Abstract
Objective
SH2B1 has been identified as an interesting candidate gene for complex obesity through genome-wide association studies. Therefore, we set out to replicate the reported association with rs7498665 in our Belgian study population and to extend our study with an additional tagSNP for the SH2B1 gene region.
Methods
We genotyped both rs7498665 and rs7201929 in a population of 1,045 obese adults and 317 healthy lean individuals. Statistical analyses were performed to evaluate the role of these polymorphisms in the development of obesity.
Results
We found that the rs7498665 minor allele increases obesity risk by 26% (ORage-sex adj = 1.26, 95% CI 1.04–1.52, nominal p = 0.016). Logistic regression showed that the rs7201929 minor allele decreases obesity risk by 24% in the population investigated (ORage-sex adj = 0.76, 95% CI 0.61–0.94, nominal p = 0.011). Conditional analyses showed that both associations represent the same association signal (rs7498665 ORadjusted for rs7201929 = 1.17, 95% CI 0.95–1.45, nominal P = 0.14; rs7201929 ORadjusted for rs7498665 = 0.82, 95% CI 0.65–1.04, nominal p = 0.10).
Methods
With the current study we were able to replicate and confirm that the SH2B1 gene locus is significantly associated with complex obesity in a Caucasian population.
Key Words: SH2B1, Obesity, Association study
Introduction
Recent genome-wide association studies (GWAS) have identified the Src homology 2 (SH2) B adaptor protein 1 (SH2B1) gene as a novel susceptibility locus for complex obesity [1, 2]. SH2B1 is a member of the SH2B protein family of adaptor proteins involved in a variety of signaling pathways. The protein was initially identified as a JAK2-interacting protein through a yeast two-hybrid screen [3]. Further investigations demonstrated that SH2B1 enhanced JAK2 activation after binding of leptin to its receptor [4, 5], demonstrating that the protein is important in the regulation of food intake by leptin. SH2B1 has also been shown to bind to the insulin receptor and enhance insulin sensitivity [6].
Further evidence implicating SH2B1 in the development of obesity comes from animal models. SH2B1 null mice demonstrate a phenotype of obesity with severe hyperphagia, hyperleptinemia, hyperinsulinemia, hyperlipidemia, hepatic steatosis, hyperglycemia, and glucose intolerance. The expression of orexigenic neuropeptide Y (NPY) and Agouti-related peptide (AgRP) was also elevated in SH2B1 null mice [7,8]. Hypothalamic restoration of SH2B1 rescues the hyperphagia, obesity, hyperglycemia, and glucose intolerance seen in knock-out (KO) mice. These SH2B1TgKO mice also showed improved leptin signaling and protection against high fat diet-induced obesity [9].
Recent genetic studies have strengthened the evidence that SH2B1 may play a key role in the development of obesity. In 2008, two GWAS identified rs7498665 (A484T) in SH2B1 as a novel susceptibility SNP for complex obesity [1, 2]. The obesity risk increased by 11% per allele of rs7498665 (OR = 1.11, 95% CI 1.06–1.17, p = 0.000022). BMI increased by 0.15 units per allele in the populations from the GIANT consortium, and 0.08% of variance in BMI was explained by rs7498665 [1]. Similar results were obtained in the deCODE GWAS [2]. Previously, the same polymorphism had already been linked to serum leptin, total fat mass, waist, and weight in a UK female twin population [10].
Furthermore, a 220 kb deletion on 16p11.2 was found in several obese individuals. This deletion encompasses the SH2B1 gene. Patients carrying the SH2B1-containing deletion have a phenotype of early-onset obesity with hyperphagia and severe insulin resistance [11].
Together these studies demonstrate that SH2B1 is a susceptibility gene for common obesity. In the present study, we aimed to replicate the previously reported association of rs7498665 with complex obesity. Furthermore, we also extended our study to include all tagSNPs for the SH2B1 region as indicated by HapMap.
Material and Methods
Population
We recruited a population of 1,045 obese individuals (486 men and 559 women, BMI ≥ 30 kg/m2; table 1) from consecutive patients consulting the outpatient obesity clinic at the Antwerp University Hospital (a tertiary referral facility). Inclusion criteria were obesity (BMI ≥ 30 kg/m2) and age 21–69 years. Exclusion criteria were pregnancy, diabetes, or impaired glucose tolerance.
317 lean control individuals (101 men and 216 women, BMI 18.5–25 kg/m2; table 1) were recruited among the university and hospital personnel and among couples seeking prenatal counseling at the Department of Medical Genetics (due to high maternal age or increased triple test). Couples seeking prenatal genetic counseling because of familial disease history were excluded. All subjects were Caucasian, and at enrolment none were involved in an ongoing weight management program. Newly diagnosed or treated diabetics and individuals with impaired glucose tolerance were excluded. The study protocol was approved by the local ethics committee; all subjects gave their written informed consent. The study protocol was in accordance to the Declaration of Helsinki.
Anthropometry
Height was measured to the nearest 0.5 cm; body weight was measured with a digital scale to the nearest 0.1 kg. BMI was calculated as weight (in kg) over height (in m) squared.
Genotyping
Genomic DNA was extracted from whole blood by a method adapted from Miller et al. [12]. We selected 2 single nucleotide polymorphisms in SH2B1 to cover genetic variation in the SH2B1 gene region. The rs7498665 polymorphism was selected because of previously reported associations with obesity in two genome-wide association studies [1, 2]. Rs7201929 was identified as tagSNP through Haploview analysis of HapMap data (HapMap Phase II Nov08, region chr16: 28772815..28797309) [13, 14]. We included the entire SH2B1 gene plus a 10 kb upstream and 3.9 kb downstream region (up to the ATP2A1 gene) in the analysis. Only SNPs with a minor allele frequency (MAF) ≥ 0.05 were included in the Tagger analysis, using aggressive tagging of 2- and 3-marker haplotypes and with r2 and LOD thresholds at 0.8 and 3.0, respectively [13]. This analysis showed that only rs7201929 was necessary as an additional SNP to cover most of the genetic variation in the SH2B1 gene and the surrounding region (rs7498665 was force-included in the analysis). TaqMan Pre-Designed Genotyping Assays (Applied Biosystems Inc., Foster City, CA, USA) were used to genotype the two selected SNPs, according to the manufacturer’s protocol, on a Lightcycler 480 Real-Time PCR System
(Roche, Penzberg, Germany). Blank samples were included as negative controls. Rs7498665 is a A/G SNP (A recoded to allele 1, G recoded to allele 2), while rs7201929 is a T/C SNP (T recoded to allele 1, C recoded to allele 2).
Statistical Analyses
Hardy-Weinberg equilibrium was checked for all individual SNPs using the HWE program from LINKUTIL [15]. Comparison of population characteristics was performed with an independent samples t-test. Logistic regression with age and sex as covariables was used to calculate odds ratios for all SNPs. To quantify the effect of a SNP on height, weight, and BMI, we performed linear regressions with age and sex as covariates. Conditional analysis to see whether rs7498665 and rs7201929 had independent effects was performed with a likelihood ratio test. All analyses were performed under an additive mode of inheritance. Significance level was set at 0.05. All statistical analyses were performed with SPSS version 15.0 (IBM, Armonk, NY, USA). Power calculations were performed using Quanto and with our population we have 80% power to detect a risk of 1.3–1.5 with a SNP with MAF of 10–35% [16]. Correction for multiple testing was performed using the false discovery rate (FDR) as described by Benjamini and Hochberg [17].
Results
In the current study we set out to replicate the previously found association between obesity and the rs7498665 SNP in the SH2B1 gene [1, 2]. Furthermore, we extended the study by also genotyping a second tagSNP for the SH2B1 gene region (rs7201929). Hardy-Weinberg equilibrium was present for both polymorphisms (rs7468665 p = 0.66; rs7201929 p = 0.06).
Both SNPs were genotyped in a population of 1,045 obese patients and 317 lean control individuals. Genotype and allele frequencies in both populations are given in table 2.
When performing logistic regression analysis to determine the obesity risk associated with the mutant allele, we found that the rs7498665 minor allele increases obesity risk by 26% (ORage-sex adj = 1.26, 95% CI 1.04–1.52, nominal p = 0.016; table 2). Each allele increases BMI by 0.89 units (95% CI 0.24–1.54, nominal page-sex adj = 0.007; table 3). Weight increased by 2.56 kg per mutant allele (95% CI 0.68–4.44, nominal page-sex adj = 0.008; table 3). Rs7498665 explained 0.5% of variance in BMI and 0.5% of variance in weight in our population.
Logistic regression showed that the rs7201929 minor allele decreases obesity risk by 24% in the population investigated (ORage-sex adj = 0.76, 95% CI 0.61–0.94, nominal p = 0.011; table 2). BMI decreased by 1.06 units per minor allele (95% CI –1.82 to –0.30, nominal page-sex adj = 0.006; table 3). No association with weight was found. Rs7201929 explained 0.5% of variance in BMI in the investigated population. All associations with rs7498665 and rs7201929 remain significant after multiple testing corrections by FDR.
Neither SNP was associated with height (rs7498665 nominal page-sex adj = 0.27; rs7201929 nominal page-sex adj = 0.34; table 3).
Conditional analyses were performed to investigate whether the effects of both SNPs were independent. These analyses showed that both associations represent the same association signal as the observed associations are no longer significant if the other SNP is included as covariate in the analysis (rs7498665 ORadjusted for rs7201929 = 1.17, 95% CI 0.95–1.45, nominal P = 0.14; rs7201929 ORadjusted for rs7498665 = 0.82, 95% CI 0.65–1.04, nominal p = 0.10).
Discussion
Since 2007, when the FTO locus was first discovered [18–20], genome-wide association studies have identified several new susceptibility loci for complex obesity [1, 2, 21–23]. The most replicated loci remain FTO [reviewed in 24] and the chromosome 18 locus near MC4R [1, 2, 23, 25–28]. However, these studies have also raised interest in other, less known genes that may be involved in the development of common obesity. One of these is the SH2B1 gene [1, 2]. This gene was immediately identified as a very good candidate gene for obesity because of prior knowledge about the obese SH2B1 knock-out mouse [7, 8]. These mice which completely lack SH2B1, are obese and display severe hyperphagia, hyperleptinemia, hyperinsulinemia, hyperlipidemia, hepatic steatosis, hyperglycemia, and glucose intolerance. Furthermore, SH2B1 is a known downstream signaling molecule in the leptin receptor signaling cascade that enhances JAK2 activation [4, 5]. Therefore, we decided to design a replication study to confirm that SH2B1 variants are associated with complex obesity. We also expanded our study to include tagSNPs covering the entire SH2B1 gene region and flanking sequences. Based on HapMap data, it was clear that we could capture most of the genetic variation in this region by genotyping the previously associated rs7498665 coding variant (A484T) and rs7201929 (3.3 kb upstream of the SH2B1 gene). We are confident that our population is suited for a replication study as we have been able to replicate the associations at the FTO [29] and near MC4R loci [30] in the past.
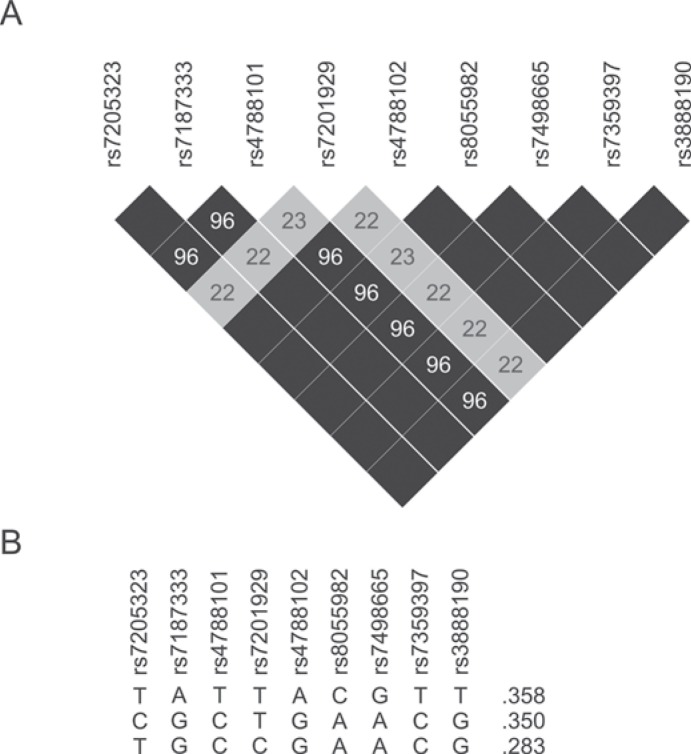
In the present study we replicated the results that rs7498665 is associated with an increased obesity risk. We also found that rs7201929 is associated with a reduction in the risk for obesity. Conditional analyses showed that both association signals are linked and can thus be considered to be the result of the same causal variant. Whether this variant is either one of the genotyped SNPs or a variant in linkage disequilibrium (LD) with them remains to be determined. However, since rs7498665 is a coding variant changing an alanine to a threonine, this SNP is the most likely candidate at the moment. This variant is also in very high LD (r2 = 0.96–1.0, HapMap data; fig. 1) with several other SNPs in or near SH2B1: rs7205323, rs7187333, rs4788101, rs4788102, rs8055982, rs7359397, and rs3888190. Only rs8055982 is also located within the SH2B1 gene, but this SNP is an intronic variant and therefore less likely to be the causal variant. Several other coupled SNPs (rs7205323, rs7187333, rs4788101, rs4788102) are located upstream from SH2B1 and could thus possibly be located in the promoter region and influence gene expression. It is clear that further investigation will be required to identify which variant may cause the observed association, although rs7498665 itself remains the best candidate at the moment.
When we compare our results to the previous studies reporting SH2B1 variants as obesity risk factors, we see some differences. In the GWAS by Willer et al. [1], rs7498665 was associated to obesity with an OR of 1.11 (95% CI 1.06–1.17), while we found an OR of 1.26 for the minor allele (95% CI 1.06–1.52). Thorleifsson et al. [2] also found a combined OR of 1.08 (95% CI 1.03–1.13) which is in line with the results from Willer et al. [1]. These differences can be attributed to differences in population structure and selection of study samples. Our patients were recruited from an outpatient obesity clinic, while the cohorts used for the GWAS were mostly population-based studies, birth cohorts or case studies recruited for other diseases (e.g. type 2 diabetes) [1, 2]. Therefore, our population may be enriched in individuals carrying the risk allele.
Recently, Speliotes et al. [31] also confirmed the involvement of SNPs in or near SH2B1 in complex obesity by demonstrating that rs7359397 is also significantly associated with BMI. Rs7359397 is in absolute LD with rs7498665 (r2 = 1; fig. 1) and thus represents the same association signal as reported by the previous GWAS and the current study [31].
With the current study we were able to replicate and confirm that the SH2B1 gene locus is significantly associated with complex obesity in a Caucasian population. We could not only replicate the association with rs7498665 but also found evidence that rs7201929 is associated with obesity risk, strengthening the evidence that SH2B1 is linked to the development of common obesity. The reported association with rs7201929, however, does require replication in an independent study group. Since the initially reported SNP rs7498665 is a non-synonymous coding variant, this polymorphism remains the best candidate to have a functional effect. Further functional studies will be necessary to study the effect this variation has on protein function. The SH2B1 gene might also be a good candidate gene for mutation analysis in patients with early-onset monogenic obesity as deletions covering this gene were already shown to be associated with an obese phenotype [11]. Therefore loss-of-function mutations in this gene may also be a direct cause of obesity and further study in this direction may be warranted.
Disclosure Statement
None declared.
Fig. 1.
Linkage disequilibrium and haplotypes for SNPs from the selected region. A Pairwise LD values (%; r2) are given for informative SNPs. Black shaded boxes indicate 100% LD. Figure based on Haploview LD plot. B Frequent haplotypes for informative SNPs from the covered region are shown (based on HapMap data). Frequency per haplotype is given. Only haplotypes with a frequency above 1% are included.
Table 1.
Description of the study groupa
| Parameter | Cases | Controls | p value |
|---|---|---|---|
| N | 1045 | 317 | – |
| Age, years | 42 ± 12 | 35 ± 7 | <0.001 |
| Male/female | 486 / 559 | 101 / 216 | – |
| Height, m | 1.71 ± 0.09 | 1.72 ± 0.10 | 0.16 |
| Weight, kg | 111.5 ± 21.8 | 65.2 ± 9.0 | <0.001 |
| BMI, kg/m2 | 38.2 ± 6.2 | 22.1 ± 1.7 | <0.001 |
Mean ± SD for age, height, weight, and BMI is given for cases and controls separately. Characteristics of cases and controls were compared using an independent samples t-test (SPSS v15.0).
Table 2.
Comparison of genotype and allele frequencies between cases and controlsa
| Genotype frequency |
Allele frequency |
OR | 95% CI | p value | ||||
|---|---|---|---|---|---|---|---|---|
| 11 | 12 | 22 | 1 | 2 | ||||
| Rs7498665 | ||||||||
| Cases (%) Controls (%) |
359 (34.4) 129 (40.7) | 505 (48.3) 143 (45.1) | 181 (17.3) 45 (14.2) | 1,223 (59) 401 (63) | 867 (41) 233 (37) | 1.26 | 1.04–1.52 | 0.016 |
| Rs7201929 | ||||||||
| Cases (%) Controls (%) |
659 (63.1) 181 (57.1) | 333 (31.9) 111 (35.0) | 53 (5.1) 25 (7.9) | 1,651 (79) 473 (75) | 439 (21) 161 (25) | 0.76 | 0.61–0.94 | 0.011 |
Genotype and allele frequencies are shown as absolute numbers (% in parentheses). OR, 95% CI and nominal p value are as calculated by logistic regression with age and sex as covariates (using SPSS 15.0).
Table 3.
Multiple regression analysis determining the change in height, weight and BMI per copy of the minor allele of each SNPa
| Rs7498665 |
Rs7201929 |
|||
|---|---|---|---|---|
| B (95% CI) | p value | B (95% CI) | p value | |
| Height, m | 0.00 (–0.00 to 0.01) | 0.27 | −0.00 (–0.01 to 0.00) | 0.34 |
| Weight, kg | 2.56 (0.68–4.44) | 0.008 | −1.90 (–4.12 to 0.32) | 0.09 |
| BMI, kg/m2 | 0.89 (0.24–1.54) | 0.007 | −1.06 (–1.82 to –0.30) | 0.006 |
Data are shown as mean effect and 95% CI. B, 95% CI and nominal p values are as calculated by linear regression with age, and sex as covariates (using SPSS 15.0).
Acknowledgements
This study was supported by a grant from the ‘Fund for Scientific Research Flanders’ (FWO-Vlaanderen) to WVH and LVG. S.B. is a postdoctoral researcher of the ‘Fund for Scientific Research Flanders’ (FWO-Vlaanderen). D.Z. holds a pre-doctoral specialization scholarship from the ‘Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen)’.
References
- 1.Willer CJ, Speliotes EK, Loos RJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, Jonsdottir T, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Jonsson F, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Lauritzen T, Aben KK, Verbeek AL, Roeleveld N, Kampman E, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Becker DM, Gulcher J, Kiemeney LA, Pedersen O, Kong A, Thorsteinsdottir U, Stefansson K. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 3.Rui L, Mathews LS, Hotta K, Gustafson TA, Carter-Su C. Identification of SH2-Bbeta as a substrate of the tyrosine kinase JAK2 involved in growth hormone signaling. Mol Cell Biol. 1997;17:6633–6644. doi: 10.1128/mcb.17.11.6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duan C, Li M, Rui L. SH2-B promotes insulin receptor substrate 1 (IRS1)- and IRS2-mediated activation of the phosphatidylinositol 3-kinase pathway in response to leptin. J Biol Chem. 2004;279:43684–43691. doi: 10.1074/jbc.M408495200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z, Zhou Y, Carter-Su C, Myers MG Jr, Rui L. SH2B1 enhances leptin signaling by both Janus kinase 2 Tyr813 phosphorylation-dependent and -independent mechanisms. Mol Endocrinol. 2007;21:2270–2281. doi: 10.1210/me.2007-0111. [DOI] [PubMed] [Google Scholar]
- 6.Morris DL, Cho KW, Zhou Y, Rui L. Sh2b1 enhances insulin sensitivity by both stimulating the insulin receptor and inhibiting tyrosine dephosphorylation of insulin receptor substrate proteins. Diabetes. 2009;58:2039–2047. doi: 10.2337/db08-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duan C, Yang H, White MF, Rui L. Disruption of the SH2-B gene causes age-dependent insulin resistance and glucose intolerance. Mol Cell Biol. 2004;24:7435–7443. doi: 10.1128/MCB.24.17.7435-7443.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ren D, Li M, Duan C, Rui L. Identification of SH2-B as a key regulator of leptin sensitivity, energy balance, and body weight in mice. Cell Metab. 2005;2:95–104. doi: 10.1016/j.cmet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Ren D, Zhou Y, Morris D, Li M, Li Z, Rui L. Neuronal SH2B1 is essential for controlling energy and glucose homeostasis. J Clin Invest. 2007;117:397–406. doi: 10.1172/JCI29417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jamshidi Y, Snieder H, Ge D, Spector TD, O’Dell SD. The SH2B gene is associated with serum leptin and body fat in normal female twins. Obesity (Silver Spring) 2007;15:5–9. doi: 10.1038/oby.2007.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bochukova EG, Huang N, Keogh J, Henning E, Purmann C, Blaszczyk K, Saeed S, Hamilton-Shield J, Clayton-Smith J, O’Rahilly S, Hurles ME, Farooqi IS. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 463:666–670. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. Nucleic Acids Res;1988:16. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 14.International HapMap Project www.hapmap.org, accessed on 14 July 2009.
- 15.Ott J. Documentation to LINKAGE UTILITY programs. http://linkage.rockefeller.edu/ott/linkutil.htm accessed on 15 December 2009.
- 16.Gauderman WJ, Morrison JM. QUANTO 1.1: A computer program for power and sample size calculations for genetic-epidemiology studies. http://hydra.usc.edu/gxe, accessed on 15 December 2009.
- 17.Benjamini Y. Hochberg Y: Controlling the False Discovery Rate – a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- 18.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CN, Doney AS, Morris AD, Smith GD, Hattersley AT, McCarthy MI. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, Najjar S, Nagaraja R, Orru M, Usala G, Dei M, Lai S, Maschio A, Busonero F, Mulas A, Ehret GB, Fink AA, Weder AB, Cooper RS, Galan P, Chakravarti A, Schlessinger D, Cao A, Lakatta E, Abecasis GR. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dina C, Meyre D, Gallina S, Durand E, Korner A, Jacobson P, Carlsson LM, Kiess W, Vatin V, Lecoeur C, Delplanque J, Vaillant E, Pattou F, Ruiz J, Weill J, Levy-Marchal C, Horber F, Potoczna N, Hercberg S, Le Stunff C, Bougneres P, Kovacs P, Marre M, Balkau B, Cauchi S, Chevre JC, Froguel P. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 21.Loos RJ, Lindgren CM, Li S, Wheeler E, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindgren CM, Heid IM, Randall JC, et al. Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS Genet. 2009;5:e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyre D, Delplanque J, Chevre JC, Lecoeur C, Lobbens S, Gallina S, Durand E, Vatin V, Degraeve F, Proenca C, Gaget S, Korner A, Kovacs P, Kiess W, Tichet J, Marre M, Hartikainen AL, Horber F, Potoczna N, Hercberg S, Levy-Marchal C, Pattou F, Heude B, Tauber M, McCarthy MI, Blakemore AI, Montpetit A, Polychronakos C, Weill J, Coin LJ, Asher J, Elliott P, Jarvelin MR, Visvikis-Siest S, Balkau B, Sladek R, Balding D, Walley A, Dina C, Froguel P. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 24.Loos RJ, Bouchard C. FTO: the first gene contributing to common forms of human obesity. Obes Rev. 2008;9:246–250. doi: 10.1111/j.1467-789X.2008.00481.x. [DOI] [PubMed] [Google Scholar]
- 25.Heard-Costa NL, Zillikens MC, Monda KL, et al. NRXN3 is a novel locus for waist circumference a genome-wide association study from the CHARGE Consortium. PLoS Genet. 2009;5:e1000539. doi: 10.1371/journal.pgen.1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zobel DP, Andreasen CH, Grarup N, Eiberg H, Sorensen TI, Sandbaek A, Lauritzen T, Borch-Johnsen K, Jorgensen T, Pedersen O, Hansen T. Variants near MC4R are associated with obesity and influence obesity-related quantitative traits in a population of middle-aged people: studies of 14,940 Danes. Diabetes. 2009;58:757–764. doi: 10.2337/db08-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Renstrom F, Payne F, Nordstrom A, Brito EC, Rolandsson O, Hallmans G, Barroso I, Nordstrom P, Franks PW. Replication and extension of genome-wide association study results for obesity in 4923 adults from northern Sweden. Hum Mol Genet. 2009;18:1489–1496. doi: 10.1093/hmg/ddp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenesa A, Campbell H, Theodoratou E, Dunlop L, Cetnarskyj R, Farrington SM, Dunlop MG. Common genetic variants at the MC4R locus are associated with obesity but not with dietary energy intake or colorectal cancer in the Scottish population. Int J Obes (Lond) 2009;33:284–288. doi: 10.1038/ijo.2008.257. [DOI] [PubMed] [Google Scholar]
- 29.Peeters A, Beckers S, Verrijken A, Roevens P, Peeters P, Van Gaal L, Van Hul W. Variants in the FTO gene are associated with common obesity in the Belgian population. Mol Genet Metab. 2008;93:481–484. doi: 10.1016/j.ymgme.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 30.Beckers S, Zegers D, de Freitas F, Mertens IL, Van Gaal LF, Van Hul W. Association study of mc4r with complex obesity and replication of the rs17782313 association signal. Mol Genet Metab. 2011;103:71–75. doi: 10.1016/j.ymgme.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 31.Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]