Abstract
Intestinal carriage of multi-drug resistant (MDR) Gram-negative bacteria including Pseudomonas aeruginosa (Psae) constitutes a pivotal prerequisite for subsequent fatal endogenous infections in patients at risk. We here addressed whether fecal microbiota transplantation (FMT) could effectively combat MDR-Psae carriage. Therefore, secondary abiotic mice were challenged with MDR-Psae by gavage. One week later, mice were subjected to peroral FMT from either murine or human donors on 3 consecutive days. Irrespective of murine or human origin of fecal transplant, intestinal MDR-Psae loads decreased as early as 24 h after the initial FMT. Remarkably, the murine FMT could lower intestinal MDR-Psae burdens by approximately 4 log orders of magnitude within 1 week. In another intervention study, mice harboring a human gut microbiota were perorally challenged with MDR-Psae and subjected to murine FMT on 3 consecutive days, 1 week later. Strikingly, within 5 days, murine FMT resulted in lower loads and carrier rates of MDR-Psae in mice with a human gut microbiota. In conclusion, FMT might be a promising antibiotics-independent option to combat intestinal MDR-Psae carriage and thus prevent from future endogenous infections of patients at risk.
Keywords: Multi-drug resistant Pseudomonas aeruginosa, fecal microbiota transplantation, antibiotics-independent intervention strategies, human microbiota associated mice, pro-inflammatory immune responses, host-pathogen-interaction, intestinal multi-drug resistant Gram-negative bacterial carriage
Introduction
Pseudomonas aeruginosa (Psae) are responsible for approximately 15% of nosocomial infections affecting up to one third of intensive care unit (ICU) patients [1]. In the United States, Psae causes over 50,000 healthcare-related cases annually posing a serious threat particularly to immunocompromised individuals infected with multidrug-resistant (MDR) strains [2]. Especially patients suffering from cystic fibrosis, post-surgery or burn wounds are at increased risk of infection [3, 4]. Complications of Psae infection include respiratory morbidities including ventilator-associated pneumonia, urinary tract infections, bacteremia, and sepsis with potentially fatal outcome [3, 4]. In addition to exerting a broad variety of metabolic functions enabling the bacteria to utilize even antimicrobial agents as substrate, Psae display a primary (i.e., natural) resistance to multiple antibiotic compounds [1, 5]. In addition, almost one third of all isolates obtained in the European Union exhibited a secondary (i.e., acquired) resistance to at least one bactericidal group out of piperacillin ± tazobactam, fluoroquinolones, ceftazidime, aminoglycosides, and carbapenems, with the highest incidence of resistance found for fluoroquinolones according to the Antimicrobial Resistance Surveillance Report 2017 published by the European Center for Disease Prevention and Control (ECDC) [6]. Various mechanisms of adaption and survival help Psae to acquire novel resistance mechanisms when encountering various classes of antibiotics, which leads to an increasing spectrum of antimicrobial resistance worldwide, especially in isolates recovered from ICU patients [7]. Bacterial evasion strategies comprize a plethora of multi-drug efflux systems, OprD loss in the outer membrane, target mutations such as changes in the penicillin-binding proteins and production of β-lactamases, carbapenemases, and 16S rRNA methylases, for instance [8]. Whereas Psae is not considered as part of the commensal intestinal microbiota, rectal carriage of the opportunistic pathogen was shown to be a predictor of Psae-associated infections in ICU patients [9]. The human gastrointestinal tract (GIT) should therefore be regarded as a potential internal Psae reservoir. Especially in conditions when the gut commensal bacterial ecosystem is disturbed upon antibiotic treatment, the compromized colonization resistance preventing from establishment of (opportunistic) pathogens [10, 11] may facilitate stable Psae colonization within the GIT [12, 13]. However, valid data regarding the immunopathological potential of intestinal Psae colonization in the vertebrate host is scarce. For the first time, our group provided evidence that MDR Psae carriage resulted in pronounced pro-infammatory immune responses in intestinal as well as extra-intestinal including systemic compartments of otherwise healthy wild-type mice [14, 15]. Moreover, pre-existing inflammatory conditions in the gut facilitated stable intestinal Psae colonization in mice which subsequently aggravated the underlying disease [16–19].
Hence, application of antimicrobial compounds for combating Psae infections provides only rather limited therapeutic (“health-benefical”) success, given the collateral damages caused by the antimicrobial compound itself resulting in depletion of the commensal microbiota in other non-infected body compartments and further by the emergence of MDR strains due to extensive antimicrobial resistance selection pressure. Furthermore, we were able to show that broad-spectrum antimicrobial treatment resulted in inhibition of secretory Immunoglobulin A (IgA) production at mucosal interfaces rendering the host susceptible to acquisition of bacterial strains [20]. Therefore, the development of alternative antimicrobial-independent approaches for prevention and therapy of infections is utmost appreciated to help out the above-mentioned and in many cases fatal dilemma.
The reintroduction of a complex microbiota into the host via fecal microbiota transplantation (FMT) is a well-known therapeutical approach dating back to the Chinese Dong-jin dynasty in the fourth century [21] and has undergone a renaissance for the treatment of recurrent and refractory Clostridium difficile toxin-induced acute necrotizing pseudo-membranous enterocolitis for instance [22–26]. In the present study, we therefore addressed whether fecal FMT from murine or human donors could effectively combat MDR Psae carriage.
Material and Methods
Generation of Secondary Abiotic Mice
Female C57BL/6j mice were bred under specific pathogen-free (SPF) conditions at the Forschungseinrichtungen für Experimentelle Medizin (Charité – University Medicine, Berlin, Germany). The murine gut microbiota was depleted as described previously [27]. In brief, 8-weeks-old mice were transferred into sterile cages and subjected to a broad-spectrum antibiotic treatment for 8 to 10 weeks by adding ampicillin plus sulbactam (1 g/L; Ratiopharm, Germany), vancomycin (500 mg/L; Cell Pharm, Germany), ciprofloxacin (200 mg/L; Bayer Vital, Germany), imipenem (250 mg/L; MSD, Germany), and metronidazole (1 g/L; Fresenius, Germany) to the drinking water (ad libitum) resulting in secondary abiotic mice. Three days before infecton, the antibiotic cocktail was replaced by autoclaved tap water to assure antibiotic washout in mice.
P. aeruginosa Infection
Secondary abiotic mice were perorally infected with 109 colony forming units (CFU) of a MDR P. aeruginosa strain by gavage in a total volume of 0.3 mL phosphate buffered saline (PBS; Gibco, life technologies, UK) on two consecutive days (days 0 and 1) as reported earlier [16]. The P. aeruginosa isolate had been initially cultured from respiratory material of a patient suffering from nosocomial pneumonia and was kindly provided by Prof. Dr. Bastian Opitz (Charité – University Medicine, Berlin, Germany). Notably, the bacterial strain displayed antimicrobial sensitivity to fosfomycin and colistin only [16].
Cultural Analysis of P. aeruginosa
In order to quantitatively survey intestinal P. aeruginosa loads, fecal pellets were homogenized in sterile PBS. Serial dilutions were streaked onto Columbia agar supplemented with 5% sheep blood (Oxoid, Germany) and cetrimid agar (Oxoid) and incubated in an aerobic atmosphere at 37 °C for at least 48 h as reported earlier [16].
Fecal Microbiota Transplantation
Seven days following the initial Psae infection, mice were subjected to peroral FMT from either human or murine donors on 3 consecutive days (i.e., days 7, 8, and 9 postinfection [p.i.]) as reported earlier [15]. In brief, fresh fecal samples, free of enteropathogenic bacteria, viruses, and parasites, were collected from 5 individual healthy human volunteers, dissolved in sterile PBS, aliquoted, and stored at –80 °C, as stated elsewhere [10, 14, 28]. Immediately before FMT, individual fecal aliquots were thawed and pooled. In addition, fresh murine fecal samples were collected from 10 age-and-sex-matched SPF control mice, pooled, dissolved in 10 mL sterile PBS, and the supernatant served as murine donor suspension. For FMT intervention studies, Psae-infected mice were subjected to peroral FMT either with 0.3 mL of the human or murine donor suspension by gavage on 3 consecutive days [10, 14, 28, 29]. Immediately before respective FMT, aliquots from each human and murine fecal donor solutions were collected for quantitative molecular analyses of main intestinal bacterial communities, as described elsewhere [10, 27, 30]. Seven days after the first out of three FMT, mice were sacrificed by isofluran inhalation (Abbott, Germany).
Murine Fecal Microbiota Transplantation in Psae-Infected Mice Harboring a Human Gut Microbiota
For murine FMT intervention studies, mice harboring a human gut microbiota were obtained by peroral human FMT of secondary abiotic mice on 3 consecutive days [28]. One week later, the thus-obtained (with respect to their gut microbiota composition) “humanized” mice were subjected to 109 CFU MDR Psae on days 0, 1, and 2 by gavage, followed by peroral murine FMT on days 7, 8, and 9 p.i. and necropsy on day 14 p.i.
Molecular Analysis of Gut Microbiota Composition
DNA was extracted from fecal samples or fecal donor suspensions as described previously [27, 31]. In brief, DNA was quantified by using Quant-iT PicoGreen reagent (Invitrogen, UK) and adjusted to 1 ng per μL. Then, the total eubacterial loads, as well as the main bacterial groups abundant in the murine and human intestinal microbiota including enterobacteria, enterococci, lactobacilli, bifidobacteria, Bacteroides/Prevotella species, Clostridium coccoides group, Clostridium leptum group, and Mouse Intestnal Bacteroides, were assessed by quantitative real-time polymerase chain reaction (qRT-PCR) with species-, genus-, or group-specific 16S rRNA gene primers (Tib MolBiol, Germany) as described previously [10, 30, 32] and numbers of 16S rRNA gene copies per ng DNA of each sample determined.
Statistical Analysis
Medians and levels of significance were determined using Mann–Whitney test (GraphPad Prism v7, USA) as indicated. Two-sided probability (p) values ≤0.05 were considered significant. Experiments were reproduced three times.
Ethical Statement
All animal experiments were conducted according to the European Guidelines for animal welfare (2010/63/EU) with approval of the commission for animal experiments headed by the “Landesamt für Gesundheit und Soziales” (LaGeSo, Berlin; registration numbers G0097/12 and G0039/15). Animal welfare was monitored twice daily by assessment of clinical conditions and weight loss of mice.
Results
Changes in Intestinal Loads of Multi-Drug Resistant P. aeruginosa Following Fecal Microbiota Transplantation in P. aeruginosa-Challenged Mice
Secondary abiotic mice were challenged with 109 CFU Psae on days 0 and 1 by gavage. Immediately before FMT on 3 consecutive days starting on day 7 p.i., the commensal microbiota composition of fecal suspensions derived from either human or murine donors were assessed by culture-independent 16S rRNA-based methods. Human fecal suspensions contained more enterobacteria, enterococci, Bacteroides/Prevotella species, and both, Clostridium coccoides and Clostridium leptum group, as compared to those from murine donors, whereas suspensions from murine donors contained more lactobacilli and Mouse Intestinal Bacteroides compared to those of human donors (Figure 1). Cultural analyses revealed that immediately before the first FMT (i.e., day 7 p.i.), mice from either cohort harbored comparably high median fecal Psae loads of approximately 108 CFU per g (Figures 2 and 3). As early as 24 h following the first FMT, however, fecal Psae loads were approximately one order of magnitude lower as compared to mock-treated mice, irrespective whether the fecal suspension had been generated from human or murine donors (p < 0.001; Figures 2 and 3). Seven days post-FMT from either origin (i.e., on day 14 p.i.), mice harbored even up to 3.5 log orders of magnitude lower Psae numbers in their intestine as compared to mock control animals (p < 0.001; Figures 2 and 3). Of note, individual mice from the FMT cohort had even completely lost the MDR Psae until day 14 p.i. (Figures 2 and 3). When compared to the respective Psae loads assessed immediately before the first FMT, mice challenged with complex microbiota from human and murine donors had lost approximately 2.5 and 4.0 log orders of magnitude of the MDR bacterial strain, respectively (p < 0.001; Figure 3). Hence, FMT (irrespective whether from human or murine fecal donors) could effectively lower the intestinal loads of MDR Psae with most prominent effect exerted by fecal microbiota of murine origin.
Figure 1.
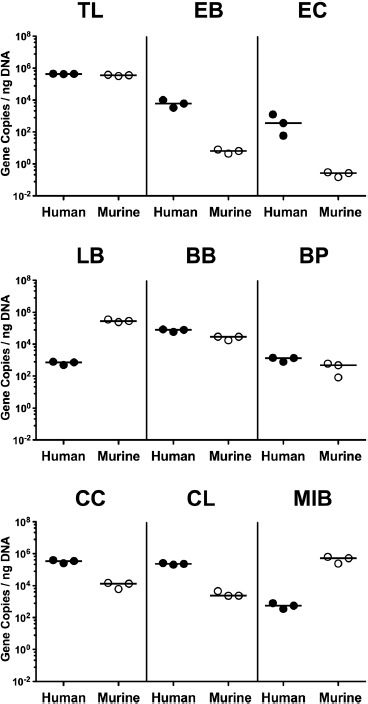
Microbiota composition of human and murine fecal suspensions used for the fecal microbiota transplantation. Immediately before fecal microbiota transplantation, the commensal microbiota composition of fecal suspensions derived from human (closed circles) and murine donors (open circles) was assessed by culture-independent 16S rRNA-based methods. The total eubacterial load (TL) and main bacterial groups including enterobacteria (EB), enterococci (EC), lactobacilli (LB), bifidobacteria (BB), Bacteroides/Prevotella species (BP), Clostridium coccoides group (CC), Clostridium leptum group (CL), and Mouse Intestinal Bacteroides (MIB) were quantitated (expressed as gene copies per ng DNA). Data shown were pooled from 3 independent suspensions from respective origin
Figure 2.
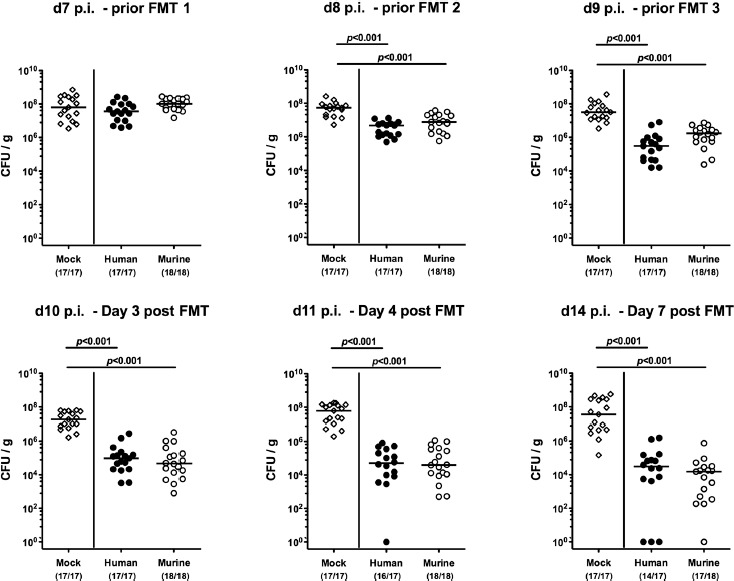
Fecal multi-drug resistant P. aeruginosa loads over time, following human or murine fecal microbiota transplantation in the infected secondary abiotic mice. Secondary abiotic mice were challenged with multi-drug resistant P. aeruginosa on day (d) 0 and d1 by gavage and subjected to peroral fecal microbiota transplantation (FMT) from either human (closed circles) or murine donors (open circles) at d7, d8, and d9 postinfection (p.i.) or to PBS (mock, open diamonds). Immediately before and at defined time points after the FMT, fecal samples were taken to assess intestinal P. aeruginosa loads by culture (expressed as colony forming units per gram, CFU/g). Medians, significance levels (p-values) determined by the Mann–Whitney U test, and the number of P. aeruginosa positive mice out of the total number of animals (in parentheses) are indicated. Data shown were pooled from 4 independent experiments
Figure 3.
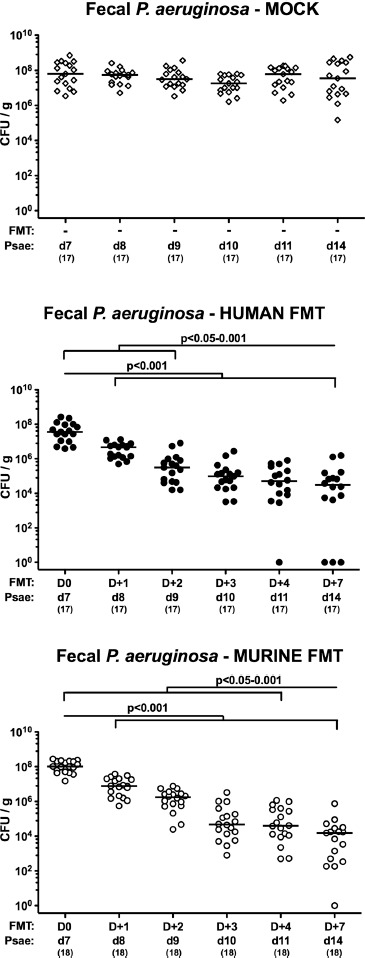
Kinetic survey of intestinal multi-drug resistant P. aeruginosa loads, following human or murine fecal microbiota transplantation in the infected secondary abiotic mice. Secondary abiotic mice were challenged with multi-drug resistant P. aeruginosa (Psae) on day (d) 0 and d1 by gavage and subjected to peroral fecal microbiota transplantation (FMT) from either human (closed circles) or murine donors (open circles) at d7, d8, and d9 postinfection (p.i.; i.e., D0, D+1 and D+2) or to PBS (mock, open diamonds). Immediately before and at defined time points (D) after the FMT, fecal samples were taken to assess intestinal P. aeruginosa loads by culture (expressed as colony forming units per gram, CFU/g). Medians, significance levels (p-values) determined by the Mann–Whitney U test, and the numbers of analyzed mice (in parentheses) are indicated. Data shown were pooled from 4 independent experiments
Changes in Intestinal Microbiota Composition Following Human or Murine FMT in Multi-Drug Resistant P. aeruginosa-Challenged Mice
We further addressed how the gut microbiota composition had changed in mice 7 days following respective FMT. Given that we applied molecular 16S rRNA-based qPCR methodology, bacterial gene copies could also be amplified from Psae-infected secondary abiotic mice (i.e., mock group) at relatively low levels (Figure 4), even though fecal samples were culture-negative for any bacteria except for Psae (not shown). Following either FMT regimen, mice displayed higher fecal loads of lactobacilli (p < 0.05), Bacteroides/Prevotella species (p < 0.01–0.001), and Clostridium leptum group (p < 0.001; Figure 4). Whereas following murine FMT only bifidobacteria and Mouse Intestinal Bacteroides had increased (p < 0.01 and p < 0.001, respectively; Figure 4), this held true for the Clostridium coccoides group following FMT from human donors (p < 0.01, Figure 4). When compared to FMT from human donors, mice that had been subjected to murine FMT harbored more bifidobacteria (p < 0.01) and Mouse Intestinal Bacteroides (p < 0.001), but less Bacteroides/Prevotella species (p < 0.05) and Clostridium coccoides group (p < 0.01) in their feces at day 14 p.i. (Figure 4). Hence, the commensal gut microbiota composition differed considerably following FMT from human and murine donors.
Figure 4.
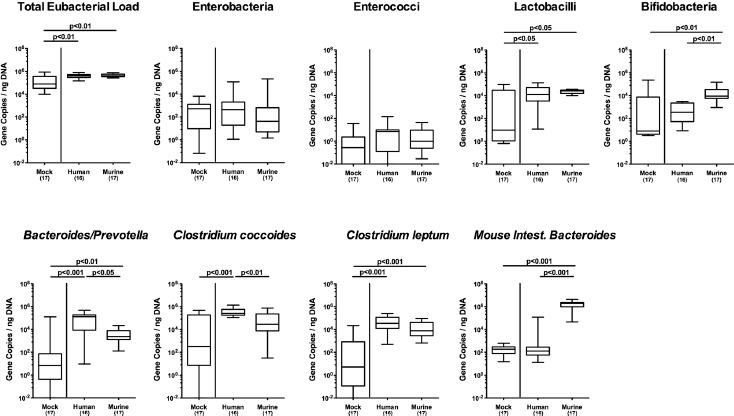
Changes in intestinal microbiota composition, following human or murine fecal microbiota transplantation in the multi-drug resistant P. aeruginosa infected mice. Secondary abiotic mice were challenged with multi-drug resistant P. aeruginosa on day (d) 0 and d1 by gavage and subjected to peroral fecal microbiota transplantation (FMT) from either human or murine donors at d7, d8, and d9 postinfection (p.i.; i.e., D0, D+1, and D+2) or to PBS (mock). Seven days later (i.e., D+7), the fecal commensal microbiota composition was assessed applying culture-independent 16S rRNA methods quantitating the total eubacterial load and main commensal bacterial groups such as enterobacteria, enterococci, lactobacilli, bifidobacteria, Bacteroides/Prevotella species, Clostridium coccoides group, Clostridium leptum group, and Mouse Intestinal Bacteroides (expressed as gene copies per ng DNA). Box plots represent the 75th and 25th percentiles of medians (black bar inside the boxes). The total range, significance levels (p-values) determined by the Mann–Whitney U, and total numbers of analyzed mice are indicated. Data were pooled from 4 independent experiments
Changes in Intestinal Multi-Drug Resistant P. aeruginosa Loads Following Murine Fecal Microbiota Transplantation in P. aeruginosa-Challenged Mice Harboring a Human Gut Microbiota
We next addressed whether murine FMT could sufficiently lower intestinal MDR Psae loads in mice harboring a human gut microbiota. Therefore, secondary abiotic mice were perorally challenged with human feces on 3 consecutive days. One week later, the thus-obtained (with respect to their gut microbiota composition) “humanized” mice were subjected to 109 CFU MDR Psae on days 0, 1, and 2 p.i. by gavage, followed by peroral murine FMT on days 7, 8, and 9 p.i. Immediately before the initial FMT (i.e., day 7 p.i.), the “humanized” mice harbored median fecal Psae loads of 104 CFU per g (Figure 5). As early as 3 days following the latest murine FMT, however, more than half of mice with a human gut microbiota were culture-negative for fecal Psae and harbored median loads of 0 CFU per g in their intestines (Figure 5). Whereas immediately before the initial murine FMT, 75.0% of the humanized mice harbored Psae in their intestines, this was true in only 45.8% of cases on day 14 p.i. (i.e., 7 days following murine FMT start; p < 0.05 vs d7 p.i.; Figure 5). Hence, murine FMT resulted in lower loads and carrier rates of MDR Psae in mice with a human gut microbiota.
Figure 5.
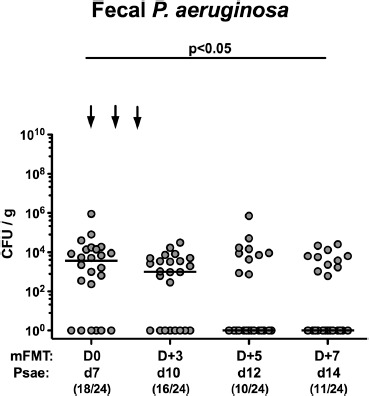
Kinetic survey of intestinal multi-drug resistant P. aeruginosa loads, following murine fecal microbiota transplantation in the infected mice harboring a human gut microbiota. Mice with a human gut microbiota were challenged with multi-drug resistant P. aeruginosa (Psae) on day (d) 0 and d1 by gavage and subjected to peroral murine fecal microbiota transplantation (mFMT) at d7, d8, and d9 postinfection (as indicated by the arrows; i.e., D0, D+1, and D+2). Immediately before and at defined time points after the FMT, fecal samples were taken to assess intestinal P. aeruginosa loads by culture (expressed as colony forming units per gram, CFU/g). Medians, significance levels (p-values) determined by the Mann–Whitney U test, and the number of P. aeruginosa positive mice out of the total number of analyzed animals (in parentheses) are indicated. Data shown were pooled from 4 independent experiments
Changes in Intestinal Microbiota Composition Following Murine FMT in Multi-Drug Resistant P. aeruginosa-Challenged Mice Harboring a Human Gut Microbiota
We next surveyed changes in the gut microbiota composition of Psae-infected “humanized” mice, 7 days following murine FMT. Whereas fecal numbers of enterobacteria (p < 0.005), enterococci (p < 0.05), Bacteroides/Prevotella species (p < 0.001), Clostridium coccoides (p < 0.05), and Clostridium leptum (p < 0.001) groups had decreased upon 3 murine FMT, higher fecal loads of lactobacilli (p < 0.001), bifidobacteria (p < 0.05) and Mouse Intestinal Bacteroides (p < 0.001) could be assessed, 7 days following the initial murine FMT (i.e., day 14 p.i.; Figure 6). Hence, within 1 week following murine FMT, the gut microbiota composition of Psae-infected “humanized” mice had substantially changed.
Figure 6.
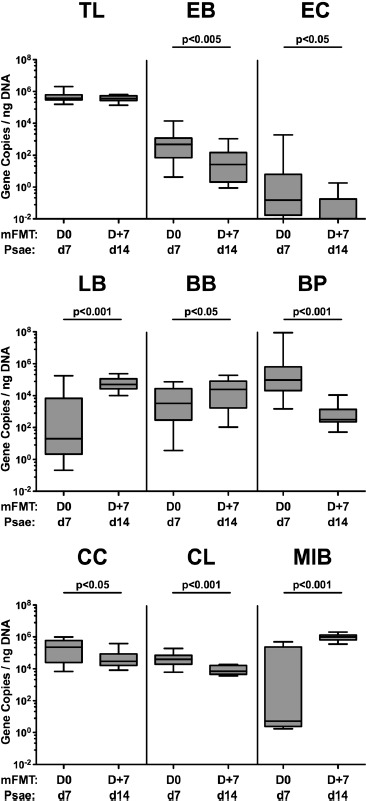
Changes in intestinal microbiota composition, following murine fecal microbiota transplantation in the multi-drug resistant P. aeruginosa infected mice harboring a human gut microbiota. Mice with a human gut microbiota were challenged with multi-drug resistant P. aeruginosa (Psae) on day (d) 0 and d1 by gavage and subjected to peroral murine fecal microbiota transplantation (mFMT) at d7, d8, and d9 postinfection (i.e., D0, D+1, and D+2) or to PBS (mock). Seven days later (i.e., D+7 and d14 p.i.), the fecal commensal microbiota composition was assessed applying culture-independent 16S rRNA methods quantitating the total eubacterial load (TL) and main commensal bacterial groups including enterobacteria (EB), enterococci (EC), lactobacilli (LB), bifidobacteria (BB), Bacteroides/Prevotella species (BP), Clostridium coccoides group (CC), Clostridium leptum group (CL), and Mouse Intestinal Bacteroides (MIB) and expressed as gene copies per ng DNA. Box plots represent the 75th and 25th percentiles of medians (black bar inside the boxes). The total range and significance levels (p-values) determined by the Mann–Whitney U are indicated. Data were pooled from 4 independent experiments (n = 24)
Discussion
In recent years, MDR bacterial strains are progressively emerging as collateral damages of inappropriate usage of broad-spectrum antimicrobial compounds leading to severe infections with high mortality rates due to limited treatment options for the often multi-morbid patients [13, 33]. This prompted the World Health Organization (WHO) to rate MDR Gram-negative bacteria including Psae as a serious global threat to human health further emphasizing the need for novel antibiotics-independent treatment strategies [34]. FMT constitutes an established antibiotics-independent way to treat gastrointestinal morbidities with a complex gut ecosystem consisting of microbes and their metabolites [21, 35]. In our present study we applied fecal samples derived from either murine and human donors for peroral FMT in mice carrying a clinical MDR Psae isolate in 2 different clinical conditions. In the first case, mice with a depleted gut microbiota had been stably colonized with up to 109 viable Psae bacteria per g feces. Remarkably, irrespective whether from human or murine donors, FMT could effectively lower intestinal MDR Psae loads by up to 4 log orders of magnitude with most prominent effects following peroral application of complex murine feces. Our comprehensive molecular survey of the gut microbiota composition post-FMT revealed substantial quantitative differences in distinct gut bacterial groups and species dependent on the host-specific fecal donors. For instance, following murine FMT, mice harbored more bifidobacteria and Mouse Intestinal Bacteroides in their feces as compared to mice upon human FMT, whereas fecal loads of Bacteroides/Prevotella species and Clostridium coccoides group were lower in the former as compared to the latter. These results are well in line with our previous results following human FMT of initially secondary abiotic mice [15].
In the other clinical scenario, we obtained, with respect to their gut microbiota, “humanized” mice that were subsequently perorally challenged with the clinical MDR Psae isolate. Following peroral murine FMT, not only the intestinal Psae loads could be lowered by up to 4 log orders of magnitude, but also the intestinal MDR bacterial carrier rates dropped below 50%. Of note, the commensal gut microbiota composition in “humanized” mice harboring MDR Psae substantially changed within 1 week following murine FMT as indicated by decreases in fecal loads of enterobacteria, enterococci, Bacteroides/Prevotella species, and Clostridia, whereas numbers of lactobacilli, bifidobacteria, and Mouse Intestinal Bacteroides were higher in the feces derived post-FMT as compared to the pre-FMT status. It is tempting to speculate that, particularly, the higher loads of lactobacilli and bifidobacteria might be involved in the MDR-Psae-clearing effect due to more pronounced production of bacteriocins and short chain fatty acids (SCFA), for instance [36, 37].
When considering transferring our results for clinical application (“from bench to bedside”), our obtained results might be promising for future clinical (i.e., therapeutic or prophylactic) application, particularly in immunocompromised and hospitalized subjects, the main patient groups at risk for acquisition of MDR bacterial infections [35, 38]. In support, a few clinical studies have reported successful intestinal decolonization of MDR bacteria including extended spectrum β-lactamase (ESBL)-producing enterobacteria, vancomycin-resistant enterococci (VRE), and methicillin-resistant Staphylococcus aureus (MRSA), following FMT from healthy human donors so far [35, 39–41].
In our previous studies, we could further prove the feasibility and reliability of murine and human FMT in secondary abiotic mice for mimicking host-specific gut microbiota conditions in different infection and inflammation models of the small as well as the large intestinal tract, which may also be used in preclinical intervention studies [10, 11, 15–17, 28, 29, 42–44]. In this context, we are currently investigating FMT as a treatment option for infections by enteropathogens such as Campylobacter jejuni.
In conclusion, FMT might be a promising antibiotics-independent option to combat intestinal MDR-Psae carriage and thus prevent from endogenous infections of patients at risk. In the future, the distinct bacterial strains and metabolites defining the gut luminal milieu, as well as the exact mechanisms that are involved in the observed MDR bacterial lowering effects of FMT need to be identified and unraveled in more detail.
Acknowledgements
We thank Alexandra Bittroff-Leben, Ines Puschendorf, Ulrike Escher, Ulrike Fiebiger, Gernot Reifenberger, and the staff of the animal research facility at Charité – University Medicine Berlin for excellent technical assistance and animal breeding.
Footnotes
Funding Sources
This work was supported from the German Federal Ministries of Education and Research (BMBF) in frame of the zoonoses research consortium PAC-Campylobacter to SB and MMH (IP7 / 01KI1725D) and from the German Research Foundation (DFG) to MMH (SFB TR84 TP A05). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Authors’ Contributions
KM analyzed the data and co-wrote the paper. SB provided advice in experimental design, critically discussed results, and co-edited the paper. MMH designed and performed the experiments, analyzed data, and wrote the paper.
Conflict of Interest
Stefan Bereswill and Markus M. Heimesaat are members of the Editorial Board.
References
- 1.Moradali MF, Ghods S, Rehm BH. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front Cell Infect Microbiol. 2017;7:39 doi: 10.3389/fcimb.2017.00039. PubMed PMID: 28261568; PubMed Central PMCID: PMCPMC5310132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shah PN, Shah KN, Smolen JA, Tagaev JA, Torrealba J, Zhou L, et al. A novel in vitro metric predicts in vivo efficacy of inhaled silver-based antimicrobials in a murine Pseudomonas aeruginosa pneumonia model. Sci Rep. 2018;8:6376 doi: 10.1038/s41598-018-24200-w. PubMed PMID: 29686296; PubMed Central PMCID: PMCPMC5913254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gellatly SL, Hancock RE. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis. 2013;67:159–73. doi: 10.1111/2049-632X.12033. PubMed PMID: 23620179. [DOI] [PubMed] [Google Scholar]
- 4.Driscoll JA, Brody SL, Kollef MH. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs. 2007;67:351–68. PubMed PMID: 17335295. [DOI] [PubMed] [Google Scholar]
- 5.Frimmersdorf E, Horatzek S, Pelnikevich A, Wiehlmann L, Schomburg D. How Pseudomonas aeruginosa adapts to various environments: a metabolomic approach. Environ Microbiol. 2010;12:1734–47. doi: 10.1111/j.1462-2920.2010.02253.x. PubMed PMID: 20553553. [DOI] [PubMed] [Google Scholar]
- 6.European Centre for Disease Prevention and Control. Surveillance of antimicrobial resistance in Europe – Annual report of the European Antimicrobial Resistance Surveillance Network (EARS-Net) 2017. Stockholm: ECDC; 2018. doi 10.2900/230516; ISBN 978-92-9498-279-7 [Google Scholar]
- 7.MacVane SH. Antimicrobial Resistance in the Intensive Care Unit: A Focus on Gram-Negative Bacterial Infections. J Intensive Care Med. 2017;32:25–37. doi: 10.1177/0885066615619895. PubMed PMID: 26772199. [DOI] [PubMed] [Google Scholar]
- 8.Wolter DJ, Lister PD. Mechanisms of beta-lactam resistance among Pseudomonas aeruginosa. Curr Pharm Des. 2013;19:209–22. PubMed PMID: 22894618. [PubMed] [Google Scholar]
- 9.Gomez-Zorrilla S, Camoez M, Tubau F, Canizares R, Periche E, Dominguez MA, et al. Prospective observational study of prior rectal colonization status as a predictor for subsequent development of Pseudomonas aeruginosa clinical infections. Antimicrob Agents Chemother. 2015;59:5213–9. doi: 10.1128/AAC.04636-14. PubMed PMID: 26077248; PubMed Central PMCID: PMCPMC4538513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bereswill S, Fischer A, Plickert R, Haag LM, Otto B, Kuhl AA, et al. Novel murine infection models provide deep insights into the “menage a trois” of Campylobacter jejuni, microbiota and host innate immunity. PloS one. 2011;6:e20953 doi: 10.1371/journal.pone.0020953. PubMed PMID: 21698299; PubMed Central PMCID: PMCPMC3115961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fiebiger U, Bereswill S, Heimesaat MM. Dissecting the Interplay Between Intestinal Microbiota and Host Immunity in Health and Disease: Lessons Learned from Germfree and Gnotobiotic Animal Models. Eur J Microbiol Immunol. 2016;6:253–71. doi: 10.1556/1886.2016.00036. PubMed PMID: 27980855; PubMed Central PMCID: PMCPMC5146645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohara T, Itoh K. Significance of Pseudomonas aeruginosa colonization of the gastrointestinal tract. Intern Med. 2003;42:1072–6. PubMed PMID: 14686744. [DOI] [PubMed] [Google Scholar]
- 13.Oliver A, Mulet X, Lopez-Causape C, Juan C. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat. 2015;21–22:41–59. doi: 10.1016/j.drup.2015.08.002. PubMed PMID: 26304792. [DOI] [PubMed] [Google Scholar]
- 14.von Klitzing E, Bereswill S, Heimesaat MM. Multidrug-Resistant Pseudomonas aeruginosa Induce Systemic Pro-Inflammatory Immune Responses in Colonized Mice. Eur J Microbiol Immunol. 2017;7:200-9. doi: 10.1556/1886.2017.00022. PubMed PMID: 29034109; PubMed Central PMCID: PMCPMC5632747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Klitzing E, Ekmekciu I, Bereswill S, Heimesaat MM. Intestinal and Systemic Immune Responses upon Multi-drug Resistant Pseudomonas aeruginosa Colonization of Mice Harboring a Human Gut Microbiota. Front Microbiol. 2017;8:2590 doi: 10.3389/fmicb.2017.02590. PubMed PMID: 29312263; PubMed Central PMCID: PMCPMC5744425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Klitzing E, Ekmekciu I, Bereswill S, Heimesaat MM. Acute ileitis facilitates infection with multidrug resistant Pseudomonas aeruginosa in human microbiota-associated mice. Gut pathogens. 2017;9:4 doi: 10.1186/s13099-017-0154-4. PubMed PMID: 28115993; PubMed Central PMCID: PMCPMC5241993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Klitzing E, Ekmekciu I, Kuhl AA, Bereswill S, Heimesaat MM. Intestinal, extra-intestinal and systemic sequelae of Toxoplasma gondii induced acute ileitis in mice harboring a human gut microbiota. PloS one. 2017;12: e0176144 doi: 10.1371/journal.pone.0176144. PubMed PMID: 28414794; PubMed Central PMCID: PMCPMC5393883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Klitzing E, Ekmekciu I, Kuhl AA, Bereswill S, Heimesaat MM. Multidrug-resistant Pseudomonas aeruginosa aggravates inflammatory responses in murine chronic colitis. Sci Rep. 2018;8 doi: ARTN 6685/10.1038/s41598-018-25034-2. PubMed PMID: WOS:000431003700011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heimesaat MM, Escher U, Grunau A, Kühl AA, Bereswill S. Multidrug-resistant Pseudomonas aeruginosa accelerate intestinal, extra-intestinal and systemic inflammatory responses in human microbiota-associated mice with subacute ileitis. Front Immunol 2019;10:49 doi: 10.3389/fimmu.2019.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robak OH, Heimesaat MM, Kruglov AA, Prepens S, Ninnemann J, Gutbier B, et al. Antibiotic treatment-induced secondary IgA deficiency enhances susceptibility to Pseudomonas aeruginosa pneumonia. J Clin Invest. 2018;128:3535-45. doi: 10.1172/JCI97065. PubMed PMID: 29771684; PubMed Central PMCID: PMCPMC6063483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang F, Luo W, Shi Y, Fan Z, Ji G. Should we standardize the 1,700-year-old fecal microbiota transplantation? Am J Gastroenterol. 2012;107:1755; author reply p -6. doi: 10.1038/ajg.2012.251. PubMed PMID: 23160295. [DOI] [PubMed] [Google Scholar]
- 22.Niel MBvan, Fauber BP, Cartwright M, Gaines S, Killen JC, Rene O, et al. A reversed sulfonamide series of selective RORc inverse agonists. Bioorg Med Chem. 2014;24:5769-76. doi: 10.1016/j.bmcl.2014.10.037. PubMed PMID: 25453817. [DOI] [PubMed] [Google Scholar]
- 23.Rohlke F, Surawicz CM, Stollman N. Fecal flora reconstitution for recurrent Clostridium difficile infection: results and methodology. J Clin Gastroenterol. 2010;44:567-70. doi: 10.1097/MCG.0b013e3181dadb10. PubMed PMID: 20485184. [DOI] [PubMed] [Google Scholar]
- 24.Brandt LJ, Aroniadis OC, Mellow M, Kanatzar A, Kelly C, Park T, et al. Long-term follow-up of colonoscopic fecal microbiota transplant for recurrent Clostridium difficile infection. Am J Gastroenterol. 2012;107:1079-87. doi: 10.1038/ajg.2012.60. PubMed PMID: 22450732. [DOI] [PubMed] [Google Scholar]
- 25.Fischer M, Kao D, Kelly C, Kuchipudi A, Jafri SM, Blumenkehl M, et al. Fecal Microbiota Transplantation is Safe and Efficacious for Recurrent or Refractory Clostridium difficile Infection in Patients with Inflammatory Bowel Disease. Inflamm Bowel Dis. 2016;22:2402-9. doi: 10.1097/MIB.0000000000000908. PubMed PMID: 27580384. [DOI] [PubMed] [Google Scholar]
- 26.Scaldaferri F, Pecere S, Petito V, Zambrano D, Fiore L, Lopetuso LR, et al. Efficacy and Mechanisms of Action of Fecal Microbiota Transplantation in Ulcerative Colitis: Pitfalls and Promises From a First Meta-Analysis. Transplant Proc. 2016;48:402-7. doi: 10.1016/j.transproceed.2015.12.040. PubMed PMID: 27109966. [DOI] [PubMed] [Google Scholar]
- 27.Heimesaat MM, Bereswill S, Fischer A, Fuchs D, Struck D, Niebergall J, et al. Gram-negative bacteria aggravate murine small intestinal Th1-type immunopathology following oral infection with Toxoplasma gondii. J. Immunol. 2006;177:8785-95. Epub 2006/12/05. doi: 177/12/8785 [pii]. PubMed PMID: 17142781. [DOI] [PubMed] [Google Scholar]
- 28.von Klitzing E, Oz F, Ekmekciu I, Escher U, Bereswill S, Heimesaat MM. Comprehensive Survey of Intestinal Microbiota Changes in Offspring of Human Microbiota-Associated Mice. Eur. J. Immunol. 2017;7:65-75. doi: 10.1556/1886.2017.00002. PubMed PMID: 28386472; PubMed Central PMCID: PMCPMC5372482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ekmekciu I, von Klitzing E, Fiebiger U, Escher U, Neumann C, Bacher P, et al. Immune Responses to Broad-Spectrum Antibiotic Treatment and Fecal Microbiota Transplantation in Mice. Front Immunol. 2017;8:397 doi: 10.3389/fimmu.2017.00397. PubMed PMID: 28469619; PubMed Central PMCID: PMCPMC5395657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heimesaat MM, Nogai A, Bereswill S, Plickert R, Fischer A, Loddenkemper C, et al. MyD88/TLR9 mediated immunopathology and gut microbiota dynamics in a novel murine model of intestinal graft-versus-host disease. Gut. 2010;59:1079-87. doi: 10.1136/gut.2009.197434. PubMed PMID: 20639251. [DOI] [PubMed] [Google Scholar]
- 31.Bereswill S, Kuhl AA, Alutis M, Fischer A, Mohle L, Struck D, et al. The impact of Toll-like-receptor-9 on intestinal microbiota composition and extra-intestinal sequelae in experimental Toxoplasma gondii induced ileitis. Gut pathogens. 2014;6:19 doi: 10.1186/1757-4749-6-19. PubMed PMID: 24932221; PubMed Central PMCID: PMC4057803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rausch S, Held J, Fischer A, Heimesaat MM, Kuhl AA, Bereswill S, et al. Small intestinal nematode infection of mice is associated with increased enterobacterial loads alongside the intestinal tract. PloS one. 2013;8:e74026 doi: 10.1371/journal.pone.0074026. PubMed PMID: 24040152; PubMed Central PMCID: PMC3769368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livermore DM. Has the era of untreatable infections arrived? J Antimicrob Chemother. 2009;64:i29-36. doi: 10.1093/jac/dkp255. PubMed PMID: 19675016. [DOI] [PubMed] [Google Scholar]
- 34.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18:318-27. doi: 10.1016/S1473-3099(17)30753-3. PubMed PMID: 29276051. [DOI] [PubMed] [Google Scholar]
- 35.Manges AR, Steiner TS, Wright AJ. Fecal microbiota transplantation for the intestinal decolonization of extensively antimicrobial-resistant opportunistic pathogens: a review. Infect Dis (Lond). 2016;48:587-92. doi: 10.1080/23744235.2016.1177199. PubMed PMID: 27194400. [DOI] [PubMed] [Google Scholar]
- 36.Corr SC, Li Y, Riedel CU, O’Toole PW, Hill C, Gahan CG. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci U S A. 2007;104:7617-21. doi: 10.1073/pnas.0700440104. PubMed PMID: 17456596; PubMed Central PMCID: PMCPMC1863472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong JM, Souza Rde, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40:235-43. PubMed PMID: 16633129. [DOI] [PubMed] [Google Scholar]
- 38.Markou P, Apidianakis Y. Pathogenesis of intestinal Pseudomonas aeruginosa infection in patients with cancer. Front Cell Infect Microbiol. 2014;3:115 doi: 10.3389/fcimb.2013.00115. PubMed PMID: 24432250; PubMed Central PMCID: PMCPMC3882663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bilinski J, Grzesiowski P, Sorensen N, Madry K, Muszynski J, Robak K, et al. Fecal Microbiota Transplantation in Patients With Blood Disorders Inhibits Gut Colonization With Antibiotic-Resistant Bacteria: Results of a Prospective, Single-Center Study. Clin Infect Dis. 2017;65:364-70. doi: 10.1093/cid/cix252. PubMed PMID: 28369341. [DOI] [PubMed] [Google Scholar]
- 40.Stalenhoef JE, Terveer EM, Knetsch CW, Van’t Hof PJ, Vlasveld IN, Keller JJ, et al. Fecal Microbiota Transfer for Multidrug-Resistant Gram-Negatives: A Clinical Success Combined With Microbiological Failure. Open Forum Infect Dis. 2017;4:ofx047 doi: 10.1093/ofid/ofx047. PubMed PMID: 28470023; PubMed Central PMCID: PMCPMC5407212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh R, Groot PFde, Geerlings SE, Hodiamont CJ, Belzer C, Berge I, et al. Fecal microbiota transplantation against intestinal colonization by extended spectrum beta-lactamase producing Enterobacteriaceae: a proof of principle study. BMC Res Notes. 2018;11:190 doi: 10.1186/s13104-018-3293-x. PubMed PMID: 29566738; PubMed Central PMCID: PMCPMC5863815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Escher U, Giladi E, Dunay IR, Bereswill S, Gozes I, Heimesaat MM. Anti-inflammatory Effects of the Octapeptide NAP in Human Microbiota-Associated Mice Suffering from Subacute Ileitis. Eur J Microbiol Immunol. 2018;8:34-40. doi: 10.1556/1886.2018.00006. PubMed PMID: 29997909; PubMed Central PMCID: PMCPMC6038539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heimesaat MM, Escher U, Grunau A, Fiebiger U, Bereswill S. Peroral Low-Dose Toxoplasma gondii Infection of Human Microbiota-Associated Mice - A Subacute Ileitis Model to Unravel Pathogen-Host Interactions. Eur J Microbiol Immunol. 2018;8:53-61. doi: 10.1556/1886.2018.00005. PubMed PMID: 29997912; PubMed Central PMCID: PMCPMC6038537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ekmekciu I, von Klitzing E, Neumann C, Bacher P, Scheffold A, Bereswill S, et al. Fecal Microbiota Transplantation, Commensal Escherichia coli and Lactobacillus johnsonii Strains Differentially Restore Intestinal and Systemic Adaptive Immune Cell Populations Following Broad-spectrum Antibiotic Treatment. Front Microbiol. 2017;8:2430 doi: 10.3389/fmicb.2017.02430. PubMed PMID: 29321764; PubMed Central PMCID: PMCPMC5732213. [DOI] [PMC free article] [PubMed] [Google Scholar]


