Abstract
Effect of miRNA-200c (miR-200c) on the proliferation, invasion and apoptosis of prostate cancer cell line LNCaP was investigated. The difference in miR-200c expression was observed using RT-qPCR in the NC group (transfected empty plasmid), simulation group (simulation sequence) and inhibition group (transferred inhibition sequence), which were established by transfecting LNCaP cells with a kit. The proliferation, invasion and apoptosis of cells after transfection were detected using the cell counting kit-8 (CCK-8) method, Transwell chamber and flow cytometry. RT-qPCR detection showed that the relative expression of miR-200c in LNCaP cells significantly increased compared with RWPE-1 cells (P<0.05). The difference was statistically significant in the relative expression of miR-200c cells among NC group, simulation group and inhibition group after transfection (P<0.05) and they significantly decreased in NC group of cells compared with the simulation group (P<0.05). CCK-8 detection showed that there were differences at the 2nd, 3rd, 4th and 5th days of growth in the NC group, simulation group and inhibition group of cells (P<0.05) and there was a difference in the proliferation ability between NC group and simulation group (P<0.05). Transwell chamber detection showed that there was a difference in the invasion ability among NC group, simulation group and inhibition group of cells (P<0.05), among which the number of passed membrane cells in inhibition group was significantly smaller than that in NC group and simulation group (P<0.05), and the difference was not statistically significant between NC group and simulation group (P>0.05). Flow cytometry detection of the apoptosis ability of each group of cells showed that there was a difference in the apoptotic rate in the NC, simulation and inhibition groups (P<0.05). The low expression of miR-200c is beneficial to inhibit the proliferation and invasion of LNCaP cells in vitro and to promote apoptosis, which may be a potential target for prostate cancer biotherapy.
Keywords: prostate cancer, miR-200c, proliferation, invasion, apoptosis, LNCaP cells
Introduction
Prostate cancer (PCa) is a common malignancy in male patients. Statistics have shown that (1) PCa is second only to lung cancer in middle-aged and elderly male malignancies. PCa patients in the United States have surpassed lung cancer patients and it has become a malignancy with the highest incidence (2). Although the incidence of PCa in developing countries is not as high as in developed countries in Europe and America, it has shown an upward trend in recent years and has gradually developed towards a younger trend (3). Data have shown that there were 1.1 million PCa patients in the world in 2012, of whom 307,000 individuals succumbed to the disease (4). At present, PCa patients are mainly elderly people, and studies have shown that the incidence of PCa in patients over the age of 60 has increased significantly (5). However, the risk factors for PCa are not completely clear except for age, race and heredity.
The prostate is an important androgen-dependent organ, and the target and signaling pathways mediated by androgen and its receptors play a key role in the growth and development of the prostate (6). Currently, the main treatment method for PCa is to inhibit the secretion of androgens in patients by surgery or drugs, thereby reducing androgen level (7). However, most PCa patients undergoing castration therapy will turn into castration-resistant PCa, which further deteriorates the patients' conditions and imposes a great burden on their families and themselves (8).
MicroRNAs (miRs) are non-coding RNAs approximately 10–22 nt in length and more than 1000 are known to exist in the human genome (9). By binding to the 3′UTR end of the downstream target gene messenger RNA, the target gene is degraded or inhibited during the translation, which has a regulatory effect on the cell growth and differentiation of the body (10). Findings have shown that miRs that participate in the occurrence and development of a variety of tumors have an important regulatory effect (11). miR-200c is a member of miR-200 and it plays a key role in the growth of various tumors (12). For example, miR-200c is highly expressed in the serum of ovarian cancer patients and is expected to become a biomarker for the diagnosis of ovarian cancer.
However, reports on miR-200c are rare in PCa. Therefore, this study evaluated the effect of miR-200c on the proliferation, invasion and apoptosis of PCa LNCaP cells using RT-qPCR. The results showed that, the low expression of miR-200c is beneficial in the inhibition of the proliferation and invasion of LNCaP cells in vitro and to promote apoptosis, which may be a potential target for prostate cancer biotherapy.
Materials and methods
Main experimental reagents, instruments and cells
PCa cell line LNCaP and normal prostate cell line RWPE-1 were purchased from the Shanghai Institute of Life Sciences. RPMI-1640 culture solution, fetal bovine serum (FBS), trypsin and penicillin-streptomycin double antibody were obtained from Gibco Co. (Grand Island, NY, USA). miR-200c primer sequences, miR-200c mimics (mimics), miR-NC control vector and miR-inhibitor (inhibitor) were all designed and synthesized by Shanghai Jema Corporation (Shanghai, China). RNA extraction TRIzol reagent and transfection kit Lipofectamine™ 2000 were purchased from Invitrogen. Thermo Fisher Scientific, Inc. (Waltham, MA, USA); Annexin V-FITC and cell counting kit-8 (CCK-8) kit were obtained from Shanghai Biyuntian Institute of Biotechnology (Shanghai, China). Transwell chamber was purchased from Corning Corporation (Corning, NY, USA); SYBR-Green PCR Master Mix kit was from Applied Biosystems (Foster City, CA, USA); the microplate reader SpectraMax M5 was from Shanghai Meigu Molecule (Shanghai, China); ABI 7900 PCR amplification instrument was purchased from Applied Biosystems; and flow cytometry CytoFLEX LX was from Beckman Coulter, Inc. (Brea, CA, USA).
Cell culture and transfection
LNCaP and RWPE-1 cells were cultured in RPMI-1640 medium (10% FBS, 1% penicillin-streptomycin double antibody) and in a thermostatic incubator at 37°C and 5% CO2 to observe cell growth. When the cells adhered to the wall and their fusion reached 80–90%, they were collected, washed with PBS, digested with 0.25% trypsin and added to RPMI-1640 culture solution (10% FBS) after digestion to culture. The LNCaP cells at logarithmic growth phase were grouped and transfected. The experiments were divided into 3 groups. miR-NC (NC group) transfected empty plasmids, miR-200c-mimics (simulation group) were transferred into the simulation sequence, and miR-200c-inhibitor (inhibition group) into the inhibition sequence. The cells were transfected according to the Lipofectamine™ 2000 manufacturer's kit instruction and collected at 48 h after transfection for subsequent experiments. The study was approved by the Ethics Committee of Shengzhou People's Hospital (Shengzhou, China).
CCK-8 detection of cell proliferation ability
The CCK-8 kit was used for detection of proliferation of each group of cells after transfection. The main steps were as follows: Cells in each group at 48 h after transfection were collected and inoculated in a 96-well plate for 24 h. Cells (5×103) were inoculated in each well. The day the cells adhered to the wall was recorded as the first day, and CCK-8 solution (20 µl/well) was added at 1st, 2nd, 3rd, 4th and 5th days, respectively. After the addition of the reagent, the cells were cultured in an incubator (37°C, 5% CO2) for 4 h. The OD values were measured using a SpectraMax M5 microplate reader at 450 nm for detection of cell proliferation, and the growth curve was plotted. The experiment was repeated three times.
Transwell chamber detection of cell invasion ability
Transwell chamber was used for detection of invasion ability of the cells in each group after transfection. The cells at 48 h after transfection were collected and inoculated in a 24-well plate. The cell density was adjusted to 5×104 cells/well (200 µl serum-free culture solution) and added to the upper chamber, and 400 µl of the culture solution containing 10% FBS PRMI-1640 to the lower chamber, and incubated at 37°C and 5% CO2 for 12 h. After the incubation, non-migrated cells on the membrane were removed with a cotton swab and then washed with PBS. The migrated cells at the bottom of the membrane were fixed with 4% paraformaldehyde solution for 10 min, washed with PBS and stained with 0.5% crystal violet for 10 min. With 95% ethanol as the solvent, it was gently agitated at room temperature for 6 h. The migrated cells were then quantified under light microscopy (Olympus Corporation, Tokyo, Japan), the cell invasion of five fields was calculated, and the average value was obtained for comparison. The experiment was repeated three times.
Flow cytometry detection of cell apoptosis
The Annexin V- FITC apoptosis detection kit was used for the detection of apoptosis of the cells in each group after transfection. The cells in each group were transfected for 48 h to make a single cell suspension and washed with PBS, and centrifuged at 3,000 × g for 8 min at 4°C to remove the supernatant, resuspended again and incubated at room temperature. It was fixed with an equal volume of ethanol (75%) for 20 min and washed with PBS. Five microliters of Annexin V-FITC and 10 ml of PI were added for incubation for 60 min at 4°C in the dark. CytoFLEX LX flow cytometry was used for detection of cell apoptosis. The experiment was repeated three times.
miR-200c expression in cells
Total RNA was extracted from untransfected LNCaP and RWPE-1 cells and each group of cells at 48 h after transfection using TRIzol reagent. A UV spectrophotometer (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used for the detection of RNA concentration and total RNA was transcribed into cDNA using the kit as 2 µl 10 mM dNTP, 0.5 µl RNase inhibitor, 0.5 µl miR reverse primer, 0.5 µl U6 reverse primer, 4 µl 5X buffer, 0.5 µl M-MLV (Promega Corporation, Madison, WI, USA). Transcription method was performed according to the manufacturer's kit. The transcribed cDNA was used for PCR amplification and its system was configured according to the manufacturer's instructions. PCR reaction conditions were: Pre-denaturation at 95°C for 10 min, denaturation at 95°C for 30 sec, annealing at 60°C for 30 sec and elongation at 74°C for 30 sec for a total of 40 cycles. The real-time fluorescence quantitative PCR detection was performed using ABI 7900 real-time PCR amplification instrument. With U6 as the internal reference, the relative expression of each group was calculated using 2−ΔCq and three reactions were performed for each sample (13). Table I shows the primer sequences used in the present study.
Table I.
Primer sequences.
| Gene | Upstream primers | Downstream primers |
|---|---|---|
| miR-200c | 5′-GGATAATACTGCCGGGT-3′ | 5′-GTGCGTGTCGTGGAGTC-3′ |
| U6 | 5′-CTCGCTTCGGCAGCACA-3′ | 5′-AACGCTTCACGAATTTGCGT-3′ |
Statistical analysis
In this study, SPSS 20.0 software package (Shanghai Kaibei) was used for statistical analysis of the collected data. GraphPad Prism 7 software was used to plot all the pictures of this experiment. The measurement data are expressed as mean ± standard deviation (mean ± SD), the Students' t-test was used for analysis of comparison between the two groups, and variance for analysis of comparison among multiple groups with LSD test. P<0.05, indicates a statistical difference.
Results
miR-200c expression in each group of cells
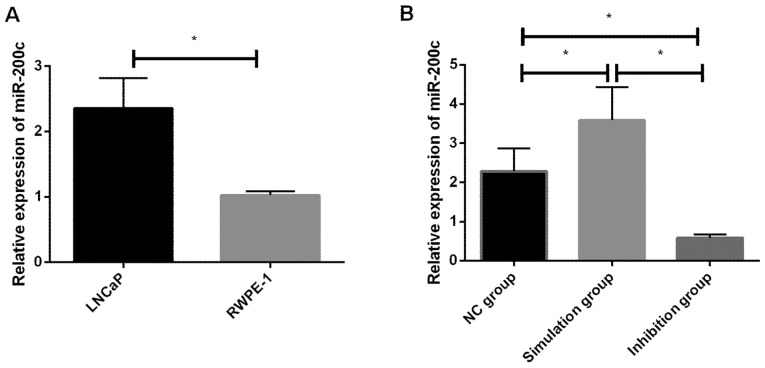
In this study, the relative expression of miR-200c in LNCaP and RWPE-1 cells before transfection and in the NC, simulation and inhibition groups of cells after transfection were detected, and it was found that the relative expression of miR-200c in LNCaP cells before transfection was significantly increased compared with RWPE-1 cells, and the difference was statistically significant (P<0.05). The difference was statistically significant in the relative expression of miR-200c cells among the NC, simulation and inhibition groups after transfection (P<0.05). In addition, there was a significant decrease in the inhibition group, as well as a difference between the inhibition, NC, and simulation groups (P<0.05). A significant decrease in the NC group of cells compared with the simulation group was also identified, which was statistically different (P<0.05) (Tables II and III and Fig. 1).
Table II.
Relative expression of miR-200c in LNCaP and RWPE-1 cells before transfection.
| Groups | LNCaP cells | RWPE-1 cells | F-value | P-value |
|---|---|---|---|---|
| Relative expression of miR-200c | 2.353±0.468 | 1.025±0.058 | 4.878 | 0.008 |
Table III.
Relative expression of miR-200c in each group of cells after transfection.
| Groups | NC group | Simulation group | Inhibition group | F-value | P-value |
|---|---|---|---|---|---|
| Relative expression of miR-200c | 2.284±0.584 | 3.584±0.845 | 0.584±0.095 | 19.143 | 0.003 |
Figure 1.
(A) Relative expression of miR-200c in LNCaP and RWPE-1 cells. RT-qPCR detection showed that the relative expression of miR-200c in LNCaP cells increased significantly, and there was a significant difference between the LNCaP cells and RWPE-1 cells (P<0.05). There is a difference between the two groups (*P<0.05). (B) miR-200c expression in each group of cells after transfection and RT-qPCR detection showed that the relative expression of miR-200c significantly decreased in the inhibition group, and there was a difference between the inhibition, NC, and simulation groups (*P<0.05), and it significantly decreased in NC group of cells compared with simulation group, indicating a difference (P<0.05). *P<0.05 indicates the difference between the two groups.
Proliferation in each group of cells following transfection
Proliferation in each group of cells after transfection was detected using CCK-8. It was found that there were differences at the 2nd, 3rd, 4th and 5th days of cell growth among the NC, simulation and inhibition groups (P<0.05), among which the growth ability in the inhibition group of cells was significantly suppressed. Additionally, there was a difference between the inhibition, NC, and simulation groups (P<0.05), and the proliferation ability in NC group significantly decreased compared with simulation group, and there was a statistical difference (P<0.05) (Fig. 2).
Figure 2.
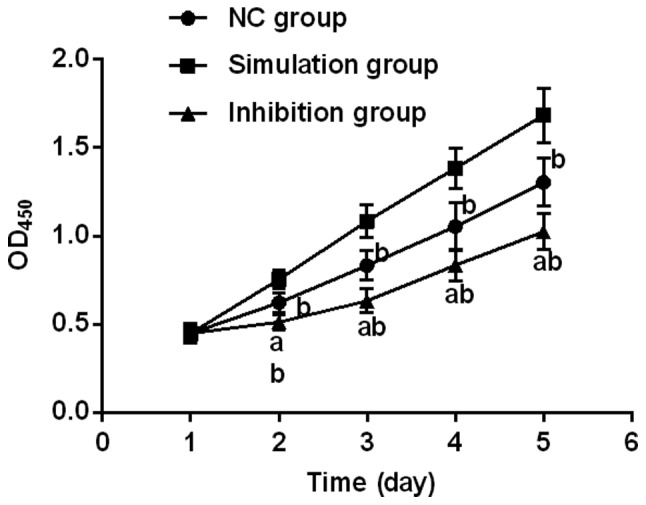
Proliferation in each group of cells after transfection. Proliferation in each group of cells after transfection was detected using CCK-8 kit. It was found that there was a difference in the proliferation ability at the 2nd day of growth between the inhibition, NC, and simulation groups (P<0.05). It was significantly decreased in NC group compared with simulation group, and there was a difference. a, indicates that there is a difference compared with NC group (P<0.05); and b, indicates that there is a difference compared with simulation group (P<0.05). CCK-8, cell counting kit-8.
Effect on cell invasion ability after transfection
The invasion ability of each group of cells after transfection was detected using Transwell chamber, and it was found that there was a difference in the three groups of cells (P<0.05), among which the number of passed membrane cells in the inhibition group was significantly smaller than that in the remaining two groups, with a statistical difference (P<0.05). Additionally, there was no significant decrease in NC group compared with simulation group (P>0.05) (Table IV and Fig. 3).
Table IV.
Number of passed membrane cells.
| Groups | No. of passed membrane cells | F-value | P-value |
|---|---|---|---|
| NC group | 97.51±10.58ab | ||
| Simulation group | 119.58±13.9a | 23.058 | 0.002 |
| Inhibition group | 58.36±8.37 |
Indicates that there is a difference compared to inhibition group (P<0.05)
indicates that there is no difference compared to simulation group (P>0.05).
Figure 3.
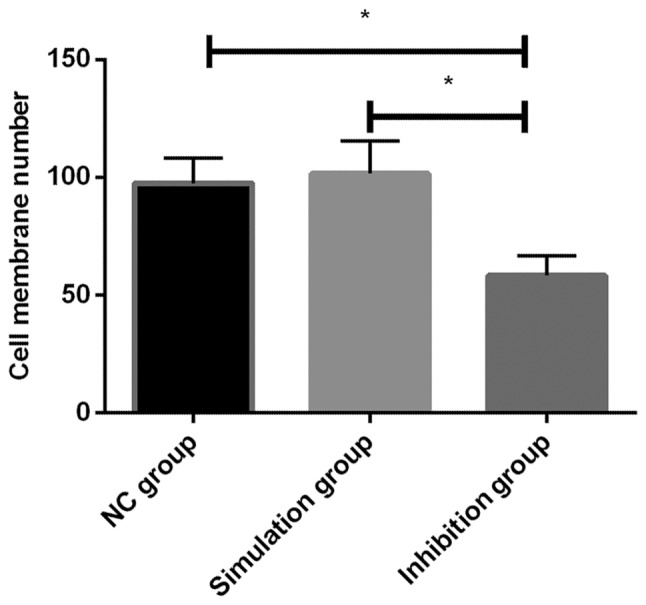
Effect on invasion ability of each group of cells after transfection. The invasion ability of each group of cells after transfection was detected by Transwell chamber, and it was found that the number of passed membrane cells in inhibition group significantly decreased, and there was a difference among the three groups (P<0.05). There was no significant decrease in NC group compared with simulation group (P>0.05). *P<0.05 indicates the difference between the two groups.
Cell apoptosis after transfection
By flow cytometry detection of the apoptosis ability of each group of cells after transfection, it was found that there was a difference in the apoptosis rate among the NC, simulation and inhibition groups (P<0.05), of which a significant increase was evident in the inhibition group. Additionally, there was a difference between the inhibition, NC, and simulation groups (P<0.05), but there was no statistical difference between the NC and simulation groups (P>0.05) (Table V and Fig. 4).
Table V.
Cell apoptosis (%).
| Groups | Apoptosis rate | F-value | P-value |
|---|---|---|---|
| NC group | 12.84±5.32ab | ||
| Simulation group | 18.35±5.94a | 6.088 | 0.036 |
| Inhibition group | 33.67±8.61 |
Compared to the inhibition group (P<0.05)
indicates that there is no difference compared to simulation group (P>0.05).
Figure 4.
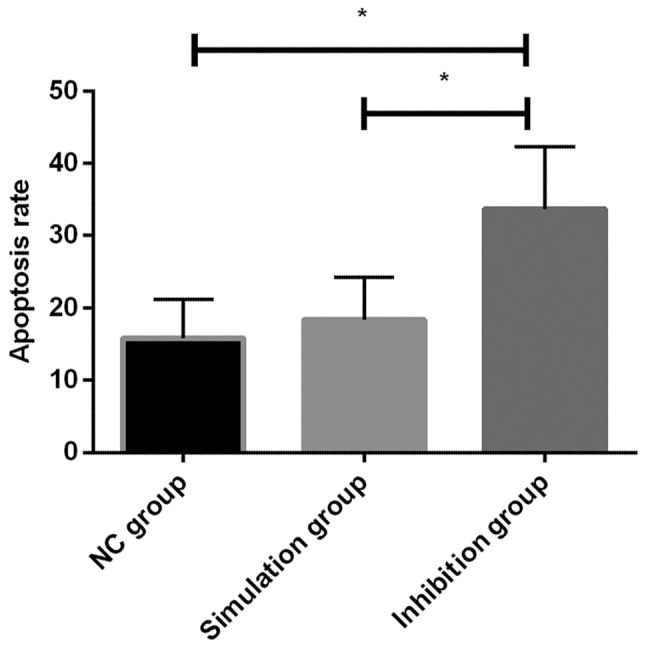
Cell apoptosis of each group after transfection. By flow cytometry detection of the apoptosis ability of each group of cells after transfection, the apoptosis rate in inhibition group was significantly higher than that in the NC and simulation groups, and the difference was significant (P<0.05). There was no statistical difference between the NC and simulation groups (P>0.05). *P<0.05 indicates the difference between the two groups.
Discussion
PCa is a global malignancy of male with a high incidence. Statistics have shown that there were approximately 180,890 new PCa patients in the United States in 2016, and approximately 26,120 patients succumbed to the disease (5). The early clinical symptoms of PCa are not obvious, and when the tumor is too large to block the urinary tract or invade to the bladder neck, there is a severe lower urinary tract infection. In the late stage of tumor, many patients experience tumor metastasis, the most common of which is bone metastasis, which is often accompanied by bone pain and fracture (14). As with other solid tumors, the main cause of death of PCa is due to cancer metastasis that is a complex physiological process, including factors such as changes in cell micro-environment, cell movement and cell growth (15).
At present, the treatment method of PCa at the middle and late stage is mainly chemotherapy, but it is found during the treatment of PCa that PCa tumor has a poor sensitivity to chemotherapy, which leads to the emergence of drug resistance during long-term chemotherapy, resulting in less than expected effect (6). The main diagnostic method of PCa is rectal examination (16) of serum PCa-specific antigen (PSA) (17). Early screening is based on rectal examination combined with PSA examination. However, the subjective nature of rectal examination requires the experience of clinicians for many years, and the early diagnosis of it before the formation of nodules is prone to missed diagnosis (16). PSA, as an important serum marker of PCa, has a high sensitivity that results in a low specificity, and it may increase in patients with prostatitis or other prostate diseases (18). Studies have shown that (19) the diagnosis rate of PCa is <30% with PSA concentration between 4–10 ng/ml, and PSA can easily lead to over-diagnosis and over-treatment of PCa. Therefore, there is a large study space for the diagnosis and treatment of PCa, and the occurrence and development of it and its molecular mechanism are still unclear, so it is of great significance for clinical treatment and diagnosis to probe into the main mechanism of PCa.
As a class of endogenous non-coding single-stranded small RNA, miRs mainly present in eukaryotes and can regulate target genes through transcription (20). Based on the binding of specific target mRNA 3′-UTR and its target gene specificity, the target gene mRNA will be degraded or the translation of it will be inhibited, so as to regulate its post-transcriptional expression (21). Increasing number of studies have proven that miRs play an important role in the occurrence and development of a variety of cancers. Studies have shown that miR-409-3p/-5p significantly increases in PCa tissues but miR-375 expression is low, suggesting that there is a difference in expression of different miRs in PCa (22,23). miR-200c, one of the five members of the miR-200 family, is an important regulator of epithelial-mesenchymal transformation. In addition to its role in normal cell phenotypic transformation, miR-200c expression is differential in many cancer cells. For example, high expression of serum miR-200c is associated with poor prognosis in lung cancer patients (24). However, there are few studies on miR-200c and PCa, and it is unclear whether miR-200c regulates PCa. Therefore, we aimed to discover the biological function of miR-200c in PCa through this investigation.
The miR-200c expression in PCa cells and normal prostate cells were detected, and it was found that they significantly increased in PCa cells compared with normal prostate cells, and the difference was significant. In study of Vrba et al (25), it was shown that miR-200c in normal prostate tissues was lower than that in cancer tissues. In study of Tao et al (26), it was shown that miR-200c was also highly expressed in PCa patient tissues, consistent with our findings. We transfected prostate cancer LNCaP cells and observed changes in cell biological function after miR-200c. Promoting cell apoptosis and inhibiting cell proliferation are currently important ideas for clinical treatment of tumors. Through detection of each group of cells after transfection, it was found that by inhibition of the miR-200c expression can effectively reduce cell proliferation and promote cell apoptosis rate. Ali et al (27), showed that curcumin inhibited the proliferation and apoptosis of PCa cells, which was significantly reduced by detection of the miR-200c expression in cells, suggesting that low expression of miR-200c can inhibit PCa proliferation and promote apoptosis. Cancer metastasis is a major cause of cancer exacerbations in the late stage. Through detection of cell invasion ability, it was found that inhibition of the miR-200c expression had a significant inhibitory effect on the invasion ability of cells. Burk et al (28), showed that inhibition of the miR-200c expression by ZEB1 inhibited the invasion ability of PCa cells, which is consistent with our findings, suggesting that down-regulating miR-200c can reduce the invasion ability of PCa cells.
However, there are still some defects in this study. First of all, the number of cells in vitro experiments was small. Secondly, more in-depth studies were not conducted, and the target genes and related pathways were not tested and validated. Therefore, in future studies, we hope to collect clinical specimens to validate our results, and test their target genes and pathways to further explore their mechanisms, laying the foundation for future clinical treatment and diagnosis.
In conclusion, the low expression of miR-200c is beneficial in inhibiting proliferation and invasion of LNCaP cells in vitro and promoting apoptosis, which may be a potential target for PCa biotherapy.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
JL drafted the manuscript. JL and YL were responsible for cell culture and transfection. XZ assisted with CCK-8 detection. QM contributed to MTT assay. LY performed the PCR. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Shengzhou People's Hospital (Shengzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Schröder FH, Hugosson J, Roobol MJ, Tammela TL, Zappa M, Nelen V, Kwiatkowski M, Lujan M, Määttänen L, Lilja H, et al. ERSPC Investigators Screening and prostate cancer mortality: Results of the European Randomised Study of Screening for Prostate Cancer (ERSPC) at 13 years of follow-up. Lancet. 2014;384:2027–2035. doi: 10.1016/S0140-6736(14)60525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Wong MC, Goggins WB, Wang HH, Fung FD, Leung C, Wong SY, Ng CF, Sung JJ. Global incidence and mortality for prostate cancer: Analysis of temporal patterns and trends in 36 countries. Eur Urol. 2016;70:862–874. doi: 10.1016/j.eururo.2016.05.043. [DOI] [PubMed] [Google Scholar]
- 4.Stewart BW, Wild CP, editors. IARC Publications; Lyon: 2014. World Cancer Report 2014. [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 6.Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15:701–711. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ceder Y, Bjartell A, Culig Z, Rubin MA, Tomlins S, Visakorpi T. The molecular evolution of castration-resistant prostate cancer. Eur Urol Focus. 2016;2:506–513. doi: 10.1016/j.euf.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 8.Nelson WG, De Marzo AM, Isaacs WB. Prostate cancer. N Engl J Med. 2003;349:366–381. doi: 10.1056/NEJMra021562. [DOI] [PubMed] [Google Scholar]
- 9.Bandara KV, Michael MZ, Gleadle JM. MicroRNA biogenesis in hypoxia. MicroRNA. 2017;6:80–96. doi: 10.2174/2211536606666170313114821. [DOI] [PubMed] [Google Scholar]
- 10.Wu HC, Lai MT, Wu CI, Chen HY, Wan L, Tsai FJ, Chen WC. E-cadherin gene 3-UTR C/T polymorphism is associated with prostate cancer. Urol Int. 2005;75:350–353. doi: 10.1159/000089173. [DOI] [PubMed] [Google Scholar]
- 11.Colden M, Dar AA, Saini S, Dahiya PV, Shahryari V, Yamamura S, Tanaka Y, Stein G, Dahiya R, Majid S. MicroRNA-466 inhibits tumor growth and bone metastasis in prostate cancer by direct regulation of osteogenic transcription factor RUNX2. Cell Death Dis. 2017;8:e2572. doi: 10.1038/cddis.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meng X, Müller V, Milde-Langosch K, Trillsch F, Pantel K, Schwarzenbach H. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget. 2016;7:16923–16935. doi: 10.18632/oncotarget.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Freitag MT, Radtke JP, Hadaschik BA, Kopp-Schneider A, Eder M, Kopka K, Haberkorn U, Roethke M, Schlemmer HP, Afshar-Oromieh A. Comparison of hybrid (68)Ga-PSMA PET/MRI and (68)Ga-PSMA PET/CT in the evaluation of lymph node and bone metastases of prostate cancer. Eur J Nucl Med Mol Imaging. 2016;43:70–83. doi: 10.1007/s00259-015-3206-3. [DOI] [PubMed] [Google Scholar]
- 15.Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JM, Papaemmanuil E, Brewer DS, Kallio HML, Högnäs G, Annala M, et al. ICGC Prostate Group The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353–357. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Catalona WJ, Richie JP, Ahmann FR, Hudson MA, Scardino PT, Flanigan RC, DeKernion JB, Ratliff TL, Kavoussi LR, Dalkin BL, et al. Comparison of digital rectal examination and serum prostate specific antigen in the early detection of prostate cancer: Results of a multicenter clinical trial of 6,630 men. J Urol. 2017;197:S200–S207. doi: 10.1016/j.juro.2016.10.073. [DOI] [PubMed] [Google Scholar]
- 17.Jemal A, Fedewa SA, Ma J, Siegel R, Lin CC, Brawley O, Ward EM. Prostate cancer incidence and PSA testing patterns in relation to USPSTF screening recommendations. JAMA. 2015;314:2054–2061. doi: 10.1001/jama.2015.14905. [DOI] [PubMed] [Google Scholar]
- 18.Abd-Alazeez M, Ahmed HU, Arya M, Charman SC, Anastasiadis E, Freeman A, Emberton M, Kirkham A. The accuracy of multiparametric MRI in men with negative biopsy and elevated PSA level - can it rule out clinically significant prostate cancer? Urol Oncol. 2014;32:45.e17–45.e22. doi: 10.1016/j.urolonc.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SW, Hosokawa K, Kim S, Jeong OC, Lilja H, Laurell T, Maeda M. A highly sensitive porous silicon (P-Si)-based human kallikrein 2 (hK2) immunoassay platform toward accurate diagnosis of prostate cancer. Sensors (Basel) 2015;15:11972–11987. doi: 10.3390/s150511972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki HI, Young RA, Sharp PA. Super-enhancer-mediated RNA processing revealed by integrative MicroRNA network analysis. Cell. 2017;168:1000–1014.e15. doi: 10.1016/j.cell.2017.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li XL, Andersen JB, Ezelle HJ, Wilson GM, Hassel BA. Post-transcriptional regulation of RNase-L expression is mediated by the 3-untranslated region of its mRNA. J Biol Chem. 2007;282:7950–7960. doi: 10.1074/jbc.M607939200. [DOI] [PubMed] [Google Scholar]
- 22.Josson S, Gururajan M, Hu P, Shao C, Chu GY, Zhau HE, Liu C, Lao K, Lu CL, Lu YT, et al. miR-409-3p/-5p promotes tumorigenesis, epithelial-to-mesenchymal transition, and bone metastasis of human prostate cancer. Clin Cancer Res. 2014;20:4636–4646. doi: 10.1158/1078-0432.CCR-14-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang X, Yuan T, Liang M, Du M, Xia S, Dittmar R, Wang D, See W, Costello BA, Quevedo F, et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol. 2015;67:33–41. doi: 10.1016/j.eururo.2014.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu XG, Zhu WY, Huang YY, Ma LN, Zhou SQ, Wang YK, Zeng F, Zhou JH, Zhang YK. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med Oncol. 2012;29:618–626. doi: 10.1007/s12032-011-9923-y. [DOI] [PubMed] [Google Scholar]
- 25.Vrba L, Jensen TJ, Garbe JC, Heimark RL, Cress AE, Dickinson S, Stampfer MR, Futscher BW. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS One. 2010;5:e8697. doi: 10.1371/journal.pone.0008697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tao T, Liu D, Liu C, Xu B, Chen S, Yin Y, Ang L, Huang Y, Zhang X, Chen M. Autoregulatory feedback loop of EZH2/miR-200c/E2F3 as a driving force for prostate cancer development. Biochim Biophys Acta. 2014;1839:858–865. doi: 10.1016/j.bbagrm.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Ali S, Ahmad A, Banerjee S, Padhye S, Dominiak K, Schaffert JM, Wang Z, Philip PA, Sarkar FH. Gemcitabine sensitivity can be induced in pancreatic cancer cells through modulation of miR-200 and miR-21 expression by curcumin or its analogue CDF. Cancer Res. 2010;70:3606–3617. doi: 10.1158/0008-5472.CAN-09-4598. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.