Abstract
Long noncoding RNAs (lncRNAs) have been reported to serve as diagnostic and prognostic biomarkers of cancers, which play vital roles in tumorigenesis and tumor progression. Several studies have been performed to explore diagnostic value of lncRNA H19 in cancer detection and diagnosis. However, there are still inconsistent results in diagnostic accuracy and reliability in individual studies. Therefore, the present study was performed to summarize the overall diagnostic performance of lncRNA H19 in cancer detection and diagnosis. A total of eight studies with 770 cases and 815 controls were included in this pooled analysis. The pooled diagnostic results were as follows: sensitivity, 0.69 (95%CI=0.62-0.76), specificity, 0.79 (95% CI=0.70-0.86), positive likelihood ratio (PLR), 3.31 (95%CI=2.29-4.78), negative likelihood (NLR), 0.39 (95%CI=0.31-0.49), diagnostic odds ratio (DOR), 8.53 (95%CI=4.99-14.60), and area under the curve (AUC), 0.79 (95%CI=0.76-0.83). Deeks' funnel plot asymmetry test (P=0.13) suggested no potential publication bias. Our results indicated that lncRNA H19 had a relatively moderate accuracy in cancer detection and diagnosis. Further comprehensive prospective studies with large sample sizes are urgently required to validate our findings.
1. Introduction
With incremental incidence and mortality in recent years, cancer has been a major public health problem all over the world [1, 2]. Although tremendous improvements have been made in therapeutic method including surgery, radiotherapy, chemotherapy, and precision therapy over past decades, the prognosis and quality of life of cancer patients remain poor, particularly in patients with advanced staged or metastatic cancers [3–5]. The lack of early diagnostic techniques contributes to the current situation [2]. Therefore, finding a potential diagnostic biomarker with good specificity and sensitivity for early cancer detection and diagnosis seems urgently needed.
Long noncoding RNAs (LncRNAs) are a subclass of regulatory ncRNAs longer than 200 nucleotides, lacking functional open reading frames (ORFs) and protein-coding capability [6, 7]. LncRNAs are widely reported to regulate gene expression at epigenetic, transcriptional, and posttranscriptional levels [8–10] and aberrant expression of lncRNAs can be involved in cancer initiation, progression, and metastasis [11–13]. Furthermore, increasing evidences suggested that lncRNAs could serve as potential biomarkers with high sensitivity and specificity in cancer detection and diagnosis [14–17].
H19, a subclass of long noncoding RNA, is a paternally imprinted gene which locates in chromosome 11p15.5 [18]. In recent years, lncRNA H19 was identified to be significantly associated with various human cancers including breast cancer, gastric cancer, thyroid cancer, and hepatic carcinoma [19–26]. Several studies indicated that lncRNA H19 could function as an oncogene in tumorigenesis and tumor progression [27–29]. Quite a few studies had explored the clinical use of lncRNA H19 in cancer detection and diagnosis. However, the diagnostic accuracy of lncRNA H19 in the individual studies is still inconsistent and controversial. For example, Zhou et al. [23] showed that lncRNA H19 can be used for diagnosis of gastric cancer with a moderate-high sensitivity and specificity of 82.9% and 72.9%, respectively, but Hashad et al. [20] revealed a low sensitivity and specificity of 68.75% and 56.67%, respectively, in gastric cancer detection. These results failed to reach the agreement due to the difference of ethnicity, study design, types of tumors, stage of cancer, and the small sample size, which made it difficult to interpret. Thus, this pooled analysis was conducted to summarize the overall diagnostic performance of lncRNA H19 in cancer detection and diagnosis and further explored its clinical value.
2. Methods
2.1. Search Strategy and Study Selection Criteria
Literature research was performed in database including PubMed, Web of Science, Wanfang library, and CNKI up to May 18, 2018, by the following searching strategy: “cancer” or “tumor” or “carcinoma” or “neoplasm” or “malignancy” or “neoplasm” and “H19” and “sensitivity” or “specificity” or “ROC curve” or “accuracy”. Three investigators (HAB, LBE, and LYH) checked the titles and abstracts of the studies and scanned the full texts to eliminate irrelevant studies with the following included criteria: (1) the diagnostic value of lncRNA H19 for detecting cancer evaluated in articles, (2) explicitly defined article population and control sources; (3) completed data for calculating sensitivity and specificity; and (4) being published in English or Chinese.
2.2. Data Extraction and Quality Assessment
For each study, the following information was extracted: first author, year of publication, country, ethnicity, sample size, specimen and cancer type, detection method, cutoff value, true positive (TP), false positive (FP), true negative (TN), and false negative (FN).The QUADAS-2 was applied to systematically evaluate the quality of the studies included in this pooled analysis. With the max QUADAS-2 score of 7, we can judge the quality of the included studies based on the results.
2.3. Statistical Analysis
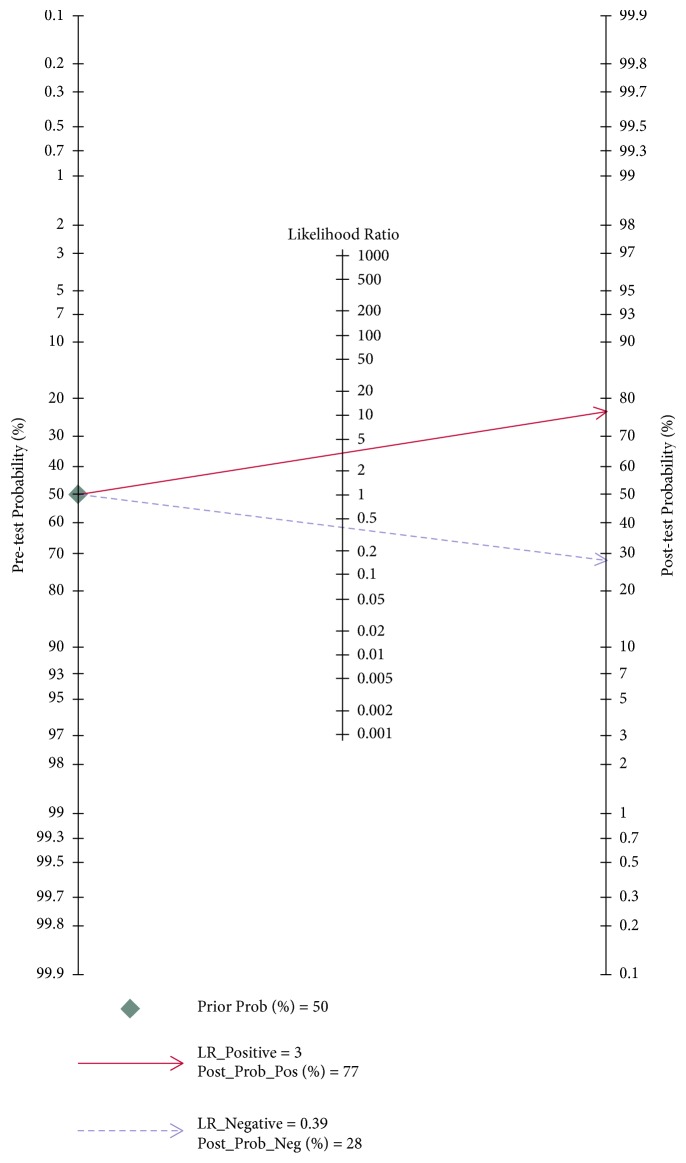
All statistical analyses were performed using Stata 14.0 (Stata, College Station, TX, USA). The pooled sensitivity, specificity, diagnostic odds ratio (DOR), positive likelihood ratio (PLR) and negative likelihood ratio (NLR) and other parameters were calculated by the bivariate meta-analysis model. Then, we performed summary receiver operator characteristic (SROC) curves analysis and calculated the area under the ROC curves (AUC) to assess the overall diagnostic value of lncRNA H19 in cancer detection and diagnosis [30]. These data were confirmed by a hierarchical summary receiver operating characteristics (HSROC) model. Spearman correlation coefficients were conducted to evaluate heterogeneity of threshold effect. Heterogeneity of nonthreshold effects was assessed by Cochran-Q and Inconsistency index (I2) test [31]. A P value less than 0.10 for the Q test or I2 value higher than 50% indicated obvious heterogeneity between the studies [32]. Moreover, Fagan's Nomogram was used to certify relationships between prior-test probability, likelihood ratio, and posttest probability. The publication bias was tested by Deeks' funnel plots [33].
3. Results
3.1. Studies Selection and Characteristics of Included Studies
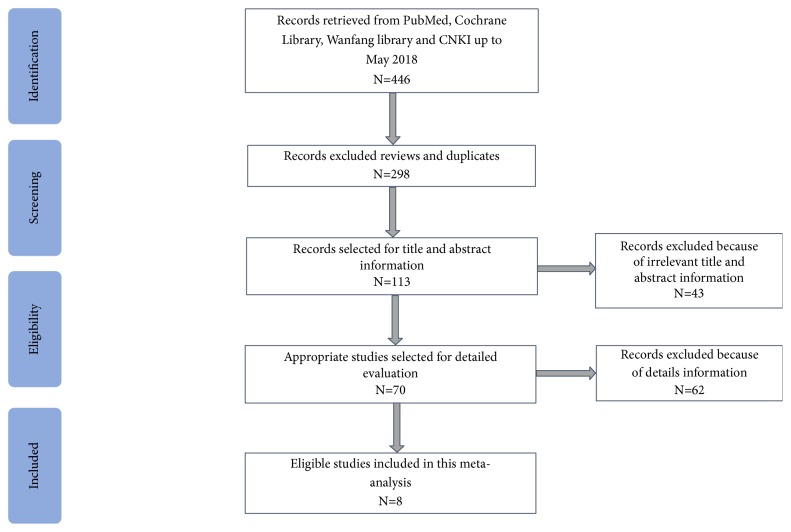
By searching PubMed, Web of Science, Wanfang, and CNKI databases, a total of 8 eligible studies [19–26] including 770 cases and 815 controls from 116 records published from 2013 to 2018 were according to inclusion and exclusion criteria (Figure 1). The main features of included articles were displayed in Table 1. In total, there were studies on breast cancer (n=3), gastric cancer (n=3), hepatic carcinoma (n=1), and thyroid cancer (n=1). Among the 8 studies tested lncRNA H19 expression using qRT-PCR methods was based on plasma (n=4), tissue (n=2), serum (n=1), and urinary (n=1).
Figure 1.
The flow diagram of the included and excluded studies.
Table 1.
Characteristics of the included studies.
| First author | Year | Country | Ethnicity | Cancer type | Normalizer | Sample type | Test method | Cutoff | Cases/ controls | TP | FP | FN | TN | QUADAS-2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Zhang | 2016 | China | Asian | BRC | β-actin | Plasma | qRT-PCR | 1.085 | 97/116 | 56 | 16 | 41 | 100 | 5 |
| Zhang | 2016 | China | Asian | BRC | β-actin | Urinary | qRT-PCR | NA | 30/42 | 22 | 14 | 8 | 28 | 4 |
| Sun | 2016 | China | Asian | HCC | GAPDH | Serum | qRT-PCR | 0. 073 | 180/211 | 140 | 16 | 40 | 195 | 4 |
| Chen | 2016 | China | Asian | GC | GAPDH | tissue | qRT-PCR | 4.615 | 128/128 | 79 | 33 | 49 | 95 | 5 |
| Hashad | 2016 | Egypt | African | GC | GAPDH | Plasma | qRT-PCR | 0.5 | 32/30 | 22 | 13 | 10 | 17 | 6 |
| Zhang | 2016 | China | Asian | BRC | β-actin | Plasma | qRT-PCR | NA | 102/96 | 58 | 13 | 44 | 83 | 4 |
| Zhou | 2015 | China | Asian | GC | GAPDH | Plasma | qRT-PCR | NA | 70/70 | 58 | 19 | 12 | 51 | 5 |
| Liu | 2017 | China | Asian | TC | GAPDH | Tissue | qRT-PCR | 3.58 | 131/122 | 95 | 30 | 36 | 92 | 5 |
BRC: breast cancer; HCC: hepatic carcinoma; GC: gastric cancer; TC: thyroid cancer; NA.: not available; TP: true positive; FP: false positive; FN: false negative; TN: true negative.
3.2. Quality Assessment
The results of the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) study quality assessment were also shown in Table 1. All of the QUADAS-2 scores for studies on diagnosis were ≥4, indicating a moderate-high quality for most of the studies.
3.3. Data Analysis
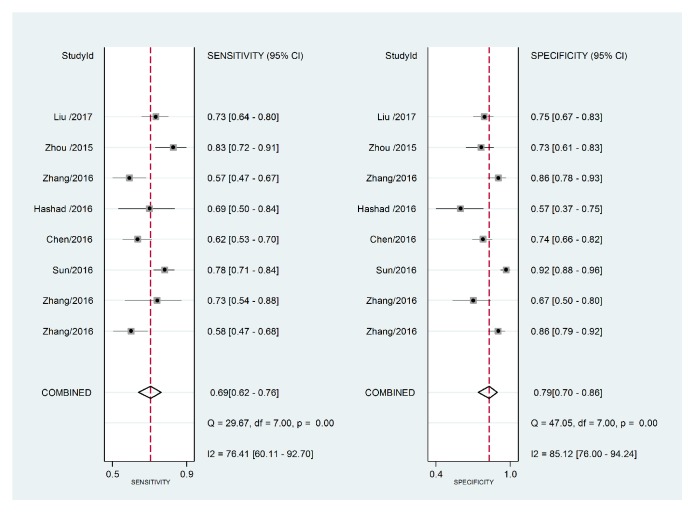
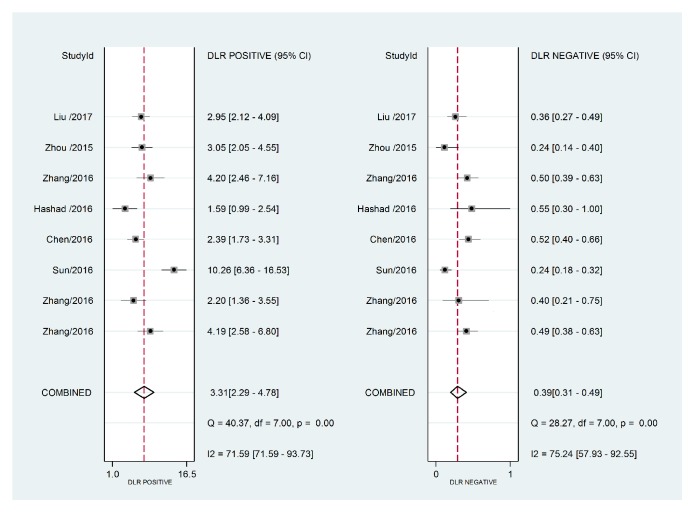
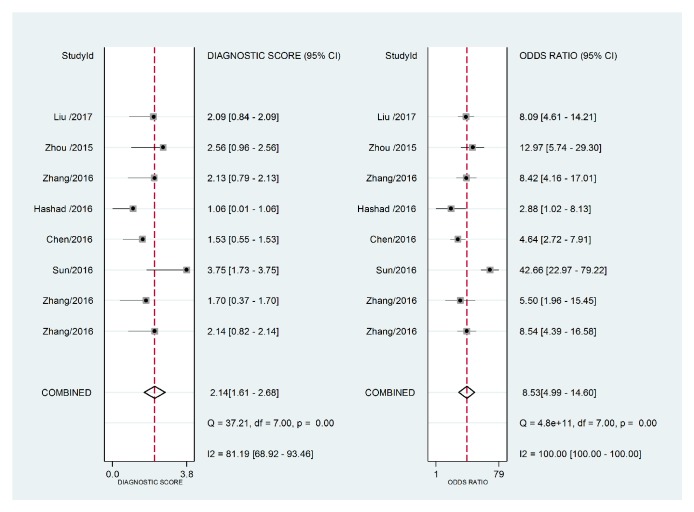
The forest plot of data from included articles on sensitivity and specificity for H19 assay in diagnosing cancer was shown in Figure 2. Overall, the sensitivity and specificity for the pooled data were 0.69 (95%CI=0.62-0.76) and 0.79 (95%CI=0.70-0.86), respectively. Significant heterogeneity was found for both sensitivity (I2=76.41%, 95%CI=60.11%-92.70%) and specificity (I2=85.12%, 95%CI=76.00%-94.24%). In addition, the pooled PLR was 3.31 (95%CI=2.29-4.78), the NLR was 0.39 (95%CI=0.31-0.49), and the DOR was 8.53 (95%CI=4.99-14.60) (Figures 3 and 4). The SROC curve for the 8 included studies is shown in Figure 5. The AUC of H19 was 0.79 (95%CI=0.76-0.83), implying a relatively moderate diagnostic value.
Figure 2.
Forest plots of pooled sensitivity and specificity of the overall 8 included publications.
Figure 3.
Forest plots of positive likelihood ratio (PLR) and negative likelihood ratio (NLR) for lncRNA H19 in the diagnosis of cancer.
Figure 4.
Forest plots of pooled diagnostic odds ratio (DOR) for lncRNA H19 in the diagnosis of cancer.
Figure 5.
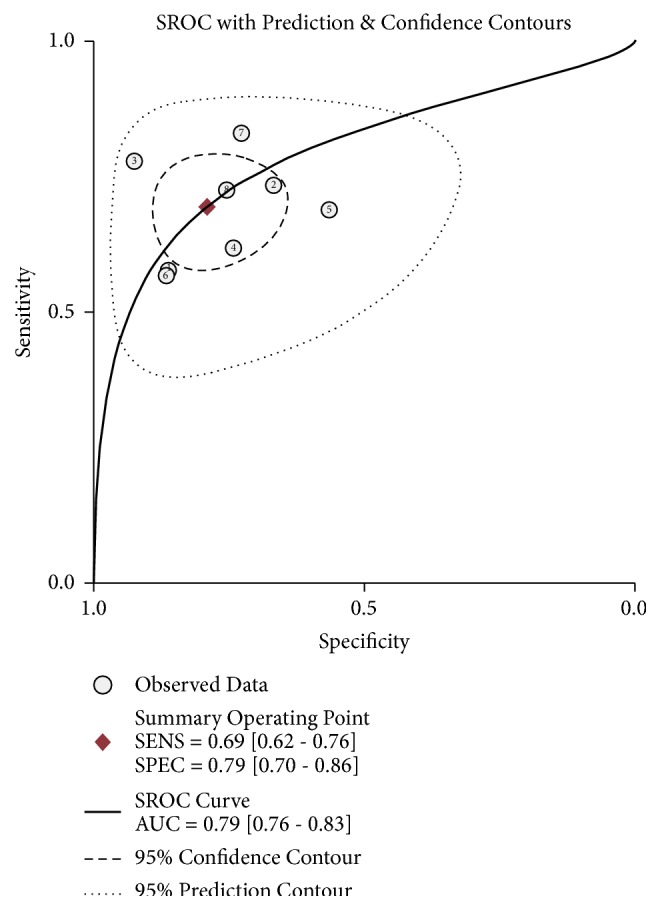
Summary receiver operating characteristic (SROC) graph of included studies.
The HSROC curve of these included studies was in line with the results from the bivariate model. The value of β was 0.47 (95%CI=0.44-1.39), and the P value was 0.314 which indicated that the HSROC was symmetrical. The value of γ was 2.08 (95%CI=1.56-2.60) (Figure 6). To evaluate the clinical utility of the index test, a Fagan's Nomogram was performed to predict the increasing inerrability about a positive diagnosis by using the value of the test and it is used for estimating posttest probabilities. As shown in Figure 7, when H19 assays were tested for all individuals with a pretest probability of 50% to have cancer, a positive result would improve posttest probability having cancer to 77%, while a negative result would drop the posttest probability to 28%. All of the results indicated that H19 had a relatively moderate accuracy in distinguishing cancer patients from all individuals.
Figure 6.
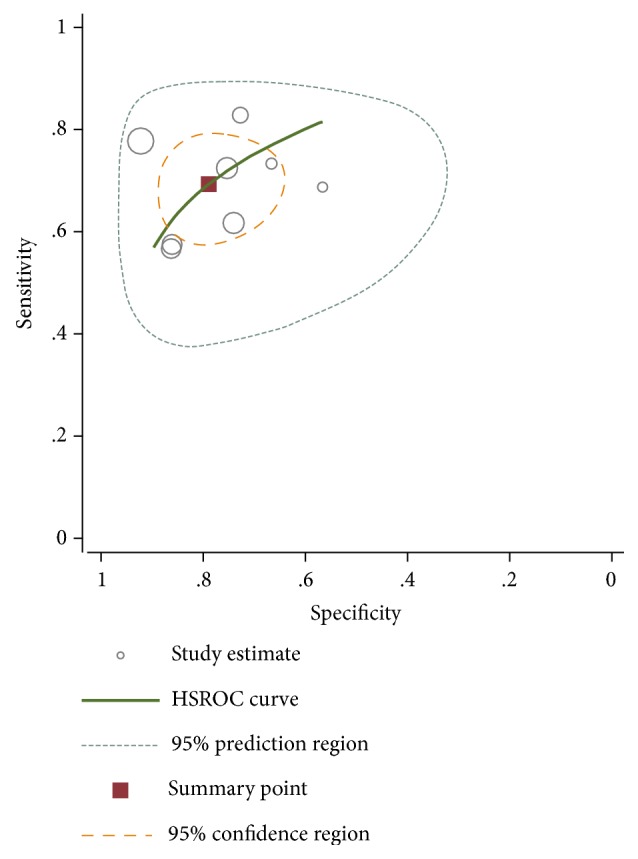
Hierarchical summary receiver operating characteristics (HSROC) curve for lncRNA H19 in the diagnosis of cancer.
Figure 7.
Fagan's Nomogram for calculation of posttest probabilities.
3.4. Influence Analysis and Robustness Tests
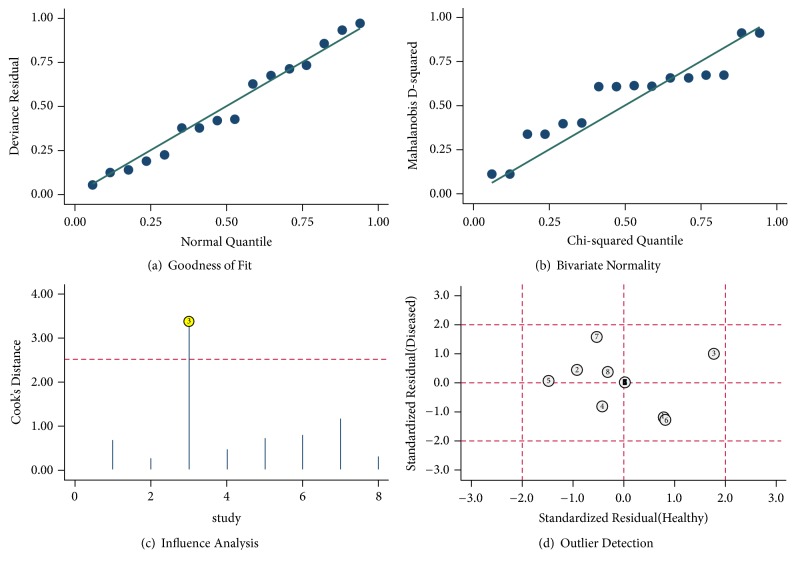
God-of-fit and bivariate normality analyses (Figures 8(a) and 8(b)) showed that the bivariate model was moderately robust. We also performed sensitivity analyses and further excluded 1 outliner found by influence analysis and outlier detection in Figures 8(c) and 8(d). After exclusion, the sensitivity dropped from 0.69 to 0.68, specificity dropped from 0.79 to 0.76, the PLR dropped from 3.3 to 2.9, the NLR increased from 0.39 to 0.42, DOR dropped from 9 to 7, and AUC decreased from 0.79 to 0.78, showing no significant change after excluding the outliner. Finally, Deeks' funnel plot asymmetry test was conducted to evaluate publication bias in this pooled analysis (Figure 9), which suggested no significant publication bias (P=0.13). The above tests confirm the robustness of our results in present meta-analysis.
Figure 8.
Graphs for sensitivity analyses: (a) goodness of fit, (b) bivariate normality, (c) influence analysis, and (d) outlier detection.
Figure 9.
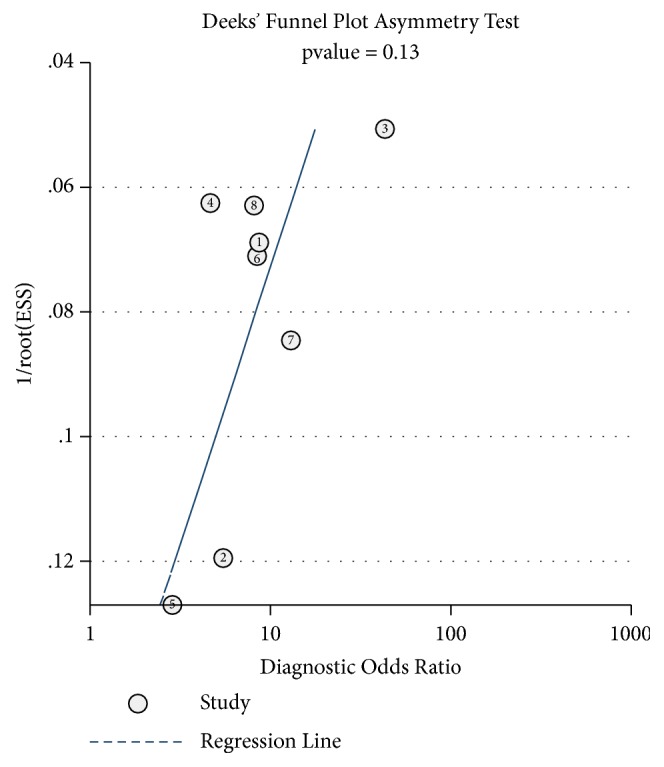
Graph of Deeks' funnel plot asymmetry test.
3.5. Threshold Effect and Heterogeneity
The I2 of the heterogeneity test was 89%, indicating significant heterogeneity. In the present study, the calculated Spearman correlation coefficient value was −0.11 with p=0.01 (P<0.05), suggesting that the threshold effect was the major source of heterogeneity. However, there were only eight articles included; metaregression analysis and subgroup analysis cannot be used.
4. Discussion
Through the next generation sequencing technology and large-scale transcriptome mapping, many lncRNAs have been reported to involve in the development of cancer as a regulator in a variety of biological processes. These lncRNAs, located in the nucleus, interact with chromatin remodeling complexes (CRCs) to regulate the genes expression locating on the same chromosome in cis or on another chromosome in trans through fine-tuning of chromatin architecture [34, 35]. Previous studies have demonstrated that lncRNAs were associated with tumor proliferation, invasion, replicative senescence, resistance to drugs and radiation by interaction with proteins, RNA, or DNA [36, 37]. Moreover, lncRNAs could serve as diagnosis and prognosis biomarkers in human cancers due to the fact that lncRNAs can be conveniently collected from body fluid, such as plasma and urine [38].
Much effort has been made to investigate the link between aberrant lncRNA expression and cancer, including lncRNA H19 [27–29, 39–41]. Emerging studies have reported that lncRNA H19 was upregulated in various cancers, such as nonsmall cell lung cancer, bladder cancer, breast cancer, and gastric cancer [42–45]. Several studies have been done to explore diagnostic value of lncRNA H19 in cancer detection and diagnosis. However, there are still inconsistent results in diagnostic accuracy and reliability in individual studies. Therefore, we performed this pooled analysis to evaluate the diagnostic value of H19 in cancer detection. The pooled results in the present study were sensitivity of 0.69 (95%CI=0.62-0.76), specificity of 0.79 (95%CI=0.70-0.86), and the AUC of0.79 (95%CI=0.76-0.83), suggesting that H19 may be a potential biomarker to discriminate cancer patients from normal people. In our study, the pooled DOR of 8.53 (95%CI=4.99-14.60) reflects a moderate level of diagnostic accuracy. Additionally, the likelihood ratio (LR) combines the stability of sensitivity and specificity to provide an omnibus index of test performance [46]. In present meta-analysis, a pooled PLR of 3.31 (95%CI=2.29-4.78) and NLR 0.39 (95%CI =0.31-0.49) suggested that patients with cancer have a 3.31-fold higher possibility of being H19 positive for patients with cancer compared with controls, and 39% of all individuals have negative results, implying that the diagnostic value of H19 is relatively moderate. From the Fagan's Nomogram, we found that when a pretest probability of 50% was specified, the posttest probability positivity would raise to 77% with a positive likelihood ratio of 3, and the posttest probability negativity would decrease to 28% with a negative likelihood ratio of 0.39. All of the results revealed that lncRNA H19 had a relatively moderate diagnostic accuracy in cancer detection and diagnosis.
Heterogeneity is an inescapable problem that can interpret the results of the meta-analysis [47]. There was still potential heterogeneity in our present study because of the existence of other confounding factors. In this study, Spearman rank correlation test was performed to analyze the threshold effect, and the Spearman correlation coefficient was -0.11 with p=0.01 (P<0.05), which indicated that threshold effect was a prime source of heterogeneity. In addition, subgroup analysis and metaregression analysis cannot be used because of the insufficient eligible articles. Thus, the possible reasons such as test method and ethnicity were not investigated as sources of heterogeneity.
Nevertheless, several defects of this pooled analysis should be emphasized. First, eight studies with a limited number of subjects were included in this study, which may weaken the reliability for determining the diagnostic value of H19 for different types of cancers. Second, our articles have a very high ratio of data in Chinese populations, which may result in inevitable publication bias. Third, research and sample size in single tumor type was relatively small; more cancer types studies with large sample size need to be included in analysis. Fourth, not all of the studies reported the cutoff values of lncRNA H19. Finally, only publications in English or Chinese were included; researches in other languages should not be missed.
In summary, all of the results indicated that H19 had a relatively moderate accuracy in distinguishing cancer patients from all individuals, suggesting that H19 could serve as a potential diagnostic biomarker for cancer detection and diagnosis. Furthermore, well-designed prospective studies with large sample sizes and different population groups must be conducted in the future to confirm our findings.
Acknowledgments
This research was supported by Health Care 3F Project of Shenzhen (Peking University First Hospital-The Second People's Hospital of Shenzhen, Academician YingluGuo's Team), the Shenzhen Key Medical Discipline Fund, Special Support Funds of Shenzhen for Introduced High-Level Medical Team, Shenzhen Foundation of Science and Technology (JCYJ20150330102720182), and Shenzhen Health and Family Planning Commission Scientific Research Project [201601025, 201606019].
Conflicts of Interest
All authors declare that there are no conflicts of interest.
Authors' Contributions
Yuhan Liu, Anbang He, Baoer Liu, and Zhengxian Huang are equal contributors.
References
- 1.Siegel R. L., Miller K. D., Jemal A. Cancer statistics. CA: A Cancer Journal for Clinicians. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2017. CA: A Cancer Journal for Clinicians. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Yang K., Park W., Jae Huh S., Bae D.-S., Kim B.-G., Lee J.-W. Clinical outcomes in patients treated with radiotherapy after surgery for cervical cancer. Radiation Oncology Journal. 2017;35(1):39–47. doi: 10.3857/roj.2016.01893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton M. B., Frommer M., Shafiq J. Role of radiotherapy in cancer control in low-income and middle-income countries. The Lancet Oncology. 2006;7(7):584–595. doi: 10.1016/S1470-2045(06)70759-8. [DOI] [PubMed] [Google Scholar]
- 5.Cree I. A., Kurbacher C. M., Lamont A., et al. A prospective randomized controlled trial of tumour chemosensitivity assay directed chemotherapy versus physician's choice in patients with recurrent platinum-resistant ovarian cancer. Anti-Cancer Drugs. 2007;18(9):1093–1101. doi: 10.1097/CAD.0b013e3281de727e. [DOI] [PubMed] [Google Scholar]
- 6.Ruan X., Li P., Cangelosi A., Yang L., Cao H. A Long Non-coding RNA, lncLGR, Regulates Hepatic Glucokinase Expression and Glycogen Storage during Fasting. Cell Reports. 2016;14(8):1867–1875. doi: 10.1016/j.celrep.2016.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Heesch S., Van Iterson M., Jacobi J., et al. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biology. 2014;15(1):p. R6. doi: 10.1186/gb-2014-15-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong C., Li Z., Ramanujan K., et al. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Developmental Cell. 2015;34(2):181–191. doi: 10.1016/j.devcel.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Chalei V., Sansom S. N., Kong L., et al. The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. eLife. 2014;3 doi: 10.7554/elife.04530.e04530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin W., Li X., Xie L., et al. A long non-coding RNA, APOA4-AS, regulates APOA4 expression depending on HuR in mice. Nucleic Acids Research. 2016;44(13):6423–6433. doi: 10.1093/nar/gkw341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kugel J. F., Goodrich J. A. Non-coding RNAs: Key regulators of mammalian transcription. Trends in Biochemical Sciences. 2012;37(4):144–151. doi: 10.1016/j.tibs.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ponting C. P., Oliver P. L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Batista P. J., Chang H. Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X., Chen X., Zhang D., et al. Prognostic and clinicopathological role of long non-coding RNA taurine upregulated 1 in various human malignancies: A systemic review and meta-analysis. Tumour biology : the journal of the international society for oncodevelopmental biology and medicine. 2017;39(7) doi: 10.1177/1010428317714361. [DOI] [PubMed] [Google Scholar]
- 15.Wang M., Dong X., Feng Y., Sun H., Shan N., Lu T. Prognostic role of the long non-coding RNA, SPRY4 Intronic Transcript 1, in patients with cancer: A meta-analysis. Oncotarget. 2017;8(20):33713–33724. doi: 10.18632/oncotarget.16735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X., Peng F., Cheng L., et al. Prognostic and clinicopathological role of long non-coding RNA UCA1 in various carcinomas. Oncotarget. 2017;8(17):28373–28384. doi: 10.18632/oncotarget.16059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian G.-W., Li N., Xin Y. Prognostic and clinicopathological significance of CCAT2 in Chinese patients with various tumors. The International Journal of Biological Markers. 2017;32(3):e344–e351. doi: 10.5301/ijbm.5000281. [DOI] [PubMed] [Google Scholar]
- 18.Gabory A., Jammes H., Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. BioEssays. 2010;32(6):473–480. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- 19.Chen J. S., Wang Y. F., Zhang X. Q., et al. H19 serves as a diagnostic biomarker and up-regulation of H19 expression contributes to poor prognosis in patients with gastric cancer. Neoplasma. 2016;63(2):223–230. doi: 10.4149/207_150821N454. [DOI] [PubMed] [Google Scholar]
- 20.Hashad D., Elbanna A., Ibrahim A., Khedr G. Evaluation of the Role of Circulating Long Non-Coding RNA H19 as a Promising Novel Biomarker in Plasma of Patients with Gastric Cancer. Journal of Clinical Laboratory Analysis. 2016;30(6):1100–1105. doi: 10.1002/jcla.21987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu N., Zhou Q., Qi Y.-H., Wang H., Yang L., Fan Q.-Y. Effects of long non-coding RNA H19 and microRNA let7a expression on thyroid cancer prognosis. Experimental and Molecular Pathology. 2017;103(1):71–77. doi: 10.1016/j.yexmp.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Zhang K., Luo Z., Zhang Y., et al. Circulating lncRNA H19 in plasma as a novel biomarker for breast cancer. Cancer Biomarkers. 2016;17(2):187–194. doi: 10.3233/CBM-160630. [DOI] [PubMed] [Google Scholar]
- 23.Zhou X., Yin C., Dang Y., Ye F., Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Scientific Reports. 2015;5 doi: 10.1038/srep11516.11516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang K., Zhang Y., Luo Z., Wu L., Zhang L., Liu J. Diagnostic value of urinary lncRNA H19 for breast cancer. Shandong Medical Journal. 2016;56(18):42–44. [Google Scholar]
- 25.Zhang Y., Luo Z., Zhang K., Wu L., Zhang L., Liu J. Potential value of plasma long non-coding RNA H19 in the diagnosis of breast cancer. Chinese Journal of Clinical Laboratoy Science. 2016;34(4):264–267. [Google Scholar]
- 26.Sun Y., Zhao F., Wang F., Wang W. Diagnostic value of long non coding RNA H19 in primary hepatic carcinoma. Chinses Journal of Health Laboratory Technology. 2016;26(3):305–310. [Google Scholar]
- 27.Luo M., Li Z., Wang W., Zeng Y., Liu Z., Qiu J. Upregulated H19 contributes to bladder cancer cell proliferation by regulating ID2 expression. FEBS Journal. 2013;280(7):1709–1716. doi: 10.1111/febs.12185. [DOI] [PubMed] [Google Scholar]
- 28.Matouk I. J., Raveh E., Abu-lail R., et al. Oncofetal H19 RNA promotes tumor metastasis. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2014;1843(7):1414–1426. doi: 10.1016/j.bbamcr.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 29.Shi Y., Wang Y., Luan W., et al. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS ONE. 2014;9(1) doi: 10.1371/journal.pone.0086295.e86295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moses L. E., Shapiro D., Littenberg B. Combining independent studies of a diagnostic test into a summary ROC curve: data-analytic approaches and some additional considerations. Statistics in Medicine. 1993;12(14):1293–1316. doi: 10.1002/sim.4780121403. [DOI] [PubMed] [Google Scholar]
- 31.Vamvakas E. C. Meta-analyses of studies of the diagnostic accuracy of laboratory tests: a review of the concepts and methods. Archives of Pathology & Laboratory Medicine. 1998;122(8):675–686. [PubMed] [Google Scholar]
- 32.Rutter C. M., Gatsonis C. A. A hierarchical regression approach to meta-analysis of diagnostic test accuracy evaluations. Statistics in Medicine. 2001;20(19):2865–2884. doi: 10.1002/sim.942. [DOI] [PubMed] [Google Scholar]
- 33.Deeks J. J., Macaskill P., Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. Journal of Clinical Epidemiology. 2005;58(9):882–893. doi: 10.1016/j.jclinepi.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 34.Djebali S., Davis C. A., Merkel A., et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Derrien T., Johnson R., Bussotti G., et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Research. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dey B. K., Mueller A. C., Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 2014;5(4) doi: 10.4161/21541272.2014.944014.e944014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li C. H., Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. The International Journal of Biochemistry & Cell Biology. 2013;45(8):1895–1910. doi: 10.1016/j.biocel.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 38.Kunej T., Obsteter J., Pogacar Z., Horvat S., Calin G. A. The decalog of long non-coding RNA involvement in cancer diagnosis and monitoring. Critical Reviews in Clinical Laboratory Sciences. 2014;51(6):344–357. doi: 10.3109/10408363.2014.944299. [DOI] [PubMed] [Google Scholar]
- 39.Chen N., Guo D., Xu Q., et al. Long non-coding RNA FEZF1-AS1 facilitates cell proliferation and migration in colorectal carcinoma. Oncotarget. 2016;7(10):11271–11283. doi: 10.18632/oncotarget.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo F., Guo L., Li Y., Zhou Q., Li Z. MALAT1 is an oncogenic long non-coding RNA associated with tumor invasion in non-small cell lung cancer regulated by DNA methylation. International Journal of Clinical and Experimental Pathology. 2015;8(12):15903–15910. [PMC free article] [PubMed] [Google Scholar]
- 41.Wu L., Jin L., Zhang W., Zhang L. Roles of long non-coding rna ccat2 in cervical cancer cell growth and apoptosis. Medical Science Monitor. 2016;22:875–879. doi: 10.12659/MSM.897754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cui J., Mo J., Luo M., et al. C-Myc-activated long non-coding RNA H19 downregulates miR-107 and promotes cell cycle progression of non-small cell lung cancer. International Journal of Clinical and Experimental Pathology. 2015;8(10):12400–12409. [PMC free article] [PubMed] [Google Scholar]
- 43.Li S., Yu Z., Chen S.-S., et al. The YAP1 oncogene contributes to bladder cancer cell proliferation and migration by regulating the H19 long noncoding RNA. Urologic Oncology: Seminars and Original Investigations. 2015;33(10):427.e1–427.e10. doi: 10.1016/j.urolonc.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 44.Barsyte-Lovejoy D., Lau S. K., Boutros P. C., et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Research. 2006;66(10):5330–5337. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- 45.Zhuang M., Gao W., Xu J., Wang P., Shu Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochemical and Biophysical Research Communications. 2014;448(3):315–322. doi: 10.1016/j.bbrc.2013.12.126. [DOI] [PubMed] [Google Scholar]
- 46.Gallagher E. J. Clinical utility of likelihood ratios. Annals of Emergency Medicine. 1998;31(3):391–397. doi: 10.1016/S0196-0644(98)70352-X. [DOI] [PubMed] [Google Scholar]
- 47.Lijmer J. G., Bossuyt P. M. M., Heisterkamp S. H. Exploring sources of heterogeneity in systematic reviews of diagnostic tests. Statistics in Medicine. 2002;21(11):1525–1537. doi: 10.1002/sim.1185. [DOI] [PubMed] [Google Scholar]