Abstract
Asthma is associated with the overproduction of leukotrienes (LTs), including LTB4. Patients with severe asthma can be highly responsive to 5-lipoxygenase (5-LO) inhibition, which blocks production of both the cysteinyl LTs and LTB4. Production of LTB4 has traditionally been ascribed to neutrophils, mononuclear phagocytes, and epithelial cells, and acts as a chemoattractant for inflammatory cells associated with asthma. The source of LTB4 is unclear, especially in eosinophilic asthma. We speculated that the benefit of 5-LO inhibition could be mediated in part by inhibition of eosinophil-derived LTB4. LTB4 concentrations were assayed in BAL fluid from patients with severe asthma characterized by isolated neutrophilic, eosinophilic, and paucigranulocytic inflammation. Expression of LTA4 hydrolase (LTA4H) by airway eosinophils was determined by immunohistochemistry (IHC). Subsequently, peripheral blood eosinophils were activated and secreted LTB4 was quantified by enzyme immunoassay. Blood eosinophil LTA4H expression was determined by flow cytometry, qPCR, and IHC. LTB4 concentrations were elevated in BAL fluid from patients with severe asthma, including those with isolated eosinophilic inflammation, and these eosinophils displayed LTA4H via IHC. LTA4H expression by blood eosinophils was confirmed by flow cytometry, IHC, and qPCR. Robust LTB4 production by blood eosinophils was observed in response to some, but not all, stimuli. We demonstrated that eosinophils express LTA4H transcripts and protein, and can be stimulated to secrete LTB4. We speculate that in many patients with asthma, eosinophil-derived LTB4 is increased, and this may contribute to the efficacy of 5-LO inhibition.
Keywords: asthma, eosinophils, leukotrienes, neutrophils
Clinical Relevance
Eosinophils were never previously thought to express LTA4 hydrolase and as such be a source of LTB4. LTB4 overexpression contributes to the severity of asthma, and the current observations support the efficacy of 5-lipoxygenase inhibitors in this disease.
Inhibition of 5-lipoxygenase (5-LO) is an effective pharmacological intervention in asthma and may be particularly beneficial in more severe presentations of asthma (1, 2). By inhibiting the synthesis of cysteinyl leukotrienes (CysLTs), 5-LO inhibitors block generation of the ligands for the CysLT receptors and, as such, share activities mediated by CysLT type 1 receptor (CysLT1R) antagonists. However, the broader efficacy associated with 5-LO inhibition suggests additional relevant biological activities. Thus, by inhibiting the generation of CysLTs, 5-LO inhibitors would also block CysLT-mediated signaling through the CysLT type 2 receptor and the more recently described leukotriene (LT) E4 receptor (3, 4). It is increasingly recognized that the production of LTB4 may also contribute to asthma severity. LTB4 is produced from the sequential metabolism of arachidonic acid by 5-LO and LTA4 hydrolase (LTA4H). LTB4 is found at high levels in the asthmatic airway (5–15), and engagement of its receptors, BLT1 and BLT2, contributes to the recruitment and activation of polymorphonuclear neutrophils (PMNs), eosinophils, and lymphocytes, in particular, CD8+ cytotoxic T cells (16–22). What is less clear, however, is the source of LTB4 in asthmatic inflammation. LTA4H is primarily associated with PMNs, inflammatory mononuclear phagocytes, and to a lesser extent epithelial cells (23). In asthma, the source(s) of LTB4 is less apparent—in particular, its source in the presentation of this disorder that is associated with alternatively activated macrophages, alveolar macrophages, and eosinophils, but few, if any PMNs or inflammatory mononuclear phagocytes. It is increasingly recognized that eosinophils have an ever-expanding arsenal of proinflammatory functions. As such, we speculated that although they are not traditionally associated with these cells, eosinophils could be a central source of LTB4 production in asthma. We therefore investigated the presence of LTB4 in eosinophilic (and noneosinophilic) presentations of severe asthma, and subsequently confirmed the expression by airway and blood eosinophils of the metabolic enzymes necessary to generate LTB4, as well as the capacity of activated eosinophils to secrete this lipid mediator.
Methods
Reagents
PerCP anti-human Siglec-8 and anti-human LTA4H were obtained from R&D Systems, and BV510 anti-human CD16b and a BD Cytofix/Cytoperm kit were obtained from BD Biosciences. Anti-human LTA4H was conjugated to PE using the Lightning-Link (R) R-PE Antibody Labeling Kit (Novus Biologicals). Carrier-free goat anti-human 5-LO peptide N-15 and rabbit anti-human LTC4 synthase peptide S-18 antibodies were obtained from Santa Cruz Biotechnology as special orders. The antibodies were conjugated with Alexa Fluor 488 and 647, respectively, by monoclonal antibody labeling kits (Invitrogen). DAPI was purchased from Roche Diagnostic Corp. Diff-Quik was obtained from Siemens Healthcare Diagnostics, Fast Green was obtained from Acros Organics, and Wright-Giemsa was obtained from Volu-Sol.
BAL Samples
BAL samples were obtained from clinically indicated bronchoscopies performed on children and adolescents with poorly controlled asthma who had been referred to the University of Virginia, as previously described (24). Briefly, all subjects underwent an initial assessment protocol (25, 26) that included 1) confirmation of the diagnosis of asthma; 2) correction of remediable factors, including poor adherence and incorrect inhaler technique; 3) evaluation of comorbidities; 4) identification of allergen sensitization with institution of appropriate interventions; and 5) evaluation of initial lung function. Baseline symptom control was measured and controller treatment was adjusted based on severity and control status in accordance with standard guidelines (27, 28) and the children were followed longitudinally. Children whose asthma remained poorly controlled despite confirmation of adherence to treatment (29) were then offered a diagnostic bronchoscopy, with additional BAL fluid (BALF) shared with our research laboratories through protocols approved by the University of Virginia Institutional Review Board. Informed consent was provided by both parents or by legal guardians when feasible, and adolescents provided consent to participate in the protocol.
Bronchial Fluid Granulocyte Patterns
The BALF samples were cytospun and stained, and differential cellular analyses were performed by counting 100 consecutive cells. BALF granulocyte patterns (neutrophilic, eosinophilic, mixed granulocytic, and paucigranulocytic) were identified based on the approach in original proof-of-concept studies performed on sputum granulocytes (30) and correlated with phenotypic features in adults by investigators in the Severe Asthma Research Program of the National Heart, Lung, and Blood Institute, National Institutes of Health (31, 32). Cutpoints for BALF eosinophilia and neutrophilia were defined according to differential counts summarized in the ERS Task Force on BAL in Children (33) and investigations in healthy children (34).
Blood Eosinophils
Eosinophils were purified from 100 ml of peripheral blood from healthy control subjects and patients with asthma. The healthy control subjects were nonallergic. Patients with asthma were defined by physician diagnosis. Subjects with a recent exacerbation or recent use of oral corticosteroids (within 60 d) were excluded from participation. Eosinophils (>99% pure as determined by flow cytometry) were enriched using Ficoll-Hypaque density gradient centrifugation followed by dextran sedimentation. Eosinophils were then separated from PMNs by negative selection using anti-CD16 magnetic affinity beads (Miltenyi Biotec), with counts ranging from 20 × 106 to 100 × 106 total eosinophils.
Flow Cytometry
Briefly, isolated eosinophils were resuspended in ice-cold staining buffer (PBS + 2% FCS) and incubated with human IgG for 5 minutes at 4°C. Subsequently, the cells were stained with PerCP anti-human Siglec-8, BV510 anti-human CD16B, or the appropriate isotype controls at 4°C for 20 minutes and then washed twice with staining buffer. The cells were then permeabilized with a BD Cytofix/Cytoperm kit according to the manufacturer’s directions. The cells were then stained with Alexa Fluor 488 anti–5-LO, PE anti-LTA4H, and Alexa Fluor 647 anti-LTC4S at 4°C for 20 minutes, after which they were washed twice and fixed with 2% paraformaldehyde. Samples were analyzed on a FACSCantoII instrument using FACSDiva software (BD Biosciences).
Immunohistochemistry
Immunohistochemistry (IHC) was performed on cytospins of the BALF and peripheral blood eosinophils, and stained for LTA4H and LTC4S. Before staining, the cells were fixed in 2% p-formaldehyde-lysine-periodate for 3 hours and equilibrated in sucrose as previously described (35). The antibodies were conjugated with Alexa Fluor 555 and 647. respectively, by monoclonal antibody labeling kits (Invitrogen) and used in cell staining at 2 μg/ml for confocal microscopy analysis on a Zeiss LSM 700 microscope assembly with ZEN software for image analysis. Alexa Fluor 555 sheep IgG and 647 rabbit IgG were used as isotype controls for staining.
qPCR
qPCR was performed on peripheral blood eosinophils for LTA4H. Blood eosinophils were enriched as described above and total RNA was extracted using TRI reagent (Sigma). Conversion of mRNA to cDNA was performed using a Taqman Reverse Transcription kit (Roche). RNA (200 ng) was added to each reaction along with oligo dT primers, 5.5 mM MgCl2, 2 mM dNTPs, RNasin, and reverse transcriptase. Reactions went through 10 minutes at 25°C, 30 minutes at 48°C, and 5 minutes at 95°C in a Bio-Rad iCycler thermocycler (Bio-Rad). The PCR mix consisted of Sensimix (Bioline), which contains the dye SYBR Green, cDNA, and 200 μM of each primer. Data were analyzed as the change in CT of the LTA4H transcript in comparison with the housekeeping gene β-actin. The primers for β-actin were as previously described (36). The primers for LTA4H were sense 5′-GCCCGAGATAGTGGATACCTG-3′ and antisense 5′-TTTTTGCTCAAAGCGATAGGA-3′ (Integrated DNA Technologies, Inc.).
Eosinophil Stimulation
Blood eosinophils were obtained as described earlier and then activated for 30 minutes at 37°C with lysine aspirin (LysASA, 10 mM; Sanofi-Aventis), platelet-activating factor (PAF) (1 μM; Sigma), trypsin (1 μM: Sigma), and a combination of PMA (33 ng/ml [low] or 100 ng/ml [high]) and ionomycin (2 μg/ml; Sigma). Supernatants were collected and LTB4 was quantified by enzyme immunoassay (EIA) as described below.
EIA
LTB4 in supernatants of BALF and of resting and activated blood eosinophils was quantified with a commercial EIA (ENZO Life Sciences) according to the manufacturer’s directions. The lower limit of detection of the assay was 11.7 pg/ml. Eosinophil-derived neurotoxin (EDN) release by activated blood eosinophils was also quantified by EIA (MBL).
Statistical Analyses
Data from unstimulated and stimulated cells were compared using the Wilcoxon rank-sum test for nonparametric data analyses or a paired t test for parametric data. Control and asthma cohorts were compared using unpaired t tests. Statistical analyses were performed using GraphPad Prism 6.
Results
LTB4 in Airways of Patients with Severe Asthma
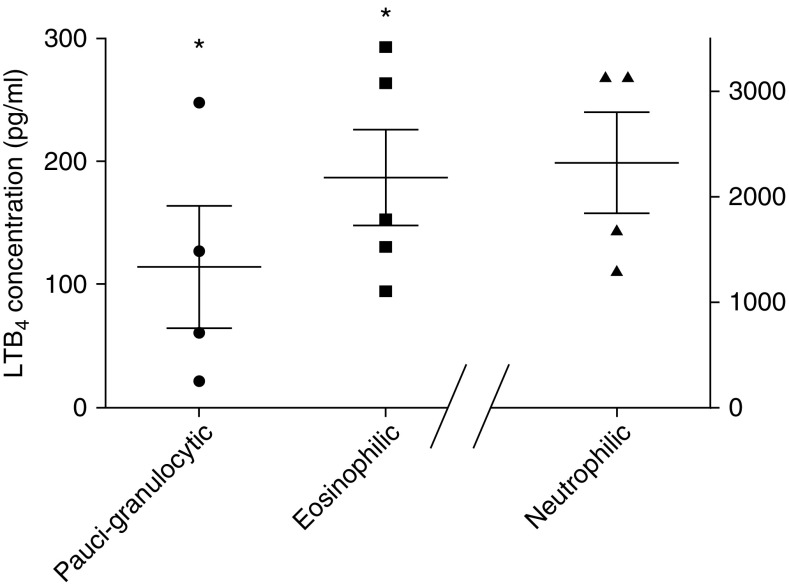
Children and adolescents with severe treatment-resistant asthma were identified and subsequently underwent a clinically indicated bronchoscopy, with additional BALF obtained under a research protocol. Distinct granulocytic patterns were identified in the BALF, and subjects with eosinophilic, neutrophilic, or paucigranulocytic patterns underwent further evaluation. As summarized in Table 1, these were patients with severe, very poorly controlled asthma, as demonstrated by poor responses on their asthma control test (ACT) despite high-dose inhaled corticosteroid and/or oral prednisone use with reduced lung function. In the eosinophilic cohort, four out of five subjects were allergic, and all demonstrated elevated blood eosinophilia (absolute eosinophil count ≥ 300/μl). As demonstrated in Figure 1, all 14 subjects demonstrated expression of LTB4 in their BALF at concentrations well exceeding the sensitivity of the EIA. Not surprisingly, LTB4 concentrations were highest among children with predominant neutrophilic disease. However, LTB4 was readily identified in the BALF of children without PMNs and instead was characterized by eosinophilic inflammation, and LTB4 concentrations were higher than those observed in the paucigranulocytic cohort.
Table 1.
Patient Characteristics
| Eosinophilic | Neutrophilic | Paucigranulocytic | |
|---|---|---|---|
| Number | 5 | 4 | 5 |
| Age (± SEM) | 11.2 ± 1.4 | 4.5 ± 1.8 | 8.3 ± 2.1 |
| Male sex, n (%) | 3 (60%) | 3 (75%) | 3 (60%) |
| Present atopy, n (%) | 4 (80%) | 2 (50%) | 2 (40%) |
| Eosinophils (%) | 4 ± 1 | 0 ± 0 | 0 ± 0 |
| PMNs (%) | 2 ± 1 | 34 ± 5 | 1 ± 1 |
| Mononuclear phagocytes (%) | 69 ± 6 | 54 ± 2 | 71 ± 10 |
| Serum IgE (median [25th–75th percentile]) | 330 (63–1,662) | 101 (18–288) | 30 (12–894) |
| AEC (median [25th–75th percentile]) | 593 (415–768) | 355 (115–835) | 180 (100–215) |
| ACT (mean ± SEM) | 13.2 ± 1.7 | 20.0 ± 3.5 | 15 ± 1.5 |
| ICU admission history (n) | 3 | 0 | 2 |
| Daily prednisone use (n) | 3 | 0 | 1 |
| ICS (fluticasone equivalents μg/d; median [25th–75th percentile]) | 800 (160–800) | 330 (160–725) | 480 (160–890) |
| Pre-FEV1/FVC (± SEM) | 0.76 ± 0.05 | 0.89* | 0.81 ± 0.05 |
| Post-FEV1/FVC (± SEM) | 0.83 ± 0.05 | ND | 0.87 ± 0.03 |
Definition of abbreviations: AEC = absolute eosinophil count; ACT = asthma control test; FEV1 = forced expiratory volume in 1 second; FVC = forced vital capacity; ICS = inhaled corticosteroid; ND = not done; PMNs = polymorphonuclear leukocytes.
Only one subject was old enough to participate in spirometry.
Figure 1.
Leukotriene B4 (LTB4) levels in BAL fluid from patients with severe asthma and different granulocyte patterns. BAL was collected during bronchoscopy of individuals with severe treatment-resistant asthma, and enzyme immunoassays were run to measure LTB4 concentrations. Samples were analyzed based on BAL granulocyte patterns and LTB4 levels (presented as pg/ml). Data are presented as individual data points along with the mean ± SEM for each group. *P < 0.005 as compared with neutrophilic.
LTA4H Protein in BALF Eosinophils
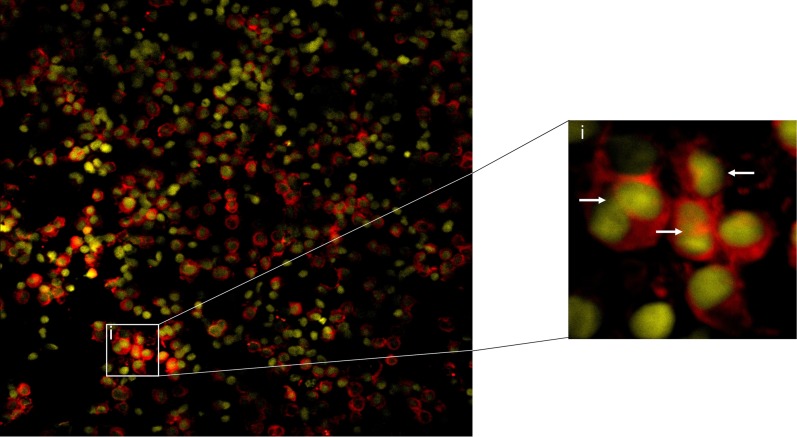
EIA of BALF—even when obtained from patients with asthma and an isolated eosinophilic granulocyte pattern—cannot exclude noneosinophils as the source of the LTB4. In support of the argument that BAL eosinophils had the potential to be a source of this lipid mediator, we evaluated their expression of LTA4H protein via IHC. BAL samples obtained from subjects with eosinophilic inflammation were cytospun and IHC was performed for LTA4H. Airway eosinophils routinely demonstrated expression of LTA4H protein (a representative sample is displayed in Figure 2).
Figure 2.
Immunofluorescence for LTA4 hydrolase (LTA4H) in BAL eosinophils. LTA4H staining of BAL eosinophils using a primary antibody directed against LTA4H and an APC-labeled secondary antibody (red) with DAPI nuclear stain (yellow). The inset (i) shows a close-up view, with white arrows indicating eosinophils.
LTA4H Expression in Resting Blood Eosinophils
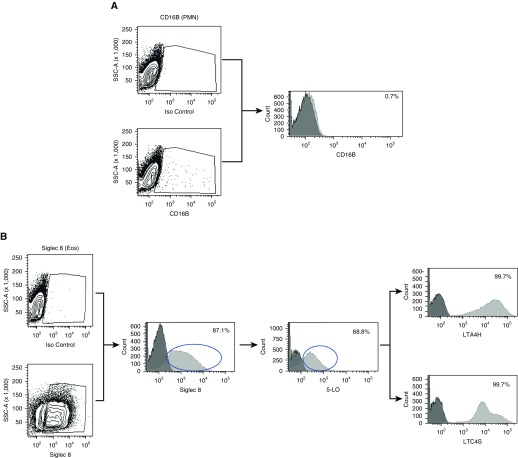
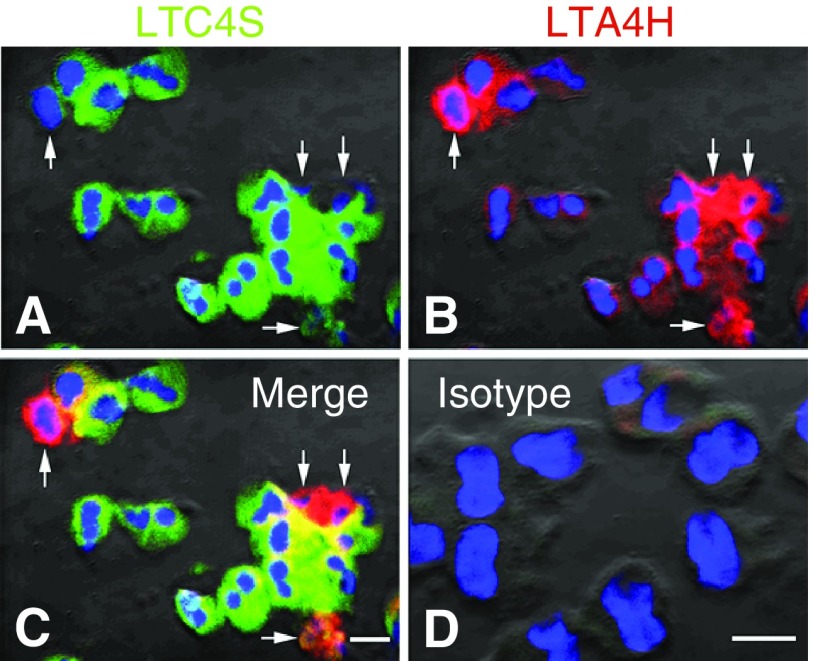
To verify the purity of the eosinophil preparation and confirm the absence of neutrophils, flow cytometry was performed to examine CD16b expression in the cell population. As shown in Figure 3A, less than 1% of the cells expressed CD16b, indicating very little neutrophil contamination (Figure 3A). Of the CD16b+ cells, 83% expressed 5-LO, and within this population 100% of the cells expressed LTA4H and LTC4S (data not shown). The purity of the eosinophil preparation was also confirmed via hematological staining of cytospun samples and again demonstrated >99% eosinophils (Figure E1 in the data supplement). We then examined LTA4H expression in the eosinophil population by flow cytometry. All of the eosinophils that were 5-LO+ (69%) expressed both LTA4H and LTC4S (Figure 3B). LTA4H protein expression in blood eosinophils was also confirmed via IHC. In line with the flow cytometry results, we identified LTA4H protein in most, but not all, blood eosinophils. This was in contrast to LTC4S, which was identified in all blood eosinophils (Figure 4).
Figure 3.
Flow cytometry for eosinophils and LTA4H. (A) Purified eosinophils were analyzed for neutrophil contamination by measuring CD16b expression by flow cytometry; results are shown in a representative histogram plot. (B) Eosinophil expression of LTA4H and LTC4 synthase (LTC4S) was determined by examining Siglec 8+ eosinophils for 5-lipoxygenase (5-LO) expression, and within that population the percentage of cells that expressed these molecules. Circles approximate the Siglec 8+ population that was gated upon to identify cells that were also positive for 5-LO and similarly the 5-LO+ cells that were analyzed for percent positive for LTA4H or LTC4S. Eos = eosinophil; Iso = isotype; PMN = polymorphonuclear neutrophil; SSC = side scatter.
Figure 4.
Immunofluorescence for LTA4H in purified blood eosinophils. Eosinophils were purified from blood by Ficoll-Hypaque density centrifugation and magnetic bead affinity column purification. Staining of LTC4S was performed using an Alexa Fluor 647–labeled antibody directed against LTC4S (green), LTA4H staining was performed using an Alexa Fluor 555–labeled antibody directed against LTA4H (red), and DAPI was used as a nuclear stain (blue). (A and B) Note that all eosinophils are positive for expression of LTC4S (A) and most, but not all, are positive for LTA4H (B). (C and D) A merged image is shown (C), and the isotype control is included to show background staining (D). Arrows represent dual positive staining eosinophils for LTC4S and LTA4H. Scale bars: 10 μM.
LTA4H Transcript in Peripheral Blood Eosinophils
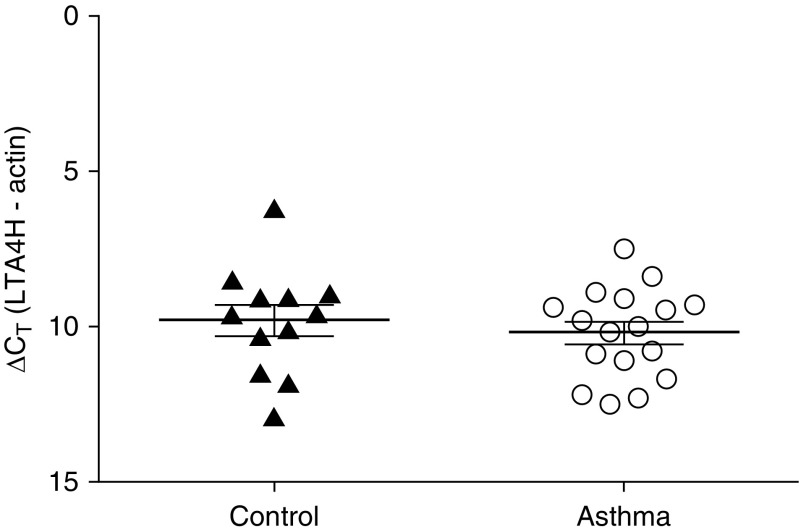
We next investigated expression of LTA4H mRNA in blood eosinophils via qPCR. We compared expression in healthy control subjects (n = 12) and patients with asthma (n = 17), with the data normalized to expression of the housekeeping gene β-actin. As demonstrated in Figure 5, LTA4H was expressed by blood eosinophils derived from both groups; however, no differences in transcript expression were observed between the two cohorts.
Figure 5.
LTA4H mRNA expression in eosinophils. After separation of blood using Ficoll-Hypaque density centrifugation and dextran sedimentation, eosinophils were enriched using magnetic affinity column purification. Transcript levels of LTA4H were quantified using PCR with SYBR Green detection. Data (mean ± SEM) reflect the relative expression of each gene in comparison with the housekeeping gene β-actin (∆CT). Control samples (n = 12) are depicted by solid triangles, and asthma samples are indicated (n = 17) by open circles.
Blood Eosinophil LTB4 Secretion
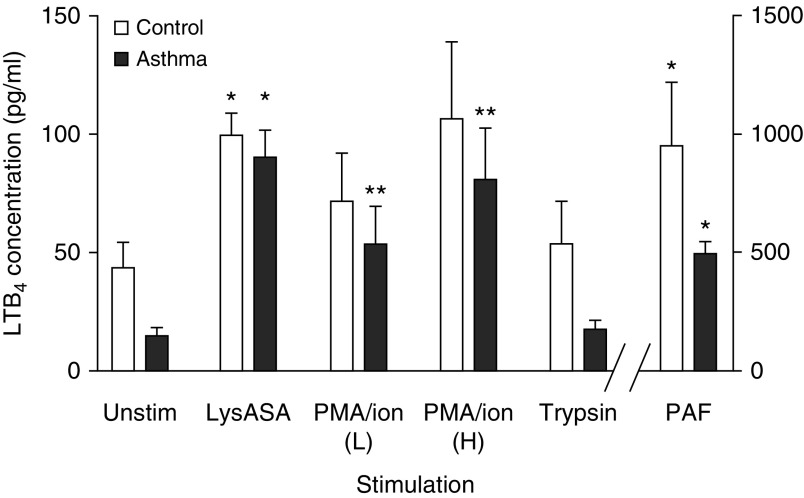
Blood eosinophils from healthy control subjects (n = 12) and patients with asthma (n = 11) were activated with LysASA, PMA/ionomycin, PAF, and trypsin, and supernatants were assayed for LTB4 and EDN by EIA. Although all of the stimuli activated eosinophils as shown by EDN release (not shown), LTB4 production was observed only in response to LysASA, PMA/ionophore, and PAF, and not with trypsin. No differences were observed in the production of LTB4 in control subjects versus patients with asthma (Figure 6).
Figure 6.
LTB4 release from aspirin-activated eosinophils. Eosinophils isolated from peripheral blood of control subjects (open bars: n = 12) and patients with asthma (solid bars: n = 11) were activated with lysine aspirin (LysASA; 10 mM), the combination of ionomycin (2 μg/ml) with low (L; 33 ng/ml) or high (H; 100 ng/ml) dose phorbol 12-myristate 13-acetate (PMA), trypsin 1 μM, or platelet-activating factor (PAF) 1 μM for 30 minutes. Supernatants were collected and LTB4 levels were quantified by enzyme immunoassay (pg/105 cells). Data are presented as mean ± SEM with *P < 0.001 or **P < 0.02 in comparison with unstimulated cells.
Discussion
In contrast to the healthy airway (8, 13), LTB4 has been found at high levels in the asthmatic airway (5–15), and this finding was confirmed in the current studies (Figure 1). LTB4 contributes to asthma severity through its ability to recruit and activate PMNs, eosinophils, and CD8+ cytotoxic lymphocytes (16–22). Although it is considered to be primarily engaged in PMN recruitment, the LTB4 receptor BLT1 is expressed on eosinophils (37), where it drives eosinophil recruitment. Although isolated blockade of LTB4 receptors has not demonstrated a striking benefit in asthma (38, 39) a contribution of LTB4 to asthma severity may underlie the improved efficacy of 5-LO inhibitors in comparison with isolated CysLT1R antagonists (1).
Although its production is primarily ascribed to PMNs, inflammatory mononuclear phagocytes, and epithelial cells, the source of LTB4 in asthma is less clear, especially in those patients with asthma who do not display prominent neutrophilia. In the current studies, we investigated the capacity of eosinophils to serve as an additional source of LTB4 in asthma. We demonstrated that eosinophils derived from the asthmatic airway and the peripheral blood express both LTA4H transcripts and protein. In addition, these cells secrete LTB4 after stimulation with LysASA, PAF, or the combination of PMA/ionomycin (Figure 6).
We appreciate that although LTB4 concentrations were high in asthmatic airways characterized by eosinophilia (and only rarely neutrophils), this does not exclude mononuclear phagocytic or other cells (or even tissue PMNs) as a source. Indeed, LTB4 was measurable even in paucigranulocytic disease (Figure 1), which may argue for “background” production by airway cells, including tissue macrophages and epithelial cells. However, LTB4 concentrations were higher in eosinophilic disease, where these cells were available as an additional source. Not surprisingly, the highest concentrations were observed in neutrophilic subjects, given the prominent role of these cells in making (and responding) to this mediator (5, 6). Of note, monocytes were the most prevalent cell observed in the BALF samples, but their numbers did not vary significantly among the three cohorts (Table 1), arguing that they could plausibly be responsible for the “background” concentrations of LTB4 observed.
We previously reported the ability of LysASA to activate eosinophils (40) and suggested that this could be relevant in patients with asthma and aspirin-exacerbated respiratory disease. In contrast, trypsin (an activator of eosinophil protease-activated receptors) did not induce LTB4 release, arguing for a distinct signaling pathway driving LTB4 secretion that can be differentially regulated from degranulation pathways (41).
Eosinophils in peripheral blood from control subjects and patients with asthma did not exhibit any difference in either their LTA4H transcript or protein expression or their ability to produce LTB4. Presumably, it would be the presence of eosinophils themselves, as well as the presence of the appropriate activating signals, that would distinguish LTB4 production in allergic inflammatory disorders.
It was also noteworthy that, unlike LTC4S, LTA4H was not universally expressed by all circulating eosinophils (Figures 3B and 4B). This suggests the possibility that unique features drive its expression during eosinophilopoiesis. (Although it is noteworthy that insofar as comparable protein levels were observed in control subjects and patients with asthma, whatever factor(s) is involved in driving transcript expression, it is unlikely to involve mechanisms specific to type 2 inflammation.)
Given the chemotactic function of LTB4 on both PMNs and eosinophils, these data cannot explain the selective recruitment of eosinophils or PMNs in these two cohorts. Presumably, these distinct granulocyte patterns must reflect the requirement for other factors, including different adhesion molecules, chemokines, and other chemotaxins, as well as all of the variable influences of factors involved in cell expression and survival (including variable responsiveness to high-dose inhaled steroids).
In summary, the current studies demonstrate the ability of eosinophils to express LTA4H and generate LTB4 in response to their activation. Eosinophil-derived LTB4 likely contributes to the presence and severity of inflammation in asthma, in particular, those presentations characterized by prominent eosinophilia (and a paucity of neutrophils).
Supplementary Material
Footnotes
Supported by National Institutes of Health grants UO1AI123337, AI057438, and AI120055.
Author Contributions: K.P. and X.F. performed and helped design the experiments. S.-S.S. designed and performed the immunofluorescence studies. M.D.B. designed and performed the flow cytometry studies. W.G.T. performed the bronchoscopies and helped with data analysis and authorship of the paper. J.W.S., Y.M.S., and L.B. designed the experiments, performed the data analyses, and authored the paper.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2018-0175OC on October 23, 2018
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Dahlén B, Nizankowska E, Szczeklik A, Zetterström O, Bochenek G, Kumlin M, et al. Benefits from adding the 5-lipoxygenase inhibitor zileuton to conventional therapy in aspirin-intolerant asthmatics. Am J Respir Crit Care Med. 1998;157:1187–1194. doi: 10.1164/ajrccm.157.4.9707089. [DOI] [PubMed] [Google Scholar]
- 2.Steinke JW, Payne SC, Borish L. Interleukin-4 in the generation of the AERD phenotype: implications for molecular mechanisms driving therapeutic benefit of aspirin desensitization. J Allergy (Cairo) 2012;2012:182090. doi: 10.1155/2012/182090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanaoka Y, Maekawa A, Austen KF. Identification of GPR99 protein as a potential third cysteinyl leukotriene receptor with a preference for leukotriene E4 ligand. J Biol Chem. 2013;288:10967–10972. doi: 10.1074/jbc.C113.453704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bankova LG, Lai J, Yoshimoto E, Boyce JA, Austen KF, Kanaoka Y, et al. Leukotriene E4 elicits respiratory epithelial cell mucin release through the G-protein-coupled receptor, GPR99. Proc Natl Acad Sci USA. 2016;113:6242–6247. doi: 10.1073/pnas.1605957113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seymour ML, Rak S, Aberg D, Riise GC, Penrose JF, Kanaoka Y, et al. Leukotriene and prostanoid pathway enzymes in bronchial biopsies of seasonal allergic asthmatics. Am J Respir Crit Care Med. 2001;164:2051–2056. doi: 10.1164/ajrccm.164.11.2008137. [DOI] [PubMed] [Google Scholar]
- 6.Zaitsu M, Hamasaki Y, Matsuo M, Ichimaru T, Fujita I, Ishii E. Leukotriene synthesis is increased by transcriptional up-regulation of 5-lipoxygenase, leukotriene A4 hydrolase, and leukotriene C4 synthase in asthmatic children. J Asthma. 2003;40:147–154. doi: 10.1081/jas-120017985. [DOI] [PubMed] [Google Scholar]
- 7.Shindo K, Fukumura M, Miyakawa K. Leukotriene B4 levels in the arterial blood of asthmatic patients and the effects of prednisolone. Eur Respir J. 1995;8:605–610. [PubMed] [Google Scholar]
- 8.Sampson AP, Castling DP, Green CP, Price JF. Persistent increase in plasma and urinary leukotrienes after acute asthma. Arch Dis Child. 1995;73:221–225. doi: 10.1136/adc.73.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wenzel SE, Trudeau JB, Kaminsky DA, Cohn J, Martin RJ, Westcott JY. Effect of 5-lipoxygenase inhibition on bronchoconstriction and airway inflammation in nocturnal asthma. Am J Respir Crit Care Med. 1995;152:897–905. doi: 10.1164/ajrccm.152.3.7663802. [DOI] [PubMed] [Google Scholar]
- 10.Zaitsu M, Hamasaki Y, Ishii K, Kita M, Hayasaki R, Muro E, et al. Direct evidence that LTC4 and LTB4 but not TXA2 are involved in asthma attacks in children. J Asthma. 1998;35:445–448. doi: 10.3109/02770909809048953. [DOI] [PubMed] [Google Scholar]
- 11.Csoma Z, Kharitonov SA, Balint B, Bush A, Wilson NM, Barnes PJ. Increased leukotrienes in exhaled breath condensate in childhood asthma. Am J Respir Crit Care Med. 2002;166:1345–1349. doi: 10.1164/rccm.200203-233OC. [DOI] [PubMed] [Google Scholar]
- 12.Montuschi P, Barnes PJ. Exhaled leukotrienes and prostaglandins in asthma. J Allergy Clin Immunol. 2002;109:615–620. doi: 10.1067/mai.2002.122461. [DOI] [PubMed] [Google Scholar]
- 13.Wardlaw AJ, Hay H, Cromwell O, Collins JV, Kay AB. Leukotrienes, LTC4 and LTB4, in bronchoalveolar lavage in bronchial asthma and other respiratory diseases. J Allergy Clin Immunol. 1989;84:19–26. doi: 10.1016/0091-6749(89)90173-5. [DOI] [PubMed] [Google Scholar]
- 14.Trischler J, Müller CM, Könitzer S, Prell E, Korten I, Unverzagt S, et al. Elevated exhaled leukotriene B4 in the small airway compartment in children with asthma. Ann Allergy Asthma Immunol. 2015;114:111–116. doi: 10.1016/j.anai.2014.11.022. [DOI] [PubMed] [Google Scholar]
- 15.Higham A, Cadden P, Southworth T, Rossall M, Kolsum U, Lea S, et al. Leukotriene B4 levels in sputum from asthma patients. ERJ Open Res. 2016;2:00088–2015. doi: 10.1183/23120541.00088-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rae SA, Smith MJ. The stimulation of lysosomal enzyme secretion from human polymorphonuclear leucocytes by leukotriene B4. J Pharm Pharmacol. 1981;33:616–617. doi: 10.1111/j.2042-7158.1981.tb13884.x. [DOI] [PubMed] [Google Scholar]
- 17.Sumimoto H, Takeshige K, Minakami S. Superoxide production of human polymorphonuclear leukocytes stimulated by leukotriene B4. Biochim Biophys Acta. 1984;803:271–277. doi: 10.1016/0167-4889(84)90117-4. [DOI] [PubMed] [Google Scholar]
- 18.Sehmi R, Wardlaw AJ, Cromwell O, Kurihara K, Waltmann P, Kay AB. Interleukin-5 selectively enhances the chemotactic response of eosinophils obtained from normal but not eosinophilic subjects. Blood. 1992;79:2952–2959. [PubMed] [Google Scholar]
- 19.Hébert MJ, Takano T, Holthöfer H, Brady HR. Sequential morphologic events during apoptosis of human neutrophils. Modulation by lipoxygenase-derived eicosanoids. J Immunol. 1996;157:3105–3115. [PubMed] [Google Scholar]
- 20.Gelfand EW, Dakhama A. CD8+ T lymphocytes and leukotriene B4: novel interactions in the persistence and progression of asthma. J Allergy Clin Immunol. 2006;117:577–582. doi: 10.1016/j.jaci.2005.12.1340. [DOI] [PubMed] [Google Scholar]
- 21.Dakhama A, Collins ML, Ohnishi H, Goleva E, Leung DY, Alam R, et al. IL-13-producing BLT1-positive CD8 cells are increased in asthma and are with airway obstruction. Allergy. 2013;68:666–673. doi: 10.1111/all.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gelfand EW. Importance of the leukotriene B4-BLT1 and LTB4-BLT2 pathways in asthma. Semin Immunol. 2017;33:44–51. doi: 10.1016/j.smim.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bigby TD, Lee DM, Minami M, Ohishi N, Shimizu T, Baker JR. Characterization of human airway epithelial cell leukotriene A4 hydrolase. Am J Respir Cell Mol Biol. 1994;11:615–624. doi: 10.1165/ajrcmb.11.5.7946391. [DOI] [PubMed] [Google Scholar]
- 24.Wisniewski JA, Muehling LM, Eccles JD, Capaldo BJ, Agrawal R, Shirley DA, et al. TH1 signatures are present in the lower airways of children with severe asthma, regardless of allergic status. J Allergy Clin Immunol. 2018;141:2048–2060.e13. doi: 10.1016/j.jaci.2017.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bush A, Saglani S. Management of severe asthma in children. Lancet. 2010;376:814–825. doi: 10.1016/S0140-6736(10)61054-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharples J, Gupta A, Fleming L, Bossley CJ, Bracken-King M, Hall P, et al. Long-term effectiveness of a staged assessment for paediatric problematic severe asthma. Eur Respir J. 2012;40:264–267. doi: 10.1183/09031936.00209511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Heart, Lung, and Blood Institute. National Asthma Education and Prevention Program. Expert Panel 3: guidelines for the diagnosis and management of asthma. Full report2007August 28, 2007 [accessed 2018 Dec 12]. Available from: http://www.nhlbi.nih.gov/guidelines/asthma/asthgdln.pdf.
- 28.Global Initiative for AsthmaGlobal Strategy for Asthma Management and Prevention; 2017. [accessed 2018 Dec 12]. Available from: https://ginasthma.org/gina-reports/
- 29.Fitzpatrick AM, Kir T, Naeher LP, Fuhrman SC, Hahn K, Teague WG. Tablet and inhaled controller medication refill frequencies in children with asthma. J Pediatr Nurs. 2009;24:81–89. doi: 10.1016/j.pedn.2008.02.027. [DOI] [PubMed] [Google Scholar]
- 30.Simpson JL, Scott R, Boyle MJ, Gibson PG. Inflammatory subtypes in asthma: assessment and identification using induced sputum. Respirology. 2006;11:54–61. doi: 10.1111/j.1440-1843.2006.00784.x. [DOI] [PubMed] [Google Scholar]
- 31.Hastie AT, Moore WC, Meyers DA, Vestal PL, Li H, Peters SP, et al. National Heart, Lung, and Blood Institute Severe Asthma Research Program. Analyses of asthma severity phenotypes and inflammatory proteins in subjects stratified by sputum granulocytes. J Allergy Clin Immunol. 2010;125:1028–1036.e13. doi: 10.1016/j.jaci.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore WC, Hastie AT, Li X, Li H, Busse WW, Jarjour NN, et al. National Heart, Lung, and Blood Institute Severe Asthma Research Program. Sputum neutrophil counts are associated with more severe asthma phenotypes using cluster analysis. J Allergy Clin Immunol. 2014;133:1557–1563.e5. doi: 10.1016/j.jaci.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Blic J, Midulla F, Barbato A, Clement A, Dab I, Eber E, et al. European Respiratory Society. Bronchoalveolar lavage in children. ERS Task Force on bronchoalveolar lavage in children. Eur Respir J. 2000;15:217–231. doi: 10.1183/09031936.00.15121700. [DOI] [PubMed] [Google Scholar]
- 34.Heaney LG, Stevenson EC, Turner G, Cadden IS, Taylor R, Shields MD, et al. Investigating paediatric airways by non-bronchoscopic lavage: normal cellular data. Clin Exp Allergy. 1996;26:799–806. [PubMed] [Google Scholar]
- 35.Sung SJ, Ge Y, Dai C, Wang H, Fu SM, Sharma R, et al. Dependence of glomerulonephritis induction on novel intraglomerular alternatively activated bone marrow-derived macrophages and Mac-1 and PD-L1 in lupus-prone NZM2328 mice. J Immunol. 2017;198:2589–2601. doi: 10.4049/jimmunol.1601565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steinke JW, Crouse CD, Bradley D, Hise K, Lynch K, Kountakis SE, et al. Characterization of interleukin-4-stimulated nasal polyp fibroblasts. Am J Respir Cell Mol Biol. 2004;30:212–219. doi: 10.1165/rcmb.2003-0071OC. [DOI] [PubMed] [Google Scholar]
- 37.Tager AM, Luster AD. BLT1 and BLT2: the leukotriene B(4) receptors. Prostaglandins Leukot Essent Fatty Acids. 2003;69:123–134. doi: 10.1016/s0952-3278(03)00073-5. [DOI] [PubMed] [Google Scholar]
- 38.Evans DJ, Barnes PJ, Spaethe SM, van Alstyne EL, Mitchell MI, O’Connor BJ. Effect of a leukotriene B4 receptor antagonist, LY293111, on allergen induced responses in asthma. Thorax. 1996;51:1178–1184. doi: 10.1136/thx.51.12.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh RK, Tandon R, Dastidar SG, Ray A. A review on leukotrienes and their receptors with reference to asthma. J Asthma. 2013;50:922–931. doi: 10.3109/02770903.2013.823447. [DOI] [PubMed] [Google Scholar]
- 40.Steinke JW, Negri J, Liu L, Payne SC, Borish L. Aspirin activation of eosinophils and mast cells: implications in the pathogenesis of aspirin-exacerbated respiratory disease. J Immunol. 2014;193:41–47. doi: 10.4049/jimmunol.1301753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miike S, McWilliam AS, Kita H. Trypsin induces activation and inflammatory mediator release from human eosinophils through protease-activated receptor-2. J Immunol. 2001;167:6615–6622. doi: 10.4049/jimmunol.167.11.6615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.