Abstract
Objective
Cystatin C, a protein coded by CST3 gene, is implicated in adipose tissue biology. Our hypothesis is that common variants in CST3 gene could play a role in the development of corpulence during lifetime.
Methods
Two tag SNPs were selected to capture all SNPs in the CST3 region. We first investigated the association of the two tag SNPs individually and combined into haplotypes with corpulence related phenotypes in 4,288 French subjects (BMI = 24.31 (3.74 kg/m2). Significant findings were replicated in five independent populations – 790 Danish lean men (BMI = 24.63 (2.30 kg/m2), 672 Danish obese men (BMI = 33.23 (2.34 kg/m2), 763 Swedish women (BMI = 21.73 (2.87 kg/m2), 1,848 Danish lean women (BMI = 22.66 (2.85 kg/m2) and 2,061 Danish obese women (BMI = 37.01 (3.59 kg/m2).
Results
Rs2424577 was associated with BMI in three independent populations – G/G carriers were less corpulent than A/A carriers in the French individuals (p = 0.045) and in the Danish lean men (p = 0.021), and they were more corpulent in the group of Swedish women (p = 0.004). This phenomenon has been described as a flip-flop phenomenon, probably caused by a multilocus effect.
Conclusion
CST3 rs2424577 is associated with BMI in a complex fashion. This association is probably caused by the interaction between several functional variants.
Keywords: Genetics, Linkage disequilibrium, Flip-flop, Cystatin C, BMI evolution
Introduction
Obesity is due to a large number of causes that can be summarized as an interaction between a ‘deleterious’ environment and a predisposing genetic background. Depending on individuals, genetic background and environmental factors are forces driving energy imbalance, and participate in energy storage in adipose tissue. There is a wide spectrum ranging from genetically determined obesity to environmentally determined obesity with most individuals containing a complex mix of these factors (i.e. many individual effects of genes, environmental influences and the complex interaction between these). While epidemiological approaches, including twin studies, showed that genetic factors could cause 57–86% of BMI variations [1], there have been more than 400 genes referenced in the ‘NCBI gene’ database (www.ncbi.nlm.nih.gov/sites/entrez) as being associated with obesity, but each of these genes individually has a small effect on BMI variance. This is very well illustrated by the results of genome wide associations on large populations which have investigated the implication of several hundreds of thousands SNPs in BMI variance [2–5]. New variants influencing BMI were discovered, nevertheless, these variants even if combined only explain a very small part of the observed BMI variations. Therefore, it seems that most of the causal variants are still to be discovered. It is most likely that most of the variance of BMI attributable to genetic factors is due to a large number of variants, each of which has a very small effect [6–8]. Nevertheless most of these studies have been limited to data that was gathered at only a single time point and therefore only few studies have investigated the potential contribution of genetic factors to the development of corpulence during lifetime [9–11]. Understanding the mechanisms that underlie development of adipose tissue would certainly provide help to identify novel genes involved in BMI and fat mass variations during life. Using a large scale transcriptomic approach in human adipose tissue, we identified Cathepsin S as a novel biomarker of adiposity [12] with a correlation between Cathepsin S gene (CTSS) expression in adipose tissue and BMI in obese subjects. Cathepsin K or L knockout mice are protected against diet induced obesity, which highlights the potential role of Cathepsin S and K in obesity [13]. Cathepsins’ enzymatic activity is regulated by endogenous inhibitors, the most powerful being cystatin C, which is coded by the CST3 gene [14]. Emerging results from mice and cellular models suggest that cathepsin family members and cystatin C are involved in adipose tissue physiology, obesity and related complications. Several epidemiological studies have reported associations between obesity or increased adiposity and cystatin C circulating levels. Positive associations of serum cystatin C with BMI were found in an elderly population [15] and in the general population [16] and with body weight in a large population of European origin [17]. In apparently healthy subjects, higher waist circumference, an index of visceral adiposity, and increased % body fat were also associated with higher serum cystatin C [18, 19]. Recently, a graded association between higher BMI and elevated serum cystatin C was reported in American adults [20]. In addition, cystatin C appears to be a biomarker associated with vascular diseases known to be strongly associated with obesity [21]. Our team confirmed the associations between circulating cystatin C and corpulence in two populations of severely and moderately obese persons. Importantly, we showed that adipose tissue contributes to this significant elevation. Obese patients had both higher expression rates in adipose tissue and higher circulating levels of cystatin C compared to lean subjects [22]. In addition we observed that cystatin C is expressed and released by human preadipocytes differentiated in vitro [23]. Based on these findings, our main aim was to evaluate if CST3 (the gene that codes for cystatin C) is one of the numerous genetic factors that causes BMI variation in individuals. In this study, we thus searched for association between common variants (SNPs) located in the CST3 gene and BMI development during time in several populations from different European countries.
Participants and Methods
Study Design
We carried out a two stage study, the first stage was a discovery stage and the second one was a replication stage – only associations that were nominally significant in the discovery stage were thus tested in the replication stage.
In the first stage we investigated the association of two selected CST3 tag SNPs individually and then combined into haplotypes with corpulence related phenotypes – BMI, waist circumference, hip circumference and waist to hip ratio in a group of 4,288 French individuals from the SUVIMAX study. In the second stage, we only tested the association of the variants that were found to be associated with corpulence related phenotypes at a 0.05 significance threshold in the first stage. These variants were tested in five independent populations of European ancestry – 763 Swedish women from the Stockholm Pregnancy And Women’s Nutrition study (SPAWN), a group of 790 Danish lean men, a group of 672 Danish obese men and two groups of Danish women from the Danish National Birth Cohort (DNBC) (1,848 lean and 2,061 obese women). In the Danish men and the Swedish women from the SPAWN cohort, BMI, waist circumference, hip circumference and waist to hip ratio were available. In the two groups of Danish women from the DNBC cohort, only BMI and waist circumference were available.
Populations
We selected these populations because BMI measures were available at several time points for each of them. BMI at each exam was calculated as weight (kg) divided by height (m) squared. The discovery population was a group of 4,288 French individuals from the SUVIMAX study [24, 25], a group representative of the French general population in many aspects. Only 4.4% of investigated participants were obese at inclusion in 1994–1995. Height of all participants was measured at inclusion. This study lasted 13 years. Participants were invited to a follow-up exam at year 2, 4, 6 and 13 of the study. At each exam, participants were weighed. Their waist and hip circumference were measured at each exam with an inelastic tape measure using standardized methods. This population is described in table 1.
The first and second replication populations were 2 samples of Danish men – 790 lean and 672 obese subjects. Both of these groups were part of a study carried out in Denmark; participants were recruited between 1943 and 1977, they underwent 3 follow-up exams – one in 1981–1983, one in 1992–1994, and one in 1998–2000. At each exam, they were weighed and measured [10]. Data concerning their height and weight during childhood were also available (from 6 to 14 years old, measured once a year). These populations are described in table 2.
The third replication population included 763 Swedish women (1.8% of whom were obese at inclusion) from the SPAWN cohort [26] described in table 2. These women were part of a longitudinal study describing the development of corpulence after pregnancy; they underwent four exams – before pregnancy (1984), at least 1 year after pregnancy, in 2000–2001 and in 2001–2002. Height and weight were measured at inclusion and participants were weighed again at each exam.
The two last replication populations were two samples of Danish pregnant women from the DNBC [27] – a group of 1,848 lean women and a group of 2,061 obese women (table 2). These women were involved in a longitudinal study addressing the impact of intrauterine exposures on the health of the offspring. Data was available at 6 time points – before pregnancy, at the 16th gestational week, at the 30th gestational week as well as 6 months, 18 months and 7 years after birth. Participants provided their height and weight during telephone interviews except for the 7-year follow-up, which was a web-based questionnaire. They also provided their waist circumference at the 2 last exams (18 months and 7 years after birth).
Phenotypes of Interest
Four phenotypes were studied in the French SUVIMAX group – BMI, waist circumference, hip circumference and waist to hip ratio. In the groups of Danish lean and obese men and in the group of Swedish women from the SPAWN cohort, BMI was available at each follow-up time point. Waist circumference, hip circumference and waist to hip ratio were also available in these populations but not at each time point. BMI was also available in the two groups of Danish women from the DNBC cohort at each time point. Waist circumference was also available in these two groups but not at each time point.
SNP Selection
Tag SNPs were selected using data from 60 unrelated individuals from the CEU HapMap population (HapMap Data Rel 27 Phase II + III, February 2009 on NCBI B36 assembly, dbSNP B126) using the tagger pairwise method in order to capture all SNPs in the CST3 gene sequence and nearby flanking sequences (chromosome 20; 20p11.2; position 23559434 to 23569433) that are referenced in HapMap and that have a minor allele frequency equal or above 5%. We required the minimum estimated r2 between the tagged and tag SNPs sets to be > 0.90, implying only a slight loss of power in typing only tags.
Genotyping
Genotypes were determined for all individuals either by Taqman semi quantitative PCR assays (Applied Biosystem) according to the manufacturer’s instructions, by SNPlex™ at INTEGRAGEN facility (Evry, France) or using the Illumina 610-QUAD chip.
LD Measure and Haplotype Reconstruction
The amount of linkage disequilibrium (LD) between tag SNPs was measured by computing the r2 correlation coefficient [28] with FAMHAP 18 [29]. Associations between haplotypes and phenotypes were tested with HAPSTAT 3.0 [30].
Statistical Analysis
All statistical analysis was performed with R 2.10.1 [31]. Hardy-Weinberg (HW) equilibrium was tested for each SNP in each population by chi-square test. This test provided exact p values (without Yates’ correction for continuity). Deviation from HW equilibrium was considered as significant for p < 0.025 in the discovery sample and for p < 0.05 in the replication samples.
Normality of the distribution of each phenotype was tested by Cramer von Misses test. Phenotypes were transformed either logarithmically or by box cox method, using R car package [32], in order to obtain a normal distribution.
Associations between phenotypes and genotypes were tested by linear mixed models assuming an additive model using R nlme package [33] as recommended in the case of longitudinal observational studies [34].
The model that was used for the linear mixed models procedure was phenotype = genotype + exam ID indicator + age + population specific covariates, random effects = Individual ID; this model only contains main effects.
All models were adjusted for age. Models were also adjusted for gender in the French SUVIMAX group, for number of children in the Swedish women group (SPAWN cohort) and for number of pregnancies and number of children in the two groups of Danish women from the DNBC cohort.
When phenotype data was only available at one time point (waist circumference, waist to hip ratio in the group of Swedish women) associations between phenotypes and genotypes were tested by linear regression with the following model: phenotype = genotype + age + number of children.
P values < 0.05 were considered significant since we carried out a discovery analysis followed by a replication stage. The significant findings were then tested assuming a dominant and recessive model in the discovery population and tested in the five independent replication groups. Importantly, a meta-analysis was performed on the estimates from the mixed models carried out in each study using standard procedures for meta-analysis using R rmeta package [35].
Results
Two tag SNPs were selected in order to capture all SNPs referenced in HapMap that are located in CST3 gene and nearby flanking sequences – rs2424577 (HapMap reference allele: G) and rs3827143 (HapMap reference allele: A). SNP rs2424577 captured no other SNP, whereas rs3827143 captured five other SNPs – rs3787499, rs3827142, rs3787498, rs35610040 and rs3827141 – all with r2 = 1. Tag SNP frequencies, haplotype frequencies, and LD between both tag SNPs (r2) are described in table 3. All genotyped SNPs were found to be in HW equilibrium except for rs3827143 which deviated moderately from HW equilibrium in the discovery sample. Association tests carried out in each population are summarized in tables 4 and 5.
Discovery Sample (French SUVIMAX Population)
All tests carried out in this population were adjusted for age and gender; all results are for genotype main effects.
Association between CST3 rs2424577 and Corpulence Related Phenotypes
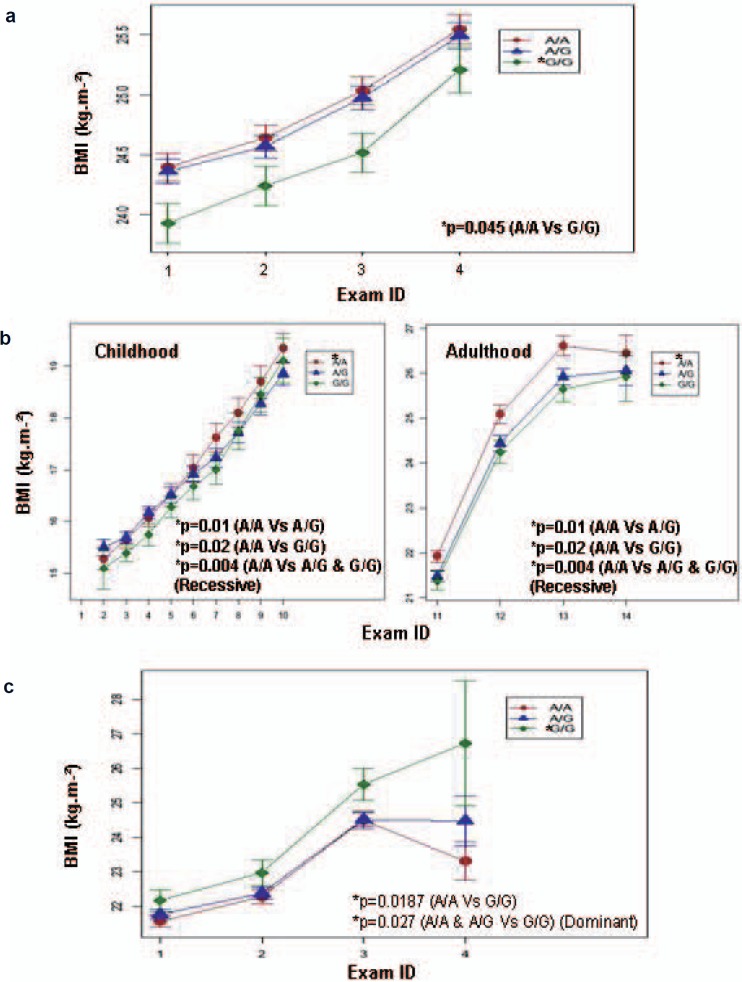
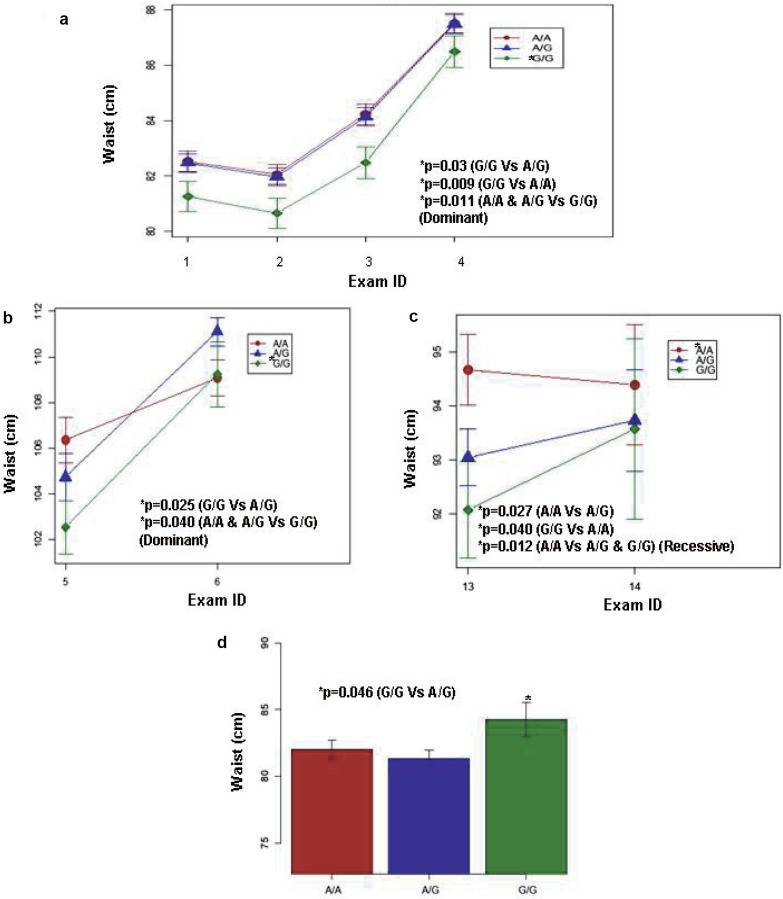
We found significant association between rs2424577 and BMI as well as waist circumference and waist to hip ratio in the SUVIMAX population. At each time point of the study, G/G carriers had significantly lower BMI (fig. 1a, table 4) than A/A carriers. G/G carriers also had lower waist circumference (fig. 2a, table 4) and waist to hip ratio (table 4) than A/A and A/G carriers. When carrying out the analysis under a dominant model (A/A and A/G vs. G/G), the association with BMI was of borderline significance and the associations with waist circumference and waist to hip ratio remained significant (table 4). No association was found when carrying out the analysis in a recessive manner (A/A vs. A/G and G/G). BMI accounts for general corpulence, whereas waist circumference, hip circumference and waist to hip ratio account for abdominal adiposity which is a risk factor for obesity associated co-morbidities. In order to investigate the specific influence of rs2424577 on adiposity while taking global corpulence into account we tested for association between rs2424577 and waist circumference as well as hip circumference and waist to hip ratio, and we included BMI as a cofactor in each model; no significant association was found; however, the association between rs2424577 and waist to hip ratio when including BMI as cofactor in the model was of borderline significance (p = 0.0534, data not shown) when carrying out the analysis in a dominant manner (G/G vs. AG and A/A). This probably suggests that the associations found with markers of body fat distribution strongly depend on global corpulence (i.e. BMI) in this population. Although G/G carriers already had a lower BMI at baseline, this difference did not reach statistical significance. Nevertheless it appeared that the association between rs2424577 and corpulence was stable during lifetime.
Association between CST3 rs3827143 and Corpulence Related Phenotypes
No significant associations between CST3 rs3827143 and any of the investigated phenotypes were found in this population.
Association between CST3 Haplotypes and Corpulence Related Phenotypes
Moderate LD was found between the two tag SNPs (table 2). Because of their ability to capture LD more informatively, testing associations with haplotypes may be more powerful than those with individual SNPs. We hence reconstructed CST3 haplotypes in order to investigate their role in the phenotypes of interest. In order to look for specific haplotype effects beyond the already identified SNP effects, we added this variant as an extra cofactor in the haplotype analysis. No association was found. Thus only CST3 rs2424577 was investigated in our five replication samples.
Replication Samples
We investigated the association between rs2424577 and three corpulence related phenotypes – BMI, waist circumference and waist to hip ratio (when available) – in the five replication populations.
Danish Men
Analysis in the group of Danish lean men revealed an association between rs2424577 and BMI (fig. 1b) in the same way as in the SUVIMAX group – G/G carriers were less corpulent than A/A carriers (table 5). In addition, we also found that A/G carriers were significantly less corpulent than A/A carriers (table 5). In this group, when carrying out the analysis under a dominant model (A/A and A/G vs. G/G), the association with BMI was no longer significant, but when the analysis was carried out under a recessive model (A/A vs. A/G and G/G), the association with BMI was significant (table 5). We also found an association between rs2424577 and both waist circumference and waist to hip ratio in the same way as in the SUVIMAX group. G/G carriers had lower waist circumference (table 5, fig. 2c) and waist to hip ratio (table 5) than A/A carriers. A/G carriers also had lower waist circumference (table 5, fig. 2c) and waist to hip ratio (table 5) than A/A carriers. When carrying out the analysis under a dominant model (A/A and A/G vs. G/G), the associations with waist circumference and waist to hip ratio were no longer significant, but when the analysis was carried out under a recessive model (A/A vs. A/G and G/G), the association with waist circumference and waist to hip ratio were significant (table 5). In contrast to lean men, we found no association between rs2424577 and BMI, waist circumference and waist to hip ratio in the group of Danish obese men (table 5).
Swedish Women (SPAWN)
In the third replication group (763 women from the SPAWN cohort), CST3 rs2424577 was significantly associated with BMI (table 5, fig. 1c) and with waist circumference (table 5, fig. 2d). Surprisingly, G/G carriers were more corpulent than A/A carriers (table 5, fig. 1c and 2d). When carrying out the analysis under a dominant model (A/A and A/G vs. G/G), only the association with BMI remained significant (table 5), but when the analysis was carried out under a recessive model (A/A vs. A/G and G/G), these associations were no longer significant.
Danish Women (DNBC)
In the two last replication cohorts, associations between rs2424577 and BMI as well as waist circumference were investigated. No significant result was found in the 1,848 lean women from the DNBC cohort (table 5). However, in the 2,091 obese women from the DNBC cohort, waist circumference was significantly associated with rs2424577 – as found in other groups, G/G carriers had a lower waist circumference than A/G carriers (table 5, fig. 2b). When carrying out the analysis under a dominant model (A/A and A/G vs. G/G), the association with waist circumference remained significant (table 5, fig. 2b).
Gene × Gender × Age Interactions
Since our results were heterogeneous, we decided to perform additional investigations. First we searched for a gender specific action of CST3 rs2424577 on BMI that could explain the different findings. We investigated the role of CST3 rs2424577 on BMI separately in men and women in our discovery sample (French SUVIMAX group). In men, G/G carriers had a lower BMI than A/A and A/G carriers (data not shown), but this difference did not reach statistical significance. In women, we found that G/G carriers had a significantly lower BMI than A/A and A/G carriers (p = 0.045 under a dominant model – A/A and A/G vs. G/G; data not shown).
Second, we searched for gene × gender, gene × age and gene × age × gender interactions in the French cohort, focusing on the association between rs2424577 (data not shown) and BMI, waist circumference and waist to hip ratio. We thus added a ‘gene × age’ or a ‘gene × gender’ or ‘gene × age × gender’ interaction term to the model described in ‘Participants and Methods’ (see above). While the ‘gene × gender’ interaction term did not reach statistical significance, the ‘gene × age’ interaction term yielded several significant results – age of the individuals apparently influenced the difference in BMI between A/G and G/G carriers (p = 0.0432), the difference in waist circumference between A/A and G/G carriers (p = 0.0358), the difference in waist circumference between A/G and G/G carriers (p = 0.0333) and the difference in waist to hip ratio between A/A and G/G carriers (p = 0.0493). The ‘gene × age × gender’ interaction term also yielded several significant results – the interaction between age and gender influenced the difference in waist circumference between A/A and G/G carriers (p = 0.0126) and between A/G and G/G carriers (p = 0.0045). In this model, gender influenced the difference in waist circumference between A/A and G/G carriers and between A/G and G/G carriers (p = 0.008 and p = 0.0039, respectively). The interaction between age and gender influenced the difference in waist to hip ratio between A/G and G/G carriers (p = 0.0294). In this model, gender also significantly influenced the difference in waist to hip ratio between A/G and G/G carriers (p = 0.0314).
Third, we tested for gene × age interactions in the association between rs2424577 and BMI, waist circumference and waist to hip ratio in our replication cohorts. In the group of Danish lean men, age also significantly influenced the difference in BMI between A/A and A/G carriers and between A/A and G/G carriers (p = 0.0007 and p = 0.022, respectively). In the group of Swedish women, age influenced the difference in BMI between A/A and G/G carriers and between A/G and G/G carriers (p = 0.0261 and p = 0.0088, respectively). Age influenced the difference in waist circumference between A/A and A/G carriers (p = 0.032) in the group of Danish lean women and the difference in waist circumference between A/A and A/G carriers (p = 0.007) and between A/A and G/G carriers (p = 0.03) in the group of Danish obese women.
Meta-Analysis of the Association of rs2424577 and BMI
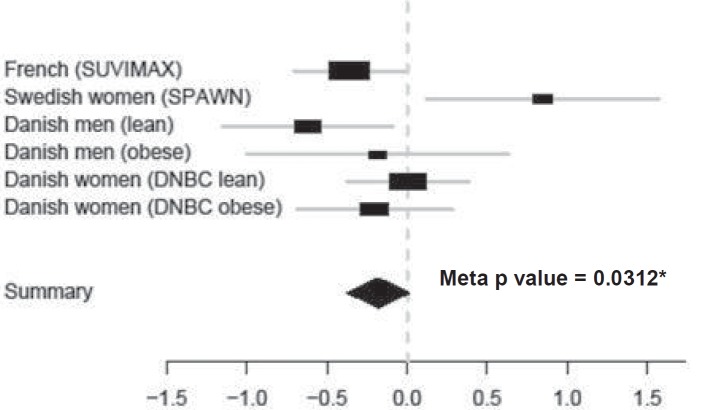
To substantiate our results, we carried out a meta-analysis on the difference in BMI between A/A and G/G carriers for rs2424577 in our six cohorts (fig. 3). The association between BMI (at each available time point in each population of our study) and rs2424577 remains significant, and the G/G genotype is associated with a lower BMI (meta p value = 0.0312, meta estimate = –0.18, 95% CI = –0.37 to 0.02).
Discussion
In our study we have identified a relationship between a variant in the CST3 gene – rs2424577 – and corpulence related phenotypes in four independent populations. Nevertheless the contribution of this SNP remains unclear since G/G genotype was associated with a protective effect in the French SUVIMAX, Danish lean men and Danish obese women groups and a deleterious effect in the Swedish female SPAWN cohort. G/G carriers had lower BMI, waist circumference and waist to hip ratio in the French SUVIMAX and the Danish lean men groups, lower waist circumference in the group of Danish obese women (DNBC), but they had higher BMI and waist circumference in the group of Swedish women (SPAWN).
This type of result is described in the literature as a ‘flip-flop phenomenon’. Lin et al. [36] demonstrated that such findings may arise from a multi-locus effect, implying a change in LD pattern between the two markers in the different studied populations. Nevertheless Zaykin et al. [37] demonstrated that such findings could simply be due to a change in haplotype frequencies in the different populations. Finally, our observations could simply be random findings. However, Clarke et al. [38] calculated the probability of obtaining such findings due to a true association or at random. They demonstrated that under the null hypothesis of no association, the probability of a significant allele flip is always α2/2 = 0.00125, whereas, in the case of a true association, the probability of such observation depends on the power of the study. In other words, it appeared that the probability to randomly obtain such findings is negligible [38]. Such flip-flop associations have already been reported for several other complex diseases including autism and late onset Alzheimer’s disease. In autism, both the long and short allele at the HTTLR locus of the serotonin transporter gene have been reported as risk alleles for autistic disorders [39–44]. In late onset Alzheimer’s disease, two independent studies report the opposite allele at the same SNP in the glutathione-S-transferase omega-1 gene (GSTO1) to be positively associated with age at onset of the disease [45, 46]. Based on these previous studies, our observation could therefore be considered as relevant. Importantly, we found significant gene × age interactions in the association with corpulence traits in the different and heterogeneous populations we explored. Age might influence the association between CST3 rs2424577 and corpulence related phenotypes. However, a gene × age interaction seems quite unlikely to explain the flip-flop phenomenon observed in this study since, when considering the age fitted values of BMI, we still retrieved the flip-flop effect, i.e. G/G carriers were still more corpulent in the group of Swedish women and less corpulent in the other groups. We carried out a meta-analysis to get an overview of the global significance of our findings. Since there is some evidence that CST3 could undergo alternate expression (see below and [47]), we decided to carry out this analysis on the results that we obtained for CST3 rs2424577 A/A versus G/G (homozygotes subjects only) and searched for an overall association with BMI in our six longitudinal studies. According to this meta-analysis, the overall association of CST3 rs2424577 and BMI was significant with G/G genotype associated to a lower BMI.
While the effect of the actual causal variant might be small, our study may nevertheless highlight an implication of CST3 – the gene that codes for cystatin C – on the level of corpulence during life time. Since cystatin C is the most powerful endogenous inhibitor of cathepsin S, K and L [14], one might speculate that the causal variant could influence corpulence by affecting the ability of cystatin C to inhibit cathepsin activity.
In our study, we used tag SNPs, which actually reflect genetic variation of the entire CTS3 region. It is now crucial to identify the variants with functional consequences in CST3 gene that are in LD with rs2424577 in order to confirm their role in corpulence related phenotypes. The contribution of CST3 gene variants has been previously described, mainly in neurological diseases and cardiovascular pathologies. The functional G73A polymorphism in CST3 gene that codes for an alanine to threonine change in position 25 was associated to Alzheimer’s disease, with homozygosity for the B haplotype (containing the A nucleotide) as a risk factor [48]. Benussi et al. [47] carried out an in vitro analysis with this SNP, and it appeared that B/B human primary epithelial fibroblasts displayed a higher intracellular retention of the protein through a less efficient cleavage of the signal peptide than A/A cells (homozygous for the G nucleotide) which is possibly due to a change in hydrophobicity in the signal peptide. However, they report that in A/B cells, while expected to have an intermediate phenotype, two out of the four analyzed cultures had A/A cells phenotype (low intracellular retention) and the two other cultures a B/B phenotype (high intracellular retention). This result suggested that only one allele of CST3 could be alternatively expressed [47]. The G73A variant was also associated with differences in cystatin C circulating levels, with the B/B genotype associated with lower cystatin C circulating levels [49, 50]. This polymorphism was also identified as associated to frontotemporal lobar degeneration [51] and to age related macular degeneration [52]. Importantly and similarly to our findings with corpulence related parameters, Crawford et al. [53] also found an association between this SNP and Alzheimer’s disease, but in the opposite direction. A/A genotype was associated with an increased risk of developing Alzheimer’s disease [53]. These findings bring evidence of association between CST3 genotypes and Alzheimer’s disease, but in different ways depending on the studied populations. The G73A variant is referenced in dbSNP (www.ncbi.nlm.nih.gov) as rs1064039. This variant has not been genotyped in any of the HapMap populations; therefore, we have no information concerning LD between this variant and the SNPs that are investigated in our study. Two other SNPs located in CST3 promoter, –82G/C and –78T/G, have been described by Eriksson et al. [54] and are associated with cystatin C circulating levels in healthy individuals and patients with recent myocardial infarctions. The authors showed that in vitro these two polymorphisms influenced the binding of nuclear factors and affected the basal rate of gene transcription in an allele specific manner. Additional SNPs such as –82G/C, –5G/A and +4A/C were also associated with cystatin C circulating levels [55]. These polymorphisms were in LD in the population studied, and individuals who carry two copies of the major haplotyp (–82G –5G +4A) had 12.4% higher circulating levels of cystatin C than non-carriers. Individuals who were heterozygous for this haplotype displayed an intermediate phenotype.
Unfortunately, none of these SNPs are referenced in HapMap. Thus no information concerning LD between these SNPs and the SNPs investigated in our study is available. We cannot exclude that other functional variants, affecting other genes located nearby CST3, might explain our findings. More explorations are necessary to understand the functional relevance of our findings.
In conclusion, we found an association between a CST3 variant and corpulence during life. The complexity of this association can be explained by a combination of variants with functional effects and by the fact that in vivo CST3 could undergo an alternative expression.
Disclosure Statement
None of the authors have a financial interest related to this work.
Fig. 1.
Evolution of BMI according to CST3 rs2424577 genotypes. Evolution (means ± SE) of BMI according to rs2424577 genotype (A/A: red circle, A/G: blue triangle and G/G: green diamond) during study period in a French individuals from the SUVIMAX group (2 to 13 years follow-up), b the group of Danish lean men, and c Swedish women from the SPAWN group (1984–2000) (exams are described in table 1 and 2). G/G and A/G carriers had a lower BMI than A/A carriers at each exam in the group of French individuals and in the group of Danish lean men. On the contrary, G/G carriers had a higher BMI at each exam than A/G and A/A carriers in the group of Swedish women. *Significant p values.
Fig. 2.
Evolution of waist circumference according to CST3 rs2424577 genotypes. Waist circumference (means ± SE) according to rs2424577 genotype (A/A: red, A/G: blue and G/G: green) in a the group of French individuals (SUVIMAX) during study period (2–13 years follow-up), b the group of Danish obese women (DNBC obese), 18 months and 7 years after birth, c the group of Danish lean men in 1992–1994 and in 1998–2000, and in D) the group of Swedish women (SPAWN) in 2001–2002 (all exams are described in table 1 and 2). G/G carriers have a lower waist circumference than A/G and A/A carriers in the group of French individuals, the group of Danish obese women and the group of Danish lean men. However, G/G carriers have higher waist circumference than A/A and A/G carriers in the group of Swedish women. *Significant p values.
Fig. 3.
Meta analysis of CST3 rs2424577 A/A versus G/G association with BMI. Estimates (black rectangles) ± 95% CI (grey lines) for CST3 rs2424577 G/G versus A/A comparison in each group and global estimate (black diamond). The overall estimate of G/G versus A/A is –0.18 (95% CI –0.37 to 0.02) and the overall p value is 0.0312. Globally, G/G carriers have a significantly lower BMI than A/A carriers.
Table 1.
Evolution of corpulence related phenotypes of French individuals (SUVIMAX, N = 4,300)
| Exam ID | Exam type | Exam description | Age, years | BMI, kg/m2 | Waist circumference, cm | Hip circumference, cm | Waist to hip ratio |
|---|---|---|---|---|---|---|---|
| 1 | follow-up | 2 years | 51.58 ± 6.24 | 24.31 ± 3.74 | 82.29 ± 12.13 | 98.84 ± 7.27 | 0.83 ± 0.089 |
| 2 | 4 years | 53.58 ± 6.24 | 24.55 ± 3.80 | 81.79 ± 12.26 | 98.49 ± 8.02 | 0.83 ± 0.094 | |
| 3 | 6 years | 55.58 ± 6.24 | 24.92 ± 3.99 | 83.88 ± 12.56 | 98.17 ± 8.55 | 0.85 ± 0.091 | |
| 4 | 13 years | 62.10 ± 6.18 | 25.47 ± 4.16 | 87.33 ± 12.50 | 98.83 ± 8.53 | 0.88 ± 0.091 |
Table 2.
Evolution of corpulence related phenotypes in replication populations
| Exam ID | Exam type | Exam description | Age, years | BMI, kg/m2 | Waist circumference, cm | Hip circumference, cm | Waist to hip ratio |
|---|---|---|---|---|---|---|---|
| Danish men (lean) n = 790 | |||||||
| 1 | school health record | birth | 0 | – | – | – | – |
| 2 | 6 years | 6 | 15.36 ± 1.05 | – | – | – | |
| 3 | 7 years | 7 | 15.62 ± 1.10 | – | – | – | |
| 4 | 8 years | 8 | 16.06 ± 1.18 | – | – | – | |
| 5 | 9 years | 9 | 16.47 ± 1.47 | – | – | – | |
| 6 | 10 years | 10 | 16.91 ± 1.65 | – | – | – | |
| 7 | 11 years | 11 | 17.32 ± 1.85 | – | – | – | |
| 8 | 12 years | 12 | 17.86 ± 2.02 | – | – | – | |
| 9 | 13 years | 13 | 18.45 ± 2.15 | – | – | – | |
| 10 | 14 years | 14 | 19.09 ± 1.90 | – | – | – | |
| 11 | follow-up | 1943–1977 | 19.98 ± 1.89 | 24.63 ± 2.30 | – | – | – |
| 12 | 1981–1983 | 37.16 ± 8.51 | 24.65 ± 3.08 | – | – | – | |
| 13 | 1992–1994 | 47.79 ± 8.55 | 26.13 ± 3.60 | 93.49 ± 10.53 | 100.26 ± 7.30 | 0.93 ± 0.07 | |
| 14 | 1998–2000 | 49.75 ± 6.13 | 26.18 ± 3.81 | 93.94 ± 10.83 | 98.08 ± 6.94 | 0.95 ± 0.06 | |
| Danish men (obese) n = 672 | |||||||
| 1 | school health record | birth | 0 | – | – | – | – |
| 2 | 6 years | 6 | 17.76 ± 1.97 | – | – | – | |
| 3 | 7 years | 7 | 18.54 ± 2.39 | – | – | – | |
| 4 | 8 years | 8 | 19.69 ± 2.51 | – | – | – | |
| 5 | 9 years | 9 | 20.63 ± 2.65 | – | – | – | |
| 6 | 10 years | 10 | 21.59 ± 2.79 | – | – | – | |
| 7 | 11 years | 11 | 22.68 ± 2.93 | – | – | – | |
| 8 | 12 years | 12 | 24.02 ± 3.08 | – | – | – | |
| 9 | 13 years | 13 | 25.09 ± 3.12 | ||||
| 10 | 14 years | 14 | 26.25 ± 3.27 | – | – | – | |
| 11 | follow-up | 1943–1977 | 19.84 ± 1.81 | 33.23 ± 2.34 | |||
| 12 | 1981–1983 | 32.52 ± 6.2 | 33.60 ± 5.06 | ||||
| 13 | 1992–1994 | 43.00 ± 6.23 | 35.61 ± 5.64 | 116.89 ± 14.08 | 114.75 ± 11.18 | 1.02 ± 0.09 | |
| 14 | 1998–2000 | 47.46 ± 5.21 | 36.01 ± 5.88 | 118.12 ± 14.34 | 114.14 ± 11.67 | 1.03 ± 0.07 | |
| Swedish women (SPAWN) n = 750 | |||||||
| 1 | before pregnancy | 1984 | 29.16 ± 4.54 | 21.73 ± 2.87 | – | – | – |
| 2 | follow-up | 1 year later | 30.16 ± 4.54 | 22.41 ± 3.28 | – | – | – |
| 3 | 2000–2001 | 46.12 ± 4.62 | 24.64 ± 4.10 | – | – | – | |
| 4 | 2001–2002 | 45.96 ± 5.20 | 24.30 ± 4.14 | 81.96 ± 10.94 | 99.53 ± 9.17 | 0.82 ± 0.07 | |
| Danish women (DNBC lean) n = 1,848 | |||||||
| 1 | before pregnancy | before pregnancy | 29.64 ± 4.21 | 22.66 ± 2.85 | – | – | – |
| 2 | during pregnancy | 12th gestational week | 29.87 ± 4.17 | 23.68 ± 2.71 | – | – | – |
| 3 | 30th gestational week | 30.24 ± 4.22 | 25.35 ± 2.49 | – | – | – | |
| 4 | after pregnancy | 6 months after birth | 31.02 ± 4.13 | 23.30 ± 3.02 | – | – | – |
| 5 | 18 months after birth | 32.16 ± 4.20 | 23.05 ± 3.09 | 79.14 ± 8.70 | – | – | |
| 6 | 7 years after birth | 36.72 ± 4.12 | 23.44 ± 2.90 | 82.42 ± 8354 | – | – | |
| Danish women (DNBC obese) n = 2,061 | |||||||
| 1 | before pregnancy | before pregnancy | 29.31 ± 4.24 | 37.01 ± 3.59 | – | – | – |
| 2 | during pregnancy | 12th gestational week | 29.55 ± 4.25 | 36.89 ± 3.69 | – | – | – |
| 3 | 30th gestational week | 29.91 ± 4.23 | 37.83 ± 4.14 | – | – | – | |
| 4 | after pregnancy | 6 months after birth | 30.72 ± 4.24 | 36.07 ± 3.83 | – | – | – |
| 5 | 18 months after birth | 31.81 ± 4.14 | 35.85 ± 3.91 | 104.88 ± 12.70 | – | – | |
| 6 | 7 years after birth | 36.53 ± 4.16 | 36.95 ± 4.79 | 110.04 ± 12.65 | – | – | |
Table 3.
Description of CST3 variability in study populations.
| French individuals (SUVIMAX) | Danish men (lean) | Danish men (obese) | Swedish women (SPAWN) | Danish women (DNBC lean) | Danish women (DNBC obese) | |
|---|---|---|---|---|---|---|
| rs2424577 | ||||||
| A | ||||||
| frequency | 0.61 | 0.61 | 0.61 | 0.62 | 0.59 | 0.60 |
| G | ||||||
| frequency | 0.39 | 0.39 | 0.39 | 0.38 | 0.41 | 0.40 |
| A/A | ||||||
| frequency | 0.38 | 0.36 | 0.35 | 0.38 | 0.35 | 0.36 |
| n | 1,600 | 289 | 236 | 288 | 641 | 744 |
| A/G | ||||||
| frequency | 0.46 | 0.49 | 0.51 | 0.49 | 0.49 | 0.48 |
| n | 1,916 | 379 | 345 | 372 | 899 | 983 |
| G/G | ||||||
| frequency | 0.16 | 0.15 | 0.14 | 0.13 | 0.17 | 0.16 |
| n | 653 | 122 | 91 | 101 | 308 | 334 |
| rs3827143 | ||||||
| A | ||||||
| frequency | 0.80 | – | – | – | – | – |
| G | ||||||
| frequency | 0.20 | – | – | – | – | – |
| A/A | ||||||
| frequency | 0.62 | – | – | – | – | – |
| n | 2,588 | – | – | – | – | – |
| A/G | ||||||
| frequency | 0.34 | – | – | – | – | – |
| n | 1,438 | – | – | – | – | – |
| G/G | ||||||
| frequency | 0.04 | – | – | – | – | – |
| n | 147 | – | – | – | – | – |
| LD | ||||||
| r2 | 0.41 | – | – | – | – | – |
| Haplotype | ||||||
| Estimated frequencies | ||||||
| AA | 0.6117 | – | – | – | – | – |
| AG | 0.0007 | – | – | – | – | – |
| GA | 0.1806 | – | – | – | – | – |
| GG | 0.2069 | – | – | – | – | – |
Table 4.
Genotype phenotype and haplotype phenotype association results in the discovery stage (significant results are shown in italics)
| CST3 variant | BMI, kg/m2 | Waist circumference, cm | Hip circumference, cm | Waist to hip ratio | |
|---|---|---|---|---|---|
| rs2424577 | |||||
| A/A vs. A/G | |||||
| p | 0.535 | 0.484 | 0.324 | 0.853 | |
| estimate | −0.096 | −0.225 | −0.262 | 0.000 | |
| 95% CI lower | −0.347 | −0.968 | −0.767 | −0.004 | |
| 95% CI upper | 0.155 | 0.518 | 0.243 | 0.003 | |
| A/A vs. G/G | |||||
| p | 0.045 | 0.009 | 0.099 | 0.009 | |
| estimate | 0.359 | 1.139 | 0.609 | 0.007 | |
| 95% CI lower | 0.015 | 0.257 | −0.086 | 0.002 | |
| 95% CI upper | 0.703 | 2.021 | 1.303 | 0.012 | |
| A/G vs. G/G | |||||
| p | 0.111 | 0.032 | 0.339 | 0.011 | |
| estimate | 0.263 | 0.912 | 0.347 | 0.006 | |
| 95% CI lower | −0.073 | 0.051 | −0.331 | 0.001 | |
| 95% CI upper | 0.599 | 1.772 | 1.024 | 0.011 | |
| A/A and A/G vs. G/G (dominant) | |||||
| p | 0.055 | 0.011 | 0.170 | 0.005 | |
| estimate | 0.307 | 1.015 | 0.466 | 0.006 | |
| 95% CI lower | −0.009 | 0.205 | −0.172 | 0.002 | |
| 95% CI upper | 0.623 | 1.825 | 1.103 | 0.011 | |
| A/A vs. A/G and G/G (recessive) | |||||
| p | 0.217 | 0.13 | 0.164 | 0.266 | |
| estimate | 0.163 | 0.458 | 0.350 | 0.002 | |
| 95% CI lower | −0.073 | −0.146 | −0.126 | −0.002 | |
| 95% CI upper | 0.399 | 1.062 | 0.825 | 0.005 | |
| rs3827143 | |||||
| A/A vs. A/G | |||||
| p | 0.780 | 0.955 | 0.959 | 0.981 | |
| estimate | 0.035 | −0.057 | −0.016 | 0.000 | |
| 95% CI lower | −0.210 | −0.685 | −0.510 | −0.004 | |
| 95% CI upper | 0.280 | 0.570 | 0.477 | 0.003 | |
| A/A vs. G/G | |||||
| p | 0.574 | 0.378 | 0.973 | 0.099 | |
| estimate | −0.180 | −0.766 | −0.015 | −0.008 | |
| 95% CI lower | −0.807 | −2.376 | −1.281 | −0.017 | |
| 95% CI upper | 0.447 | 0.843 | 1.251 | 0.001 | |
| A/G vs. G/G | |||||
| p | 0.511 | 0.400 | 0.958 | 0.108 | |
| estimate | 0.215 | 0.709 | −0.001 | 0.008 | |
| 95% CI lower | −0.426 | −0.936 | −1.295 | −0.002 | |
| 95% CI upper | 0.856 | 2.354 | 1.292 | 0.017 | |
| Haplotypes | |||||
| AA | |||||
| p | 0.552 | 0.859 | 0.681 | 0.857 | |
| estimate | −0.231 | 0.175 | 0.329 | −0.001 | |
| 95% CI lower | −0.993 | −1.755 | −1.239 | −0.012 | |
| 95% CI upper | 0.530 | 2.105 | 1.897 | 0.010 | |
| AG | |||||
| p estimate | – | – | – | – | |
| 95% CI lower | – | – | – | – | |
| 95% CI upper | – | – | – | – | |
| GA | |||||
| p | 0.260 | 0.351 | 0.208 | 0.741 | |
| estimate | −0.151 | −0.321 | −0.342 | −0.001 | |
| 95% CI lower | −0.414 | −0.995 | −0.875 | −0.005 | |
| 95% CI upper | 0.112 | 0.354 | 0.190 | 0.003 | |
| GG | |||||
| p | 0.192 | 0.393 | 0.271 | 0.697 | |
| estimate | 0.172 | 0.288 | 0.294 | 0.001 | |
| 95% CI lower | −0.086 | −0.373 | −0.229 | −0.003 | |
| 95% CI upper | 0.431 | 0.950 | 0.817 | 0.004 | |
Table 5.
Genotype phenotype association results in the replication stage (significant results are shown in italics)
| Population and phenotype | CST3 rs2424577 |
||||
|---|---|---|---|---|---|
| A/A vs. A/G | A/A vs. G/G | A/G vs. G/G | A/A and A/G vs. G/G (dominant) | A/A vs. A/G and G/G (recessive) | |
| Danish men (lean) BMI | |||||
| p | 0.010 | 0.022 | 0.670 | 0.181 | 0.004 |
| estimate | −0.510 | −0.621 | 0.111 | −0.332 | −0.537 |
| 95% CI lower | −0.895 | −1.153 | −0.402 | −0.819 | −0.901 |
| 95% CI upper | −0.125 | −0.089 | 0.624 | 0.155 | −0.173 |
| Danish men (lean) waist circumference | |||||
| p | 0.027 | 0.040 | 0.634 | 0.211 | 0.012 |
| estimate | −1.784 | −2.298 | 0.515 | −1.283 | −1.907 |
| 95% CI lower | −0.206 | −0.100 | 2.633 | 0.729 | −0.413 |
| 95% CI upper | −3.361 | −4.497 | −1.604 | −3.294 | −3.400 |
| Danish men (lean) waist to hip ratio | |||||
| p | 0.025 | 0.027 | 0.535 | 0.158 | 0.010 |
| estimate | −0.011 | −0.015 | 0.004 | −0.009 | −0.012 |
| 95% CI lower | −0.001 | −0.002 | 0.017 | 0.003 | −0.003 |
| 95% CI upper | −0.021 | −0.028 | −0.009 | −0.021 | −0.021 |
| Danish men (obese) BMI | |||||
| p | 0.355 | 0.655 | 0.260 | 0.370 | 0.534 |
| estimate | 0.263 | −0.186 | 0.449 | −0.341 | 0.169 |
| 95% CI lower | 0.822 | 0.631 | 1.231 | 0.406 | 0.703 |
| 95% CI upper | −0.295 | −1.003 | −0.333 | −1.089 | −0.365 |
| Danish men (obese) waist circumference | |||||
| p | 0.635 | 0.605 | 0.377 | 0.435 | 0.820 |
| estimate | 0.561 | −0.895 | 1.456 | −1.230 | 0.258 |
| 95% CI lower | 2.886 | 2.497 | 4.691 | 1.864 | 2.482 |
| 95% CI upper | −1.763 | −4.286 | −1.779 | −4.323 | −1.966 |
| Danish men (obese) waist/hip ratio | |||||
| p | 0.946 | 0.461 | 0.411 | 0.401 | 0.860 |
| estimate | 0.000 | −0.007 | 0.007 | −0.007 | −0.001 |
| 95% CI lower | 0.013 | 0.012 | 0.025 | 0.010 | 0.011 |
| 95% CI upper | −0.012 | −0.026 | −0.010 | −0.024 | −0.013 |
| Swedish women BMI | |||||
| p | 0.297 | 0.019 | 0.090 | 0.027 | 0.165 |
| estimate | 0.143 | 0.846 | −0.704 | 0.766 | 0.293 |
| 95% CI lower | −0.350 | 0.119 | −1.409 | 0.094 | −0.178 |
| 95% CI upper | 0.636 | 1.574 | 0.002 | 1.437 | 0.763 |
| Swedish women waist circumference | |||||
| p | 0.514 | 0.131 | 0.046 | 0.058 | 0.945 |
| estimate | −0.623 | 2.186 | −2.808 | 2.540 | −0.063 |
| 95% CI lower | −2.495 | −0.654 | −5.562 | −0.091 | −1.857 |
| 95% CI upper | 1.250 | 5.025 | −0.055 | 5.171 | 1.732 |
| Swedish women waist to hip ratio | |||||
| p | 0.164 | 0.363 | 0.993 | 0.676 | 0.146 |
| estimate | −0.008 | −0.008 | 0.000 | −0.003 | −0.008 |
| 95% CI lower | −0.020 | −0.025 | −0.024 | −0.020 | −0.019 |
| 95% CI upper | 0.003 | 0.009 | −0.017 | 0.013 | 0.003 |
| Danish women (DNBC lean) BMI | |||||
| p | 0.425 | 0.989 | 0.537 | 0.711 | 0.532 |
| estimate | 0.117 | 0.003 | 0.114 | −0.064 | 0.086 |
| 95% CI lower | −0.171 | −0.376 | −0.248 | −0.405 | −0.184 |
| 95% CI upper | 0.404 | 0.382 | 0.476 | 0.276 | 0.357 |
| Danish women (DNBC lean) waist circumference | |||||
| P | 0.957 | 0.900 | 0.863 | 0.870 | 0.995 |
| estimate | −0.032 | 0.097 | −0.129 | 0.115 | 0.003 |
| 95% CI lower | −1.190 | −1.425 | −1.595 | −1.260 | −1.083 |
| 95% CI upper | 1.126 | 1.619 | 1.337 | 1.490 | 1.089 |
| Danish women (DNBC obese) BMI | |||||
| p | 0.666 | 0.413 | 0.236 | 0.270 | 0.961 |
| estimate | 0.078 | −0.202 | 0.280 | −0.247 | 0.008 |
| 95% CI lower | −0.277 | −0.686 | −0.184 | −0.686 | −0.328 |
| 95% CI upper | 0.434 | 0.282 | 0.744 | 0.192 | 0.345 |
| Danish women (DNBC obese) waist | circumference | ||||
| p | 0.994 | 0.140 | 0.025 | 0.040 | 0.908 |
| estimate | 0.893 | −1.914 | 2.807 | −2.411 | 0.107 |
| 95% CI lower | −1.058 | −4.455 | 0.353 | −4.708 | −1.723 |
| 95% CI upper | 2.845 | 0.628 | 5.262 | −0.114 | 1.937 |
Acknowledgments
We would like to thank Dr Audrey Sabbagh for her precious advice and re-read of the manuscript.
This work was supported by a Programme Hospitalier de Recherche Clinique, Assistance Publique des Hôpitaux de Paris (AOR 02076), two grants from the French National Agency of Research (OBCAT, program ANR-05-PCOD-026–01, and COMPALIMAGE), a prize DANONE/Fondation pour la Recherche Médicale to KC, AFA insurance company (Sweden), by the Commission of the European Communities (Collaborative Project ADAPT, contract number HEALTH-F262008–201100). Support was obtained by region Ile de France and by the EU FP6 program HEPADIP. The Danish National Research Foundation established the Danish Epidemiology Science Centre, which initiated and created the Danish National Birth Cohort. The cohort is furthermore a result of a major grant from this Foundation. Additional support for the Danish National Birth Cohort was obtained from the Pharmacy Foundation, the Egmont Foundation, the March of Dimes Birth Defects Foundation and the Augustinus Foundation.
References
- 1.Silventoinen K, Kaprio J. Genetics of tracking of body mass index from birth to late middle age: evidence from twin and family studies. Obes Facts. 2009;2:196–202. doi: 10.1159/000219675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willer CJ, Speliotes EK, Loos RJF, Li S, Lindgren CM, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, Jonsdottir T, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Jonsson F, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Lauritzen T, Aben KK, Verbeek ALM, Roeleveld N, Kampman E, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Becker DM, Gulcher J, Kiemeney LA, Pedersen O, Kong A, Thorsteinsdottir U, Stefansson K. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 4.Lindgren CM, Heid IM, Randall JC, Lamina C, Steinthorsdottir V, et al. Genome-wide association scan meta-analysis identifies three loci influencing adiposity and fat distribution. PLoS Genet. 2009;5:e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naukkarinen J, Surakka I, Pietiläinen KH, Rissanen A, Salomaa V, Ripatti S, Yki-Järvinen H, van Duijn CM, Wichmann H, Kaprio J, Taskinen M, Peltonen L. Use of genome-wide expression data to mine the 'gray zone' of GWA studies leads to novel candidate obesity genes. PLoS Genetics. 2010;6:e1000976. doi: 10.1371/journal.pgen.1000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, Guo Y, Zhang L, Pei Y, Yu N, Yu P, Papasian CJ, Deng H. Biological pathway-based genome-wide association analysis identified the vasoactive intestinal peptide (VIP) pathway important for obesity. Obesity (Silver Spring.) 2010;18:2339–2346. doi: 10.1038/oby.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moonesinghe R, Liu T, Khoury MJ. Evaluation of the discriminative accuracy of genomic profiling in the prediction of common complex diseases. Eur J Hum Genet. 2010;18:485–489. doi: 10.1038/ejhg.2009.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van den Berg SW, Dollé MET, Imholz S, van der A DL, van't Slot R, Wijmenga C, Verschuren WMM, Strien C, Siezen CLE, Hoebee B, Feskens EJM, Boer JMA. Genetic variations in regulatory pathways of fatty acid and glucose metabolism are associated with Obesity phenotypes a population-based cohort study. Int J Obes. 2009;33:1143–1152. doi: 10.1038/ijo.2009.152. [DOI] [PubMed] [Google Scholar]
- 10.Jess T, Zimmermann E, Kring SII, Berentzen T, Holst C, Toubro S, Astrup A, Hansen T, Pedersen O, Sørensen TIA. Impact on weight dynamics and general growth of the common FTO rs9939609: a longitudinal Danish cohort study. Int J Obes. 2008;32:1388–1394. doi: 10.1038/ijo.2008.110. [DOI] [PubMed] [Google Scholar]
- 11.Luan J, Kerner B, Zhao J, Loos RJ, Sharp SJ, Muthén BO, Wareham NJ. A multilevel linear mixed model of the association between candidate genes and weight and body mass index using the Framingham longitudinal family data. BMC Proc. 2009;3((suppl 7)):S115. doi: 10.1186/1753-6561-3-s7-s115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taleb S, Lacasa D, Bastard J, Poitou C, Cancello R, Pelloux V, Viguerie N, Benis A, Zucker J, Bouillot J, Coussieu C, Basdevant A, Langin D, Clement K, Cathepsin S. a novel biomarker of adiposity: relevance to atherogenesis. FASEB J. 2005;19:1540–1542. doi: 10.1096/fj.05-3673fje. [DOI] [PubMed] [Google Scholar]
- 13.Lafarge J, Naour N, Clément K, Guerre-Millo M. Cathepsins and cystatin C in atherosclerosis and obesity. Biochimie. 2010;92:1580–1586. doi: 10.1016/j.biochi.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 14.Turk V, Bode W. The cystatins: protein inhibitors of cysteine proteinases. FEBS Lett. 1991;285:213–219. doi: 10.1016/0014-5793(91)80804-c. [DOI] [PubMed] [Google Scholar]
- 15.Wasén E, Isoaho R, Mattila K, Vahlberg T, Kivelä S, Irjala K. Serum cystatin C in the aged: relationships with health status. Am J Kidney Dis. 2003;42:36–43. doi: 10.1016/s0272-6386(03)00406-2. [DOI] [PubMed] [Google Scholar]
- 16.Luc G, Bard J, Lesueur C, Arveiler D, Evans A, Amouyel P, Ferrieres J, Juhan-Vague I, Fruchart J, Ducimetiere P. Plasma cystatin-C and development of coronary heart disease: the PRIME study. Atherosclerosis. 2006;185:375–380. doi: 10.1016/j.atherosclerosis.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 17.Knight EL, Verhave JC, Spiegelman D, Hillege HL, de Zeeuw D, Curhan GC, de Jong PE. Factors influencing serum cystatin C levels other than renal function and the impact on renal function measurement. Kidney Int. 2004;65:1416–1421. doi: 10.1111/j.1523-1755.2004.00517.x. [DOI] [PubMed] [Google Scholar]
- 18.Ognibene A, Mannucci E, Caldini A, Terreni A, Brogi M, Bardini G, Sposato I, Mosconi V, Salvadori B, Rotella CM, Messeri G. Cystatin C reference values and aging. Clin Biochem. 2006;39:658–661. doi: 10.1016/j.clinbiochem.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 19.Ichihara K, Saito K, Itoh Y. Sources of variation and reference intervals for serum cystatin C in a healthy Japanese adult population. Clin Chem Lab Med. 2007;45:1232–1236. doi: 10.1515/CCLM.2007.504. [DOI] [PubMed] [Google Scholar]
- 20.Muntner P, Winston J, Uribarri J, Mann D, Fox CS. Overweight, obesity, and elevated serum cystatin C levels in adults in the United States. Am J Med. 2008;121:341–348. doi: 10.1016/j.amjmed.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taglieri N, Koenig W, Kaski JC. Cystatin C and cardiovascular risk. Clin Chem. 2009;55:1932–1943. doi: 10.1373/clinchem.2009.128397. [DOI] [PubMed] [Google Scholar]
- 22.Naour N, Fellahi S, Renucci J, Poitou C, Rouault C, Basdevant A, Dutour A, Alessi M, Bastard J, Clément K, Guerre-Millo M. Potential contribution of adipose tissue to elevated serum cystatin C in human obesity. Obesity (Silver Spring) 2009;17:2121–2126. doi: 10.1038/oby.2009.96. [DOI] [PubMed] [Google Scholar]
- 23.Taleb S, Cancello R, Clément K, Lacasa D. Cathepsin S promotes human preadipocyte differentiation: possible involvement of fibronectin degradation. Endocrinology. 2006;147:4950–4959. doi: 10.1210/en.2006-0386. [DOI] [PubMed] [Google Scholar]
- 24.Hercberg S, Preziosi P, Briançon S, Galan P, Triol I, Malvy D, Roussel AM, Favier A. A primary prevention trial using nutritional doses of antioxidant vitamins and minerals in cardiovascular diseases and cancers in a general population: the SU. VI.MAX study - design, methods, and participant characteristics. SUpplementation en VItamines et Minéraux AntioXydants. Control Clin Trials. 1998;19:336–351. doi: 10.1016/s0197-2456(98)00015-4. [DOI] [PubMed] [Google Scholar]
- 25.Dolley G, Bertrais S, Frochot V, Bebel J, Guerre-Millo M, Tores F, Rousseau F, Hager J, Basdevant A, Hercberg S, Galan P, Oppert J, Lacorte J, Clément K. Promoter adiponectin polymorphisms and waist/hip ratio variation in a prospective French adults study. Int J Obes. 2008;32:669–675. doi: 10.1038/sj.ijo.0803773. [DOI] [PubMed] [Google Scholar]
- 26.Linné Y, Rössner S. Interrelationships between weight development and weight retention in subsequent pregnancies: the SPAWN study. Acta Obstet Gynecol Scand. 2003;82:318–325. doi: 10.1080/j.1600-0412.2003.00150.x. [DOI] [PubMed] [Google Scholar]
- 27.Olsen J, Melbye M, Olsen SF, Sørensen TI, Aaby P, Andersen AM, Taxbøl D, Hansen KD, Juhl M, Schow TB, Sørensen HT, Andresen J, Mortensen EL, Olesen AW, Søndergaard C. The Danish national birth cohort - its background, structure and aim. Scand J Public Health. 2001;29:300–307. doi: 10.1177/14034948010290040201. [DOI] [PubMed] [Google Scholar]
- 28.Devlin B, Risch N. A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics. 1995;29:311–322. doi: 10.1006/geno.1995.9003. [DOI] [PubMed] [Google Scholar]
- 29.Becker T, Knapp M. Maximum-likelihood estimation of haplotype frequencies in nuclear families. Genet Epidemiol. 2004;27:21–32. doi: 10.1002/gepi.10323. [DOI] [PubMed] [Google Scholar]
- 30.Lin DY, Huang BE. The use of inferred haplotypes in downstream analyses. Am J Hum Genet. 2007;80:577–579. doi: 10.1086/512201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.R Development Core Team: R A language and environment for statistical computing. 2008 available from www.R-project.org. [Google Scholar]
- 32.Fox J. car: Companion to Applied Regression. 2009 available from www.r-project.org http://socserv.socsci.mcmaster.ca/jfox/ [Google Scholar]
- 33.33 J, Bates D, DebRoy S, Deepayan Sarkar D, R Core team: nlme. Linear and Nonlinear Mixed Effects Models. 2008 available from www.r-project.org. [Google Scholar]
- 34.34 MJ. West Sussex: Wiley and Sons; 2007. The R Book. [Google Scholar]
- 35.35 T. Rmeta: meta-analysis. R package version 2.16. available from www.r-project.org. [Google Scholar]
- 36.Lin P, Vance JM, Pericak-Vance MA, Martin ER, No gene is an island. the flip-flop phenomenon. Am J Hum Genet. 2007;80:531–538. doi: 10.1086/512133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaykin DV, Shibata K. Genetic flip-flop without an accompanying change in linkage disequilibrium. Am J Hum Genet. 2008;82:794–796. doi: 10.1016/j.ajhg.2008.02.001. author reply 796–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clarke GM, Cardon LR. Aspects of observing and claiming allele flips in association studies. Genet Epidemiol. 2010;34:266–274. doi: 10.1002/gepi.20458. [DOI] [PubMed] [Google Scholar]
- 39.Devlin B, Cook EH, Coon H, Dawson G, Grigorenko EL, McMahon W, Minshew N, Pauls D, Smith M, Spence MA, Rodier PM, Stodgell C, Schellenberg GD, Autism and the serotonin transporter. the long and short of it. Mol Psychiatry. 2005;10:1110–1116. doi: 10.1038/sj.mp.4001724. [DOI] [PubMed] [Google Scholar]
- 40.Betancur C, Corbex M, Spielewoy C, Philippe A, Laplanche JL, Launay JM, Gillberg C, Mouren-Siméoni MC, Hamon M, Giros B, Nosten-Bertrand M, Leboyer M. Serotonin transporter gene polymorphisms and hyperserotonemia in autistic disorder. Mol Psychiatry. 2002;7:67–71. doi: 10.1038/sj.mp.4001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maestrini E, Lai C, Marlow A, Matthews N, Wallace S, Bailey A, Cook EH, Weeks DE, Monaco AP. Serotonin transporter (5-HTT) and gamma-aminobutyric acid receptor subunit beta3 (GABRB3) gene polymorphisms are not associated with autism in the IMGSA families. The International Molecular Genetic Study of Autism Consortium. Am J Med Gene. 1999;88:492–496. doi: 10.1002/(sici)1096-8628(19991015)88:5<492::aid-ajmg11>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 42.McCauley JL, Olson LM, Dowd M, Amin T, Steele A, Blakely RD, Folstein SE, Haines JL, Sutcliffe JS. Linkage and association analysis at the serotonin transporter (SLC6A4) locus in a rigid-compulsive subset of autism. Am J Med Genet B Neuropsychiatr Genet. 2004;127B:104–112. doi: 10.1002/ajmg.b.20151. [DOI] [PubMed] [Google Scholar]
- 43.Tordjman S, Gutknecht L, Carlier M, Spitz E, Antoine C, Slama F, Carsalade V, Cohen DJ, Ferrari P, Roubertoux PL, Anderson GM. Role of the serotonin transporter gene in the behavioral expression of autism. Mol Psychiatry. 2001;6:434–439. doi: 10.1038/sj.mp.4000873. [DOI] [PubMed] [Google Scholar]
- 44.Yirmiya N, Pilowsky T, Nemanov L, Arbelle S, Feinsilver T, Fried I, Ebstein RP. Evidence for an association with the serotonin transporter promoter region polymorphism and autism. Am J Med Genet. 2001;105:381–386. doi: 10.1002/ajmg.1365. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, Oliveira SA, Xu P, Martin ER, Stenger JE, Scherzer CR, Hauser MA, Scott WK, Small GW, Nance MA, Watts RL, Hubble JP, Koller WC, Pahwa R, Stern MB, Hiner BC, Jankovic J, Goetz CG, Mastaglia F, Middleton LT, Roses AD, Saunders AM, Schmechel DE, Gullans SR, Haines JL, Gilbert JR, Vance JM, Pericak-Vance MA, Hulette C, Welsh-Bohmer KA. Glutathione S-transferase omega-1 modifies age-at-onset of Alzheimer disease and Parkinson disease. Hum Mol Genet. 2003;12:3259–3267. doi: 10.1093/hmg/ddg357. [DOI] [PubMed] [Google Scholar]
- 46.Kölsch H, Linnebank M, Lütjohann D, Jessen F, Wüllner U, Harbrecht U, Thelen KM, Kreis M, Hentschel F, Schulz A, von Bergmann K, Maier W, Heun R. Polymorphisms in glutathione S-transferase omega-1 and AD vascular dementia, and stroke. Neurology. 2004;63:2255–2260. doi: 10.1212/01.wnl.0000147294.29309.47. [DOI] [PubMed] [Google Scholar]
- 47.Benussi L, Ghidoni R, Steinhoff T, Alberici A, Villa A, Mazzoli F, Nicosia F, Barbiero L, Broglio L, Feudatari E, Signorini S, Finckh U, Nitsch RM, Binetti G. Alzheimer disease-associated cystatin C variant undergoes impaired secretion. Neurobiol Dis. 2003;13:15–21. doi: 10.1016/s0969-9961(03)00012-3. [DOI] [PubMed] [Google Scholar]
- 48.Finckh U, von der Kammer H, Velden J, Michel T, Andresen B, Deng A, Zhang J, Müller-Thomsen T, Zuchowski K, Menzer G, Mann U, Papassotiropoulos A, Heun R, Zurdel J, Holst F, Benussi L, Stoppe G, Reiss J, Miserez AR, Staehelin HB, Rebeck GW, Hyman BT, Binetti G, Hock C, Growdon JH, Nitsch RM. Genetic association of a cystatin C gene polymorphism with late-onset Alzheimer disease. Arch Neurol. 2000;57:1579–1583. doi: 10.1001/archneur.57.11.1579. [DOI] [PubMed] [Google Scholar]
- 49.Noto D, Cefalu' AB, Barbagallo CM, Pace A, Rizzo M, Marino G, Caldarella R, Castello A, Pernice V, Notarbartolo A, Averna MR, Cystatin c levels are decreased in acute myocardial infarction. effect of cystatin c g73a gene polymorphism on plasma levels. Int J Cardiol. 2005;101:213–217. doi: 10.1016/j.ijcard.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 50.Chuo L, Sheu WHH, Pai M, Kuo Y. Genotype and plasma concentration of cystatin C in patients with late-onset Alzheimer disease. Dement Geriatr Cogn Disord. 2007;23:251–257. doi: 10.1159/000100021. [DOI] [PubMed] [Google Scholar]
- 51.Benussi L, Ghidoni R, Galimberti D, Boccardi M, Fenoglio C, Scarpini E, Frisoni GB, Binetti G. The cst3 b haplotype is associated with frontotemporal lobar degeneration. Eur J Neurol. 2010;17:143–146. doi: 10.1111/j.1468-1331.2009.02767.x. [DOI] [PubMed] [Google Scholar]
- 52.Zurdel J, Finckh U, Menzer G, Nitsch RM, Richard G. CST3 genotype associated with exudative age related macular degeneration. Br J Ophthalmol. 2002;86:214–219. doi: 10.1136/bjo.86.2.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crawford FC, Freeman MJ, Schinka JA, Abdullah LI, Gold M, Hartman R, Krivian K, Morris MD, Richards D, Duara R, Anand R, Mullan MJ. A polymorphism in the cystatin C gene is a novel risk factor for late-onset Alzheimer's disease. Neurology. 2000;55:763–768. doi: 10.1212/wnl.55.6.763. [DOI] [PubMed] [Google Scholar]
- 54.Eriksson P, Deguchi H, Samnegård A, Lundman P, Boquist S, Tornvall P, Ericsson C, Bergstrand L, Hansson L, Ye S, Hamsten A. Human evidence that the cystatin C gene is implicated in focal progression of coronary artery disease. Arterioscler Thromb Vasc Biol. 2004;24:551–557. doi: 10.1161/01.ATV.0000117180.57731.36. [DOI] [PubMed] [Google Scholar]
- 55.Loew M, Hoffmann MM, Koenig W, Brenner H, Rothenbacher D. Genotype and plasma concentration of cystatin C in patients with coronary heart disease and risk for secondary cardiovascular events. Arterioscler Thromb Vasc Biol. 2005;25:1470–1474. doi: 10.1161/01.ATV.0000168416.74206.62. [DOI] [PubMed] [Google Scholar]