Abstract
Background
Breast cancer stem cells have self-renewal capability and are resistant to conventional chemotherapy. PD-L1 could promote the expression of stemness markers (OCT4 and Nanog) in breast cancer stem cells. However, the mechanisms by which PD-L1 regulates the stemness of breast cancer cells and PD-L1 is regulated in breast cancer cells are still unclear.
Methods
Lentivirus infection was used to construct stable cell lines. The correlation between PD-L1 and stemness markers expression was evaluated in clinical samples. Additionally, luciferase reporter assay combined with RNA-Fluorescence in situ hybridization (RNA-FISH) and RNA-binding protein immunoprecipitation (RIP) assays were used to verify the direct binding of miR-873 on PD-L1. Furthermore, flow cytometry, mammosphere formation combined with nude mouse tumor xenograft model were carried out to examine the effects of miR-873/PD-L1 axis on the stemness of breast cancer cells. Finally, MTT assay was performed to determine the effects of miR-873/PD-L1 axis on drug resistance.
Findings
PD-L1 expression was positively correlated with the expression of stemness markers, and overexpression of PD-L1 contributed to chemoresistance and stemness-like properties in breast cancer cells via activating PI3K/Akt and ERK1/2 pathways. Mechanistically, miR-873 inhibited PD-L1 expression through directly binding to its 3′-untranslated region (UTR), and miR-873 attenuated the stemness and chemoresistance of breast cancer cells which was dependent on PD-L1 and the downstream PI3K/Akt and ERK1/2 signaling. Notably, the promotion of PD-L1 on the stemness and chemoresistance was enhanced by recombinant PD-1 (rPD-1), this effect was attenuated by PD-1/PD-L1 inhibitor.
Interpretation
miR-873/PD-L1 regulatory axis might serve as a therapeutic target to enhance the chemo-sensitivity and eliminate the stemness of breast cancer cells.
Fund
This work was supported by the National Nature Science Foundation of China, No. 81702957, China Postdoctoral Science Foundation, No. 2017M620230, the Postdoctoral Research Funding Scheme of Jiangsu Province (2017), No. 1701197B, and the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions.
Keywords: miR-873, PD-L1, Cancer stem cells, Drug resistance, PI3K/Akt, ERK1/2
Highlights
-
•
A critical role of PD-L1 in stemness and chemoresistance is proposed.
-
•
A negative miR-873/PD-L1 interaction was identified in breast cancer cells.
-
•
The mechanisms of miR-873/PD-L1 in stemness and chemoresistance were studied.
-
•
The results provide new insights for breast cancer progression and treatment.
Research in context.
Evidence before this study
PD-L1 is associated with epithelial to mesenchymal transition and PD-L1 could promote OCT4 and Nanog expression in breast cancer stem cells. Moreover, PD-L1 expression can be promoted in cells and tissue following chemotherapy. Previous study has demonstrated that miR-873 could attenuate tamoxifen resistance in ERalpha-positive breast cancer.
Added value of this study
We firstly clarified that PD-L1 was a direct target of miR-873 in breast cancer, which could facilitate the understanding of the mechanisms by which PD-L1 was regulated, and future works could be performed to explore the effects of combined miR-873 agonist with PD-L1 antibody on breast cancer progression.
Implications of all the available evidence
This study provided evidence suggesting a targeting strategy involving miR-873 together with chemo-therapy or immune checkpoint blockage to treat breast cancer.
Alt-text: Unlabelled Box
1. Introduction
The main treatments of breast cancer are surgery, targeting therapy, radiotherapy, and chemotherapy, especially for triple-negative breast cancer, chemotherapy is the only option. However, chemotherapy induces tumor heterogeneity derived from both normal and cancer cells, this effect could lead to chemoresistance and disease progression [1,2]. Cancer stem cells (CSCs) hold the ability to self-renew and differentiate into the heterogeneous lineages of cancer cells in response to chemotherapeutic agents, and are considered as the mediators of cancer metastasis, drug resistance and cancer relapse [[3], [4], [5]]. Although successful cancer therapy could kill the proliferating tumor cells, a subset of remaining CSCs can survive [6]. Therefore, it is important to reveal the mechanisms underlying CSCs formation.
Programmed cell death ligand 1 (PD-L1/B7-H1/CD274), an immune checkpoint molecule, is the ligand of PD-1 [7]. Currently, the launch of an anti-PD-L1 antibody has been represented as a significant breakthrough for patients with advanced solid tumors [8], as PD-L1 is overexpressed in solid cancers [9]. Interestingly, PD-L1 expression can be promoted following chemotherapeutic treatment, which is recognized as a signal of poor prognosis in patients with NSCLC [10]. Meanwhile, PD-L1 expression is associated with epithelial to mesenchymal transition (EMT) process [11], this process could be resulted from CSCs [12]; and PD-L1 could promote the expression of stemness markers (OCT4 and Nanog) [13]. Additionally, PD-L1 is frequently overexpressed in basal type of breast cancer, which exhibits a relative stronger stemness [14,15]. These effects suggest that PD-L1 might promote the stemness of breast cancer cells. Notably, the mechanisms by which PD-L1 is regulated are not well defined in breast cancer.
MicroRNAs (miRNAs) are a class of small noncoding RNA molecules that post-transcriptionally modulate gene expression by binding to the 3′-untranslated region (3′-UTR) of target genes [16]. Notably, PD-L1 has been identified as the target of various miRNAs [[17], [18], [19]]. In addition, recent studies have shown that miRNAs could regulate cancer stemness and drug resistance in breast cancer [[20], [21], [22]]. Previous studies have demonstrated that miR-873 acts as a tumor suppressor via suppressing IGF2BP1 expression in glioblastoma [23] and by targeting differentiated embryonic chondrocyte expressed gene 2 (DEC2) in esophageal cancer [24], respectively. Moreover, miR-873 attenuates tamoxifen resistance via regulating ERα transcriptional activity through targeting CDK3 in breast cancer cells [25]. However, the roles and related mechanisms of miR-873 in regulating the stemness and chemoresistance remain unclear in breast cancer.
Here, we found that PD-L1 expression was increased in breast cancer tissues and cells with adriamycin resistance, and enhanced the stemness of breast cancer cells via activating PI3K/Akt and ERK1/2 signaling, this effect was strengthened by recombinant PD-1 (rPD-1). In addition, miR-873 attenuated the stemness and chemoresistance via directly targeting PD-L1 and thus inactivating the downstream PI3K/Akt and ERK1/2 signaling. Therefore, miR-873/PD-L1 regulatory axis might serve as a therapeutic target to enhance the chemo-sensitivity and eliminate the stemness of breast cancer cells.
2. Materials and methods
2.1. Cell culture and clinical samples
Human breast cancer cell lines MCF-7 and MDA-MB-231 cells were obtained from ATCC (Manassas, Virginia, USA), and respectively cultured in high glucose DMEM (Gibco, Grand Island) and L15 medium (Gibco). Adriamycin resistant MCF-7/ADR cells were purchased from KeyGen BioTECH (Nanjing, China), and cultured in RPMI-1640 medium (Gibco). HEK-293 T cells were preserved in our laboratory and cultured in high glucose DMEM. The three media were supplemented with 10% fetal bovine serum (FBS, Gibco, Grand Island), 80 U/ml penicillin (Sangon Biotech, China), and 0.08 mg/ml streptomycin (Sangon Biotech, China) at 37 °C under humidified atmosphere with 5% CO2. MCF-7/ADR cells were cultured with 1 μM adriamycin to maintain adriamycin resistance. PI3K inhibitor (LY-294002) and ERK1/2 inhibitor (VX-11e) were purchased from APExBIO. Adriamycin and human recombinant PD-1 (rPD-1) were purchased from the Zhongda Hospital Southeast University and BioVision, respectively. BMS-202 (synonyms: PD-1/PD-L1 inhibitor 2) and Human IFN-gamma were purchased from the MedChemExpress and Pepro Tech, respectively.
Four pairs of fresh breast cancer and adjacent mammary gland epithelial tissue samples were randomly selected from the Tumor Hospital of Jiangsu Province. Written informed consent from all patients and approval of the hospital ethics review committees were obtained.
2.2. Transfection of plasmids, miRNA, siRNA
The detailed procedure for transfection was referred our previous work [26]. The coding sequence of PD-L1, and the 1000 bp of pri-miR-873 were inserted into the plvx-IRES-ZsGreen1 vector, and denoted as plvx-PD-L1 and plvx-873, respectively. ShRNA against PD-L1 was cloned into Plko.1, and named as Plko.1-PD-L1. PD-L1 3′ UTR (WT) which contains binding site of miR-873 was inserted into pMIR-Report or pcDNA3.1(+), and PD-L1 3′ UTR (MUT) which contains mutated binding sites for miR-873 were constructed using Fast Mutagenesis Kit V2 (Vazamy, China) following the manufacturer's instruction, and all the constructs were verified by DNA sequencing. The sequences of siRNAs, miR-873 (mimics or inhibitor) were shown in Supplementary Table 1. All the primer sequences for plasmids construction were listed in Supplementary Table 2.
2.3. Construction of stable cell lines
Cells with miR-873 stable overexpression, PD-L1 stable knockdown or miR-873 stable overexpression together with PD-L1 stable overexpression were obtained by lentivirus infection. The detailed procedure was mentioned in our previous study [27]. Briefly, HEK-293T cells were co-transfected with Plko.1-PD-L1, plvx-873 or plvx-PD-L1, and packaging vectors psPAX2 and pMD2.G using Lentifectin (Abm, USA). Cells were infected with the virus in the presence of 2 μg/ml Polybrene. Cells infected with Plko.1-PD-L1 were selected with puromycin (Sigma, 2 μg/ml) for 2 weeks, and cells infected with plvx-873 were screened by fluorescent cell sorting, after which qRT-PCR analysis was used for verification.
2.4. Western blot
Total protein from cells was harvested using RIPA lysis buffer (Beyotime, China), and total protein from tissues was extracted using total protein extraction kit (invent, USA). About 10–20 μg protein was fractionated by 10% SDS/PAGE, and then transferred onto polyvinylidene difluoride membranes, and incubated with the primary antibody. The primary antibodies were listed in Supplementary Table 3. After incubating with secondary antibodies (Jackson ImmunoResearch, USA), ECL system (Thermo Fisher, USA) were performed to determine protein expression by density analysis.
2.5. Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted using Trizol (TransGen Biotech, China). The first-strand cDNA was reversely transcribed with M-MLV (Vazyme Biotech, China) following the standard protocols. qRT-PCR was carried out to determine mRNA expression with SYBR Green master mix (Vazyme Biotech, China) on an ABI Prism 7500 Sequence Detector (Applied Biosystems, USA). MiRNA/U6 snRNA RT primer mix and PCR specific primer set (GenePharma, China) were used to detect and quantify miRNA expression. Primer sequences of P-gp were described in our previous study [26]. Other Primer sequences used were mentioned in Supplementary Table 4. mRNA and miRNA levels were normalized to GAPDH or U6 sRNA, respectively. The relative expression was calculated using 2−∆∆Ct method.
2.6. Luciferase reporter assay
HEK-293T cells were co-transfected with pMIR-PD-L1 3′UTR (WT or MUT), miR-873 mimics or mimics NC and β-gal reporter control plasmid. 72 h later, the luciferase activity was measured by POLARstar Omega multimode microplate reader according to manufacturer's protocol and normalized to β-gal activity.
2.7. Flow cytometry analysis
MDA-MB-231, MCF-7/ADR and MCF-7 cells were seeded into a 6-well plate and transfected with miR-873 mimics or inhibitor, or si-PD-L1, or plvx-PD-L1, and followed by detecting CD44+/CD24− population via flow cytometry analysis according to the procedure described in our recent work [27]. To assay the surface expression of PD-L1, cells at a density of 105 cells/mL were harvested, stained with APC-labeled anti-PD-L1 Ab or an isotype control, and PD-L1 expression was measured by flow cytometry.
2.8. MTT assay
MTT (Amresco's, USA) assay was used to measure cell viability. MCF-7/ADR and MCF-7 cells were seeded in 96-well plates at the density of 3000/well, and treated with different concentrations of adriamycin for MCF-7/ADR (0, 0.08, 2.5, 5, 8.75, 10, 17.5, 20, 35 μM) and MCF-7 (0, 0.078125, 0.15625, 0.3125, 0.625, 1.25, 2.5, 5, 10 μM) for 24 h, 48 h, 72 h, respectively. MTT was added to the medium to a final concentration of 0.5 mg/ml, and incubated for 4 h. After medium was removed, the formazan crystals were dissolved in 150 μl dimethyl-sulfoxide (DMSO) in room temperature for 15 min. Finally, the absorbance was measured at 490 nm Biorad iMark by a microplate reader. The inhibition percentage was calculated by the formula: inhibition percentage = (1 − adriamycin treated group OD/no adriamycin treated group OD) ∗ 100.
2.9. RNA-binding protein immunoprecipitation assay (RIP)
RIP assay was performed using the Protein A/G Agarose Resin (YEASEN, China) following the manufacturer's protocol. The detailed procedure was described in our previous studies [28,29]. MCF-7 cells were transfected with pcDNA3.1(+) PD-L1 3′UTR (WT) and pcDNA3.1(+) PD-L1 3′UTR (MUT), and lysed by NP-40 lysis buffer (Beyotime, China). Ago2-RNA complex was dissociated from the beads. qRT-PCR was performed to detect miR-873 level.
2.10. Mammosphere formation assay
Cells were grown in the MammoCult medium (Stem Cell Technologies, Vancouver, Canada) supplemented with MammoCult Proliferation Supplements (Stem Cell Technologies, Vancouver, Canada) and plated in 24-wells plates with ultra-low attachment at a density of 10,000 viable cells/ml and grown for 10 days. Mammospheres were counted and photographed.
2.11. RNA-Fluorescence in situ hybridization (RNA-FISH)
Fluorescence in situ hybridization for miR-873 and PD-L1 was performed on frozen slice. The frozen slice was rehydrated in citrate buffer. Proteinase K digestion was used to treat fixed tissues at 37 °C for 20 min. After digestion, slice was hydrated through a graded series of alcohol to tap water after immersing in RNase-free water for 3 min and then air dried. Hybridization was carried out overnight at 37 °C using hybridization buffer supplemented with denatured 5′ CY3-labeled Locked Nucleic Acid (LNA) probes directed against miR-873 and 5′ FAM-labeled Locked Nucleic Acid (LNA) probes directed against PD-L1 (GenePharma, China). After washing, the sections were counterstained with 4,6-diamidino-2-phenylindole (DAPI) for 20 min and observed with the confocal microscopy.
2.12. Nude mouse tumor xenograft model
All animal experiments were performed with the approval of Ethics Committee for Animal Experimentation of China Pharmaceutical University. 4–6 weeks male athymic BALB/c nude mice were purchased from Model Animal Research Center of Nanjing University, housed and fed in standard pathogen-free conditions. For tumor-limiting dilution assays, 1 × 106 and 1 × 105 and 1 × 104 MDA-MB-231 cells with miR-873 overexpression, PD-L1 knockdown or not were mixed with Matrigel matrix (1:1, BD Biosciences) and orthotopically implanted in the inguinal mammary gland of mice. After 8 days, all mice were euthanized, and tumor tissues were collected. The stem cell frequencies were calculated using an ELDA (http://bioinf.wehi.edu.au/software/elda/).
Additionally, 1 × 106 MCF-7/ADR cells were implanted in the inguinal mammary gland of mice. Fourteen days later saline or lentivirus gene therapy vectors were administered into the tumor using the following doses: plvx-873, Plko.1-PD-L1 or plvx-873 together with plvx-PD-L1: 0.40 × 108 pfu/site. When the tumors reached the volume of 100 mm3, we randomly allocated the mice to groups in which they received adriamycin (0.5 mg/kg, Shenzhen Main Luck Pharmaceuticals Inc. China) or saline. Mice were euthanized 18 days after the inoculation.
2.13. Statistical analysis
Results were presented as the mean ± SD (standard deviation) and statistical analyses compared using unpaired Student's t-test. Values of P < 0.05 or less were considered statistically significant. (*P < 0.05, **P < 0.01,***p < 0.001); ns indicates no significant differences from control.
3. Results
3.1. PD-L1 positively correlated with the expression of stemness markers in breast cancer tissues
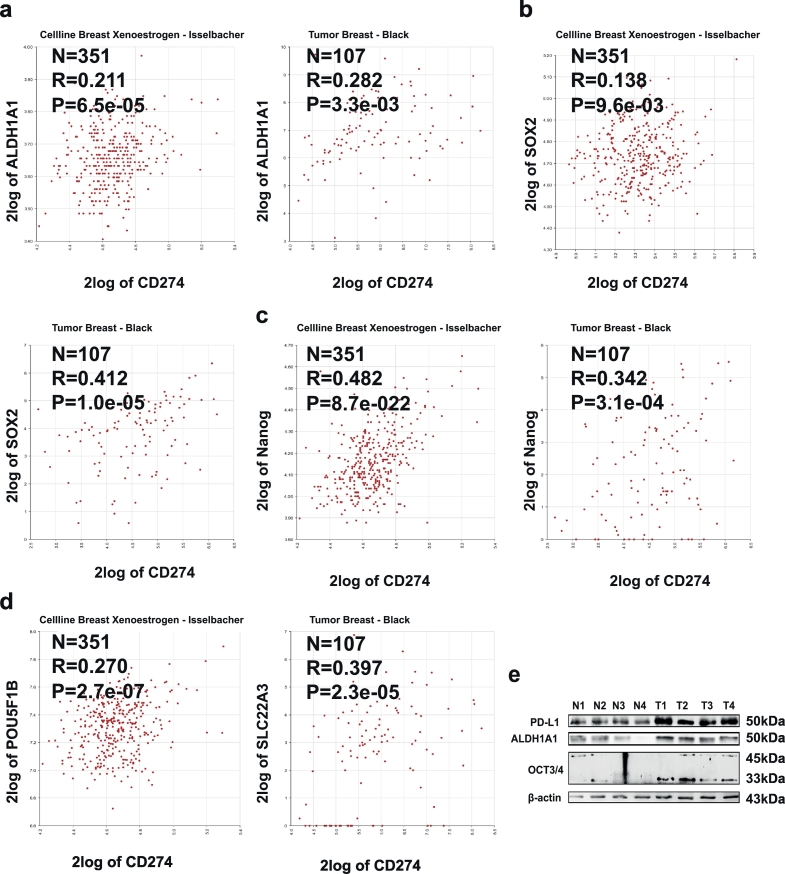
First, we assessed the expression correlation between stemness markers (ALDH1A1, SOX2, Nanog, POU5F1B) and PD-L1 across cellline breast xenoestrogen using R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl), and found that the expression of PD-L1 and stemness markers displayed a positive correlation in cellline breast xenoestrogen (Transcriptome analysis of MCF-7 cells exposed for 48 h to various concentrations of xenoestrogen chemicals) and tumor breast (Fig. 1a–d). Furthermore, PD-L1 expression was significantly increased in breast cancer tissues, and the expression of PD-L1 and stemness markers (ALDH1A1 and OCT3/4) exhibited a similar pattern in clinical samples as higher in tumor samples and lower in normal samples (Fig. 1e). These results indicate that PD-L1 might regulate the stemness of breast cancer cells.
Fig. 1.
PD-L1 was positively correlated with the expression of stemness markers in breast cancer tissues and cells. (a–d) PD-L1 expression is positively correlated with the expression of stemness markers in cellline breast xenoestrogen or tumor breast. (e) The expression of PD-L1 and stemness signatures (OCT3/4, ALDH1A1) was detected in breast cancer and normal mammary tissues via western blot.
3.2. miR-873 directly targeted PD-L1 3′UTR
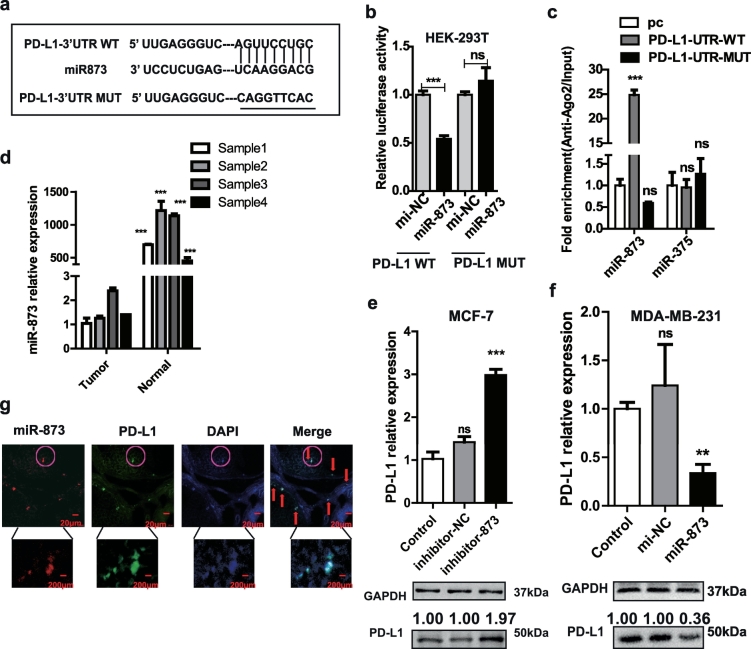
miRWalk2.0 (a comprehensive atlas of predicted and validated miRNA-target interactions) showed that PD-L1 is a potential direct target of miR-873 (Fig. 2a), which had been confirmed to inhibit breast cancer proliferation by our group and others [25,26]. Luciferase reporter assay revealed that the activity of pMIR-PD-L1 3′ UTR WT was remarkably inhibited by miR-873, while no significant difference in pMIR-PD-L1-3′ UTR MUT (Fig. 2b). In addition, miRNAs conjugated in RNA-induced silencing complex (RISC) were pulled down by Ago2 and detected by qRT-PCR. Results showed that miR-873 was enriched in cells with PD-L1-UTR-WT overexpression, but not in PD-L1-UTR-MUT, and miR-375 served as a negative control without binding sites on PD-L1 (Fig. 2c). Moreover, miR-873 expression was significantly decreased in tumor tissues (Fig. 2d). Since PD-L1 expression is higher in MDA-MB-231 cells than MCF-7 cells [15], which is opposite to miR-873 expression in these two cell lines (Fig. S1a and b). miR-873 was chosen for overexpression in MDA-MB-231 and inhibition in MCF-7 cells, respectively. As shown in Fig. 2e, f, and Fig. S1e, f, PD-L1 expression was promoted by inhibitor-873 in MCF-7 cells and suppressed by miR-873 in MDA-MB-231 cells, respectively. The transfection efficiency of miR-873 and inhibitor-873 was tested by qRT-PCR (Fig. S1c and d). Finally, FISH (Fluorescence in situ hybridization) assay revealed a clear overlap between miR-873 and PD-L1 in breast cancer tissues (Fig. 2g). Taken together, these findings indicate that miR-873 could directly target PD-L1 in breast cancer.
Fig. 2.
miR-873 directly targeted PD-L1 3′UTR. (a) Binding sites among miR-873 in PD-L1-3′ UTR and PD-L1-3′ UTR-MUT were shown. (b) Luciferase reporter assay confirmed the direct binding of miR-873 on PD-L1. Luciferase activity was measured and normalized to β-galactosidase activity. (***p < 0.001 vs Control group). (c) RIP assay was further verified for direct association between miR-873 and PD-L1, followed by qRT-PCR analysis for miR-873. miR-375 was used as negative control. (***p < 0.001 vs pcDNA3.1(+) empty vector). (d) The expression of miR-873 was detected in breast cancer and normal mammary tissues via qRT-PCR. (e, f) Effects of miR-873 or inhibitor-873 on PD-L1 expression in MDA-MB-231 or MCF-7 cells. (***p < 0.001 vs Control). (g) RNA-FISH of location between miR-873 and PD-L1 in breast cancer tissues. (Data were presented as the mean ± SD, n = 3).
3.3. miR-873 attenuated the stemness of breast cancer cells through PD-L1
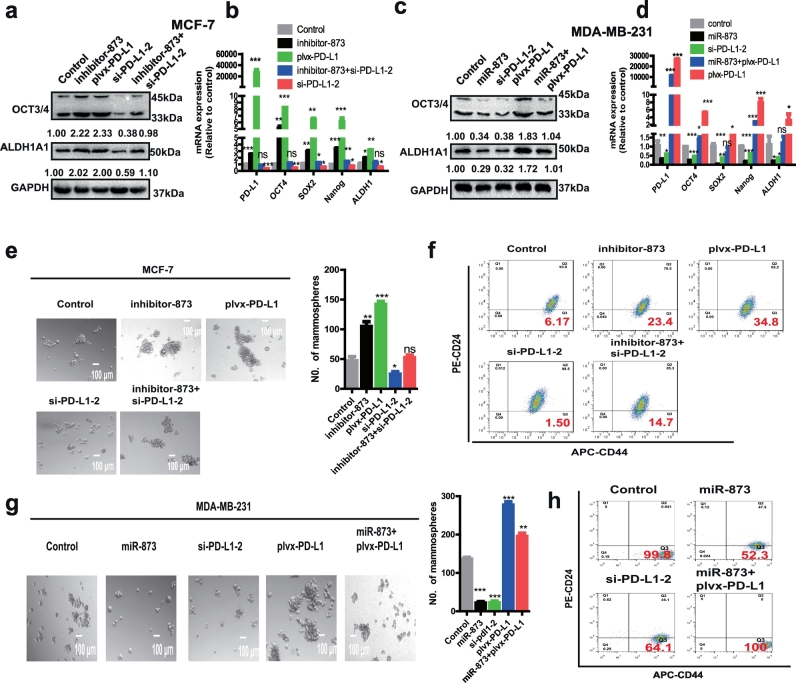
Since PD-L1 expression was statistically highly associated with stemness-correlated genes in breast cancer tissues and it could sustain their expression in breast cancer stem cells [13], we inferred that miR-873/PD-L1 regulatory axis regulates the stemness of breast cancer cells. Firstly, we selected si-PD-L1-2 for knocking down PD-L1 as it displayed the highest knockdown efficiency in both MCF-7 and MDA-MB-231 cells (Fig. S2a and b). Moreover, PD-L1 overexpression reversed the knockdown effects of si-PD-L1, excluding the off-target effects of PD-L1 siRNA (Fig. S2c). The expression of several pluripotent transcription factors (OCT4, ALDH1A1, Nanog and SOX2) was increased in MCF-7 cells with inhibitor-873 or plvx-PD-L1 transfection, while co-transfection with inhibitor-873 and si-PD-L1-2 attenuated or even reversed the promoting effects induced by inhibitor-873 (Fig. 3a and b). In contrast, the expression of stemness markers was inhibited in MDA-MB-231 cells with miR-873 overexpression or PD-L1 knockdown, and the suppressive effects mediated by miR-873 were attenuated by PD-L1 overexpression (Fig. 3c and d). The transfection efficiency was confirmed and denoted in Fig. S2d–f. Furthermore, transfection of inhibitor-873 or plvx-PD-L1 increased the CD44+/CD24− population or cell spheroid formation in MCF-7 cells, which could characterize the stemness of breast cancer cells [30], and inhibitor-873-mediated promotion was attenuated by PD-L1 knockdown (Fig. 3e and f). In addition, consistent results were obtained in MDA-MB-231 cells (Fig. 3g and h). Overall, these findings indicate that the miR-873/PD-L1 axis negatively regulates the stemness of breast cancer cells in vitro.
Fig. 3.
miR-873 attenuated the stemness of breast cancer cells through PD-L1. (a, b) The expression of stemness signatures (OCT3/4, ALDH1A1) was detected in MCF-7 cells with miR-873 knockdown or PD-L1 overexpression, or miR-873 knockdown plus PD-L1 knockdown. (c, d) Expression of several pluripotent transcription factors (OCT4, SOX2, Nanog, ALDH1) was examined in MDA-MB-231 cells with miR-873 overexpression or PD-L1 knockdown, or miR-873 overexpression plus PD-L1 overexpression. (e–h) Spheroid formation or CD44+/CD24- population was measured in cells depicted in (a and c). (Data were presented as the mean ± SD, n = 3. *p < 0.05, **p < 0.01, ***p < 0.001 vs Control group).
3.4. miR-873/PD-L1 regulatory axis inhibited the tumor initiating ability of breast cancer cells in vivo
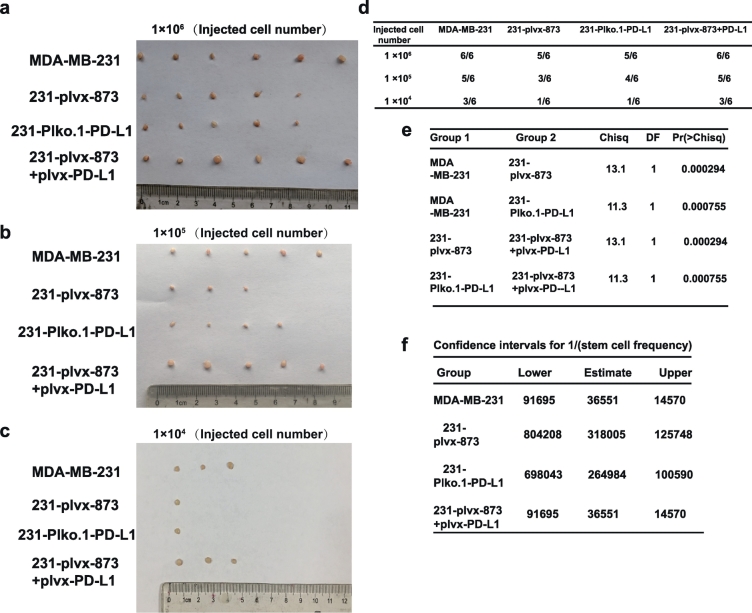
To further investigate whether the miR-873/PD-L1 regulatory axis could inhibit tumor-initiating potential of breast cancer cells in vivo, we constructed MDA-MB-231 cells with miR-873 (plvx-873) stable overexpression, PD-L1 stable knockdown (Plko.1-PD-L1), or miR-873 stable overexpression together with PD-L1 overexpression (plvx-873 and plvx-PD-L1) to seed tumors at limiting dilutions. The stable expression of miR-873 and PD-L1 was verified by qRT-PCR (Fig. S3a and b). We noticed that overexpression of miR-873 or knockdown of PD-L1 remarkably attenuated the tumor-initiating potential of MDA-MB-231 cells (Fig. 4a–f). Importantly, the attenuated tumor-initiating potential of miR-873 overexpression was rescued by PD-L1 overexpression (Fig. 4a–f). Therefore, our results suggest that miR-873 could suppress the tumor-initiating ability through PD-L1.
Fig. 4.
miR-873/PD-L1 axis inhibited the tumor initiating ability of breast cancer cells in vivo. (a–f) Images of tumors and index of tumors harvested when serially diluted MDA-MB-231 cells with Plko.1-PD-L1, plvx-873 alone or together with plvx-PD-L1 were planted.
3.5. miR-873 suppressed the stemness through inactivating PI3K/Akt and ERK1/2 pathways in a PD-L1 dependent manner
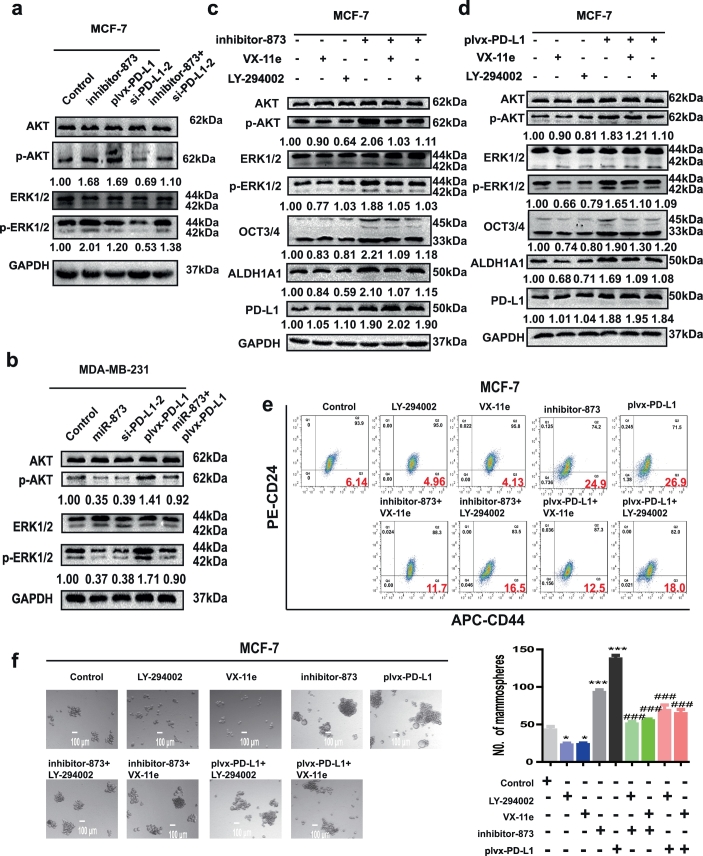
Previous studies have shown that PD-L1 could activate PI3K/AKT and ERK pathways in breast cancer [13,31]. Thus, we inferred whether miR-873 attenuated the stemness of breast cancer cells through these two pathways. As expected, transfection with inhibitor-873 increased p-AKT and p-ERK levels as well as PD-L1 overexpression in MCF-7 cells, and knockdown of PD-L1 reversed the effects mediated by inhibitor-873 (Fig. 5a). Consistently, p-AKT and p-ERK levels were downregulated in MDA-MB-231 cells with miR-873 overexpression or PD-L1 knockdown, and the effects of miR-873 overexpression were reversed by PD-L1 overexpression (Fig. 5b). Furthermore, the increased expression of OCT3/4 and ALDH1A1 induced by both inhibitor-873 and plvx-PD-L1 was attenuated by PI3K/AKT inhibitor (LY-294002) or ERK1/2 inhibitor (VX-11e) (Fig. 5c and d). Additionally, consistent results were acquired in regulating CD44+/CD24− sub-population and spheroid formation (Fig. 5e and f). Collectively, these results demonstrate that miR-873/PD-L1 axis regulates the stemness of breast cancer cells through PI3K/Akt and ERK1/2 pathways.
Fig. 5.
miR-873 suppressed the stemness through inactivating PI3K/Akt and ERK1/2 pathways in a PD-L1 dependent manner. (a, b) p-AKT and p-ERK levels were detected in MCF-7 and MDA-MB-231 cells with different treatment shown in labels. (c, d) The expression levels of p-AKT/p-ERK and stemness markers were examined in MCF-7 cells transfected with inhibitor-873 plus PI3K/AKT inhibitor (LY-294002) or ERK1/2 inhibitor (VX-11e), or plvx-PD-L1 plus LY-294002 or VX-11e, or not. (e, f) Spheroid formation or CD44+/CD24- sub-population was measured in MCF-7 cells depicted in (c). (***p < 0.001 vs Control group. ###p < 0.001 vs plvx-PD-L1 or inhibitor-873). (Data were presented as the mean ± SD, n = 3).
Interferon-γ (IFN-γ) is a key inducer of PD-L1 expression and is among the cytokines present in the microenvironment of some breast cancer and other tumors [[32], [33], [34]]. MCF-7 cells were incubated with IFN-γ for 24 h, and PD-L1 expression was induced by IFN-γ stimulation (Fig. S4a and b). Furthermore, PI3K/Akt and ERK1/2 pathways were activated, and the expression of stemness markers was increased by IFN-γ treatment, these effects were attenuated by PD-L1 knockdown. Additionally, consistent results were observed upon detecting the CD44+/CD24- population and spheroid formation (Fig. S4c and d).
3.6. miR-873/PD-L1 regulatory axis regulated the sensitivity of adriamycin via modulating the stemness of breast cancer cells
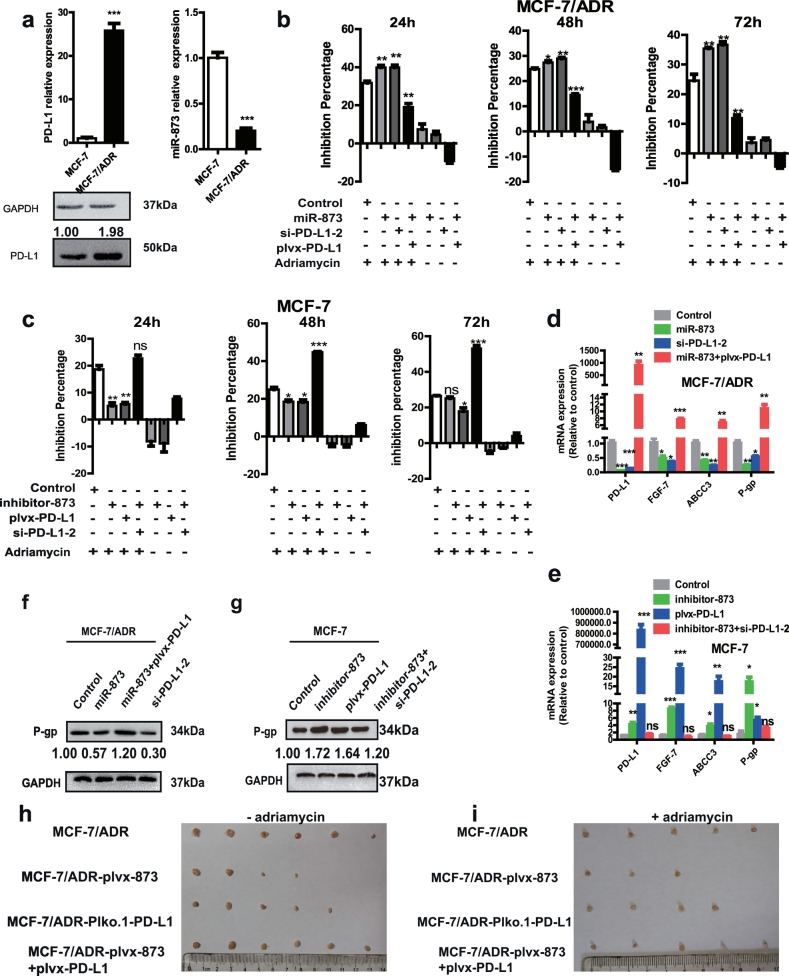
Since CSCs could contribute to chemoresistance in cancers [35], we attempted to explore the potential involvement of miR-873/PD-L1 axis in regulating chemoresistance of breast cancer cells. Firstly, we found that PD-L1 expression was significantly increased in MCF-7/ADR cells, while miR-873 exhibited a lower level (Fig. 6a). We selected si-PD-L1-2 as a candidate of siRNA-mediated knockdown of PD-L1 for MCF-7/ADR as it displayed the strongest knockdown efficiency in MCF-7/ADR cells (Fig. S5a). Additionally, MCF-7/ADR cells displayed a stronger stemness relative to MCF-7 cells, characterized as the higher expression of stemness markers (Fig. S5b and c), stronger capacity of spheroids formation (Fig. S5d), and higher proportion of CD44+/CD24− phenotype (Fig. S5e). A consistent hyper-activation of PI3K/Akt and ERK1/2 pathways was observed in MCF-7/ADR cells relative to MCF-7 cells (Fig. S5c). These effects concluded that MCF-7/ADR cells held a stronger stemness than MCF-7 cells. Then we further investigated whether miR-873/PD-L1 axis held the similar effects in MCF-7/ADR cells with that in MDA-MB-231 cells. As expected, an obvious downregulation of pluripotent transcription factors expression, cell spheroid formation and CD44+/CD24− sub-population by si-PD-L1-2 and miR-873 transfection was observed in MCF-7/ADR cells, and miR-873-mediated inhibition was indeed attenuated or even reversed by PD-L1 overexpression (Fig. S5b–e).
Fig. 6.
miR-873/PD-L1 regulatory axis regulated the sensitivity of adriamycin via modulating the stemness of breast cancer cells. (a) PD-L1 and miR-873 relative expression in MCF7/ADR and MCF-7 cells was detected by qRT-PCR and western blot. (b, c) MTT assay was performed to detect inhibition rate of proliferation of adriamycin in MCF-7/ADR cells with miR-873 overexpression or PD-L1 knockdown, or miR-873 overexpression plus PD-L1 overexpression (b), and MCF-7 cells with transfection of inhibitor-873 or plvx-PD-L1, or inhibitor-873 plus si-PD-L1-2 (c). (d, e) Expression of drug resistance genes (FGF-7, ABCC3, P-gp) and PD-L1 was detected by qRT-PCR in MCF-7/ADR cells depicted in (b), and MCF-7 cells depicted in (c). (f, g) Western blot was performed to test P-gp expression in MCF-7 and MCF-7/ADR cells depicted in (b and c). (h, i) Images of tumors harvested when nude mice received MCF-7/ADR cells 2 weeks, after then mice were received saline or gene therapy vectors (plvx-873, Plko.1-PD-L1, plvx-873 with plvx-PD-L1) before adriamycin treatment or not. (Data were presented as the mean ± SD, n = 3. *p < 0.05, **p < 0.01, ***p < 0.001 vs Control group).
After MCF-7/ADR cells and MCF-7 cells were treated with different concentrations of adriamycin, MCF-7/ADR and MCF-7 cells possessed around 20% inhibition at 8.75 μM and 0.08 μM, respectively (Fig. S6a and b), which could be a favorable concentration to test the sensitivity as it was not sufficient to create an irreversible damage. As expected, there was a higher inhibition percentage with miR-873 overexpression or PD-L1 knockdown in MCF-7/ADR cells, overexpression of PD-L1 reversed the effects of miR-873 (Fig. 6b). Similarly, transfection with inhibitor-873 or PD-L1 overexpression displayed a lower inhibition relative to control, whereas PD-L1 knockdown reversed the effect of inhibitor-873 (Fig. 6c). Additionally, transfection alone without additional drug treatment could exert similar but weaker (Fig. 6b) or stronger (Fig. 6c) effects than groups with drug treatment. These effects demonstrated the combined effects of miR-873/PD-L1 axis with adriamycin. The transfection efficiency and miR-873/PD-L1 interaction were confirmed in MCF-7/ADR cells (Fig. S7a and b). Similarly, the off-target effect was excluded (Fig. S7c).
Furthermore, MCF-7/ADR cells with miR-873 overexpression or PD-L1 knockdown displayed reduced mRNA levels of the drug-resistant genes [36], and PD-L1 overexpression reversed miR-873-mediated effects (Fig. 6d). Moreover, the expression of drug resistance genes was increased in MCF-7 cells with transfection of inhibitor-873 or plvx-PD-L1, and inhibitor-873-mediated effects were rescued by si-PD-L1-2 (Fig. 6e). Consistent results were obtained in P-gp protein level (Fig. 6f and g). Importantly, PD-L1 knockdown or miR-873 overexpression impaired the tumor-initiating potential of MCF-7/ADR cells, and overexpression of PD-L1 reversed plvx-873-mediated attenuation (Fig. 6h), these effects were enhanced by adriamycin treatment together (Fig. 6i). The transfection efficiency of plvx-873, Plko.1-PD-L1, plvx-873 with plvx-PD-L1 were confirmed in MCF-7/ADR cells (Fig. S7d and e). Thus, these results demonstrated that miR-873/PD-L1 axis could attenuate adriamycin resistance via reducing the stemness of breast cancer cells.
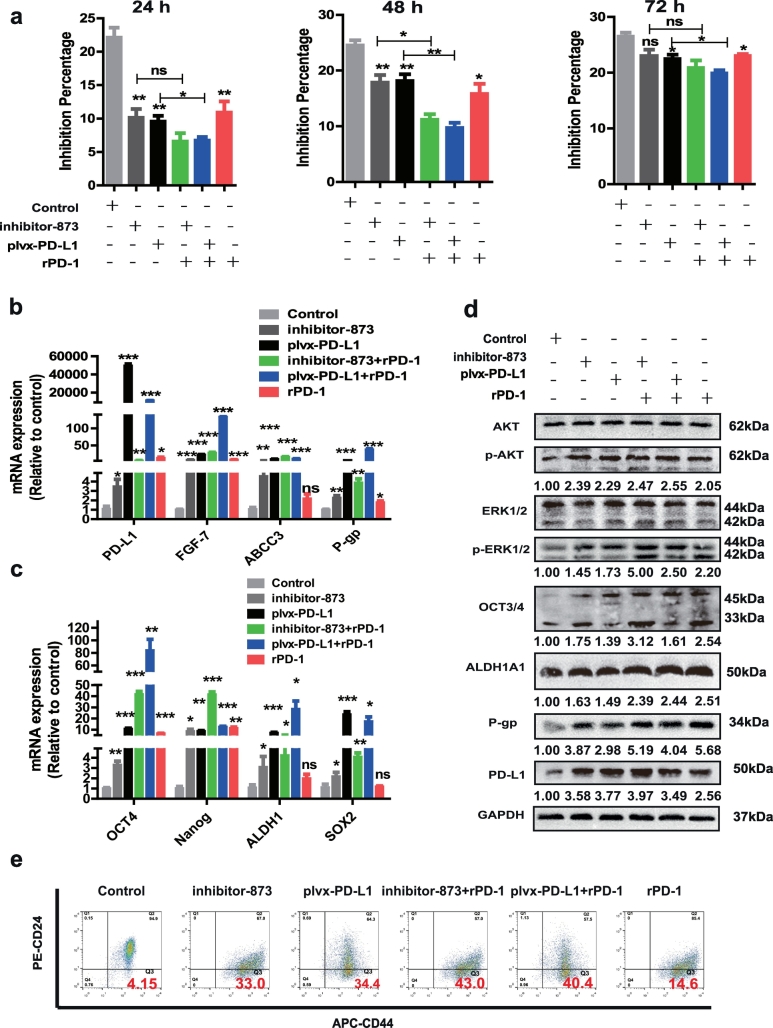
3.7. rPD-1 attenuated the inhibitory effect of miR-873/PD-L1 axis on the stemness and drug resistance
Eventually, we explored whether PD-1/PD-L1 axis is involved in miR-873/PD-L1 axis-mediated effects on the stemness and drug resistance in breast cancer cells. MCF-7 cells were incubated with rPD-1 for 24 h prior to exposure to adriamycin. Interestingly, rPD-1 possessed similar effects with inhibitor-873 or plvx-PD-L1, and enhanced inhibitor-873 or plvx-PD-L1-mediated inhibition on adriamycin sensitivity (Fig. 7a). Additionally, inhibitor-873- or plvx-PD-L1-induced promoting effects on the expression of drug resistance genes (Fig. 7b) and stemness markers, and PI3K/Akt and ERK1/2 pathways activation (Fig. 7c and d) were strengthened by rPD-1. Moreover, rPD-1 with inhibitor-873 or plvx-PD-L1 exerted an additive promotion on the CD44+/CD24- population (Fig. 7e). Finally, the effects of rPD-1 on the CD44+/CD24- population were attenuated by si-PD-L1-2 or BMS-202 (PD-1/PD-L1 inhibitor 2), respectively (Fig. S8a and b). Thus, these results suggest that PD-1 could interact with PD-L1 to enhance the promotion of PD-L1 on the stemness and drug resistance, and attenuate miR-873-induced inhibition.
Fig. 7.
rPD-1 attenuated the inhibitory effect of miR-873/PD-L1 axis on the stemness and drug resistance. (a) Inhibition percentage of inhibitor-873 and plvx-PD-L1 after exposure to rPD-1 (0.5 μg/ml) 24 h before adriamycin in MCF-7 cells. (b, c) Expression of drug resistance and stemness markers genes was measured in cells depicted in (a) by qRT-PCR. (d) The activation of PI3K/Akt and ERK1/2 pathways, and stemness properties were examined in MCF-7 cells depicted in (a). (e) CD44+/CD24- populations in MCF-7 cells depicted in (a) was evaluated. (Data were presented as the mean ± SD, n = 3. *p < 0.05, **p < 0.01, ***p < 0.001 vs Control group).
4. Discussion
In this study, analysis of clinical samples showed that PD-L1 expression was positively correlated with the expression of stemness markers. And PD-L1 exhibited a higher level, while miR-873 displayed a lower level in MCF-7/ADR cells, which held stronger stemness relative to MCF-7 cells. Luciferase reporter assay combined with RNA-FISH and RIP verified miR-873 targeting on PD-L1. Furthermore, we demonstrated that the inhibitory roles of miR-873/PD-L1 axis in regulating drug resistance and stemness were through inactivating PI3K/Akt and ERK1/2 pathways. Notably, rPD-1 counteracted the inhibitory effects of miR-873/PD-L1 axis on the stemness of breast cancer cells.
Among all immune checkpoints, the PD-1/PD-L1 pathway is prominent for its proven results as a therapeutic target in a variety of cancers [37]. Recently, antibodies targeting the PD-1/PD-L1 axis have been approved for some kinds of cancers, however, they were not approved in breast cancer although PD-L1 correlates with poor prognosis in breast cancer patients [14,38], this presumably due to the confused mechanisms by which PD-L1 is regulated or functions exerted by PD-L1 alone. Compelling evidences have shown that PD-L1 protein levels are regulated by miRNAs, such as miR-513 [39], miR-138-5p in colorectal cancer [19], miR-200 in NSCLC [40] and miR-15a in malignant pleural mesothelioma [41]. We firstly clarified that PD-L1 was directly regulated by miR-873 in breast cancer, which could facilitate the understanding of the mechanisms by which PD-L1 was regulated, and future works could be performed to explore the effects of combined miR-873 agonist with PD-L1 antibody in breast cancer. Additionally, we found that PD-L1 promotes the stemness of breast cancer alone, this effect could be enhanced by rPD-1, demonstrating that PD-L1 could function independent on PD-1/PD-L1 axis. However, it was not elucidated whether miR-873 promoted immunotherapy in our research. Since miRNAs could influence antitumor immune responses by affecting the expression of immune modulatory molecules in tumor and immune cells [42], and PD-L1/PD-1 immune checkpoint pathway could inhibit the T-cell antitumor immune response [43], miR-873 might exhibit immune modulation in breast cancer cells via regulating PD-L1.
CSCs contribute to drug resistance. Recent study has demonstrated an upregulation of PD-L1 expression in cancer cells by chemo-preventive agents and a resulting decrease in tumor-specific T-cell activity that potentially promotes immune evasion [17]. Here, we detected that drug resistance cells MCF-7/ADR possessed higher PD-L1 expression than MCF-7, indicating that PD-L1 overexpression might promote immune evasion in MCF-7/ADR cells. Notably, PD-L1 overexpression promoted the stemness of breast cancer cells, suggesting that CSCs might escape from the immune surveillance, this could be explored in the future work. Additionally, ectopic inactivation of miR-873/PD-L1 signaling reduced adriamycin sensitivity, while activation of this signaling attenuated adriamycin resistance, supporting that PD-1/PD-L1 antibodies could be combined with chemotherapy in breast cancer [44]. Moreover, as anti-PD-1 has been widely used as immune-therapy in multiple cancer types, such treatments may become more effective if the PD-L1 positive CSCs can be rendered simultaneously with miR-873 agonist. Notably, the effects of miR-873/PD-L1 axis and rPD-1 seem to progressively decrease over time in Fig. 7a, we think this could be due to the fact that the transient transfection has a limited timeless and validity.
The Ras/Raf/MEK/ERK and RAS/PI3K/PTEN/mTOR pathways are well known regulators of cell proliferation and apoptosis which ultimately lead to drug resistance in various cancers including breast cancer [[45], [46], [47]]. Other researchers indicated that the PD-1/PD-L1-induced MDR1/P-gp expression was mediated by activating PI3K/AKT and MAPK/ERK pathways [31]; and PD-1/PD-L1 axis led to chemoresistance of tumor cells [48], this is consistent with our results that PD-1/PD-L1 axis contributed to both stemness and drug resistance via activating PI3K/Akt and ERK1/2 pathways. Notably, as the modulation of miR-873/PD-L1 axis on stemness is attenuated but not abrogated by AKT/ERK inhibitors, suggesting the presence of other pathways involved. Conventional chemotherapies seemed to have reached a therapeutic puzzle in the treatment of solid tumors and many metastatic diseases are still incurable. Immunotherapy has raised interests in advanced cancer therapy, however this strategy needs to face this new challenge exploring its efficacy in cure CSCs, and its potential against CSCs is a novel field of research. Importantly, to date, mechanistic studies of PD-1 have been largely focused on its role on T cells as PD-1 in tumor micro-environment could facilitate the immune evasion of tumor cells, however, PD-1 has also been found to be expressed on tumor cells [49]. Here, we showed that rPD-1 promoted the stemness of breast cancer cells, indicating that endogenous PD-1 in tumor cells might help CSCs cells escaping from immune-surveillance and thus leading to drug resistance. To the best of our knowledge, this is the first study indicating PD-1 roles and related mechanism in regulating the stemness of breast cancer cells. Notably, we must admit the deficiency of this work, such as, 8 days from tumor implantation may not be a sufficient time to appreciate differences in tumorigenicity and to perform limiting-dilution essay (LDA). More days on this exploration could strengthen the current conclusions. Meanwhile, we should further confirm that the miR-873/PD-L1 axis and its modulatory effects could hold true even in cases where endogenous PD-L1 induced by IFN-γ on stemness not only in tumor cases with constitutive expression of PD-L1.
In conclusion, we identified a mechanism that miR-873 influenced the PD-L1/PD-1 immune checkpoint signaling pathways responsible for drug resistance and the stemness in breast cancer cells (Fig. 8). This study provided compelling evidence that suggests a targeting strategy involving miR-873 together with chemo-therapy or immune blockage to treat breast cancer.
Fig. 8.
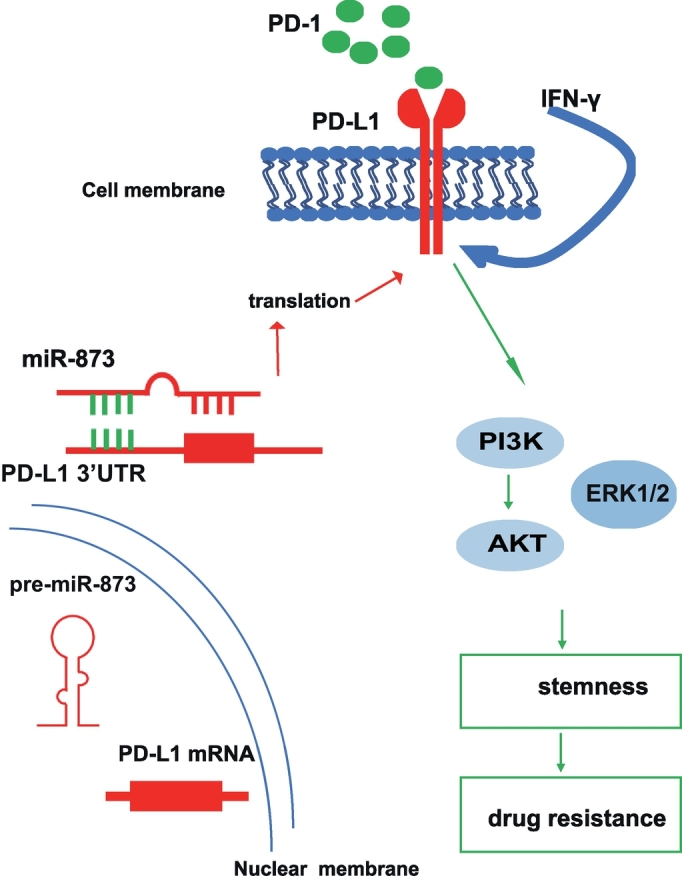
Proposed model that miR-873 inhibits the stemness of breast cancer cells and thus drug resistance. miR-873 could suppress the stemness of breast cancer cells through inactivating PI3K/Akt and ERK1/2 pathways via directly targeting PD-L1 3′UTR, this effect was attenuated by rPD-L1.
Acknowledgments
Acknowledgements
This work was supported by the National Nature Science Foundation of China, No. 81702957, China Postdoctoral Science Foundation, No. 2017M620230, the Postdoctoral Research Funding Scheme of Jiangsu Province (2017), No. 1701197B, and the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions.
Conflicts of interest
The authors declare no potential conflicts of interest.
Author contributions
Tao Xi and Lufeng Zheng designed the research and analyzed the data. Lanlan Gao, Qianqian Guo, Xiaoman Li, Xuan Yang, Haiwei Ni, Ting Wang, Qiong Zhao performed the research and analyzed the data. Hai Liu and Yingying Xing provided additional help. Lanlan Gao and Lufeng Zheng wrote the paper. All authors read and approved the final manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.02.034.
Contributor Information
Tao Xi, Email: Xitao18@hotmail.com.
Lufeng Zheng, Email: zhlf@cpu.edu.cn.
Appendix A. Supplementary data
Supplementary material
References
- 1.Reid P.A., Wilson P., Li Y., Marcu L.G., Bezak E. Current understanding of cancer stem cells: review of their radiobiology and role in head and neck cancers. Head Neck. 2017;39(9):1920–1932. doi: 10.1002/hed.24848. [DOI] [PubMed] [Google Scholar]
- 2.Gottesman M.M. Mechanisms of cancer drug resistance. Annu Rev Med. 2002;53(1):615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 3.Alison M.R., Lim S.M., Nicholson L.J. Cancer stem cells: problems for therapy? J Pathol. 2011;223(2):147–161. doi: 10.1002/path.2793. [DOI] [PubMed] [Google Scholar]
- 4.Ou Y., Guo X.L. Tumor stem cells and drug resistance. Sheng Li Ke Xue Jin Zhan. 2007;38(2):115–119. [PubMed] [Google Scholar]
- 5.Visvader J.E., Lindeman G.J. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8(10):755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 6.Yang Z.J., Wechsler-Reya R.J. Hit 'em where they live: targeting the cancer stem cell niche. Cancer Cell. 2007;11(1):3–5. doi: 10.1016/j.ccr.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Ishibashi M., Tamura H., Sunakawa M., Kondo-Onodera A., Okuyama N., Hamada Y. Myeloma drug resistance induced by binding of myeloma B7-H1 (PD-L1) to PD-1. Cancer Immunol Res. 2016;4(9):779–788. doi: 10.1158/2326-6066.CIR-15-0296. [DOI] [PubMed] [Google Scholar]
- 8.Brahmer J.R., Tykodi S.S., Chow L.Q., Hwu W.J., Topalian S.L., Hwu P. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel S.P., Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther. 2015;14(4):847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 10.Fujita Y., Yagishita S., Hagiwara K., Yoshioka Y., Kosaka N., Takeshita F. The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Mol Ther. 2015;23(4):717–727. doi: 10.1038/mt.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alsuliman A., Colak D., Al-Harazi O., Fitwi H., Tulbah A., Al-Tweigeri T. Bidirectional crosstalk between PD-L1 expression and epithelial to mesenchymal transition: significance in claudin-low breast cancer cells. Mol Cancer. 2015;14:149. doi: 10.1186/s12943-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mukherjee S., Manna A., Bhattacharjee P., Mazumdar M., Saha S., Chakraborty S. Non-migratory tumorigenic intrinsic cancer stem cells ensure breast cancer metastasis by generation of CXCR4 migrating cancer stem cells. Oncogene. 2016;35(37):4937–4948. doi: 10.1038/onc.2016.26. [DOI] [PubMed] [Google Scholar]
- 13.Almozyan S., Colak D., Mansour F., Alaiya A., Al-Harazi O., Qattan A. PD-L1 promotes OCT4 and Nanog expression in breast cancer stem cells by sustaining PI3K/AKT pathway activation. Int J Cancer. 2017;141(7):1402–1412. doi: 10.1002/ijc.30834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghebeh H., Mohammed S., Al-Omair A., Qattan A., Lehe C., Al-Qudaihi G. The B7-H1 (PD-L1) T lymphocyte-inhibitory molecule is expressed in breast cancer patients with infiltrating ductal carcinoma: correlation with important high-risk prognosticfactors. Neoplasia. 2006;8(3):190–198. doi: 10.1593/neo.05733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soliman H., Khalil F., Antonia S. PD-L1 expression is increased in a subset of basal type breast cancer cells. PLoS One. 2014;9(2) doi: 10.1371/journal.pone.0088557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felekkis K., Touvana E., Stefanou C., Deltas C. microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia. 2010;14(4):236–240. [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X., Li J., Dong K., Lin F., Long M., Ouyang Y. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell Signal. 2015;27(3):443–452. doi: 10.1016/j.cellsig.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Xu S., Tao Z., Hai B., Liang H., Shi Y., Wang T. miR-424(322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat Commun. 2016;7 doi: 10.1038/ncomms11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao L., Yu H., Yi S., Peng X., Su P., Xiao Z. The tumor suppressor miR-138-5p targets PD-L1 in colorectal cancer. Oncotarget. 2016;7(29):45370–45384. doi: 10.18632/oncotarget.9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park E.Y., Chang E., Lee E.J., Lee H.W., Kang H.G., Chun K.H. Targeting of miR34a-NOTCH1 axis reduced breast cancer stemness and chemoresistance. Cancer Res. 2014;74(24):7573–7582. doi: 10.1158/0008-5472.CAN-14-1140. [DOI] [PubMed] [Google Scholar]
- 21.Zhu Y., Yu F., Jiao Y., Feng J., Tang W., Yao H. Reduced miR-128 in breast tumor-initiating cells induces chemotherapeutic resistance via Bmi-1 and ABCC5. Clin Cancer Res. 2011;17(22):7105–7115. doi: 10.1158/1078-0432.CCR-11-0071. [DOI] [PubMed] [Google Scholar]
- 22.Celia-Terrassa T., Liu D.D., Choudhury A., Hang X., Wei Y., Zamalloa J. Normal and cancerous mammary stem cells evade interferon-induced constraint through the miR-199a-LCOR axis. Nat Cell Biol. 2017;19(6):711–723. doi: 10.1038/ncb3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang R.J., Li J.W., Bao B.H., Wu H.C., Du Z.H., Su J.L. MicroRNA-873 (miRNA-873) inhibits glioblastoma tumorigenesis and metastasis by suppressing the expression of IGF2BP1. J Biol Chem. 2015;290(14):8938–8948. doi: 10.1074/jbc.M114.624700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang Y., Zhang P., Li S., Li H., Song S., Lu B. MicroRNA-873 acts as a tumor suppressor in esophageal cancer by inhibiting differentiated embryonic chondrocyte expressed gene 2. Biomed Pharmacother. 2018;105:582–589. doi: 10.1016/j.biopha.2018.05.152. [DOI] [PubMed] [Google Scholar]
- 25.Cui J., Yang Y., Li H., Leng Y., Qian K., Huang Q. MiR-873 regulates ERalpha transcriptional activity and tamoxifen resistance via targeting CDK3 in breast cancer cells. Oncogene. 2015;34(30):3895–3907. doi: 10.1038/onc.2014.430. [DOI] [PubMed] [Google Scholar]
- 26.Zheng L., Meng X., Li X., Zhang Y., Li C., Xiang C. miR-125a-3p inhibits ERalpha transactivation and overrides tamoxifen resistance by targeting CDK3 in estrogen receptor-positive breast cancer. FASEB J. 2018;32(2):588–600. doi: 10.1096/fj.201700461RR. [DOI] [PubMed] [Google Scholar]
- 27.Zheng L., Xiang C., Li X., Guo Q., Gao L., Ni H. STARD13-correlated ceRNA network-directed inhibition on YAP/TAZ activity suppresses stemness of breast cancer via co-regulating Hippo and Rho-GTPase/F-actin signaling. J Hematol Oncol. 2018;11(1):72. doi: 10.1186/s13045-018-0613-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo X., Xiang C., Zhang Z., Zhang F., Xi T. Displacement of Bax by BMF mediates STARD13 3'UTR-induced breast cancer cells apoptosis in an miRNA-depedent manner. Mol Pharm. 2018;15(1):63–71. doi: 10.1021/acs.molpharmaceut.7b00727. [DOI] [PubMed] [Google Scholar]
- 29.Zheng L., Zhang Z., Zhang S., Guo Q., Zhang F., Gao L. RNA binding protein RNPC1 inhibits breast cancer cell metastasis via activating STARD13-correlated ceRNA network. Mol Pharm. 2018;15(6):2123–2132. doi: 10.1021/acs.molpharmaceut.7b01123. [DOI] [PubMed] [Google Scholar]
- 30.Liu S., Clouthier S.G., Wicha M.S. Role of microRNAs in the regulation of breast cancer stem cells. J Mammary Gland Biol Neoplasia. 2012;17(1):15–21. doi: 10.1007/s10911-012-9242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu S., Chen S., Yuan W., Wang H., Chen K., Li D. PD-1/PD-L1 interaction up-regulates MDR1/P-gp expression in breast cancer cells via PI3K/AKT and MAPK/ERK pathways. Oncotarget. 2017;8(59):99901–99912. doi: 10.18632/oncotarget.21914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Oliveira K.B., Guembarovski R.L., Guembarovski A.M., da Silva do Amaral Herrera A.C., Sobrinho W.J., Ariza C.B. CXCL12, CXCR4 and IFNgamma genes expression: implications for proinflammatory microenvironment of breast cancer. Clin Exp Med. 2013;13(3):211–219. doi: 10.1007/s10238-012-0194-5. [DOI] [PubMed] [Google Scholar]
- 33.Abiko K., Matsumura N., Hamanishi J., Horikawa N., Murakami R., Yamaguchi K. IFN-gamma from lymphocytes induces PD-L1 expression and promotes progression of ovarian cancer. Br J Cancer. 2015;112(9):1501–1509. doi: 10.1038/bjc.2015.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dorand R.D., Nthale J., Myers J.T., Barkauskas D.S., Avril S., Chirieleison S.M. Cdk5 disruption attenuates tumor PD-L1 expression and promotes antitumor immunity. Science. 2016;353(6297):399–403. doi: 10.1126/science.aae0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu F., Yao H., Zhu P., Zhang X., Pan Q., Gong C. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131(6):1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 36.AlHossiny M., Luo L., Frazier W.R., Steiner N., Gusev Y., Kallakury B. Ly6E/K signaling to TGFbeta promotes breast cancer progression, immune escape, and drug resistance. Cancer Res. 2016;76(11):3376–3386. doi: 10.1158/0008-5472.CAN-15-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun C., Mezzadra R., Schumacher T.N. Regulation and function of the PD-L1 checkpoint. Immunity. 2018;48(3):434–452. doi: 10.1016/j.immuni.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin T., Zeng Y.D., Qin G., Xu F., Lu J.B., Fang W.F. High PD-L1 expression was associated with poor prognosis in 870 Chinese patients with breast cancer. Oncotarget. 2015;6(32):33972–33981. doi: 10.18632/oncotarget.5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gong A.Y., Zhou R., Hu G., Li X., Splinter P.L., O'Hara S.P. MicroRNA-513 regulates B7-H1 translation and is involved in IFN-gamma-induced B7-H1 expression in cholangiocytes. J Immunol. 2009;182(3):1325–1333. doi: 10.4049/jimmunol.182.3.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen L., Gibbons D.L., Goswami S., Cortez M.A., Ahn Y.H., Byers L.A. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun. 2014;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kao S.C., Cheng Y.Y., Williams M., Kirschner M.B., Madore J., Lum T. Tumor suppressor microRNAs contribute to the regulation of PD-L1 expression in malignant pleural mesothelioma. J Thorac Oncol. 2017;12(9):1421–1433. doi: 10.1016/j.jtho.2017.05.024. [DOI] [PubMed] [Google Scholar]
- 42.Eichmuller S.B., Osen W., Mandelboim O., Seliger B. Immune modulatory microRNAs involved in tumor attack and tumor immune escape. J Natl Cancer Inst. 2017;109(10) doi: 10.1093/jnci/djx034. [DOI] [PubMed] [Google Scholar]
- 43.Xu C., Fillmore C., Koyama S., Wu H., Zhao Y., Chen Z. Loss of Lkb1 and Pten leads to lung squamous cell carcinoma with elevated PD-L1 expression. Cancer Cell. 2014;25(5):590–604. doi: 10.1016/j.ccr.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Postow M.A., Callahan M.K., Wolchok J.D. Immune checkpoint blockade in cancer therapy. J Clin Oncol. 2015;33(17):1974–1982. doi: 10.1200/JCO.2014.59.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis J.M., Navolanic P.M., Weinstein-Oppenheimer C.R., Steelman L.S., Hu W., Konopleva M. Raf-1 and Bcl-2 induce distinct and common pathways that contribute to breast cancer drug resistance. Clin Cancer Res. 2003;9(3):1161–1170. [PubMed] [Google Scholar]
- 46.McCubrey J.A., Abrams S.L., Stadelman K., Chappell W.H., Lahair M., Ferland R.A. Targeting signal transduction pathways to eliminate chemotherapeutic drug resistance and cancer stem cells. Adv Enzyme Regul. 2010;50(1):285–307. doi: 10.1016/j.advenzreg.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steelman L.S., Navolanic P.M., Sokolosky M.L., Taylor J.R., Lehmann B.D., Chappell W.H. Suppression of PTEN function increases breast cancer chemotherapeutic drug resistance while conferring sensitivity to mTOR inhibitors. Oncogene. 2008;27(29):4086–4095. doi: 10.1038/onc.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Black M., Barsoum I.B., Truesdell P., Cotechini T., Macdonald-Goodfellow S.K., Petroff M. Activation of the PD-1/PD-L1 immune checkpoint confers tumor cell chemoresistance associated with increased metastasis. Oncotarget. 2016;7(9):10557–10567. doi: 10.18632/oncotarget.7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao Y., Harrison D.L., Song Y., Ji J., Huang J., Hui E. Antigen-presenting cell-intrinsic PD-1 neutralizes PD-L1 in cis to attenuate PD-1 signaling in T cells. Cell Rep. 2018;24(2):379–390. doi: 10.1016/j.celrep.2018.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material