Abstract
Cross-talk between competing endogenous RNAs (ceRNAs) play key roles in tumor development. In this study, we performed exon-level expression profiling on 26 glioblastomas (GBMs) and 6 controls to identify long non-coding RNAs (lncRNAs) of GBM initiation and progression using lncRNA-mediated ceRNA network (LMCN). The mRNA and lncRNA expression data, as well as miRNA-target interactions were firstly collected. Then, we used hypergeometric test to detect the lncRNA-mRNA interactions, followed by the construction of LMCN based on Pearson correlation coefficient. With the goal of investigation of the network organization, degree distribution of LMCN was performed. Next, the synergistic, competing lncRNA modules were identified using jActiveModule plug-in of Cytoscape. Moreover, we implemented the pathway analysis for its mRNAs in the module to explore the functions of significant lncRNAs. Using the criteria of degrees >50, 8 hub genes were identified, including EPB41L4A-AS1, ZRANB2-AS2, XIST, HOTAIR, TRAF3IP2-AS1, TPT1-AS1, PVT1 and DLG1-AS1. Furthermore, 1 synergistic, competitive module was identified. In this module, lncRNAs XIST and PVT1 were also the hubs in the synergistic, competing lncRNA module. Functional annotation demonstrated that 5 pathways were identified, including cytokine-cytokine receptor interaction, neuroactive ligand-receptor interaction, and mTOR signaling pathway. We have successfully identified several hubs (such as XIST and PVT1) and significant pathways (for instance, cytokine-cytokine receptor interaction, and neuroactive ligand-receptor interactions) for GBM via establishing the LMCN. These findings might offer potential biomarkers to early diagnose, and predict GBM prognosis in the future.
Keywords: glioblastomas, long non-coding RNA-mediated competing endogenous RNA network, pathway
Introduction
Glioblastomas (GBM), one of the most aggressive and lethal types of brain tumor, accounts for more than half of all gliomas (1). The GBM prognosis is still very poor, although the standard of care has improved survival (2,3). The dismal outcome makes GBM an urgent subject of cancer research. Thus, detecting suitable biomarkers is urgently needed for GBM diagnosis as well as prognosis.
Well-defined risks for GBM are radiation exposure and certain genetic syndromes (4). For example, mutation of PTEN, EGFR, TP53, PIK3CA, IDH1, NF1, RB1 and PDGFRA have been identified in GBM genome, whereas the functional consequences of many of these alterations are still unknown (5–7). With the development of high-througput technology, it has been well accepted that at least 90% of genome sequences is transcribed into noncoding RNAs (ncRNAs), but <2% of the genome encode proteins (8,9). It is increasingly recognized that the roles of ncRNAs constitute an important component in the most biological processes, in part due to the findings that most of the transcripts do not encode for proteins (10,11). According to the size of ncRNA, they are classfied into small ncRNAs as well as long ncRNAs (lncRNAs). Growing evidence has demonstrated that lncRNAs participate in many kinds of biological pathways, for example, neural lineage commitment, and immune response (12–14). Furthermore, lncRNAs have been demonstrated to be important regulators of disease processes (15,16). Significantly, lncRNAs have been revealed to regulate tumor growth and metastasis, including GBM. For example, Pastori et al indicated that lncRNA HOTAIR plays important roles in glioblastoma through promoting cell proliferation (17). Another study has indicated that the oncogenic lncRNA MALAT1 is related to survival and metastasis in GBM (18). However, the overall study of lncRNAs in brain tumors has lagged behind (19,20). Therefore, the extraction of lncRNA signatures is challenging.
Significantly, competing endogenous RNAs (ceRNAs) have been reported to be crucial in illuminating how lncRNAs regulate the expression of coding genes (21,22). For instance, a previous study has reported that the ceRNA interaction network for GBM reveals oncogenic pathways (23). Moreover, Zhang et al (24) have proposed the biological functions of lncRNA associated-ceRNAs in GBM. Collectively, the dysregulated ceRNA network might offer new hope for revealing the pathogenesis underlying GBM.
Herein, in order to better understand the pathogenic processes of GBM, we sought to identify the candidate lncRNA signatures through establishing a functional lncRNA-mediated ceRNA network (LMCN) for GBM.
Materials and methods
The specific steps of our study were divided into the following procedures: Firstly, mRNA as well as lncRNA expression data of GBM were obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo/) based on the platform of exon-array data on the Affymetrix Human 1.0 ST Array. Simultaneously, miRNA-target interactions were downloaded from starBase v2.0 (25). Then, a hypergeometric test was applied to identify the competing lncRNA-mRNA interactions, following by co-expression analysis to extract the co-expressed lncRNA-mRNA pairs to further establish the LMCN. Functional analyses for the mRNAs in the LMCN to reveal the biological roles of lncRNAs.
Collecting miRNA-target interactions and affymetrix microarray
The mRNA as well as lncRNA data of GBM (accession number GSE9385) (26) were received from the GEO database, which is on the basis of the (GPL5188) platform of GeneChip Human Exon 1.0 ST Array. The GSE9385 included 26 GBMs, 22 oligodendrogliomas (ODs) as well as 6 control samples. In our study, with the goal of better understanding the molecular mechanisms of GBM, we only selected 26 GBMs and 6 control samples for subsequent analysis. After the probes were mapped to the gene symbols, we obtained a total of 35,367 genes for the following study.
Identification of miRNA-target interactions
starBase v2.0 provides high-quality and experimentally validated miRNA-target interactions. In the present study, the experimentally confirmed lncRNA-miRNA as well as miRNA-mRNA interactions were obtained from starBase 2.0. Then, we aligned the obtained 35,367 genes to the lncRNA-miRNA as well as miRNA-mRNA interactions obtained from starBase 2.0, and we derived the new expression profiles of 11,423 genes (including 212 lncRNAs and 11,211 mRNAs). Moreover, we obtained the interactions covering any genes of 212 lncRNAs and 11,211 mRNAs from the lncRNA-miRNA and miRNA-mRNA interactions, respectively. Ultimately, 2551 lncRNA-miRNA interactions and 352,647 miRNA-mRNA interactions were obtained.
Identifying potential ceRNA interactions
Herein, to get the competing lncRNA-mRNA interactions, the hypergeometric test was used. In an attempt to measure the enrichment significance of the common miRNAs, we computed the P-values. Subsequently, false discovery rate (FDR) was utilized to correct the original P-values based on Benjamini & Hochberg method (27). The interactions with FDR <0.01 was considered as the significant and competing interactions.
Co-expression analysis
With an attempt of measuring the co-expression probability of lncRNA-mRNA interactions, we calculated the PCC relying on the expression of the potential lncRNA-mRNAs pair. In this study, the PCC absolute value of an edge was determined as the weight values, and only edges having weight value >1.15 were reserved to establish the LMCN based on Pearson correlation coefficient. Cytoscape (28) was employed to illustrate the highly competitive LMCN.
Topological analysis for LMCN
As documented, topological centralities (degree, closeness as well as betweenness) are broadly utilized to reveal the properties of network (29). Among these centralities, degree is the simplest index. Herein, we analyzed the degree distribution of all nodes in LMCN and we determined the nodes having degrees greater than 50 as hubs.
Mining synergistic, competing lncRNA modules from the LMCN
Although LMCN can supply an overall view of all lncRNA-mRNA interactions, the sub-networks or clusters reveal a more detailed picture of how lncRNAs synergized with the competing mRNAs. Thus, in this study, the jActiveModule plug-in of Cytoscape was employed to further screened out the synergistic competing modules from the LMCN as previously described (30).
Analysis of functional annotation of synergistic, competing lncRNA modules
DAVID is widely used to analyze genes obtained from the genomic experiments. In the present study, we utilized the DAVID to extract significant pathway terms enriched in genes in the module based on the ‘guilt by association’ strategy, thereby revealing the underlying biological process of lncRNAs. We used Fisher's exact test to detect the significant pathways using the criteria of FDR <0.001.
Results
Constructing a highly competitive LMCN
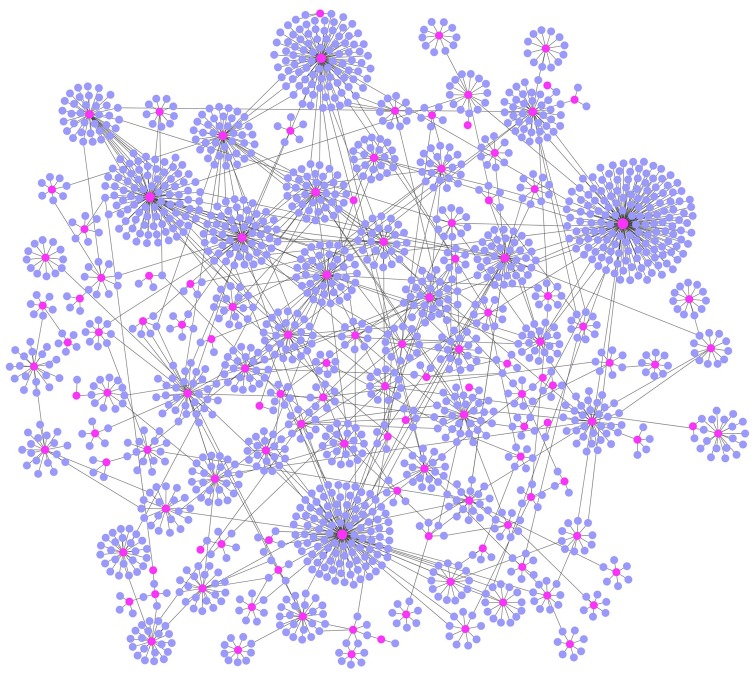
To establish a highly competitive LMCN, the hypergeometric test was first utilized to detect the candidate lncRNA-mRNA interactions via measuring the significance of the common miRNAs among each lncRNA-mRNA interaction. Eventually, using the criteria of FDR <0.01, we identified 212 lncRNAs, 11,199 mRNAs, as well as 218,696 ceRNA interactions. Based on a weight value >1.15, a highly competive LMCN was built, which included 199 lncRNAs, 2052 mRNAs, and 2715 ceRNA interactions. This LMCN is shown in Fig. 1. We observed that the lncRNAs were distributed in the central area of the LMCN, while the mRNAs were distributed in the outside layer (Fig. 1).
Figure 1.
Establishment of a highly competitive LMCN. The LMCN included 199 lncRNAs, 2052 mRNAs, 2715 ceRNA interactions. Pink represents lncRNAs, blue stands for mRNAs. lncRNA, long non-coding RNA; LMCN, lncRNA-mediated competing endogenous RNA network.
Degree distribution of the LMCN
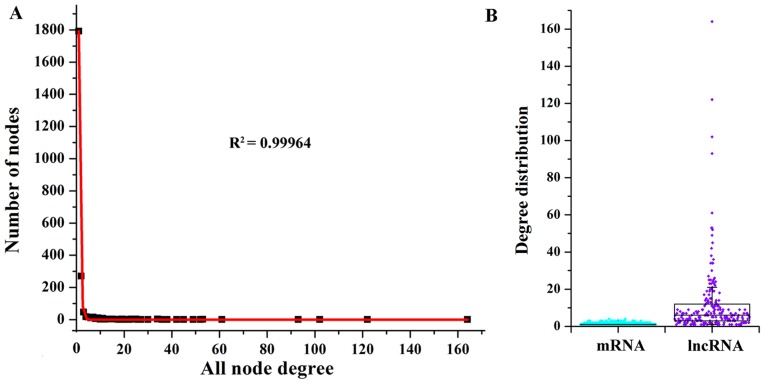
Then, we investigated the degree properties of LMCN to reveal the organization of the LMCN in GBM patients. The degree distribution of the LMCN (R2=0.99964) demonstrated power law distributions (Fig. 2A). In general, the node with a higher degree demonstrated a hub, which regulated more ceRNA interactions. As displayed in Fig. 2B, the degree distribution of the mRNAs was smaller than that of the lncRNAs, which suggested that though lncRNAs did not encode for proteins, these lncRNAs showed more degree distribution than mRNAs within the LMCN. On the basis of the traits of the lncRNAs of the LMCN, some hub nodes were considered as risk lncRNAs for GBM. With the goal of identifying the hub nodes of the LMCN, the degrees of all nodes in the entire network were ordered in a descending rank on the basis of their degree distribution. Using the degrees >50 as the cut-off threshold, there were 8 hub genes, including EPB41L4A-AS1 (degree=164), ZRANB2-AS2 (degree=122), XIST (degree=102), HOTAIR (degree=93), TRAF3IP2-AS1 (degree=61), TPT1-AS1 (degree=53), PVT1 (degree=53), and DLG1-AS1 (degree=52).
Figure 2.
Degree analysis of the LMCN. (A) Degree distributions of the LMCN reflected a power-law distribution. (B) Box-type scatterplot.
Identifying synergistic, competing lncRNA modules from the LMCN
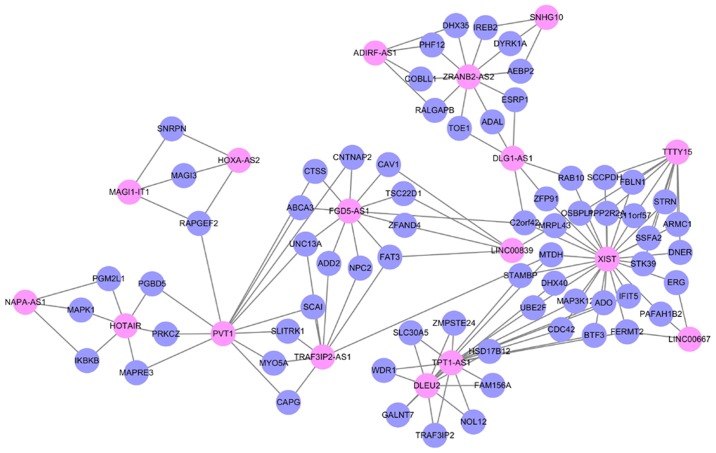
To explore the modularity features of the LMCN, the jActiveModule plug-in of Cytoscape was employed. One synergistic, competitive module was identified, which contained 83 nodes, as listed in Fig. 3. Similar to the LMCN, synergistic, competitive module possessed a scale-free characteristics with power law degree distribution (Fig. 4, R2=0.99431). Based on the network organization, we found that lncRNA XIST competed with 26 mRNAs and 6 lncRNAs (LINC00667, TTTY15, TPT1-AS1, DLG1-AS1, DLEU2 and LINC00839) in the module (Fig. 3), suggesting its significant roles in GBM. Of note, three lncRNAs XIST, TPT1-AS1 and PVT1 in the LMCN were also the hubs in the synergistic, competing lncRNA module.
Figure 3.
A synergistic, competing lncRNA module identified using jActiveModule plug-in of Cytoscape. LMCN, lncRNA-mediated ceRNA network.
Figure 4.
Degree analysis revealing the specific properties of the synergistic, competing lncRNA module. lncRNA, long non-coding RNA.
Analysis of functional annotation
To further investigate the biological significance of lncRNAs in GBM, pathway enrichment analyses of mRNAs in the synergistic, competing lncRNA module were conducted. According to the FDR <0.001, 5 pathways enriched by the mRNAs of the module were identified, including cytokine-cytokine receptor interaction, neuroactive ligand-receptor interaction, and mTOR signaling pathway. Specific information is shown in Table I.
Table I.
The significant pathways relying on FDR<0.001.
| Pathway terms | FDR values |
|---|---|
| Neuroactive ligand-receptor interaction | 4.28×10−12 |
| Cytokine-cytokine receptor interaction | 4.38×10−05 |
| mTOR signaling pathway | 1.99×10−04 |
| Oocyte meiosis | 2.40×10−04 |
| Spliceosome | 3.63×10−04 |
FDR, false discovery rate.
Discussion
The ceRNAs have been demonstrated to regulate the expression based on the competing mechanisms, which play important roles in diverse tumor pathological as well as physiological processes. The ‘guilt by association’ method is typically, utilized to explore lncRNA roles (31). Thus, we decided to use a more comprehensive method to establish a functional LMCN, module extraction, and functional analysis. The results suggested that the GBM-related LMCN can be applied to accelerate biosignature identification and therapeutic development for GBM.
In the present study, hub lncRNAs XIST, and PVT1 in the LMCN were also the hubs in the synergistic, competing lncRNA module. The lncRNA XIST is the main regulator of X inactivation in mammals (32). Previous studies have indicated that XIST exerts critical functions in cell proliferation, cell differentiation, and genome maintenance (33). Significantly, a former study has implicated that XIST is overexpressed in glioma tissues and glioblastoma stem cells, and knockdown of XIST plays tumor-suppressive roles via decreasing cell proliferation, and migration as well as inducing apoptosis (34). PVT1 is an oncogene as well as a Myc protein target known to be upregulated in transformed cells (35). As reported, lncRNA PVT1 is related to cell proliferation, lymph node invasion, cell apoptosis, and metastasis (36,37). The aberrant expression of PVT1 has been identified in cancers, including gastric cancer, prostate cancer, and breast cancer (38). However, no knowledge on the relationship between PVT1 and GBM has been reported. Demonstrated here, we infer that regulating the expression of XIST and PVT1 might be of therapeutic benefit in GBM patients.
Neuroactive ligand-receptor interaction was the most significant pathway in this study. Neuroactive ligand-receptor interactions involve in signaling molecules and interactions which exert critical functions in many cellular processes, for example, apoptosis and cell proliferation. Moreover, apoptosis and cell proliferation are key processes for cancer onset and progression (39,40). Cytokine-cytokine receptor interaction is another significant pathway. Cytokines are crucial regulators involved in cell growth, and angiogenesis (41). As reported, cytokines can induce inflammatory responses which are induced through binding to the corresponding receptors of cytokines (42). Significantly, cell growth, angiogenesis, and inflammation contribute to cancer development and progression (43,44). Importantly, cytokine-cytokine receptor interaction has been indicated to play key roles in cancer (45). Remarkably, neuroactive ligand-receptor interactions and cytokine-cytokine receptor interactions have been indicated to be highly associated with GBM (41).
Therefore, we suggested that altered expression of neuroactive ligand-receptor interactions and cytokine-cytokine receptor interactions caused GBM occurrence.
In the present study, computational method of LMCN network was used to identify the potential lncRNAs and significant pathways. This approach potentially helps us move closer to understanding the biological phenomena of GBM. However, some limitations still remained. To begin with, samples were limited. In addition, study was performed using bioinformatics method, but was not validated using experiments. Moreover, the data used in our study were retrieved from the public database, not produced by us. Therefore, it is urgent to develop chip data on GBM and to do animal experiments to confirm our obtained findings.
In short, we identified several hub lncRNAs (such as XIST and PVT1) and significant pathways (for instance neuroactive ligand-receptor interactions and cytokine-cytokine receptor interactions) for GBM based on the establishment of LMCN. These findings might offer potential biomarkers to early diagnose, and predict GBM prognosis.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XYZ conceived the study, analyzed the data and drafted the manuscript; LL conceived the study and revised the manuscript. Both authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Liang Y, Diehn M, Watson N, Bollen AW, Aldape KD, Nicholas MK, Lamborn KR, Berger MS, Botstein D, Brown PO, et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci USA. 2005;102:5814–5819. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuan J, Xiao G, Peng G, Liu D, Wang Z, Liao Y, Liu Q, Wu M, Yuan X. MiRNA-125a-5p inhibits glioblastoma cell proliferation and promotes cell differentiation by targeting TAZ. Biochem Biophys Res Commun. 2015;457:171–176. doi: 10.1016/j.bbrc.2014.12.078. [DOI] [PubMed] [Google Scholar]
- 3.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 4.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: A clinical review. JAMA. 2013;310:1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 5.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al. TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frattini V, Trifonov V, Chan JM, Castano A, Lia M, Abate F, Keir ST, Ji AX, Zoppoli P, Niola F, et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45:1141–1149. doi: 10.1038/ng.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 9.He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW, Li J. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344:20–27. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 10.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. FANTOM Consortium Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 11.Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Children's Hospital Oakland Research Institute Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell. 2013;152:743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramos AD, Andersen RE, Liu SJ, Nowakowski TJ, Hong SJ, Gertz C, Salinas RD, Zarabi H, Kriegstein AR, Lim DA. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell. 2015;16:439–447. doi: 10.1016/j.stem.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aprea J, Prenninger S, Dori M, Ghosh T, Monasor LS, Wessendorf E, Zocher S, Massalini S, Alexopoulou D, Lesche M, et al. Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J. 2013;32:3145–3160. doi: 10.1038/emboj.2013.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao X, Tang Z, Zhang H, Atianjoh FE, Zhao JY, Liang L, Wang W, Guan X, Kao SC, Tiwari V, et al. A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat Neurosci. 2013;16:1024–1031. doi: 10.1038/nn.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerin T, Ramanathan A, Rivas K, Grepo N, Coetzee GA, Campbell DB. A noncoding RNA antisense to moesin at 5p14.1 in autism. Sci Transl Med. 2012;4:128ra40. doi: 10.1126/scitranslmed.3003479. [DOI] [PubMed] [Google Scholar]
- 17.Pastori C, Daniel M, Penas C, Volmar CH, Johnstone AL, Brothers SP, Graham RM, Allen B, Sarkaria JN, Komotar RJ, et al. BET bromodomain proteins are required for glioblastoma cell proliferation. Epigenetics. 2014;9:611–620. doi: 10.4161/epi.27906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Wang Y, Li J, Zhang Y, Yin H, Han B. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Lett. 2015;367:122–128. doi: 10.1016/j.canlet.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Kiang KM, Zhang GP, Leung GK. Long non-coding RNAs dysregulation and function in glioblastoma stem cells. Noncoding RNA. 2015;1:69–86. doi: 10.3390/ncrna1010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ulitsky I, Bartel DP. lincRNAs: Genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lü M, Tang B, Zeng S, Hu C, Xie R, Wu Y, Wang S, He F, Yang S. Long noncoding RNA BC032469, a novel competing endogenous RNA, upregulates hTERT expression by sponging miR-1207-5p and promotes proliferation in gastric cancer. Oncogene. 2016;35:3524–34. doi: 10.1038/onc.2015.413. [DOI] [PubMed] [Google Scholar]
- 23.Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang K, Qi L, Kang X, Wang Y, Wang S. Identification and functional characterization of lncRNAs acting as ceRNA involved in the malignant progression of glioblastoma multiforme. Oncol Rep. 2016;36:2911–2925. doi: 10.3892/or.2016.5070. [DOI] [PubMed] [Google Scholar]
- 25.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.French PJ, Peeters J, Horsman S, Duijm E, Siccama I, van den Bent MJ, Luider TM, Kros JM, van der Spek P, Sillevis Smitt PA. Identification of differentially regulated splice variants and novel exons in glial brain tumors using exon expression arrays. Cancer Res. 2007;67:5635–5642. doi: 10.1158/0008-5472.CAN-06-2869. [DOI] [PubMed] [Google Scholar]
- 27.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 28.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osareh F, Khademi R, Rostami MK, Shirazi MS. Co-authorship Network Structure Analysis of Iranian Researchers' scientific outputs from 1991 to 2013 based on the Social Science Citation Index (SSCI) Collnet J Scientometrics Inf Manage. 2014;8:263–271. doi: 10.1080/09737766.2014.1015301. [DOI] [Google Scholar]
- 30.Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol. 2011;696:291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- 31.Guo Q, Cheng Y, Liang T, He Y, Ren C, Sun L, Zhang G. Comprehensive analysis of lncRNA-mRNA co-expression patterns identifies immune-associated lncRNA biomarkers in ovarian cancer malignant progression. Sci Rep. 2015;5:17683–17683. doi: 10.1038/srep17683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 33.Weakley SM, Wang H, Yao Q, Chen C. Expression and function of a large non-coding RNA gene XIST in human cancer. World J Surg. 2011;35:1751–1756. doi: 10.1007/s00268-010-0951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J, Chen L, Xi Z, Teng H, Wang Z, et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015;359:75–86. doi: 10.1016/j.canlet.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 35.Carramusa L, Contino F, Ferro A, Minafra L, Perconti G, Giallongo A, Feo S. The PVT-1 oncogene is a Myc protein target that is overexpressed in transformed cells. J Cell Physiol. 2007;213:511–518. doi: 10.1002/jcp.21133. [DOI] [PubMed] [Google Scholar]
- 36.Yang YR, Zang SZ, Zhong CL, Li YX, Zhao SS, Feng XJ. Increased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7:6929–6935. [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi Y, Sawada G, Kurashige J, Uchi R, Matsumura T, Ueo H, Takano Y, Eguchi H, Sudo T, Sugimachi K, et al. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. Br J Cancer. 2014;110:164–171. doi: 10.1038/bjc.2013.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer KB, Maia AT, O'Reilly M, Ghoussaini M, Prathalingam R, Porter-Gill P, Ambs S, Prokunina-Olsson L, Carroll J, Ponder BA. A functional variant at a prostate cancer predisposition locus at 8q24 is associated with PVT1 expression. PLoS Genet. 2011;7:e1002165. doi: 10.1371/journal.pgen.1002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lorenz HM, Herrmann M, Winkler T, Gaipl U, Kalden JR. Role of apoptosis in autoimmunity. Apoptosis. 2000;5:443–449. doi: 10.1023/A:1009692902805. [DOI] [PubMed] [Google Scholar]
- 40.Urrego D, Tomczak AP, Zahed F, Stühmer W, Pardo LA. Potassium channels in cell cycle and cell proliferation. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130094. doi: 10.1098/rstb.2013.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu H, Li L. Biological pathway selection through nonlinear dimension reduction. Biostatistics. 2011;12:429–444. doi: 10.1093/biostatistics/kxq081. [DOI] [PubMed] [Google Scholar]
- 42.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao D, Johnson RS. Hypoxia: A key regulator of angiogenesis in cancer. Cancer Metastasis Rev. 2007;26:281–290. doi: 10.1007/s10555-007-9066-y. [DOI] [PubMed] [Google Scholar]
- 45.Weidle UH, Klostermann S, Eggle D, Krüger A. Interleukin 6/interleukin 6 receptor interaction and its role as a therapeutic target for treatment of cachexia and cancer. Cancer Genomics Proteomics. 2010;7:287–302. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.