Abstract
Background
The therapeutic potential of faecal microbiota transplantation (FMT) is under investigation for a range of inflammatory conditions. While mechanisms of benefit are poorly understood, most models rely on the viability of transplanted microbes. We hypothesised that protocols commonly used in the preparation of faecal transplants will substantially reduce the number, diversity and functional potential of viable microbes.
Methods
Stools from eight screened donors were processed under strict anaerobic conditions, in ambient air, and freeze-thawed. Propidium monoazide (PMA) sample treatment was combined with quantitative PCR, 16S rRNA gene amplicon sequencing and short-chain fatty acid (SCFA) analysis to define the viable microbiota composition and functional potential.
Findings
Approximately 50% of bacterial content of stool processed immediately under strict anaerobic conditions was non-viable. Homogenisation in ambient air or freeze-thaw reduced viability to 19% and 23% respectively. Processing of samples in ambient air resulted in up to 12-fold reductions in the abundance of important commensal taxa, including the highly butyrogenic species Faecalibacterium prausnitzii, Subdoligranulum variable, and Eubacterium hallii. The adverse impact of atmospheric oxygen exposure on the capacity of the transplanted microbiota to support SCFA biosynthesis was demonstrated by significantly reduced butyrate and acetate production by faecal slurries processed in ambient air. In contrast, while reducing overall levels of viable bacteria, freeze-thaw did not significantly alter viable microbiota composition.
Interpretation
The practice of preparing material for faecal transplantation in ambient air profoundly affects viable microbial content, disproportionately reducing the abundance of anaerobic commensals and the capacity for biosynthesis of important anti-inflammatory metabolites.
Fund
This work was supported by the South Australian Health and Medical Research Institute. LP is supported by a scholarship from the Flinders Foundation. GR is supported by a Matthew Flinders Research Fellowship.
Keywords: Bacterial viability, Fecal microbiota transplantation, qPCR, Propidium monoazide
Research in context.
Evidence before this study
Stool from healthy donors is processed into faecal transplant material with the expectation that viable bacteria from the donor will be transplanted into the gut of the recipient. Despite the prominence of FMT in the clinical trials literature, few studies have attempted to assess the viability of bacteria in FMT material. We searched Pubmed for the terms “viability fecal microbiota”, “freezing fecal microbiota” and “PMA fecal microbiota” from 1980–2018 and also looked at citations of relevant articles. We were able to identify only three studies that have attempted to assess the viability of commensals in human faecal material. Using standard culture methods, Costello et al assessed the viability of Escherichia coli, Bifidobacterium and Lactobacilli in faecal microbiota transplant (FMT) material that had been processed in anaerobic conditions and then frozen for up to 6 months, reporting no significant effect of freezing when glycerol was used as a cryoprotective agent. Fouhy et al examined faecal material from 7 healthy donors before and after freezing using 16S rRNA amplicon gene sequencing and with culture evaluating total aerobes, total anaerobes and Bifidobaterium. Fouhy et al also found no significant effect overall of freezing using either culture-based methods or 16S rRNA amplicon sequencing. However, both approaches used in these studies have significant limitations. Culture-based assessment used by Costello et al is that only a small fraction of species present in stool can be readily cultured. Although molecular methods have the advantage of identifying the broad range of bacteria present in stool, standard sequencing methodology is ineffective at assessing viability as these methods will amplify DNA from both live and dead microorganisms. Only one publication, by Chu et al, combined propidium monoazide (PMA) based exclusion of non-viable DNA with 16S rRNA amplicon sequencing to assess the effect of freezing and ambient air processing on FMT material. This study had the limitation of assessing only one donor and methodological problems in applying PMA to stool samples. In the process of validating the PMA based method used in this study, we demonstrated that if using undiluted faecal slurry with PMA, as done by Chu et al, the method is not effective.
Added value of this study
We have performed the first comprehensive analysis of bacterial viability in faecal slurries used for FMT. Unlike previous publications, we were able to show a significant effect of freezing on the overall bacterial viability in FMT material, despite the use of glycerol. However, our findings in relation to the effects of freezing on specific bacterial taxa confirm the results shown by Costello et al’s culture-based viability study, as we also show no significant effect of freezing on the viability of Bifidobacterium and E. coli. We show that processing faecal material in ambient air profoundly affects the viability of several important bacterial taxa, with Faecalibacterium prausnitzii being the most affected. F. prausnitzii was also the species whose relative abundance was found by Chu et al to be most decreased after aerobic processing (this effect was observed regardless of PMA treatment). We report that the commensal organisms most affected by oxygen exposure are collectively major producers of butyrate in the gut, a metabolite which has been shown to be an important signalling molecule which influences inflammatory and metabolic pathways. Direct assessment of the levels of an enzyme central to bacterial butyrate biosynthesis, and measurement of butyrate levels by gas chromatography following in-vitro fermentation with pre-biotic starch supports the conclusion that ability of the microbiota as a whole to produce butyrate is significantly reduced by oxygen exposure.
Implications of all the available evidence
Following the publication of several high impact trials, such as that of van Nood et al published in 2013 (NEJM), showing the benefit faecal microbiota transplantation (FMT) for recurrent Clostridium difficile colitis, FMT has become an established treatment for this condition. FMT trials using stool processed by a variety of methods, including ambient air homogenisation, freezing, and freeze-drying, have consistently demonstrated benefit in C. difficile colitis. However, it cannot be assumed that these methods, which we show profoundly affect the viability of important commensal species, will not adversely affect the ability of FMT to produce benefit or increase the risk of harming the recipient, particularly when used for conditions other than C. difficile colitis. FMT is increasingly being investigated as a therapeutic intervention for a wide range of other conditions through numerous clinical trials. Many, particularly those targeting inflammatory and metabolic disorders, hypothesize that viable beneficial microbiota delivered via FMT will mediate their effects via the production of beneficial metabolites. Our finding that the ability of microbiota to produce short-chain fatty acids is significantly reduced by homogenisation of stool in ambient air is particularly relevant in these contexts. Little consideration is given in most clinical trials to the method of preparing FMT and how this might impact the safety and efficacy of FMT material. We believe an evidence based and standardized approach to preparing FMT as well as routine assessment of donor stool composition and viability is required in order minimize risks to the patient and ultimately understand how FMT might be able to produce clinical benefit.
Alt-text: Unlabelled Box
1. Introduction
In recent years, following major advances in nucleic acid sequencing technology, there has been a dramatic increase in research linking gut microbiota not only to intestinal pathologies such as inflammatory bowel disease [[1], [2], [3], [4]] or colorectal cancer [5], but also to a broad range of other medical conditions that include metabolic disorders and even mental health conditions [[6], [7], [8]]. This has led to the hypothesis that modification of gut microbiota via faecal microbiota transplantation (FMT) could have a therapeutic role in a diverse range of diseases.
FMT is a therapeutic intervention in which stool from one or more healthy donors is processed into a faecal slurry (FS) and delivered into the intestinal tract of the recipient. Although FMT is best established as a therapy for recurrent Clostridium difficile infection [9,10], there is increasing evidence for the use of FMT in inflammatory disorders, particularly ulcerative colitis [11].
Despite its increasing use, there is neither standardisation of donor screening nor standardisation of stool processing [12]. This lack of standardization extends to the use of FMT in clinical trials, of which there are over 200 registered on clinicaltrials.gov, making comparison of outcomes difficult. Worryingly, FMT is also often performed in private clinics, or even by patients themselves, for unproven indications and in a completely unregulated fashion [13].
Current guidelines for processing stool for FMT are intended for the treatment of C. difficile colitis and are based on expert opinion in the absence of evidence [[14], [15], [16]]. The exact mechanism by which FMT results in clearance of C. difficile from stool is not known, and variations in stool processing protocols appear to have little impact on the efficacy of FMT for this indication [14,17,18]. This has resulted in protocols designed for use in C. difficile infection being adopted in trials using FMT for other indications, where mechanisms of action are likely to be different. These protocols commonly involve the homogenisation of stool in ambient air, despite oxygen exposure being known to cause the rapid death of many obligate anaerobic bacterial commensals [19].
At present there is little evidence available to guide clinicians in selecting a stool processing methodology. Characterisation of the microbiome composition in processed FMT donor material is often not attempted, and where performed, typically involves high throughput sequencing of extracted faecal DNA [20]. Such an approach will detect DNA derived from both viable and non-viable organisms and therefore have a limited capacity to indicate which bacteria are viable and capable of replicating in the recipient. Culture methods readily isolate only a small subset of the total gut microbiota and are therefore unsuitable for characterising the impact of processing on many of the commensal anaerobic species present.
A strategy to overcome these challenges is combine molecular techniques, such as next-generation sequencing and targeted quantitative PCR assays, with propidium monoazide sample treatment (PMA-qPCR) [21]. PMA is a red fluorescent dye which selectively enters cells with compromised cell membranes. Upon exposure to light PMA will covalently bind to DNA in these cells, inhibiting PCR amplification [22]. In this way, PMA treatment of sample material allows the selective amplification of DNA from only viable cells in the sample [22]. The PMA-qPCR methodology applied in this study was specifically optimised for use in faecal slurries for transplantation and validated in comparison to culture methods [23].
We report the effects of anaerobic homogenisation, aerobic homogenisation and freeze-thaw on the viability and functional capacity of bacteria in donor stools processed for faecal microbiota transplantation.
2. Methods
2.1. FMT faecal slurry (FS) processing
Stool was collected with informed consent from healthy participants being screened as FMT donors for a clinical trial [24] with approval from the Queen Elizabeth Hospital Human Research Ethics Committee (HREC/16/TQEH/32). All donors had passed a screening questionnaire (see appendix) used to identify potential FMT donors. Stool was collected on site and processed within 15 min of passage. Stool was divided into two aliquots weighing at least 30 g. Each aliquot was blended (in a 240 V Waring SS515 laboratory blender at 22,000 rpm) with normal saline (NS) and glycerol to produce a faecal slurry (FS) consisting of 25% (wt/vol) stool, 65% NS, and 10% glycerol, as previously described [12]. In the first aliquot (ANO2) stool blending and PMA treatment were performed under anaerobic conditions within an anaerobic chamber. In the second aliquot (O2), the same procedure was performed in ambient air. The resultant FS was frozen at −80 oC in 50 mL centrifuge tubes. To assess the effects of freeze-thaw, a 50 mL aliquot of anaerobically processed FS was stored at −80 oC for 48 h and then allowed to thaw at room temperature within the anaerobic chamber. The freeze-thawed specimens were not exposed to oxygen during the processing or PMA treatment. Heat killing was performed by subjecting a 1 mL aliquot of thawed FS to 99 oC for 30 min.
2.2. Dilution and PMA treatment of fresh, frozen and thawed, and heat-killed FS
Immediately following the processing described above, neat FS (25% stool content) was diluted 100-fold in phosphate buffered saline (PBS) and divided into six 95 μL aliquots in clear RNase-free 1.5 mL tubes (Ambion, Thermo Fisher Scientific, Waltham MA, USA). Diluted samples were treated with and without PMA in triplicate, as described above. Samples were stored at −80 oC prior to extraction. PMA-treatment was performed using a protocol specifically developed and validated for faecal slurries [23]. A stock PMA solution was prepared by dissolving 1 mg of PMA (Biotium Inc., Fremont, CA, USA) in 1 mL of 20% dimethyl sulfoxide. For PMA treatment, 5 μL of PMA stock was added to 95 μL of sample to achieve 100 μM final concentration of PMA. Following a 30 min incubation at room temperature in the dark, samples were exposed to an LED light (Aqua Zonic, Singapore) for 20 min. In non-PMA treated control aliquots, 5 μL of PBS was added instead of PMA. Control samples underwent identical incubation and light-exposure as the matching PMA treated samples.
2.3. Determination of microbiota composition
DNA was extracted from the entire (100 μL) unspun sample. DNA was extracted using the PowerLyzer PowerSoil DNA Isolation Kit (MO BIO Laboratories, Carslbad, CA, USA) in accordance with the manufacturer’s instructions and stored at −20 oC.
The microbial composition of the faecal slurry specimens was determined by paired-end sequencing of the V4 hypervariable region of the bacterial 16S rRNA gene. Amplicon sequencing was performed on an Illumina MiSeq platform as described previously. Paired-end reads were merged, demultiplexed and analysed using Quantitative Insights in to Microbial Ecology (QIIME) software (v1.9.1) using a previously described bioinformatics pipeline [25]. Sequences were assigned to operational taxonomic units (OTUs) using an open reference approach against the SILVA 16S rRNA reference database (release 128) clustered at 97% similarity. Subsampling was performed on all samples to a depth of 6188 sequence reads. The alpha diversity metrics for determining taxa richness (observed species) and diversity (PD whole tree) was computed using QIIME. Sequence data was submitted to the National Center for Biotechnology Information SRA database with accession number PRJNA491383.
Taxa with zero counts were normalised to a single count across all samples. Changes in taxon relative abundance were determined by calculating the log2 values of fold change in relative abundance against the anaerobically processed matching controls across all taxa. Taxon relative abundance average fold change was computed based on the inverse logarithm of the sum of log2 fold change divided by the number of donors in which the taxa was detected. Only bacterial taxa that were present (sequence count ≥2) in at least one comparison group within each donor and were present in at least 7 of 8 donors were included in the analysis.
2.4. Species-specific bacterial enumeration
Levels of total bacteria, butyryl-CoA:acetate CoA-transferase gene, Anaerostipes hadrus, Faecalibacterium prausnitzii, Eubacterium hallii, Roseburia spp/Eubacterium rectale, Bifidobacterium spp., Alistipes putredinis, and Bacteriodes spp. were determined using previously described qPCR assays (detailed in Supplementary Table 1) using a QuantStudio 6 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). The Subdoligranulum variable qPCR assay was developed as part of this study (see Supplementary Methods). Escherichia coli DNA was amplified using a probe-based assay using KAPPA PROBE FAST ROX Low Master Mix reagents (Kapa Biosystems, Cape Town, South Africa). All other PCR assays were performed using SYBR green fluorophore reagents (PowerUp SYBR Green Master Mix, Applied Biosystems). Quantitative PCR assays for the butyryl-CoA:acetate CoA-transferase gene, fastidious bacteria, including A. hadrus, E. hallii, Roseburia/E. rectale, and S. variable, a 10-fold dilution series of DNA extracted from donor faecal slurry was used as the positive qPCR standard.
The proportion of bacterial cells in each sample that were viable was determined by dividing the quantity of cells amplified in the presence of PMA by the quantity of cells amplified in matching untreated controls. To determine the proportion viable in heat-killed specimens, the quantity of cells amplified in PMA-treated heat-killed samples was divided by the quantity of cells amplified in the control sample prior to heat killing. Heat-killed specimens served as negative controls, representing levels of amplification expected in non-viable specimens.
2.5. Assessment of metabolomic functional capacity
We assessed the functional capacity of differentially processed faecal slurries using an in vitro fermentation model, as described previously [26]. This method assesses the ability of microbiota in the sample to produce SCFA when incubated with a fermentation substrate, high amylose maize starch (HAMS). Briefly, stored frozen faecal slurry samples from FMT donors (n=8) that had been processed ANO2 or O2 as described earlier were thawed and incubated under strict anaerobic conditions with HAMS. Heat-killed faecal slurry (n=2) and HAMS only (n=2) were used as negative controls. SCFA levels were determined by gas chromatography with flame ionisation detection (Hewlett-Packard6890; Palo Alto, CA, USA). Acetate, butyrate and propionate levels were measured pre-incubation and after 24 h of anaerobic incubation at 37 °C with shaking. Results were normalised using 4-methylvaleric acid (Sigma-Aldrich) as an internal standard. SCFA production in the two groups were compared using the Wilcoxon matched pairs signed rank test.
2.6. Statistical analysis
Statistical analysis was performed using GraphPad Prism 7.03 software. Significance (p-value <0.05) was determined using paired t-tests for parametric data and the Wilcoxon matched-pairs signed rank test for non-parametric data.
3. Results
3.1. Donor characteristics
Donors median age was 28.5 years (range 21–41) and with an equal male to female ratio. An equal number of donors had South East Asian and European heritage (Supplementary Table 2).
3.2. Impact of processing methodology on bacteria viability in faecal slurries
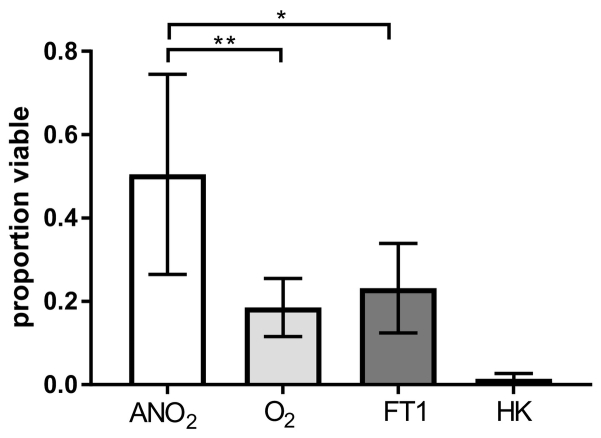
With immediate anaerobic processing, the mean proportion of bacteria in FS that was viable was 0.50 ± 0.24 (mean ± SD). After processing in ambient air, this proportion fell to 0.19 ± 0.07, and after anaerobic processing followed by one freeze-thaw cycle it was 0.23 ± 0.11 (Fig. 1). A significant reduction in the proportion of bacterial cells that were viable was observed following processing in ambient air (p = 0.007) and after freeze-thawing (p = 0.027), as compared to anaerobic processing alone. The proportion of viable bacteria in all processing methods was greater than detected in heat-killed specimens (p > 0.001).
Fig. 1.
Proportion of bacteria determined to be viable using 16S rRNA gene qPCR in conjunction with PMA treatment. Proportion of viable cells was determined by dividing viable cells amplified in PMA-treated samples over total number of cells amplified in non-PMA treated control samples. Bacterial viability in faecal slurry was assessed after processing in fresh anerobic conditions (ANO2), fresh aerobic conditions (O2), after one cycle of freezing and thawing in anaerobically processed specimens (FT1) or after heat-killing (HK). (Bars depict mean ± SD of 8 donor faecal slurry samples *= p<0·05, **=p<0·01; paired t-test).
3.3. Diversity of microbiota
The diversity of viable taxa detected in specimens processed under anaerobic conditions after PMA treatment was significantly lower than that in specimens processed using standard (without PMA) methods (Fig. 2; taxa richness: p <0.001; PD whole tree: p = 0.007). Observed viable taxon richness was significantly lower after processing in ambient air (O2) as compared to either ANO2 processed (p = 0.023) or FT specimens (p = 0.023). No difference in diversity was observed between ANO2 and FT groups (Fig. 2). Differences in PD whole tree diversity between O2 and ANO2 and between O2 and FT groups were not significant.
Fig. 2.
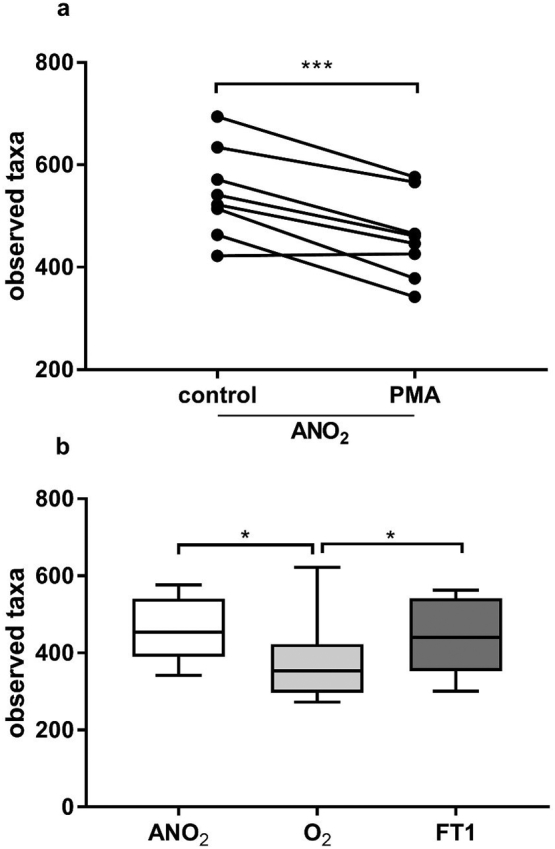
Taxa richness of faecal microbiota transplant material from 8 donors as assessed by 16S rRNA gene amplicon sequencing with and without PMA treatment. Viable diversity (PMA treated group) was significantly lower than diversity observed in control specimens, even in samples processed immediately in anaerobic conditions (Fig. 2a ***=p<0·001, paired t-test). When comparing only viable diversity between samples processed in anaerobic conditions (ANO2), in ambient air (O2), or after one cycle of freezing and thawing in anaerobically processed specimens (FT1) there are significantly lower observed species in specimens processed O2, whereas freeze-thawing of specimens did not significantly reduce diversity (Fig. 2b, box plot depicts median and IQR and error bars depict minimum to maximum values; *= p<0·05; Wilcoxon matched-pairs signed rank test).
3.4. Viable microbiota composition
The five taxa (OTUs) that displayed the greatest relative abundance across all samples belonged to the genera Bacteroides, Prevotella, Bifidobacterium, Faecalibacterium, and the family Lachnospiraceae (0.21, 0.09, 0.05, 0.05 and 0.05 mean relative abundance, respectively). To determine which taxa were affected most by processing in ambient oxygen or freeze-thawing, taxa were ranked by relative abundance fold-change (Fig. 3). Nineteen taxa displayed a 2.5-fold or greater drop in relative abundance after processing in ambient air, as compared to 2 taxa in the freeze-thaw group. The taxa most affected by processing in ambient air included Faecalibacterium, Subdoligranulum, Eubacterium hallii, Eubacterium rectale, Roseburia and Anaerostipes, representing major butyrate producing taxa in healthy human gut microbiota [27]. Escherichia-Shigella and Alistipes were the only taxa to show a 2.5-fold or greater increase in relative abundance processing in ambient oxygen or freeze-thawing and this was observed only following processing in ambient air.
Fig. 3.
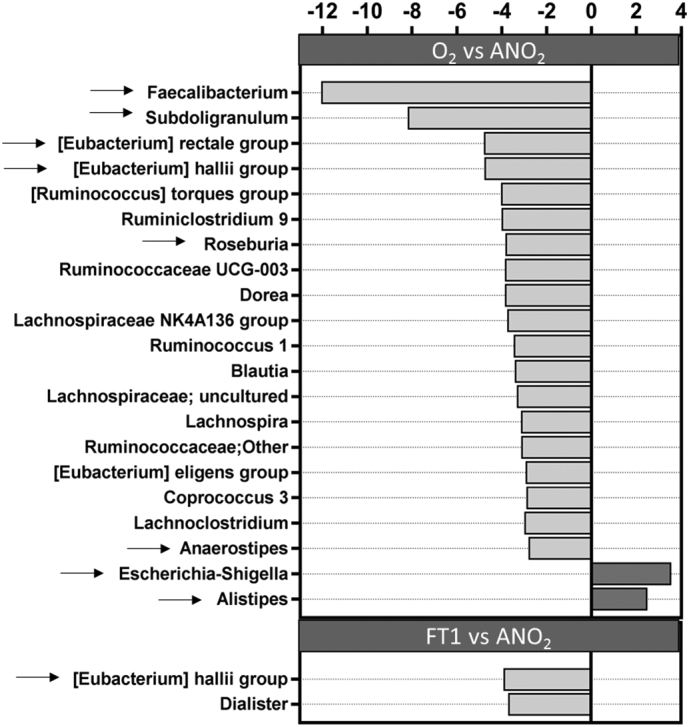
Change in the relative abundance of viable taxa after processing in ambient air (O2 vs ANO2) or after freeze-thawing (FT1 vs ANO2). Light grey bars represent decreased relative abundance and dark grey bars represent increased relative abundance. Selected bacterial taxa were further assessed by qPCR (arrows). Only taxa with at least 2·5-fold change in relative abundance are depicted.
To corroborate changes observed in taxon relative abundance, absolute levels of the taxa listed above that were substantially affected by processing were determined by quantitative PCR. This included an assessment of all major butyrate producers that displayed a ≥2.5-fold in relative abundance, as well as those taxa displaying the greatest overall prevalence (Bacteriodes/Prevetolla and Bifidobacterium spp). Changes in relative abundance of viable bacteria (PMA-treated) in the anaerobically processed group, compared to standard analysis of the same group, are shown in Supplementary Fig. 1.
3.5. Targeted amplification of viable microbial DNA
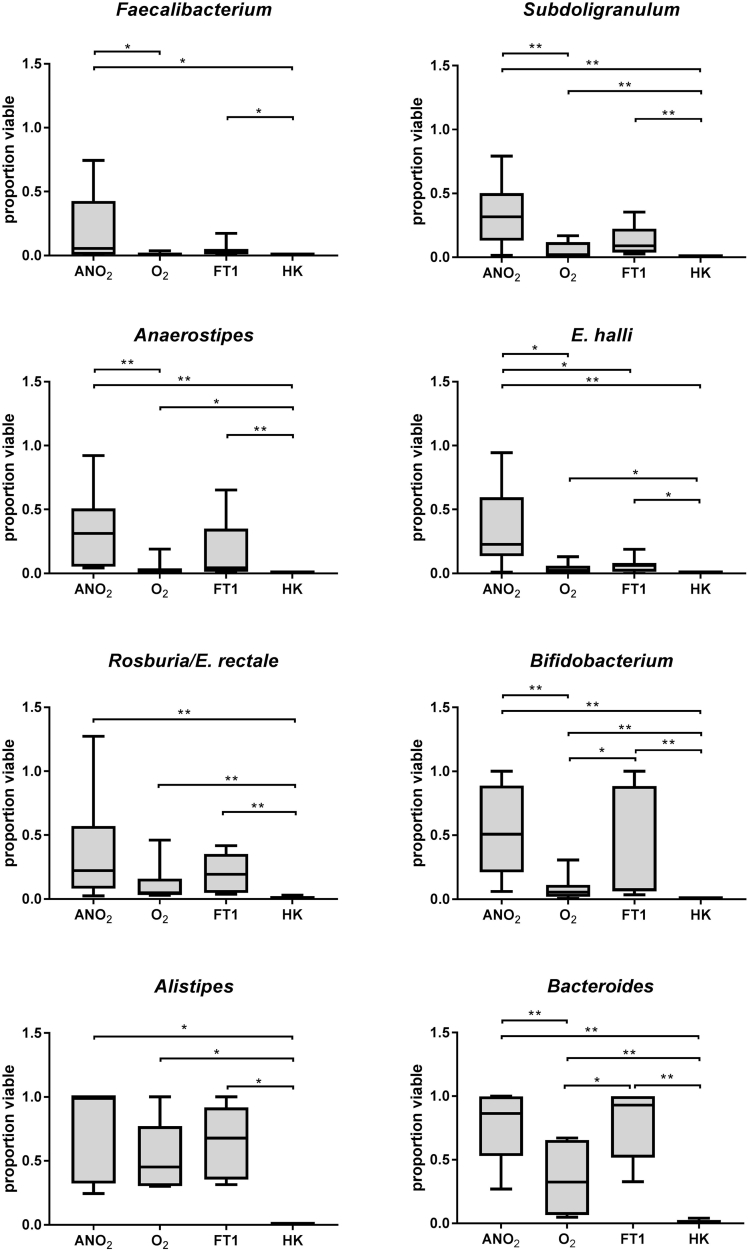
The use of targeted quantitative PCR assays allowed the proportion of viable cells belonging to particular bacterial species to be determined (Fig. 4). Assays for F. prausnitzii, S. variable, A. hadrus, E. hallii, and Roseburia/E. rectale were included as these taxa showed >2.5 fold or greater decrease in relative abundance following processing in ambient air. Assays for Bacteriodes/Prevotella spp. and Bifidobacterium spp. were also performed as these were the genera with the highest relative abundance in our cohort. Results for individual donors for each processing condition are shown in Supplemental Tables 3a–c.
Fig. 4.
Proportion of bacteria determined to be viable by specific qPCR assays. Proportion of viable cells was determined by dividing viable cells amplified in PMA-treated samples over total number of cells amplified in non-PMA treated control samples. Bacterial viability in faecal slurry was assessed after processing in fresh anaerobic conditions (ANO2), fresh aerobic conditions (O2), after one cycle of freezing and thawing in anaerobically processed specimens (FT1) or after heat-killing (HK). Box plot depicts median and IQR and error bars depict minimum to maximum values of faecal slurry samples from 8 individual donors. All significant comparisons are indicated by stars (*= p<0·05; **= p<0·01; ***= p<0·001; Wilcoxon matched-pairs signed rank test).
Significant reductions in levels of viable bacteria following ambient air processing were observed for all taxa except the Roseburia/E. rectale (for which no significant effects of processing were observed). In the case of F. prausnitzii, the proportion viable bacteria in the O2 group was not significantly different from heat-killed aliquots. E. hallii was the only species that showed a significant reduction in absolute viable levels following freeze-thaw.
An assessment of E. coli absolute abundance was performed as this taxon showed the greatest increase in relative abundance after processing in ambient air. The absolute amplification of E. coli in the donors was below the threshold of quantification in all but one individual, and there was no significant difference between the different processing methods (Supplemental Tables 3a–c). Alistipes also showed an increase in relative abundance following processing in ambient air, but there was no increase in absolute abundance.
A semi-quantitative PCR method was used to estimate the carriage in viable bacterial cells of the gene encoding the butyryl-CoA:acetate CoA-transferase gene, the terminal enzyme in the dominant pathway of butyrate biosynthesis (this was employed as a surrogate measure of overall butyrate biosynthesis capacity) [27]. When comparing amplification from viable cells processed in ambient air, levels of this enzyme are significantly lower than that detected in viable cells from anaerobically processed samples (p = 0.012) or freeze-thawed samples (p = 0.001). The level the butyryl-CoA:acetate CoA-transferase gene detected within viable cells processed in ambient air was equivalent to samples that had been heat-killed (Fig. 5).
Fig. 5.
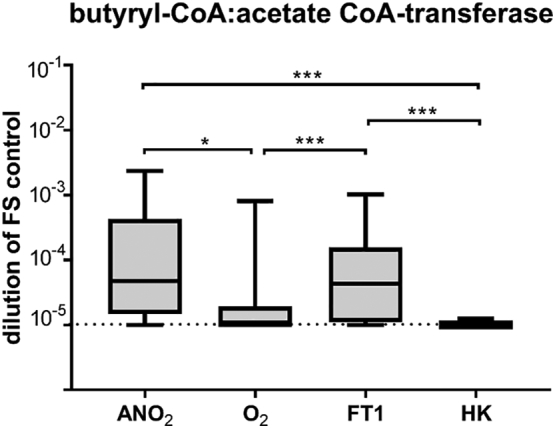
Amplification of the butyryl-CoA:acetate CoA-transferase gene, the terminal enzyme of the central butyrate synthesis pathway of human gut microbiota, in fresh anaerobic conditions (ANO2), fresh aerobic conditions (O2), after one cycle of freezing and thawing in anaerobically processed specimens (FT1) or after heat-killing (HK) in PMA treated samples. Butyryl-coenzyme A(CoA) CoA transferase gene levels were measured relative to amplification in a 10-fold dilution series of neat faecal slurry (FS control). The dotted line represents limit of quantification of the assay. Box plots depict median and IQR and error bars depict minimum to maximum values of faecal slurry samples from 8 individual donors. Significant comparisons are indicated by stars. (*= p<0·05; **= p<0·01; ***= p<0·001; Wilcoxon matched-pairs signed rank test).
3.6. SCFA biosynthesis
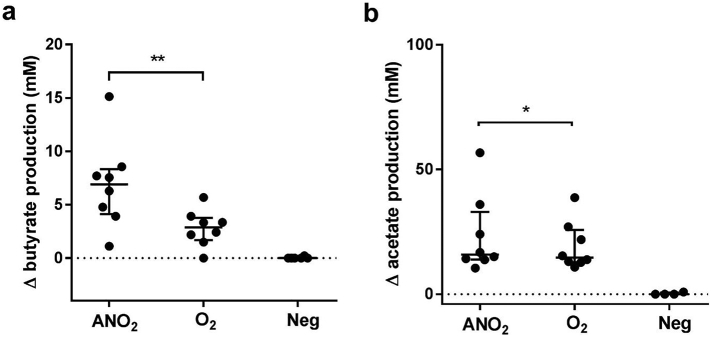
Paired comparisons of post-fermentation SCFA levels demonstrated microbial butyrogenic and acetogenic capacity to be significantly reduced when donor stool was processed in ambient air (O2 vs ANO2, p = 0.008 and p = 0.016 respectively, Fig. 6). In contrast, no significant change in propionate biosynthesis was associated with oxygen exposure.
Fig. 6.
Net production of butyrate (panel a) and acetate (panel b) following in-vitro fermentation of faecal slurries for FMT with high-amylose maize starch. Matching samples (n=8) were processed either under anaerobic conditions (ANO2), or under aerobic conditions (O2). Significant comparisons are indicated by stars. (*= p<0·05; **= p<0·01; Wilcoxon matched-pairs signed rank test).
4. Discussion
At present there is little evidence to guide clinicians using FMT on how to best ensure that the viability of donor microbiota is preserved in faecal transplant material. Analysis of microbiota in donor material is not routine, and when performed, the methods used do not assess viability. PMA based methodology is able to overcome many challenges in assessing the viability of the complex community of fastidious bacteria in stool. However, the main limitation of PMA methodology is that it is prone to over-estimating the number of live bacteria in a sample [22]. This means the number of viable cells could be lower than we report. Furthermore, the dilution of specimens required for PMA treatment means that very rare taxa are excluded from this type of analysis.
Despite these limitations we show that current methods of reporting the microbiota present in FMT material significantly overestimate the number of live bacteria transplanted. On average, only half of bacteria in faecal transplants in our study were still viable after immediate processing in strict anaerobic conditions. The use of PMA sample treatment also revealed that the diversity of bacteria in these transplants is significantly less than what would reported using standard sequencing methods.
We observed substantial inter-donor variation in the impact of sample processing. Such differences can be explained by individual variation in microbiome composition, resulting in microbiota with different vulnerabilities to oxygen exposure and freezing, and indicate the need for viability assessments to be performed on all donor material.
Our study revealed that homogenization by blending stool in ambient air has a profound impact on its viable bacterial composition. Ambient air processing is the default practice in most clinical trials and is described in American [28], British [16] and European consensus guidelines [14], although in many protocols stools are homogenized manually and not blended as in this study. The increased air flow produced during high-speed blending may result increased oxygen-exposure and be more detrimental to oxygen sensitive species than manual homogenization.
Obligate anaerobic gut commensal species that are most affected by oxygen exposure, including Faecalibacterium prausnitzii, Subdoligranulum variable, Eubacterium rectale, Eubacterium hallii and Anaerostipes hadrus, are major contributors to the biosynthesis of butyrate [27]. Butyrate is a short-chain fatty acid produced from the fermentation by the intestinal microbiota. Apart from being the major energy source of colonocytes, butyrate and has both anti-inflammatory and anti-carcinogenic properties [29,30]. Reduced luminal butyrate concentration or butyrate utilisation is associated with enterocyte adenosine triphosphate depletion, loss of tight junctions, reduced mucus production and resultant colonic barrier disruption with inflammatory and immunological consequences [30].
We observed the proportion of viable F. prausnitzii and A. hadrus in the majority of donors, as well as the levels of the butyryl-CoA:acetate CoA-transferase gene, to be reduced to levels detected in heat-killed specimens when specimens were processed in the presence of oxygen. The relative reduction of butyrate producing bacteria, F. prausnitzii in particular, within the gut has been associated with the presence of a diverse range of chronic diseases including depression [8], obesity [7], type 2 diabetes [6] and inflammatory bowel disease [1,3,4]. These findings suggest that processing faecal material in ambient air may negatively influence the outcome of efforts to achieve anti-inflammatory or immunomodulatory outcomes using FMT.
Bifidobacterium spp and Bacteroides spp, are major producers of the short-chain fatty acids, acetate and propionate respectively, and like butyrate are important energy and signalling molecules [29]. Although, their beneficial effects are less well-established than those of butyrate, these metabolites have also been found to be important immune and metabolic regulators [29]. Members of these genera were also significantly reduced in abundance following oxygen exposure. However, significant residual viable populations remained, particularly in the case of Bacteroides spp., suggesting the potential for re-expansion in the gut of the recipient.
Although freezing did reduce the overall viability of the transplant material (reducing overall viability to around 25%) the viable microbiota composition was not significantly different to that in fresh, anaerobically processed, specimens. Only one bacterial species, Eubacterium hallii, was found to be significantly reduced in freeze-thawed specimens. While E. hallii is a major butyrate-producer [27], this function is also performed by several other bacterial species. The potential clinical implications of the loss of this single species is uncertain.
Beyond the depletion of beneficial commensal bacteria in faecal material through processing in ambient air, the relative abundance of potentially pathogenic species, such E. coli and other oxygen-tolerant Gram-negative bacteria, will increase proportionally. In our donor cohort levels of E. coli were very low in fresh stool, with no evidence of an increase in absolute abundance during processing. However, delays in sample processing that result in prolonged periods at room temperature could result in substantial increases in the abundance of opportunistic pathogens. The loss of butyrate-producing anaerobes combined with the overgrowth of oxygen-tolerant species could potentially transform faecal transplant material from healthy donors into faecal material with a microbiota profile more closely resembling those linked to inflammatory bowel disease [2,19], type 2 diabetes [6] and colorectal cancer [5].
Processing FMT material in an anaerobic chamber achieves optimal preservation of important commensal species. In the context of C. difficile colitis, aerobic processing does not appear to adversely influence clinical outcomes. However optimal preservation of commensals should be attempted when FMT is being investigated for other indications where it is not yet known how the loss of commensal anaerobes could influence clinical outcomes. A limitation of our study was that the effect of delays in processing were not analyzed. Although stool specimens were processed within 15 min in this study, this is not achievable in most clinical settings, and a delay in processing of several hours is more typical. Such delays in processing may result in changes in viable microbiota composition. A number of previous studies employing molecular strategies, have reported delays in stool analysis to be associated with shifts in microbiota composition and viability [25,[31], [32], [33]]. Chu et al assessed changes in FMT material in one donor after processing delays of incremental periods up to 7 hours and saw a trend towards a change in microbiome composition over this time [21]. These reports are supported by a number of culture-based studies that have demonstrated reduced recovery of anaerobes when processing is delayed [34], particularly at room temperature [35] and when samples were not stored in anaerobic conditions [36]. Until further research clarifies the effects or processing delays, we suggest that periods between sample collection and processing are as short as possible.
Following processing, faecal material should be frozen promptly at −80 °C. The ability to freeze samples prior to instillation is important as it provides an opportunity to complete systematic testing for pathogens, including viruses and parasites [12]. Freezing also allows stool to be available on demand for use in urgent clinical situations. Our analysis suggests that, while freeze-thaw does impact the viable composition of stool, in general this effect is relatively limited. However, there is variability in the individual donor microbiota response to freezing. In this study, the overall bacterial viability in two donors dropped below levels seen after ambient air processing after freeze-thaw alone.
The role that stool processing plays in changing the composition of faecal transplants has been widely overlooked in the design of FMT clinical trials. Adherence to strict anaerobic stool processing protocols is likely to result in increased benefit from FMT in some clinical settings. Other factors, such as delays in stool processing and storage conditions, might have as great an impact on bacterial viability as anaerobic processing, but were not assessed in this study. It would be beneficial for future trials to assess the composition of donor transplant material with a viability assay to ensure that the microbiota composition includes a broad range of viable bacteria some of which may be crucial in mediating the therapeutic effects of FMT. A detailed analysis of the types and numbers of viable bacteria transplanted is critical to understanding of the mechanisms by which FMT produces, or fails to produce, therapeutic effects.
Acknowledgments/funding
We would like to acknowledge Ge Liu from the South Australian Health and Medical Research Institute for performing SCFA gas chromatography. This work was supported by the South Australian Health and Medical Research Institute. LP is supported by a scholarship from the Flinders Foundation. GR is supported by a Matthew Flinders Research Fellowship. The funders of this study did not play a role in study design, data collection, data analysis, interpretation of data, writing of the report, or the decision to submit the paper for publication.
Declaration of interests
SC has received speaking fees from Pfizer, Janssen and Shire and has been paid as a consultant for Microbiotica. All other authors have nothing to declare.
Author contributions
LP and GR conceived of the study. LP, GR, SC, DG and SW had input into the design of the study. JC and LP performed the microbiome analysis. LP, JC, YW and LL contributed to PCR assay development, statistical analysis and PMA method validation. LP and YW performed in-vitro fermentation experiments. LP, GR, SC and JC interpreted the results and contributed to writing the manuscript. All authors critically reviewed the manuscript and approved he final version of the report.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.02.023.
Appendix A. Supplementary data
Supplementary material
References
- 1.Fuentes S., Rossen N.G., van der Spek M.J. Microbial shifts and signatures of long-term remission in ulcerative colitis after faecal microbiota transplantation. ISME J. 2017;11(8):1877–1889. doi: 10.1038/ismej.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frank D.N., St Amand A.L., Feldman R.A., Boedeker E.C., Harpaz N., Pace N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104(34):13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kostic A.D., Xavier R.J., Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146(6):1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sokol H., Pigneur B., Watterlot L. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105(43):16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang T., Cai G., Qiu Y. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6(2):320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin J., Li Y., Cai Z. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 7.Remely M., Aumueller E., Merold C. Effects of short chain fatty acid producing bacteria on epigenetic regulation of FFAR3 in type 2 diabetes and obesity. Gene. 2014;537(1):85–92. doi: 10.1016/j.gene.2013.11.081. [DOI] [PubMed] [Google Scholar]
- 8.Jiang H., Ling Z., Zhang Y. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun. 2015;48:186–194. doi: 10.1016/j.bbi.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 9.van Nood E., Vrieze A., Nieuwdorp M. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368(5):407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 10.Cammarota G., Masucci L., Ianiro G. Randomised clinical trial: faecal microbiota transplantation by colonoscopy vs. vancomycin for the treatment of recurrent Clostridium difficile infection. Aliment Pharmacol Ther. 2015;41(9):835–843. doi: 10.1111/apt.13144. [DOI] [PubMed] [Google Scholar]
- 11.Costello S.P., Soo W., Bryant R.V., Jairath V., Hart A.L., Andrews J.M. Systematic review with meta-analysis: faecal microbiota transplantation for the induction of remission for active ulcerative colitis. Aliment Pharmacol Ther. 2017;46(3):213–224. doi: 10.1111/apt.14173. [DOI] [PubMed] [Google Scholar]
- 12.Costello S.P., Tucker E.C., La Brooy J., Schoeman M.N., Andrews J.M. Establishing a fecal microbiota transplant service for the treatment of Clostridium difficile infection. Clin Infect Dis. 2016;62(7):908–914. doi: 10.1093/cid/civ994. [DOI] [PubMed] [Google Scholar]
- 13.Segal J.P., Abbasi F., Kanagasundaram C., Hart A. Does the internet promote the unregulated use of fecal microbiota transplantation: a potential public health issue? Clin Exp Gastroenterol. 2018;11:179–183. doi: 10.2147/CEG.S159609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cammarota G., Ianiro G., Tilg H. European consensus conference on faecal microbiota transplantation in clinical practice. Gut. 2017;66(4):569–580. doi: 10.1136/gutjnl-2016-313017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trubiano J.A., Cheng A.C., Korman T.M. Australasian Society of Infectious Diseases updated guidelines for the management of Clostridium difficile infection in adults and children in Australia and New Zealand. Intern Med J. 2016;46(4):479–493. doi: 10.1111/imj.13027. [DOI] [PubMed] [Google Scholar]
- 16.Mullish B.H., Quraishi M.N., Segal J.P. The use of faecal microbiota transplant as treatment for recurrent or refractory Clostridium difficile infection and other potential indications: joint British Society of Gastroenterology (BSG) and Healthcare Infection Society (HIS) guidelines. Gut. 2018;67(11):1920–1941. doi: 10.1136/gutjnl-2018-316818. [DOI] [PubMed] [Google Scholar]
- 17.Lee C.H., Steiner T., Petrof E.O. Frozen vs Fresh fecal microbiota transplantation and clinical resolution of diarrhea in patients with recurrent Clostridium difficile infection: a randomized clinical trial. JAMA. 2016;315(2):142–149. doi: 10.1001/jama.2015.18098. [DOI] [PubMed] [Google Scholar]
- 18.Youngster I., Russell G.H., Pindar C., Ziv-Baran T., Sauk J., Hohmann E.L. Oral, capsulized, frozen fecal microbiota transplantation for relapsing Clostridium difficile infection. JAMA. 2014;312(17):1772–1778. doi: 10.1001/jama.2014.13875. [DOI] [PubMed] [Google Scholar]
- 19.Rigottier-Gois L. Dysbiosis in inflammatory bowel diseases: the oxygen hypothesis. ISME J. 2013;7(7):1256–1261. doi: 10.1038/ismej.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fouhy F., Deane J., Rea M.C. The effects of freezing on faecal microbiota as determined using MiSeq sequencing and culture-based investigations. PLoS One. 2015;10(3):e0119355. doi: 10.1371/journal.pone.0119355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu N.D., Smith M.B., Perrotta A.R., Kassam Z., Alm E.J. Profiling living bacteria informs preparation of fecal microbiota transplantations. PLoS One. 2017;12(1):e0170922. doi: 10.1371/journal.pone.0170922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nocker A., Sossa-Fernandez P., Burr M.D., Camper A.K. Use of propidium monoazide for live/dead distinction in microbial ecology. Appl Environ Microbiol. 2007;73(16):5111–5117. doi: 10.1128/AEM.02987-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papanicolas L.E., Wang Y., Choo J.M., Gordon D.L., Wesselingh S.L., Rogers G.B. Optimisation of a propidium monoazide based method to determine the viability of microbes in faecal slurries for transplantation. J Microbiol Methods. 2019;156:40–45. doi: 10.1016/j.mimet.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Australia New Zealand Clinical Trials Registry Gastrointestinal eradication of multi-resistant gram negative bacteria by faecal microbiota transplantation (FMT) 2018. https://www.anzctr.org.au/Trial/Registration/TrialReview.aspx?id=371671&isReview=true
- 25.Choo J.M., Leong L.E., Rogers G.B. Sample storage conditions significantly influence faecal microbiome profiles. Sci Rep. 2015;5:16350. doi: 10.1038/srep16350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y., Leong L.E.X., Keating R.L. Opportunistic bacteria confer the ability to ferment prebiotic starch in the adult cystic fibrosis gut. Gut Microbes. 2018:1–15. doi: 10.1080/19490976.2018.1534512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Louis P., Young P., Holtrop G., Flint H.J. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ Microbiol. 2010;12(2):304–314. doi: 10.1111/j.1462-2920.2009.02066.x. [DOI] [PubMed] [Google Scholar]
- 28.IDSA . 2013. Fecal Microbiota Transplant (FMT) protocol by enema for patients with recurrent Clostridium difficile infection. [Google Scholar]
- 29.Koh A., De Vadder F., Kovatcheva-Datchary P., Backhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165(6):1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 30.McNabney S.M., Henagan T.M. Short chain fatty acids in the colon and peripheral tissues: a focus on butyrate, colon cancer, obesity and insulin resistance. Nutrients. 2017;9(12) doi: 10.3390/nu9121348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dominianni C., Wu J., Hayes R.B., Ahn J. Comparison of methods for fecal microbiome biospecimen collection. BMC Microbiol. 2014;14:103. doi: 10.1186/1471-2180-14-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flores R., Shi J., Yu G. Collection media and delayed freezing effects on microbial composition of human stool. Microbiome. 2015;3:33. doi: 10.1186/s40168-015-0092-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tedjo D.I., Jonkers D.M., Savelkoul P.H. The effect of sampling and storage on the fecal microbiota composition in healthy and diseased subjects. PLoS One. 2015;10(5):e0126685. doi: 10.1371/journal.pone.0126685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Helstad A.G., Kimball J.L., Maki D.G. Recovery of anaerobic, facultative, and aerobic bacteria from clinical specimens in three anaerobic transport systems. J Clin Microbiol. 1977;5(6):564–569. doi: 10.1128/jcm.5.6.564-569.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Justesen T., Jensen A.M., Hoffmann S. The survival of anaerobic bacteria at 4 degrees C and 22 degrees C on swabs in three transport systems. Statistical evaluation by application of a variance component model. Acta Pathol Microbiol Immunol Scand B. 1983;91(1):17–22. doi: 10.1111/j.1699-0463.1983.tb00003.x. [DOI] [PubMed] [Google Scholar]
- 36.Hill G.B. Effects of storage in an anaerobic transport system on bacteria in known polymicrobial mixtures and in clinical specimens. J Clin Microbiol. 1978;8(6):680–688. doi: 10.1128/jcm.8.6.680-688.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material