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 3.Schuurbiers O.C., Meijer T.W., Kaanders J.H., Looijen-Salamon M.G., de Geus-Oei L.F., van der Drift M.A. Glucose metabolism in NSCLC is histology-specific and diverges the prognostic potential of 18FDG-PET for adenocarcinoma and squamous cell carcinoma. J Thorac Oncol. 2014;9(10):1485–1493. doi: 10.1097/JTO.0000000000000286. [DOI] [PubMed] [Google Scholar]
- 4.Upadhyay M., Samal J., Kandpal M., Singh O.V., Vivekanandan P. The Warburg effect: insights from the past decade. Pharmacol Ther. 2013;137(3):318–330. doi: 10.1016/j.pharmthera.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Koppenol W.H., Bounds P.L., Dang C.V. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 6.Le A., Lane A.N., Hamaker M., Bose S., Gouw A., Barbi J. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15(1):110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan T., Gao L., Wu G., Shen G., Xie S., Wen H. Elevated expression of glutaminase confers glucose utilization via glutaminolysis in prostate cancer. Biochem Biophys Res Commun. 2015;456(1):452–458. doi: 10.1016/j.bbrc.2014.11.105. [DOI] [PubMed] [Google Scholar]
- 8.Metallo C.M., Vander Heiden M.G. Understanding metabolic regulation and its influence on cell physiology. Mol Cell. 2013;49(3):388–398. doi: 10.1016/j.molcel.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ward P.S., Thompson C.B. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wise D.R., Thompson C.B. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35(8):427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hensley C.T., Wasti A.T., DeBerardinis R.J. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123(9):3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Son J., Lyssiotis C.A., Ying H., Wang X., Hua S., Ligorio M. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496(7443):101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren P., Yue M., Xiao D., Xiu R., Gan L., Liu H. ATF4 and N-Myc coordinate glutamine metabolism in MYCN-amplified neuroblastoma cells through ASCT2 activation. J Pathol. 2015;235(1):90–100. doi: 10.1002/path.4429. [DOI] [PubMed] [Google Scholar]
- 14.Yuneva M.O., Fan T.W., Allen T.D., Higashi R.M., Ferraris D.V., Tsukamoto T. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 2012;15(2):157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kastenhuber E.R., Lowe S.W. Putting p53 in Context. Cell. 2017;170(6):1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gnanapradeepan K., Basu S., Barnoud T., Budina-Kolomets A., Kung C.P., Murphy M.E. The p53 tumor suppressor in the control of metabolism and ferroptosis. Front Endocrinol (Lausanne) 2018;9:124. doi: 10.3389/fendo.2018.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin A., Li C., Xing Z., Hu Q., Liang K., Han L. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18(2):213–224. doi: 10.1038/ncb3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hung C.L., Wang L.Y., Yu Y.L., Chen H.W., Srivastava S., Petrovics G. A long noncoding RNA connects c-Myc to tumor metabolism. Proc Natl Acad Sci U S A. 2014;111(52):18697–18702. doi: 10.1073/pnas.1415669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redis R.S., Vela L.E., Lu W., Ferreira de Oliveira J., Ivan C., Rodriguez-Aguayo C. Allele-specific reprogramming of cancer metabolism by the long non-coding RNA CCAT2. Mol Cell. 2016;61(4):520–534. doi: 10.1016/j.molcel.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song J., Wu X., Liu F., Li M., Sun Y., Wang Y. Long non-coding RNA PVT1 promotes glycolysis and tumor progression by regulating miR-497/HK2 axis in osteosarcoma. Biochem Biophys Res Commun. 2017;490(2):217–224. doi: 10.1016/j.bbrc.2017.06.024. [DOI] [PubMed] [Google Scholar]
- 21.Huarte M., Guttman M., Feldser D., Garber M., Koziol M.J., Kenzelmann-Broz D. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang F., Zhang H., Mei Y., Wu M. Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol Cell. 2014;53(1):88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Barsotti A.M., Beckerman R., Laptenko O., Huppi K., Caplen N.J., Prives C. p53-dependent induction of PVT1 and miR-1204. J Biol Chem. 2012;287(4):2509–2519. doi: 10.1074/jbc.M111.322875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marechal R., Demetter P., Nagy N., Berton A., Decaestecker C., Polus M. High expression of CXCR4 may predict poor survival in resected pancreatic adenocarcinoma. Br J Cancer. 2009;100(9):1444–1451. doi: 10.1038/sj.bjc.6605020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yabuta N., Onda H., Watanabe M., Yoshioka N., Nagamori I., Funatsu T. Isolation and characterization of the TIGA genes, whose transcripts are induced by growth arrest. Nucleic Acids Res. 2006;34(17):4878–4892. doi: 10.1093/nar/gkl651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lange P.S., Chavez J.C., Pinto J.T., Coppola G., Sun C.W., Townes T.M. ATF4 is an oxidative stress-inducible, prodeath transcription factor in neurons in vitro and in vivo. J Exp Med. 2008;205(5):1227–1242. doi: 10.1084/jem.20071460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suragani R.N., Zachariah R.S., Velazquez J.G., Liu S., Sun C.W., Townes T.M. Heme-regulated eIF2alpha kinase activated Atf4 signaling pathway in oxidative stress and erythropoiesis. Blood. 2012;119(22):5276–5284. doi: 10.1182/blood-2011-10-388132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee P.C., Wildt D.E., Comizzoli P. Nucleolar translocation of histone deacetylase 2 is involved in regulation of transcriptional silencing in the cat germinal vesicle. Biol Reprod. 2015;93(2):33. doi: 10.1095/biolreprod.115.129106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maldonado E.N., Patnaik J., Mullins M.R., Lemasters J.J. Free tubulin modulates mitochondrial membrane potential in cancer cells. Cancer Res. 2010;70(24):10192–10201. doi: 10.1158/0008-5472.CAN-10-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maldonado E.N. VDAC-tubulin, an anti-Warburg pro-oxidant switch. Front Oncol. 2017;7:4. doi: 10.3389/fonc.2017.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gurnev P.A., Rostovtseva T.K., Bezrukov S.M. Tubulin-blocked state of VDAC studied by polymer and ATP partitioning. FEBS Lett. 2011;585(14):2363–2366. doi: 10.1016/j.febslet.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emerling B.M., Platanias L.C., Black E., Nebreda A.R., Davis R.J., Chandel N.S. Mitochondrial reactive oxygen species activation of p38 mitogen-activated protein kinase is required for hypoxia signaling. Mol Cell Biol. 2005;25(12):4853–4862. doi: 10.1128/MCB.25.12.4853-4862.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barabutis N., Schally A.V., Siejka A. P53, GHRH, inflammation and cancer. EBioMedicine. 2018;37:557–562. doi: 10.1016/j.ebiom.2018.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanikawa C., Zhang Y.Z., Yamamoto R., Tsuda Y., Tanaka M., Funauchi Y. The transcriptional landscape of p53 signalling pathway. EBioMedicine. 2017;20:109–119. doi: 10.1016/j.ebiom.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwartzenberg-Bar-Yoseph F., Armoni M., Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64(7):2627–2633. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- 36.Zhang C., Liu J., Wu R., Liang Y., Lin M., Liu J. Tumor suppressor p53 negatively regulates glycolysis stimulated by hypoxia through its target RRAD. Oncotarget. 2014;5(14):5535–5546. doi: 10.18632/oncotarget.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bensaad K., Tsuruta A., Selak M.A., Vidal M.N., Nakano K., Bartrons R. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126(1):107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 38.Green D.R., Chipuk J.E. p53 and metabolism: inside the TIGAR. Cell. 2006;126(1):30–32. doi: 10.1016/j.cell.2006.06.032. [DOI] [PubMed] [Google Scholar]
- 39.Jiang P., Du W., Wang X., Gao X., Wu M., Yang X. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol. 2011;13(3):310–316. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu W., Zhang C., Wu R., Sun Y., Levine A., Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci U S A. 2010;107(16):7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eriksson M., Ambroise G., Ouchida A.T., Lima Queiroz A., Smith D., Gimenez-Cassina A. Effect of mutant p53 proteins on glycolysis and mitochondrial metabolism. Mol Cell Biol. 2017;37(24) doi: 10.1128/MCB.00328-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang C., Liu J., Liang Y., Wu R., Zhao Y., Hong X. Tumour-associated mutant p53 drives the Warburg effect. Nat Commun. 2013;4:2935. doi: 10.1038/ncomms3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abu-Hamad S., Sivan S., Shoshan-Barmatz V. The expression level of the voltage-dependent anion channel controls life and death of the cell. Proc Natl Acad Sci U S A. 2006;103(15):5787–5792. doi: 10.1073/pnas.0600103103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keinan N., Tyomkin D., Shoshan-Barmatz V. Oligomerization of the mitochondrial protein voltage-dependent anion channel is coupled to the induction of apoptosis. Mol Cell Biol. 2010;30(24):5698–5709. doi: 10.1128/MCB.00165-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tikunov A., Johnson C.B., Pediaditakis P., Markevich N., Macdonald J.M., Lemasters J.J. Closure of VDAC causes oxidative stress and accelerates the Ca(2+)-induced mitochondrial permeability transition in rat liver mitochondria. Arch Biochem Biophys. 2010;495(2):174–181. doi: 10.1016/j.abb.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wortel I.M.N., van der Meer L.T., Kilberg M.S., van Leeuwen F.N. Surviving stress: modulation of ATF4-mediated stress responses in normal and malignant cells. Trends Endocrinol Metab. 2017;28(11):794–806. doi: 10.1016/j.tem.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang H., Cai F., Zhang Y., Xue M., Liu L., Yang A. Early-stage autophagy protects nucleus pulposus cells from glucose deprivation-induced degeneration via the p-eIF2alpha/ATF4 pathway. Biomed Pharmacother. 2017;89:529–535. doi: 10.1016/j.biopha.2017.02.074. [DOI] [PubMed] [Google Scholar]
- 48.Zheng Q., Ye J., Cao J. Translational regulator eIF2alpha in tumor. Tumour Biol. 2014;35(7):6255–6264. doi: 10.1007/s13277-014-1789-0. [DOI] [PubMed] [Google Scholar]
- 49.Comito G., Calvani M., Giannoni E., Bianchini F., Calorini L., Torre E. HIF-1alpha stabilization by mitochondrial ROS promotes Met-dependent invasive growth and vasculogenic mimicry in melanoma cells. Free Radic Biol Med. 2011;51(4):893–904. doi: 10.1016/j.freeradbiomed.2011.05.042. [DOI] [PubMed] [Google Scholar]
- 50.Lu H., Dalgard C.L., Mohyeldin A., McFate T., Tait A.S., Verma A. Reversible inactivation of HIF-1 prolyl hydroxylases allows cell metabolism to control basal HIF-1. J Biol Chem. 2005;280(51):41928–41939. doi: 10.1074/jbc.M508718200. [DOI] [PubMed] [Google Scholar]
- 51.Zepeda A.B., Pessoa A., Jr., Castillo R.L., Figueroa C.A., Pulgar V.M., Farias J.G. Cellular and molecular mechanisms in the hypoxic tissue: role of HIF-1 and ROS. Cell Biochem Funct. 2013;31(6):451–459. doi: 10.1002/cbf.2985. [DOI] [PubMed] [Google Scholar]
- 52.Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23(1):27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McBrian M.A., Behbahan I.S., Ferrari R., Su T., Huang T.W., Li K. Histone acetylation regulates intracellular pH. Mol Cell. 2013;49(2):310–321. doi: 10.1016/j.molcel.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xia M., Liu J., Liu S., Chen K., Lin H., Jiang M. Ash1l and lnc-Smad3 coordinate Smad3 locus accessibility to modulate iTreg polarization and T cell autoimmunity. Nat Commun. 2017;8 doi: 10.1038/ncomms15818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L., Bu P., Ai Y., Srinivasan T., Chen H.J., Xiang K. A long non-coding RNA targets microRNA miR-34a to regulate colon cancer stem cell asymmetric division. Elife. 2016;5 doi: 10.7554/eLife.14620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Y., Santoro R., Grummt I. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 2002;21(17):4632–4640. doi: 10.1093/emboj/cdf460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1-S6
Table S1: Genes alteration in EBP41L4A-AS1 knockdown
Table S2: TIGA1-interating proteins by MS