Abstract
Background
A growing number of studies clearly demonstrate a substantial association between chronic obstructive pulmonary disease (COPD) and cardiovascular diseases (CVD), although little is known about the shared genetics that contribute to this association.
Methods
We conducted a large-scale cross-trait genome-wide association study to investigate genetic overlap between COPD (Ncase = 12,550, Ncontrol = 46,368) from the International COPD Genetics Consortium and four primary cardiac traits: resting heart rate (RHR) (N = 458,969), high blood pressure (HBP) (Ncase = 144,793, Ncontrol = 313,761), coronary artery disease (CAD)(Ncase = 60,801, Ncontrol = 123,504), and stroke (Ncase = 40,585, Ncontrol = 406,111) from UK Biobank, CARDIoGRAMplusC4D Consortium, and International Stroke Genetics Consortium data.
Results
RHR and HBP had modest genetic correlation, and CAD had borderline evidence with COPD at a genome-wide level. We found evidence of local genetic correlation with particular regions of the genome. Cross-trait meta-analysis of COPD identified 21 loci jointly associated with RHR, 22 loci with HBP, and 3 loci with CAD. Functional analysis revealed that shared genes were enriched in smoking-related pathways and in cardiovascular, nervous, and immune system tissues. An examination of smoking-related genetic variants identified SNPs located in 15q25.1 region associated with cigarettes per day, with effects on RHR and CAD. A Mendelian randomization analysis showed a significant positive causal effect of COPD on RHR (causal estimate = 0.1374, P = 0.008).
Conclusion
In a set of large-scale GWAS, we identify evidence of shared genetics between COPD and cardiac traits.
Electronic supplementary material
The online version of this article (10.1186/s12931-019-1036-8) contains supplementary material, which is available to authorized users.
Keywords: Chronic obstructive pulmonary disease, Cardiovascular diseases, Genetic overlap
Background
Chronic obstructive pulmonary disease (COPD) is a chronic inflammatory disease of the lungs that is the fourth leading cause of death in the world, accounting for more than 3 million deaths each year [1]. There is now considerable evidence of an association between COPD and cardiovascular disease (CVD). Several population-based studies have shown that COPD and airflow limitation is a predictor of cardiovascular risk [2]. The SUMMIT randomized clinical trial reported that exacerbations of COPD confer an increased risk of subsequent CVD [3, 4]. The Lung Health Study reported that for every 10% decrease in forced expiratory volume in 1 s (FEV1), there is a 28% increase in fatal coronary events among subjects with mild to moderate COPD [5]. In addition, CVD is a leading cause of death in patients with COPD, with a 5-year mortality of up to 25% due to a cardiovascular event [5, 6], such as high resting heart rate (RHR), systemic hypertension, coronary artery disease (CAD), or stroke [7–10].
We and colleagues recently identified shared genetic architecture between COPD and lung function/pulmonary fibrosis [11], asthma and allergic diseases [12], Alzheimer’s disease and metabolic disorders [13], psychiatric disorders [14], indicating potential pleiotropic effects among these diseases. COPD and CVD are both highly heritable traits [11, 15]. Parallel epidemic trends worldwide suggest shared genetic and environmental components for both conditions. However, there is little knowledge about shared genetic components between COPD and CVD. Although a previous study identified some genetic loci that influencing both lung function and CAD [16], the findings were not genome-wide in scale and were limited by small sample size. Therefore, it remains largely unknown to what extent the phenotypic association between COPD and CVD is due to shared genetic and biologic effects.
Therefore, we investigated the genetic correlation between COPD and cardiac traits and attempted to describe the specific shared genetic loci and biological pathways between traits. We conducted a large-scale, genome-wide association study (GWAS) cross-trait analysis of COPD from the International COPD Genetics Consortium (ICGC) and 4 cardiac traits from UK Biobank, CARDIoGRAMplusC4D Consortium, and International Stroke Genetics Consortium (ISGC) data, including RHR, high blood pressure (HBP), CAD [17], and stroke [18].
Methods
Study populations
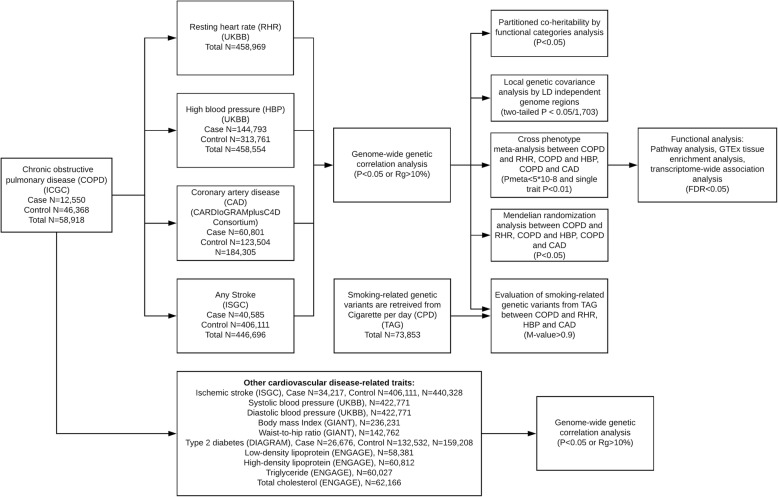
We included 4 major data sources—ICGC, UK Biobank, CARDIoGRAMplusC4D Consortium, and ISGC—in the overall study design (Fig. 1). Previous reports have detailed disease definition and baseline characteristics of the ICGC study cohorts [11] and UK Biobank cohort [19]. In brief, the ICGC defined COPD by GOLD criteria based on pre-bronchodilator spirometry: FEV1 of < 80% and FEV1 to forced vital capacity (FVC) ratio of < 0.7 for cases; or FEV1 of > 80% and FEV1/FVC of > 0.7 for controls, and adjusted for age, sex, pack-years, and smoking status. In UK Biobank, we used both data field 102 and 95 for RHR and data field 6150 for HBP. RHR was assessed via two methods: automated reading during blood pressure measurement (in 501,340 participants); and pulse waveform obtained from the finger with an infrared sensor during arterial stiffness measurement (in 193,472 participants). RHR was averaged if multiple measurements were available for one individual [20]. HBP was assessed by touch screen questionnaire of participants’ HBP diagnosis by doctor. We retrieved summary statistics from publicly available GWAS studies: CAD (Ncase/control = 60,801/123,504) from CARDIoGRAMplusC4D Consortium [17], and stroke (Ncase/control = 40,585/406,111) from ISGC [18]. CAD diagnoses in CARDIoGRAMplusC4D was defined by an inclusive CAD diagnosis (e.g. myocardial infarction (MI), acute coronary syndrome, chronic stable angina, or coronary stenosis > 50%) [17]. The ISGC defined stroke by an inclusive stroke diagnosis (e.g. ischemic stroke, large artery stroke, cardioembolic stroke and small vessel stroke). We standardized GWAS summary data to minimize potential bias due to quality control procedures. Indels and rare/low frequency variants with a minor allele frequency of < 1% were excluded. In addition, we restricted analysis to autosomal chromosomes. Aside from RHR and HBP, both tested in Biobank, we are not aware of specific sample overlap between COPD and 4 major cardiovascular traits in this study, including RHR, HBP, CAD and stroke. Details of each dataset can be found in Additional file 1: Table S1. All subjects consent to participate the study by the time of data analysis.
Fig. 1.
Overall study design. Multiple GWAS data sources were first retrieved. We first conducted genome-wide genetic correlation between COPD and 4 major cardiovascular disease (CVD) traits. For CVD traits that were shown genetic correlation with COPD, we conducted further post-GWAS analyses to investigate genetic overlap between them (variant/region/functional levels, smoking effect and causal inference). We also evaluated the genetic correlation between COPD and other CVD related traits. Abbreviations: ICGC: International COPD Genomic Consortium; UKBB: UK Biobank; ISGC: International Stroke Genetics Consortium; GIANT: The Genetic Investigation of ANthropometric Traits (GIANT) consortium; DIAGRAM: DIAbetes Genetics Replication And Meta-analysis consortium; ENGAGE: European Network for Genetic and Genomic Epidemiology consortium; TAG: Tobacco and Genetics Consortium
GWAS analysis in UK biobank
We performed GWAS analysis on RHR and HBP using a linear mixed model (LMM) method [21] based on European ancestry. See the Additional file 2: Supplemental Note for additional information.
LD score regression (LDSC) analysis
We conducted post-GWAS genetic correlation analysis with LDSC, which estimates genetic correlation between true causal effects of two traits (genetic correlation estimate Rg ranging from − 1 to 1) [22]. Cardiac traits showing genome-wide genetic correlation with COPD were further studied in the downstream analysis. See the Additional file 2: Supplemental Note for additional information.
In addition, we performed genetic correlation analysis between COPD and ischemic stroke subtypes, and metabolic traits (lipids, obesity, and glucose).
Partitioned genetic correlation
To characterize genetic overlap at the level of functional categories, we estimated genetic correlation between COPD and cardiac traits in 11 annotation categories using LDSC. These annotations included transcribed regions, transcription factor binding sites, super-enhancers, introns, DNaseI digital genomic footprinting (DGF) regions, DNaseI hypersensitivity sites (DHSs), fetalDHSs, and histone marks h3k9ac, h3k4me1, h3k4me3, and h3k27ac [23]. For each annotation, we re-calculated LD scores for SNPs assigned to that particular category and then used annotation-specific LD scores to estimate the COPD–cardiac trait genetic correlation.
Local genetic correlation
To identify local genetic correlations between COPD and cardiac traits, we performed ρ-HESS to estimate local genetic correlation between a pair of traits at each LD-independent region in the genome [24]. Approximately 1703 independent LD blocks of 1.5 Mb were used to calculate local genetic heritability and covariance. All GWAS data were restricted to European ancestry, and Bonferroni correction was used to adjust multiple testing (two-tailed P < 0.05/1703) according to the original method description [24].
Cross-trait meta-analysis
After assessing genetic correlations among all traits, we applied 2 cross-trait GWAS meta-analysis methods to combine binary or continuous traits [25]. We used association analysis based on SubSETs (ASSET) to combine association evidence for COPD with HBP and CAD at individual variants because it is designed for meta-analysis of binary traits [26]. We also applied another cross-trait GWAS meta-analysis method, cross phenotype association (CPASSOC), to combine association evidence for COPD with RHR at individual variants, since this method allows meta-analysis of continuous traits [27]. See the Additional file 2: Supplemental Note for additional information.
We applied PLINK [28] clumping function (parameters: --clump-p1 5e-8 --clump-p2 1e-5 --clump-r2 0.2 --clump-kb 500) to determine top loci that were independent from one another (i.e., variants with P < 1 × 10− 5, r2 > 0.2, and < 500 kb away from a peak). The variant with the lowest p-value was defined as the sentinel variant. Putative genes for each variant were considered to be those within the clump. We used Variant Effect Predictor based on Ensembl/GENCODE basic transcripts database for detailed variant annotation [29].
Fine-mapping of credible sets
To identify the 99% credible set of variants within each 500-kb sentinel variant, we identified a credible set of causal variants at each shared locus that met cross-trait meta-analysis criteria using the Bayesian likelihood fine-mapping algorithm [30]. The algorithm maps primary signal and uses a flat prior with steepest descent approximation.
Pathway and GTEx tissue enrichment analysis
To gain biological insights for shared genes, we used the WebGestalt tool [31] to assess enrichment of the identified shared gene set in the Gene Ontology (GO) biological process. We conducted GTEx tissue enrichment analysis using functional mapping and annotation (FUMA) [32] with 53 tissue types from GTEx version 7 [33]. Both analyses were based on shared genes that were identified from cross-trait meta-analysis.
Transcriptome-wide association study (TWAS)
To identify shared COPD and cardiac trait gene expression associations in specific tissues, we conducted TWAS using the FUSION software package based on 43 GTEx (version 6) tissue expression weights [34]. Multiple testing correction was applied for each trait’s gene–tissue pairs on TWAS P-values using false discovery rate (FDR) Benjamini-Hochberg procedure (FDR < 0.05).
Evaluation of effect of smoking-related genetic variants between COPD and cardiac traits
To evaluate the potential effect of smoking-related genetic variants between COPD and cardiac traits, we retrieved 129 genome-wide significant SNPs for cigarette per day (CPD) from the Tobacco and Genetics Consortium (TAG) [35]. We also looked up GWAS results for 2 other smoking related traits from TAG, ever vs never smoked and current vs former smoker, however no SNPs reached genome-wide significance. Thus, we merged 129 SNPs with COPD and CVD traits (RHR, HBP and CAD) and identified 45 SNPs in common for all traits. We used M-value posterior probability [36] to evaluate if the CPD genetic variant effect exists among COPD and CVD traits. A M-value > 0.9 was considered evidence that the SNP had an effect on the trait.
Mendelian randomization (MR) analysis
Finally, we performed MR analysis using Mendelian Randomization Pleiotropy RESidual Sum and Outlier (MR-PRESSO) [37] in order to infer putative causal relationships between COPD and 3 cardiac traits (RHR, HBP, CAD). MR-PRESSO estimates effect of exposure on outcome using SNPs significantly associated with exposure and allows for the evaluation of horizontal pleiotropy in multi-instrument Mendelian Randomization utilizing GWAS summary association statistics. We constructed instruments using genome-wide significant LD-independent SNPs with P-value less than 5 × 10− 8. Prior to running MR-PRESSO, we removed strand-ambiguous SNPs and SNPs in the MHC region (chr6:25-34 M).
Results
Genome-wide association and SNP-based heritability
The phenotype–genotype association test was carried out on ~ 460,000 samples and ~ 5.26 million SNPs from UK Biobank data after quality control. The genomic inflation factor (λgc) from LDSC for RHR and HBP were 1.8405 [LDSC intercept: 1.1256, standard error (SE): 0.0502; Additional file 3: Figure S1] and 1.7648 (LDSC intercept: 1.1061, SE: 0.0244; Additional file 3: Figure S2), respectively; these values suggest that much of the inflation is due to polygenic inheritance [38]. Estimates of SNP-based heritability on the observed scale using GWAS summary statistics were 20.11% (SE: 2.61%) for COPD, 15.19% (SE: 1.28%) for RHR, 12.80% (SE: 0.58%) for HBP, 6.71% (SE: 0.52%) for CAD, and 1.21% (SE: 0.14%) for stroke (Additional file 1: Table S2).
Genome-wide genetic correlation
We evaluated the genetic correlation of COPD and cardiac traits using cross-trait LDSC. Nominally significant genetic correlation with COPD was found for both RHR (Rg = 0.0722; P = 0.0434) and HBP (Rg = 0.0751; P = 0.0467) (Table 1). Genetic correlation for COPD and CAD was approximately 10%, but this value did not reach statistical significance; we did not observe significant genetic correlation between COPD and stroke (Table 1), or additional blood pressure traits, such as systolic blood pressure, diastolic blood pressure (Additional file 1: Table S3). In addition, we did not find evidence of genetic correlation between COPD and ischemic stroke subtype or any CVD related metabolic traits (Additional file 1: Table S3).
Table 1.
Genome-wide genetic correlation between COPD and cardiac traits
| Phenotype 1 | Phenotype 2 | Rg | Rg_SE | P |
|---|---|---|---|---|
| COPD | Resting heart rate | 0.0722 | 0.0357 | 0.0434 |
| COPD | High blood pressure | 0.0751 | 0.0378 | 0.0467 |
| COPD | Coronary artery disease | 0.1015 | 0.0528 | 0.0548 |
| COPD | Stroke | 0.0226 | 0.0689 | 0.7428 |
COPD chronic obstructive pulmonary disease, Rg genetic correlation estimate, SE standard error
Partitioned genetic correlation
In partitioned LDSC analysis, we used 11 functional annotations to evaluate genetic correlations between COPD and cardiac traits by specific functional category. The highest magnitude of significant genetic correlation between COPD and HBP was in introns (Rg = 0.1711; P = 0.0233) and h3k9ac (Rg = 0.1428; P = 0.033) (Additional file 3: Figure S3, Additional file 1: Table S4). Super enhancers had the highest magnitude of genetic correlation between COPD and RHR (Rg = 0.1259; P = 0.0173).
Local genetic correlation
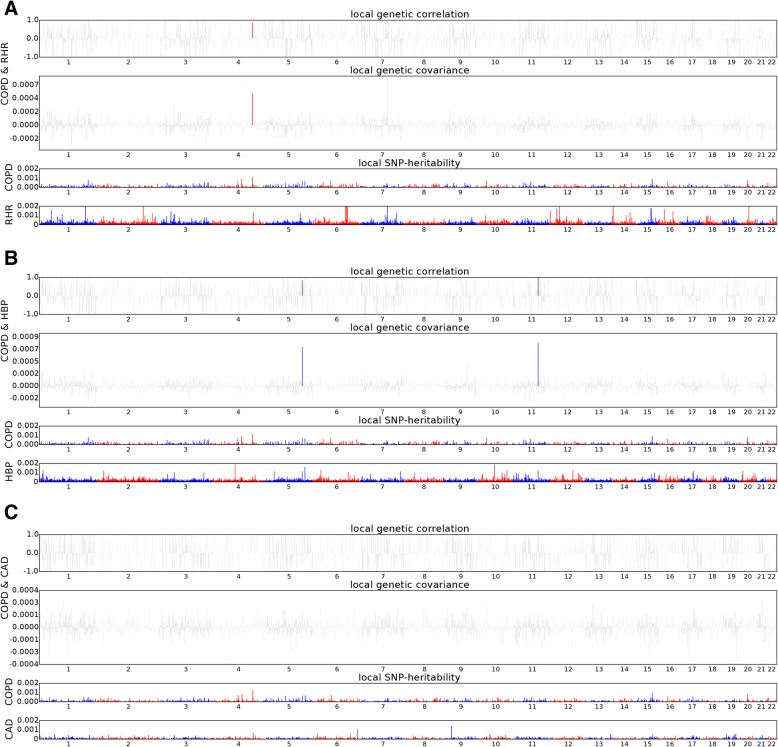
We performed ρ-HESS to investigate whether specific regions of the genome had genetic correlation between COPD and cardiac traits. Analysis of the COPD/RHR trait pair showed that the 4q31 region (chromosome 4: 143443265–146,178,187) had strong local genetic correlations (P = 7.42 × 10− 7) (Fig. 2 and Additional file 1: Table S5). Analysis of the COPD/HBP trait pair showed strong local genetic correlations in 11q22 (chromosome 11: 100417169–101,331,121; P = 6.31 × 10− 7) and 5q32 (chromosome 5: 147181998–148,662,624; P = 3.98 × 10− 6) regions (Fig. 2 and Additional file 1: Table S6). We did not observe any significant local genetic correlations for the COPD/CAD trait pair (Fig. 2 and Additional file 1: Table S7).
Fig. 2.
Plots depicting local genetic correlation (top), genetic covariance (middle), and SNP heritability (bottom) for COPD and RHR (a), COPD and HBP (b), and COPD and CAD (c). Blue or red highlights indicate significant local genetic correlation and covariance after multiple testing correction
Cross-trait meta-analysis between COPD and cardiac traits
ASSET and CPASSOC were applied for genome-wide meta-analysis to identify genetic loci associated with COPD and cardiac traits (meta-analysis P < 5 × 10− 8; trait-specific P < 0.01). After pruning, we found 21 loci significantly associated with COPD and RHR (Table 2 and Additional file 1: Table S8). The most significant SNP was rs7655625 (Pmeta = 1.92 × 10− 19), located at the HHIP locus. The second most significant locus (sentinel SNP: rs59985166, Pmeta = 4.08 × 10− 18) was located at the CASZ1 locus [39].
Table 2.
Genome-wide significant loci by cross-trait meta-analysis at sentinel SNPs associated with COPD and RHR (Pmeta < 5 × 10− 8; single trait P < 0.01)
| Sentinel SNP | CHR | N | Position | P RHR | P COPD | P META | Variant annotation | Genes within clumping region |
|---|---|---|---|---|---|---|---|---|
| rs3738676 | 1 | 162 | chr1:39552963–40,088,043 | 2.30 × 10− 10 | 1.51 × 10− 3 | 3.07 × 10− 11 | 3′ UTR | BMP8A, KIAA0754, MACF1, PABPC4, PPIEL, SNORA55 |
| rs59985166 | 1 | 47 | chr1:10653622–10,753,094 | 2.50 × 10−16 | 2.92 × 10− 3 | 4.08 × 10−18 | Intron | CASZ1, PEX14 |
| rs13409705 | 2 | 17 | chr2:28820641–29,115,712 | 2.80 × 10− 10 | 6.57 × 10− 4 | 8.61 × 10− 12 | Intron | PLB1, PPP1CB, SPDYA, TRMT61B |
| rs3791979 | 2 | 21 | chr2:218667372–218,704,894 | 2.40 × 10− 9 | 4.15 × 10− 3 | 2.0 × 10− 9 | 3′ UTR | TNS1 |
| rs2955083 | 3 | 119 | chr3:127715196–128,067,275 | 6.90 × 10− 4 | 2.0 × 10−8 | 3.56 × 10− 8 | Intron | EEFSEC, RUVBL1, SEC61A1 |
| rs9859058 | 3 | 78 | chr3:156353161–156,695,011 | 3.60 × 10−10 | 6.57 × 10−3 | 4.01 × 10− 11 | Intron | LEKR1, LINC00886, PA2G4P4, TIPARP, TIPARP-AS1 |
| rs7655625 | 4 | 140 | chr4:145196359–145,974,688 | 2.70 × 10−5 | 3.02 × 10−19 | 1.92 × 10− 19 | Intergenic | ANAPC10, HHIP, HHIP-AS1 |
| rs7449334 | 5 | 43 | chr5:90309787–90,418,617 | 1.0 × 10− 10 | 3.59 × 10− 3 | 1.62 × 10−11 | Intron | GPR98 |
| rs1158317 | 6 | 5 | chr6:120518685–121,035,654 | 6.50 × 10−11 | 7.45 × 10−3 | 1.19 × 10− 11 | Intergenic | C6orf170* |
| rs12173787 | 6 | 144 | chr6:33350201–33,789,899 | 1.20 × 10−15 | 5.4 × 10−3 | 2.79 × 10− 15 | Downstream gene | BAK1, CUTA, GGNBP1, IP6K3, ITPR3, KIFC1, LEMD2, LINC00336, MIR5004, MLN, PHF1, SYNGAP1, UQCC2, ZBTB9 |
| rs57942103 | 8 | 11 | chr8:106513461–106,589,409 | 1.90 × 10−10 | 5.06 × 10−3 | 3.10 × 10− 11 | Intron | ZFPM2 |
| rs10883944 | 10 | 21 | chr10:105522875–105,667,110 | 7.80 × 10−9 | 2.11 × 10−4 | 2.33 × 10−10 | Intron | OBFC1, SH3PXD2A |
| rs4746139 | 10 | 92 | chr10:75404300–75,692,923 | 4.50 × 10−10 | 9.94 × 10−3 | 1.60 × 10− 10 | Synonymous | AGAP5, BMS1P4, C10orf55, CAMK2G, CHCHD1, FUT11, GLUD1P3, NDST2, PLAU, SEC24C, SYNPO2L, ZSWIM8, ZSWIM8-AS1 |
| rs2512519 | 11 | 68 | chr11:77924870–78,286,462 | 2.10 × 10− 10 | 2.32 × 10−3 | 4.39 × 10−12 | Intron | GAB2, NARS2, USP35 |
| rs8756 | 12 | 12 | chr12:66306441–66,389,968 | 2.20 × 10−11 | 3.22 × 10−3 | 2.57 × 10− 12 | 3′ UTR | HMGA2 |
| rs56386186 | 14 | 77 | chr14:102418380–102,784,274 | 1.70 × 10−10 | 2.79 × 10−3 | 1.22 × 10−11 | Intron | DYNC1H1, HSP90AA1, MOK, WDR20, ZNF839 |
| rs7143026 | 14 | 17 | chr14:65825854–66,272,664 | 1.90 × 10−6 | 1.22 × 10−4 | 2.54 × 10−8 | Intergenic | FUT8, FUT8-AS1 |
| rs754388 | 14 | 34 | chr14:93068516–93,118,229 | 3.0 × 10−4 | 7.07 × 10−12 | 1.03 × 10−11 | Intron | RIN3 |
| rs17186681 | 15 | 138 | chr15:63800237–64,148,582 | 2.50 × 10−11 | 6.57 × 10−3 | 1.50 × 10−12 | Intron | FBXL22, HERC1, USP3, USP3-AS1 |
| rs17431820 | 16 | 10 | chr16:64939145–65,128,819 | 5.30 × 10−12 | 9.78 × 10−3 | 2.38 × 10− 12 | Intron | CDH11 |
| rs12709669 | 18 | 14 | chr18:19780858–19,826,742 | 4.10 × 10−9 | 4.80 × 10−3 | 8.11 × 10−10 | Intron | GATA6 |
SNP single nucleotide polymorphisms, CHR chromosome, N number of SNPs clumped with peak variant, RHR resting heart rate, COPD chronic obstructive pulmonary disease
In addition, we found 22 loci significantly associated with both COPD and HBP (Table 3 and Additional file 1: Table S9). The top significant locus was near ARHGAP42 (sentinel SNP: rs633185, Pmeta = 1.80 × 10− 47) [40], Notably, rs7655625, the most significant SNP for COPH/RHR, also had strong correlation with COPD/HBP (Pmeta = 9.69 × 10− 19).
Table 3.
Genome-wide significant loci by cross-trait meta-analysis at sentinel SNPs associated with COPD and HBP (Pmeta < 5 × 10− 8; single trait P < 0.01)
| Sentinel SNP | CHR | N | Position | P HBP | P COPD | P META | Variant annotation | Genes within clumping region |
|---|---|---|---|---|---|---|---|---|
| rs12759054 | 1 | 72 | chr1:234091759–234,196,884 | 1.2 × 10−6 | 5.78 × 10−3 | 4.88 × 10−8 | Intron | SLC35F3 |
| rs301819 | 1 | 99 | chr1:8440643–8,895,970 | 1.3 × 10−10 | 5.47 × 10−3 | 9.93 × 10−12 | Intron | RERE |
| rs305221 | 1 | 115 | chr1:88899115–89,440,896 | 3.7 × 10−10 | 5.32 × 10−4 | 2.91 × 10−12 | Intron | CCBL2, GTF2B, LOC101927891, PKN2 |
| rs61781370 | 1 | 97 | chr1:39554034–40,060,025 | 9.0 × 10−8 | 6.66 × 10−3 | 7.47 × 10−9 | Upstream gene | BMP8A, KIAA0754, MACF1, PABPC4, PPIEL, SNORA55 |
| rs71636784 | 1 | 198 | chr1:26994245–27,298,564 | 3.60 × 10−10 | 0.002705 | 6.41 × 10−12 | Intron | ARID1A, GPATCH3, GPN2, KDF1, NR0B2, NUDC, PIGV, SFN, ZDHHC18 |
| rs2293947 | 3 | 153 | chr3:127620467–128,349,376 | 1.0 × 10−8 | 1.96 × 10−3 | 9.09 × 10−10 | Intron | C3orf27, DNAJB8, DNAJB8-AS1, EEFSEC, GATA2, KBTBD12, LOC90246, RPN1, RUVBL1, SEC61A1 |
| rs6799272 | 3 | 163 | chr3:157977793–158,421,824 | 2.0 × 10−8 | 2.55 × 10−3 | 1.22 × 10−9 | Intron | GFM1, LOC100996447, LXN, MLF1, RARRES1, RSRC1 |
| rs2869966 | 4 | 135 | chr4:89750361–90,028,653 | 2.20 × 10−3 | 1.11 × 10−14 | 2.73 × 10−14 | Intron | FAM13A |
| rs7655625 | 4 | 111 | chr4:145228728–145,974,688 | 7.10 × 10−3 | 3.02 × 10−19 | 9.69 × 10−19 | Intergenic | ANAPC10, HHIP, HHIP-AS1 |
| rs7733088 | 5 | 66 | chr5:147682118–147,856,522 | 1.50 × 10−3 | 4.41 × 10−14 | 1.26 × 10−13 | Intron | FBXO38, HTR4, LOC102546294, SPINK7, SPINK9 |
| rs9399401 | 6 | 15 | chr6:142652344–142,865,106 | 4.20 × 10−3 | 3.59 × 10−10 | 9.38 × 10−10 | Intron | GPR126, LOC153910 |
| rs11771259 | 7 | 73 | chr7:7174042–7,348,633 | 1.60 × 10−15 | 6.16 × 10−4 | 6.66 × 10−18 | Intron | C1GALT1, LOC101927354 |
| rs36044436 | 7 | 40 | chr7:74027839–74,140,925 | 7.40 × 10−10 | 3.77 × 10−3 | 4.87 × 10−11 | Intron | GTF2I, LOC101926943 |
| rs633185 | 11 | 256 | chr11:100421331–100,713,227 | 1.90 × 10−48 | 1.63 × 10− 4 | 1.80 × 10− 47 | Intron | ARHGAP42 |
| rs11168245 | 12 | 125 | chr12:47981942–48,212,719 | 1.20 × 10−9 | 6.23 × 10−3 | 7.26 × 10−11 | Intron | ENDOU, HDAC7, RAPGEF3, RPAP3, SLC48A1 |
| rs1549306 | 16 | 95 | chr16:75304623–75,491,327 | 4.0 × 10−9 | 7.61 × 10−4 | 3.36 × 10−11 | Intron | CFDP1, TMEM170A |
| rs200528 | 16 | 58 | chr16:24699511–24,879,963 | 1.70 × 10−7 | 1.86 × 10−3 | 1.37 × 10−8 | Intron | SLC5A11, TNRC6A |
| rs4787486 | 16 | 26 | chr16:29958216–30,093,779 | 1.10 × 10−8 | 8.61 × 10−3 | 7.10 × 10−9 | Intron | ALDOA, C16orf92, DOC2A, FAM57B, HIRIP3, INO80E, PPP4C, TAOK2, TMEM219 |
| rs55804009 | 18 | 72 | chr18:1840658–1,902,417 | 9.0 × 10−9 | 7.36 × 10−3 | 6.62 × 10−10 | Intergenic | LINC00470* |
| rs13040716 | 20 | 44 | chr20:30660621–31,035,129 | 1.20 × 10−5 | 3.36 × 10−5 | 4.0 × 10−8 | Downstream gene | ASXL1, HCK, KIF3B, NOL4L, PLAGL2, POFUT1, TM9SF4, TSPY26P |
| rs12627514 | 21 | 29 | chr21:44740327–44,824,964 | 1.70 × 10−12 | 7.55 × 10−3 | 2.82 × 10−14 | Intergenic | LINC00322 |
| rs229340 | 21 | 173 | chr21:44914815–45,133,868 | 6.10 × 10−11 | 9.87 × 10−3 | 1.57 × 10−11 | Intron | HSF2BP, MIR6070, RRP1B |
SNP single nucleotide polymorphisms, CHR chromosome, HBP high blood pressure, COPD chronic obstructive pulmonary disease
In addition to rs7655625 in HHIP, we also observed two more overlapping significant loci in meta-analyses of COPD/RHR and COPD/HBP. The first locus was EEFSEC (sentinel SNP: rs2955083, Pmeta = 3.56 × 10− 8 for COPD/RHR; sentinel SNP: rs2293947, Pmeta = 9.09 × 10− 10 for COPD/HBP) [11, 41]. The other locus was BMP8A (sentinel SNP: rs3738676, Pmeta = 3.07 × 10− 11 for COPD/RHR; sentinel SNP: rs61781370, Pmeta = 7.47 × 10− 9 for COPD/HBP) [42, 43]. Finally, we identified 3 loci significantly associated with COPD and CAD (Table 4 and Additional file 1: Table S10). The first locus (sentinel SNP: rs2128739, Pmeta = 3.17 × 10− 12) is a transcript of long non-coding RNA gene RP11-563P16.1. The second locus represented by rs8108474 (Pmeta = 1.49 × 10− 8) was mapped to DMPK [44]. The third locus (sentinel SNP: rs8046697, Pmeta = 3.80 × 10− 8), was mapped to BCAR1 [45]. Detailed annotation for each sentinel variant is shown in Additional file 1: Table S11.
Table 4.
Genome-wide significant loci by cross-trait meta-analysis at sentinel SNPs associated with COPD and CAD (Pmeta < 5 × 10−8; single trait P < 0.01)
| Sentinel SNP | CHR | N | Position | P CAD | P COPD | P META | Variant annotation | Genes within clumping region |
|---|---|---|---|---|---|---|---|---|
| rs2128739 | 11 | 13 | chr11:103660567–103,718,660 | 7.05 × 10−11 | 3.69 × 10−3 | 3.17 × 10−12 | Intergenic | RP11-563P16.1 |
| rs8046697 | 16 | 164 | chr16:75236763–75,516,534 | 3.24 × 10−6 | 8.1 × 10−4 | 3.80 × 10−8 | Intron | BCAR1, CFDP1, CHST6, CTRB1, CTRB2, LOC100506281, TMEM170A |
| rs8108474 | 19 | 27 | chr19:46190268–46,370,381 | 7.51 × 10−6 | 5.62 × 10−5 | 1.49 × 10−8 | Intron | DMPK, DMWD, FBXO46, FOXA3, LOC388553, QPCTL, RSPH6A, SIX5, SNRPD2, SYMPK |
SNP single nucleotide polymorphisms, CHR chromosome, CAD coronary artery disease, COPD chronic obstructive pulmonary disease
Identification of causal variants
We identified a credible set of causal SNPs using Bayesian fine-mapping at each shared loci meeting significance criteria in the COPD–cardiac traits meta-analysis. The credible set of variants at each locus were 99% likely to contain the causal variant. A list of credible sets of SNPs for each locus is provided in Additional file 1: Tables S11–S14.
We found 5 loci (in MACF1, SYNPO2L, RIN3, TNS1, and MLN) for COPD/RHR (Additional file 1: Table S15), 4 loci (NR0B2, C1orf172, MAFC1, and TNRC6A) for COPD/HBP (Additional file 1: Table S16), and 7 loci (CD3EAP, C19orf83, GIPR, FBXO46, AC074212.3, SIX5, and DMPK) for COPD/CAD (Additional file 1: Table S17) in which the credible set included exonic missense polymorphisms. However, most variants in credible sets at each locus were either intronic or intergenic, which is consistent with prior studies showing most variants detected by GWAS involve gene regulatory effects, rather than protein structure changes [46].
Biological pathway, tissue enrichment, and TWAS
We performed pathway analyses to identify biological pathways enriched for shared loci related to COPD and cardiac traits based on significant cross-trait meta-analysis results. COPD and RHR response to nicotine was present only at a liberal FDR (FDR = 0.198) (Additional file 1: Table S18). COPD shared pathways of detection of chemical stimulus involved in sensory perception of smell with HBP (FDR = 1.06 × 10− 10) (Additional file 1: Table S19). No biological pathways were significantly shared by COPD and CAD (Additional file 1: Table S20).
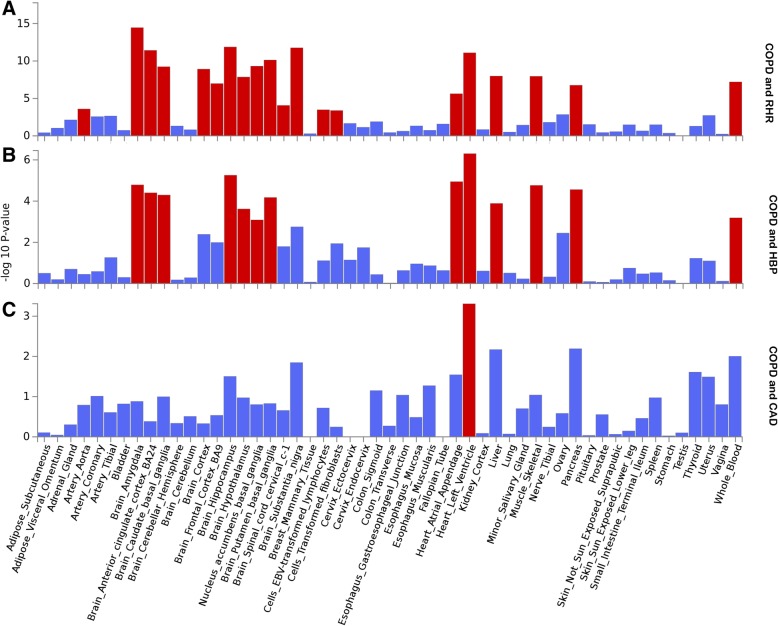
GTEx enrichment analysis identified 20 independent tissues that were significantly enriched (after Benjamin-Hochberg correction) for expression of cross-trait-associated genes for COPD and RHR traits, the top of which was brain amygdala (Fig. 3). In addition, all 13 independent tissues enriched for COPD and HBP trait expression overlapped with COPD and RHR traits. COPD and CAD trait expression only showed one significantly enriched tissue, heart left ventricle.
Fig. 3.
GTEx tissue enrichment analysis for expression of cross-trait-associated genes for COPD and RHR (a), COPD and HBP (b), or COPD and CAD (c). Red represents significant tissue enrichment after Benjamin-Hochberg correction
To identify associations between COPD and cardiac traits with gene expression in specific tissues, we conducted TWAS analysis in 44 GTEx tissues. A total of 231 gene–tissue pairs were significantly associated with COPD, in addition to 8504 gene–tissue pairs with RHR, 8272 gene–tissue pairs with HBP, and 805 gene–tissue pairs with CAD. Most associations were found in heart, vascular system, and lung tissues. Notably, 18 COPD-associated gene–tissue pairs were shared with RHR, 16 pairs were shared with HBP, and 2 pairs were shared with CAD (Additional file 1: Table S21).
Effect of smoking-related genetic variants between COPD and cardiac traits
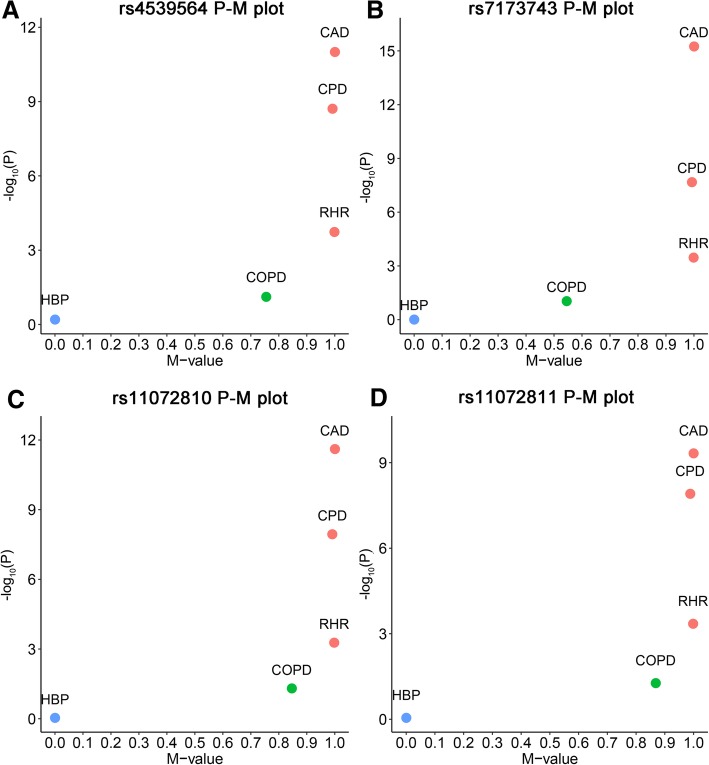
In the GWAS cross-trait subset effect analysis of smoking-related genetic variants, four SNPs located in the 15q25.1 region (rs4539564, rs11072810, rs11072811 and rs7173743) with CPD genetic effect, were also identified to be associated with RHR and CAD traits. These SNPs also had a moderate effect in COPD with M-values more than 0.5 (Fig. 4 and Additional file 1: Table S22).
Fig. 4.
PM plots of 4 smoking related SNPs from Tobacco and Genetics Consortium that also have an effect on at least one CVD trait. a rs4539564, (b) rs7173743, (c) rs11072810, (d) rs11072811. Red dot represents the SNP has an effect on certain traits (M-value> 0.9); green dot represents the SNP may have an effect on certain traits (0.1 ≤ M-value≤0.9), but the effect is ambiguous; blue dot represents the SNP does not have an effect on certain traits (M-value< 0.1)
Causal inference
We identified a significant positive causal effect of COPD on RHR (causal estimate = 0.1374, P = 0.008), but not on HBP (causal estimate = 0.007, P = 0.35) or CAD (causal estimate = 0.004, P = 0.40) (Additional file 1: Table S23).
Discussion
To our knowledge, this study is the first large-scale genome-wide analysis to investigate genetic overlap between COPD and cardiac traits. We found significant positive genome-wide genetic correlation of COPD with RHR or HBP, and a positive correlation between COPD and CAD, although this latter association failed to reach statistical significance. In the analysis of functional partitioned LDSC, we observed positive genetic correlations between COPD and cardiac traits in most annotated regions of the genome. Among them, introns, h3k9ac, and super enhancers had the highest magnitude and significance.
GWAS most frequently detects non-coding variants, and variants affecting gene expression have been shown to have pervasive effects on most diseases [46]. Histone markers like h3k9ac and h3k4me3 are some of the most essential modification markers involved in arterial pressure [47] and development of bronchial epithelial cells influencing COPD [48]. Super enhancer regions have multiple enhancers that drive transcription of genes involved in cell identity in diseases and heart development [49]. In local genetic correlation analysis, we identified multiple novel regions that have strong local genetic correlation between COPD and cardiac traits, such as the 4q31 region shared by COPD and RHR, and 11q22 and 5q32 regions shared by COPD and HBP. The 4q31 region was previously reported to have an independent association with COPD and RHR [20, 50], although it has not been identified as a shared region. By contrast, we did not observe any significant local genetic correlation between COPD and CAD.
We also discovered 21 shared loci between COPD and RHR, 22 shared loci between COPD and HBP, and 3 shared loci between COPD and CAD using cross-trait meta-analysis. Among them, we highlight the novel association of HHIP, EEFSEC, RIN3, SIX5, and DMPK with COPD and cardiac traits due to their potentially interesting functions.
First, the top sentinel variant for both COPD/RHR and COPD/HBP was rs7655625 near HHIP, known to be associated with COPD susceptibility by influencing crucial lung development signaling pathway [51]. HHIP is also downregulated during angiogenesis and under oxidative stress [52], and its knockdown in late endothelial progenitor cells improves endothelial angiogenesis, promoting vascular repair [53]. Another top association common to the COPD/RHR and COPD/HBP meta-analysis was with variants near EEFSEC, however the two analyses identified different sentinel variants. EEFSEC encodes a translation factor necessary for incorporation of selenocysteine into proteins associated with COPD [11] and cardiovascular events [41]. DMPK encodes a myotonic dystrophy protein kinase that is involved in heart cells, and SIX5, encodes a homeodomain-containing transcription factor that appears to function in the regulation of organogenesis [44]. Fine-mapping analysis identified multiple missense variants. For example, in meta-analysis of COPD and RHR only, we identified RIN3 as a significant locus. Fine-mapping analysis found that rs117068593 is a missense variant in which the effect allele T results in mutation R279C in RIN3. Also, several missense variants were found in SIX5 and DMPK, which are associated with COPD and CAD. However, we stress that the causal genes in these and other associated regions cannot be determined without further study.
Post-GWAS functional analyses provided biological insights to the shared genes between COPD and cardiac traits. GTEx tissue enrichment analysis identified shared genes that were significantly enriched in several tissues, including cardiovascular, nervous, and immune systems. Our findings of cardiovascular system genetic enrichment could eventually have therapeutic implications for managing COPD patients through exploration of shared mechanisms in genes such as HHIP [53].
Although the association between COPD/CVD and the nervous system may initially seem counterintuitive, further exploring their genetic link may provide functional and molecular understanding of their etiologies. Impaired brain function is a complication of COPD and CVD [54], which can be due to systemic inflammation, induced stress, and neurochemical abnormalities [55]. Further, stimulation of nicotinic cholinergic receptors releases a variety of neurotransmitters in the brain, which have adverse effects [55]. Nicotine-related functions in both diseases were also highlighted in our biological pathway analysis.
In TWAS analysis, we integrated data from GWAS and GTEx tissue expression to identify shared mechanistic hypotheses between COPD and cardiac traits on a tissue–gene pair level. We found 231 unique gene–tissue pairs with transcriptome-wide significant associations with COPD, in addition to 8504 with RHR, 8272 with HBP, and 805 with CAD. Most were associated with heart, vascular system, and lung tissues. Notably, 18 COPD-associated gene–tissue pairs were shared with RHR, 16 pairs were shared with HBP, and 2 pairs were shared with CAD, thus implicating specific shared regulatory features for functional follow-up.
In addition to genetic contributions to COPD and CVD, environmental, behavioral, and clinical factors also play important roles in their comorbidity. Notably, smoking is a major common environmental risk factor for both COPD and CVD. One possible mechanism linking COPD and CVD is systemic inflammation due to smoking [9]. Thus the impact of controlling such modifiable risk factor can be large. Several interventions, such as smoking cessation, exercise, drug use (e.g., statins), increased awareness of the connection between COPD and CVD, and improved collaboration between pulmonary and cardiovascular clinicians, have been shown to improve COPD and CVD and currently represent the most hopeful approaches to disease prevention and treatment [56]. While we adjusted for cigarette smoking in our ICGC COPD GWAS, other GWAS did not, and accurate measurement of exposure is challenging. Some loci such as 15q25.1 are clearly related to cigarette smoking, which is also a risk factor for CVD. Previous studies have suggested that the 15q25.1 region played a role in nicotine, alcohol, and cocaine dependence [57]. This region has been reported related to multiple diseases, such as COPD [11]. In our cross-trait subset effect analysis, we also found 4 variants in 15q25.1 region have an effect with RHR and CAD. However, interestingly, these variants were not related to COPD, suggesting that the genetic effect of cigarette smoking between COPD and CVD is complex, and not necessarily based on the same genetic variants in 15q25.1 region.
Finally, our MR analysis suggested a significant positive causal effect of COPD on RHR. One possible causal pathway example is genetic variation leading to COPD could exacerbate right ventricular diastolic dysfunction and alterations in heart rate [8]. However, our MR results should be taken with caution as other potential confounders may bias the causal relationship. For example, COPD is also known to be associated with cardiovascular autonomic neuropathy resulting in decreased parasympathetic and increased sympathetic activity, which can alter the heart rate [58]. In addition, medication use (bronchodilators) or stimulants (such as cigarettes and caffeine) may also contribute to elevated RHR in COPD patients [7].
We also acknowledge other potential limitations in this study. First, additional GWAS cohorts are not available to replicate our findings. However, we used the largest datasets available at the time of our study to perform our analyses. Genome-wide genetic correlation results were relatively weak, and did not reach significance level after multiple testing correction. However, we found a strong local genetic correlation between COPD and RHR at 4q31, between COPD and HBP at 11q22 and 5q32 regions after multiple testing correction, which highlights the genetic overlap between COPD and CVD at regional level. In addition, we identified a credible-set of SNPs that contains potential causal variants. Further functional experiments are needed to investigate the causal variants or genes. Finally, the current study was limited to assessing shared genetic factors between COPD and CVD. Future studies on shared environmental factors between COPD and CVD are needed.
Conclusions
Understanding the genetic overlap between COPD and CVD is important for disease prevention, timely diagnosis and treatment of both diseases. Our study shows evidence of significant positive genetic correlations between COPD and cardiac traits. Shared genetic variants were fine-mapped to improve resolution and identify potential shared causal variants with exonic missense polymorphisms. We also found multiple common biological pathways and tissue enrichments, such as nicotine response, cardiovascular, brain, and immune-related tissues, which can further our understanding of the connection between these diseases. Such shared genes and pathways might serve as common drug targets in both COPD and CVD.
Additional files
Table S1. Summary of GWAS data. Table S2. SNP based heritability and genomic inflation factor estimated by LDSC. Table S3. Evaluation of genetic correlation between COPD and CVD related metabolic traits. Table S4. Partitioned genetic correlation between COPD and 3 cardiac traits. Table S5. Local genetic covariance analysis between COPD and RHR (only P < 0.01 shown in this table). Table S6. Local genetic coveriance analysis between COPD and HBP (only P < 0.01 shown in this table). Table S7. Local genetic covariance analysis between COPD and CAD. Table S8. Genome-wide significant loci by cross-trait meta-analysis at sentinel SNPs. Table S9. Genome-wide significant loci by cross-trait meta-analysis at sentinel SNPs. Table S10. Genome-wide significant loci by cross-trait meta-analysis at sentinel SNPs. Table S11. Detailed annotation of cross-trait meta-analysis genome-wide significant SNPs. Table S12. Fine-mapping credible set analysis for 21 top loci. Table S13. Fine-mapping credible set analysis for 22 top loci. Table S14. Fine-mapping credible set analysis for 3 top loci. Table S15. Missense variants in 99% credible set. Table S16. Missense variants in 99% credible-set. Table S17. Missense variantsin 99% credible-set. Table S18. GO biological process pathway analysis for COPD and RHR. Table S19. GO biological process pathway analysis for COPD and HBP. Table S20. GO biological process pathway analysis for COPD and CAD. Table S21. Significant overlap transcriptome-wide association analysis results. Table S22. Characterization of trait-specific association for the smoking related. Table S23. Mendelian randomization analysis between COPD and cardiac traits. (XLSX 240 kb)
Online Data Supplemental Text. (DOCX 129 kb)
Figure S1. QQ plot of resting heart rate. Figure S2. QQ plot of high blood pressure. Figure S3. Genetic Correlation between COPD and Cardiac Traits by Functional Category. (PDF 360 kb)
Acknowledgements
This research has been conducted using the UK Biobank Resource under application number 16549. We would like to thank participants and researchers from the UK Biobank who significantly contributed or collected data. We are grateful to all participants from International COPD Genetics Consortium, CARDIoGRAMplusC4D Consortium, International Stroke Genetics Consortium, GIANT consortium, DIAGRAM consortium and ENGAGE consortium for their significant contributions to share data.
Funding
This study is supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health and National Institute of Environmental Health Sciences (Grant R01HL060710 and R56HL134356 to D.C.C, R01HL113264, R01HL137927, R01HL135142 to M.H.C, and P30ES000002 to Z.Z).
Availability of data and materials
UK Biobank summary GWAS statistics will be available at the GWAS catalog (https://www.ebi.ac.uk/gwas/downloads/summary-statistics).
Abbreviations
- ASSET
Association analysis based on SubSETs
- CARDIoGRAMplusC4D
Coronary ARtery DIsease Genome wide Replication and Meta-analysis (CARDIoGRAM) plus The Coronary Artery Disease (C4D) consortium
- COPD
Chronic Obstructive Pulmonary Disease
- CPASSOC
Cross-Phenotype Association
- DIAGRAM
DIAbetes Genetics Replication And Meta-analysis consortium
- ENGAGE
European Network for Genetic and Genomic Epidemiology consortium
- FDR
False Discovery Rate
- GIANT
The Genetic Investigation of ANthropometric Traits (GIANT) consortium
- GO
Gene Ontology
- GOLD
The Global Initiative for Chronic Obstructive Lung Disease
- GTEx
The Genotype-Tissue Expression
- HESS
Heritability Estimation from Summary Statistics
- ICGC
International COPD Genomic Consortium
- ISGC
International Stroke Genetics Consortium
- LDSC
LD Score Regression
- MR-PRESSO
Mendelian Randomization Pleiotropy RESidual Sum and Outlier
- M-value
Posterior probability to evaluate if the genetic variant effect exists among traits
- TAG
Tobacco and Genetics Consortium.
- TWAS
Transcriptome-Wide Association Study
- UKBB
UK Biobank
- ρ-HESS
A tool to quantify the correlation between pairs of traits due to genetic variation at a small region in the genome
Authors’ contributions
ZZ, XW, CLL, HMB, MHC, and DCC designed the study. ZZ, XL and BHH performed statistical analysis. ZZ, YL, XW, KH, and CAC first drafted the manuscript. All authors helped interpret data, reviewed and edited the final paper, and approved the submission.
Ethics approval and consent to participate
This research has been approved by UK Biobank (application number 16549). The institutional review boards of the UK Biobank approved use of UK Biobank resources. All UK Biobank participants involved in this study consented to participate.
Consent for publication
Not applicable
Competing interests
Dr. Michael H. Cho has received grant support from GSK. The other authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Raherison C, Girodet PO. Epidemiology of COPD. Eur Respir Rev. 2009;18:213–221. doi: 10.1183/09059180.00003609. [DOI] [PubMed] [Google Scholar]
- 2.Macnee W, Maclay J, McAllister D. Cardiovascular injury and repair in chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2008;5:824–833. doi: 10.1513/pats.200807-071TH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kunisaki KM, Dransfield MT, Anderson JA, Brook RD, Calverley PMA, Celli BR, Crim C, Hartley BF, Martinez FJ, Newby DE, et al. Exacerbations of chronic obstructive pulmonary disease and cardiac events. A post hoc cohort analysis from the SUMMIT randomized clinical trial. Am J Respir Crit Care Med. 2018;198:51–57. doi: 10.1164/rccm.201711-2239OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hurst JR, Sin DD. Chronic obstructive pulmonary disease as a risk factor for cardiovascular disease. A view from the SUMMIT. Am J Respir Crit Care Med. 2018;198:2–4. doi: 10.1164/rccm.201802-0347ED. [DOI] [PubMed] [Google Scholar]
- 5.Anthonisen NR, Connett JE, Enright PL, Manfreda J. Lung health study research G: hospitalizations and mortality in the lung health study. Am J Respir Crit Care Med. 2002;166:333–339. doi: 10.1164/rccm.2110093. [DOI] [PubMed] [Google Scholar]
- 6.Grosdidier S, Ferrer A, Faner R, Pinero J, Roca J, Cosio B, Agusti A, Gea J, Sanz F, Furlong LI. Network medicine analysis of COPD multimorbidities. Respir Res. 2014;15:111. doi: 10.1186/s12931-014-0111-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen MT, Marott JL, Lange P, Vestbo J, Schnohr P, Nielsen OW, Jensen JS, Jensen GB. Resting heart rate is a predictor of mortality in COPD. Eur Respir J. 2013;42:341–349. doi: 10.1183/09031936.00072212. [DOI] [PubMed] [Google Scholar]
- 8.Falk JA, Kadiev S, Criner GJ, Scharf SM, Minai OA, Diaz P. Cardiac disease in chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2008;5:543–548. doi: 10.1513/pats.200708-142ET. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boschetto P, Beghe B, Fabbri LM, Ceconi C. Link between chronic obstructive pulmonary disease and coronary artery disease: implication for clinical practice. Respirology. 2012;17:422–431. doi: 10.1111/j.1440-1843.2011.02118.x. [DOI] [PubMed] [Google Scholar]
- 10.Austin V, Crack PJ, Bozinovski S, Miller AA, Vlahos R. COPD and stroke: are systemic inflammation and oxidative stress the missing links? Clin Sci (Lond) 2016;130:1039–1050. doi: 10.1042/CS20160043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hobbs BD, de Jong K, Lamontagne M, Bosse Y, Shrine N, Artigas MS, Wain LV, Hall IP, Jackson VE, Wyss AB, et al. Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nat Genet. 2017;49:426–432. doi: 10.1038/ng.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Z, Lee PH, Chaffin MD, Chung W, Loh PR, Lu Q, Christiani DC, Liang L. A genome-wide cross-trait analysis from UK biobank highlights the shared genetic architecture of asthma and allergic diseases. Nat Genet. 2018;50:857–864. doi: 10.1038/s41588-018-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Z, Lin Y, Li X, Driver JA, Liang L. Shared genetic architecture between metabolic traits and Alzheimer's disease: a large-scale genome-wide cross-trait analysis. Hum Genet. 2019;138:271–85. [DOI] [PMC free article] [PubMed]
- 14.Lee PH, Anttila V, Won H, Feng Y-CA, Rosenthal J, Zhu Z, Tucker-Drob EM, Nivard MG, Grotzinger AD, Posthuma D, et al. Genome wide meta-analysis identifies genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. bioRxiv. 2019:528117. 10.1101/528117.
- 15.Kathiresan S, Srivastava D. Genetics of human cardiovascular disease. Cell. 2012;148:1242–1257. doi: 10.1016/j.cell.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sabater-Lleal M, Malarstig A, Folkersen L, Soler Artigas M, Baldassarre D, Kavousi M, Almgren P, Veglia F, Brusselle G, Hofman A, et al. Common genetic determinants of lung function, subclinical atherosclerosis and risk of coronary artery disease. PLoS One. 2014;9:e104082. doi: 10.1371/journal.pone.0104082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, et al. A comprehensive 1,000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, Rutten-Jacobs L, Giese AK, van der Laan SW, Gretarsdottir S, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50:524–537. doi: 10.1038/s41588-018-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eppinga RN, Hagemeijer Y, Burgess S, Hinds DA, Stefansson K, Gudbjartsson DF, van Veldhuisen DJ, Munroe PB, Verweij N, van der Harst P. Identification of genomic loci associated with resting heart rate and shared genetic predictors with all-cause mortality. Nat Genet. 2016;48:1557–1563. doi: 10.1038/ng.3708. [DOI] [PubMed] [Google Scholar]
- 21.Loh PR, Tucker G, Bulik-Sullivan BK, Vilhjalmsson BJ, Finucane HK, Salem RM, Chasman DI, Ridker PM, Neale BM, Berger B, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47:284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, ReproGen C, Psychiatric Genomics C. Genetic Consortium for anorexia nervosa of the Wellcome Trust case control C, Duncan L, et al: an atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR, Anttila V, Xu H, Zang C, Farh K, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi H, Mancuso N, Spendlove S, Pasaniuc B. Local genetic correlation gives insights into the shared genetic architecture of complex traits. Am J Hum Genet. 2017;101:737–751. doi: 10.1016/j.ajhg.2017.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Z, Anttila V, Smoller JW, Lee PH. Statistical power and utility of meta-analysis methods for cross-phenotype genome-wide association studies. PLoS One. 2018;13:e0193256. doi: 10.1371/journal.pone.0193256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhattacharjee S, Rajaraman P, Jacobs KB, Wheeler WA, Melin BS, Hartge P, GliomaScan C, Yeager M, Chung CC, Chanock SJ, Chatterjee N. A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. Am J Hum Genet. 2012;90:821–835. doi: 10.1016/j.ajhg.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu X, Feng T, Tayo BO, Liang J, Young JH, Franceschini N, Smith JA, Yanek LR, Sun YV, Edwards TL, et al. Meta-analysis of correlated traits via summary statistics from GWASs with an application in hypertension. Am J Hum Genet. 2015;96:21–36. doi: 10.1016/j.ajhg.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Giron CG, et al. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang H, Fang M, Jostins L, Umicevic Mirkov M, Boucher G, Anderson CA, Andersen V, Cleynen I, Cortes A, Crins F, et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature. 2017;547:173–178. doi: 10.1038/nature22969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Consortium GT, Laboratory DA. Coordinating center -analysis working G, statistical methods groups-analysis working G, enhancing gg, fund NIHC, Nih/Nci, Nih/Nhgri, Nih/Nimh, Nih/Nida, et al: genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Consortium GT. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tobacco, Genetics C Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han B, Eskin E. Interpreting meta-analyses of genome-wide association studies. PLoS Genet. 2012;8:e1002555. doi: 10.1371/journal.pgen.1002555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J, Weedon MN, Purcell S, Lettre G, Estrada K, Willer CJ, Smith AV, Ingelsson E, O'Connell JR, Mangino M, et al. Genomic inflation factors under polygenic inheritance. Eur J Hum Genet. 2011;19:807–812. doi: 10.1038/ejhg.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.International Consortium for Blood Pressure Genome-Wide Association S. Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, et al. genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carney EF. Hypertension: role of ARHGAP42 in hypertension. Nat Rev Nephrol. 2017;13:134. doi: 10.1038/nrneph.2017.13. [DOI] [PubMed] [Google Scholar]
- 41.Terao C, Yoshifuji H, Kimura A, Matsumura T, Ohmura K, Takahashi M, Shimizu M, Kawaguchi T, Chen Z, Naruse TK, et al. Two susceptibility loci to Takayasu arteritis reveal a synergistic role of the IL12B and HLA-B regions in a Japanese population. Am J Hum Genet. 2013;93:289–297. doi: 10.1016/j.ajhg.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aguilera-Aguirre L, Hosoki K, Bacsi A, Radak Z, Sur S, Hegde ML, Tian B, Saavedra-Molina A, Brasier AR, Ba X, Boldogh I. Whole transcriptome analysis reveals a role for OGG1-initiated DNA repair signaling in airway remodeling. Free Radic Biol Med. 2015;89:20–33. doi: 10.1016/j.freeradbiomed.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen QM, Maltagliati AJ. Nrf2 at the heart of oxidative stress and cardiac protection. Physiol Genomics. 2018;50:77–97. doi: 10.1152/physiolgenomics.00041.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, Chu A, Chakroun I, Islam U, Blais A. Cooperation between myogenic regulatory factors and SIX family transcription factors is important for myoblast differentiation. Nucleic Acids Res. 2010;38:6857–6871. doi: 10.1093/nar/gkq585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boardman-Pretty F, Smith AJ, Cooper J, Palmen J, Folkersen L, Hamsten A, Catapano AL, Melander O, Price JF, Kumari M, et al. Functional analysis of a carotid intima-media thickness locus implicates BCAR1 and suggests a causal variant. Circ Cardiovasc Genet. 2015;8:696–706. doi: 10.1161/CIRCGENETICS.115.001062. [DOI] [PubMed] [Google Scholar]
- 46.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kresovich JK, Zhang Z, Fang F, Zheng Y, Sanchez-Guerra M, Joyce BT, Zhong J, Chervona Y, Wang S, Chang D, et al. Histone 3 modifications and blood pressure in the Beijing truck Driver air pollution study. Biomarkers. 2017;22:584–593. doi: 10.1080/1354750X.2017.1347961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leclercq B, Platel A, Antherieu S, Alleman LY, Hardy EM, Perdrix E, Grova N, Riffault V, Appenzeller BM, Happillon M, et al. Genetic and epigenetic alterations in normal and sensitive COPD-diseased human bronchial epithelial cells repeatedly exposed to air pollution-derived PM2.5. Environ Pollut. 2017;230:163–177. doi: 10.1016/j.envpol.2017.06.028. [DOI] [PubMed] [Google Scholar]
- 49.Anderson KM, Anderson DM, McAnally JR, Shelton JM, Bassel-Duby R, Olson EN. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature. 2016;539:433–436. doi: 10.1038/nature20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilk JB, Chen TH, Gottlieb DJ, Walter RE, Nagle MW, Brandler BJ, Myers RH, Borecki IB, Silverman EK, Weiss ST, O'Connor GT. A genome-wide association study of pulmonary function measures in the Framingham heart study. PLoS Genet. 2009;5:e1000429. doi: 10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou X, Qiu W, Sathirapongsasuti JF, Cho MH, Mancini JD, Lao T, Thibault DM, Litonjua AA, Bakke PS, Gulsvik A, et al. Gene expression analysis uncovers novel hedgehog interacting protein (HHIP) effects in human bronchial epithelial cells. Genomics. 2013;101:263–272. doi: 10.1016/j.ygeno.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lao T, Jiang Z, Yun J, Qiu W, Guo F, Huang C, Mancini JD, Gupta K, Laucho-Contreras ME, Naing ZZ, et al. Hhip haploinsufficiency sensitizes mice to age-related emphysema. Proc Natl Acad Sci U S A. 2016;113:E4681–E4687. doi: 10.1073/pnas.1602342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee BNR, Son YS, Lee D, Choi YJ, Kwon SM, Chang HK, Kim PH, Cho JY. Hedgehog-interacting protein (HIP) regulates apoptosis evasion and Angiogenic function of late endothelial progenitor cells. Sci Rep. 2017;7:12449. doi: 10.1038/s41598-017-12571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goto T, Shimada YJ, Faridi MK, Camargo CA, Jr, Hasegawa K. Incidence of acute cardiovascular event after acute exacerbation of COPD. J Gen Intern Med. 2018;33(9):1461–1468. doi: 10.1007/s11606-018-4518-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benowitz NL. Nicotine addiction. N Engl J Med. 2010;362:2295–2303. doi: 10.1056/NEJMra0809890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hunninghake DB. Cardiovascular disease in chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2005;2:44–49. doi: 10.1513/pats.200410-050SF. [DOI] [PubMed] [Google Scholar]
- 57.Erlich PM, Hoffman SN, Rukstalis M, Han JJ, Chu X, Linda Kao WH, Gerhard GS, Stewart WF, Boscarino JA. Nicotinic acetylcholine receptor genes on chromosome 15q25.1 are associated with nicotine and opioid dependence severity. Hum Genet. 2010;128:491–499. doi: 10.1007/s00439-010-0876-6. [DOI] [PubMed] [Google Scholar]
- 58.Chhabra SK, De S. Cardiovascular autonomic neuropathy in chronic obstructive pulmonary disease. Respir Med. 2005;99:126–133. doi: 10.1016/j.rmed.2004.06.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Summary of GWAS data. Table S2. SNP based heritability and genomic inflation factor estimated by LDSC. Table S3. Evaluation of genetic correlation between COPD and CVD related metabolic traits. Table S4. Partitioned genetic correlation between COPD and 3 cardiac traits. Table S5. Local genetic covariance analysis between COPD and RHR (only P < 0.01 shown in this table). Table S6. Local genetic coveriance analysis between COPD and HBP (only P < 0.01 shown in this table). Table S7. Local genetic covariance analysis between COPD and CAD. Table S8. Genome-wide significant loci by cross-trait meta-analysis at sentinel SNPs. Table S9. Genome-wide significant loci by cross-trait meta-analysis at sentinel SNPs. Table S10. Genome-wide significant loci by cross-trait meta-analysis at sentinel SNPs. Table S11. Detailed annotation of cross-trait meta-analysis genome-wide significant SNPs. Table S12. Fine-mapping credible set analysis for 21 top loci. Table S13. Fine-mapping credible set analysis for 22 top loci. Table S14. Fine-mapping credible set analysis for 3 top loci. Table S15. Missense variants in 99% credible set. Table S16. Missense variants in 99% credible-set. Table S17. Missense variantsin 99% credible-set. Table S18. GO biological process pathway analysis for COPD and RHR. Table S19. GO biological process pathway analysis for COPD and HBP. Table S20. GO biological process pathway analysis for COPD and CAD. Table S21. Significant overlap transcriptome-wide association analysis results. Table S22. Characterization of trait-specific association for the smoking related. Table S23. Mendelian randomization analysis between COPD and cardiac traits. (XLSX 240 kb)
Online Data Supplemental Text. (DOCX 129 kb)
Figure S1. QQ plot of resting heart rate. Figure S2. QQ plot of high blood pressure. Figure S3. Genetic Correlation between COPD and Cardiac Traits by Functional Category. (PDF 360 kb)
Data Availability Statement
UK Biobank summary GWAS statistics will be available at the GWAS catalog (https://www.ebi.ac.uk/gwas/downloads/summary-statistics).