Abstract
Background & Aims
RET, the receptor for the glial cell line–derived neurotrophic factor (GDNF) family ligands, is the most frequently mutated gene in congenital aganglionic megacolon or Hirschsprung’s disease (HSCR). The leading cause of mortality in HSCR is HSCR-associated enterocolitis (HAEC), which is characterized by altered mucin composition, mucin retention, bacterial adhesion to enterocytes, and epithelial damage, although the order of these events is obscure. In mice, loss of GDNF signaling leads to a severely underdeveloped enteric nervous system and neonatally fatal kidney agenesis, thereby precluding the use of these mice for modeling postnatal HSCR and HAEC. Our aim was to generate a postnatally viable mouse model for HSCR/HAEC and analyze HAEC etiology.
Methods
GDNF family receptor alpha-1 (GFRa1) hypomorphic mice were generated by placing a selectable marker gene in the sixth intron of the Gfra1 locus using gene targeting in mouse embryonic stem cells.
Results
We report that 70%–80% reduction in GDNF co-receptor GFRa1 expression levels in mice results in HSCR and HAEC, leading to death within the first 25 postnatal days. These mice mirror the disease progression and histopathologic findings in children with untreated HSCR/HAEC.
Conclusions
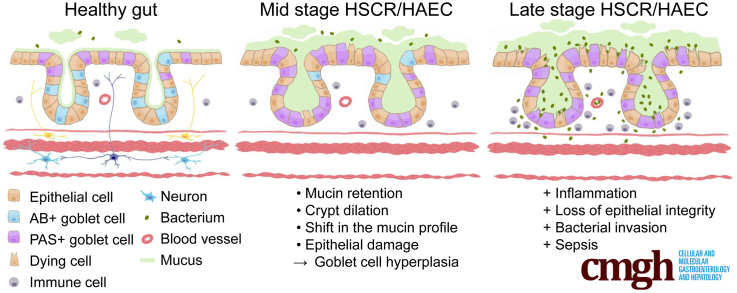
In GFRa1 hypomorphic mice, HAEC proceeds from goblet cell dysplasia, with abnormal mucin production and retention, to epithelial damage. Microbial enterocyte adherence and tissue invasion are late events and therefore unlikely to be the primary cause of HAEC. These results suggest that goblet cells may be a potential target for preventative treatment and that reduced expression of GFRa1 may contribute to HSCR susceptibility.
Keywords: Aganglionosis, Megacolon, Transgenic Mouse Model
Abbreviations used in this paper: AB, Alcian Blue; cDNA, complementary DNA; E, embryonic day; ELISA, enzyme-linked immunosorbent assay; ENCC, enteric neural crest-derived cell; ENS, enteric nervous system; GDNF, glial cell line–derived neurotrophic factor; GFRa1, glial cell line–derived neurotrophic factor family receptor alpha-1; GI, gastrointestinal; HAEC, Hirschsprung’s disease–associated enterocolitis; HPF, high-power field; HSCR, Hirschsprung’s disease; IL, interleukin; KO, knockout; LMMP, longitudinal muscle/myenteric plexus preparation; mRNA, messenger RNA; NADPH, reduced nicotinamide adenine dinucleotide phosphate; P, postnatal day; PAS, periodic acid–Schiff; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; qPCR, quantitative polymerase chain reaction; RET, rearranged during transfection; TBS, Tris-buffered saline; TNF, tumor necrosis factor; WT, wild type
Graphical abstract
Summary.
A 70% to 80% underexpression of glial cell line–derived neurotrophic factor family receptor alpha-1 results in Hirschsprung’s disease and an associated enterocolitis phenotype in a new mouse model. Enterocolitis proceeds from goblet cell dysplasia, with mucin abnormalities, to epithelial damage. Reduced expression of glial cell line–derived neurotrophic factor family receptor a1 may contribute to Hirschsprung disease susceptibility.
Hirschsprung’s disease (HSCR, also known as congenital intestinal aganglionosis) is a congenital malformation characterized by a lack of enteric ganglia in the distal part of the gut leading to constipation, gut distention, and associated failure to thrive. The underlying pathogenic etiology is widely accepted to be based on defects in craniocaudal migration, proliferation, differentiation, and survival of neuroblasts that originate from the neural crest in the early stage of pregnancy.1, 2 The prevalence of HSCR is 1 in 5000 live births, and the disease generally is subdivided into short-segment and long-segment types based on the point at which histologically characterized aganglionosis begins. Currently, HSCR routinely is treated by surgical removal of the aganglionic gut segment. The most life-threatening HSCR-associated complication is HSCR-associated enterocolitis (HAEC), which can occur preoperatively and/or postoperatively.3 HAEC develops in approximately 30% of patients with short-segment and 50% of patients with long-segment HSRC.4, 5 The incidence of HAEC suggests either a multigenic and/or environmental contribution,4, 6 but the pathogenesis of HAEC remains poorly understood. Mucosal barrier dysfunction as a predisposing factor is one of the several proposed drivers for HAEC.4, 5, 6 HAEC is clinically characterized by fever, abdominal distention, diarrhea, and sepsis. Histopathologic features in the colon include crypt dilatation, mucin retention, enterocyte adherence of bacteria, a shift from acidic toward neutral mucin production, epithelial damage, leukocyte infiltration, ulceration, and, in the terminal stages, transmural necrosis and perforation.4, 5, 6, 7 However, at least in part owing to a lack of suitable animal models, the order in which these HAEC-associated clinical features develop is currently poorly understood, which has hampered advances in the treatment and prevention of HAEC.
The most common genetic cause of HSCR is the presence of inactivating mutations in the receptor tyrosine kinase rearranged during transfection (RET), the signaling receptor for glial cell line–derived neurotrophic factor (GDNF); approximately 50% of cases are familial, and 15%–35% are sporadic cases.2, 8, 9, 10, 11 In addition, at least 13 other HSCR susceptibility genes have been identified, including GDNF, NRTN, EDNRB, EDN3, ECE1, SOX10, PHOX2B, KIF1BP, ZEB2, TCF4, and TTF1; together, mutations in these genes account for approximately 20% of all HSCR cases.2, 9, 12, 13 HSCR penetrance also is known to be influenced by gene expression levels because copy number variants of MAPK10, ZFHX1B, and SOX2 loci were associated with HSCR in a pilot study on 67 candidate genes.12 Currently, the underlying genetic cause is unknown for approximately half of all sporadic cases of HSCR.2, 9
RET is activated upon binding with a complex comprising GDNF and GDNF family receptor alpha-1 (GFRa1), a glycosylphosphatidylinositol-anchored protein.14 Studies using transgenic mice have shed light on how GDNF signaling regulates the development of the enteric nervous system (ENS). In the developing mouse gut, ENS progenitor cells called enteric neural crest‒derived cells (ENCCs) enter the foregut on embryonic day 9–9.5 (E9-E9.5). ENCCs migrate rostrocaudally along the gut toward a GDNF source, reach the proximal colon at E12, and colon colonization is complete by E14.5.15, 16 In addition to regulating ENCC migration, GDNF signaling also controls the proliferation, differentiation, and survival of ENCCs.17, 18, 19 Mice that lack expression of genes encoding GDNF, GFRa1, or RET have a common phenotype that includes kidney agenesis and a lack of enteric ganglia distal to the stomach.20, 21, 22 However, heterozygous Gdnf, Gfra1, or Ret null-allele mice with a reduced gene dose have a relatively mild reduction in enteric neuron numbers and do not develop the clinical features of childhood HSCR or HAEC.23 Mouse studies using knock-in or timed conditional deletion alleles for Gfra1 or Ret support the importance of GFRa1/RET signaling in ENS development and survival through postnatal day 1 (P1).18, 24, 25 However, animal models with defective GDNF/GFRa1/RET signaling that phenocopy postnatal HSCR and/or HAEC are currently not available.
Mutations in the RET gene are the most common genetic cause of HSCR, however, causative mutations in GFRa1 have not been found. Here, we report the generation of GFRa1 hypomorphic mice with reduced expression of Gfra1 at the endogenous locus. These mice were generated by inserting a selectable marker gene within intron 6 of the Gfra1 gene. Homozygous GFRa1 hypomorphic (Gfra1hypo/hypo) mice have a 70%–80% reduction in Gfra1 expression in the developing gut and kidney. Although the kidneys develop normally, Gfra1hypo/hypo mice have congenital features reminiscent of childhood long-segment HSCR, with early postnatal onset of symptoms and accompanying progressive HAEC; these mice generally die at P7–P25. Our further analysis also suggests a possible explanation why reduced gene expression in heterozygous Gdnf, Gfra1, and Ret null-allele mice does not result in HSCR, whereas a more severe reduction in Gfra1 expression in Gfra1hypo/hypo mice does lead to HSCR. In addition, our analysis of Gfra1hypo/hypo mice has enabled us to better define the sequence of events in the pathogenesis of HAEC. Specifically, our results allow us to exclude bacterial enterocyte adherence as the primary cause of HAEC in our model, and suggest that goblet cell dysplasia is an early, ubiquitous event that precedes the epithelial changes, including crypt dilatation, mucin retention, and epithelial damage. Together, these findings suggest that goblet cells could be a candidate target for developing preventative therapies for HAEC and that reduced expression of GFRa1 may represent a novel susceptibility trait for HSCR.
Results
Generation and Primary Characterization of Gfra1hypo/hypo Mice
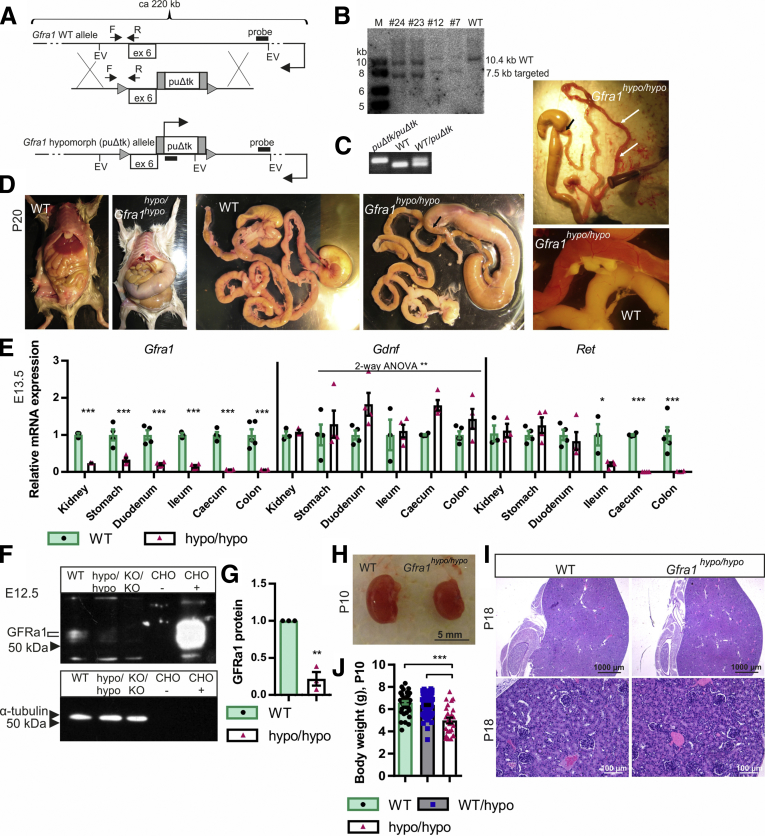
Insertion of a marker gene into an intron of a target gene often interferes with expression of the target, yielding a hypomorphic allele.26, 27, 28, 29, 30 We inserted a puΔtk cassette31 into intron 6 of the Gfra1 gene (Figure 1A–C) and analyzed the phenotype of homozygous (Gfra1puΔtk/puΔtk) mice generated by crossing 2 heterozygous animals. Compared with their expected Mendelian inheritance, homozygous mice were under-represented by approximately 3- to 4-fold at the time of weaning (P18–P25); moreover, homozygous mice developed severe abdominal swelling, failed to thrive, showed increasing inactivity, and other signs of discomfort. Of the 8 homozygous mice that were monitored beginning at P15, none survived beyond P27; because of ethical considerations, we did not proceed with subsequent survival studies. During further experiments, animals that showed obvious signs of discomfort, including abdominal swelling, cachexia, and inactivity, were euthanized. One or more of those signs usually became evident by P4, with gradual progression until the end stage/euthanasia, usually between P7 and P25. Necropsy performed at P18–P25 showed marked caudal gut distention (Figure 1D) starting between the distal ileum and the midcolon. Four of 8 homozygous animals analyzed at P18–P25 had acute erythema/hyperemia in the gastrointestinal (GI) tract, indicative of acute inflammation (Figure 1D, right panel). Upon further inspection, narrowed, contracted gut that was distal to the distention was less flexible and stiff, a feature commonly reported for aganglionic gut segments in HSCR patients.
Figure 1.
Generation of GFRa1 hypomorphic mice and analysis of Gfra1 expression. (A) The Gfra1 hypomorphic allele was generated by inserting a puΔtk cassette31 into intron 6 in the Gfra1 gene. (B) Representative Southern blot analysis of EcoRV-digested genomic DNA isolated from embryonic stem cells, confirming correct targeting of Gfra1 via homologous recombination (expected sizes: targeted, 7564 bp; WT, 10397 bp). (C) PCR-based genotyping of mice using the primers indicated in panel A by F and R (expected sizes: WT, 88 bp; Gfra1puΔtk/puΔtk, 100 bp). (D) Gfra1hypo/hypo mice develop megacolon, constipation, and abdominal distention, and they fail to thrive (note that the Gfra1hypo/hypo mouse is smaller than the WT mouse). The point of dilation is indicated by black arrows. At P18–P25, 50% of Gfra1hypo/hypo mice, sacrificed due to obvious discomfort due to abdominal swelling, have clearly visible acute erythema/hyperemia in the gastrointestinal tract (white arrows) indicative of enterocolitis; representative images are shown. (E) Relative Gfra1, Gdnf, and Ret mRNA levels at E13.5 in the indicated organs measured using qPCR (N = 4). (F and G) Western blot analysis of GFRa1 protein in the gut segment spanning from the duodenum to the ileum in E12.5 embryos. (F) Representative Western blot (samples obtained from E12.5 Gfra1 KO mice were included as a negative control); (G) normalized average GFRa1 intensity (N = 3 independent experiments). (H) At P10, the kidneys in Gfra1hypo/hypo mice appear anatomically normal. (J) Gfra1hypo/hypo mice are smaller than WT and heterozygous littermates at P10 (P < .001). The slight reduction in kidney size in the Gfra1hypo/hypo mice likely reflects an overall failure of the mice to thrive. (I) Compared with WT littermates, kidney histology is grossly normal in P18 Gfra1hypo/hypo mice. F, forward primer; R, reverse primer; EV, EcoRV site; ex 6, exon 6; puΔtk/puΔtk, Gfra1puΔtk/puΔtk; KO/KO, Gfra1KO/KO; CHO, Chinese hamster ovary cells transfected with a plasmid encoding GFP (-) or Gfra1 (+). All summary data are presented as means ± SEM. *P < .05, **P < .01, and ***P < .001. Statistical analyses were performed using the Student t test, unless indicated otherwise.
Analysis of Gfra1, Ret, and Gdnf Expression
Quantitative real-time polymerase chain reaction (PCR) analysis performed at E13.5 showed that Gfra1 messenger RNA (mRNA) levels were reduced by 75%–80% in the kidneys, stomach, and duodenum of Gfra1hypo/hypo mice; expression levels decreased further toward the colon, where Gfra1 mRNA levels were reduced by approximately 90% compared with control animals (Figure 1E). Ret mRNA levels were normal in the stomach and duodenum, but sharply decreased in the ileum of Gfra1hypo/hypo mice. In the cecum and colon, Ret mRNA was not detectable in our analysis (Figure 1E). This was expected because in the developing gut Ret is expressed exclusively in ENCCs, whereas Gfra1 is expressed both in ENCCs and in the surrounding mesenchyme.16, 32, 33, 34 As our parallel analysis showed, ENCCs in Gfra1hypo/hypo mice were missing distal from the midileum (see later).
Gdnf mRNA levels were slightly—albeit significantly—increased throughout the GI tract, possibly reflecting mechanistic compensation for the reduced expression of GDNF receptors (Figure 1E). Western blot analysis showed that the amount of GFRa1 protein in the gut segment from the duodenum to the ileum of Gfra1hypo/hypo mice was reduced by 75%–80% at E13.5 (Figure 1F and G). In the cecum and colon, GFRa1 protein was not detectable in our analysis. Kidney development in Gfra1hypo/hypo mice was generally normal (Figure 1H and I). Given the hypomorphic Gfra1 mRNA and protein levels measured in these mice, we renamed the Gfra1puΔtk allele the Gfra1 hypomorphic (Gfra1hypo) allele.
Homozygous Gfra1hypo/hypo Mice Have Impaired Neuronal Progenitor Colonization and Develop Aganglionosis and Hypertrophic Fibers in the Colon
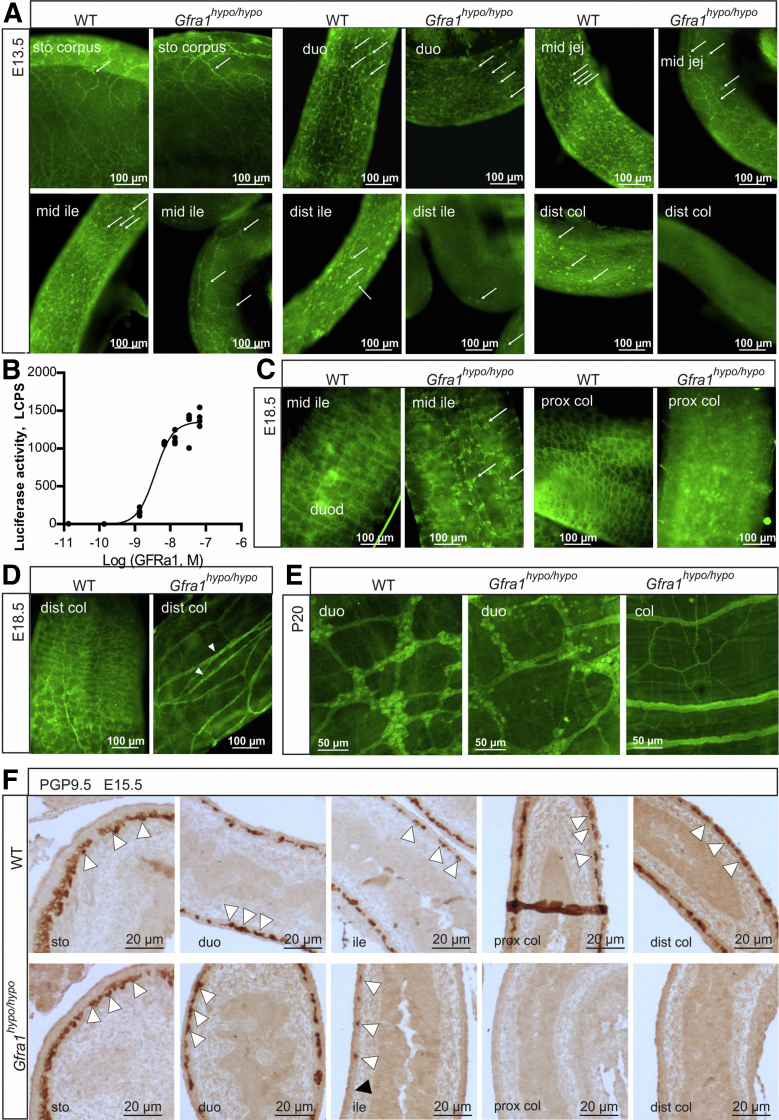
Mice that lack Gdnf, Gfra1, or Ret expression lack enteric nerves distal to the stomach.20, 21, 22 Moreover, experiments in which Gfra1 or Ret expression was deleted under temporal control in the developing gut showed that GDNF/GFRa1/RET signaling regulates the migration, proliferation, differentiation, and survival of ENCCs, particularly in the distal gut, in a gene dose-dependent manner.18, 25 Consistent with these findings, innervation of the stomach in Gfra1hypo/hypo mice appeared normal at E13.5, and the number of neuronal progenitors decreased gradually toward the distal gut (Figure 2A). At E13.5, Gfra1hypo/hypo embryos lacked neuronal progenitors from the midileum onward (Figure 2A). In the developing gut, Ret is expressed exclusively in ENCCs, whereas Gfra1 is expressed both in ENCCs and in the surrounding mesenchyme.16, 32, 33, 34 As shown in Figure 2A, neuronal progenitors in the distal gut in Gfra1hypo/hypo embryos are missing, explaining the observed craniocaudal reduction in Gfra1 mRNA and the absence of Ret mRNA in the distal gut (Figure 1E). Similar innervation defects were observed at E15.5 (ie, 3–4 days before birth in mice) (Figure 2F).
Figure 2.
Impaired enteric neuronal progenitor colonization and hypertrophic nerve fibers in the distal gut of GFRa1 hypomorphic mice. (A) Immunohistochemistry using the pan-neuronal marker ubiquitin C-terminal hydrolase L1 in E13.5 WT and Gfra1hypo/hypo mice whole-mount GI tract preparations. Note the reduced numbers of neuronal progenitors (white arrows) beginning at the midjejunum of Gfra1hypo/hypo mice and the absence of neuronal progenitors from the distal and GDNF ileum onward; representative images are shown (N = 3–4 animals per genotype). (B) Dose-dependent activation of mitogen-activated protein kinase signaling by soluble GFRa1. MG87 fibroblasts stably expressing RET and the PathDetect Elk-1 luciferase system were treated with the indicated concentration of soluble GFRa1 (N = 4 biological replicates per experiment; N = 2 experiments). (C and D) ubiquitin C-terminal hydrolase L1 immunohistochemistry was performed in E18.5 WT and Gfra1hypo/hypo mice from whole-mount GI tract preparations, showing reduced and scattered innervation in the midileum, an absence of innervation in the proximal colon, and hypertrophic fibers in the distal colon (white arrowheads, a hallmark feature of HSCR); the white arrows indicate neuronal somas. Representative images are shown (N = 3–4 animals per genotype). (E) ubiquitin C-terminal hydrolase L1 immunohistochemistry performed on P20 whole-mount myenteric plexus preparations showing normal innervation in the duodenum, an absence of neuronal cell bodies in the colon, and hypertrophic fibers in the colon of Gfra1hypo/hypo mice; representative images are shown (N = 2–3 animals per genotype). (F) The ENCC colonization of the mouse hindgut is complete by E14.5. Representative image of sagittal gut sections immunostained with the pan-neuronal marker ubiquitin C-terminal hydrolase L1 in E15.5 WT and Gfra1hypo/hypo mice, showing that the enteric neuronal progenitors have reached the distal colon in the WT mice (white arrowheads); in contrast, enteric neuronal progenitors reached the proximal ileum (white arrowheads) in E15.5 Gfra1hypo/hypo mice, but are absent from the distal ileum, cecum, and colon. The point at which aganglionosis begins is indicated by a black arrowhead; representative images from WT and Gfra1hypo/hypo mice are shown. dist, distal; duo, duodenum; ile, ileum; jej, jejunum; prox, proximal.
Heterozygous Gfra1 null-allele (Gfra1wild type [WT]/knockout [KO]) mice, which had an approximate 50% reduction in GFRa1 expression, had reduced enteric neuron fiber density and defects in gut function measured ex vivo; however, they did not have reduced numbers of enteric neurons, and they did not develop symptoms of either HSCR or HAEC.20, 23, 35 Our finding that a 70%–80% reduction in Gfra1 expression caused long-segment HSCR suggests a threshold effect in GFRa1-mediated downstream signaling. In the developing gut, 2 forms of the GFRa1 protein are present—the membrane-associated glycosylphosphatidylinositol-anchored form and the extracellular soluble form; these 2 forms can function cooperatively in mediating GDNF signaling in ENCCs.34 Because the concentration of a solute decreases steeply as a function of distance from the source, signaling via the soluble form of GFRa1 is likely to be particularly sensitive to decreased expression. We analyzed the concentration dependence of soluble GFRa1 on downstream signaling in an MG87 fibroblast model system that stably expressed both Ret and a luciferase gene that reflected the activity of mitogen-activated protein kinases,36 a downstream target of RET signaling. We stimulated these cells with GDNF together with increasing concentrations of soluble GFRa1 and measured a sharp response in RET downstream signaling within a relatively narrow GFRa1 concentration range of 1.35–6.75 nmol/L (Figure 2B). These results provide a possible explanation for the phenotype differences observed between Gfra1WT/KO and Gfra1hypo/hypo mice. In Gfra1WT/KO mice, the GFRa1 levels were within the effective range, whereas GFRa1 levels fell below the effective range in Gfra1hypo/hypo mice.
Long-segment HSCR is defined as aganglionosis proximal to the splenic flexure.37 Another hallmark clinical feature of HSCR is the presence of extrinsic hypertrophic nerve fibers in the distal colon.38, 39, 40 At E18.5, Gfra1hypo/hypo mice had varying-length aganglionic segments proceeding from the midcolon (long-segment HSCR) or even from the distal ileum (total colonic aganglionosis), and they had hypertrophic nerve fibers in the distal colon (Figure 2C and D). Because the presence of hypertrophic fibers in the colon of HSCR patients is diagnosed postnatally, we analyzed the colon in postnatal (P20) Gfra1hypo/hypo mice and found that the hypertrophic fibers still were present (Figure 2E).
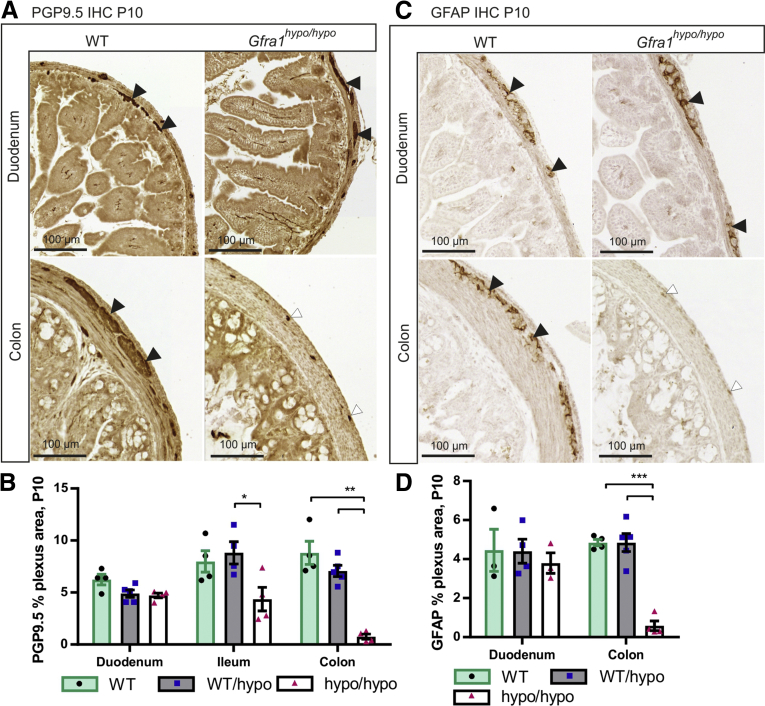
Gfra1hypo/hypo mice are born with a normal appearance; however, they then develop symptoms that resemble HSCR, including constipation, distention of the gut, and a failure to thrive (Figure 1J and see later), with 100% penetrance by P10. Consistent with early mortality, the genotypes in the animal cohort were at the expected Mendelian ratios at E18.5, but not at P10 or P15–P23 (Table 1). Immunohistologic analysis of the ENS in Gfra1hypo/hypo mice at P10 using the pan-neuronal marker ubiquitin C-terminal hydrolase L1 showed normal ganglia in the duodenum, reduced ganglia size in the distal ileum, and absent or rudimentary ubiquitin C-terminal hydrolase L1-positive structures in the colon, reflecting hypertrophic fibers in the colon (Figure 3A and B). Similar results were obtained using the glial fibrillary acidic protein marker (Figure 3C and D). Collectively, results presented in Figures 2 and 3 are consistent with previous results on timed deletion of GFRa1 and RET until E18.5,18, 25 and suggest that reduced Gfra1 expression in Gfra1hypo/hypo mice causes impaired craniocaudal colonization of the gut by enteric neuronal progenitors, with subsequent aganglionosis of the distal gut and the occurrence of hypertrophic nerve fibers in the distal colon.
Table 1.
Distribution of Offspring Genotypes Born From Breedings Between Gfra1wt/hypo Animals
| Distribution, number of animals (% total) | Expected, number of animals (% total) | ||
|---|---|---|---|
| E18.5 | WT | 27 (25.23) | 26.75 (25) |
| P = .308 | WT/hypo | 59 (55.14) | 53.5 (50) |
| Hypo/hypo | 21 (19.63) | 26.75 (25) | |
| P10 | WT | 39 (24.38) | 35.75 (25) |
| P = .018 | WT/hypo | 96 (60.00) | 71.5 (50) |
| Hypo/hypo | 25 (15.63) | 35.75 (25) | |
| P15–P23 | WT | 37 (35.24) | 26.25 (25) |
| P < .001 | WT/hypo | 60 (57.14) | 52.5 (50) |
| Hypo/hypo | 8 (7.62) | 26.25 (25) |
NOTE. At E18.5, Gfra1hypo/hypo mice are present at the expected Mendelian ratio, but not at P10 (P < .05) or at P15–P23 (P < .001), chi-square analysis.
Figure 3.
Analysis of the enteric nervous system in P10 Gfra1 hypomorphic mice. (A) Representative images of the pan-neuronal marker ubiquitin C-terminal hydrolase L1 immunohistochemistry in coronal gut sections prepared from P10 mice. (B) Quantification of the ganglionated plexus area (black arrowheads in A) in P10 WT and Gfra1hypo/hypo coronal gut sections; the residual staining in the Gfra1hypo/hypo mice colon reflects hypertrophic fibers or transition zone (white arrowheads in A). P < .05 Gfra1WT/hypo vs Gfra1hypo/hypo in ileum and P < .01 WT and Gfra1WT/hypo vs Gfra1hypo/hypo in colon, N = 4–5 mice per genotype. Analysis of variance and the Tukey multiple comparisons tests were used for statistical analysis. (C) Immunostaining for the glial marker, glial fibrillary acidic protein (GFAP), in the duodenum of P10 mice is similar between genotypes (N = 3; representative images are shown). Black arrowheads indicate ganglia stained with GFAP, white arrowheads mark residual staining from hypertrophic fibers of the transition zone. Reminiscent of the results obtained using ubiquitin C-terminal hydrolase L1 at the same age (A and B) in the colon, Gfra1hypo/hypo mice have a smaller GFAP-positive plexus area in the myenteric plexus relative to littermate controls quantified in panel D; N = 4–5 animals per genotype; P < .001, analysis of variance and the Tukey multiple comparisons test (data are presented as means ± SEM). All summary data are presented as means ± SEM. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001.
Hypertrophic Fibers in the Colon of Gfra1hypo/hypo Mice Are Cholinergic
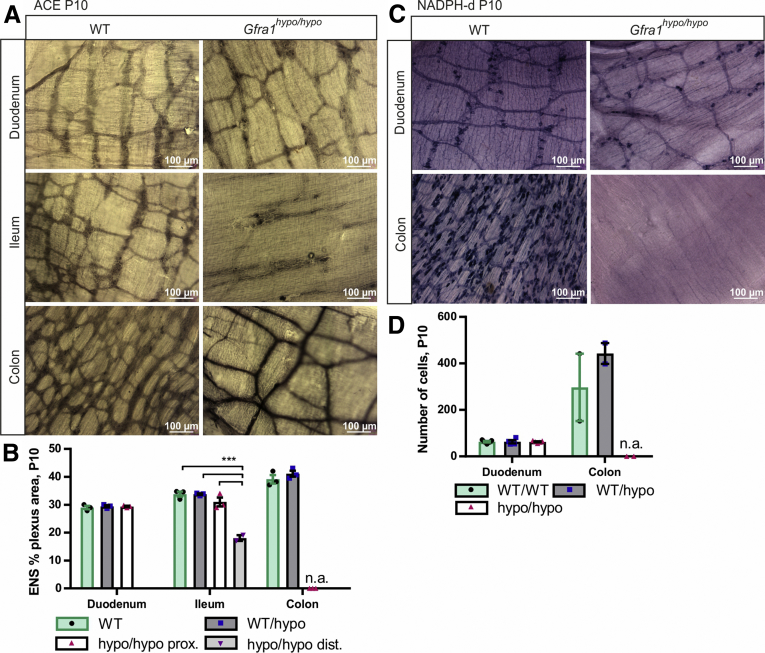
Hypertrophic nerve fibers in the distal colon of HSCR patients are known to be cholinergic.40, 41 The reduced flexibility of the aganglionic gut segment in HSCR patients is believed to result from excess extrinsic excitatory cholinergic stimuli and a lack of intrinsic inhibitory nitrergic stimuli.42, 43 To examine the properties of the myenteric plexus in Gfra1hypo/hypo mice, we stained for acetylcholinesterase. Our analysis showed that Gfra1hypo/hypo mice have normal cholinergic innervation in the duodenum and proximal ileum, reduced cholinergic innervation in the distal ileum, a lack of cholinergic innervation in the midcolon, and hypertrophic cholinergic fibers in the distal colon (Figure 4A and B). Next, we stained the myenteric plexus for reduced nicotinamide adenine dinucleotide phosphate (NADPH) diaphorase, a marker of nitric oxide synthase‒positive nitrergic enteric neurons. Our analysis showed that Gfra1hypo/hypo mice have normal numbers of NADPH-diaphorase‒positive neurons in the duodenum, but a complete lack of NADPH-diaphorase‒positive cells in the distal colon (Figure 4C and D), reminiscent of the histopathologic findings in patients with HSCR.42, 43
Figure 4.
The hypertrophic nerve fibers in the distal gut of GFRa1 hypomorphic mice are cholinergic. (A) Acetylcholinesterase histochemistry at P10 showing similar innervation in the duodenum, as well as cholinergic hypertrophic nerve fibers characteristic of HSCR in the colon of Gfra1hypo/hypo mice. In the ileum, an image distal of the point of dilatation is shown (N = 3 animals per genotype). (B) Quantification of the acetylcholinesterase-positive plexus area in P10 WT and Gfra1hypo/hypo mice (N = 3 animals per genotype, ***P < .001). (C) NADPH diaphorase histochemistry for nitric oxide synthase (NOS)-positive neurons in P10 mice showing similar innervation in the duodenum and an absence of NOS-positive innervation in the colon of Gfra1hypo/hypo mice; N = 4–5 animals per genotype (N = 2 animals per genotype for the colon). (D) Quantification of NOS-positive plexus area in P10 WT and Gfra1hypo/hypo mice; N = 4–5 animals per genotype (N = 2 animals per genotype for the colon). Analysis of variance and the Tukey multiple comparisons tests were used for statistical analysis. n.a., not applicable for Gfra1hypo/hypo mice. ACE, acetylcholinesterase; dist, distal; prox, proximal (data are presented as means ± SEM). All summary data are presented as means ± SEM. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001.
Histopathologic Characterization of HSCR/HAEC in Gfra1hypo/hypo Mice
Improving our understanding of the histopathologic changes that occur in HAEC may eventually help in developing more effective treatments. We used the histopathologic grading system for HAEC developed by Teitelbaum et al7 to analyze Gfra1hypo/hypo mice. Teitelbaum et al7 scored the histopathologic findings in HAEC in patients as follows: 0, no abnormalities; I, crypt dilatation and mucin retention; II, cryptitis or 2 crypt abscesses per high-power field; III, more than 2 crypt abscesses per high-power field; IV, fibrinopurulent debris and mucosal ulceration; and V, transluminal necrosis or perforation. However, compared with patients, mice with HAEC had milder infiltration of inflammatory cells into the crypts (milder cryptitis and crypt abscesses),44 although the epithelial pathologies were similar (see later); therefore, we modified the grading system to primarily reflect epithelial pathology (see the Materials and Methods section). Another characteristic feature associated with HAEC is an overproduction of mucus and the retention of mucus in the colon, as well as a shift from producing acidic mucins to producing neutral mucins.5, 7 Mucins are produced primarily by goblet cells and form a protective barrier preventing bacterial enterocyte adherence.45 Goblet cell hyperplasia46 and altered goblet cell function47 have been reported in HSCR.
Histopathologic characterization of HSCR/HAEC in E18.5, P4, and P10 Gfra1hypo/hypo mice
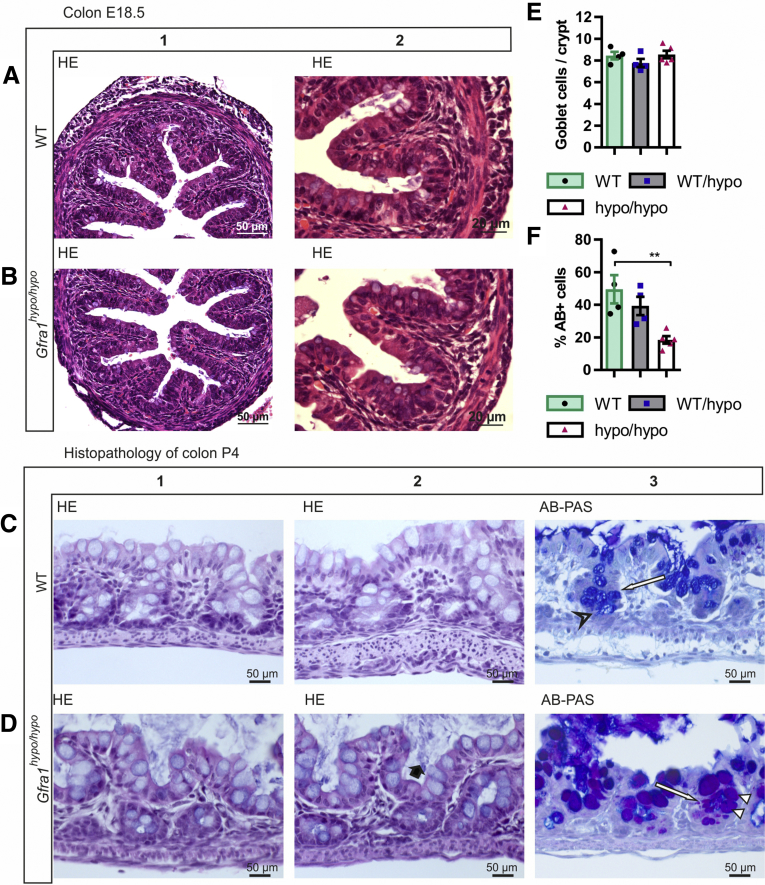
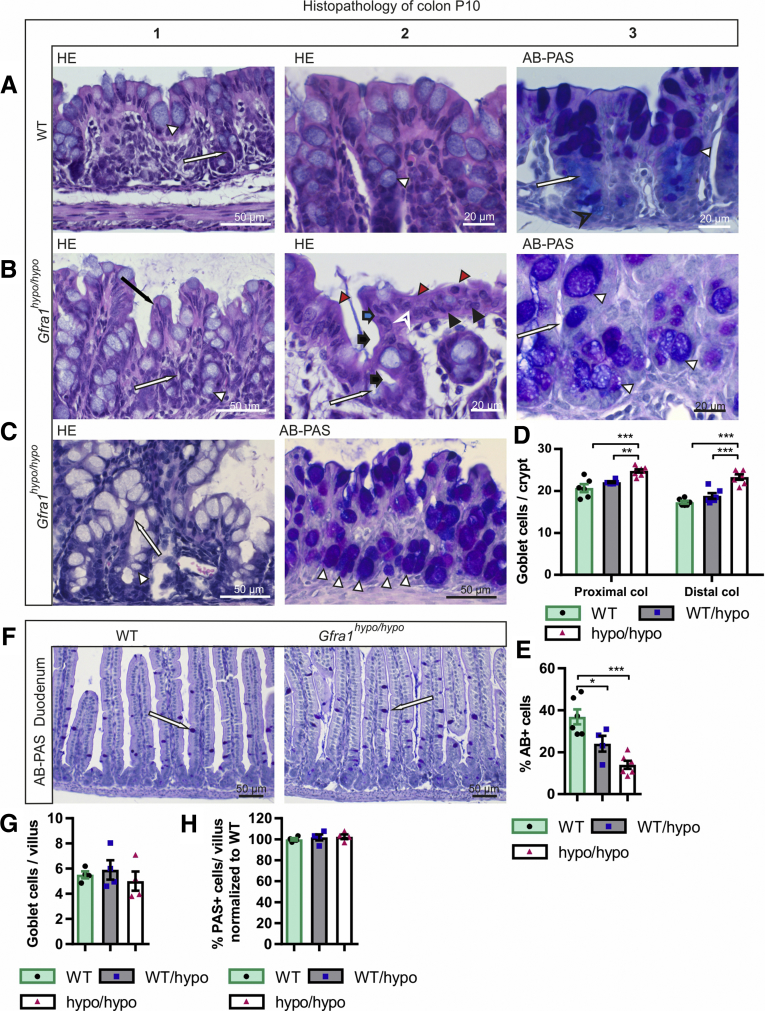
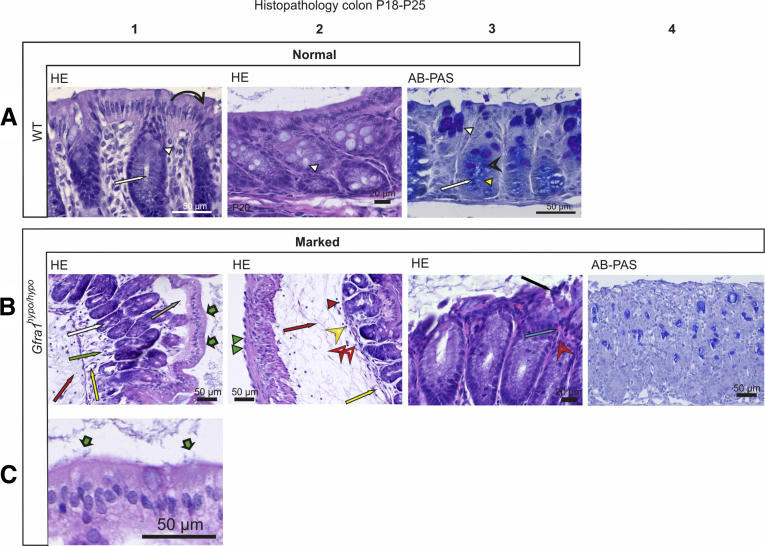
Histologic analysis of the colon in E18.5 Gfra1hypo/hypo mice showed no overt changes compared with control mice (Figure 5A and B). However, at P4, histologic analysis using Alcian blue (AB), periodic acid‒Schiff (PAS), H&E, and Gram staining showed from minimal to moderate epithelial damage accompanied by crypt dilatation and a mucin shift from acidic to neutral with mucus retention in the goblet cells (Figure 5C–F, Table 2). At P10 the differences were more advanced: slight to mild multifocal to diffuse epithelial hyperplasia, as well as dysplasia (ie, disorganization) accompanied by degeneration of surface colonocytes in Gfra1hypo/hypo mice (Figures 6A and B) was observed, indicating mild surface epithelial damage. In the hyperplastic and degenerated areas of the colon, the epithelial cells grew in disorganized, undulating rows with partly overlapping nuclei (Figure 6B and C). In addition, the crypts in the colon were mildly to moderately dilated with various degrees of mucin retention (Figure 6B and C), indicating an overproduction and retention of mucus in the colon. We also observed increased numbers of intraepithelial apoptotic cell remnants in 5 of 8 Gfra1hypo/hypo mice, indicating a degenerative process (Figure 6B). Importantly, the severity of these features varied between animals (the histopathologic data from each animal are presented in Table 3).
Figure 5.
Histopathology of colon at E18.5 and P4. (A and B) Representative H&E-stained coronal sections of the colon at E18.5. No gross difference was observed between genotypes (N = 3–4 animals per genotype). (C and D). H&E staining from WT and Gfra1hypo/hypo colon at P4. Gfra1hypo/hypo animals typically show mild crypt dilatation accompanied with mucus retention (D2, black bold arrow). AB-PAS staining shows more acidic mucins in WT animals at the basal crypts (C3, open arrowhead) and more neutral mucins in Gfra1hypo/hypo animals (white arrowheads) quantified in F. **P < .01 (N = 3–4). (E) The number of goblet cells (C3 and D3, white arrows) was not changed at P4 (N = 4–5). Analysis of variance and the Tukey multiple comparisons test, data are presented as means ± SEM. All summary data are presented as means ± SEM.
Table 2.
Summary of Histopathologic Findings at P4
| P4 | ID | Weight, g | Sex | Applied Teitelbaum et al7 grade | Surface epithelial injury | Crypt dilation | Intraepithelial apoptotic remnants | Goblet cell hyperplasia | Mucins in crypts |
Bacteria in crypts | Enterocyte adherence | Leukocytes |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mucin shift to PAS | Basal | Surface | Mucus retention | Granulocytes | Lymphocytes | |||||||||||
| WT | 7615-2 | 3.02 | F | 0 | - | - | - | No | No | AB | PAS | - | - | No | - | - |
| WT | 7640-3 | 2.96 | F | 0 | - | - | - | No | No | AB | PAS | - | - | No | - | - |
| WT | 7640-4 | 3.16 | M | 0 | - | - | - | No | No | AB | PAS | - | - | No | - | - |
| WT | 7640-4a | 2.92 | M | 0 | - | - | - | No | No | AB | PAS | - | - | No | - | - |
| WT/hypo | 7615-4 | 3.15 | F | 0 | - | - | - | No | No | AB | PAS | - | - | No | - | + |
| WT/hypo | 7615-1 | 3.21 | F | 0 | - | - | - | No | No | AB | PAS | - | - | No | - | - |
| WT/hypo | 7615-5 | 3.06 | M | 0 | - | - | - | No | No | AB | PAS | - | - | No | - | - |
| WT/hypo | 7640-5a | 2.78 | M | 0 | - | - | - | No | No | AB | PAS | - | - | No | - | - |
| Hypo/hypo | 7615-3 | 2.60 | F | I | Mild | ++ | - | No | Yes | PAS | PAS | ++ | - | No | - | - |
| Hypo/hypo | 7640-1 | 2.38 | F | I | Mild | ++ | - | No | Yes | PAS | PAS | ++ | - | No | - | - |
| Hypo/hypo | 7640-1a | 2.34 | M | I | Mild | +++ | - | No | Yes | PAS | PAS | ++ | - | No | - | - |
| Hypo/hypo | 7640-3a | 2.44 | M | I | Minimal | + | - | No | Yes | PAS | PAS | ++ | - | No | - | - |
| Hypo/hypo | 7640-2 | 2.51 | M | II | Moderate | ++ | + | No | Yes | PAS | PAS | ++ | - | No | - | - |
| Minimal | Mild + | Mild + | Mild + | Mild + | Normal - (<4/HPF) | |||||||||||
| 0–V | Mild | Moderate ++ | Moderate ++ | Yes/no | Yes/no | Moderate ++ | Moderate ++ | Yes/no | Mild + (5–15/HPF) | |||||||
| Moderate | Marked +++ | Marked +++ | Marked +++ | Moderate ++ (20–30/HPF) | ||||||||||||
| Marked | ||||||||||||||||
NOTE. See the Materials and Methods section for details regarding the histopathologic grading.
Figure 6.
Mild epithelial damage, mucin retention, mucin type changes, and goblet cell hyperplasia in P10 Gfra1hypo/hypomice. (A1–2) H&E and AB-PAS (A3) staining of coronal colon sections from P10 WT mice. In WT mice, the mucosa is thin and compact with an evenly palisading, regular surface epithelium. The crypts of Lieberkühn (A1 and A3, white arrow) in the colon are shallow and straight. Goblet cells (A1–3, white arrowhead) are abundant in surface epithelium and within the proximal third of the crypts, but less abundant at the crypt bases (A3). The goblet cells in the surface epithelium and in the proximal parts of the crypts contain PAS-positive neutral mucins, staining deep purple (A3, white arrowhead). The goblet cells in the basal parts of the crypts primarily contain AB-positive acidic droplets, staining light blue (A3, black open arrowhead). (B and C) In Gfra1hypo/hypo mice, the colon crypts (B1, B3, and C1, white arrow) are mildly to moderately dilated, with variable levels of mucin retention (B2, black bold arrow). The surface of the mucosa in the colon of Gfra1hypo/hypo mice is bumpy in appearance (compare A1 with B1), and epithelial cells grow in disorganized, undulating rows with partly overlapping nuclei (B1, black arrow). The height variation of the epithelial cells (B2, blue bold arrow), the round to oval bland nuclei (B2, black arrowhead), increased eosinophilia in the apical cytoplasm (B2, red arrowheads), and the indistinct cell borders (B2, white open arrowhead) are indicative of degeneration and mild epithelial damage. Unlike in WT mice (A3), the goblet cells in Gfra1hypo/hypo mice often are abundant in the basal parts of the crypts (C2, white arrowheads) and show a shift toward the production of PAS-positive (ie, neutral) mucins (compare C2 with A3). (D) Quantification of goblet cells in WT, Gfra1WT/hypo, and Gfra1hypo/hypo mice, for which the number of goblet cells is increased both in proximal (P < .001 and P < .01) and distal colons (P < .001) compared with both heterozygous and WT animals; N = 6–8 animals per genotype. (E) Gfra1hypo/hypo mice and Gfra1WT/hypo have fewer AB-positive cells per crypt compared with WT colon (P < .001 and P < .05, N = 4–5). (F) AB-PAS staining from WT and hypomorphic duodenum at P10. (G) Quantification of goblet cells (F, white arrows) in WT, Gfra1WT/hypo, and Gfra1hypo/hypo mice in P10 duodenum (N = 4 per genotype). (H) Quantification of PAS-positive goblet cells, AB-positive goblet cells were absent in duodenum (N = 4 mice per genotype). *P < .05, **P < .01, and ***P < .001. col, colon. Analysis of variance and the Tukey multiple comparisons test, data are presented as means ± SEM. All summary data are presented as means ± SEM.
Table 3.
Summary of Histopathologic Findings at P10
| P10 | ID | Weight, g | Sex | Applied Teitelbaum et al7 grade | Surface epithelial damage | Crypt dilation | Intraepithelial apoptotic remnants | Goblet cell hyperplasia | Mucins in crypts |
Bacteria in crypts | Enterocyte adherence | Leukocytes |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mucin shift to PAS | Basal | Surface | Mucus retention | Granulocytes | Lymphocytes | |||||||||||
| WT | 10 | 6.8 | F | 0 | - | - | - | No | No | AB | PAS | - | - | No | - | - |
| WT | 23 | 5.3 | M | 0 | - | - | - | No | No | AB | PAS | - | - | No | - | - |
| WT | 6280-1 | 8 | M | 0 | - | - | - | No | No | AB | PAS | - | - | No | - | - |
| WT | 31 | 4.12 | M | 0 | - | - | - | No | No | AB/PAS | PAS | - | - | No | - | - |
| WT/hypo | 22 | 6.09 | M | 0 | - | - | - | No | No | AB/PAS | PAS | - | - | No | - | - |
| WT/hypo | 24 | 6.42 | F | 0 | - | - | - | No | No | AB | PAS | - | - | No | - | -/+ |
| WT/hypo | 6777-3 | 5.18 | M | 0 | - | - | - | No | No | AB | PAS | - | - | No | - | - |
| WT/hypo | 6280-2 | 7.84 | M | 0 | - | - | - | No | No | AB | PAS | - | - | No | - | - |
| WT/hypo | 12 | 6.42 | M | 0-I | No/minimal | - | - | No | No | AB/PAS | PAS | - | - | No | - | - |
| Hypo/hypo | 11 | 3.61 | M | I | Minimal | - | - | Yes | Yes | PAS | PAS | - | - | No | - | - |
| Hypo/hypo | 27 | 7.57 | M | II | Mild | - | + | Yes | Yes | PAS | PAS | - | - | No | - | - |
| Hypo/hypo | 6277-7 | 3.6 | F | II | Mild | +/++ | - | Yes | Yes | PAS | PAS | + | - | No | - | - |
| Hypo/hypo | 21 | 4.93 | F | III | Moderate | + | + | Yes | Yes | PAS | PAS | ++ | - | No | - | + |
| Hypo/hypo | 6224-2 | 4.15 | M | II | Mild | + | + | Yes | Yes | PAS | PAS + AB | + | - | No | - | - |
| Hypo/hypo | 6224-7 | 5.93 | F | III | Moderate | ++ | + | Yes | Yes | PAS | PAS + AB | + | - | No | ++ | - |
| Hypo/hypo | 7310-1 | 4.53 | M | III | Moderate | ++ | + | Yes | Yes | PAS/ foamyAB | PAS/ foamyAB | ++ | - | No | - | - |
| Hypo/hypo | 7310-2 | 3.62 | M | II | Mild | ++/+++ | - | Yes | Yes | PAS/ foamyAB | PAS/ foamyAB | ++ | - | No | - | - |
| Minimal | Mild + | Mild + | Mild + | Mild + | Normal - (<4/HPF) | |||||||||||
| 0–V | Mild | Moderate ++ | Moderate ++ | Yes/no | Yes/no | Moderate ++ | Moderate ++ | Yes/no | Mild + (5–15/HPF) | |||||||
| Moderate | Marked +++ | Marked +++ | Marked +++ | Moderate ++ (20–30/HPF) | ||||||||||||
| Marked | ||||||||||||||||
NOTE. See the Materials and Methods section for details regarding the histopathologic grading.
At P10, goblet cell pathology was more advanced, with a further shift in mucin production from acidic to neutral now also accompanied by goblet cell hyperplasia in all mutant mice. Notably, the earlier-described goblet cell pathology appeared in all mice regardless of the extent of the aforementioned epithelial pathology (Figure 6C, Table 3). More specifically, we observed that PAS-positive goblet cells, which are normally found in the apical and central parts of the crypts, were present at the base of the crypts in the Gfra1hypo/hypo colon (Figure 6B and C). On the other hand, AB, which stains acidic mucins, showed that AB-positive goblet cells, which are normally found at the base of crypts, are either absent or filled with a foamy mixture of both neutral and acidic mucins in the Gfra1hypo/hypo colon (Figure 6B and C; Table 3). To gain insight into whether altered goblet cell physiology in the Gfra1hypo/hypo colon results from a deficit in the ENS in the colon and distal intestine, or from a reduction in GFRa1 levels in fetal mesenchyme in the whole intestine, we also analyzed goblet cells in the duodenum at P10 using AB-PAS staining. The number of goblet cells and the mucin profile were unchanged in the duodenum at P10 (Figure 6F–H).
Finally, both Gram and H&E staining showed no evidence of bacterial enterocyte adherence or microbial infection in the Gfra1hypo/hypo colon at P10 (Table 3). Based on our histopathologic grading system of HAEC, 3 of the 8 Gfra1hypo/hypo mice at P10 had a grade III HAEC score, 4 mice had a grade II HAEC score, and 1 mouse had a grade I HAEC score.
Taken together, these results suggest that a mucin shift from acidic to neutral in goblet cells at P4 is further enhanced and accompanied with hyperplasia at P10 and that those are generally ubiquitous early events in Gfra1hypo/hypo mice. On the other hand, the interanimal variations in epithelial damage, epithelial degeneration, crypt dilatation, and mucin retention (Table 3) suggest that these changes may be secondary to goblet cell dysfunction and appear later in HAEC.
Histopathologic characterization of HSCR/HAEC in P18–P25 Gfra1hypo/hypo mice
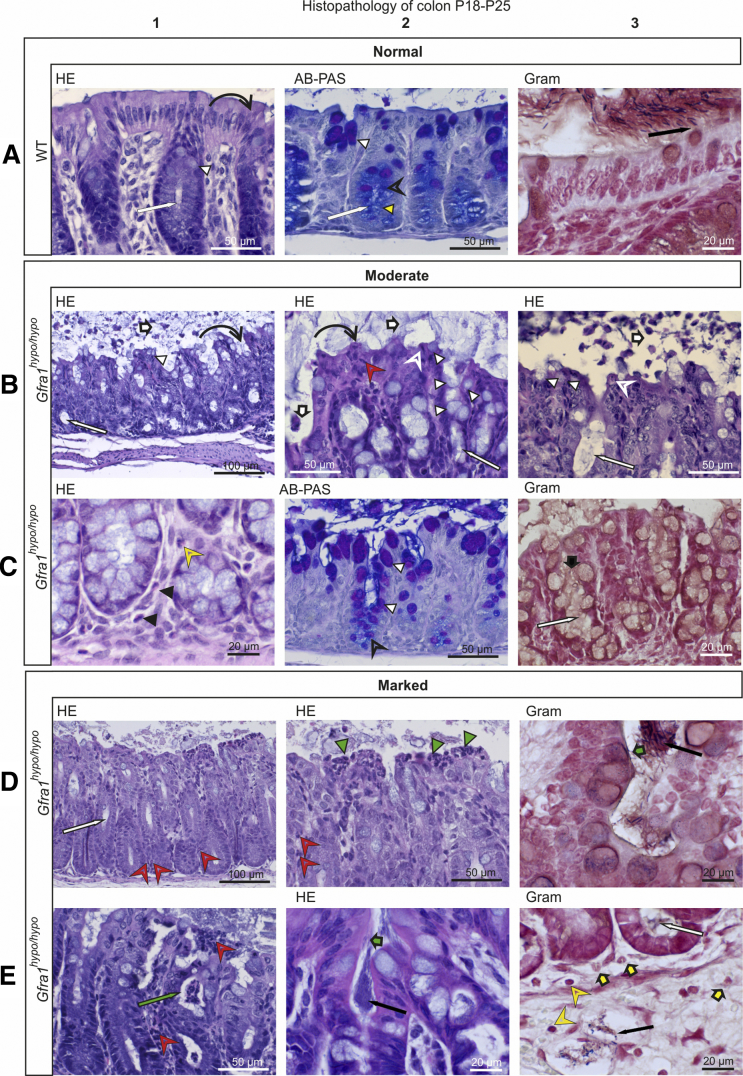
To analyze the features of advanced HAEC in our Gfra1hypo/hypo mice, we examined P18–P25 mice that showed obvious signs of discomfort, including cachexia, abdominal swelling, and reduced spontaneous activity. In these mice, we observed histopathologic features that are reminiscent of advanced HAEC in HSCR patients5, 6, 7 (Figures 7 and 8; Table 4). Compared with P10 mice, in all animals analyzed the P18–P25 mice had more advanced levels of leukocyte accumulation, epithelial damage (including the presence of apoptotic cell remnants), mucus accumulation, and crypt dilatation‒associated edema (Figure 6, Figure 7, Figure 8; Tables 3 and 4). The shift from acidic to neutral mucin production observed at P10 also clearly was evident at P18–P25 (Figure 7). Moreover, goblet cell hyperplasia (Figure 7B and C) was present only in mice with moderate epithelial damage; in contrast, mice with marked epithelial damage had either degenerated goblet cells or a combined phenotype of goblet cell hyperplasia and goblet cell degeneration (Figures 7C and 8B; Table 4). A variety of other features that are typical in advanced HAEC, including ulceration, necrotic purulent colitis, crypt abscesses, and/or the presence of focal bacterial aggregates, were observed in 3 of the 4 animals investigated (Figure 7D and E). Bacterial enterocyte adherence on the other hand was observed in only 2 of the 3 mice with severe epithelial damage (Table 4); these same 2 mice also had bacteria within the colon crypts and blood vessels (Figures 7D and E and 8B and C), indicating sepsis. The histopathologic findings of each mouse examined at P4, P10, or P18–P25 are summarized in Table 2, Table 3, and 4, respectively.
Figure 7.
Histopathologic analysis of moderate and marked HSCR/HAEC in P18–P25 Gfra1hypo/hypomice. (A–C) Exacerbated mucin type changes, epithelial damage, scarce inflammation in the lamina propria, and a lack of bacterial invasion in P18–P25 Gfra1hypo/hypo mice with moderate-grade HAEC. (A) In WT mice, the crypts in the colon (A1 and A2, white arrow) are relatively shallow and straight, and the surface epithelium is even and regularly palisading (A1, curved black arrow). The goblet cells in the surface epithelium and in the proximal parts of the crypts contain PAS-positive (ie, neutral) mucins, staining deep purple (A2, white arrowhead). In the basal parts of the crypts, apical mucin droplets in the colonocytes and the mucus in the goblet cells stain primarily with AB, staining light blue (mucin droplets; A2, yellow arrowhead) to dark blue (goblet cells; A2, open arrowhead). In A3, gram-positive (blue to black) and gram-negative (intense red) bacteria are isolated from the surface epithelium by a layer of mucus (A3, black arrow). (B) The intestinal lumens of Gfra1hypo/hypo mice contain fecal material and copious amounts of mucus with eosinophilic debris of shed epithelial cells (B1–B3, white bold arrows). The crypts were markedly dilated with retained mucin (B1–B3, white arrow). The surface epithelium had diffuse degeneration (B1 and B2, curved black arrow) and an increased number of goblet cells (B1–B3, white arrowhead). Multifocal necrotic and apoptotic colonocytes (B2 and B3, open white arrowhead) and scattered intraepithelial neutrophils (B2, red open arrowhead). (C) The basal lamina propria contains moderately increased numbers of eosinophils (C1, black arrowhead) and mildly increased numbers of lymphocytes (C1, yellow open arrowhead), indicative of an inflammatory process. Note the reduction in AB-stained (ie, acidic) mucin (C2, black open arrowhead) in the cytoplasm of basal crypt colonocytes. Gram staining shows copious amounts of mucus (C3, black bold arrow) and a lack of bacteria in mildly dilated crypts (C3, white arrow). (D and E) Marked epithelial damage at the surface epithelium, crypt abscesses, bacterial invasion, and inflammation in P18–P25 Gfra1hypo/hypo mice with marked HAEC score. (D) In Gfra1hypo/hypo mice, crypt dilatation (D1, white arrow) along with sparse neutrophil infiltrates in basal (D1, red open arrowheads) and apical (D2, red open arrowheads) crypt epithelium. The superficial epithelium shows multifocal necrotic areas and degenerative changes accompanied by superficial dense infiltrates of neutrophils (ie, microabscesses; D2, green arrowheads). Crypt abscesses (E1, green arrow), dense bacterial aggregates (D3, black arrow), and enterocyte-attaching bacteria (D3 and E2, green bold arrow) also are indicated. Bacteria inside a submucosal blood vessel (E3, black arrow) and a bacterial aggregate in mildly dilated crypt base (E3, white arrow) are present. Single granulocytes (E3, yellow bold arrows) and lymphocytes (E3, yellow arrowheads) are present in the lamina propria and submucosa, indicative of inflammation.
Figure 8.
Histopathologic analysis of HSCR/HAEC in P18–P25 Gfra1hypo/hypomice. (A) A WT colon. (B and C) Focal epithelial ulceration, variable colon wall changes after loss of mucosal integrity, and bacterial invasion in P18–P25 Gfra1hypo/hypo mice. Marked subepithelial edema (B1, grey arrow) and basal edema (B1, green arrow) in the lamina propria, and copious edema in the submucosa (B1, red arrow). Vacuole formation, indicative of acute degenerative changes (B1 and B2, yellow arrow) and mild dilatation of crypts in the colon (B1, white arrow) also are present. Copious numbers of bacteria are attached to the surface epithelium (C1, green bold arrows). Reactive mesothelial cells (B2, green arrowheads) and thin infiltrates of eosinophils (B2, red arrowhead), neutrophils (B2, red open arrowheads), and lymphocytes in the submucosa (B2, yellow open arrowhead) are present. Focal marked epithelial damage with epithelial degeneration and necrosis (B3, black arrow) are present. Subepithelial small hemorrhage (B3, blue arrow) and infiltrating neutrophils (B3, red open arrowheads) are present. Also evident is a relative absence of AB-PAS‒positive cells (compare with panels A2 and C2 in Figure 7), which indicate goblet cell degeneration (B4).
Table 4.
Summary of Histopathologic Findings at P18–P25
| P18–P25 | ID | Weight, g | Sex | Applied Teitelbaum et al7 grade | Surface epithelial injury | Crypt dilation | Intraepithelial apoptotic remnants | Goblet cell hyperplasia | Mucins in crypts |
Bacteria in crypts | Enterocyte adherence | Leukocytes |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mucin shift to PAS | Basal | Surface | Mucus retention | Granulocytes | Lymphocytes | |||||||||||
| WT | 29.12.8-9 | ND | ND | 0 | - | - | - | No | No | AB | PAS | - | - | No | - | - |
| WT | 23.3-9-5 | 11.1 | F | 0 | - | - | - | No | No | AB | PAS | - | - | No | - | - |
| WT | 22.9.10CA-2 | 10.1 | F | 0 | - | - | - | No | No | AB | PAS | - | - | No | - | - |
| WT | 5.1.9-9 | 12.23 | M | 0 | - | - | - | No | No | AB | PAS | - | - | No | - | - |
| WT/hypo | 29.12.8-5 | ND | ND | 0 | - | - | - | No | No | AB | PAS | - | - | No | - | + |
| WT/hypo | 23.3-9-6 | 12.1 | F | 0 | - | - | - | No | No | AB | PAS | - | - | No | - | - |
| WT/hypo | 22.9.10CA-7 | 84 | F | 0 | - | - | - | No | No | AB | PAS | - | - | No | - | - |
| WT/hypo | 22.9.10 CA-8 | 8.9 | F | 0 | - | - | - | No | No | AB | PAS | - | - | No | - | - |
| Hypo/hypo | 29.12.8-8 | ND | ND | III/IV | Marked | ++ | ++ | Degenerated | Yes | PAS | AB | + | + | Yes | ++ | - |
| Hypo/hypo | 23.3-9-4 | 9.4 | M | II | Moderate | ++ | + | Yes | Yes | PAS/ foamy AB | PAS/ foamy AB | ++ | - | No | - | + |
| Hypo/hypo | 22.9.10CA-3 | 8.5 | M | III/IV | Marked | +/++ | ++ | Yes + degenerated | Degenerated | degenerated | Atypical AB | + | - | No | ++ | + |
| Hypo/hypo | 22.9.10 CA-10 | 7.8 | F | II | Moderate | ++/+++ | ++ | Yes | Yes | PAS/ foamy AB | PAS/ foamy AB | ++ | - | No | + | + |
| Hypo/hypo | 5.1.9-7 | 8.8 | M | IV/V | Marked | ++/+++ | +++ | Yes + degenerated | Yes | PAS/ foamy AB | PAS/ foamy AB | +/++ | ++ | Yes | + | + |
| Minimal | Mild + | Mild + | Mild + | Mild + | Normal - (<4/HPF) | |||||||||||
| 0–V | Mild | Moderate ++ | Moderate ++ | Yes/no | Yes/no | Moderate ++ | Moderate ++ | Yes/no | Mild + (5–15/HPF) | |||||||
| Moderate | Marked +++ | Marked +++ | Marked +++ | Moderate ++ (20–30/HPF) | ||||||||||||
| Marked | ||||||||||||||||
NOTE. See the Materials and Methods section for details regarding histopathologic grading.
ND, not determined.
Taken together, these data suggest that Gfra1hypo/hypo animals recapitulate both the early and advanced histopathologic findings present in patients with HAEC.4, 5, 7
Characterization of Cytokine mRNA Expression and Serum Protein Levels in Gfra1hypo/hypo Mice
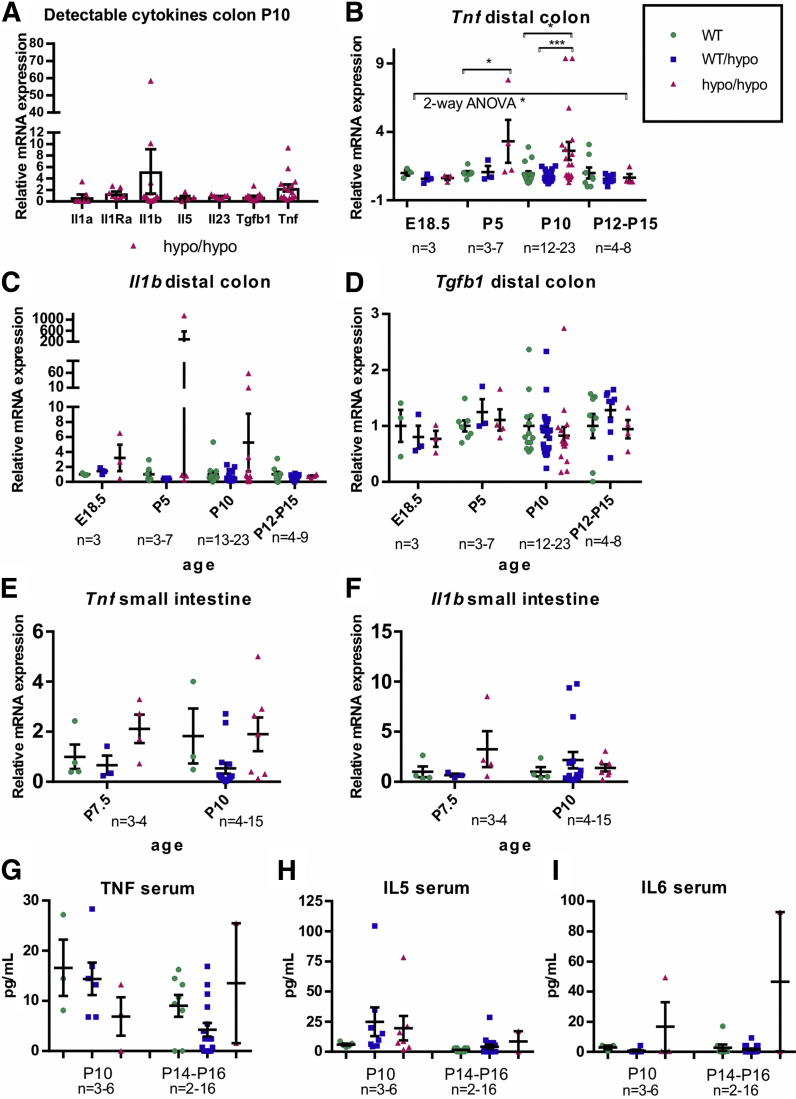
A mouse long-segment HSCR/HAEC model may allow identification of diagnostic markers and further explore the relationship between immune response and HAEC. As a first step in this direction we analyzed the expression of mRNAs for Il1a, Il1b, Il1Ra, Il2, Il4, Il5, Il6, Il10, Il13, Il23, Tgfb1, Tnf, and Ifng selected based on their presumed or hypothetical involvement in GI tract inflammation. First, we performed a pilot study on mRNA expression in the colon at P10, when the HSCR/HAEC phenotype is readily detectable but less variable than at the end stage of the disease. Of 13 selected mRNAs, 7 were detectably expressed at P10 (Figure 9A). Based on those preliminary results Tnf, Il1b, and Tgfb1 mRNA were selected for further analysis at E18,5, P5, and P12–P15, while the n number of animals analyzed at P10 was increased. We observed moderate 2- to 3-fold up-regulation of Tnf at P5 and P10 in the colon of Gfra1hypo/hypo animals (Figure 9B). Il1b mRNA expression showed increased expression in Gfra1hypo/hypo mice with remarkable interanimal variation (Figure 9C). Expression levels of Tgfb1 were similar between genotypes (Figure 9D). The downward trend in Tnf and Il1b mRNA expression in older animals may reflect the fact that more severely affected individuals die before P10 (Table 1). The observed peak in Tnf and Il1b mRNA expression in a few animals therefore may reflect the end stage of the disease, which is associated with acute bacterial enterocyte attachment, tissue invasion, and sepsis.
Figure 9.
Analysis of cytokine mRNA and protein levels. (A–D) qPCR analysis of cytokine mRNA levels in the colon of Gfra1hypo/hypo and control mice at indicated ages. Tnf mRNA levels are up-regulated in P5 colon (P < .05, analysis of variance and Tukey multiple comparisons) and in P10 colon (P < .05, analysis of variance and Tukey multiple comparisons). (E and F) Tnf and Il1b mRNA levels are not up-regulated in the small intestine (a segment spanning the duodenum–jejunum was analyzed). (G–I) Measurements of serum cytokine protein levels by ELISA. In analysis of Gfra1hypo/hypo and control mice at the indicated ages, no significant differences between the genotypes were observed. The number of animals is given for each time point in the Figure panels (qPCR: 2–3 biological replicates, 1–2 independent experiments per animal; ELISA: 2 biological replicates, 1 experiment per animal). All summary data are presented as means ± SEM. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001.
In the small intestine we were unable to detect significant differences in Tnf and Il1b mRNA expression (Figure 9E and F).
We also analyzed cytokine interferon γ, interleukin (IL)12, IL4, IL5, IL6, tumor necrosis factor (TNF), IL1b, and IL9 protein levels in the serum using commercially available enzyme-linked immunosorbent assay (ELISA) analysis tools at P10 and P14–P16. We were able to detect TNF, IL5, and IL6 with no clear genotype–phenotype correlation (Figure 9H and I). We hypothesized that systemic immune activation with notable serum cytokine changes in the Gfra1hypo/hypo HSCR/HAEC mouse model was a late event that was associated with bacterial invasion and sepsis at the end stage.
Discussion
Here, we report that reducing GFRa1 expression in mice does not affect renal development, but is sufficient to cause a long-segment HSCR-like phenotype. Why is ENS development in heterozygous Gfra1WT/KO mice grossly normal, whereas homozygous Gfra1hypo/hypo mice, which have only a 20%–30% further reduction in GFRa1 levels compared with Gfra1WT/KO mice, develop HSCR? Our results using a reporter cell line suggest that GFRa1 levels need to exceed a specific threshold before downstream signaling is activated. It is possible that during ENS development this threshold level of GFRa1 in Gfra1hypo/hypo mice is not reached. Previous attempts to alter GDNF/GFRa1/RET signaling by introducing knock-in alleles for Ret led to defects in both the ENS and kidneys.24 It is possible that the level of RET phosphorylation required for normal development is higher in the developing ENS compared with the developing kidneys. GDNF and GFRa1 also can promote synapse formation in a RET-independent manner via a process called ligand-induced cell adhesion, which also may influence cell migration,48 and therefore synapse formation could be another process in which a reduction in GFRa1 levels may impact ENS development differently from the kidneys. Unfortunately, methods for quantifying RET phosphorylation in vivo are not currently available, thus precluding the ability to quantify RET signaling to discriminate between RET-dependent from RET-independent effects in ENS and kidney development.
Our Gfra1hypo/hypo mouse provides an opportunity to model childhood HSCR and HAEC arising from a defect in the GDNF/GFRa1/RET signaling axis. At 10 days of age, Gfra1hypo/hypo mice recapitulate the principal features of childhood long-segment HSCR and the early features of HAEC, including a failure to thrive, long-segment aganglionosis of the colon, the presence of hypertrophic cholinergic fibers in the distal colon, hyperplasia of goblet cells in the colon, mucus retention, a shift from acidic to neutral mucin production, crypt dilatation, and surface epithelial damage. Epithelial damage included focal epithelial necrosis and modest neutrophil infiltration, corresponding to mild cryptitis in human beings. Importantly, although the presence of most of these pathologic changes varied among the Gfra1hypo/hypo mouse colon samples at P4 and P10, 100% of the Gfra1hypo/hypo mice at both ages had a shift from acidic to neutral mucin production. By P10, 100% of Gfra1hypo/hypo mice also showed similar levels of goblet cell hyperplasia, whereas bacterial enterocyte adherence was not observed in any mice at this age. These results suggest that goblet cell dysfunction in the colon precedes the other pathologic changes, and that bacterial enterocyte adherence is not an initiating factor in HAEC. Gfra1hypo/hypo mice also provided us with an opportunity to study whether the observed altered goblet cell physiology results from the lack of the ENS in the colon and distal parts of the small intestine or developmental reduction in GFRa1 in the mesenchyme of the whole fetal intestine. We found that in the duodenum of Gfra1hypo/hypo mice both the goblet cell numbers and the mucin profile are normal, suggesting that colonic goblet cell hyperplasia results from the lack of ENS.
At a later age (P18–P25), all of the Gfra1hypo/hypo mice studied showed previously known features of HAEC, including copious mucus retention, epithelial damage in the colon, goblet cell hyperplasia mixed with goblet cell degeneration, and crypt dilatation and inflammation. Importantly, enterocyte adherence and bacterial invasion were present in only 2 of the 3 mice that had the most advanced epithelial damage, suggesting that enterocyte adherence and bacterial invasion are late events that mark the end stage of the disease. Taken together, our results suggest that HAEC may be associated with goblet cell dysfunction, which manifests first as a shift from acidic toward neutral mucin production in the early stage, followed by hyperplasia and a continuous mucin shift, and mucin overproduction during the later stages.5 Over time, these features lead to mucus retention and crypt dilation, which in turn contribute to progressive dysfunction of the intestinal barrier, including progressive epithelial damage and ulceration; at the end stage of the disease, these pathologic changes facilitate bacterial enterocyte adherence, bacterial invasion, and, ultimately, sepsis.6, 7 Thus, our results shed light on the sequence of events in HAEC and suggest that addressing goblet cell dysplasia, which could remain in the proximal colon or transition zone after surgical removal of the aganglionic gut segment, may be beneficial in treating or preventing HAEC.
Together, mutations in endothelin-related genes that encode EDN3, EDNRB, and ECE1 account for approximately 5% of all HSCR cases.1 Various animal models of HSCR with defective endothelin signaling have been generated, and substantial progress has been made using those models toward understanding short-segment HSCR and HAEC. A careful comparison between these endothelin-defective models and our Gfra1hypo/hypo mice is an important future objective. Such a study likely would show similarities—and probably also differences—between the 2 genotypes and might pave the way for the development of gene-specific treatment strategies.
To date, more than 10 HSCR susceptibility genes have been identified.1, 2, 9, 11, 49 However, together these genes account for approximately 50% of all HSCR cases.1, 9, 49 Thus, other genetic mutations and/or traits likely underlie HSCR. At present, neither GFRa1 protein nor GFRa1 mRNA levels are routinely measured in colon samples resected from HSCR patients. One study using semiquantitative PCR reported reduced levels of GFRa1 mRNA in 3 of 13 HSCR patients analyzed.33 Given that our results show that reducing GFRa1 levels is sufficient to cause HSCR in mice, measuring GFRa1 levels in a larger cohort of HSCR patients in conjunction with epigenetic and genomic sequencing analysis is an important future objective.
Materials and Methods
Animals
Gfra1hypo mice (registered in the Mouse Genome Database as the 129Ola/ICR/C57BL6-Gfra1tm1Joao mouse line) were generated by inserting a puΔtk cassette31 into intron 6 of the Gfra1 gene using gene targeting via homologous recombination in embryonic stem cells (Figure 1A–C). Embryonic stem cell clones that had undergone homologous recombination in 1 Gfra1 allele were identified by performing Southern blot analysis on EcoRV-digested genomic DNA using the probe indicated in Figure 1A (expected sizes: targeted, 7564 bp; WT, 10397 bp). Routine PCR-based genotyping of the offspring was performed using the primers indicated in Figure 1A (GfraloxF forward: 5’-cattggccaggtgaaagaca; Gfra1loxR reverse: 5’-agaaagagagatgatcacagtacacatg). The mice were maintained on a 129Ola/ICR/C57BL6 mixed genetic background, housed under a 12-hour/12-hour light/dark cycle at 20°C–22°C, with 1 mother and litter per cage; standard chow and water were available ad libitum. All animal experiments were approved by the National Animal Experiment Board of Finland.
Tissue Processing
Paraffin-embedded sections, whole-mount GI tracts (E13.5), and myenteric plexuses (P10 or P18–P25) were used for histologic preparations. Comparable samples obtained from the duodenum, ileum, and colon were analyzed from all genotypes, using littermates as controls. The samples were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 24 hours at room temperature. Automated dehydration of the tissues and subsequent paraffin embedding were performed using an ASP300 S tissue processor (Leica, Wetzlar, Germany). Paraffin blocks were cut to prepare 5-μm sections using a Tissue-Tek microtome (Sakura, Alphen aan den Rijn, The Netherlands). For whole-mount histology of P10 and P18–P25 tissues, the mice were perfused intracardially with PBS and 4% paraformaldehyde. The longitudinal muscle/myenteric plexus (LMMP) preparations were isolated under a dissection microscope by peeling off the outer muscle layer of the GI tract and postfixed for 15 minutes in paraformaldehyde.
Histopathology
Paraffin sections of the duodenum and colon were stained with H&E, Gram stain, or a combination of AB (pH 2.5) and PAS using standard protocols. LMMPs from the small and large intestine were stained for acetylcholinesterase as previously described.50 Briefly, LMMP was washed in PBS for 1 hour, 100 mmol/L maleic acid buffer pH 6.0 for 5 minutes, pre-incubated for 30 minutes in 65 mmol/L maleic acid buffer including 0.5 mol/L potassium ferricyanide, 4 mmol/L copper sulfate, and 5 mmol/L sodium citrate, and reacted for 30–45 minutes in 5 mmol/L acetylthiocholine iodide in 100 mmol/L maleic acid buffer. NADPH diaphorase was used to visualize nitrergic neurons in LMMPs; briefly, LMMPs were washed 2 times 1 hour in PBS and reacted 1 hour at +37°C in a solution prepared right before use containing 0.3% Triton (Sigma) X-100, 0.01% nitro blue tetrazolium chloride, and 0.1% NADPH in PBS. Finally, the LMMP preparations were washed 6 times for 5 minutes in PBS and mounted in glycerol for imaging.
Histopathology Grading
The criteria for histopathologic grading are defined later. HAEC-associated inflammatory cell infiltration of the crypts (cryptitis and crypt abscesses) was milder in mice than in human beings, whereas epithelial pathology characteristic of HAEC was recapitulated well in mice. Therefore, we modified the grading system reported by Teitelbaum et al7 to primarily reflect epithelial pathology.
Surface epithelial damage
Epithelial damage was graded as minimal, mild, moderate, or marked.
Minimal surface epithelial damage
The surface epithelium consists of mild disorganization and slight degeneration of the surface epithelium. In contrast to regular palisading, colonocytes are clustered multifocally in disorganized, undulating rows with partially overlapping nuclei. Apical eosinophilia and indistinct cell borders are also visible focally.
Mild surface epithelial damage
The surface epithelium has mild to moderate disorganization and mild degenerative changes. The size and shape of the epithelial cells and nuclei vary, and they grow in disorganized, undulating rows with partially overlapping nuclei. Apical cytoplasms are generally homogenously eosinophilic or vacuolar, and the cell borders are indistinct; cells are occasionally low columnar to cuboidal in shape. Scattered intraepithelial apoptotic/necrotic cell remnants are present.
Moderate surface epithelial damage
The lumen of the intestine contains copious amounts of mucus and deeply eosinophilic debris of shed epithelial cells. The surface epithelium has multifocal to diffuse moderate degeneration and cuboidal to flattened cells with indistinct cell borders and intense staining. Increased numbers of apoptotic/necrotic cells or cell remnants are present, as well as clusters of apoptotic/necrotic cells. Scattered intraepithelial neutrophils (1–5 per high-power field [HPF]) may be present in the lamina propria. Mildly increased numbers of lymphocytes in the basal lamina propria (indicative of mild chronic colitis) were detected in 1 mouse; however, in accordance with a previous report on HAEC in mice,44 inflammatory activity is generally very low.
Marked surface epithelial damage
The surface epithelium has generally diffuse degeneration, and multiple necrotic areas accompanied by small superficial infiltrates of neutrophils (microabscesses) are present. The apical parts of the crypts of Lieberkühn contain few to several intraepithelial neutrophils with sporadic crypt abscesses. The goblet cells are degenerated. The lamina propria contains mildly to moderately increased numbers of eosinophils and neutrophils in the basal areas and between the crypts. The inflammatory reaction in the intestinal wall is mild.
Loss of integrity of the gut wall/bacterial invasion
The surface epithelium shows gross changes of varying degrees. The lamina propria has mild to moderate edema and mildly to moderately increased numbers of eosinophils, neutrophils, and lymphocytes. Profuse bacterial accumulates are present in some of the moderately to markedly dilated crypts of Lieberkühn. The submucosa has marked edema, sparse infiltrates of leukocytes, and small numbers of bacteria. The circular and longitudinal muscle layers have marked diffuse degeneration and moderate neutrophilic infiltrates. Mesothelial cells are reactive.
Crypt dilatation
Crypt dilatation was graded as minimal to mild (+), moderate (++), or marked (+++) to quantify the extent of the changes. In cases in which large segmental variation was present, both grades were reported.
Mild crypt dilatation
The dilatation is present primarily at the base of the crypts, and any from a few to approximately half of the crypts are affected.
Moderate crypt dilatation
Most of the crypts are affected.
Marked crypt dilatation
The apical areas of the crypts also are affected, and most of the crypts are affected.
Leukocyte numbers in the mucosa
A 3-tier system was used to grade leukocyte numbers as minimal to mild (+), moderate (++), and marked (+++).
Lymphocytes
Lymphocyte number is considered to be normal if 5 or fewer cells are generally present in the lamina propria between the crypts per HPF. A minimal to mild increase in lymphocytes is scored if they fill the intercryptal region, and they may be increased basally below the crypt bases. There is no increase in separation of the crypts. A moderate increase is scored if the lymphocytes increase the separation of the crypts; a marked increase is scored if there is a diffuse distribution of lymphocytes with distortion of the cryptal architecture. In accordance with a previous report of HAEC in mice,44 none of our samples contained moderate or marked increases in lymphocytes. Accordingly, extremely few plasma cells were detected, and plasma cells therefore were excluded from grading.
Granulocytes
Eosinophils and neutrophils were graded as a single entity. Neutrophils related to surface epithelial damage (eg, crypt abscesses, microabscesses in the epithelium, and epithelial neutrophils) were omitted from this section and were graded under surface epithelial damage.
Granulocyte number is considered to be normal if no more than 3–4 granulocytes typically are present per HPF in the lamina propria. A minimal to mild increase is scored if approximately 15 granulocytes are present per HPF in the lamina propria. A moderate increase is scored if the infiltrate contains 20–30 granulocytes per HPF; this may be accompanied by macrophages. A marked increase is scored if granulocytes are the dominant population and are not easily counted per HPF.
Quantification of Goblet Cells
Goblet cells were counted in the distal third of the colon from the basal half of the crypts. Twenty to 30 crypts were counted per sample. Goblet cells that primarily were stained light blue were counted as AB+ and goblet cells stained primarily magenta or purple were counted as PAS+. Goblet cells fitting neither category were counted as AB-PAS+. Sample genotypes were blinded for the counter.
Immunohistochemistry
Sections were deparaffinized through serial washes with xylene, alcohol, and water. Antigen retrieval was performed by boiling the samples for 10 minutes in citrate buffer (10 mmol/L sodium citrate, 0.05% Tween 20, pH 6.0). Endogenous peroxidase activity was quenched in 1:53 H2O2 standard solution in Tris-buffered saline (TBS) for 30 minutes at room temperature. After washing with TBS containing 0.1% Tween 20, the tissue was blocked in 1.5% normal goat serum in TBS containing 0.1% Tween 20 for 30 minutes at room temperature. The sections were incubated overnight at 4°C in primary antibody solution containing rabbit anti-ubiquitin C-terminal hydrolase L1 (1:250, BML-PG9500; Enzo Life Sciences) or rabbit anti–glial fibrillary acidic protein (1:200, Ab-4 RB-087-A; Thermo-Scientific). After washing, the sections were incubated for 90 minutes at room temperature in secondary antibody solution containing biotinylated anti-rabbit antibody (1:200; Vector Laboratories) or goat anti-rabbit Alexa 488 antibody (1:400, A11034; Invitrogen). The biotinylated secondary antibody signal was enhanced using the ABC reaction kit (PK-4001, Vector Laboratories) and visualized using 3,3′-diaminobenzidine (SK-4100, Vector Laboratories). Samples were dehydrated through serial washes with water, alcohol, and xylene, and mounted using Depex (Science Services GmbH, München, Germany). Fluorescence-labeled, whole-mount samples were mounted in glycerol and imaged using an Olympus BX-61 microscope. As negative controls, either the primary or secondary antibody was omitted.
ELISA
ELISA was performed with commercial kits according to the manufacturer’s instructions (Invitrogen ProcartaPlex multiplex interferon γ, IL12, IL4, IL5, IL6, TNF, EPX060-20831-90, IL1B Mouse ProcartaPlex Simplex Kit, EPX01A-26002-901, and IL9 Mouse ProcartaPlex Simplex Kit, EPX01A-26041-901).
Quantitative PCR
RNA isolation
RNA was isolated from snap-frozen tissues using the TRIzol reagent (Invitrogen) for samples collected from E18.5 on or the RNAqueous Micro-kit (AM1931; Life Technologies) in accordance with the manufacturer’s instructions. DNase I was supplied in the same kit.
Reverse transcription
First-strand complementary DNA (cDNA) was synthesized from 50 to 300 ng of RNA (with equal amounts of starting RNA used in each experiment) using random hexamer primers in a final volume of 20 μL (Transcriptor First-Strand cDNA synthesis kit; Roche). In brief, 2 μL of random hexamer primers was mixed with 11 μL of RNA sample diluted in nuclease-free water and incubated at 65°C for 10 minutes. Next, 7 μL of a mixture containing 4 μL of 5× reverse transcriptase buffer, 2 μL of 100 mmol/L deoxynucleoside triphosphate, 0.5 μL of RNase inhibitor, and 0.5 μL of Transcriptor reverse transcriptase (Transcriptor First-Strand cDNA synthesis kit; Roche) were added, mixed gently, and incubated at 25°C for 10 minutes, 55°C for 30 minutes, and 85°C for 5 minutes. A control sample containing no reverse transcriptase was included in each experiment. The cDNA was cooled on ice, diluted 1:15 in water, and either stored at -20°C or used immediately for quantitative PCR (qPCR).
Real-time qPCR
qPCR was performed using the LightCycler 480 real-time PCR system (Roche Diagnostics) with the LightCycler 480 SYBR Green I Master mix and 2.5 pmol of primers at a final volume of 10 μL. The reactions were run on white 384-well plates sealed with adhesive plate sealers (04729749001; Roche Diagnostics). Each reaction contained 2.5 μL of diluted cDNA. Each qPCR run contained 2–3 replicates of each reaction. The following qPCR program was used: pre-incubation: 10 minutes at 95°C; amplification: 10 seconds at 95°C, 15 seconds at 60°C, and 15 seconds at 72°C for 45 cycles; melting curve: 5 seconds at 95°C, 30 seconds at 55°C, continuous acquisition mode at 95°C with 2 acquisitions per degree Celsius; and cooling: 10 seconds at 40°C. The data were analyzed using LightCycler 480 Software Release 1.5.0 SP1, with the Absolute Quantification/2nd Derivative Max calculation. Actb was amplified as a reference gene. The following mus musculus (m) primer sets were used in this study: mActb: 5’-ctaaggccaaccgtgaaaag and 5’-accagaggcatacagggaca; mGdnf ex2–3: 5’-cgctgaccagtgactccaatatgc and 5’-tgccgcttgtttatctggtgacc; mGfra1: 5’-ttcccacacacgttttacca and 5’-gcccgatacattggatttca; mIfng: 5’-ttcttcagcaacagcaaggc and 5’-tcagcagcgactccttttcc; mIl1b: 5’-agttgacggaccccaaaag and 5’-agctggatgctctcatcagg; mIl1a: 5’-gtcggcaaagaaatcaagatg and 5’-gtcttcgttttcactgtaacag; mIl1Ra: 5’-aaccagctcattgctgggtactta and 5’-gcccaagaacacactatgaaggtc; mIl2: 5’-ttgtgctccttgtcaacagc and 5’-ctggggagtttcaggttcct; mIl4: 5’-aacgaggtcacaggagaagg and 5’-tctgcagctccatgagaaca; mIl5: 5’-accgagctctgttgacaag and 5’-tcctcgccacacttctcttt; mIl6: 5’-accacttcacaagtcggagg and 5’-tgcaagtgcatcatcgttgt; mIl10: 5’-ataactgcacccacttccca and 5’-cttggcaacccaagtaaccc; mIl13: 5’-gcagcatggtatggagtgtg and 5’-tggcgaaacagttgctttgt; mIl23: 5’-gctgtgcctaggagtagcag and 5’-tggctgttgtccttgagtcc; mRet t2 : 5’-tcccttccacatggattga and 5’-atcggctctcgtgagtggta; mTgfb1: 5’-tggagcaacatgtggaactc and 5’-gtcagcagccggttacca; and mTnf: 5’-gatcggtccccaaagggatg and 5’-tgagggtctgggccatagaa.
Western Blot Analysis
Snap-frozen E12.5 tissue was homogenized in a homogenization solution containing 0.3 mol/L sucrose, 10 mmol/L HEPES, 1 mmol/L EDTA (pH 7.2–7.4), and a proteinase inhibitor (Complete Mini-Tabs Cocktail Set; Roche); the protein concentration was measured using the Lowry method (Bio-Rad). A total of 5–10 μg of protein was mixed with Laemmli sample buffer and incubated at 95°C for 5 minutes. The proteins were separated on an 8% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane (Amersham Protran, GE Healthcare). After washing and blocking at room temperature for 60 minutes in 5% milk, the membranes were incubated in goat anti-Gfra1 antibody (1:1000, GT15004; Neuromics). After washing, the membranes were incubated at room temperature for 2 hours in horseradish peroxidase–conjugated anti-goat antibody (1:2000, P0449; Dako). The protein bands were visualized using enhanced chemiluminescence substrate (Pierce). As a loading control, the membranes then were stripped and reprobed using anti–α-tubulin antibody (1:60,000, T9026; Sigma), followed by horseradish peroxidase–conjugated donkey anti-mouse antibody (1:2000, P0449; Dako). Three independent Western blots were quantified with ImageJ 1.50a software (National Institutes of Health, Bethesda, MD) and an unpaired Student t test was performed as a statistical analysis using GraphPad Prism software.
Luciferase Assay
RET-dependent activation of mitogen-activated protein kinase by soluble GFRa1 was measured using MG87 fibroblasts that stably express RET (MG87RET) and the PathDetect-Elk1 system (219005; Stratagene). One day before the assay, cells were plated in a 96-well plate (20,000 cells/well) in 100 μL/well Dulbecco's modified Eagle medium containing 10% fetal bovine serum and 100 μg/mL normocin (ant-nr-1; InvivoGEN) and cultured overnight in an incubator with 5% CO2. Soluble GFRa1 (1–5000 ng/mL, 560-GR; R&D Systems) and GDNF (100 ng/mL 450-10; Peprotech) were added to wells in quadruplicate per GFRa1 concentration. The plates then were cultured for an additional 22–24 hours for luciferase expression, after which the culture media was discarded and the cells were incubated with NeoLite reagent (6016711; Perkin Elmer) for 10 minutes. Luciferase activity was measured using a MicroBeta 2 (PerkinElmer) plate counter. Each experiment was performed twice, and the results were analyzed using GraphPad Prism software.
Acknowledgments
L. Lauriina Porokuokka and Heikki T. Virtanen are currently affiliated with the Department of Pharmacology, Faculty of Medicine & Helsinki Institute of Life Science, University of Helsinki, Helsinki, Finland, P.O. Box 63, 00014 Helsinki, Finland.
Jaan-Olle Andressoo is currently affiliated with the Department of Pharmacology, Faculty of Medicine & Helsinki Institute of Life Science, University of Helsinki, Helsinki, Finland, P.O. Box 63, 00014 Helsinki, Finland and Division of Neurogeriatrics, Department of Neurobiology, Care Sciences and Society, Karolinska Institutet SE-171 77 Stockholm, Sweden.
The authors would like to thank Ms Susanne Dahlström, Ms Jenni Montonen, and personnel of the Finnish Centre for Laboratory Animal Pathology for help with experimental procedures, Ms Ana Montaño-Rodríguez and Ms Sonja Huovila for help with the artwork, and Mr Mark C. Cowlishaw for English language editing.
Footnotes
Author contributions L. Lauriina Porokuokka acquired, analyzed, and interpreted the data, drafted the manuscript, and performed the statistical analysis; Heikki T. Virtanen acquired, analyzed, and interpreted the data; Jere Linden acquired, analyzed, and interpreted the histopathologic data and drafted the manuscript; Yulia A. Sidorova acquired, analyzed, and interpreted the data and drafted the manuscript; Tatiana Danilova acquired, analyzed, and interpreted the data; Maria Lindahl analyzed and interpreted the data, was responsible for the study design, and critically read the manuscript; Mart Saarma initiated the in vivo GFRa1 studies, critically read the manuscript, and provided funding; and Jaan-Olle Andressoo designed and generated glial cell line–derived neurotrophic factor family receptor a1 hypomorphic mice, acquired and interpreted the data, was responsible for the study design, drafted the manuscript, and provided funding.
Conflicts of interest The authors disclose no conflicts.
Funding Supported by the Integrative Life Science doctoral program (L.P.); Academy of Finland grant 117044 (M.L.); grants from the Sigrid Juselius Foundation, grant 1186236 from the Academy of Finland, grant R21-AT008265 from the National Institutes of Health, and the Institute of Biotechnology, University of Helsinki (M.S.); by Academy of Finland grant 297727, Sigrid Juselius Foundation, Faculty of Medicine at the University of Helsinki, the Helsinki Institute of Life Science Fellow grant, and by the European Research Council under the European Union’s Horizon 2020 research and innovation program, grant agreement 724922 (J.O.A.).
References
- 1.Amiel J., Sproat-Emison E., Garcia-Barcelo M., Lantieri F., Burzynski G., Borrego S., Pelet A., Arnold S., Miao X., Griseri P., Brooks A.S., Antinolo G., de Pontual L., Clement-Ziza M., Munnich A., Kashuk C., West K., Wong K.K., Lyonnet S., Chakravarti A., Tam P.K., Ceccherini I., Hofstra R.M., Fernandez R., Hirschsprung Disease C. Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet. 2008;45:1–14. doi: 10.1136/jmg.2007.053959. [DOI] [PubMed] [Google Scholar]
- 2.Butler Tjaden N.E., Trainor P.A. The developmental etiology and pathogenesis of Hirschsprung disease. Transl Res. 2013;162:1–15. doi: 10.1016/j.trsl.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lister J., Tam P.K.H. Hirschsprung's disease. In: Lister J., Irving I.M., editors. Neonatal surgery. Butterworth; London: 1990. pp. 523–546. [Google Scholar]
- 4.Frykman P.K., Short S.S. Hirschsprung-associated enterocolitis: prevention and therapy. Semin Pediatr Surg. 2012;21:328–335. doi: 10.1053/j.sempedsurg.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy F., Puri P. New insights into the pathogenesis of Hirschsprung's associated enterocolitis. Pediatr Surg Int. 2005;21:773–779. doi: 10.1007/s00383-005-1551-1. [DOI] [PubMed] [Google Scholar]
- 6.Demehri F.R., Halaweish I.F., Coran A.G., Teitelbaum D.H. Hirschsprung-associated enterocolitis: pathogenesis, treatment and prevention. Pediatr Surg Int. 2013;29:873–881. doi: 10.1007/s00383-013-3353-1. [DOI] [PubMed] [Google Scholar]
- 7.Teitelbaum D.H., Caniano D.A., Qualman S.J. The pathophysiology of Hirschsprung's-associated enterocolitis: importance of histologic correlates. J Pediatr Surg. 1989;24:1271–1277. doi: 10.1016/s0022-3468(89)80566-4. [DOI] [PubMed] [Google Scholar]
- 8.Burzynski G.M., Nolte I.M., Bronda A., Bos K.K., Osinga J., Plaza Menacho I., Twigt B., Maas S., Brooks A.S., Verheij J.B., Buys C.H., Hofstra R.M. Identifying candidate Hirschsprung disease-associated RET variants. Am J Human Genet. 2005;76:850–858. doi: 10.1086/429589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldstein A.M., Hofstra R.M., Burns A.J. Building a brain in the gut: development of the enteric nervous system. Clin Genet. 2013;83:307–316. doi: 10.1111/cge.12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kenny S.E., Tam P.K., Garcia-Barcelo M. Hirschsprung's disease. Semin Pediatr Surg. 2010;19:194–200. doi: 10.1053/j.sempedsurg.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Plaza-Menacho I., Burzynski G.M., de Groot J.W., Eggen B.J., Hofstra R.M. Current concepts in RET-related genetics, signaling and therapeutics. Trends Genet. 2006;22:627–636. doi: 10.1016/j.tig.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Jiang Q., Ho Y.Y., Hao L., Nichols Berrios C., Chakravarti A. Copy number variants in candidate genes are genetic modifiers of Hirschsprung disease. PLoS One. 2011;6:e21219. doi: 10.1371/journal.pone.0021219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallace A.S., Anderson R.B. Genetic interactions and modifier genes in Hirschsprung's disease. World J Gastroenterol. 2011;17:4937–4944. doi: 10.3748/wjg.v17.i45.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Airaksinen M.S., Saarma M. The GDNF family: signalling, biological functions and therapeutic value. Nat Rev Neurosci. 2002;3:383–394. doi: 10.1038/nrn812. [DOI] [PubMed] [Google Scholar]
- 15.Sasselli V., Pachnis V., Burns A.J. The enteric nervous system. Dev Biol. 2012;366:64–73. doi: 10.1016/j.ydbio.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 16.Young H.M., Hearn C.J., Ciampoli D., Southwell B.R., Brunet J.F., Newgreen D.F. A single rostrocaudal colonization of the rodent intestine by enteric neuron precursors is revealed by the expression of Phox2b, Ret, and p75 and by explants grown under the kidney capsule or in organ culture. Dev Biol. 1998;202:67–84. doi: 10.1006/dbio.1998.8987. [DOI] [PubMed] [Google Scholar]
- 17.Heanue T.A., Pachnis V. Enteric nervous system development and Hirschsprung's disease: advances in genetic and stem cell studies. Nat Rev Neurosci. 2007;8:466–479. doi: 10.1038/nrn2137. [DOI] [PubMed] [Google Scholar]
- 18.Uesaka T., Nagashimada M., Yonemura S., Enomoto H. Diminished Ret expression compromises neuronal survival in the colon and causes intestinal aganglionosis in mice. J Clin Invest. 2008;118:1890–1898. doi: 10.1172/JCI34425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uesaka T., Nagashimada M., Enomoto H. GDNF signaling levels control migration and neuronal differentiation of enteric ganglion precursors. J Neurosci. 2013;33:16372–16382. doi: 10.1523/JNEUROSCI.2079-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enomoto H., Araki T., Jackman A., Heuckeroth R.O., Snider W.D., Johnson E.M., Jr., Milbrandt J. GFR alpha1-deficient mice have deficits in the enteric nervous system and kidneys. Neuron. 1998;21:317–324. doi: 10.1016/s0896-6273(00)80541-3. [DOI] [PubMed] [Google Scholar]
- 21.Pichel J.G., Shen L., Sheng H.Z., Granholm A.C., Drago J., Grinberg A., Lee E.J., Huang S.P., Saarma M., Hoffer B.J., Sariola H., Westphal H. Defects in enteric innervation and kidney development in mice lacking GDNF. Nature. 1996;382:73–76. doi: 10.1038/382073a0. [DOI] [PubMed] [Google Scholar]
- 22.Schuchardt A., D'Agati V., Larsson-Blomberg L., Costantini F., Pachnis V. Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature. 1994;367:380–383. doi: 10.1038/367380a0. [DOI] [PubMed] [Google Scholar]
- 23.Gianino S., Grider J.R., Cresswell J., Enomoto H., Heuckeroth R.O. GDNF availability determines enteric neuron number by controlling precursor proliferation. Development. 2003;130:2187–2198. doi: 10.1242/dev.00433. [DOI] [PubMed] [Google Scholar]
- 24.de Graaff E., Srinivas S., Kilkenny C., D'Agati V., Mankoo B.S., Costantini F., Pachnis V. Differential activities of the RET tyrosine kinase receptor isoforms during mammalian embryogenesis. Genes Dev. 2001;15:2433–2444. doi: 10.1101/gad.205001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uesaka T., Jain S., Yonemura S., Uchiyama Y., Milbrandt J., Enomoto H. Conditional ablation of GFRalpha1 in postmigratory enteric neurons triggers unconventional neuronal death in the colon and causes a Hirschsprung's disease phenotype. Development. 2007;134:2171–2181. doi: 10.1242/dev.001388. [DOI] [PubMed] [Google Scholar]
- 26.Wu L., Gu J., Cui H., Zhang Q.Y., Behr M., Fang C., Weng Y., Kluetzman K., Swiatek P.J., Yang W., Kaminsky L., Ding X. Transgenic mice with a hypomorphic NADPH-cytochrome P450 reductase gene: effects on development, reproduction, and microsomal cytochrome P450. J Pharmacol Exp Ther. 2005;312:35–43. doi: 10.1124/jpet.104.073353. [DOI] [PubMed] [Google Scholar]
- 27.Meyers E.N., Lewandoski M., Martin G.R. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- 28.Nagy A., Moens C., Ivanyi E., Pawling J., Gertsenstein M., Hadjantonakis A.K., Pirity M., Rossant J. Dissecting the role of N-myc in development using a single targeting vector to generate a series of alleles. Curr Biol. 1998;8:661–664. doi: 10.1016/s0960-9822(98)70254-4. [DOI] [PubMed] [Google Scholar]
- 29.Wei Y., Zhou X., Fang C., Li L., Kluetzman K., Yang W., Zhang Q.Y., Ding X. Generation of a mouse model with a reversible hypomorphic cytochrome P450 reductase gene: utility for tissue-specific rescue of the reductase expression, and insights from a resultant mouse model with global suppression of P450 reductase expression in extrahepatic tissues. J Pharmacol Exp Ther. 2010;334:69–77. doi: 10.1124/jpet.110.167411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang B., He L., Dong H., Dalton T.P., Nebert D.W. Generation of a Slc39a8 hypomorph mouse: markedly decreased ZIP8 Zn(2)(+)/(HCO(3)(-))(2) transporter expression. Biochem Biophys Res Commun. 2011;410:289–294. doi: 10.1016/j.bbrc.2011.05.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y.T., Bradley A. A new positive/negative selectable marker, puDeltatk, for use in embryonic stem cells. Genesis. 2000;28:31–35. doi: 10.1002/1526-968x(200009)28:1<31::aid-gene40>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 32.Durbec P.L., Larsson-Blomberg L.B., Schuchardt A., Costantini F., Pachnis V. Common origin and developmental dependence on c-ret of subsets of enteric and sympathetic neuroblasts. Development. 1996;122:349–358. doi: 10.1242/dev.122.1.349. [DOI] [PubMed] [Google Scholar]
- 33.Lui V.C., Samy E.T., Sham M.H., Mulligan L.M., Tam P.K. Glial cell line-derived neurotrophic factor family receptors are abnormally expressed in aganglionic bowel of a subpopulation of patients with Hirschsprung's disease. Lab Invest. 2002;82:703–712. doi: 10.1097/01.lab.0000017364.13014.ae. [DOI] [PubMed] [Google Scholar]
- 34.Worley D.S., Pisano J.M., Choi E.D., Walus L., Hession C.A., Cate R.L., Sanicola M., Birren S.J. Developmental regulation of GDNF response and receptor expression in the enteric nervous system. Development. 2000;127:4383–4393. doi: 10.1242/dev.127.20.4383. [DOI] [PubMed] [Google Scholar]
- 35.Cacalano G., Farinas I., Wang L.C., Hagler K., Forgie A., Moore M., Armanini M., Phillips H., Ryan A.M., Reichardt L.F., Hynes M., Davies A., Rosenthal A. GFRalpha1 is an essential receptor component for GDNF in the developing nervous system and kidney. Neuron. 1998;21:53–62. doi: 10.1016/s0896-6273(00)80514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sidorova Y.A., Matlik K., Paveliev M., Lindahl M., Piranen E., Milbrandt J., Arumae U., Saarma M., Bespalov M.M. Persephin signaling through GFRalpha1: the potential for the treatment of Parkinson's disease. Mol Cell Neurosci. 2010;44:223–232. doi: 10.1016/j.mcn.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 37.Ryan E.T., Ecker J.L., Christakis N.A., Folkman J. Hirschsprung's disease: associated abnormalities and demography. J Pediatr Surg. 1992;27:76–81. doi: 10.1016/0022-3468(92)90111-j. [DOI] [PubMed] [Google Scholar]
- 38.Payette R.F., Tennyson V.M., Pham T.D., Mawe G.M., Pomeranz H.D., Rothman T.P., Gershon M.D. Origin and morphology of nerve fibers in the aganglionic colon of the lethal spotted (ls/ls) mutant mouse. J Comp Neurol. 1987;257:237–252. doi: 10.1002/cne.902570209. [DOI] [PubMed] [Google Scholar]
- 39.Tam P.K., Boyd G.P. Origin, course, and endings of abnormal enteric nerve fibres in Hirschsprung's disease defined by whole-mount immunohistochemistry. J Pediatr Surg. 1990;25:457–461. doi: 10.1016/0022-3468(90)90394-o. [DOI] [PubMed] [Google Scholar]
- 40.Niemi M., Kouvalainen K., Hjelt L. Cholinesterases and monoamine oxidase in congenital megacolon. J Pathol Bacteriol. 1961;82:363–366. doi: 10.1002/path.1700820214. [DOI] [PubMed] [Google Scholar]
- 41.Kamijo K., Hiatt R.B., Koelle G.B. Congenital megacolon; a comparison of the spastic and hypertrophied segments with respect to cholinesterase activities and sensitivities to acetylcholine, DFP and the barium ion. Gastroenterology. 1953;24:173–185. [PubMed] [Google Scholar]
- 42.Bealer J.F., Natuzzi E.S., Buscher C., Ursell P.C., Flake A.W., Adzick N.S., Harrison M.R. Nitric oxide synthase is deficient in the aganglionic colon of patients with Hirschsprung's disease. Pediatrics. 1994;93:647–651. [PubMed] [Google Scholar]
- 43.Vanderwinden J.M., De Laet M.H., Schiffmann S.N., Mailleux P., Lowenstein C.J., Snyder S.H., Vanderhaeghen J.J. Nitric oxide synthase distribution in the enteric nervous system of Hirschsprung's disease. Gastroenterology. 1993;105:969–973. doi: 10.1016/0016-5085(93)90938-9. [DOI] [PubMed] [Google Scholar]
- 44.Cheng Z., Dhall D., Zhao L., Wang H.L., Doherty T.M., Bresee C., Frykman P.K. Murine model of Hirschsprung-associated enterocolitis. I: phenotypic characterization with development of a histopathologic grading system. J Pediatr Surg. 2010;45:475–482. doi: 10.1016/j.jpedsurg.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pelaseyed T., Bergstrom J.H., Gustafsson J.K., Ermund A., Birchenough G.M., Schutte A., van der Post S., Svensson F., Rodriguez-Pineiro A.M., Nystrom E.E., Wising C., Johansson M.E., Hansson G.C. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev. 2014;260:8–20. doi: 10.1111/imr.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiagarajah J.R., Yildiz H., Carlson T., Thomas A.R., Steiger C., Pieretti A., Zukerberg L.R., Carrier R.L., Goldstein A.M. Altered goblet cell differentiation and surface mucus properties in Hirschsprung disease. PLoS One. 2014;9:e99944. doi: 10.1371/journal.pone.0099944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakamura H., Tomuschat C., Coyle D., O’Donnel A.-M., Lim T., Puri P. Altered goblet cell function in Hirschsprung’s disease. Pediatr Surg Int. 2018;34:121–128. doi: 10.1007/s00383-017-4178-0. [DOI] [PubMed] [Google Scholar]
- 48.Ledda F., Paratcha G., Sandoval-Guzman T., Ibanez C.F. GDNF and GFRalpha1 promote formation of neuronal synapses by ligand-induced cell adhesion. Nat Neurosci. 2007;10:293–300. doi: 10.1038/nn1855. [DOI] [PubMed] [Google Scholar]
- 49.Kapur R.P. Practical pathology and genetics of Hirschsprung's disease. Semin Pediatr Surg. 2009;18:212–223. doi: 10.1053/j.sempedsurg.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 50.Vincent S.R. Histochemistry of endogenous enzymes. In: Bolam J.P., editor. Experimental neuroanatomy: a practical approach. IRL Press; Oxford: 1992. pp. 154–155. [Google Scholar]