Abstract
MIR143 is pathologically downregulated and may function as a tumor suppressor in prostate cancer. Likewise, the urokinase plasminogen activator receptor (UPAR) is overexpressed in prostate carcinoma, representing a negative prognostic marker and putative therapeutic target gene. In this paper, we establish UPAR as a new direct target of MIR143. Luciferase reporter gene constructs identify one of the two in silico-predicted binding sites as functionally relevant for direct MIR143 binding to the 3′ UTR, and, concomitantly, transfection of MIR143 reduces UPAR protein levels in prostate carcinoma cells in vitro. Inhibitory effects on cell proliferation and colony formation, spheroid growth and integrity, and cell viability are extensively analyzed, and they are compared to direct small interfering RNA (siRNA)-mediated uPAR knockdown or combined microRNA (miRNA)-siRNA treatment. Switching to a therapeutically more relevant in vivo model, we demonstrate tumor-inhibitory effects of MIR143 replacement therapy by systemic treatment of mice bearing subcutaneous PC-3 tumor xenografts with MIR143 formulated in polymeric nanoparticles. This efficient, nanoparticle-mediated delivery of intact MIR143 mediates the marked downregulation of uPAR protein, but not mRNA levels, thus indicating translational inhibition rather than mRNA degradation. In summary, we identify UPAR as a direct target gene of MIR143, and we establish the therapeutic anti-tumor potential of nanoparticle-based MIR143 replacement in prostate cancer.
Keywords: MIR143, urokinase plasminogen activator receptor, prostate cancer, miRNA replacement, PEI nanoparticles, xenograft
Introduction
Through repression of translation or cleavage of RNA transcripts in a sequence-specific manner, microRNAs (miRNAs) are able to (patho-)physiologically regulate the expression of their respective target genes. When aberrantly downregulated in the tumor context, certain miRNAs may thus contribute to cancer development, progression, and metastasis, by de-repressing oncogene expression. Levels of the human miRNA MIR143 have been found reduced in several tumor entities, and its function as a tumor-inhibitory miRNA (anti-oncomiR) has been described in various cancers, including prostate carcinoma1 and colorectal,2, 3, 4, 5, 6 gastric,7 bladder,8, 9 cervical,10 breast,11 and nasopharyngeal cancers12 as well as chronic lymphocytic leukemia and B cell lymphoma.13 Tumor-inhibitory effects of MIR143 may be based on regulating different tumor-related pathways upon binding of MIR143 to the 3′ UTRs of already validated target genes (see, e.g., miRTarBase at http://mirtarbase.mbc.nctu.edu.tw/php/index.php for a summary). Previously, we and others reported decreased MIR143 levels in prostate cancer compared to normal tissue.14, 15, 16, 17, 18 In accordance with this finding, the re-introduction of MIR143 in prostate cancer cell lines decreased tumor cell proliferation and cell migration and invasion,14 and MIR143 and MIR145 inhibited stem cell characteristics of PC-3 prostate cancer cells.19
Recently, our in silico analysis of novel putative MIR143 target genes identified the urokinase plasminogen activator (uPA) receptor (UPAR, or PLAUR) as a potential target of MIR143. This may be of particular importance since UPAR has been found overexpressed in tumors and can modulate various (patho-)physiological processes, such as cell adhesion, migration, and invasion, which are also critically important in cancers.20 In fact, UPAR may play a central role in solid tumors and exert various functions in different cancer entities, including prostate cancer.21, 22 Supporting its clinicopathological significance, we and others demonstrated a correlation between increased UPAR tumor and/or serum levels and a poor prognosis for prostate cancer patients. Taken together, UPAR may serve as a negative prognostic marker and putative target gene in prostate cancer therapy.23, 24, 25, 26, 27 Based on our in silico analysis, the aberrant overexpression of UPAR in prostate carcinoma may, at least in part, be mediated by reduced MIR143 levels.
In this study, we analyzed this putative MIR143-UPAR axis in prostate carcinoma, and indeed we identified UPAR as a direct MIR143 target. In vitro as well as in a therapeutic in vivo model in mice, we demonstrate tumor-inhibitory effects of MIR143 replacement through affecting UPAR expression.
Results
UPAR Is a Target Gene of MIR143
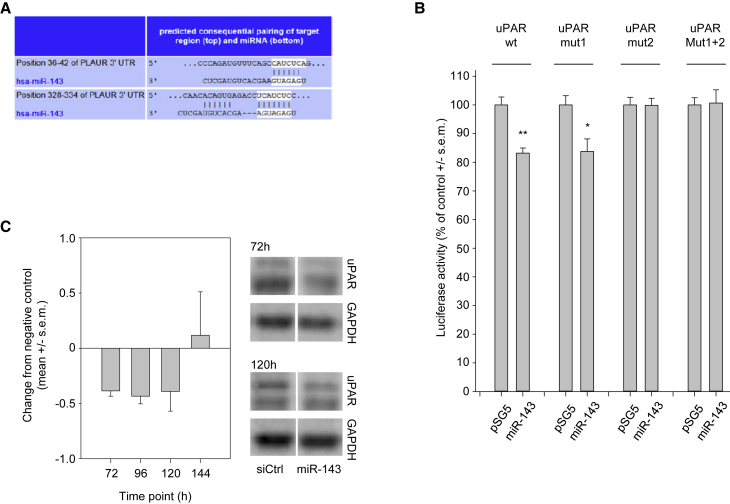
In silico analyses predicted UPAR as a potential target gene of MIR143, with two putative binding sites in the 3′ UTR (positions 35–41 and 327–333 of the UPAR 3′ UTR; http://www.targetscan.org/vert_71/: release 5.2; Figure 1A). To test for this, we generated a reporter gene construct with the luciferase gene under the regulatory control of the uPAR 3′ UTR. As shown in Figure 1B, the simultaneous transfection of the wild-type (WT) reporter gene plasmid with a MIR143 expression vector into HEK293T cells indeed led to a significant ∼15% reduction in reporter gene activity, which is well in the range of effects to be expected from miRNAs. To identify the active binding site, we mutated the first (Mut I), the second (Mut II), or both predicted MIR143-binding sites (Mut I + II) by site-directed mutagenesis. The mutation of both binding sites, or of the second binding site alone, completely abrogated the inhibitory effect of MIR143 on the reporter gene (Figure 1B). In contrast, mutation of the first binding site had no effect on the regulation of the reporter gene by MIR143. This result demonstrates that the 3′ UTR of the UPAR gene has one direct, functionally active interaction site for MIR143 (nucleotides 327–333 of the 3′ UTR).
Figure 1.
Regulation of UPAR by MIR143
(A) Two putative MIR143-binding sites in the 3′ UTR of the uPAR-PLAUR gene are predicted by TargetScan (http://www.targetscan.org/vert_71/), located at 35–41 bp and 327–333 bp, respectively. (B) Luciferase reporter gene analysis. HEK293T cells were co-transfected with reporter gene constructs (top) and with the MIR143 expression vector or the empty vector (bottom). Firefly luciferase activity was normalized against the activity of Renilla luciferase. pMIR-UPAR-WT, vector with non-modified UPAR 3′ UTR; pMIR-UPAR MUT I, MUT II, and MUT I + II, vector containing the UPAR 3′ UTR with MIR143-binding sites I, II, and I + II, respectively, mutated by site-directed mutagenesis. (C) MIR143-mediated regulation of UPAR protein expression in vitro. PC-3 cells were transfected with synthetic MIR143 mimics or non-targeting control oligonucleotides for the indicated time. The expression of UPAR was analyzed by western blotting. The uPAR protein expression of each sample was normalized to GAPDH as the loading control. Bars represent changes from negative control-transfected cells; right, representative western blots. Data are presented as mean ± SEM; *p < 0.05; **p < 0.03.
To study the effects of MIR143 on endogenous UPAR protein expression, PC-3 prostate carcinoma cells were transfected with synthetic MIR143 mimics, and, at different time points, the amounts of UPAR protein were measured by western blotting. Notably, UPAR protein exhibited a rather prolonged stability. The complete inhibition of protein synthesis by cycloheximide indicated that more than 72 h was necessary to detect a substantial decrease in UPAR protein (data not shown). Therefore, the time frame after transfection was extended up to 7 days. Indeed, 72 h after MIR143 transfection of PC-3 cells, reduced UPAR protein levels were detected as compared to transfection with negative control RNA, recovering to normal levels only after 144 h (Figure 1C).
While this finding confirms the direct regulation of UPAR by MIR143, it also suggests that pathologically decreased MIR143 levels in prostate carcinoma may account for UPAR upregulation. To test for a correlation between UPAR and MIR143 levels in prostate carcinoma, 26 primary prostate carcinoma tissues were analyzed by UPAR ELISA and qRT-PCR. Indeed, an inverse correlation between MIR143 and UPAR protein levels was observed (rs = −0.443; p = 0.027, Spearman’s test; Figure S1A).
Effects of MIR143 and UPAR Knockdown on Cell Proliferation and Cell Viability
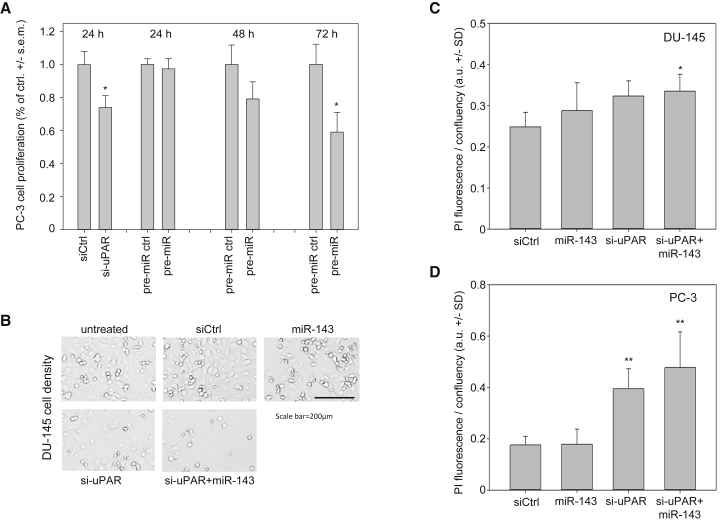
Next, we studied the effect of transfection of MIR143 on PC-3 or DU-145 cell proliferation and cell viability, in direct comparison to small interfering RNA (siRNA) directed against UPAR. In PC-3 cells, treatment with siRNA against UPAR led to a statistically significant reduction in cell proliferation compared to cells treated with control siRNA, with this effect starting already at 24 h after transfection (Figure 2A, left). A similar decrease in cell proliferation was observed upon transfection of pre-MIR143; however, this inhibition set in only after 48 h, with a subsequent further decrease in cell proliferation to approximately 58% of control miRNA-treated cells at 72 h (Figure 2A, middle and right). This time frame is consistent with the delayed effects of MIR143 on UPAR protein levels (see above).
Figure 2.
Biological Effects of MIR143 Replacement or UPAR Knockdown In Vitro
(A) Inhibition of PC-3 cell proliferation upon MIR143 replacement or UPAR knockdown. PC-3 cells were transfected with pre-MIR143, siRNA against UPAR, or the appropriate negative controls. At the indicated time points, cell proliferation was determined by BrdU incorporation. (B) Effects on cell density upon single or combined transfection of MIR143 and/or si-UPAR in DU-145 cells. Scale bar, 200 μm. Decreased numbers of cells were also associated with increased percentages of dead cells in (C) DU-145 and (D) PC-3 cells, as determined by propidium iodide staining in Celigo Imaging Cytometry. Data are presented as mean ± SEM (A) or as mean ± SD (C and D); *p < 0.05; **p < 0.03.
Inhibitory effects on tumor cell growth were also observed in DU-145 cells, with a marked reduction of cell density at 72 h after transfection with MIR143 or si-UPAR, respectively (Figure 2B). While the inhibition was slightly more profound after UPAR knockdown as compared to miRNA replacement, the strongest inhibitory effects on DU-145 tumor cell growth were observed upon double transfection (Figure 2B, bottom). Beyond reduced viable cell density, a slight increase in the percentage of dead cells was observed, especially upon si-UPAR transfection, as determined by propidium iodide (PI) staining using Celigo Imaging Cytometry (Figure 2C; Figure S2A). Again, effects showed a very slight trend toward further increased PI signals upon double transfection, with differences to negative control-transfected cells then reaching statistical significance. In PC-3 cells, the transfection of 10 nM MIR143 did not yet yield an increase in PI-positive cells, while 5 nM si-UPAR was already sufficient to induce a 2-fold upregulation of PI positivity. The combined transfection with both siRNA and miRNA, however, led to a further increase in PI-positive cells (Figure 2D; Figure S2B).
Notably, here and in all other experiments, equal amounts of total siRNA and miRNA were employed to exclude non-specific effects of small RNA transfection, i.e., where applicable, negative control siRNA was added to the different amounts of specific siRNA and miRNA to yield a total of 15 nM in each setting. Additionally, the parallel transfection of cells with a scramble control RNA, for direct comparison with the negative control siLuc, revealed no differences between the two negative controls with regard to induction of cell death or effects on proliferation, proving that both negative control RNAs do appropriately control for non-specific effects (data not shown). Thus, differences in PI induction must be considered exclusively as specific effects. Interestingly, when doubling the RNA amounts used for transfection, a different picture was seen, with MIR143 gaining cell death-inducing potential, as indicated by an ∼2-fold upregulation of PI signal (Figure S3). While this was even slightly above 10 nM si-UPAR effects, no further increase in PI positivity was observed upon double treatment (Figure S3, right).
Effects of MIR143 and UPAR Knockdown on Colony Formation and 3D Cell Growth
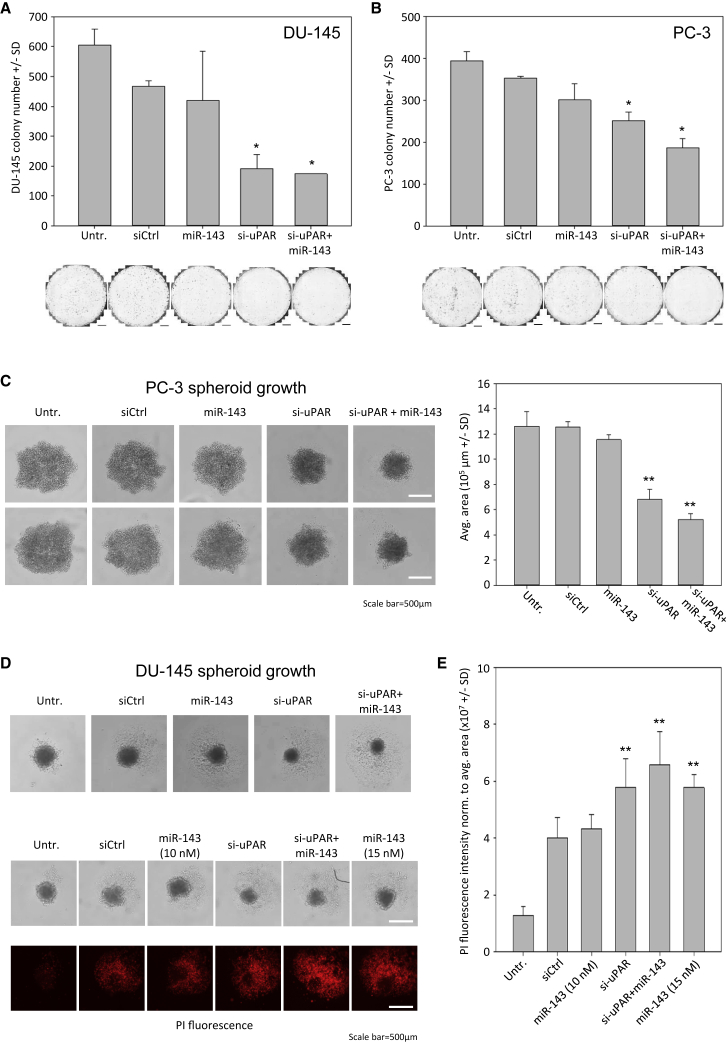
To further explore MIR143 and/or si-UPAR-mediated tumor cell inhibitory effects, studies were extended toward other in vitro models. DU-145 colony formation was mainly impaired by siRNA-mediated UPAR knockdown, while MIR143 transfection exerted little effects over negative control transfection (Figure 3A). This is in line with the results on anchorage-dependent cell proliferation and on induction of cell death (see Figures 2B–2D), and it was also observed in PC-3 cells. Again, si-UPAR effects were found more pronounced as compared to MIR143 transfection. In this cell line, however, a further decrease in colony formation was observed upon double transfection (Figure 3B, right). Notably, since again identical amounts of total RNA were used for transfection (with negative control siRNA being used for filling up, where appropriate), enhanced inhibition of cell proliferation upon combined treatment cannot be explained by non-specific transfection effects.
Figure 3.
Effects of MIR143 and si-UPAR Transfection on Colony Formation and Spheroid Growth and Viability
Inhibition of colony formation in (A) DU-145 and (B) PC-3 cells, as determined by colony numbers. (C) Reduction of PC-3 cell spheroid growth (left, original pictures; scale bars, 500 μm; right, quantitation of average spheroid areas). (D) Slightly decreased DU-145 spheroid growth upon single or combined MIR143 and si-UPAR transfection (top). Inhibitory MIR143 effects are dose dependent (middle), especially with regard to cell death induction in the spheroid, as determined by PI staining of dead cells (bottom). (E) Quantitation of PI staining intensity. Data are presented as mean ±SD; *p < 0.05; **p < 0.03.
Spheroid assays in the same cell line revealed a profound >50% reduction of spheroid size upon combined transfection (Figure 3C). This effect again mainly relied on siRNA-mediated UPAR knockdown rather than MIR143 replacement. Less pronounced effects were observed on DU-145 cell spheroid growth (Figure 3D, top). Here, however, the considerable miRNA dose dependence was confirmed: a 1.5-fold increase in miRNA, while keeping the total RNA used for transfection again constant, already led to increased effects on DU-145 spheroid viability (Figure 3D, bottom). This was particularly seen when spheroids were analyzed for PI-positive cells, indicating the induction of cell death. While the lower miRNA amounts yielded a signal only at the level of the negative control, the 1.5-fold higher dosage (15 nM instead of 10 nM) led to a marked increase in PI-positive cells that was comparable to si-UPAR. An only slight increase was observed upon double transfection (Figure 3E). Taken together, this indicates a dose-dependent inhibitory effect of MIR143 on prostate carcinoma cells, which, at sufficient amounts, reaches results on tumor cell inhibition induced by siRNA-mediated UPAR knockdown.
Systemic Application of MIR143 Decreases Tumor Xenograft Growth in Mice
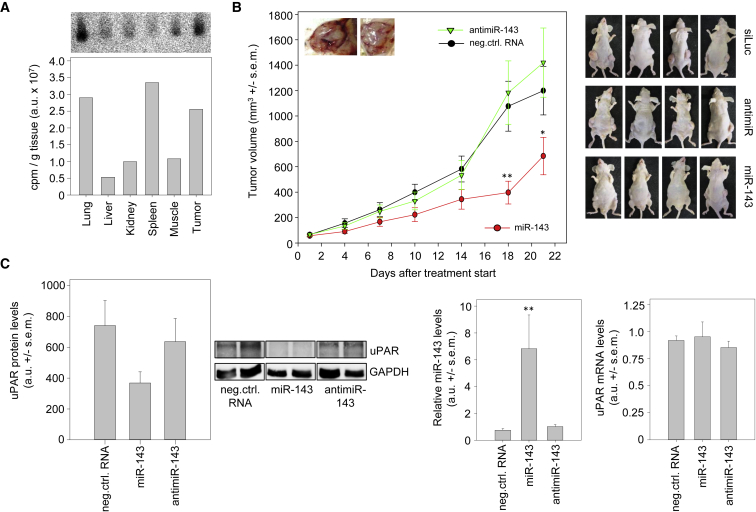
To explore the effects of miRNA replacement and double treatment in a therapeutically more relevant in vivo model, we generated subcutaneous xenograft tumors originating from PC-3 cells in athymic nude mice. To represent the clinical setting of treating only well-established tumors in the growing state, we started the treatment when the xenografts had reached 50–75 mm3, and we performed systemic injection of the mice rather than local administration into the tumor. An initial biodistribution experiment confirmed the nanoparticle-mediated delivery of intact full-length miRNA to the tumors. More specifically, the mice were treated once with complexes containing [32P]-end-labeled miRNAs, and, 6 h after a single intraperitoneal (i.p.) injection, various organs detailed in Figure 4A were analyzed for miRNA levels. Gel electrophoresis and autoradiography revealed strong miRNA signals in the tumor and lung, while weaker bands were observed in other organs. Additionally, normalization for tissue amount revealed rather profound uptake into the spleen. The presence of non-degraded bands with the radioactive miRNA end label confirmed the delivery of intact, full-length miRNAs into the respective tissue (Figure 4A).
Figure 4.
Tumor Inhibition upon Therapeutic MIR143 Replacement In Vivo
(A) Biodistribution and tissue uptake of radioactively labeled MIR143 upon PEI F25-LMW complexation and single i.p. injection. Total RNA from various mouse organs and tumor tissue was analyzed by gel electrophoresis and autoradiography (top), with bands representing intact full-length miRNA. Bottom: quantitation of miRNA levels was normalized to tissue amounts. (B) Inhibition of xenograft tumor growth upon nanoparticle-based MIR143 replacement therapy. Athymic nude mice with established PC-3 xenograft tumors were systemically treated with PEI F25-LMW-complexes containing the RNA specified in the figure. The determination of tumor volumes at the indicated time points reveals profound inhibition of tumor growth in the specific treatment group. Right: pictures show representative mice at the time point of termination of the experiment. (C) Analysis of the tumor xenograft tissues upon termination of the treatment study. Left: UPAR protein levels were normalized to GAPDH (representative examples of the western blots and a bar diagram quantitating all samples are shown; center, miRNA levels; right, UPAR mRNA levels, both normalized to reference genes). Data are presented as mean ± SEM; *p < 0.05; **p < 0.03.
For the therapy study, mice were randomized into treatment and negative control groups, prior to systemic administration by i.p. injection of the respective complexes three times per week. The complexes contained 10 μg of either MIR143 or negative control siRNA. Additionally, an anti-MIR143 treatment group was included in this experiment to address the question of whether MIR143 inhibition may lead to even enhanced tumor growth in this in vivo setting.
Tumor volumes were monitored every 3–4 days until the mice were sacrificed at day 21 after the start of therapy. During the entire treatment period, none of the mice showed any treatment-related side effects. The tumors of the mice in the MIR143 treatment group exhibited marked inhibition of tumor growth, leading to significantly reduced tumor volumes compared to their polyethylenimine (PEI)-siRNA negative control-treated counterparts (Figure 4B). The differences were most pronounced at day 18, when the volumes of the xenograft tumors of the mice treated with MIR143 were only approximately 35% of the control tumor volumes. In contrast, after 18 days of treatment, the tumors of the antimiR-treated group were not significantly different in their xenograft tumor growth compared to the control group (Figure 4B).
Tumor Xenograft-Inhibitory MIR143 Effects Are Paralleled by Decreased UPAR Protein Levels
Upon termination of the single treatment experiment at day 21 of the treatment scheme (Figure 4B), the mice were sacrificed and the xenograft tumors were surgically removed for further analyses. The determination of UPAR protein levels from the tumor xenograft lysates revealed an almost 50% reduction of UPAR expression in the MIR143 treatment group compared to the negative control or the antimiR group (Figure 4C, left). In the latter case of MIR143 inhibition, however, no further increase in UPAR protein levels was observed, which was consistent with the largely unaltered growth kinetics between the two groups (see Figure 4B). Notably, however, in the case of PEI-MIR143 treatment, the decrease in UPAR protein was even more pronounced than in our previous in vitro experiments. Consistent with this finding, an approximately 9-fold increase in MIR143 tissue levels upon treatment was observed, confirming the efficient nanoparticle-mediated delivery of the miRNA (Figure 4C, middle). A downregulation of ERK5 as an already well-established target of MIR143 was observed in the tumors as well (Figure S4). As expected, no effects of the antimiR treatment on MIR143 levels were found, because the antimiRs would not degrade the miRNAs. Interestingly, the abundance of the UPAR mRNA transcript was not influenced by either MIR143 or anti-MIR143 molecules (Figure 4C, right), indicating that the pronounced MIR143 effects on UPAR protein levels (see Figure 4C, left) result from the inhibition of translation rather than mRNA degradation.
Discussion
With miRNAs acting simultaneously on several molecular targets, the approach of miRNA replacement therapy addresses the notion of cancer as a pathway disease.28, 29 Recently, efforts and first successes of miRNA-based therapies mostly focusing on miR-34 have been reviewed,30 but they have also been reported to meet unexpected toxicity. Thus, this approach probably requires the identification of (all) major relevant target molecules; the dissection of the major contributors to tumor-suppressive effects of a given miRNA by extensive functional analyses; and solutions to issues regarding the delivery of the drug, namely, the miRNA.
MIR143 has been well established as an anti-oncogenic (tumor-suppressive) miRNA in different cancers. Direct target genes and pathways include G-protein/mitogen-activated protein kinase (MAPK)-AKT signal transduction by inhibiting KRAS, HRAS, BRAF, RREB1, MAPK7-ERK5, and AKT1;9, 12, 14, 31, 32, 33, 34, 35 epithelial-mesenchymal transition (EMT) by regulating MACC1, MAPK7-ERK5, and FNDC3B;14, 36, 37, 38, 39 and TP53-apoptosis pathways by repressing BCL2, CD44, and MDM2.34, 40, 41 In addition, MIR143 has also been described to negatively affect extracellular matrix degradation by binding to SERPINE1-PAI-1 and MMP13,42, 43 in this way repressing metastasis pathways44 and inhibiting changes in the cytoskeleton through the repression of MYO6 and FSCN1.16, 45
Here we establish for the first time the UPAR as a direct target of MIR143. The tumor cell-inhibitory effects upon MIR143 transfection, or si-UPAR transfection for comparison, are in line with previous papers, which showed that UPAR is well known to increase cell proliferation in different cancers, including prostate cancer.21 Therefore, our finding of UPAR as a target gene of MIR143 also fits into the line of cell proliferation-associated target genes of MIR143, including, e.g., ERK5, PTGS2-COX2, KDM5B, IGFR1, and ERBB3.14, 46, 47, 48, 49 As opposed to UPAR single knockdown, however, the approach of miRNA replacement may lead to broader molecular effects, potentially avoiding a cellular escape through using redundant pathways and/or preventing the emergence of secondary resistance.
Furthermore, UPAR belongs to a MIR143-regulated group of proteins (PAI-1, MMP13, VCAN, SDCN1, and FNDC3B) that are involved either structurally or as regulators in extracellular matrix (ECM) assembly or degradation, cell adhesion, migration, invasion, and metastasis.37, 50, 51, 52, 53, 54, 55, 56 Interestingly, however, the uPA inhibitor (PAI-1 or SERPINE1), a member of the urokinase-type plasminogen activator (UPA or PLAU) system, has been identified in bladder cancer as a target of MIR143 as well.43 While the uPA-UPAR system is thought to play an important role in tumorigenesis and tumor progression, the role of the uPA inhibitor PAI-1 in cancer is rather ambiguous,57 with at least partially counteracting UPA-UPAR effects. This also highlights that the entirety of molecular and cellular effects of a single inhibition or knockdown, and even more so the effects of a miRNA replacement approach, needs to be analyzed in great detail in the context of the given tumor entity.
In previous studies, we found that MIR143 was downregulated in prostate cancer tissue compared to corresponding, adjacent normal tissue,15, 16, 18 providing a solid basis for miRNA replacement therapy. Thus, moving into a therapeutically relevant preclinical in vivo setting, we explored nanoparticle-mediated MIR143 replacement in a PC-3-xenotransplant mouse model. We found a significant reduction in UPAR protein expression and a significant reduction of tumor growth in the MIR143 treatment group. Since experiments with siRNA against UPAR in PC-3 xenotransplant models have shown comparable reductions in UPAR protein levels and tumor growth,58, 59 we suggest the direct effect of MIR143 on uPAR, leading to the decreased uPAR protein levels observed in our study, as at least one major mechanism of MIR143-mediated tumor inhibition. Our results are consistent with data on other cancers, where, upon delivery of MIR143 or MIR145, an inhibition of pancreatic, breast, or colon carcinoma growth in mice models can be achieved.48, 60, 61 Recently, Niu et al.62 showed that injection of stably MIR143-transduced PC-3 cells into mice generated smaller xenograft tumors. Our in vivo model goes one step further toward a therapeutic scenario, by exploring miRNA replacement therapy in fully established tumors, thus resembling more closely the clinical situation.
Altogether, our data emphasize the putative relevance of miRNA replacement as a novel therapeutic concept in tumor treatment and the usefulness and efficacy of polymeric nanoparticles for systemic miRNA administration.62, 63, 64, 65 This notion is supported by at least 12 ongoing clinical studies that apply nanoparticles to deliver therapeutics to prostate cancer (https://clinicaltrials.gov/; accessed December 2018). Likewise, clinical studies on miRNA treatment, i.e., either inhibition of an oncogenic miRNA (e.g., antimiR-155) or delivery of tumor suppressor miRNAs (e.g., miR-16-mimic and miR-34-mimic) have been performed or are underway, for example, for the treatment of cutaneous T cell lymphoma, mesothelioma, non-small-cell lung cancer, and multiple solid tumors (e.g., ClinicalTrials.gov: NCT02580552, NCT02369198, and NCT01829971; reviewed in Rupaimoole and Slack66).
Interestingly, Niu et al.62 recently showed that insulin-like growth factor I can induce chemoresistance against docetaxel by inhibiting MIR143 in human prostate cancer cells in vitro. On the other hand, overexpression of MIR143 abolished IGF-I-induced chemoresistance to docetaxel treatment. This result indicates that miRNA replacement therapy could be able to interfere with tumor characteristics on multiple levels. Thus, while RNA-centered therapies may not be strong enough as single drugs, their combination with existing therapeutic regimens could be particularly attractive for sensitizing tumor cells. Beyond aiming at increased efficacy, this may also allow for decreasing side effects by being able to reduce drug dosages. The somewhat broader specificities of miRNAs can be beneficial when considering cancer as a pathway disease, with the simultaneous action of a given miRNA on multiple relevant targets increasing its efficacy and avoiding the development of resistance.
In summary, we identify UPAR as a direct target gene of MIR143, and we establish the therapeutic relevance of nanoparticle-based MIR143 replacement in a prostate cancer mouse model. Beyond direct tumor inhibition, MIR143, by regulating UPAR and other target genes that encode proteins involved in extracellular matrix formation and degradation, may also play a major role against tumor cell dissemination and metastasis.
Materials and Methods
Cell Culture
The prostate cancer cell lines PC-3 and DU-145 and the normal cell line HEK293T were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany) or from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in DMEM (Sigma, Taufkirchen, Germany) supplemented with 10% heat-inactivated fetal bovine serum, 1 nmol/L L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C in a humidified atmosphere of 5% CO2.
PCa Tissue
Prostate cancer (PCa) patients and PCa tissues used for expression analyses have been described previously.16 The study was approved by the local ethical review board and was performed according to the Declaration of Helsinki. Upon informed consent, prostate cancer tissues were obtained after radical prostatectomy from so far treatment-naive prostate cancer patients.
Oligonucleotides
For in vitro modulation of MIR143, we used a synthetic precursor MIR143 (PM10883), a synthetic miRNA (see below), as well as a synthetic MIR143 inhibitor (AM10883) along with appropriate negative control oligonucleotides (Thermo Fisher Scientific, Waltham, MA, USA). The synthetic MIR143 was also used for the in vivo studies and prepared as an RNA-RNA duplex with 3′ and 5′ overhangs and intramolecule nucleotide mismatches, as reported in miRBase (version 21; http://www.mirbase.org/). More specifically, RNA oligonucleotides 5′-GGUGCAGUGCUGCAUCUCUGGU-3′ and 5′-UGAGAUGAAGCACUGUAGCUC-3′ were synthesized and annealed to an RNA-RNA duplex by Integrated DNA Technologies (IDT, Leuven, Belgium). The synthetic anti-MIR143 inhibitor for in vivo application was designed and produced by Exiqon (Vedbaek, Denmark). Knockdown of UPAR was accomplished with a pool of siRNAs (L006388, ON-TARGETplus PLAUR siRNA, Dharmacon, Lafayette, CO, USA). As a negative control RNA, a non-targeting siRNA directed against the luciferase gene was used. The siRNA molecule 5′-CUUACGCUGAGUACUUCGA-dTdT-3′ and 5′-dTdT-GAAUGCGACUCAUGAAGCU-3′ was purchased from MWG (Ebersberg, Germany).
RNA and Protein Isolation
Total RNA and protein were isolated using TRIzol (Invitrogen, Darmstadt, Germany), according to the manufacturer’s instructions. In the case of cultivated cell lines, TRIzol reagent was added directly to the cells after washing with PBS. Tissue samples were mechanically disrupted in TRIzol reagent prior to RNA and protein isolation. RNA preparations were treated with recombinant DNaseI (Sigma-Aldrich, Taufkirchen, Germany) before use. For luciferase reporter gene analyses, a different extraction protocol was used. Briefly, cells were lysed with radioimmunoprecipitation assay (RIPA) lysis buffer (25 mM Tris-HCl [pH 8.0], 137 mM NaCl, 10% glycerol, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP40, 2 mM EDTA [pH 8.0], 1 mM sodium vanadate, and 1.5 mM sodium fluoride) for 15 min, and cell lysates were collected by centrifugation.
Reverse Transcription and qPCR
To quantify the mRNA levels, 1 μg total RNA was reverse transcribed using the Dynamo cDNA synthesis system (Thermo Scientific, Darmstadt, Germany), according to the manufacturer’s recommendations. qPCR was performed in a StepOne plus real-time PCR system (Thermo Scientific, Applied Biosystems, Darmstadt, Germany) in a final volume of 10 μL containing 1× gene expression master mix, 1× gene-specific primers and detection probe, and cDNA corresponding to 25 ng total RNA. All of the reactions were performed in triplicate. The gene-specific primer-probe combinations used were HPRT1 (Hs99999909_m1, Applied Biosystems) and PLAUR (Hs00182181_m1, Applied Biosystems).
For the quantification of miRNAs, 10 ng total RNA was reverse transcribed using miRNA reverse transcription reagents (Applied Biosystems) and miRNA-specific reverse transcription primers. Quantification was performed in a StepOne plus real-time PCR system (Thermo Scientific, Applied Biosystems) in a final volume of 10 μL containing 1× gene expression master mix, 1× miRNA-specific primers and detection probe, and miRNA-specific primed cDNA corresponding to 330 pg total RNA. All of the reactions were performed in triplicate. The miRNA-specific primer-probe combinations used were RNU6b (001093, Applied Biosystems) and hsa-MIR143 (002249, Applied Biosystems). Relative gene expression was calculated using the ΔΔCt method using the reference gene HPRT1 or RNU6b.67
MIR143 Reporter Luciferase Assays
The pSG5-MIR143 expression construct was generated by PCR amplification using primers MIR143-EcoRI 5′-CCGAATTCTGCTCAAATGGCAGGCCACAGAC-3′ and MIR143-BamHI 5′-CCGGATCCTCGTGAAGCAGATCGTGGCACCA-3′ (MWG-Eurofins, Ebersberg, Germany) and inserted into the pSG5 expression plasmid (Stratagene, Heidelberg, Germany). The dual luciferase reporter plasmid pMIR-RL has been described elsewhere.16 Nucleotides 21–336 of the 3′ UTR of UPAR (GenBank: NM_001005376.2) were cloned via PCR amplification from testis cDNA and inserted via the SpeI and NaeI restriction sites of pMIR-RL. The oligonucleotide primers used were UPAR WT forward 5′-GGACTAGTCCCAGATGTTTCAGCCATCTCAGCCCAGGCACCAGAC-3′ and UPAR WT reverse 5′-GCCGGCGTGGAGATGAGGTCTCACTG-3′. For the targeted mutation of the predicted MIR143-binding sites, we designed alternative oligonucleotide primers that, when used instead of the original primers, substituted the first, the second, or both MIR143-binding sites with restriction site sequences. More specifically, UPAR MUT I forward 5′-GGACTAGTCCCAGATGTTTCAGCTCGCGAAGCCCAGGCACCAGAC-3′ substituted the first MIR143-binding site with a NruI restriction site, and UPAR MUT II reverse 5′-GCCGGCGTGTCACGTGTGTCTCACTGTGTTGCCCAGGCTGATC-3′ substituted the second MIR143-binding site with a PmlI restriction site.
HEK293T cells were cultivated in 24-well plates and transfected with 0.2 μg reporter construct and 0.8 μg miRNA expression plasmid using the Nanofectin transfection reagent (PAA, Cölbe, Germany). The luciferase assays were performed 48 h after transfection using a Dual-Luciferase Reporter Assay System, according to the manufacturer’s instructions (Promega, Mannheim, Germany).
Western Blotting
Equal amounts of total protein lysates were separated by SDS-PAGE and transferred to nitrocellulose membranes (GE Healthcare, Freiburg, Germany) by electroblotting. The primary antibodies used were polyclonal rabbit anti-uPA receptor antibody (ab103791; Abcam Milton, UK; 1:100) and monoclonal rabbit anti-GAPDH (clone 14C10, NEB Cell Signaling, Frankfurt, Germany; 1:1,000). Secondary anti-rabbit antibody conjugated with horseradish peroxidase was purchased from Jackson ImmunoResearch Laboratories (Suffolk, UK) and used at a concentration of 1:5,000. Protein bands were revealed by enhanced chemiluminescence in a LAS-4000 chemiluminescence detection system (GE Healthcare, Munich, Germany). Densitometry of western blot pictures was performed using ImageJ software, and the expression of UPAR protein was normalized to GAPDH as the housekeeping control protein.
Determination of UPAR Antigen Levels by ELISA
The UPAR antigen level in tissue extracts of PCa patients was determined using a commercially available ELISA kit (IMUBIND UPAR ELISA 893; American Diagnostica/Sekisui Diagnostics, Stamford, CT, USA), according to the manufacturer’s instructions and as previously described.68 Antigen concentrations in tissue extracts are given as nanogram analyte per milligram total protein.
Cell Transfection and Measurement of Cell Proliferation
PC-3 and DU-145 cells were transfected with negative control siRNA (siCtrl), MIR143, si-UPAR, or the combination of si-UPAR and MIR143 at a final concentration of 15 nM, using INTERFERin transfection reagent (Polyplus Transfection, Illkirch, France). Single transfections with MIR143 or si-UPAR at concentrations below 15 nM were filled up with negative control to reach equal amounts of nucleic acid in all wells. For the transfection of pre-miR (Figure 2A), siPORTNeoFX transfection agent (Thermo Fisher Scientific) and a final concentration of 50 nM were used.
Cell proliferation was quantitated with the bromodeoxyuridine (BrdU) cell proliferation ELISA Kit (Roche Applied Science, Mannheim, Germany), according to the manufacturer’s instructions. At 24, 48, and 72 h after transfection, BrdU incorporation was measured in an automated microplate reader. The mean absorbance of control cells was set to 100%, and cell proliferation upon specific transfection is given as percent of control.
Spheroid and Colony Formation Assays
Cells were transfected using INTERFERin as described above. For spheroid assays, cells were trypsinized after 48 h and reseeded in Nexcelom3D 96-well round-bottom ultra-low attachment plates (Nexcelom Bioscience, Lawrence, MA) at a density of 5,000 cells/well. The morphology as well as the average area (μm2) was measured on day 7 after seeding, using a Celigo Imaging Cytometer (Nexcelom). Subsequently, the dead cell staining reagent propidium iodide (Carl Roth, Karlsruhe, Germany) was added to the spheroids at a concentration of 1 μg/mL/well. After 30-min incubation, the absolute red fluorescence intensity was measured using the Celigo Imaging Cytometer.
For colony formation assays, cells were trypsinized 48 h after transfection and reseeded in 6-well plates at a density of 1,000 cells/well. Each treatment was seeded in duplicates and cultured for 8 days. Subsequently, the colonies were fixed with methanol, stained with 0.5% crystal violet solution (Carl Roth), and counted and imaged automatically using the Celigo Imaging Cytometer Software.
Confluency and PI Assay
PC-3 or DU-145 cells were seeded in a Greiner CELLSTAR 96-well-plate (Sigma-Aldrich, Munich, Germany) at a density of 2,000 cells/well. At 1 day after seeding, cells were transfected as described above. Propidium iodide was added 72 h after transfection at a concentration of 1 μg/mL/well. The red fluorescence intensity signal relative to the confluence per well was analyzed using the Celigo Imaging Cytometer Software.
Mouse Xenograft Model and Nanoparticle Therapy
Athymic nude mice (Hsd:Athymic Nude-Foxn1nu, 6–8 weeks of age) were obtained from the animal core facility of the University of Leipzig (Medizinisch-Experimentelles Zentrum [MEZ]) and kept at 23°C in a humidified atmosphere and a 12-h-light and dark cycle, with standard rodent chow and water ad libitum. Experiments were performed according to the national regulations and approved by the local authorities of the State of Saxony, Germany. For tumor establishment, 3 × 106 PC-3 cells in 150 μL PBS were injected subcutaneously (s.c.) into both flanks of the mice. When the solid tumors reached a volume of approximately 50–75 mm3, the mice were randomized into specific treatment, negative control treatment, and non-treatment groups (n = 18–23 tumors per group).
miRNAs, antimiRs, and negative control siRNAs were complexed with the PEI F25-LMW essentially as described previously.63, 69 Briefly, 10 μg miRNA, antimiR, or siRNA was dissolved in 75 μL 10 mM HEPES, 150 mM NaCl (pH 7.4), and incubated for 10 min. Then, 50 μg PEI F25-LMW70 was dissolved in the same buffer for 10 min and mixed with the miRNA-antimiR-siRNA solution, prior to complexation for 15 min. The complexes were aliquoted and stored frozen.71 Prior to use, they were thawed and allowed to incubate for 30–60 min at room temperature (RT).
To assess the biodistribution, miRNAs were radioactively end labeled before complexation, using [32P]-ATP as described previously.69 At 6 h after i.p. injection of the PEI-miRNA complexes, the mice were sacrificed and total RNA was prepared from various organs, run on a 1.6% agarose gel, and blotted, prior to analyzing the blots by autoradiography.
For studying therapeutic effects, 10 μg PEI F25-LMW-complexed miRNAs, antimiRs, or negative control siRNAs were systemically administered three times per week by i.p. injection, described previously as optimal.69 Tumor volumes were monitored every 3–4 days at the time points indicated in Figure 4B, by measuring all three dimensions. For termination of the experiment, the mice were sacrificed 1 day after the last treatment with PEI complexes, and the tumors were immediately removed. Pieces of the tumor tissue were snap frozen in TRIzol for future RNA and protein preparation.
Statistical Analysis
All experiments were run in triplicates unless stated otherwise. Statistical analyses were performed by one-way ANOVA or Student’s t test using SigmaPlot 10 (Systat, Chicago, IL, USA), with *p < 0.05 and **p < 0.03 compared to negative control transfection.
Author Contributions
A.A., H.T., and S.W. designed the study. K.W., S.L., E.N., O.A.-J., R.S., J.G., V.L., A.H., and B.W. acquired the clinical samples and patient information. A.H. and J.G. performed the pathological review of all cases. A.A., H.T., M.B., and S.W. made the statistical analyses and M.B., H.B., K.W., S.L., E.N., O.A.-J., M.H., F.G., R.J., S.H., A.P., and C.S. performed the experimental work. A.A., H.T., M.B., and S.W. prepared tables and figures. A.A., H.T., B.W., S.W., and A.H. wrote the main manuscript. All authors reviewed the manuscript.
Conflicts of Interest
All authors declare no competing interests.
Acknowledgments
The authors are grateful to Markus Böhlmann and Kathrin Krause for expert mouse maintenance and help with the animal experiments. The project was supported by the Wilhelm Sander-Stiftung (2015.171.1) and the Johannes and Frieda Marohn-Stiftung (FWN/Zo-Tau/2012). Funding was also provided by the Deutsche Krebshilfe (Az. 111616, to A.A.) and by the FAZIT Stiftung (stipend to H.B.). In addition, H.T. and E.N. were supported by the Rudolf und Irmgard Kleinknecht-Stiftung and the Movember Foundation/Förderverein Hilfe beim Prostatakrebs e.V. (MOV-2013-05).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.02.020.
Supplemental Information
References
- 1.Goto Y., Kurozumi A., Enokida H., Ichikawa T., Seki N. Functional significance of aberrantly expressed microRNAs in prostate cancer. Int. J. Urol. 2015;22:242–252. doi: 10.1111/iju.12700. [DOI] [PubMed] [Google Scholar]
- 2.Akao Y., Nakagawa Y., Naoe T. MicroRNA-143 and -145 in colon cancer. DNA Cell Biol. 2007;26:311–320. doi: 10.1089/dna.2006.0550. [DOI] [PubMed] [Google Scholar]
- 3.Michael M.Z., O’ Connor S.M., van Holst Pellekaan N.G., Young G.P., James R.J. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 4.Motoyama K., Inoue H., Takatsuno Y., Tanaka F., Mimori K., Uetake H., Sugihara K., Mori M. Over- and under-expressed microRNAs in human colorectal cancer. Int. J. Oncol. 2009;34:1069–1075. doi: 10.3892/ijo_00000233. [DOI] [PubMed] [Google Scholar]
- 5.Slaby O., Svoboda M., Fabian P., Smerdova T., Knoflickova D., Bednarikova M., Nenutil R., Vyzula R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 6.Wang C.J., Zhou Z.G., Wang L., Yang L., Zhou B., Gu J., Chen H.Y., Sun X.F. Clinicopathological significance of microRNA-31, -143 and -145 expression in colorectal cancer. Dis. Markers. 2009;26:27–34. doi: 10.3233/DMA-2009-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takagi T., Iio A., Nakagawa Y., Naoe T., Tanigawa N., Akao Y. Decreased expression of microRNA-143 and -145 in human gastric cancers. Oncology. 2009;77:12–21. doi: 10.1159/000218166. [DOI] [PubMed] [Google Scholar]
- 8.Dyrskjøt L., Ostenfeld M.S., Bramsen J.B., Silahtaroglu A.N., Lamy P., Ramanathan R., Fristrup N., Jensen J.L., Andersen C.L., Zieger K. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69:4851–4860. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- 9.Lin T., Dong W., Huang J., Pan Q., Fan X., Zhang C., Huang L. MicroRNA-143 as a tumor suppressor for bladder cancer. J. Urol. 2009;181:1372–1380. doi: 10.1016/j.juro.2008.10.149. [DOI] [PubMed] [Google Scholar]
- 10.Wang X., Tang S., Le S.Y., Lu R., Rader J.S., Meyers C., Zheng Z.M. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS ONE. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng E.K., Li R., Shin V.Y., Siu J.M., Ma E.S., Kwong A. MicroRNA-143 is downregulated in breast cancer and regulates DNA methyltransferases 3A in breast cancer cells. Tumour Biol. 2014;35:2591–2598. doi: 10.1007/s13277-013-1341-7. [DOI] [PubMed] [Google Scholar]
- 12.Chen H.C., Chen G.H., Chen Y.H., Liao W.L., Liu C.Y., Chang K.P., Chang Y.S., Chen S.J. MicroRNA deregulation and pathway alterations in nasopharyngeal carcinoma. Br. J. Cancer. 2009;100:1002–1011. doi: 10.1038/sj.bjc.6604948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akao Y., Nakagawa Y., Kitade Y., Kinoshita T., Naoe T. Downregulation of microRNAs-143 and -145 in B-cell malignancies. Cancer Sci. 2007;98:1914–1920. doi: 10.1111/j.1349-7006.2007.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clapé C., Fritz V., Henriquet C., Apparailly F., Fernandez P.L., Iborra F., Avancès C., Villalba M., Culine S., Fajas L. miR-143 interferes with ERK5 signaling, and abrogates prostate cancer progression in mice. PLoS ONE. 2009;4:e7542. doi: 10.1371/journal.pone.0007542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hart M., Nolte E., Wach S., Szczyrba J., Taubert H., Rau T.T., Hartmann A., Grässer F.A., Wullich B. Comparative microRNA profiling of prostate carcinomas with increasing tumor stage by deep sequencing. Mol. Cancer Res. 2014;12:250–263. doi: 10.1158/1541-7786.MCR-13-0230. [DOI] [PubMed] [Google Scholar]
- 16.Szczyrba J., Löprich E., Wach S., Jung V., Unteregger G., Barth S., Grobholz R., Wieland W., Stöhr R., Hartmann A. The microRNA profile of prostate carcinoma obtained by deep sequencing. Mol. Cancer Res. 2010;8:529–538. doi: 10.1158/1541-7786.MCR-09-0443. [DOI] [PubMed] [Google Scholar]
- 17.Tong A.W., Fulgham P., Jay C., Chen P., Khalil I., Liu S., Senzer N., Eklund A.C., Han J., Nemunaitis J. MicroRNA profile analysis of human prostate cancers. Cancer Gene Ther. 2009;16:206–216. doi: 10.1038/cgt.2008.77. [DOI] [PubMed] [Google Scholar]
- 18.Wach S., Nolte E., Szczyrba J., Stöhr R., Hartmann A., Ørntoft T., Dyrskjøt L., Eltze E., Wieland W., Keck B. MicroRNA profiles of prostate carcinoma detected by multiplatform microRNA screening. Int. J. Cancer. 2012;130:611–621. doi: 10.1002/ijc.26064. [DOI] [PubMed] [Google Scholar]
- 19.Huang S., Guo W., Tang Y., Ren D., Zou X., Peng X. miR-143 and miR-145 inhibit stem cell characteristics of PC-3 prostate cancer cells. Oncol. Rep. 2012;28:1831–1837. doi: 10.3892/or.2012.2015. [DOI] [PubMed] [Google Scholar]
- 20.Smith H.W., Marshall C.J. Regulation of cell signalling by uPAR. Nat. Rev. Mol. Cell Biol. 2010;11:23–36. doi: 10.1038/nrm2821. [DOI] [PubMed] [Google Scholar]
- 21.Mekkawy A.H., Pourgholami M.H., Morris D.L. Involvement of urokinase-type plasminogen activator system in cancer: an overview. Med. Res. Rev. 2014;34:918–956. doi: 10.1002/med.21308. [DOI] [PubMed] [Google Scholar]
- 22.Mahmood N., Mihalcioiu C., Rabbani S.A. Multifaceted Role of the Urokinase-Type Plasminogen Activator (uPA) and Its Receptor (uPAR): Diagnostic, Prognostic, and Therapeutic Applications. Front. Oncol. 2018;8:24. doi: 10.3389/fonc.2018.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Janabi O., Taubert H., Lohse-Fischer A., Fröhner M., Wach S., Stöhr R., Keck B., Burger M., Wieland W., Erdmann K. Association of tissue mRNA and serum antigen levels of members of the urokinase-type plasminogen activator system with clinical and prognostic parameters in prostate cancer. BioMed Res. Int. 2014;2014:972587. doi: 10.1155/2014/972587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Almasi C.E., Brasso K., Iversen P., Pappot H., Høyer-Hansen G., Danø K., Christensen I.J. Prognostic and predictive value of intact and cleaved forms of the urokinase plasminogen activator receptor in metastatic prostate cancer. Prostate. 2011;71:899–907. doi: 10.1002/pros.21306. [DOI] [PubMed] [Google Scholar]
- 25.Kumano M., Miyake H., Muramaki M., Furukawa J., Takenaka A., Fujisawa M. Expression of urokinase-type plasminogen activator system in prostate cancer: correlation with clinicopathological outcomes in patients undergoing radical prostatectomy. Urol. Oncol. 2009;27:180–186. doi: 10.1016/j.urolonc.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Shariat S.F., Roehrborn C.G., McConnell J.D., Park S., Alam N., Wheeler T.M., Slawin K.M. Association of the circulating levels of the urokinase system of plasminogen activation with the presence of prostate cancer and invasion, progression, and metastasis. J. Clin. Oncol. 2007;25:349–355. doi: 10.1200/JCO.2006.05.6853. [DOI] [PubMed] [Google Scholar]
- 27.Wach S., Al-Janabi O., Weigelt K., Fischer K., Greither T., Marcou M., Theil G., Nolte E., Holzhausen H.J., Stöhr R. The combined serum levels of miR-375 and urokinase plasminogen activator receptor are suggested as diagnostic and prognostic biomarkers in prostate cancer. Int. J. Cancer. 2015;137:1406–1416. doi: 10.1002/ijc.29505. [DOI] [PubMed] [Google Scholar]
- 28.Bader A.G., Brown D., Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70:7027–7030. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Check Hayden E. Cancer complexity slows quest for cure. Nature. 2008;455:148. doi: 10.1038/455148a. [DOI] [PubMed] [Google Scholar]
- 30.Pichler M., Calin G.A. MicroRNAs in cancer: from developmental genes in worms to their clinical application in patients. Br. J. Cancer. 2015;113:569–573. doi: 10.1038/bjc.2015.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akao Y., Nakagawa Y., Naoe T. MicroRNAs 143 and 145 are possible common onco-microRNAs in human cancers. Oncol. Rep. 2006;16:845–850. [PubMed] [Google Scholar]
- 32.Kent O.A., Fox-Talbot K., Halushka M.K. RREB1 repressed miR-143/145 modulates KRAS signaling through downregulation of multiple targets. Oncogene. 2013;32:2576–2585. doi: 10.1038/onc.2012.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noguchi S., Yasui Y., Iwasaki J., Kumazaki M., Yamada N., Naito S., Akao Y. Replacement treatment with microRNA-143 and -145 induces synergistic inhibition of the growth of human bladder cancer cells by regulating PI3K/Akt and MAPK signaling pathways. Cancer Lett. 2013;328:353–361. doi: 10.1016/j.canlet.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 34.Pagliuca A., Valvo C., Fabrizi E., di Martino S., Biffoni M., Runci D., Forte S., De Maria R., Ricci-Vitiani L. Analysis of the combined action of miR-143 and miR-145 on oncogenic pathways in colorectal cancer cells reveals a coordinate program of gene repression. Oncogene. 2013;32:4806–4813. doi: 10.1038/onc.2012.495. [DOI] [PubMed] [Google Scholar]
- 35.Xu B., Niu X., Zhang X., Tao J., Wu D., Wang Z., Li P., Zhang W., Wu H., Feng N. miR-143 decreases prostate cancer cells proliferation and migration and enhances their sensitivity to docetaxel through suppression of KRAS. Mol. Cell. Biochem. 2011;350:207–213. doi: 10.1007/s11010-010-0700-6. [DOI] [PubMed] [Google Scholar]
- 36.Peng X., Guo W., Liu T., Wang X., Tu X., Xiong D., Chen S., Lai Y., Du H., Chen G. Identification of miRs-143 and -145 that is associated with bone metastasis of prostate cancer and involved in the regulation of EMT. PLoS ONE. 2011;6:e20341. doi: 10.1371/journal.pone.0020341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai C., Rajaram M., Zhou X., Liu Q., Marchica J., Li J., Powers R.S. Activation of multiple cancer pathways and tumor maintenance function of the 3q amplified oncogene FNDC3B. Cell Cycle. 2012;11:1773–1781. doi: 10.4161/cc.20121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan X., Chen X., Deng W., Zhong G., Cai Q., Lin T. Up-regulated microRNA-143 in cancer stem cells differentiation promotes prostate cancer cells metastasis by modulating FNDC3B expression. BMC Cancer. 2013;13:61. doi: 10.1186/1471-2407-13-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y., Wang Z., Chen M., Peng L., Wang X., Ma Q., Ma F., Jiang B. MicroRNA-143 targets MACC1 to inhibit cell invasion and migration in colorectal cancer. Mol. Cancer. 2012;11:23. doi: 10.1186/1476-4598-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borralho P.M., Kren B.T., Castro R.E., da Silva I.B., Steer C.J., Rodrigues C.M. MicroRNA-143 reduces viability and increases sensitivity to 5-fluorouracil in HCT116 human colorectal cancer cells. FEBS J. 2009;276:6689–6700. doi: 10.1111/j.1742-4658.2009.07383.x. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J., Sun Q., Zhang Z., Ge S., Han Z.G., Chen W.T. Loss of microRNA-143/145 disturbs cellular growth and apoptosis of human epithelial cancers by impairing the MDM2-p53 feedback loop. Oncogene. 2013;32:61–69. doi: 10.1038/onc.2012.28. [DOI] [PubMed] [Google Scholar]
- 42.Osaki M., Takeshita F., Sugimoto Y., Kosaka N., Yamamoto Y., Yoshioka Y., Kobayashi E., Yamada T., Kawai A., Inoue T. MicroRNA-143 regulates human osteosarcoma metastasis by regulating matrix metalloprotease-13 expression. Mol. Ther. 2011;19:1123–1130. doi: 10.1038/mt.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villadsen S.B., Bramsen J.B., Ostenfeld M.S., Wiklund E.D., Fristrup N., Gao S., Hansen T.B., Jensen T.I., Borre M., Ørntoft T.F. The miR-143/-145 cluster regulates plasminogen activator inhibitor-1 in bladder cancer. Br. J. Cancer. 2012;106:366–374. doi: 10.1038/bjc.2011.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu D., Huang P., Wang L., Zhou Y., Pan H., Qu P. MicroRNA-143 inhibits cell migration and invasion by targeting matrix metalloproteinase 13 in prostate cancer. Mol. Med. Rep. 2013;8:626–630. doi: 10.3892/mmr.2013.1501. [DOI] [PubMed] [Google Scholar]
- 45.Liu R., Liao J., Yang M., Sheng J., Yang H., Wang Y., Pan E., Guo W., Pu Y., Kim S.J., Yin L. The cluster of miR-143 and miR-145 affects the risk for esophageal squamous cell carcinoma through co-regulating fascin homolog 1. PLoS ONE. 2012;7:e33987. doi: 10.1371/journal.pone.0033987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song T., Zhang X., Wang C., Wu Y., Dong J., Gao J., Cai W., Hong B. Expression of miR-143 reduces growth and migration of human bladder carcinoma cells by targeting cyclooxygenase-2. Asian Pac. J. Cancer Prev. 2011;12:929–933. [PubMed] [Google Scholar]
- 47.Li J., Wan X., Qiang W., Li T., Huang W., Huang S., Wu D., Li Y. MiR-29a suppresses prostate cell proliferation and induces apoptosis via KDM5B protein regulation. Int. J. Clin. Exp. Med. 2015;8:5329–5339. [PMC free article] [PubMed] [Google Scholar]
- 48.Yan X., Chen X., Liang H., Deng T., Chen W., Zhang S., Liu M., Gao X., Liu Y., Zhao C. miR-143 and miR-145 synergistically regulate ERBB3 to suppress cell proliferation and invasion in breast cancer. Mol. Cancer. 2014;13:220. doi: 10.1186/1476-4598-13-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhuang M., Shi Q., Zhang X., Ding Y., Shan L., Shan X., Qian J., Zhou X., Huang Z., Zhu W. Involvement of miR-143 in cisplatin resistance of gastric cancer cells via targeting IGF1R and BCL2. Tumour Biol. 2015;36:2737–2745. doi: 10.1007/s13277-014-2898-5. [DOI] [PubMed] [Google Scholar]
- 50.PLOS ONE Editors Retraction: MicroRNA-143 targets Syndecan-1 to repress cell growth in melanoma. PLoS ONE. 2018;13:e0205767. doi: 10.1371/journal.pone.0205767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X., Hu G., Zhou J. Repression of versican expression by microRNA-143. J. Biol. Chem. 2010;285:23241–23250. doi: 10.1074/jbc.M109.084673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Balsara R.D., Ploplis V.A. Plasminogen activator inhibitor-1: the double-edged sword in apoptosis. Thromb. Haemost. 2008;100:1029–1036. [PMC free article] [PubMed] [Google Scholar]
- 53.Durand M.K., Bødker J.S., Christensen A., Dupont D.M., Hansen M., Jensen J.K., Kjelgaard S., Mathiasen L., Pedersen K.E., Skeldal S. Plasminogen activator inhibitor-I and tumour growth, invasion, and metastasis. Thromb. Haemost. 2004;91:438–449. doi: 10.1160/TH03-12-0784. [DOI] [PubMed] [Google Scholar]
- 54.Leeman M.F., Curran S., Murray G.I. The structure, regulation, and function of human matrix metalloproteinase-13. Crit. Rev. Biochem. Mol. Biol. 2002;37:149–166. doi: 10.1080/10409230290771483. [DOI] [PubMed] [Google Scholar]
- 55.Ricciardelli C., Sakko A.J., Ween M.P., Russell D.L., Horsfall D.J. The biological role and regulation of versican levels in cancer. Cancer Metastasis Rev. 2009;28:233–245. doi: 10.1007/s10555-009-9182-y. [DOI] [PubMed] [Google Scholar]
- 56.Szatmári T., Ötvös R., Hjerpe A., Dobra K. Syndecan-1 in Cancer: Implications for Cell Signaling, Differentiation, and Prognostication. Dis. Markers. 2015;2015:796052. doi: 10.1155/2015/796052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dass K., Ahmad A., Azmi A.S., Sarkar S.H., Sarkar F.H. Evolving role of uPA/uPAR system in human cancers. Cancer Treat. Rev. 2008;34:122–136. doi: 10.1016/j.ctrv.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 58.Nalla A.K., Gorantla B., Gondi C.S., Lakka S.S., Rao J.S. Targeting MMP-9, uPAR, and cathepsin B inhibits invasion, migration and activates apoptosis in prostate cancer cells. Cancer Gene Ther. 2010;17:599–613. doi: 10.1038/cgt.2010.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pulukuri S.M., Gondi C.S., Lakka S.S., Jutla A., Estes N., Gujrati M., Rao J.S. RNA interference-directed knockdown of urokinase plasminogen activator and urokinase plasminogen activator receptor inhibits prostate cancer cell invasion, survival, and tumorigenicity in vivo. J. Biol. Chem. 2005;280:36529–36540. doi: 10.1074/jbc.M503111200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Akao Y., Nakagawa Y., Hirata I., Iio A., Itoh T., Kojima K., Nakashima R., Kitade Y., Naoe T. Role of anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer Gene Ther. 2010;17:398–408. doi: 10.1038/cgt.2009.88. [DOI] [PubMed] [Google Scholar]
- 61.Pramanik D., Campbell N.R., Karikari C., Chivukula R., Kent O.A., Mendell J.T., Maitra A. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol. Cancer Ther. 2011;10:1470–1480. doi: 10.1158/1535-7163.MCT-11-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niu X.B., Fu G.B., Wang L., Ge X., Liu W.T., Wen Y.Y., Sun H.R., Liu L.Z., Wang Z.J., Jiang B.H. Insulin-like growth factor-I induces chemoresistence to docetaxel by inhibiting miR-143 in human prostate cancer. Oncotarget. 2017;8:107157–107166. doi: 10.18632/oncotarget.22362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ibrahim A.F., Weirauch U., Thomas M., Grünweller A., Hartmann R.K., Aigner A. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res. 2011;71:5214–5224. doi: 10.1158/0008-5472.CAN-10-4645. [DOI] [PubMed] [Google Scholar]
- 64.Aigner A., Fischer D. Nanoparticle-mediated delivery of small RNA molecules in tumor therapy. Pharmazie. 2016;71:27–34. [PubMed] [Google Scholar]
- 65.Höbel S., Aigner A. Polyethylenimines for siRNA and miRNA delivery in vivo. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2013;5:484–501. doi: 10.1002/wnan.1228. [DOI] [PubMed] [Google Scholar]
- 66.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 67.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 68.Fuessel S., Erdmann K., Taubert H., Lohse-Fischer A., Zastrow S., Meinhardt M., Bluemke K., Hofbauer L., Fornara P., Wullich B. Prognostic impact of urokinase-type plasminogen activator system components in clear cell renal cell carcinoma patients without distant metastasis. BMC Cancer. 2014;14:974. doi: 10.1186/1471-2407-14-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Höbel S., Koburger I., John M., Czubayko F., Hadwiger P., Vornlocher H.P., Aigner A. Polyethylenimine/small interfering RNA-mediated knockdown of vascular endothelial growth factor in vivo exerts anti-tumor effects synergistically with Bevacizumab. J. Gene Med. 2010;12:287–300. doi: 10.1002/jgm.1431. [DOI] [PubMed] [Google Scholar]
- 70.Werth S., Urban-Klein B., Dai L., Höbel S., Grzelinski M., Bakowsky U., Czubayko F., Aigner A. A low molecular weight fraction of polyethylenimine (PEI) displays increased transfection efficiency of DNA and siRNA in fresh or lyophilized complexes. J. Control. Release. 2006;112:257–270. doi: 10.1016/j.jconrel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 71.Höbel S., Prinz R., Malek A., Urban-Klein B., Sitterberg J., Bakowsky U., Czubayko F., Aigner A. Polyethylenimine PEI F25-LMW allows the long-term storage of frozen complexes as fully active reagents in siRNA-mediated gene targeting and DNA delivery. Eur. J. Pharm. Biopharm. 2008;70:29–41. doi: 10.1016/j.ejpb.2008.03.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.