Abstract
Introduction:
Glucose utilization and lactate release are 2 important indicators of cancer metabolism. Most tumors consume glucose and release lactate at a higher rate than normal tissues due to enhanced aerobic glycolysis. However, these 2 indicators of metabolism have not previously been studied on a single-cell level, in the same cell.
Objective:
To develop and characterize a novel droplet microfluidic device for multiplexed measurements of glucose uptake (via its analog 18F-fluorodeoxyglucose) and lactate release, in single live cells encapsulated in an array of water-in-oil droplets.
Results:
Surprisingly, 18F-fluorodeoxyglucose uptake and lactate release were only marginally correlated at the single-cell level, even when assayed in a standard cell line (MDA-MB-231). While 18F-fluorodeoxyglucose-avid cells released substantial amounts of lactate, the reverse was not true, and many cells released high amounts of lactate without taking up 18F-fluorodeoxyglucose.
Discussion:
These results confirm that cancer cells rely on multiple metabolic pathways in addition to aerobic glycolysis and that the use of these pathways is highly heterogeneous, even under controlled culture conditions. Clinically, the large cell-to-cell variability suggests that positron emission tomography measurements of 18F-fluorodeoxyglucose uptake represent metabolic flux only in an aggregate sense, not for individual cancer cells within the tumor.
Keywords: fluorodeoxyglucose, lactate, cancer metabolism, droplet microfluidics, microscopy
Introduction
Aberrant metabolism is a hallmark of cancer as many metabolic pathways are dysregulated in tumors.1 In particular, the upregulation of glycolysis promotes increased glucose uptake and lactate release in tumors2,3 and provides important clinical diagnostic and therapeutic targets.4 For instance, the retention of the radiolabeled glucose analogue 18F-fluorodeoxyglucose (FDG) in tissues is used routinely in positron emission tomography (PET) scans to visualize malignant tumors for cancer diagnosis, staging, and monitoring.5 However, many cancers display significant intratumoral heterogeneity, both genetically and metabolically, and there can be a significant discrepancy between bulk metabolic rates measured with PET and actual metabolism of individual cells.6 Fluorescence methods, such as flow cytometry and microscopy, are often used to study the biological properties of single cells, but their limitation is that most small molecules lack intrinsic fluorescence and cannot be fluorescently labeled without interfering with their biochemical activity.7 This limitation led us to develop radioluminescence microscopy (RLM), a method for microscopic imaging of cells using clinically relevant PET tracers for metabolism and other biological processes.8
Radioluminescence microscopy is based on detecting the scintillation of individual radionuclide decays, within a fluorescence microscopy environment. Previously, the method has been used to measure glycolysis in single cells using FDG as a tracer. The FDG uptake is directly related to the expression and activity of hexokinase, which is a key regulatory enzyme in the glycolysis pathway.9 However, cancer cells have metabolic plasticity: They can use a variety of anabolic and catabolic pathways to adapt to energetic needs and availability of nutrients10; thus, FDG uptake alone is not sufficient to fully characterize the metabolic state of cancer cells. Cancer metabolism involves a variety of fuels (eg, glucose, glutamine, lactate, and fatty acids) that feed specific molecular pathways.11 Although most cancers are characterized by a high rate of glycolysis, many cancers rely on alternative metabolic pathways.12 Hence, the metabolic state of cancer cells is a multidimensional quantity, not well defined by any single readout. Glycolysis, as measured by single-cell FDG uptake, may not fully represent the diversity of metabolic programs in a given cell population.
To address this issue, we became interested in developing a multiplexed approach to characterize the metabolic profile of individual cancer cells using 2 different indicators of cell metabolism, FDG uptake and lactate release. The assay combines 2 previous technologies, FDG-RLM and single-cell droplet microfluidics, to simultaneously measure glucose uptake and lactate secretion in single cells. We have previously demonstrated that RLM can quantitatively measure the uptake of a radiolabeled molecule by single cells individually encapsulated in small droplets13; furthermore, we have measured lactate release from single cells inside similar droplets using a fluorescent sensor.14 We show here a significant extension of this technique by multiplexing the 2 approaches and jointly measuring single-cell FDG uptake and lactate release by the same cells.
The approach has several advantages. First, the encapsulation of cells in droplets permits easy manipulation of single cells as microfluidic technology allows for the optimal arraying of droplets for higher throughput. Second, as cell secretion remains contained within the small droplet volume, the technique allows the measurement of lactate release for single cells. Finally, this technique can image radionuclides inside an optical microscope for the sensitive detection of metabolic substrates, without the need for bulky fluorophores. Using the technique, we are able to quantitatively measure glucose uptake and lactate release in the same live cells. These measurements can reveal the potential connection between the energy source (glucose) and the product (lactate) of aerobic glycolysis for individual cells.
In the current study, we use this multiplexed system to measure FDG uptake and lactate release in the MDA-MB-231 cell line, which is derived from a triple negative human breast adenocarcinoma. We demonstrate that, on a single-cell level, the metabolic state of cells varies significantly, even under homogeneous conditions within a clonal cell population.
Materials and Methods
Cells
MDA-MB-231 human breast cancer cells were purchased from the American Type Culture Collection (Manassas, Virginia) and cultured in Dulbecco’s modified Eagle medium (DMEM; Gibco, Waltham, Massachusetts) medium supplemented with 10% fetal bovine serum. For inhibitor experiments, α-cyano-4-hydroxycinnamic acid (αCHC) was dissolved in dimethyl sulfoxide. Cells were incubated with the inhibitor at a concentration of 3 mM for 24 hours at 37°C and 5% CO2 prior to experimentation. For droplet-based radioluminescence experiments, cells were incubated with 250 µCi/1 mL of FDG for 30 minutes, washed with phosphate-buffered saline 3 times, and resuspended in glucose-free media. Immediately before introduction into the microfluidic device, pelleted cells were resuspended in DMEM. To make the final cell solution, working reagent comprised of the individual reagents in the Enzyfluo l-lactate assay kit (EFFLC-100; BioAssay systems) was added to the resuspended cells in a 1:1 ratio (vol/vol). The kit’s working reagent was prepared as specified immediately prior to use and consisted of solutions containing buffer, NAD+, probe, and enzymes including lactate dehydrogenase and was added to the cell suspension just before droplet encapsulation.
Microfluidic Device
Microfluidic chips with channel depth modulations were made of polydimethylsiloxane (PDMS) using dry-film photoresist soft lithography technique15 that enables rapid prototyping of multilevel structures. The PDMS chips were plasma bonded to a 1 cm × 1 cm cadmium tungstate (CdWO4; MTI Inc, Richmond, California; 0.5 mm thickness, both sides polished) scintillator. To render the channel surface hydrophobic, Novec 1720 electronic grade coating (3 M, Maplewood, Minnesota) was flowed into the microchannel and the device was heated for 30 minutes at 150°C. This surface treatment prevented wetting and contact of the aqueous droplets with the channel walls.
Droplet Generation
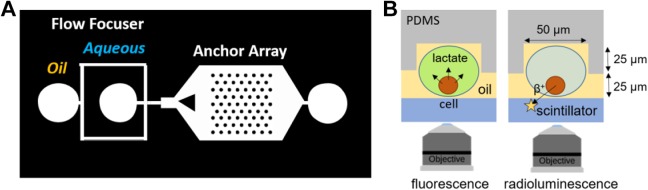
Droplets were formed using a flow focuser,16 and droplets flowed into a 2-mm-wide channel containing an array of 10 × 18 anchors. The channel height was 25 µm. The circular anchors had a diameter of 50 µm and a depth of 25 µm and were spaced 150 µm apart. The aqueous and oil flow rates were controlled to produce droplet of diameter 50 µm, the same size as the anchors. The device was used with 2% (wt/wt) of 008-fluorosurfactant (Ran Biotechnologies, Beverly, Massachusetts) in Novec 7500 (3 M, Maplewood, Minnesota) as the external oil phase.
For this combination of channel depth, anchor depth, and fluids, droplets would remain in the anchors for external oil flows of less than about 100 µL/min. Fluid flow was controlled using computer-controlled syringe pumps (Nemesys; Cetoni, Korbussen, Germany). With this design, the number of anchors occupied by droplets was greater than 160 (90% loading efficiency). Cell concentration was adjusted so that 25% to 30% of droplets contained single cells (Poisson loading statistics).
Radioluminescence Image Acquisition and Quantitation
Radioactive decay of FDG inside cells produces a β particle (positron), which can travel to the scintillator underneath the droplet array and produce a flash of light detectable within an optical microscope.13,17 Radioluminescence images were taken with an inverted bioluminescence microscope (LV200, Olympus, Tokyo, Japan) equipped with a ×20, 0.75 NA objective (Olympus UPLSAPO20X). Radioluminescence images were generated by “optical reconstruction of the β-ionization track” (ORBIT), a method described in detail in a previous publication.17 This method uses an EM-CCD camera (ImageEM C9100-14, Hamamatsu, Hamamatsu City, Japan) operating at maximum gain to acquire images of individual ionization tracks at a high frame rate (50-200 milliseconds integration time). With this exposure time, each frame contained about 10 radioactive counts. The frame was then filtered (Gaussian kernel) to reduce shot noise and segmented using a constant threshold set above the noise floor to identify radioactive decay events. The final ORBIT image was reconstructed by computing the center of mass of the light distribution for each detected track and aggregating these locations.
The final uptake of FDG is quantified as the number of FDG molecules per cell. Here, this number refers to the number of FDG molecules present in the cells at the beginning of the experiment, computed based on the half-life (109 minutes) of the radiotracer and the number of decays measured over the integration time. It should be noted that the number of FDG molecules is not equal to the number of glucose molecules taken up by the cell, but it is related to it via a “lumped constant,” which depends on the kinetic parameters of glucose transport by the cell.18
Fluorescence Imaging and Quantitation of Lactate Release
The rate of lactate release is determined using a fluorescence lactate kit (Enzyfluo, EFFLC-100; BioAssay Systems, Hayward, California) adapted for use with single cells in droplets as described previously.14 Briefly, lactate released from single cells is oxidized to pyruvate via lactate dehydrogenase present in the droplet, while NAD+ is reduced to NADH. In turn, NADH reduces a fluorescent substrate into a fluorescent probe, increasing the fluorescence of the droplet.
To quantify the rate of lactate release, a fluorescence image time series was obtained of the droplet array. Images were acquired every 30 seconds with an excitation wavelength of 460/50 nm and an emission wavelength of 535/40 nm. The time series was started no later than 2.5 minutes from the formation of the first droplets to capture the initial rise in droplet fluorescence.
Fluorescence images were processed and analyzed with Matlab (version R2015b). A dark image was first subtracted from all fluorescence images in the time series. The determination of the location of individual droplets within the array was automated using a Canny edge detector applied to the bright-field image. Cell occupancy in droplets was determined manually from bright-field images. Due to the use of a nonstandard tube lens in the LV200, fluorescence illumination and collection was not uniform across the imaging field, with higher fluorescence observed near the center of the image. A flat-field correction curve was estimated by fitting the fluorescence of an array of identical droplets to a 2-dimensional polynomial of third order. This method corrected variations in fluorescence due to spatial position in the field of view. We verified that, after correction, there was no correlation between the position of the droplet in the array and the amount of lactate measured in the droplet (data not shown). A new correction curve was produced for each day of experiments.
The fluorescence intensity of the droplets was determined from the average fluorescence near the droplet center. This droplet fluorescence intensity was used to determine the lactate release rate according to a method described in detail previously.14 Briefly, the droplet fluorescence is first corrected by subtracting the average fluorescence of empty droplets in the same array. The remaining fluorescence signal is modeled according to a polynomial of the form at 2 + c. The pre-exponential coefficient (a) is then used to determine the single-cell lactate release rate (L’) in femtomoles per minute according to the equation:
where V is the droplet volume, n is the number of cells in the droplet, and k is the slope of the calibration curve. The calibration curve was obtained from a droplet array with similar reagents as the cell experiments but with known lactate concentration. The droplets had a diameter of 50 µm corresponding to a volume of 65 pL. Droplets containing multiple cells were excluded from the analysis. The model assumes a constant release of lactate by the cells and no efflux out of the hermetic droplet.
Cluster Analysis
Single-cell measurements were analyzed using the Ward linkage clustering method. In the Ward minimum variance method, the distance between 2 clusters is the analysis of variance sum of squares between the 2 clusters added up over all the variables. At each generation, the within-cluster sum of squares is minimized over all partitions obtainable by merging 2 clusters from the previous generation. A cubic clustering criterion was employed to determine the optimal number of clusters. Other clustering metrics were used as well. In the end, these different results were summarized by manually drawing straight lines to separate the 2-D data into 4 clusters.
Results
Relationship Between Lactate Transport and FDG Uptake
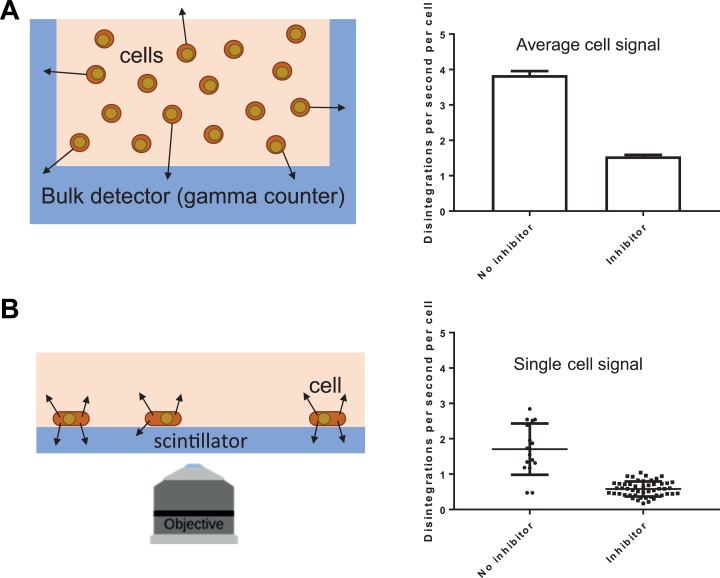
We first demonstrate that radiotracer uptake presents different levels of heterogeneity when quantified through bulk measurements and single-cell RLM measurements (Figure 1). We incubate MDA-MB-231 cells with (and without) the known MCT1 lactate transport inhibitor, αCHC. This inhibitor was found effective in our previous study where lactate release was measured at the single-cell level.14 As seen from Figure 1A, conventional γ counting (left panel) can assay tens of thousands of cells per run to report the average number of atomic disintegrations per second (DPS) per vial, which is proportional to the amount of FDG in the sample. Using this method, the average FDG uptake per cell is 3.84 ± 0.07 DPS/cell without the inhibitor and 1.54 ± 0.02 DPS/cell with the inhibitor, a 2-fold difference.
Figure 1.
Bulk and single-cell measurements of FDG uptake. A, Bulk radionuclide counting of cells using a γ counter (schematic) showing the detection of γ rays (arrows) from a suspension of cells inside the γ counter. The FDG uptake in MDA-MB-231 cells is ≈2 times lower in cells treated with αCHC, a lactate export inhibitor. B, Radionuclide counting of single cells using RLM (schematic). Here, the arrows represent β particles emitted following radioactive decay of FDG. As in the bulk experiment, mean FDG uptake is 2 times lower in cells pretreated with αCHC; in addition, quantification of single-cell FDG uptake shows lower heterogeneity when cells are treated with the inhibitor. αCHC, α-cyano-4-hydroxycinnamic acid; FDG, 18F-fluorodeoxyglucose; RLM, radioluminescence microscopy.
When we use RLM to assay FDG uptake on a single-cell level (Figure 1B), we observe that, while cell measurements congregate around an average FDG concentration, there is large cell-to-cell variability. For cells incubated without the inhibitor, the average FDG uptake per cell is 1.7 DPS/cell. Notably, we find not only a few cells with almost no detectable FDG uptake but also cells that might be considered hypermetabolic, in that they take up a very high amount of FDG. Similar to the bulk experiment, when the αCHC inhibitor is added, FDG uptake drops over 2-fold to 0.59 DPS/cell.
These 2 data sets show that γ counting and RLM are both able to quantify uptake of a radiotracer in live cells. The relative decrease induced by the inhibitor is consistent between both experiments. In addition, RLM can quantify the variance in tracer uptake within the cell population. We computed the standard deviation of the single-cell measurements and found it to be 55% ± 10% of the average uptake value for the control cells and 47% ± 5% for the cells incubated with the inhibitor, suggesting that inhibition of lactate export tends to decrease heterogeneity in FDG uptake.
Figure 1 therefore demonstrates that bulk data do not necessarily represent the behavior of individual cells. It is important to note that the variability observed between multiple γ counting replicates is due to unavoidable experimental variability, not biological heterogeneity. More importantly, the results from the αCHC inhibitor study highlight a strong association between FDG uptake and lactate release. Intuitively, as a product of glycolysis, lactate release is expected to mirror FDG uptake. Here, the results suggest that lactate release may also have a feedback effect on glucose uptake: Forcing lactate to accumulate in the cell (by blocking the efflux transporter) causes FDG uptake to decrease.
This relationship between lactate release and FDG uptake suggests a possible relationship between FDG uptake and lactate release at the single-cell level. To investigate this question, we analyze the metabolic profile of MDA-MB-231 cells using a multiplexed single-cell approach. Specifically, we combine 2 existing assays to measure FDG uptake and lactate release in the same cells. Because the inhibitor decreases metabolic heterogeneity and blocks lactate export, the remaining experiments are performed in unperturbed MDA-MB-231 cells where lactate transport is not blocked.
Multiplexed Detection of FDG and Lactate
Prior to analysis, cells are incubated with FDG for 30 minutes, washed 3 times, trypsinized, washed again, suspended in equal volumes of glucose-free DMEM and lactate assay kit, and finally introduced into the microfluidic device. Figure 2 shows the channel geometry and a cross-sectional view of the device. The microfluidic device is made of PDMS directly bonded to a CdWO4 scintillator substrate. Flow focusers are used to encapsulate single cells in water-in-oil droplets.19 A triangle-shaped obstacle spreads the flowing droplets throughout the entire width of the channel. Because the assay requires the same cells to be monitored for an extended period of time, we use a technique called “Rails and Anchors”20 to trap droplets into a static array. As demonstrated in Figure 2A, the droplets are initially squeezed by the top and bottom of the channel; as they flow into an array of microfabricated well, they are able to expand and reduce their surface energy, and they become anchored to the microwells (Figure 2B). The droplets remain stationary throughout the experiment (approximately 45 minutes for the combined measurement of FDG and lactate), even when oil is flowing. After data acquisition, the flow of oil is increased to eject the droplets from their anchors and flush the chip for subsequent experiments.
Figure 2.
Diagram of the microfluidic device. A, Device mask showing flow focuser for generating water-in-oil droplets and anchor array for imaging them. B, Cross-sectional schematic of the device and single-cell imaging techniques. Radioactive cells are encapsulated in water-in-oil droplets, which are anchored within the PDMS device for sequential analysis, in the same cells, of lactate release (by fluorescence; left) and FDG uptake (by radioluminescence; right). FDG, 18F-fluorodeoxyglucose; PDMS, polydimethylsiloxane.
The multiplexed detection of FDG uptake and lactate release combines 2 distinct techniques, implemented within the same microfluidic device. The FDG uptake is measured using RLM to image radiotracer decay. The radioactive decay releases a positron that traverses the scintillator substrate and produces a flash of light that is detected by a high numerical aperture objective (Figure 2B, left). Lactate release is quantified via an enzymatic fluorescence assay performed inside single-cell droplets (Figure 2B, right). By performing the 2 measurements sequentially, the technique allows us to correlate 2 different facets of cell metabolism, glucose transport and hexokinase activity (as measured by FDG uptake), and lactate secretion.
Figure 3 shows representative images from one of the experiments. Figure 3A is a bright-field image of the droplet array. Droplets are of similar size as the anchors (50 µm). A few cells are indicated by arrows in the image. Figure 3B is a representative fluorescence image obtained at the end of the fluorescence time series, 3 minutes after the formation of the droplets. At this time, droplets containing cells are clearly more fluorescent than unoccupied droplets. Fluorescence brightness increases over time as more lactate is released into the droplet. The rate of fluorescence increase is used to estimate the rate of lactate release. Figure 3C is a reconstructed radioluminescence image showing FDG uptake in individual cells. The intensity of the image is proportional to the number of decay events detected within each image pixel. While fluorescence and radioluminescence are acquired on the same cell, it is important to note that the 2 processes do not interfere with each other. No fluorescent light is emitted during radioluminescence imaging because the illumination source is turned off. Conversely, radioluminescence is not visible during fluorescence imaging because radioluminescence is 3 orders of magnitude weaker than fluorescence. Radioluminescence tracks are only visible when the camera EM gain is set to ×1200, whereas fluorescence imaging does not require EM gain. Figure 3D combines the 2 signals and shows that, for some cells, high FDG appears to correlate with high lactate. However, this correlation breaks down quite often. This behavior is highlighted by 2 cells indicated by arrows: The cell on the left displays both high lactate release (Figure 3B) and FDG uptake (Figure 3C), but the one on the right shows high lactate but only modest FDG uptake.
Figure 3.
Representative images from one experiment. A, Bright-field image of droplets trapped within the anchor array. Two droplets containing single cells are identified by arrows. B, Fluorescent signal (3 minutes after cell encapsulation) due to lactate release from individual cells. C, Radioluminescence microscopy image representing distribution of FDG molecules inside individual cells. D, Overlay showing lactate and FDG from previous 2 images. E, Raw fluorescence time curves for the 2 droplets identified by arrows. Time is measured from initial droplet formation. F, Radioactive event count rate for the 2 droplets identified by arrows (after decay correction). Time is measured from beginning of RLM acquisition. FDG indicates 18F-fluorodeoxyglucose; RLM, radioluminescence microscopy.
The time-dependent signals measured by fluorescence and RLM are illustrated in Figure 3E and F, respectively. Droplet fluorescence increases over time as lactate reacts with the reagents of the lactate detection kit. As this reaction depletes the substrate, droplet fluorescence eventually reaches a plateau. For this reason, lactate release is estimated from early time points, while the detection substrate is in excess. For RLM, the rate of radioactive decay for each cell is constant over time because FDG is trapped in the droplet (Figure 3F; after decay correction).
Multiparametric Analysis
The correlation between the 2 measurements is presented in Figure 4, which plots lactate release versus FDG uptake for 3 consecutive experiments (n = 127 cells in total, measured during 3 consecutive runs). Droplets containing single cells are shown as colored dots. Empty droplets are included as controls (black dots). This multiparametric analysis allows us to analyze the heterogeneity of the cell population according to 2 indicators of metabolism. The average FDG uptake over a 30-minute incubation period is 1500 ± 200 molecules/cell, and the average lactate secretion rate is 13 (2) fmol/min/cell (standard error of the mean computed from 3 experimental replicates). Monodimensional histograms are included along the x- and y-axes to highlight the univariate distribution of FDG uptake and lactate release, respectively.
Figure 4.
Scatter plot showing FDG and lactate release rate for single cells. The 2 curves along the axes are univariate histograms of FDG uptake (top) and lactate release (right) for droplets containing single cells. Empty droplets (controls) are shown as black dots. The 4 divisions represent clusters of cells with distinct metabolic properties. The 3 colors correspond to 3 separate runs of this experiment. FDG indicates 18F-fluorodeoxyglucose.
To characterize the relationship between glucose uptake and lactate release in various subgroups of cells, we clustered the data according to the Ward linkage method. Based on the cubic clustering criterion, we found the heterogeneity of the data to be best represented by 4 clusters. It should be noted that these clusters should not be interpreted as discrete subpopulation of cells; rather, they help us describe the continuum of cell phenotypes. The clusters are also not unique and different sets of clusters could be obtained. Various clustering analyses suggested the clusters represented in Figure 4 by solid lines. The first cluster (25% of the cells) is made up of cells that have both low FDG uptake and low lactate release. The second cluster (55% of the cells) describes cells that have low to intermediate FDG uptake and intermediate lactate release. Finally, the third and fourth clusters (9% and 11% of the cells, respectively) represent cells that have high lactate release or high FDG uptake, but not both simultaneously.
Finally, to evaluate the correlation between FDG uptake and lactate, we compute the Pearson and Spearman correlation coefficients. Both numbers point to weak but statistically significant correlation between FDG uptake and lactate release. Specifically, the Pearson correlation is 0.4 (P < 10−5) and the Spearman correlation is 0.6 (P < 10−5). However, if we exclude the nonmetabolic cells from cluster 1, both correlation coefficients drop to 0.2 (P < .03), which barely meets the threshold for statistical significance. Therefore, for metabolic cells, we find only marginal correlation between FDG uptake and lactate release.
Discussion
The metabolic reprograming of cancer cells is known to increase the uptake of glucose (or its analogue FDG) and the production of lactate.3 A simplified view of tumor metabolism is that cancer cells turn glucose into lactate via aerobic glycolysis; therefore, lactate release should track glucose consumption. Our results paint a more complex picture of cancer metabolism, with significant heterogeneity and little correlation between FDG uptake and lactate release. For instance, we observe cells with high lactate release but low FDG uptake. The fact that FDG uptake is nearly independent of lactate release suggests that cells have great flexibility to use multiple catabolic and anabolic pathways, even when cultured under homogenous conditions.
Within this data set, we find a cluster of cells (25% of the population) that take up little FDG and secrete little lactate. Low lactate release (<5 fmol/min) strongly predicts low FDG uptake (<1000 FDG molecules). Based on this definition, 94% of cells with low lactate release are found to have low FDG uptake. These cells may not be significantly metabolically active, may exist in a state of quiescence, or may use metabolic fuels and pathways unrelated to lactate or glucose. In a typical bulk measurement, this subset of cells would be missed because of their low signal. Small populations of cancerous cells that are dormant can cause tumor recurrence in many different types of cancers.21 What would be considered noise in a large cell population may yet have important clinical implications. We also note that the reverse is false: Cells with low FDG uptake do not follow a specific pattern of lactate release.
In the second cluster, which contains 55% of the cells, we observe cells taking up various amounts of FDG while secreting significant quantities of lactate. Although no correlation is observed in this population, we hypothesize that these cells are cycling and metabolically active and may at least partially utilize aerobic glycolysis. Interestingly, nearly all the cells in this cluster released lactate at a rate of at least 5 fmol/min, but some of the cells in this cluster had no detectable FDG uptake. This suggests that while all metabolically active cells release some lactate, not all take up glucose.
The third cluster contains cells that take up similar amounts of FDG as cluster 2 but secrete even higher amounts of lactate. These cells may rely on glutaminolysis for their energetic and biosynthetic needs and thus may be able to produce lactate without a significant input of glucose.22 They may also rely on stores of glycogen to drive glycolysis.
Finally, in fourth cluster, we see cells that take up FDG avidly but produce only moderate amounts of lactate. It is possible (although not proven by our data) that these hypermetabolic cells take up glucose mainly for biosynthesis. Alternately, pyruvate may be consumed through the citric acid cycle (oxidative phosphorylation) rather than being converted to lactate.
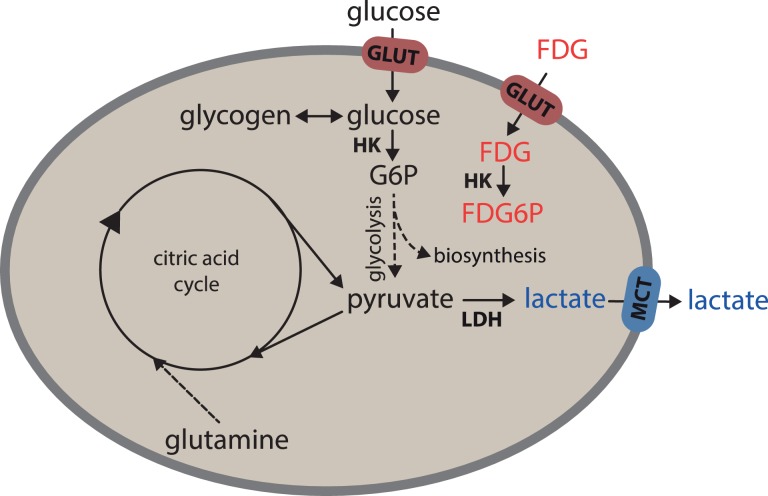
The primary byproduct of aerobic glycolysis is commonly considered to be lactate. If cells are undergoing aerobic glycolysis as their primary mechanism of metabolism, we would expect a high degree of correlation between FDG uptake and lactate production. Our data suggest that aerobic glycolysis may not be the only metabolic pathway employed by the MDA-MB-231 cells. Multiple other pathways are available for a single cell to produce adenosine triphosphate (ATP), including oxidative phosphorylation, glutaminolysis, fatty acid metabolism, and others. Pathways that are related to glucose and lactate are outlined in Figure 5. Furthermore, many studies now highlight the importance of lactate as a metabolic fuel, which has the ability to replace glucose in many cells of the body and which is constantly equilibrating with local lactate concentrations.23 Notably, in one instance, it has been suggested that glucose and glutamine only comprise 40% of the fuels used by cells to generate ATP, and up to 60% of the cell’s energy is derived from additional sources.11 In addition, it is also known that glycolysis intermediates may be diverted to energy storage (glycogen) and for anabolic purposes including lipid synthesis24 and other biosynthesis regulated by pyruvate kinase M2.25 Furthermore, lactic acid release can cause intracellular acidification and inhibit glucose uptake; thus, high lactate production and glucose uptake may not occur simultaneously.26 The complexity and redundancy inherent in metabolic and anabolic pathways makes it impossible to fully characterize individual cell metabolism based on FDG uptake and lactate secretion alone.
Figure 5.
A simplified schematic of the different energetic pathways related to FDG uptake (red) and lactate release (blue). Unlike glucose-6-phosphate, FDG-6-phosphate is not further metabolized. FDG indicates 18F-fluorodeoxyglucose; FDG6P, FDG-6-phosphate; G6P, glucose-6-phosphate; GLUT, glucose transporter; HK, hexokinase; LDH, lactate dehydrogenase; MCT, monocarboxylate transporter.
In addition, it should also be noted that the measurements of FDG uptake and lactate release represent snapshots of dynamic processes. In our experiments, FDG uptake represents the avidity of cells for glucose averaged over a 30-minute-long incubation phase (before droplet encapsulation). Once FDG is taken up by the cell, it remains trapped within the droplet for the remainder of the experiment. Lactate release is measured a few minutes after droplet encapsulation. The 2 measurements are therefore taken approximately 30 minutes apart. Variations between cells could be explained by temporal, unsynchronized fluctuation in glycolysis due to cell cycle and other factors. High-frequency oscillations in the amount of lactate dehydrogenase have been reported in cultured cells.27 This type of oscillation suggests the presence of a dynamic metabolic loop driven by intracellular lactate concentration. It has similarly been demonstrated that cells undergo an oscillating lactate switch that prevents high lactate and high glucose uptake from occurring at the same time.28 Dynamic measurements of FDG uptake and lactate release in the same cells may provide more insight on these processes, but a different technology would have to be used as our droplet technology does not enable repeated measurements of the same cells. Specifically, the position of the cells in the graph of Figure 4 may change over time, which could explain the high cell-to-cell variability. While cell lines such as MDA-MB-231 are typically used as simple models of in vivo processes, these data strikingly demonstrate that, even within a cell line, the metabolic pathways employed by cells are complex. In an in vivo tumor environment, the number of variables is even greater because of subclonal diversity and heterogeneous microenvironmental conditions such as glucose, pH, and oxygen availability. Finally, heterogeneity could be explained by chromosomal instability, which is modest in the MDA-MB-231 cell line,29 and by epigenetic heterogeneity.30
Conclusions
In conclusion, we have shown that, for MDA-MB-231 cells, FDG uptake is only a weak predictor of lactate release when measured on a single-cell level. We obtained this result using a novel droplet-based multiplexed assay of cell metabolism. The combination of RLM and droplet microfluidics allows us to better characterize the multidimensional metabolic profile of live MDA-MB-231 cells. Our data show a lack of correlation between FDG uptake and lactate release, thus highlighting a complex and heterogeneous picture of metabolism even in homogeneous cell lines. Our single-cell data also point to the existence of cancer cell populations that produce lactate in significant quantities but do not take up FDG and thus may not be observed as cancerous in diagnostic techniques such as FDG-PET. This result underscores the need to expand metabolism-based cancer screening methods, not just to rely on the Warburg effect but also to consider alternative metabolic pathways.
Acknowledgments
The authors would like to thank International Electronic Components Inc for their generous donation of dry photoresist films. They also thank Prashanth Asuri of Santa Clara University for providing cell culture facilities for device optimization.
Abbreviations
- αCHC
α-cyano-4-hydroxycinnamic acid
- ATP
adenosine triphosphate
- DMEM
Dulbecco’s modified Eagle medium
- DPS
disintegrations per second
- FDG
18F-fluorodeoxyglucose
- PDMS
polydimethylsiloxane
- PET
positron emission tomography
- ORBIT
optical reconstruction of the β-ionization track
- RLM
radioluminescence microscopy
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Guillem Pratx receives support for this project from the National Institutes of Health under grants 5R01CA186275 and 5R21CA193001, and he is a Damon Runyon-Rachleff Innovator supported (in part) by the Damon Runyon Cancer Research Foundation (DRR-36-15). Paul Abbyad is supported for this project by a National Science Foundation Career Award (grant number 1751861) and the National Institutes of Health under grant 1R15GM129674. Tae Jin Kim is a Stanford Molecular Imaging Scholar under NIH grant T32CA1186810.
ORCID iD: Guillem Pratx, PhD  https://orcid.org/0000-0002-0247-6470
https://orcid.org/0000-0002-0247-6470
References
- 1. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. [DOI] [PubMed] [Google Scholar]
- 2. Doherty JR, Cleveland JL. Targeting lactate metabolism for cancer therapeutics. J Clin invest. 2013;123(9):3685–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4(11):891–899. [DOI] [PubMed] [Google Scholar]
- 4. Zhao Y, Butler EB, Tan M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis. 2013;4:e532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Surasi DS, Bhambhvani P, Baldwin JA, Almodovar SE, O’Malley JP . 18F-FDG PET and PET/CT patient preparation: a review of the literature. J Nucl Med Technol. 2014;42(1):5–13. [DOI] [PubMed] [Google Scholar]
- 6. Sengupta D, Pratx G. Imaging metabolic heterogeneity in cancer. Mol Cancer. 2016;15:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wei L, Hu F, Shen Y, et al. Live-cell imaging of alkyne-tagged small biomolecules by stimulated Raman scattering. Nat Meth. 2014;11(4):410–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim TJ, Turkcan S, Pratx G. Modular low-light microscope for imaging cellular bioluminescence and radioluminescence. Nat Protoc. 2017;12(5):1055–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marín-Hernández A, Rodríguez-Enríquez S, Vital-González PA, et al. Determining and understanding the control of glycolysis in fast-growth tumor cells: flux control by an over-expressed but strongly product-inhibited hexokinase. FEBS J. 2006;273(9):1975–1988. [DOI] [PubMed] [Google Scholar]
- 10. DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. 2016;2(5):e1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guppy M, Leedman P, Zu X, Russell V. Contribution by different fuels and metabolic pathways to the total ATP turnover of proliferating MCF-7 breast cancer cells. Biochem J. 2002;364(pt 1):309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mazurek S, Michel A, Eigenbrodt E. Effect of extracellular AMP on cell proliferation and metabolism of breast cancer cell lines with high and low glycolytic rates. J Biol Chem. 1997;272(8):4941–4952. [DOI] [PubMed] [Google Scholar]
- 13. Tu¨rkcan S, Nguyen J, Vilalta M, et al. Single-cell analysis of [18F] fluorodeoxyglucose uptake by droplet radiofluidics. Anal Chem. 2015;87(13):6667–6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mongersun A, Smeenk I, Pratx G, Asuri P, Abbyad P. Droplet microfluidic platform for the determination of single-cell lactate release. Anal Chem. 2016;88(6):3257–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stephan K, Pittet P, Renaud L, et al. Fast prototyping using a dry film photoresist: microfabrication of soft-lithography masters for microfluidic structures. J Micromech Microeng. 2007;17(10): N69–N74. [Google Scholar]
- 16. Anna SL, Bontoux N, Stone HA. Formation of dispersions using “flow focusing” in microchannels. Appl Phys Lett. 2003;82:364–366. [Google Scholar]
- 17. Pratx G, Chen K, Sun C, et al. High-resolution radioluminescence microscopy of 18F-FDG uptake by reconstructing the β-ionization track. J Nucl Med. 2013;54(10);1841–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reivich M, Kuhl D, Wolf A, et al. The [18F] fluorodeoxyglucose method for the measurement of local cerebral glucose utilization in man. Circ Res. 1979;44(1):127–137. [DOI] [PubMed] [Google Scholar]
- 19. Ignatov SG, Ferguson JA, Walt DR. A fiber-optic lactate sensor based on bacterial cytoplasmic membranes. Biosens Bioelectron. 2001;16(1-2):109–113. [DOI] [PubMed] [Google Scholar]
- 20. Abbyad P, Dangla R, Alexandrou A, Baroud CN. Rails and anchors: guiding and trapping droplet microreactors in two dimensions. Lab Chip. 2011;11(5):813–821. [DOI] [PubMed] [Google Scholar]
- 21. Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23(10):1124–1134. [DOI] [PubMed] [Google Scholar]
- 22. Jin L, Alesi GN, Kang S. Glutaminolysis as a target for cancer therapy. Oncogene. 2016;35(28):3619–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goodwin ML, Gladden LB, Nijsten MW, Jones KB. Lactate and cancer: revisiting the Warburg effect in an era of lactate shuttling. Front Nutr. 2015;1:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lane AN, Tan J, Wang Y, Yan J, Higashi RM, Fan TW. Probing the metabolic phenotype of breast cancer cells by multiple tracer stable isotope resolved metabolomics. Metab Eng. 2017;43(pt B):125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Witney TH, James ML, Shen B, et al. PET imaging of tumor glycolysis downstream of hexokinase through noninvasive measurement of pyruvate kinase M2. Sci Transl Med. 2015;7(310):310ra169. [DOI] [PubMed] [Google Scholar]
- 26. Xie J, Wu H, Dai C, et al. Beyond Warburg effect—dual metabolic nature of cancer cells. Sci Rep. 2014;4:4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ferreira G, Hammond KD, Gilbert DA. Independent high-frequency oscillations in the amounts of individual isozymes of lactate dehydrogenase in Hl60 cells. Cell Biol Int. 1996;20(9):607–611. [DOI] [PubMed] [Google Scholar]
- 28. Mulukutla BC, Yongky A, Grimm S, Daoutidis P, Hu WS. Multiplicity of steady states in glycolysis and shift of metabolic state in cultured mammalian cells. PLoS One. 2015;10(3):e0121561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yoon DS, Wersto RP, Zhou W, et al. Variable levels of chromosomal instability and mitotic spindle checkpoint defects in breast cancer. Am J Pathol. 2002;161(2):391–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501(7467):328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]