Abstract
A level of epigenetic programming, encoded by complex sets of chemical marks on DNA and histones, and by context-specific DNA, RNA, protein interactions, that all regulate the structure, organization, and function of the genome, is critical to establish both normal and neoplastic cell identities and functions. This structure–function relationship of the genome encoded by the epigenetic programming can be thought of as an epigenetic cityscape that is built on the underlying genetic landscape. Alterations in the epigenetic cityscape of prostate cancer cells compared with normal prostate tissues have a complex interplay with genetic alterations to drive prostate cancer initiation and progression. Indeed, mutations in genes encoding epigenetic enzymes are often observed in human cancers including prostate cancer. Interestingly, alterations in the prostate cancer epigenetic cityscape can be highly recurrent, a facet that can be exploited for development of biomarkers and potentially as therapeutic targets.
Epigenetics can be defined as the study of processes that mediate potentially heritable changes in gene regulation without a change in the genetic sequence. Here, heritable refers to passing down information from parent cells to daughter cells rather than from parents to children (although the intergenerational transmission of epigenetic encoding is a fascinating and emerging area of research, such inheritance as a factor affecting prostate cancer risk has not been explored significantly). Epigenetic processes therefore mediate the level of programming that establishes distinct cell identities and functions across cell types and over time in a given individual in whom the genetic sequence would be largely unchanging except for stochastic accumulation of rare errors in a given cell and programmed genetic alterations in cells of the adaptive immune system.
The syntax of the epigenetic code is comprised of chemical modifications on the DNA, on histones in nucleosomes, and by binding interactions between DNA, RNA, proteins, and other macromolecules that signal changes to the organization and function of the genome. These epigenetic processes can therefore be thought of as a cell-intrinsic epigenetic code that establishes cellular phenotypes by modulating the structure–function dynamics of the genome. It is therefore apt to consider the epigenetic code as a cityscape architecture built on the underlying genomic landscape. An elaborate epigenetic machinery of proteins and other macromolecules (including noncoding RNAs) construct, maintain, and dynamically remodel this epigenetic cityscape (Jenuwein and Allis 2001; Wang et al. 2004; Chi et al. 2010; Chen and Dent 2014). This epigenetic machinery has been broadly classified by their functions in “writing,” “erasing,” and “reading” the epigenetic code; and here we expand this lexicon to include two additional critical functions of the epigenetic machinery, “preserving,” and “organizing/remodeling” (Table 1). This epigenetic machinery can establish and maintain the cellular epigenetic programing or can dynamically alter it. How cells choose to maintain or dynamically alter their epigenetic program during division and other processes is not fully understood.
Table 1.
Overview of the epigenetic machinery
| Writing | Erasing | Reading | Preserving | Remodeling | |
|---|---|---|---|---|---|
| Histone acetylation | Histone acetyl-transferases (e.g., KAT6B,a p300/CBP) | Histone deacetylase complexes (e.g., ARID4A,a HDAC1, HDAC2, HDAC3, HDAC4, HDAC6) | BET bromodomain proteins (e.g., BRD4, BRD2, BRD3) | Unknown | SWI/SNF remodeling complexes (e.g., CHD1,a CHD3,a CHD8,a ARID1A,a ARID2,a SMARCA1,a SMARCAD1,a ATRXa) |
| Histone methylation | Histone methyl-transferases (e.g., KMT2C,a KMT2D,a KMT2A,a SETD2,a SETDB1,a EZH2, DOT1L) | Histone demethylases (e.g., KDM6A,a KDM1A, KDM4B, KDM4C, KDM2A) | Chromodomain, PHD finger proteins | UHRF1, unknown | Mi2-NuRD remodeling complex (CHD3,a CHD4) |
| DNA methylation | DNA methyl-transferases (e.g., DNMT1, DNMT3A, DNMT3B) | TET enzymes, BER machinery, and modifiers (e.g., IDH1,a,b TET1, TET2,a TET3, TDG) | MBD, zinc finger proteins (e.g., MBD1,a MBD2, MECP2, ZBTB33) | UHRF1, DNMT1 | Variant histones (e.g., H2A.x, H2A.z, H3.3) |
Multiple epigenetic machinery proteins have recurrent driver gene mutations in prostate cancer.
aGenes recurrently harboring driver gene mutations in prostate cancer. Most of the mutations are putative loss-of-function, with the exception of IDH1. Genes that are frequently altered in expression in prostate cancer are shown in italics alone (without superscript).
bIDH1 is not directly involved in DNA demethylation activity, but recurrent hotspot mutations redirect the activity of IDH1 to generate the metabolite 2-hydroxyglutarate, which can inhibit α-ketoglutarate-dependent oxygenases, including the TET family of enzymes involved in DNA demethylation.
Remarkably, large-scale cancer genome sequencing studies have shown that genes encoding these epigenetic machinery components are often enriched in the set of mutated “driver” genes in many human cancer types, including prostate cancer. The consequences of these mutations on the epigenetic code and their mechanistic role in cancer initiation and progression are just beginning to be unraveled. Even independent of these mutations in the epigenetic machinery, alterations to the epigenetic code, seen as changes in DNA methylation and other epigenetic marks, are universal in human cancers, including prostate cancer (Fig. 1). The patterns of epigenetic alterations in a given cancer may reveal key targets for development of cancer biomarkers and therapeutic strategies. Here, we will examine the basic mechanisms and principles of the epigenetic code, summarize our understanding of epigenetic alterations in human prostate cancer, and discuss the translation of this understanding to develop strategies for prostate cancer detection, risk stratification, and therapy.
Figure 1.
DNA methylation cityscapes of lethal metastatic prostate cancer. DNA hypermethylation (top) and DNA hypomethylation (bottom) alterations in multiple anatomically distinct metastatic deposits obtained by rapid autopsy from each of 13 men that died of metastatic prostate cancer are shown. In these cityscape plots, each chromosome is folded into a two-dimensional neighborhood demarcated by light gray lines, following a Hilbert curve. In the cityscapes, each structure represents a region harboring a DNA methylation alteration in at least one tumor compared with a series of benign prostate tissues. The height of the structures represents the number of tumors having the alteration at that region. Tall structures, skyscrapers in the cityscape, are nearly universally altered in methylation in all metastases studied, and short structures (huts) are infrequently altered. The color represents the degree of maintenance of each alteration across all metastases within each subject normalized to the overall variability (R2). Hypermethylated promoter regions of cancer-related genes from the NCI Cancer Gene Index that were in the upper 10th percentile for frequency (height) or maintenance (color) are labeled on the cityscape. A region on chromosome 5, harboring the APC promoter, is magnified in the “blowout” showing the Hilbert curve indicated in the white path and shows multiple clustered hypermethylated regions with varying frequency and maintenance in this region. Although DNA hypomethylation was pervasive in large blocks across the genome, DNA hypermethylation was much more focal and often highly maintained across all metastases within subjects even when the overall frequency of these alterations was low across subjects. Highly frequent (skyscrapers) and/or highly maintained (orange to red) alterations in the hypermethylation cityscape were enriched for (1) overlap with gene promoters, particularly those associated with development/differentiation and cancer-related genes, and (2) inverse correlation with cis gene expression. Thus, in the backdrop of a general tendency for losing methylation across the genome, hypermethylation alterations were highly maintained, and appeared to be selected for across metastatic dissemination. (From Aryee et al. 2013; reprinted, with permission, from the American Association for the Advancement of Science ©2013.)
THE MAJOR EPIGENETIC MARKS AND THE MACHINERY RESPONSIBLE FOR THEIR REGULATION
DNA Methylation
Methylation of cytosine in DNA at the 5-position (5mC) is an important and common epigenetic mark, with conserved regulation and function across vertebrate species (plants and other eukaryotes and some prokaryotes also use cytosine methylation as an epigenetic mark). This mark is usually found in the palindromic cytosine-phospho-guanine (CpG) dinucleotides. Other sequence contexts can also harbor 5mC, particularly observed in embryonic stem cells and some neuronal tissues; typically, 5mC in non-CpG contexts accounts for <1% of the overall 5mC content in DNA in the majority of adult cell types but may still have some functional roles (Schultz et al. 2015). Although ∼80% of CpGs are methylated in normal cells, 5mC is usually not found in CpG-dense “CpG islands” that often surround gene transcriptional start sites and other regulatory regions (Jones 2012). In contrast, in cancer cells, CpG islands and surrounding genomic sequences referred to as “CpG island shores” can become hypermethylated, often leading to repression of the nearby gene (Jones and Baylin 2007; Irizarry et al. 2009). Such DNA methylation-mediated epigenetic repression is enriched at cancer-protective genes and in genes involved in development and differentiation (Jones 2012; Aryee et al. 2013). The epigenetic repression mediated by this CpG island hypermethylation can be very stable (Aryee et al. 2013; Vandiver et al. 2015), resembling the inactivation of genes through loss-of-function mutations and deletions. Concomitant with such DNA hypermethylation, cancer cells paradoxically also develop progressive DNA hypomethylation, often occurring in large genomic blocks spanning many megabases (Feinberg et al. 2006; Yegnasubramanian et al. 2008; Hansen et al. 2011; Aryee et al. 2013). Although such hypomethylation does not seem to directly regulate the expression of the encompassed genes in any consistent manner (Aryee et al. 2013), this loss of methylation may promote an open chromatin that is permissive of genomic instability (Ehrlich 2002), including by activation of endogenous retroviral elements.
DNA methylation marks are “written” by DNA methyltransferase enzymes, encoded by three genes, DNMT1, DNMT3A, and DNMT3B, in human cells (Jurkowska et al. 2011). All of these enzymes have preferential activity at methylating cytosines in CpG dinucleotides compared with any other cytosines. Another gene, DNMT3L, can assist in the activity of DNMT3A and DNMT3B, but does not have its own catalytic activity. DNMT1 is largely responsible for the maintenance of DNA methylation during DNA replication by tracking with the replication fork (Okano et al. 1998; Pradhan et al. 1999; Jeltsch 2002). Compared with completely unmethylated DNA, DNMT1 catalytic activity is significantly enhanced at hemimethylated DNA, as would be present when the palindromic CpG dinucleotide in the parental DNA is methylated with the daughter strand being unmethylated. DNMT1 and DNMT3 isoforms can also cooperate to maintain methylation at repetitive sequences (Liang et al. 2002). DNMT3A and DNMT3B can methylate completely unmethylated DNA with catalytic activity similar to that for hemimethylated DNA (Okano et al. 1998, 1999), and have thus been considered “de novo” methyltransferases. DNMT3A is particularly active in embryonic stem cells and neuronal cells and can mediate non-CpG methylation (Lister et al. 2009; Guo et al. 2014). DNMT3B is important for establishing and maintaining methylation at satellite sequences and in gene bodies (Liang et al. 2002; Tao et al. 2002; Yang et al. 2014). If the three DNMT enzymes fail to maintain methylation during replication, a passive demethylation ensues.
Apart from such passive loss of DNA methylation during replication, a complex sequence of enzymatic processes can also actively “erase” DNA methylation, allowing plasticity and dynamic regulation of this epigenetic mark even in nonreplicating cells such as postmitotic neurons. A family of ten-eleven translocation (TET) proteins, encoded by the TET1, TET2, and TET3 genes, has been shown to oxidize 5-methylcytosine to 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxycytosine (Kriaucionis and Heintz 2009; Tahiliani et al. 2009; He et al. 2011; Ito et al. 2011). These oxidized forms of 5mC can then lead to base excision repair processes that can remove the once-methylated cytosine and repair with an unmethylated cytosine for active demethylation (Guo et al. 2011; He et al. 2011). Compared with normal terminally differentiated cells in diverse tissue types, the amount of 5-hydroxymethylcytosine, the major oxidized product of TET-mediated oxidation of 5-methylcytosine, is profoundly reduced in stem/progenitor cells and in human cancers, including prostate cancer (Haffner et al. 2011; Lian et al. 2012; Orr et al. 2012; Ahsan et al. 2014; Munari et al. 2016).
DNA methylation can signal epigenetic repression through the action of “reader” proteins that selectively bind methylated DNA and orchestrate recruitment of corepressor and remodeling complexes, such as Mi2-NuRD, to establish repressive chromatin conformations (Bird and Wolffe 1999; Klose and Bird 2006). The methyl-binding domain proteins, encoded by MBD1, MBD2, MBD3, MBD4, and MECP2, have both overlapping and complementary roles in this capacity (Bird and Wolffe 1999). MBD1, MBD2, and MECP2 have high affinity for methylated CpGs and are associated with distinct corepressor complexes (Hendrich and Bird 1998; Bird and Wolffe 1999; Klose and Bird 2006). Despite its high degree of homology, particularly in the methyl-binding domain, to MBD2, MBD3 does not bind methylated DNA to any appreciable extent but participates in corepressor complexes found in repressed chromatin. Although MBD3 has been implicated in binding hydroxymethylated DNA (Yildirim et al. 2011), this has been controversial. More recently, MECP2, and not MBD3, has been shown to bind hydroxymethylated DNA (Mellen et al. 2012), suggesting that such hydroxymethylcytosine may not simply be an intermediate toward demethylation, but may have epigenetic signaling functions of its own. MBD4 is a glycosylase and has been implicated in the base excision repair of T:G mismatches that would arise from deamination of 5mC (Hendrich et al. 1999). Apart from the MBD family of proteins, recent work has shown that the presence of 5mC in the transcription factor target sequences can greatly change their binding to that target sequence (Hu et al. 2013; Wan et al. 2017; Yin et al. 2017). KLF4 and HOXB13, for example, have been suggested to bind a specific target consensus sequence only when the CpG within it is methylated apart from their specificity for binding their cognate unmethylated target sequences (Hu et al. 2013; Wan et al. 2017; Yin et al. 2017). Such observations may shed light on the confusing observations that methylation of some sequences leads to activation, rather than repression, of the nearby gene. It has been suggested that such transcription factors may have a “pioneer” function in converting otherwise repressed chromatin to active chromatin in critical developmental/differentiation states. The extent to which cancer cells take advantage of such alterations to alter their epigenetic programming is unclear and an area of ongoing work.
Our understanding of the “preservers” of DNA methylation as a heritable mark that can be maintained across DNA replication is the most mature, relative to that of other epigenetic modifications. DNMT1 itself is an important “preserver,” as discussed above, because it has preferential catalytic activity for hemimethylated DNA and can track with the replication fork. In addition, its ability to track with the replication fork is heavily dependent on UHRF1, which shows high expression in S phase, and can bind preferentially to hemimethylated DNA at the replication fork, and can recruit DNA methyltransferases to those sites as a means to enforce the maintenance of methylation after replication (Bostick et al. 2007; Sharif et al. 2007; Arita et al. 2008; Avvakumov et al. 2008; Hashimoto et al. 2008). Interestingly, UHRF1 is an important factor for maintaining histone methylation as well; it can recruit the G9A histone methyltransferase, and links H3K9 methylation to DNA methylation at sites of repressive chromatin (Kim et al. 2009; Unoki et al. 2009). Such mechanisms may be critical to establishment of cross talk between the different epigenetic marks to regulate chromatin states.
DNA methylation can also signal cross talk to chromatin “remodelers” capable of altering the large-scale conformations of chromatin. SWI/SNF ATP-dependent chromatin remodeling proteins such as ATRX, for example, have been shown to bind and regulate variant histone occupancy at nucleosomes surrounding methylated repetitive DNA elements, and subtelomeric sequences (Gibbons et al. 2000; Dyer et al. 2017). The impact of alterations in the DNA methylation of these regions in prostate cancer cells, and of mutations in these chromatin remodeling factors and associated alterations in DNA methylation, are still poorly understood; but such cross talk between DNA methylation and chromatin remodeling complexes appears critical to establishment and maintenance of the epigenetic chromatin organization, mediating normal and cancer cell identities.
Histone Modifications and DNA–RNA–Protein Interactions Governing Chromatin Structure and Organization
DNA is packaged through interactions with proteins and RNA into complex quaternary structures in chromatin. Epigenetic marks on the DNA-binding proteins also help constitute the epigenetic programming of DNA function. The primary level of DNA organization occurs in nucleosome subunits, composed of DNA wrapped around a histone octamer barrel-like structure containing dimers of the four histone subunits, H2A, H2B, H3, and H4. These histone subunits have long tails that protrude outward from the tight core nucleosome structure, at which numerous posttranslational modifications, including phosphorylation, methylation, acetylation, ADP-ribosylation, ubiquitination, and several others can establish an “epigenetic code” regulating the surrounding chromatin function. Repressive, active, or poised (bivalent) chromatin states are associated with specific combinations of modifications on the histone tails (Jenuwein and Allis 2001; Bernstein et al. 2006). Given the numerous histone modifications and the fact that such combinations can occur in countless combinations, the resulting epigenetic code is profoundly more complex than the genetic code. To add to this complexity, a significant fraction of the products of protein-coding and noncoding RNA genes, including linker histones, transcription factors, coactivator complexes, corepressor complexes, insulators such as CTCF (Ong and Corces 2014), and long noncoding RNAs (Fatica and Bozzoni 2014), can bind to chromatin and act as trans regulators.
Histone methylation and acetylation are among the most studied of the histone modifications. Although histone tail acetylation is uniformly associated with open or active chromatin, histone methylation can be associated with either open or closed chromatin, depending on which amino acid on the histone tail is methylated and the number of methyl groups (Jenuwein and Allis 2001; Rice and Allis 2001; Bernstein et al. 2006). For example, methylation of H3K9 (the lysine at the ninth position of H3) and H3K27, which is mediated by the Polycomb complexes and EZH2 methyltransferase, is typically associated with repressed chromatin, whereas H3K4 methylation, which can be mediated by MLL methyltransferases, is typically an active or open chromatin mark. Some regions may harbor both open and closed chromatin marks, a “poised” chromatin state observed at key differentiation regions in embryonic and tissue stem cells (Bernstein et al. 2006). Differentiation of the stem cells can progress with transition of the poised bivalent state to more committed active or closed marks alone.
These histone modifications are regulated by specific “writers,” “erasers,” and “readers.” The writers of histone methylation and acetylation are the histone methyltransferases (HMTs) and histone acetyltransferases (HATs), respectively; the “erasers” are the histone demethylases (HDMs) and histone deacetylases (HDACs), respectively. The “readers” of these marks include proteins with specific protein folds, including bromodomains (histone acetylation readers), PHD domains, tudor domains, chromodomains, among several others. One of the most studied of these of late, as a result of the development of powerful pharmacological probes, are the bromodomains, particularly the BET-bromodomain family of histone lysine acetylation readers, which can bind acetylated lysines and promote chromatin activation (Filippakopoulos et al. 2010; Loven et al. 2013). In particular, BRD4 has been identified as a critical member of the BET-bromodomain family and has been implicated in activation of “super-enhancers” controlling the expression of dominant transcriptional programs in cancer cells, including those mediated by the MYC oncogene and AR, both of which are key targets in prostate cancer, as discussed in detail in other reviews in this collection.
The readers of histone modifications can signal chromatin “remodeling” through the recruitment and action of ATP-dependent enzymes that can sculpt the positioning of nucleosomes and moderate larger-scale looping interactions of chromatin to establish the final 3D organization of the genome. The precise mechanisms governing the maintenance of histone modifications across cell division are not fully understood, but as described above, the best-known mechanisms feature cross talk between DNA methylation and histone modification (e.g., through specialized preserver proteins such as UHRF1).
Understanding the epigenetic encoding mediated through the profoundly complex combinatorial histone modifications and their signaling will require novel approaches to measure the combinations of marks present at each region of the genome and across different functional states. A comprehensive decoding of the histone code can hopefully be fueled by the ongoing revolution in genomics, proteomics, and artificial intelligence/machine learning and computational systems biology.
EPIGENETIC ALTERATIONS IN PROSTATE CANCER
Although the mechanisms behind the establishment, maintenance, and plasticity of epigenetic programming in normal and cancer cells are not fully understood, it is clear that alterations in the epigenetic programming are nearly universal in human cancers, including prostate cancer. Such epigenetic alterations are part of the causal chain that allow cancer cells to evade the rules governing normal cells, allowing the establishment of a caricaturized cell identity that enables invasion, survival, and self-renewal.
Driver Mutations in Epigenetic Machinery Proteins in Prostate Cancer
The importance of these epigenetic alterations in driving carcinogenesis and disease progression has been highlighted by the observation that many proteins in the epigenetic machinery are mutated in prostate cancer. Although each of these proteins is mutated infrequently, as a class, epigenetic machinery proteins are among the most frequently mutated in prostate cancer, and in many other cancers. Recent studies compiling large-scale genome sequencing data from more than a thousand prostate cancer specimens have estimated that ∼15%–20% of prostate cancers harbor driver genetic mutations in genes encoding epigenetic machinery components (Table 1) (Grasso et al. 2012; Baca et al. 2013; Haffner et al. 2013a; Cancer Genome Atlas Research Network 2015; Beltran et al. 2016; Armenia et al. 2018). Such mutations are mostly inactivating, including point mutations, insertions and deletions, and genomic structural alterations, and have been observed in genes encoding epigenetic machinery regulating DNA methylation (IDH1, TET2, MBD1), histone acetylation (KAT6B, ARID4A), and histone methylation (MLL methyltransferases (KMT2C, KMT2D, KMT2A), SETD2, SETDB1, KDM6A), and chromatin remodelers (CHD1, CHD3, CHD8, ARID1A, ARID2, SMARCA1, SMARCAD1, ATRX). Although each gene is relatively infrequently mutated (the most frequently mutated is KMT2C at ∼5%–8% of cases), this class of genes is among the most frequently mutated in prostate cancer. The epigenetic consequences of such mutations and their precise role in driving cancer initiation or progression are still largely unknown and is the focus of much ongoing research.
DNA Methylation Alterations in Prostate Cancer
DNA methylation alterations are the best-understood of the epigenetic alterations in prostate cancer and have been examined throughout carcinogenesis and disease progression. DNA hypermethylation of promoters of key tumor suppressor genes such as GSTP1, RASSF1A, and APC (Nakayama et al. 2003; Yegnasubramanian et al. 2004) have been observed in the main known precursor lesion to prostate cancer, high-grade prostatic intraepithelial neoplasia (PIN), and at least GSTP1 promoter hypermethylation may occur in even earlier stages of disease initiation, in some cases during the transition from benign prostate glands to inflammatory lesions called proliferative inflammatory atrophy (PIA) (Nakayama et al. 2003). In overt invasive carcinoma, there appears to be a DNA hypermethylation catastrophe, involving hundreds to thousands of CpG islands and shores, in the prostate cancer genome (Yegnasubramanian et al. 2004, 2011; Aryee et al. 2013; Cancer Genome Atlas Research Network 2015). Whether such a catastophe may have already begun in the precursor PIN lesions is not yet known and is the subject of much study. Such alterations include some hypermethylation events that are perhaps the most recurrent somatic genome alterations known in prostate cancer, even more frequent than the recurrent rearrangements involving ETS transcription factors (Tomlins et al. 2005). The hypermethylation events are enriched at promoter regions, particularly of development/differentiation genes, and are associated with gene repression (Yegnasubramanian et al. 2011; Aryee et al. 2013). Interestingly, hypermethylation alterations are also enriched near transcriptional termination sites, intron–exon junctions, and exons, and may regulate antisense transcription, poly(A)-site selection, alternative transcriptional start selection, and alternative splicing (Yegnasubramanian et al. 2011). Additionally, hypermethylation alterations are enriched at highly conserved intergenic sequences, including conserved transcription factor binding sites, and may regulate gene expression at a distance (e.g., through modulation of enhancer and insulator sequences). A subset of such intergenic hypermethylation alterations may directly repress expression of previously unknown noncoding genes in cis (Yegnasubramanian et al. 2011). Large-scale efforts to map and delineate the functional elements in the genome (ENCODE Project Consortium 2012; Roadmap Epigenomics Consortium 2015) will provide important insights into the significance of such intergenic hypermethylation alterations.
After an initial hypermethyltaion catastrophe accompanying the early stages of carcinogenesis, it is likely that multiple waves of hypermethylation alterations occur during prostate cancer progression (Nelson et al. 2009). This is apparent through studies identifying numerous DNA methylation alterations that are associated with high grade and/or stage disease, and disease recurrence after primary therapy (Yegnasubramanian et al. 2004; Stott-Miller et al. 2014; Bhasin et al. 2015). Recent studies have also identified DNA hypermethylation alterations associated with lethal metastatic prostate cancer and neuroendocrine prostate cancer (Aryee et al. 2013; Beltran et al. 2016). It is possible that a subset of such alterations are involved in development of resistance to treatment with first- and second-generation anti-androgens and chemotherapeutic agents.
Prostate cancers also harbor DNA hypomethylation, that is, the undermethylation of genomic regions that are universally methylated in normal prostate tissues (Florl et al. 2004; Yegnasubramanian et al. 2008; Aryee et al. 2013). Such hypomethylation alterations appear to arise later in the disease progression than the initial hypermethylation catastrophe and are most prominent in metastases. The hypomethylation alterations are widespread in the genome and can affect large genomic blocks spanning hundreds to thousands of kilobases, interrupted by regions of hypermethylation that are just a few hundred base pairs (Aryee et al. 2013). This hypomethylation is also readily observed at repetitive elements including L1 and Alu elements (Florl et al. 2004; Yegnasubramanian et al. 2008). Interestingly, the hypomethylation alterations do not appear to have any consistent cis regulation of gene expression, except for a class of cancer-testis antigen genes that are observed to have variable hypomethylation of their promoter regions associated with overexpression (Yegnasubramanian et al. 2008; Aryee et al. 2013). Nonetheless, the hypomethylation of large genomic regions can potentially result in a higher propensity for genomic instability that can drive cancer progression.
To visualize these complex alterations in DNA methylation in prostate cancer, we recently created genome-scale DNA methylation “cityscapes” of lethal metastatic prostate cancer (Aryee et al. 2013). These cityscapes depicted DNA methylation alterations in multiple metastatic deposits from men dying of metastatic prostate cancer compared with normal prostate tissues. Each metastatic deposit in a subject represents a separate clonal outgrowth, and thus the degree to which DNA methylation alterations in any one metastasis was maintained across all other metastases within the same individual would allow assessment of which DNA methylation alterations were maintained across metastatic dissemination. Although the degree of similarity in the methylation patterns overall within a subject were much more consistently maintained than the degree of consistency in methylation alterations across individuals, many DNA methylation alterations were not highly maintained across metastases within subjects. Such variably methylated alterations across metastases within subjects were typically hypomethylated regions, which were widespread across the genome as described above. Against this tendency for widespread and variable hypomethylation, the DNA hypermethylation alterations were sparse across the genome, but the majority were highly maintained across metastases within individuals. Taken together, the high degree of maintenance of hypermethylation in the backdrop of a pervasive tendency for hypomethylation suggest that the hypermethylation alterations were selected for and represent driver alterations that are part of the causal chain of the malignant phenotype in these subjects. In support of this notion, these consistently maintained hypermethylation events were significantly associated with repression of gene expression in cis, and were enriched at differentiation/developmental gene sets, the majority of which were already methylated in primary prostate cancer. Collectively, these observations suggest that DNA hypermethylation alterations are highly maintained and selected for longitudinally through disease progression and metastatic dissemination.
Complementary to this staunch maintenance of DNA hypermethylation alterations, prostate cancers also exhibit a profound loss of hydroxymethylation content in the genome (Haffner et al. 2011). This may indicate a reduced level of active demethylation and plasticity, and greater potential for maintenance and selection of driver hypermethylation events.
Alterations in Histone Modifications in Prostate Cancer
As technologies for measurement of histone modifications in tissues are advancing, so too is our understanding of histone modifications in human prostate cancer tissues. This understanding began with early efforts using in vitro experiments in cell lines studied with chromatin immunoprecipitation and microarrays or next-generation sequencing (ChIP-chip and ChIP-seq). One important theme that emerged from such studies was an “epigenetic switch,” in which Polycomb-mediated H3K27 methylation associated repression switched to a more stable and non-inducible H3K9 methylation and DNA hypermethylation (Gal-Yam et al. 2008). This was similar to observations in many cancer systems in which regions of bivalent chromatin marks (H3K4me and H3K27me) in embryonic stem cells that transition to H3K27me in tissue stem cells, can undergo a switch to DNA hypermethylation and loss of the Polycomb mark in the cancer cells—a phenomenon described as a DNA hypermethylation module in the Polycomb stem-cell signature of cancer cells (Easwaran et al. 2012). More recently, these ChIP-seq studies have been adapted for lower input materials and multiple studies have been able to apply such methodologies to human prostate tissues to measure the occupancy of transcription factors and chromatin marks genome-wide. These studies are poised to greatly improve our understanding of the heterogeneity of such epigenetic marks in human prostate cancer and observe their association with cancer initiation, progression, and response to therapy (Adli and Bernstein 2011; Pomerantz et al. 2015).
In the meanwhile, much has also been learned from studies examining changes in the global levels of histone modifications, using in situ immunohistochemistry and immunofluorescence microscopy in prostate cancer tissues. Alterations in the patterns in the global levels of histone acetylation at H3K9, H3K18, and H4K12, and histone dimethylation at H3R3 andH3K4 fell into distinct states within the cancers analyzed (Seligson et al. 2005). A more recent study has shown that prostate cancers show a significant global reduction in H3K27me3 Polycomb-mediated histone methylation marks, occurring in early stages and continues through the more advanced stages of the disease (Pellakuru et al. 2012). This reduction in the global levels of H3K27me3 was highly correlated, even at an individual cell level, with the global reduction in hydroxymethylation of prostate cancer tissues as described above (Haffner et al. 2013b). This correlation is highly suggestive of a direct coupling between Polycomb-mediated histone methylation and DNA methylation. Paradoxically, EZH2, the catalytic component of the Polycomb repressor complex PRC2, is consistently up-regulated in prostate cancer, especially in more aggressive and neuroendocrine prostate cancers (Varambally et al. 2002; Koh et al. 2011; Beltran et al. 2016). The precise mechanisms behind this increase in EZH2 expression despite the global reduction in the H3K27me mark that it is responsible for “writing” is still not clear. However, one recent study has suggested that EZH2 catalytic activity may be involved in mediating PRC2-independent functions modulating androgen receptor transcriptional programs in prostate cancer (Xu et al. 2012).
CLINICAL TRANSLATION OF PROSTATE CANCER EPIGENETICS
Even as our knowledge of the epigenetic mechanisms driving prostate cancer initiation and progression advances, we can opportunistically take forward consistent and recurrent epigenetic alterations in prostate cancer as biomarkers that can inform prostate cancer management decisions. Moreover, our ripening knowledge of epigenetic mechanisms in prostate cancer is leading to development of novel therapeutic strategies.
Epigenetic Alterations as Prostate Cancer Biomarkers
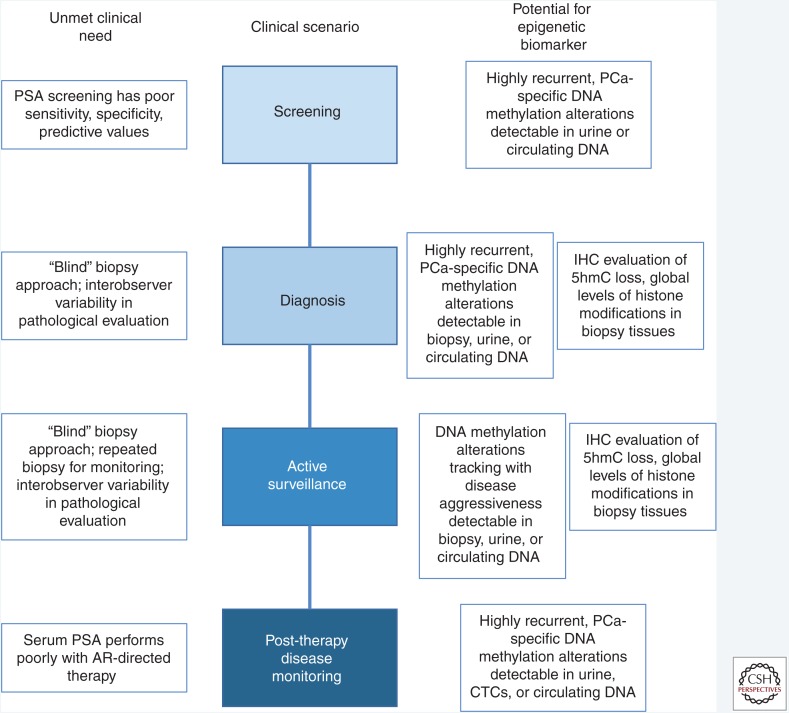
The current management of prostate cancer is challenged by multiple unmet clinical needs where development of novel biomarkers to help guide clinical decision making would be of tremendous value. Biomarkers that are developed based on our current understanding of epigenetic alterations in prostate cancer may be impactful in multiple settings of prostate cancer management (Fig. 2) including (1) screening, (2) diagnosis, (3) risk stratification at the time of diagnosis, and (4) monitoring treatment response and disease burden, especially when the therapy involves targeting the androgen receptor such as in high risk primary prostate cancer or in advanced metastatic disease.
Figure 2.
Utility of epigenetic alterations as prostate cancer biomarkers. Novel prostate cancer biomarkers are critically needed in multiple clinical contexts, including screening, diagnosis, active surveillance, and treatment response monitoring. Multiple types of epigenetic biomarkers, as shown on the right, may have utility to address these unmet needs. PSA, Prostate-specific antigen; AR, androgen receptor; PCa, prostate cancer; IHC, immunohistochemistry; CTCs, circulating tumor cells.
DNA-based biomarkers can have extraordinary analytical sensitivity because advances in molecular biology have allowed robust near single-molecule detection using elegant amplification strategies. Among DNA-based biomarkers, genetic alterations, such as mutations and genomic rearrangements can be extraordinarily specific for cancer and are being pursued aggressively. However, in prostate cancer, such alterations can be highly heterogeneous, with even the most frequent alterations, such as TMPRSS2-ERG rearrangements, occurring in 50% of cases. In contrast, many DNA methylation alterations are highly frequent across cases, with many dozens of such alterations occurring in 70% or more of prostate cancer cases but not in normal prostate. These alterations may therefore be particularly useful as stand-alone biomarkers or in combination with genetic alterations to augment current clinical decision making.
Epigenetic Biomarkers for Prostate Cancer Screening and Diagnosis
As a screening tool, measurement of serum PSA is highly controversial (Strope and Andriole 2010), in large part because of its known limitations in sensitivity, specificity, and positive and negative predictive values. The development of novel markers that can greatly add to the imperfect screening assessments based on serum PSA and other clinical parameters could greatly enhance prostate cancer screening (Nelson et al. 2009). As discussed above, many DNA methylation alterations are highly prostate cancer specific and highly recurrent in prostate cancers, including CpG island promoter methylation of GSTP1, APC, RARB, RASSF1, and PTGS2, among dozens to hundreds of others (Yegnasubramanian et al. 2004; Nelson et al. 2009; Aryee et al. 2013); if such alterations could be detected in blood or urine of asymptomatic individuals, they may serve as a valuable biomarker for prostate cancer screening. In this regard, numerous recent approaches to detect cancer DNA alterations, including mutations and DNA methylation alterations, in circulating tumor DNA and/or urine, provide proof-of-principle that such a DNA methylation informed screening tool may be feasible for prostate cancer.
In prostate cancer diagnosis, a major current problem is the use of “blind” biopsies that arbitrarily sample the prostate gland, even with the typical 12-core sampling. Although prostate magnetic resonance imaging (MRI) and other imaging advances are poised to guide biopsy toward the relevant prostate regions, such approaches are still not standard of care and have limitations in the size and type of lesions that can be detected. Given the current “blind” biopsy approach, a negative biopsy result does not rule out presence of cancer in a region that was not sampled by the biopsy. One approach to address this issue involves detection of CpG island promoter methylation of GSTP1, APC, and RASSF1 in DNA extracted from biopsy materials through a clinically used test to assess whether there was molecular evidence of cancer (Trock et al. 2012; Partin et al. 2014), even when morphological evidence was not evident in the pathological examination of the biopsy tissue. In one clinical study, a positive promoter methylation test was associated with a 3.17-fold increased risk of prostate cancer on a subsequent biopsy, whereas a negative test had a 90% negative predictive value for a subsequent biopsy procedure yielding a cancer diagnosis; a second study largely confirmed these findings (Stewart et al. 2013; Partin et al. 2014). Efforts to see if such assessments could be further improved by detecting prostate cancer DNA methylation alterations in blood or urine, as a potential way to sample the prostate gland more systematically, are also under way. Finally, apart from DNA methylation alterations, immunohistochemical or immunofluorescence approaches to detecting global alterations in the levels of histone marks or hydroxymethylcytosine content may prove useful in aiding the tissue based diagnosis of prostate cancer, particularly if conventional morphological and IHC features are inconclusive (Seligson et al. 2005; Haffner et al. 2011; Pellakuru et al. 2012).
Epigenetic Biomarkers of Risk Stratification at the Time of Diagnosis and Monitoring during Active Surveillance
Recent evidence has shown a systematic tendency to overdiagnosis and overtreatment of prostate cancer with current medical practices. Although prostate cancer claims nearly 30,000 lives every year, most of the nearly 165,000 men that will be diagnosed with prostate cancer in 2018 in the United States will likely have indolent disease that may never become life-threatening even without radical therapy (Guileyardo et al. 1980; Sakr et al. 1993; Holmberg et al. 2012; Wilt et al. 2012; Siegel et al. 2018). It has been estimated that somewhere between 5 and 48 men would need to be detected and potentially treated with definitive management of primary disease under current clinical practices to prevent one death from prostate cancer (Barry 2009; Schroder et al. 2009, 2012). This means that the majority of men diagnosed with prostate cancer are at risk for complications arising from radical prostatectomy or radiation therapy, including urinary incontinence and sexual dysfunction, but may not receive significant benefit (Michaelson et al. 2008). Thus, there is a pressing need to develop better biomarkers to distinguish men with aggressive disease that would benefit from primary therapy from men with indolent disease for whom active surveillance may be more appropriate. If such biomarkers could be noninvasively assessed, they may also aid in the monitoring during active surveillance.
Current strategies for risk stratification to guide decisions of active surveillance versus definitive management with radical prostatectomy or radiation therapy involve use of multiple clinical and pathological parameters. Perhaps the most important of these is the Gleason score. When assessed by expert pathologists of radical prostatectomy specimens, Gleason score is an extremely effective tool for prognostication. In a recent study, of >2500 men that had surgical treatment of Gleason 6 or lower prostate cancer, none died of their disease (Hernandez et al. 2008). The problem with this measure again relates to the use of “blind” biopsy during diagnosis; the threat of incorrect classification of Gleason grade and the possibility of undetected progression during active surveillance have been major barriers to effective decision making. Even with the use of 12 core biopsies, prostate biopsies sample such a small volume of the prostate that it is still very likely to miss aggressive cancers that are only seen after removal of the entire gland. Multiple studies have shown that the biopsy Gleason score and radical prostatectomy Gleason score match in only half of all cases (Steinberg et al. 1997), with a high fraction of cases being “upgraded” after surgery, which in most cases occurs because regions of higher grade cancer were not sampled in the biopsy. Finally, the need to use repetitive biopsies in the active surveillance setting can cause significant morbidity, including infection and even death. Thus, noninvasive monitoring tools would be greatly beneficial.
DNA methylation alterations that are associated with Gleason grade, and cancer aggressiveness more generally may serve as an effective biomarker to address this unmet need. Multiple previous studies have identified DNA methylation alterations associated with Gleason grade, with metastasis, and with disease recurrence after therapy for primary prostate cancer (e.g., Yegnasubramanian et al. 2004; Stott-Miller et al. 2014; Bhasin et al. 2015). If validated in larger studies, and if detection of such biomarkers could be performed with noninvasive sampling of tumor DNA in blood or urine, such DNA methylation biomarkers would hold tremendous promise for this major area of unmet clinical need. Furthermore, global levels of histone marks detected in situ in prostate cancer tissues also appear to be associated with disease aggressiveness and recurrence and may be useful for risk stratification of biopsy materials (Seligson et al. 2005).
Epigenetic Biomarkers for Monitoring Disease Burden and Treatment Response
Molecular assessment of disease burden and treatment response monitoring in prostate cancer is currently performed by serial determinations of serum PSA. Although this strategy is particularly effective in evaluating response and recurrence after radical prostatectomy, it is much less effective in other settings of prostate cancer therapy, particularly with androgen receptor targeted therapies (Eisenberger and Nelson 1996; Scher et al. 2004, 2008). First, because PSA is itself an AR target gene, inhibition of the AR axis could modulate PSA expression even if overall tumor burden did not change. Multiple attempts to validate various serum PSA parameters (e.g., percent decline, doubling time, slope, etc.) for following response to experimental or existing therapies for advanced metastatic prostate cancer have been unsuccessful (Scher et al. 2008). Finally, many compounds may affect PSA expression independent of antitumor activity or even AR signaling.
DNA-based biomarkers of prostate cancer may stoichiometrically reflect tumor burden more so than protein or RNA based markers that are affected by cellular signaling. DNA-based markers measured in cell-free cancer DNA or circulating cancer cells have already been in development for measurement of mutations and rearrangements in cancer cells, and a recent report has suggested that combinations of DNA-based markers with other modalities such as protein markers can be effective in detecting presence of cancer (Leary et al. 2010, 2012; Bettegowda et al. 2014; Newman et al. 2014; Cohen et al. 2018). DNA hypermethylation alterations are also highly promising for tracking tumor burden and treatment response because they tend to be stably maintained longitudinally, are highly cancer-specific, and can occur with very high frequency in prostate cancer. The same types of DNA methylation alterations described above as having utility for prostate cancer screening and diagnosis could also have utility in the space of prostate cancer treatment response and disease burden monitoring.
DNA Methylation Detection Technologies
The feasibility of measuring prostate cancer DNA methtylation biomarkers in liquid biopsy, including urine and blood (plasma or serum) and in prostate secretions is becoming increasingly established (Bastian et al. 2005, 2007, 2008; Rogers et al. 2006; Baden et al. 2009). A recent meta-analysis has shown that measurement of GSTP1 promoter methylation in plasma and urine using conventional DNA methylation detection assays such as methylation-specific PCR (MSP) and methylation-sensitive restriction enzyme PCR achieved sensitivities of ∼75% with specificities around 95% (Wu et al. 2011).
Further improvements in DNA methylation detection technologies could potentially improve the clinical performance of such biomarkers even further. Currently, most DNA methylation detection assays use sodium bisulfite conversion of cytosine, but not methylcytosine, to uracil. Assays such as MSP and its derivative technologies, and bisulfite sequencing/pyrosequencing/next-generation sequencing, are commonly used tools (Clark et al. 1994; Herman et al. 1996; Eads et al. 2000; Dupont et al. 2004). However, bisulfite conversion can be injurious to DNA, potentially compromising assay sensitivity. Methods that do not rely on bisulfite conversion include use of methylation-sensitive restriction enzymes to digest unmethylated DNA, while leaving methylated DNA intact for detection by quantitative PCR (qPCR). Such methods can be highly sensitive, but specificity can suffer from incomplete digestion of unmethylated DNA (Yegnasubramanian et al. 2006). Another bisulfite-free method features use of selective affinity enrichment of methylated DNA using methyl-binding domain polypeptides (MBD) or 5-methylcytosine-specific antibodies, followed by qPCR. However, background binding of unmethylated DNA or incomplete capture of methylated DNA can potentially compromise specificity and sensitivity, respectively (Yegnasubramanian et al. 2006). Compellingly, using a combination of methylation-sensitive restriction enzymes and affinity enrichment of methylated DNA (COMPARE-MS) for highly selective enrichment of methylated DNA followed by qPCR or other nucleic acid detection methods has shown very high assay-sensitivity and specificity while avoiding the weaknesses of bisulfite conversion (Yegnasubramanian et al. 2006). Whether this approach will have clinical use in the contexts described above for prostate cancer management must be rigorously evaluated. Finally, novel approaches for direct sequencing based detection of DNA methylation using nanopore sequencing or other single molecule real time sequencing approaches are being developed and may find utility for DNA methylation biomarker detection (Simpson et al. 2017; Kelleher et al. 2018).
Targeting Prostate Cancer Epigenetics for Therapy
Our increasing understanding of the epigenetic mechanisms driving prostate cancer initiation and progression is fueling development of novel therapeutic strategies that can target these epigenetic processes.
Given the highly frequent and stably maintained DNA hypermethylation alterations in the prostate cancer hypermethylation cityscape that appear to regulate key differentiation/development and tumor suppressor pathways, it would stand to reason that pharmacological modulation of DNA methylation may have therapeutic benefit in prostate cancer. Currently available DNA methyltransferase inhibitors include the nucleoside analog drugs decitabine, azacitidine, and a new decitabine prodrug called guadecitabine. Decitabine and azacitidine are Food and Drug Administration (FDA) approved for treatment of myelodysplastic syndrome (Derissen et al. 2013; Issa et al. 2015), but these agents are being investigated in numerous disease settings. For prostate cancer, and many other solid organ cancers, these agents have not been effective as single agents. This may be in part because of poor bioavailability, metabolism, or other pharmacological properties, conferring a primary resistance to these agents. One known factor in this regard is the activity of cytidine deaminase, which can inactivate decitabine and azacitidine (Qin et al. 2011; Gnyszka et al. 2013). Multiple novel nucleoside analog DNMT inhibitors that are resistant to the effects of cytidine deaminase are in development, including guadecitabine (SGI-110) (Sato et al. 2017) and ASTX-727, a combination of cytidine deaminase inhibitor with decitabine. Other nucleoside metabolizing enzymes may also confer cancer cell intrinsic primary resistance to these agents, and combinations that can overcome such resistance mechanisms are under intense investigation. In prostate cancers, the low proliferative index in many cancers (except in very late stage disease and in the relatively rare small cell neuroendocrine carcinomas) may also be a major factor, because the nucleoside analog DNMT inhibitors require cells to be replicating their DNA to allow incorporation into the genome and exert their mechanism of action in covalently trapping the DNA methyltransferase enzymes. Thus, approaches to modulate the DNA methylation machinery without requirement of high proliferation rates may be more fruitful, and such non-nucleoside DNA methyltransferase inhibitors are under investigation with some limited success (Lee et al. 2005; Lin et al. 2011; Schweizer et al. 2013).
Histone alterations are also highly pervasive in prostate cancer as discussed above. Multiple HDAC inhibitors are FDA approved for treatment of peripheral/cutaneous T-cell lymphomas and in a few other select cancer treatment settings including panabinostat for multiple myeloma (Mann et al. 2007; San-Miguel et al. 2016). Like DNMT inhibitors, these agents have also not been effective as single agents in most solid organ cancers.
However, in one recent clinical trial, the combination of the DNMT inhibitor azacitidine and the HDAC inhibitor entinostat displayed some modest response rates for treatment of advanced non-small-cell lung cancer (Juergens et al. 2011). Most intriguingly, when the patients on this trial went on to receive subsequent therapies, including chemotherapy, targeted therapies, and immunotherapy, they often had more pronounced responses than would have been expected for those agents or for patients receiving up to fourth or fifth line therapy. These anecdotal observations suggested a hypothesis that epigenetic drugs, even if only modestly effective on their own, may sensitize cancers to other therapeutic agents. Multiple mechanistic studies have suggested that epigenetic agents, including DNMT inhibitors and HDAC inhibitors, can modulate the immunogenicity and immune response of/to cancer cells, leading to a greater tendency to respond to immunotherapies. These hypotheses are now being tested in multiple cancer types, including prostate cancer.
Several new classes of epigenetic drugs are in development. For prostate cancer, given the frequent up-regulation of EZH2 particularly in aggressive and neuroendocrine prostate cancer, there is significant enthusiasm around testing novel EZH2 inhibitors (e.g., tazemetostat, CPI-1205) in extensive preclinical and clinical studies (Knutson et al. 2012; McCabe et al. 2012). CPI-1205 (Constellation Pharmaceuticals), in particular, is being tested in combination with anti-androgen therapies in the setting of metastatic castration-resistant prostate cancer. Another exciting new class of drugs are those targeting the BET-bromodomain “readers” of histone acetylation marks, including BRD4 (Filippakopoulos et al. 2010; Loven et al. 2013; Asangani et al. 2014). Because BRD4 has been shown to have critical importance in the regulation of MYC and AR, both of which have critical roles in prostate cancer initiation and progression, inhibitors of BRD4, including the first-in-class JQ-1 and several new drugs in this class, are being aggressively investigated for prostate cancer therapy in pre-clinical and clinical settings. There are also multiple efforts to target HATs (Bowers et al. 2010; Yan et al. 2013), which would be predicted to have similar effects to that of targeting BRD4, by antagonizing the “writing” of histone acetylation marks that would then be “read” by BRD4. Finally, several new programs to target various “writers” (e.g., HMTs) (Daigle et al. 2011), “erasers” (e.g., HMDs) (Hashizume et al. 2014), and “readers” (e.g., MBD proteins) (Wyhs et al. 2014), of epigenetic marks are in rapid development, but are mostly still in early stages. Another interesting approach has been to target enzymes in classical metabolic pathways, whose products can directly modulate the epigenetic machinery. The most striking example of this concept has been the development of mutant IDH1 and IDH2 inhibitors, including ivosidenib and edasidenib, which is now FDA approved for treatment of relapsed/refractory, IDH2 mutant, AML (Stein et al. 2017; Popovici-Muller et al. 2018). Mutant IDH1 and IDH2 proteins catalyze the conversion of α-ketoglutarate to 2-hydroxyglutarate (Dang et al. 2009), a known inhibitor of α-ketoglutarate-dependent oxygenase enzymes, including the TET family and other histone demethylases, and can lead to hypermethylator phenotypes in human cancers (Figueroa et al. 2010; Turcan et al. 2012). Whether these agents will have utility in IDH mutant prostate cancer is not yet known. Because such epigenetic processes targeted by these new agents are critically important in normal and cancer cells, whether such drug development programs will yield treatments with favorable therapeutic index and clinical utility will be the focus of intense research.
CONCLUDING REMARKS
Our increasing understanding of epigenetic mechanisms in health and disease has shown that epigenetic alterations are a vital part of the causal chain driving cancer initiation and progression. The assessment of whether these potentially plastic and dynamic alterations in the epigenetic cityscape are maintained and selected for during cancer progression can provide clues as to whether such alterations are truly part of the causal chain or simply passenger events. This increasing understanding is fueling the ability to target these epigenetic mechanisms for development of clinically relevant biomarkers and therapeutic strategies in prostate cancer.
ACKNOWLEDGMENTS
This work was supported by Prostate Cancer Foundation Challenge Awards (to S.Y. and W.G.N.), National Institutes of Health (NIH)/National Cancer Institute (NCI) Grants U01CA196390, P30CA006973, P50CA058236, and R01CA070196. Conflicts of interest: S.Y., W.G.N., and A.M.D.M. are paid consultants to Cepheid LLC, with whom they are developing epigenetic tests for prostate cancer. S.Y. has received sponsored research support from Cepheid for development and testing of prostate cancer epigenetic biomarkers. This arrangement has been reviewed and approved by the Johns Hopkins University in accordance with its conflict of interest policies.
Footnotes
Editors: Michael M. Shen and Mark A. Rubin
Additional Perspectives on Prostate Cancer available at www.perspectivesinmedicine.org
REFERENCES
- Adli M, Bernstein BE. 2011. Whole-genome chromatin profiling from limited numbers of cells using nano-ChIP-seq. Nat Protoc 6: 1656–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahsan S, Raabe EH, Haffner MC, Vaghasia A, Warren KE, Quezado M, Ballester LY, Nazarian J, Eberhart CG, Rodriguez FJ. 2014. Increased 5-hydroxymethylcytosine and decreased 5-methylcytosine are indicators of global epigenetic dysregulation in diffuse intrinsic pontine glioma. Acta Neuropathol Commun 2: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. 2008. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 455: 818–821. [DOI] [PubMed] [Google Scholar]
- Armenia J, Wankowicz SAM, Liu D, Gao J, Kundra R, Reznik E, Chatila WK, Chakravarty D, Han GC, Coleman I, et al. 2018. The long tail of oncogenic drivers in prostate cancer. Nat Genet 50: 645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryee MJ, Liu W, Engelmann JC, Nuhn P, Gurel M, Haffner MC, Esopi D, Irizarry RA, Getzenberg RH, Nelson WG, et al. 2013. DNA methylation alterations exhibit intraindividual stability and interindividual heterogeneity in prostate cancer metastases. Sci Transl Med 5: 169ra10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C, et al. 2014. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 510: 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. 2008. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 455: 822–825. [DOI] [PubMed] [Google Scholar]
- Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, MacDonald TY, Ghandi M, et al. 2013. Punctuated evolution of prostate cancer genomes. Cell 153: 666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden J, Green G, Painter J, Curtin K, Markiewicz J, Jones J, Astacio T, Canning S, Quijano J, Guinto W, et al. 2009. Multicenter evaluation of an investigational prostate cancer methylation assay. J Urol 182: 1186–1193. [DOI] [PubMed] [Google Scholar]
- Barry MJ. 2009. Screening for prostate cancer—The controversy that refuses to die. N Engl J Med 360: 1351–1354. [DOI] [PubMed] [Google Scholar]
- Bastian PJ, Palapattu GS, Lin X, Yegnasubramanian S, Mangold LA, Trock B, Eisenberger MA, Partin AW, Nelson WG. 2005. Preoperative serum DNA GSTP1 CpG island hypermethylation and the risk of early prostate-specific antigen recurrence following radical prostatectomy. Clin Cancer Res 11: 4037–4043. [DOI] [PubMed] [Google Scholar]
- Bastian PJ, Palapattu GS, Yegnasubramanian S, Lin X, Rogers CG, Mangold LA, Trock B, Eisenberger M, Partin AW, Nelson WG. 2007. Prognostic value of preoperative serum cell-free circulating DNA in men with prostate cancer undergoing radical prostatectomy. Clin Cancer Res 13: 5361–5367. [DOI] [PubMed] [Google Scholar]
- Bastian PJ, Palapattu GS, Yegnasubramanian S, Rogers CG, Lin X, Mangold LA, Trock B, Eisenberger MA, Partin AW, Nelson WG. 2008. CpG island hypermethylation profile in the serum of men with clinically localized and hormone refractory metastatic prostate cancer. J Urol 179: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV, Varambally S, et al. 2016. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 22: 298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326. [DOI] [PubMed] [Google Scholar]
- Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al. 2014. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 6: 224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasin JM, Lee BH, Matkin L, Taylor MG, Hu B, Xu Y, Magi-Galluzzi C, Klein EA, Ting AH. 2015. Methylome-wide sequencing detects DNA hypermethylation distinguishing indolent from aggressive prostate cancer. Cell Rep 13: 2135–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP. 1999. Methylation-induced repression—Belts, braces, and chromatin. Cell 99: 451–454. [DOI] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. 2007. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317: 1760–1764. [DOI] [PubMed] [Google Scholar]
- Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, Crump NT, Hazzalin CA, Liszczak G, Yuan H, et al. 2010. Virtual ligand screening of the p300/CBP histone acetyltransferase: Identification of a selective small molecule inhibitor. Chem Biol 17: 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. 2015. The molecular taxonomy of primary prostate cancer. Cell 163: 1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Dent SY. 2014. Chromatin modifiers and remodellers: Regulators of cellular differentiation. Nat Rev Genet 15: 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG. 2010. Covalent histone modifications—Miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 10: 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Harrison J, Paul CL, Frommer M. 1994. High sensitivity mapping of methylated cytosines. Nucleic Acids Res 22: 2990–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, Douville C, Javed AA, Wong F, Mattox A, et al. 2018. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359: 926–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, Johnston LD, Scott MP, Smith JJ, Xiao Y, et al. 2011. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 20: 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. 2009. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462: 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derissen EJ, Beijnen JH, Schellens JH. 2013. Concise drug review: Azacitidine and decitabine. Oncologist 18: 619–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont JM, Tost J, Jammes H, Gut IG. 2004. De novo quantitative bisulfite sequencing using the pyrosequencing technology. Anal Biochem 333: 119–127. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Qadeer ZA, Valle-Garcia D, Bernstein E. 2017. ATRX and DAXX: Mechanisms and mutations. Cold Spring Harb Perspect Med 7: a026567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. 2000. MethyLight: A high-throughput assay to measure DNA methylation. Nucleic Acids Res 28: E32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easwaran H, Johnstone SE, Van Neste L, Ohm J, Mosbruger T, Wang Q, Aryee MJ, Joyce P, Ahuja N, Weisenberger D, et al. 2012. A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome Res 22: 837–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M. 2002. DNA methylation in cancer: Too much, but also too little. Oncogene 21: 5400–5413. [DOI] [PubMed] [Google Scholar]
- Eisenberger MA, Nelson WG. 1996. How much can we rely on the level of prostate-specific antigen as an end point for evaluation of clinical trials? A word of caution! J Natl Cancer Inst 88: 779–781. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Bozzoni I. 2014. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet 15: 7–21. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Ohlsson R, Henikoff S. 2006. The epigenetic progenitor origin of human cancer. Nat Rev Genet 7: 21–33. [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. 2010. Selective inhibition of BET bromodomains. Nature 468: 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florl AR, Steinhoff C, Muller M, Seifert HH, Hader C, Engers R, Ackermann R, Schulz WA. 2004. Coordinate hypermethylation at specific genes in prostate carcinoma precedes LINE-1 hypomethylation. Br J Cancer 91: 985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Yam EN, Egger G, Iniguez L, Holster H, Einarsson S, Zhang X, Lin JC, Liang G, Jones PA, Tanay A. 2008. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci 105: 12979–12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons RJ, McDowell TL, Raman S, O’Rourke DM, Garrick D, Ayyub H, Higgs DR. 2000. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat Genet 24: 368–371. [DOI] [PubMed] [Google Scholar]
- Gnyszka A, Jastrzebski Z, Flis S. 2013. DNA methyltransferase inhibitors and their emerging role in epigenetic therapy of cancer. Anticancer Res 33: 2989–2996. [PubMed] [Google Scholar]
- Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. 2012. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487: 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guileyardo JM, Johnson WD, Welsh RA, Akazaki K, Correa P. 1980. Prevalence of latent prostate carcinoma in two U.S. populations. J Natl Cancer Inst 65: 311–316. [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. 2011. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145: 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Shin JH, Shin J, Li H, Xie B, Zhong C, Hu S, Le T, Fan G, et al. 2014. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci 17: 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner MC, Chaux A, Meeker AK, Esopi DM, Gerber J, Pellakuru LG, Toubaji A, Argani P, Iacobuzio-Donahue C, Nelson WG, et al. 2011. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget 2: 627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner MC, Mosbruger T, Esopi DM, Fedor H, Heaphy CM, Walker DA, Adejola N, Gurel M, Hicks J, Meeker AK, et al. 2013a. Tracking the clonal origin of lethal prostate cancer. J Clin Invest 123: 4918–4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner MC, Pellakuru LG, Ghosh S, Lotan TL, Nelson WG, De Marzo AM, Yegnasubramanian S. 2013b. Tight correlation of 5-hydroxymethylcytosine and Polycomb marks in health and disease. Cell Cycle 12: 1835–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, et al. 2011. Increased methylation variation in epigenetic domains across cancer types. Nat Genet 43: 768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. 2008. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 455: 826–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume R, Andor N, Ihara Y, Lerner R, Gan H, Chen X, Fang D, Huang X, Tom MW, Ngo V, et al. 2014. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med 20: 1394–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Bird A. 1998. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol 18: 6538–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. 1999. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature 401: 301–304. [DOI] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. 1996. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci 93: 9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez DJ, Nielsen ME, Han M, Trock BJ, Partin AW, Walsh PC, Epstein JI. 2008. Natural history of pathologically organ-confined (pT2), Gleason score 6 or less, prostate cancer after radical prostatectomy. Urology 72: 172–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg L, Bill-Axelson A, Steineck G, Garmo H, Palmgren J, Johansson E, Adami HO, Johansson JE. 2012. Results from the Scandinavian Prostate Cancer Group Trial Number 4: A randomized controlled trial of radical prostatectomy versus watchful waiting. J Natl Cancer Inst Monogr 2012: 230–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Wan J, Su Y, Song Q, Zeng Y, Nguyen HN, Shin J, Cox E, Rho HS, Woodard C, et al. 2013. DNA methylation presents distinct binding sites for human transcription factors. eLife 2: e00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. 2009. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 41: 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa JP, Roboz G, Rizzieri D, Jabbour E, Stock W, O’Connell C, Yee K, Tibes R, Griffiths EA, Walsh K, et al. 2015. Safety and tolerability of guadecitabine (SGI-110) in patients with myelodysplastic syndrome and acute myeloid leukaemia: A multicentre, randomised, dose-escalation phase 1 study. Lancet Oncol 16: 1099–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. 2011. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333: 1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeltsch A. 2002. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem 3: 274–293. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. 2001. Translating the histone code. Science 293: 1074–1080. [DOI] [PubMed] [Google Scholar]
- Jones PA. 2012. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat Rev Genet 13: 484–492. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. 2007. The epigenomics of cancer. Cell 128: 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B, Sebree R, Rodgers K, Hooker CM, Franco N, et al. 2011. Combination epigenetic therapy has efficacy in patients with refractory advanced non–small cell lung cancer. Cancer Discov 1: 598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkowska RZ, Jurkowski TP, Jeltsch A. 2011. Structure and function of mammalian DNA methyltransferases. Chembiochem 12: 206–222. [DOI] [PubMed] [Google Scholar]
- Kelleher P, Murphy J, Mahony J, van Sinderen D. 2018. Identification of DNA base modifications by means of Pacific biosciences RS sequencing technology. Methods Mol Biol 1681: 127–137. [DOI] [PubMed] [Google Scholar]
- Kim JK, Esteve PO, Jacobsen SE, Pradhan S. 2009. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res 37: 493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. 2006. Genomic DNA methylation: The mark and its mediators. Trends Biochem Sci 31: 89–97. [DOI] [PubMed] [Google Scholar]
- Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, Sacks JD, Raimondi A, Majer CR, Song J, et al. 2012. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol 8: 890–896. [DOI] [PubMed] [Google Scholar]
- Koh CM, Iwata T, Zheng Q, Bethel C, Yegnasubramanian S, De Marzo AM. 2011. Myc enforces overexpression of EZH2 in early prostatic neoplasia via transcriptional and post-transcriptional mechanisms. Oncotarget 2: 669–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. 2009. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324: 929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, Antipova A, Lee C, McKernan K, De La Vega FM, et al. 2010. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med 2: 20ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, O’Shaughnessy J, Kinzler KW, Parmigiani G, Vogelstein B, et al. 2012. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med 4: 162ra154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Yegnasubramanian S, Lin X, Nelson WG. 2005. Procainamide is a specific inhibitor of DNA methyltransferase 1. J Biol Chem 280: 40749–40756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q, et al. 2012. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 150: 1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Chan MF, Tomigahara Y, Tsai YC, Gonzales FA, Li E, Laird PW, Jones PA. 2002. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol Cell Biol 22: 480–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Haffner MC, Zhang Y, Lee BH, Brennen WN, Britton J, Kachhap SK, Shim JS, Liu JO, Nelson WG, et al. 2011. Disulfiram is a DNA demethylating agent and inhibits prostate cancer cell growth. Prostate 71: 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153: 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann BS, Johnson JR, He K, Sridhara R, Abraham S, Booth BP, Verbois L, Morse DE, Jee JM, Pope S, et al. 2007. Vorinostat for treatment of cutaneous manifestations of advanced primary cutaneous T-cell lymphoma. Clin Cancer Res 13: 2318–2322. [DOI] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A III, Diaz E, et al. 2012. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492: 108–112. [DOI] [PubMed] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. 2012. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151: 1417–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson MD, Cotter SE, Gargollo PC, Zietman AL, Dahl DM, Smith MR. 2008. Management of complications of prostate cancer treatment. CA Cancer J Clin 58: 196–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munari E, Chaux A, Vaghasia AM, Taheri D, Karram S, Bezerra SM, Gonzalez Roibon N, Nelson WG, Yegnasubramanian S, Netto GJ, et al. 2016. Global 5-hydroxymethylcytosine levels are profoundly reduced in multiple genitourinary malignancies. PLoS ONE 11: e0146302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M, Bennett CJ, Hicks JL, Epstein JI, Platz EA, Nelson WG, De Marzo AM. 2003. Hypermethylation of the human glutathione S-transferase-π gene (GSTP1) CpG island is present in a subset of proliferative inflammatory atrophy lesions but not in normal or hyperplastic epithelium of the prostate: A detailed study using laser-capture microdissection. Am J Pathol 163: 923–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WG, De Marzo AM, Yegnasubramanian S. 2009. Epigenetic alterations in human prostate cancers. Endocrinology 150: 3991–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE, et al. 2014. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 20: 548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E. 1998. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet 19: 219–220. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257. [DOI] [PubMed] [Google Scholar]
- Ong CT, Corces VG. 2014. CTCF: An architectural protein bridging genome topology and function. Nat Rev Genet 15: 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr BA, Haffner MC, Nelson WG, Yegnasubramanian S, Eberhart CG. 2012. Decreased 5-hydroxymethylcytosine is associated with neural progenitor phenotype in normal brain and shorter survival in malignant glioma. PLoS ONE 7: e41036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partin AW, Van Neste L, Klein EA, Marks LS, Gee JR, Troyer DA, Rieger-Christ K, Jones JS, Magi-Galluzzi C, Mangold LA, et al. 2014. Clinical validation of an epigenetic assay to predict negative histopathological results in repeat prostate biopsies. J Urol 192: 1081–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellakuru LG, Iwata T, Gurel B, Schultz D, Hicks J, Bethel C, Yegnasubramanian S, De Marzo AM. 2012. Global levels of H3K27me3 track with differentiation in vivo and are deregulated by MYC in prostate cancer. Am J Pathol 181: 560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz MM, Li F, Takeda DY, Lenci R, Chonkar A, Chabot M, Cejas P, Vazquez F, Cook J, Shivdasani RA, et al. 2015. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat Genet 47: 1346–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovici-Muller J, Lemieux RM, Artin E, Saunders JO, Salituro FG, Travins J, Cianchetta G, Cai Z, Zhou D, Cui D, et al. 2018. Discovery of AG-120 (Ivosidenib): A first-in-class mutant IDH1 inhibitor for the treatment of IDH1 mutant cancers. ACS Med Chem Lett 9: 300–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan S, Bacolla A, Wells RD, Roberts RJ. 1999. Recombinant human DNA (cytosine-5) methyltransferase. I: Expression, purification, and comparison of de novo and maintenance methylation. J Biol Chem 274: 33002–33010. [DOI] [PubMed] [Google Scholar]
- Qin T, Castoro R, El Ahdab S, Jelinek J, Wang X, Si J, Shu J, He R, Zhang N, Chung W, et al. 2011. Mechanisms of resistance to decitabine in the myelodysplastic syndrome. PLoS ONE 6: e23372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JC, Allis CD. 2001. Histone methylation versus histone acetylation: New insights into epigenetic regulation. Curr Opin Cell Biol 13: 263–273. [DOI] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium; Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CG, Gonzalgo ML, Yan G, Bastian PJ, Chan DY, Nelson WG, Pavlovich CP. 2006. High concordance of gene methylation in post-digital rectal examination and post-biopsy urine samples for prostate cancer detection. J Urol 176: 2280–2284. [DOI] [PubMed] [Google Scholar]
- Sakr WA, Haas GP, Cassin BF, Pontes JE, Crissman JD. 1993. The frequency of carcinoma and intraepithelial neoplasia of the prostate in young male patients. J Urol 150: 379–385. [DOI] [PubMed] [Google Scholar]
- San-Miguel JF, Hungria VT, Yoon SS, Beksac M, Dimopoulos MA, Elghandour A, Jedrzejczak WW, Gunther A, Nakorn TN, Siritanaratkul N, et al. 2016. Overall survival of patients with relapsed multiple myeloma treated with panobinostat or placebo plus bortezomib and dexamethasone (the PANORAMA 1 trial): A randomised, placebo-controlled, phase 3 trial. Lancet Haematol 3: e515. [DOI] [PubMed] [Google Scholar]
- Sato T, Issa JJ, Kropf P. 2017. DNA hypomethylating drugs in cancer therapy. Cold Spring Harb Perspect Med 7: a026948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher HI, Eisenberger M, D’Amico AV, Halabi S, Small EJ, Morris M, Kattan MW, Roach M, Kantoff P, Pienta KJ, et al. 2004. Eligibility and outcomes reporting guidelines for clinical trials for patients in the state of a rising prostate-specific antigen: Recommendations from the Prostate-Specific Antigen Working Group. J Clin Oncol 22: 537–556. [DOI] [PubMed] [Google Scholar]
- Scher HI, Halabi S, Tannock I, Morris M, Sternberg CN, Carducci MA, Eisenberger MA, Higano C, Bubley GJ, Dreicer R, et al. 2008. Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: Recommendations of the Prostate Cancer Clinical Trials Working Group. J Clin Oncol 26: 1148–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, Kwiatkowski M, Lujan M, Lilja H, Zappa M, et al. 2009. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med 360: 1320–1328. [DOI] [PubMed] [Google Scholar]
- Schroder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, Kwiatkowski M, Lujan M, Lilja H, Zappa M, et al. 2012. Prostate-cancer mortality at 11 years of follow-up. N Engl J Med 366: 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MD, He Y, Whitaker JW, Hariharan M, Mukamel EA, Leung D, Rajagopal N, Nery JR, Urich MA, Chen H, et al. 2015. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 523: 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer MT, Lin J, Blackford A, Bardia A, King S, Armstrong AJ, Rudek MA, Yegnasubramanian S, Carducci MA. 2013. Pharmacodynamic study of disulfiram in men with non-metastatic recurrent prostate cancer. Prostate Cancer Prostatic Dis 16: 357–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. 2005. Global histone modification patterns predict risk of prostate cancer recurrence. Nature 435: 1262–1266. [DOI] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. 2007. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450: 908–912. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. 2018. Cancer statistics, 2018. CA Cancer J Clin 68: 7–30. [DOI] [PubMed] [Google Scholar]
- Simpson JT, Workman RE, Zuzarte PC, David M, Dursi LJ, Timp W. 2017. Detecting DNA cytosine methylation using nanopore sequencing. Nat Methods 14: 407–410. [DOI] [PubMed] [Google Scholar]
- Stein EM, DiNardo CD, Pollyea DA, Fathi AT, Roboz GJ, Altman JK, Stone RM, DeAngelo DJ, Levine RL, Flinn IW, et al. 2017. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 130: 722–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg DM, Sauvageot J, Piantadosi S, Epstein JI. 1997. Correlation of prostate needle biopsy and radical prostatectomy Gleason grade in academic and community settings. Am J Surg Pathol 21: 566–576. [DOI] [PubMed] [Google Scholar]
- Stewart GD, Van Neste L, Delvenne P, Delree P, Delga A, McNeill SA, O’Donnell M, Clark J, Van Criekinge W, Bigley J, et al. 2013. Clinical utility of an epigenetic assay to detect occult prostate cancer in histopathologically negative biopsies: Results of the MATLOC study. J Urol 189: 1110–1116. [DOI] [PubMed] [Google Scholar]
- Stott-Miller M, Zhao S, Wright JL, Kolb S, Bibikova M, Klotzle B, Ostrander EA, Fan JB, Feng Z, Stanford JL. 2014. Validation study of genes with hypermethylated promoter regions associated with prostate cancer recurrence. Cancer Epidemiol Biomarkers Prev 23: 1331–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strope SA, Andriole GL. 2010. Prostate cancer screening: Current status and future perspectives. Nat Rev Urol 7: 487–493. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Q, Huang H, Geiman TM, Lim CY, Fu L, Qiu GH, Robertson KD. 2002. Defective de novo methylation of viral and cellular DNA sequences in ICF syndrome cells. Hum Mol Genet 11: 2091–2102. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. 2005. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310: 644–648. [DOI] [PubMed] [Google Scholar]
- Trock BJ, Brotzman MJ, Mangold LA, Bigley JW, Epstein JI, McLeod D, Klein EA, Jones JS, Wang S, McAskill T, et al. 2012. Evaluation of GSTP1 and APC methylation as indicators for repeat biopsy in a high-risk cohort of men with negative initial prostate biopsies. BJU Int 110: 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. 2012. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483: 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unoki M, Brunet J, Mousli M. 2009. Drug discovery targeting epigenetic codes: The great potential of UHRF1, which links DNA methylation and histone modifications, as a drug target in cancers and toxoplasmosis. Biochem Pharmacol 78: 1279–1288. [DOI] [PubMed] [Google Scholar]
- Vandiver AR, Idrizi A, Rizzardi L, Feinberg AP, Hansen KD. 2015. DNA methylation is stable during replication and cell cycle arrest. Sci Rep 5: 17911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. 2002. The Polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419: 624–629. [DOI] [PubMed] [Google Scholar]
- Wan J, Su Y, Song Q, Tung B, Oyinlade O, Liu S, Ying M, Ming GL, Song H, Qian J, et al. 2017. Methylated cis-regulatory elements mediate KLF4-dependent gene transactivation and cell migration. eLife 6: e20068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Fischle W, Cheung W, Jacobs S, Khorasanizadeh S, Allis CD. 2004. Beyond the double helix: Writing and reading the histone code. Novartis Found Symp 259: 3–17. [PubMed] [Google Scholar]
- Wilt TJ, Brawer MK, Jones KM, Barry MJ, Aronson WJ, Fox S, Gingrich JR, Wei JT, Gilhooly P, Grob BM, et al. 2012. Radical prostatectomy versus observation for localized prostate cancer. N Engl J Med 367: 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Giovannucci E, Welge J, Mallick P, Tang WY, Ho SM. 2011. Measurement of GSTP1 promoter methylation in body fluids may complement PSA screening: A meta-analysis. Br J Cancer 105: 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyhs N, Walker D, Giovinazzo H, Yegnasubramanian S, Nelson WG. 2014. Time-resolved fluorescence resonance energy transfer assay for discovery of small-molecule inhibitors of methyl-CpG binding domain protein 2. J Biomol Screen 19: 1060–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, Wu X, Stack EC, Loda M, Liu T, et al. 2012. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 338: 1465–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G, Eller MS, Elm C, Larocca CA, Ryu B, Panova IP, Dancy BM, Bowers EM, Meyers D, Lareau L, et al. 2013. Selective inhibition of p300 HAT blocks cell cycle progression, induces cellular senescence, and inhibits the DNA damage response in melanoma cells. J Invest Dermatol 133: 2444–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. 2014. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell 26: 577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yegnasubramanian S, Kowalski J, Gonzalgo ML, Zahurak M, Piantadosi S, Walsh PC, Bova GS, De Marzo AM, Isaacs WB, Nelson WG. 2004. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res 64: 1975–1986. [DOI] [PubMed] [Google Scholar]
- Yegnasubramanian S, Lin X, Haffner MC, DeMarzo AM, Nelson WG. 2006. Combination of methylated-DNA precipitation and methylation-sensitive restriction enzymes (COMPARE-MS) for the rapid, sensitive and quantitative detection of DNA methylation. Nucleic Acids Res 34: e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yegnasubramanian S, Haffner MC, Zhang Y, Gurel B, Cornish TC, Wu Z, Irizarry RA, Morgan J, Hicks J, DeWeese TL, et al. 2008. DNA hypomethylation arises later in prostate cancer progression than CpG island hypermethylation and contributes to metastatic tumor heterogeneity. Cancer Res 68: 8954–8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yegnasubramanian S, Wu Z, Haffner MC, Esopi D, Aryee MJ, Badrinath R, He TL, Morgan JD, Carvalho B, Zheng Q, et al. 2011. Chromosome-wide mapping of DNA methylation patterns in normal and malignant prostate cells reveals pervasive methylation of gene-associated and conserved intergenic sequences. BMC Genomics 12: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. 2011. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 147: 1498–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, Das PK, Kivioja T, Dave K, Zhong F, et al. 2017. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356: eaaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]