Abstract
Traumatic brain injury (TBI) is a major cause of injury-related death throughout the world and lacks effective treatment. Surviving TBI patients often develop neuropsychiatric symptoms, and the molecular mechanisms underlying the neuronal damage and recovery following TBI are not well understood. Extracellular vesicles (EVs) are membranous nanoparticles that are divided into exosomes (originating in the endosomal/multi-vesicular body [MVB] system) and microvesicles (larger EVs produced through budding of the plasma membrane). Both types of EVs are generated by all cells and are secreted into the extracellular environment, and participate in cell-to-cell communication and protein and RNA delivery. EVs enriched for neuronal origin can be harvested from peripheral blood samples and their contents quantitatively examined as a window to follow potential changes occurring in brain. Recent studies suggest that the levels of exosomal proteins and microRNAs (miRNAs) may represent novel biomarkers to support the clinical diagnosis and potential response to treatment for neurological disorders. In this review, we focus on the biogenesis of EVs, their molecular composition, and recent advances in research of their contents as potential diagnostic tools for TBI.
Keywords: extracellular vesicles (exosomes) and biomarkers, traumatic brain injury (TBI)
Introduction
Traumatic brain injury (TBI) is the leading cause of death and long-term disability in the developed world. Annually, in excess of 10 million people suffer a TBI event worldwide.1,2 Projections reveal that TBI will comprise the third largest portion of the total global disease burden by the year 2020.1 Within the United States, an estimated 1.7 million people per year incur a TBI, and approximately 5.3 million people live with a TBI-induced disability.3,4 By far the majority of sustained TBIs are mild to moderate in nature and account for some 80–95% of incidents, with severe TBI comprising the balance.5 This stratification of injury is based on the widely used Glasgow Coma Scale (GCS) for determining the severity of neurological injury in TBI patients by evaluating three functions: motor responsiveness, verbal performance, and eye opening. A score ranging from 1 to 6 points is assigned to each function, and their sum indicates the TBI grade: mild TBI (GCS 13–15), moderate TBI (GCS 9–12) and severe (GCS ≤8).
With improvements in survival rate following initial injury, TBI can give rise to substantial and lifelong cognitive, physical, and behavioral impairments that necessitate long-term access to health care and disability services.5,6 Particularly vulnerable are the elderly, in which the same insult leads to greater disability and can result in a dramatic rise in the risk for neurodegenerative7,8 and neuropsychiatric disorders.9 TBI symptoms can sporadically resolve within the first year following injury, but 70–90% of patients continue to exhibit protracted and often unending neurocognitive dysfunction. It is now established that TBI represents a time-dependent process, rather than a single event. Emerging evidence reveals that this process may in some individuals lead to early dementia onset,7,8 but predicting which individuals and the sequence of TBI-induced cellular cascades that underpin this has been difficult to determine in the absence of easy to sample, well-validated biomarkers. Finding and evaluating the utility of such biomarkers has become critical as, clinically, TBI is one of the most powerful environmental risk factors for development of Alzheimer's disease (AD). Recent gene expression studies have identified the upregulation of pathways leading to AD and Parkinson's disease that are triggered by mild TBI (mTBI), let alone moderate or severe forms of TBI, in animal models.10–12 In light of the lack of any available therapeutic options,13 it is imperative to understand the mechanisms that underlie head injury and the ensuing neuronal dysfunction and loss, as well as triggered degenerative pathways to support the development of effective therapeutic strategies. These mechanisms are time-dependent, and biomarker technologies to accurately and quantitatively follow them to select subjects that might best respond to a particular treatment or to optimize a treatment are sorely needed.
TBI-associated brain damage can be classified into two key phases. First, an initial primary damage phase occurs at the moment of insult, and includes contusion and laceration, diffuse axonal injury, and intracranial hemorrhage that can result in instantaneous (necrotic) cell death.10,14 This is followed by a protracted second phase that encompassess cascades of biological processes, initiated at the time of injury, which may persist over much longer times consequent to ischemia, neuroinflammation, glutamate toxicity, astrocyte reactivity, and apoptosis.15–18 As this secondary brain injury may be reversible, it is crucial to understand the biological cascades that drive the delayed secondary phase that occurs following TBI.10,13
Routinely used neuroimaging techniques, such as single photon emission computed tomography (CT) scanning and magnetic resonance imaging (MRI), are available to assess TBI, but, in broad terms, have not proved effective in the diagnosis of brain injury in relation to the true intensity, cell death, blood–brain barrier breakdown, neuroinflammation, and excitotoxicity that ensues. MRI scans are relatively costly and cannot be performed in a repeated manner over short intervals owing to their potential adverse effects in humans. Single-photon emission CT scanning has proved useful in the diagnosis of regional blood flow abnormalities, but although relatively cost-effective to apply has not been of utility in detecting TBI-associated structural damage.
Protein biomarkers potentially available in biological fluids, such as blood, plasma, and serum as well as cerebrospinal fluid (CSF; if available), can be sampled frequently and inexpensively, and offer the promise of providing information about TBI severity. How such damage time-dependently progresses, and whether predictive signatures can be found in relation to the development and progression of TBI-induced disorders, or, more positively, to gain treatment-induced improvements and drug target information would be hugely useful. Clearly, biomarkers that are linked to the extent of injury, as well as to molecular pathways associated with either degenerative or regenerative processes would be of great benefit to patients and clinicians to aid appropriate decision making regarding treatment options—particularly, if these were early biomarkers that showed substantial and predictable changes.
The present review focuses on identifying and monitoring potential protein biomarkers in biological fluids, such as blood, plasma, and serum, with the promise to aid in understanding and better treating TBI.
Biomarkers of mTBI Diagnosis
Diagnostic markers are indicative of disease status, progression, severity, and potential therapeutic interventions.19 Although several biomarkers exist for TBI diagnosis, there is no single clinical diagnostic marker to estimate injury severity.20 Whereas the GCS rating system is often used to assess the severity of TBI,21 it is unable to provide insight into the origin of TBI (i.e., by alcohol, drug use, or polytrauma).22
Bodily fluids including blood, CSF, and urine as reservoirs for diagnostic markers may offer the potential to evaluate neurological deficits in a specific, time-dependent, and sensitive manner. Markers in bodily fluids hold the advantage of being an objective and quantitative measure of biological changes.23 Currently, multiple available markers are under investigation to better understand the biological mechanisms via which mTBI occurs, as well as to assess the severity of the disease.
In this regard, numerous potential markers have been evaluated for their ability to diagnose mTBI, including some with moderate success (Table 1). S100B, a low affinity calcium binding marker expressed in Schwann and glial cells within 6 h of mTBI, is one such candidate.24–27 S100B expression is found to be elevated in the serum of TBI patients; however, the fact that other pathologies that often accompany TBI also drive the expression of S100B, calls into question the protein's specificity as a marker.28–32 Recent studies indicate S100B and glial fibrillary acidic protein (GFAP) levels are increased in TBI patients, and could potentially differentiate mTBI patients from those with severe TBI.33,34 Indeed, GFAP as a potentially promising marker holds the advantage of specific expression in nervous tissue, and can be correlated with CT or MRI images.35 The disadvantages of utilizing GFAP expression include the inability to predict its levels 6 months post-TBI, as well as that GFAP cannot be used to assess the pathophysiology or mechanism underpinning a TBI due to its inability to identify axonal and glial damage.
Table 1.
TBI Biomarkers
| TBI biomarker | Function | References |
|---|---|---|
| GFAP | Serum glial fibrillary acidic protein breakdown products in mild and moderate TBI are associated with intracranial lesions and neurosurgical intervention. | 141, 142 |
| S100β | S100β is a 21 kDa calcium-binding protein (Ca2+), found mainly in the cytosol of astroglial cells and Schwann cells. S100B may be a reliable marker of brain damage in TBI without multiple trauma at 24 h. | 143–145 |
| Myelin-basic protein (MBP) | TBI-induced axonal injury could cause damage to the myelin structure, resulting in secondary myelin sheath instability and demyelination, which may increase the vulnerability of the axons. | 146 |
| NSE | Enolase is a glycolytic enzyme and NSE is one of its five isoenzymes. NSE levels are particularly elevated in severe TBI. | 36, 142 |
| NFH (neurofilament heavy chain) | NFH is an axonal injury marker. Serum NFH levels are significantly elevated in diffuse axonal injury (DAI). | 37, 147 |
| Tau | Microtubule-associated structural protein in axons. Shearing of axons leads to the disruption of Tau binding to tubulin, and subsequent Tau hyperphosphorylation can lead to the formation of Tau oligomers. These can cause self-propagating Tau pathology under specific conditions. | 148 |
| Tumor necrosis factor-alpha (TNF-α) | Pro-inflammatory cytokine that plays an essential role in the immune response observed in patients post-TBI. When TNFα levels are unregulated and chronically elevated, they can drive neurodegenerative cascades. | 149 |
| Interleukin-6 | Pro-inflammatory cytokine that has been implicated in TBI patients and, when elevated levels are not regulated, then can drive neurodegenerative processes. | 150 |
| Ubiquitin C-terminal hydrolase-L1 (UCH-L1) | The addition or removal of ubiquitin from proteins that are destined for metabolism; this is a key part in the removal of excessive, oxidized or misfolded proteins during both normal and pathological conditions in neurons. It is abundant in neurons and has been thought of as a possible biomarker for TBI. It has previously been associated with severe TBI, an increased mortality rate at 6 weeks, and a poor 6-month GOS score. | 151–153 |
| Alpha II-spectrin (SBDPs) | Principally found in neurons and is in plentiful supply in axons and pre-synaptic terminals. The protein is processed into breakdown products (SBDPs) - specifically, SBDP150 (150 kDa size) and SBDP145 (145 kDa) by calpain, and into SBDP120 (120 kDa) by caspase-3. SBDPs are found across cellular and animal models of TBI, and levels are elevated in the CSF of humans particularly after severe TBI. SBDP150, −145, and −120 show a different temporal pattern after TBI, particularly in differentiating survivors from non-survivors. | 50, 154 |
| Estradiol and testosterone | Female TBI patients, in general, recover better than male TBI patients – postulating that differential levels of progesterone and other sex hormones may act as neuroprotective agents. However, increased oestradiol and testosterane levels over time have been associated with increased mortality and worse global outcome for both men and women. | 155, 156 |
CSF, cerebrospinal fluid; GFAP, glial fibrillary acidic protein; GOS, Glasgow Outcome Scale; NSE, neuron-specific enolase; TBI, traumatic brain injury.
Currently available markers include neuron-specific enolase (NSE), myelin basic protein (MBP), and hyperphosphorylated neurofilament heavy chain (NFH). MBP has been found to be a more specific TBI marker than NSE36 primarily because NSE can also be elevated in CSF due to lysis of erythrocytes. NFH is found to be expressed in TBI patients at 2 to 4 days post-injury, but its expression levels drastically alter.37
Tau's phosphorylated form is a well-established CSF biomarker for AD patients38; however, phosphorylated tau (P-tau) is also reported in CSF of patients suffering from chronic neurodegenerative disorders, such as Pick disease and progressive supranuclear palsy.39–41 Tau is a central nervous system (CNS)-specific protein, and a reliable marker found to be elevated 24 h following hypoxic brain injury, with delayed elevations observed up to 48 h. Ultra-sensitive immunoassays are currently available, and have the potential to measure less than 10 pg of P-tau protein, potentially allowing correlations with outcome measures.42 Additional research on P-tau protein levels, and in particular specific types of the protein (e.g., the cis vs. trans form43,44), are warranted to evaluate its potential as a reliable marker for TBI.
Spectrin is a non-erythrocytic protein found in neurons, axons, and presynaptic terminals.45 Calpain and caspase-3 are expressed in TBI and during neuronal apoptosis, and promote the breakdown of αII spectrin.45–47 Spectrin's breakdown products are increased in severe TBI, and can potentially be correlated with patient outcome.48–50
Ubiquitin carboxy-terminal hydrolase isoenzyme L1 (UCH-L1), a deubiquinatase that is highly expressed in neurons and a tissue distribution that is almost exclusively restricted to brain, is another potential biomarker for TBI.51 Research indicates UCH-L1, in tandem with spectrin levels could provide prognostic information in TBI.49,50 S100B and GFAP levels increase in a manner similar to UCH-L1 and spectrin breakdown, and similarly could be used to assess the severity of TBI using peripheral blood.48,50 UCH-L1 was detected in serum within 1 h of injury, and was found to be correlated with GCS score and CT-assessed lesions in a 96-patient TBI study.52
In synopsis, recent investigations have identified sets of interesting potential biomarkers that can be linked with and provide insight into pathobiological processes instigated by TBI (e.g., caspase-3 and calpain-mediated spectrin breakdown,48 and/or brain structural elements (as epitomized by UCH-L1 and GFAP), that are symbolic of trauma-induced brain injury. The evaluation of and combined use of such markers, when related to a comprehensive physical examination may be useful in determining the full extent of injury and possible outcome scenarios.
Extracellular Vesicles
Extracellular vesicles (EVs) are membranous nanoparticles that are found in all biological fluids investigated to date including amniotic fluid, blood, urine, saliva, breast milk, CSF, and ascetic fluid. EVs are divided into exosomes (smaller EVs in the range of 30–150 nm originating in the endosomal/multi-vesicular body [MVB] system) and microvesicles (larger EVs in the range of 100–300 nm that are produced through budding of the plasma membrane). Both types of EVs are secreted by a variety of cell types including lymphocytes, mast cells, platelets, endothelial cells, neurons, and dendritic cells via direct release from the plasma membrane.53 Crucial discoveries regarding exosome structure were made in 1992. The finding that exosomes contain the transferrin receptor, allowing them to be segregated from other membrane proteins and externalized, shed light on the role that exosomes play in intercellular signaling.
EVs are lined by a lipid bilayer, and contain numerous types of proteins and lipids. (For specifics, the website www.microvesicles.org provides a detailed catalog of proteins, RNAs, and lipids associated with EVs.) The additional website Vesiclepedia (formerly ExoCarta; http://microvesicles.org/index.html) lists 92,897 proteins, 27,642 messenger RNAs (mRNAs), 4934 microRNAs (miRNAs), and 584 lipids associated with EVs. The protein and lipid profiles of EV reveal a great deal about their functional role. Notable proteins in EVs include tetraspanins (CD9, CD63, CD81, and CD82), membrane transport and fusion proteins (GTPases, annexins, and flotillin), and heat shock proteins (Hsc70 and Hsp90).54 Proteins of the Rab family, annexins, and heat shock proteins play a key role in intracellular assembly and trafficking of EVs. The tetraspanins that are frequently found in exosomes mediate cell migration, fusion, cell–cell adhesion, and signaling. Integrins, also found abundantly in exosomes, regulate the adherence of vesicles to their target cells.53 Likewise, many further proteins are associated with EV formation and aid in cargo for cell–cell communication, and when widely expressed in EVs have the potential to be used as markers.
Lipids such as phosphatidylcholine, phosphatidylethanolamine, sphingomyelin, phosphatidylserine, phosphatidylinositol, and monosialotetrahexosylganglioside (GM3), are abundantly present in the EV lipid bilayer.55,56 Phosphatidylserine, in particular, is involved in the signaling and fusion of EVs to the plasma membrane, and docking the proteins expressed on the EV membrane by acting through different phospholipid transportation enzymes.57 GM3 and sphingomyelin are reported to be involved in the rigidity of EVs,58 and ceramide, cholesterol, and phosphoglycerides, along with saturated fatty-acid chains appear also to be present in EVs.
Biological Functions of Extracellular Vesicles
EVs are secreted by cells and via biological fluids that include the blood, CSF, synovial fluid, urine, and amniotic fluid, act as key role players in implementing intercellular communication and initiating physiological responses. They express major histocompatibility complex (MHC) class I and II molecules on their cell surface, which are secreted from antigen-presenting cells and aid in triggering specific immune responses by activating CD8+ and CD4+ T cells.59–62 EVs also carry nucleic acids, mRNA, and miRNA that can potentially be transferred to recipient cells. As an example, in the case of EVs deriving from glioblastoma cells, their mRNA and miRNA have the potential to trigger angiogenesis.63
EVs have been described as impacting a broad array of biological processes that include cellular waste removal, the maturation of erythrocytes,64 inflammation,65 coagulation,66 angiogenesis,63,67 and immune responses.68 They play a key role in cell to cell communication in a target-specific manner, and are important in the promotion of thrombosis, tumor proliferation, and angiogenesis through transfer of mRNA and KRAS protein.69
In addition to potential beneficial and physiological roles, EVs are likely involved in the spread of disease within organ systems. This is exemplified by the role EVs play in Parkinson's disease pathogenesis through the transport of misfolded proteins.70 Likewise, potential roles of EVs in other disorders such as cardiovascular disease and infectious diseases such as human immunodeficiency virus (HIV) have been reported.
Biogenesis of Extracellular Vesicles
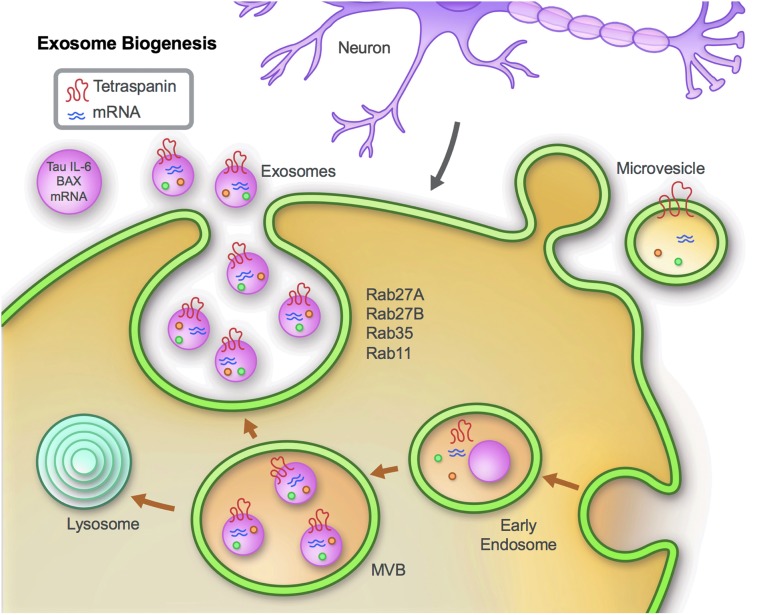
EVs display a round or cup-shaped morphology, and are released from a diverse group of both non-neuronal cells, exemplified by monocytes and lymphocytes,71,72 and neuronal cells including microglia,73 neurons,74 and astrocytes.75 EVs are generated primarily through one of two routes: via endocytic vesicles from the plasma membrane that form early endosomes and subsequently MVBs by inward budding of endosomal vesicles, which contain intraluminal vesicles that are released as exosomes by the fusion of MVBs with plasma membranes; and by budding of the plasma membrane (Fig. 1). Studies have indicated the involvement of SNARE proteins and Rab GTPases such as Rab27, 35, 11 in the regulation of EV secretion. These proteins aid in the process of tethering, docking, and the fusion of EVs at the plasma membrane.76,77 Although mechanisms such as endocytosis, receptor ligand binding, and fusion of EVs with plasma membranes have been proposed as mechanisms by which target cells receive EVs, the definitive mechanisms of interaction and secretion have yet to be fully elucidated.53,78,79 Recent research evidence does, however, support the concept that the internal cargo of EVs contributes to the health and pathology of cells within the CNS.80
FIG. 1.
EV (exosome) biogenesis. EVs are generated in late endosomes or multi-vesicular bodies (MVB). The components of the endosomal sorting complex required for transport (ESCRT) are involved in MVB and EV biogenesis, and aid in cargo loading and vesicle release.157 RabGTPases, such as Rab27, 35, 11 and the SNARE proteins are involved in tethering and fusion to the plasma membrane.76,77,158,159 The cargo of EVs contains proteins and RNAs reflective of ongoing intracellular processes, and thereby provide a window to cellular events.
Extracellular Vesicle Isolation
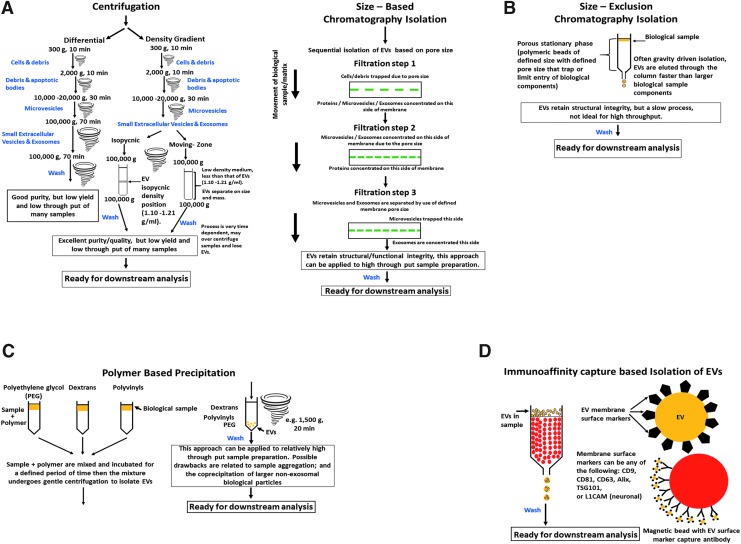
Multiple methods have been developed to support the isolation of EVs from biological fluids by exploiting their physical properties in relation to other consituents within the same sample. Such methods include centrifugation, chromatography-filtration, polymer-based precipitation, and immunological separation techniques to permit relatively consistent EV isolation (Fig. 2). These methods have assorted advantages and disadvantages of relevance to pre-clinical and clinical research. Notably, they can be combined in various manners to enrich EV populations deriving from select cell types.
FIG. 2.
Commonly used methods to isolate exosomes (EVs) from biological samples. (A) The stepwise process of centrifugation-based EV isolation is shown. Centrifugation can utilize differential or density gradient-based methods. (B) Size-based chromatography-filtration isolates EVs based on the size differences of the different biomolecules present in the samples. (C) Polymer-based precipitation of EVs is accomplished by exploiting protein–protein interactions of the EVs and the polymers. (D) Immunoaffinity capture-based isolation of EVs takes advantage of the presence of EV surface marker proteins that are selective for global EVs or even cell type-specifc surface markers, such as L1CAM (a neuronal cell-type marker).
Centrifugation-based isolation methods
Differential centrifugation is a common method used to isolate EVs from biological fluids and media. The basis of differential centrifugation is the application of successive centrifugation steps of increasing centrifugal force and duration to sequentially isolate smaller from larger objects. The larger particles sediment faster and, by leaving the majority of smaller ones in the supernatant, can be progressively removed in the initial centrifugation steps (Fig. 2A). Subsequent successive rounds of centrifugation ultimately provide the particulate of interest. For most common EV isolation protocols, there are four consecutive centrifugation steps: low speed (10 min at 300g) is initially used to sediment cells and debris, (10 min at 2000g) to remove further debris and apoptotic bodies, (30 min at 10,000 to 20,000g) to isolate microvesicles (generally with diameters in excess of 100–150 nm), followed by pelleting of smaller EVs with predominance of exosomes (100,000g for 70 min). Thereafter, washing and a repeat 100,000g centrifugation step of the re-suspended pellet is commonly applied to further purify the EV sample from any free proteins.81
Essentially, the method provides a reasonable purity of EVs but at a moderately low yield. The primary challenge for this purification technique is the separation of EVs from smaller microvesicles consequent to their close size similarity. Additionally, the method is not particularly efficient for handling large numbers of biological fluids, such as would be expected with time-dependent serum or plasma samples deriving from a clinical trial for EV analysis of potential biomarkers.
Density gradient centrifugation, a variation of ultracentrifugation, described above, is the use of a density gradient during the centrifugation process.82 There are essentially two types, isopycnic and moving-zone (Fig. 2A). The use of the former, density gradient ultracentrifugation, is being increasingly applied to the isolation of EVs where separation is achieved based on their size, mass, and density within a selected density gradient medium of increasingly higher density from top to bottom. The biological sample is added as a narrow band onto the top of the density gradient, and subjected to ultracentrifugation to allow centrifugal force to move the assorted components within the sample, including EVs, as different discrete zones depending on their specific sedimentation rate. These zones can then be collected and cleaned by re-suspending them in physiological buffered saline (PBS) and ultracentrifuged (100,000g) to provide EVs for further analysis. In the isopycnic process, a density gradient medium is selected to cover the entire range of densities of sample constituents, and thus zone separation depends on density differences between EVs versus other components, with each sedimenting along the density gradient that matches its own, the isopycnic position—which, in general, lies between 1.10 and 1.21 g/mL for EVs.83
In moving-zone ultracentrifugation, the biological sample is added as a thin layer to the top of a prepared gradient density medium of lower density than any of the sample's components. EVs hence gradually separate from other elements based on their different size and mass, rather than their density difference (as in isopycnic ultracentrifugation), and thus the moving-zone process permits the separation of EVs with similar densities but diverse sizes. When centrifugal force is applied, the separation process is dynamic for the moving-zone procedure as, over sufficient time, all components will eventally pellet together at the bottom of the centrifuge tube, and hence time and the application of centrifugal force need to be optimized to maximize EV separation. This contrasts with the isopycnic procedure, in which static zones are ultimately achieved at the isopycnic position of the alike density components.
In synopsis, when applied optimally, density gradient ultracentrifugation procedures can be advantageouly applied to differential ultracentrifugation to improve the quantity and purity of the EVs harvested, particularly when there is an expected heterogeneity in the EV population and an overlap in size in relation to other elements within the biological sample.84 A disadvantage is that such techniques are relatively labor-intense and cannot readily be applied to large numbers of samples.
Size-based chromatography and size-exclusion chromatography isolation
Size-based EV isolation techniques are widely used for blood and urine samples,85,86 particularly the application of ultrafiltration. In this, EVs can be separated by sequentially passing them through a series of membrane filters of specific molecular weight or size exclusion limits,84,87 (Fig. 2B). In several procedures, a low force is applied to speed up the EV isolation process, which has to be selected to avoid breaking or deforming larger vesicles as this may detrimentally impact later analyses.88 Several commercial EV isolation kits have been developed for plasma, serum, urine, and CSF samples.84 Such consecutive filtration permits EV isolation with relatively high purity and functional integrity. The combination of this technique with ultracentrifugation procedures has been employed to isolate therapeutic EVs for clinical studies, and appears to be scalable to handle large sample numbers.89,90
An additional size-based separation technique utilized to harvest EVs is size-exclusion chromatography (SEC). In SEC, a porous stationary phase (often polymeric beads) within a column is applied to isolate components according to their size (Fig. 2B). Components with a smaller hydrodynamic radius are capable of passing into and through the bead pores, and thus take a longer time to elute. In contrast, those with a greater hydrodynamic radius (i.e., EVs) are unable to transfuse through as many pores, and hence elute from the column more quickly, permitting their isolation. As this is often achieved using gravity flow, EV structure and integrity are retained, albeit long run times ensue that impact the scalability of SEC techniques.88,91 For both forms of isolation the eluted EV samples are washed and prepared for downstream analysis or biological assays.
Polymer-based precipitation
The basis of polymer-based precipitation is the use of polymers, such as polyethylene glycol (PEG), dextrans, or polyvinyls that, when added to a biological sample, attract water molecules away from the solvation layer around proteins. This increases protein–protein interactions and enhances precipitation92 (Fig. 2C). Taking advantage of this phenomena, several commercial isolations kits are available that use polymers to permit EVs to be pelleted under a low centrifugal force, producing a relatively high yield of useable EVs both quickly and in a scalable manner to support evaluation of large sample numbers. Available commercial kits include ExoQuick, ExoSpin, and the Invitrogen Total Exosome Purification Kit, or can be potentially replaced by use of the ExtraPEG method (8% PEG + wash).93,94 In synopsis, a small volume of precipitant is added to a biological sample, which is then incubated (for between 30 min and 12 h, 4°C, depending on the manufacturer). Thereafter, the EV fraction is then pelleted by low-speed centrifugation (1500 × g, 20 min, 4°C). This pellet is then washed and re-suspended in distilled water with a protease inhibitor cocktail and phosphatase inhibitor cocktail. Potential disadvantages of such techniques, are that polymer-based precipitation solutions can result in aggregates, the co-precipitation of larger non-exosomal elements, and precipitants deriving from the kits.95 Recent studies have demonstrated the utility of this EV isolation technique, particularly in relation to clinical sample analyses.96–99
Immunoaffinity capture-based isolation
The presence of numerous proteins and receptors within the membrane of EVs provides the opportunity to isolate EVs based on immunoaffinity capture-based techniques focused on interactions between these exposed surface proteins (antigens) and their specific antibodies (Fig. 2D). Clearly, such antigens should ideally be expressed in a highly enriched and abundant manner on EVs, versus other components within the same biological samples, be membrane-bound and lack soluble counterparts.84 Magnetic bead technology, together with other techniques, have been applied to further increase EV purity. Notably, such techniques can be advantageously combined with other isolation procedures to enrich for EVs deriving from select tissues, such as neurons, astrocytes, and/or capillary endothelial cells.82,98,100 After elution of EVs the samples are washed and prepared for downstream analysis of biological assays.
Extracellular Vesicles Characterization
EVs are classified as exosomes, microvesicles, and apoptotic bodies based on their size or origin.101,102 Exosomes are small in size and derived from the endosomal system103–106; they bear surface markers such as tetraspanins (CD9, CD81, and CD63), Alix, or TSG101107 (Fig. 1). Microvesicles (also previously named as ectosomes) are generated by evagination of plasma membranes into the extracellular space and carry cytoplasmic contents (Fig. 1). Microvesicles do not appear to arise from the endosomal pathway. The size and number of EVs are normally determined by nanoparticle tracking analysis (NTA) or tunable resistive pulse sensing (TRPS).108 Exosomes and microvesicles can be distinguished to some extent based on size, although significant overlap occurs, but cannot be easily distinguished based on protein markers on the vesicle membrane. Tetraspanins, which are commonly used to define EVs, are enriched in both exosomes and microvesicles.109,110 Apoptotic bodies are diverse in size ranging from 50 nm to 5000 nm and are released from dying cells undergoing programmed cell death. More accurate clues are required to distinguish all these EVs; even these are classified based on size and origin.
Extracellular Vesicles Involved in Neurological Diseases
EVs are secreted by cells throughout the CNS, including astrocytes, neurons, microglia, and oligodendrocytes, and their involvement in regeneration, the modulation of synaptic function, and neuronal development has been described.111–113 Similar to other organ systems, CNS EVs appear to be key players in intercellular communication and the initiation of physiological responses regulating the immune response, in eliminating cellular waste, and in communication between neural cells.65,114,115 Notable among recent studies, is that EVs appear to be involved in promoting communication between glia and neurons.116 For example, EVs from human CSF or derived from N2a cells can mitigate the synaptic plasticity disruption caused by both synthetic and AD brain-derived Aβ.117 In vitro and in vivo studies have reported that microglia EVs can promote the spread of tau protein, and inhibiting EV synthesis appears to reduce such tau propagation.113
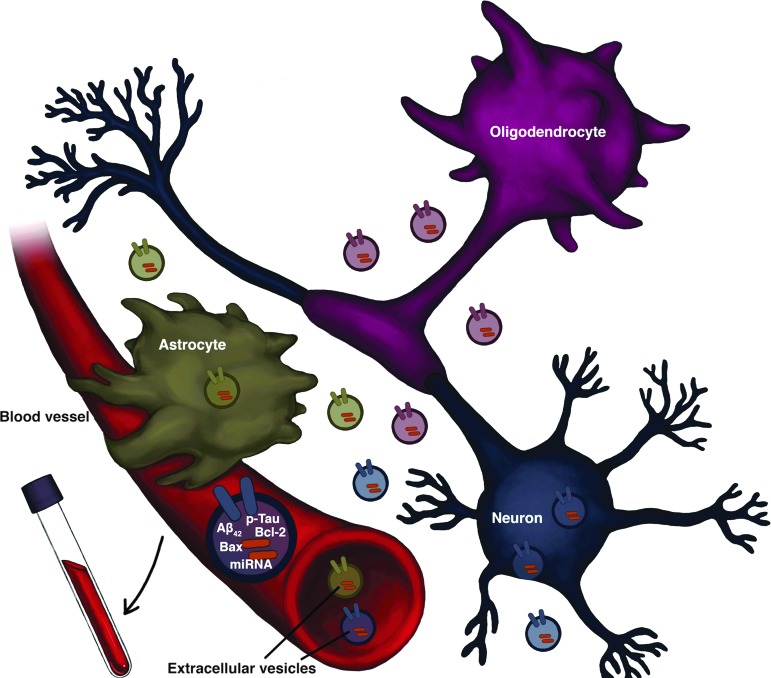
Recent studies suggest that EVs can be used as biomarkers for diagnosing neurodegenerative disorders such as Parkinson's disease, multiple sclerosis, Huntington's disease, amyotrophic lateral sclerosis, and AD. These disorders are associated with the occurrence of misfolded proteins that can be found and quantified in EVs deriving from the CNS.118 An increasing number of studies are hence evaluating the utility of EVs as diagnostic tools to characterize disease progression.119 Their ability to cross the blood–brain barrier allows their time-dependent collection and enrichment from the systemic circulation, saliva, and urine, making their contents an attractive target to quantify for diagnostic applications (Fig. 3).
FIG. 3.
Plasma extracellular vesicles (EVs) enriched for neuronal origin, astrocyte, or oligodendrocyte origin. Schematic showing the assembly and release of EVs from cells within the brain. EVs are generated by the inward budding of endosomal membranes, with the associated recruitment and internalization of cellular protein and RNA cargo. Following their cellular release EVs travel within the interstitial fluid to neighboring cells or, via the circulation, to distant targets to potentially provide modulatory actions. EVs are found in and can be isolated from plasma/serum samples from animal models as well as humans. Enrichment for their cellular origin (whether neuronal, astrocyte, or a different cell type—such as oligodendrocytces) can be achieved by immunoprecipitation with biotinylated antibodies against surface markers to isolate sub-populations from different central nervous system (CNS) cell types. The quantitative evaluation of their protein and RNA content provides a platform for their use as biomarkers to changes in neurological function, disease state, and treatment conditions.
Extracellular Vesicles as a Tool for Diagnosis and Therapy
As noted, the relative ease of collecting EVs from biological fluids, and enriching them for cellular origin, such as neuronal or astrocytic, make them a useful tool for non-invasive disease diagnosis.79 As an example among several, a study of the classical AD markers Aβ42 and P-tau (phosphorylated at the S396 or T181 sites) found them significantly increased in plasma derived EVs enriched for neuronal origin (enrichment was obtained by use of the neuronal cell surface marker L1CAM) that allowed the differentiation of AD patients from age-matched controls.120 Notably, these markers appeared to be significantly higher as early as 10 years prior to AD clinical diagnosis.121 Recent analyses of other proteins within the cargo of plasma derived, L1CAM neuronally enriched EVs are providing insight into mechanisms involved in AD pathogenesis, with the occurrence of heightened protein profiles involved in brain insulin resistance122 and declines in the levels of cellular survival factors123 and synaptic proteins.124 Notably, these protein profiles can be likewise interrogated in TBI, as there appear to be shared mechanisms that underpin AD and TBI. Indeed, elevations in P-tau, Tau, Aβ42, and IL-10 within neural-enriched EVs have recently been reported in TBI subjects.125,126 Such processes, together with measures of neuroinflammation by evaluating astrocyte-derived127 and/or microglial-derived EVs128 may provide insight into time-dependent mechanisms occurring in TBI, as well as the factors involved in TBI progression to later AD, and responses to interventions.
Growing experimental evidence suggests that EVs hold a potentially useful role in therapeutic interventions. It has become increasingly clear over the last decade that EVs provide an efficient mechanism for cell–cell communication by delivery of RNA and proteins both within the local microenvironment of their release as well as at a distance, by trafficking their contents through the systemic circulation. Interaction and binding to recipient cells can occur via multiple processes, including receptor–ligand interactions and antigen presentation. Additionally, EVs may attach and fuse to their target-cell membrane or become internalized by endocytosis. Whatever the mechanism, it has become clear that the transferred EV contents are functional at their new location. As an example, mRNA within mouse EVs has been demonstrated to be taken up and successfully translated within human cells.129 siRNA containing EVs have been used to successfully deliver short interference (siRNA) to target tissues when injected into mice.130 Plasmacytoma-derived EVs have been reported to successfully reduce the inflammatory and autoimmune responses in arthritis patients and prevent tumor development.79,131 Finally, recent studies have demonstrated that EVs derived from mesenchymal stem cells can augment functional recovery, mitigate pattern separation and spatial learning impairments, enhance neurovascular remodeling through actions on neurogenesis and angiogenesis, and diminish neuroinflammation in animal models of TBI.132–134 Such research hence opens many windows in relation to new diagnostic and therapeutic approaches. For the former, by allowing the time-dependent collection and evaluation of the contents of EVs generated in cells (neurons and/or astrocytes) impacted by physiological/environmental challenges and/or disease states, and for the latter by aiding the transport and delivery of specific proteins (as well as pharmaceutical and biotherapeutics) to recipient cells to support specific regenerative functions.
Conclusion
TBI symptoms can occasionally resolve within the first year after injury and in many cases largely do so, but 70–90% of patients continue to manifest prolonged and sometimes permanent neurocognitive dysfunction. Albeit a majority of subjects that experience a single mTBI recover completely with sufficient time, some 25% largely fail to do so.134,135 Those experiencing repetitive concussive mTBIs or blast impact mTBIs are less likely to fully recover.136,137 Perhaps more important still, how and in whom these single and repetitive forms of TBI increase the vulnerability of the recipient to later neurodegenerative disorders that manifest as AD or Parkinson's disease remain largely unknown. Under such circumstances, readily accessible biomarkers are clearly needed for identifying underlying pathobiological changes, and could additionally support early diagnosis and optimize treatment for TBI. Recent studies indicate EV protein profiles contain injury-specific biomarkers.138
Whereas advances have been made in the identification and characterization of a selection of blood and CSF biomarkers that appear well-replicated in acute and more severe TBI, there remains an insufficient understanding of the dynamic changes that transpire during the hours, days, weeks, and months that follow different forms of head injury. Elucidation of these dynamic changes would clearly improve our understanding of the pathobiologies that result from acute TBIs. They may define windows of opportunity for therapeutic interventions, define mechanisms of drug action that might best be suited as treatment options within these windows, and also define which subjects might best respond and, importantly, when its safe for them to return to work or play.139 In this regard, neuronal- and astrocyte-enriched plasma EVs appear particularly promising for identifying blood-based biomarkers140 that are more specific to CNS pathobiology as the contents of these stable, cell-derived, small phospholipid bilayer-enclosed vesicles largely mirror the attributes of their parent cells; they can cross the blood–brain barrier and are available for time-dependent sampling from biological fluids, such as blood, saliva, and urine in addition to CSF. Current methodologies allowing the relatively rapid isolation of plasma EVs enriched for CNS origin by the targeting of CNS-specific cellular markers available on their surface, such as the neural adhesion proteins NCAM and L1CAM (CD171) for neurons or glutamine aspartate transporter (GLAST) for astrocytes, permit their immunoprecipitation and collection to enable analysis of their protein and RNA contents. The current availability of technologies to reproducibly quantify within a single sample multiple proteins and/or RNAs derived from focused biochemical cascades permits the interrogation of mechanisms theoretically underpinning TBI, host homeostatic responses to TBI, and the potential of therapeutic options in a time-dependent manner. How these correlate to the more classical markers of TBI sampled from plasma and/or CSF, described in Table 1, remains to be elucidated. Such prior markers have often been evaluated at a single time-point that is often different between studies, and thereby offer only a snapshot of events arising following a head injury. These same markers can be time-dependently evaluated within EV samples alongside gateway proteins/RNAs regulating mechanistic pathways. Such studies in well-orchestrated cellular, animal, and exploratory clinical trials have the ability to provide the field truly useful biomarkers for TBI and other neurodegenerative disorders.
Acknowledgments
We thank Marc Raley and Lauren Brick for generation of Figures 1 and 3, respectively. This work was supported in part by the Intramural Research Program, National Institute on Aging, National Institutes of Health.
Author Disclosure Statement
No competing financial interests exist.
References
- 1. Hyder A.A., Wunderlich C.A., Puvanachandra P., Gururaj G., and Kobusingye O.C. (2007). The impact of traumatic brain injuries: a global perspective. NeuroRehabilitation 22, 341–353 [PubMed] [Google Scholar]
- 2. Ruff R.L., Riechers R.G., Wang X.F., Piero T., and Ruff S.S. (2012). A case-control study examining whether neurological deficits and PTSD in combat veterans are related to episodes of mild TBI. BMJ Open 18, e000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Langlois J.A., Rutland-Brown W., and Wald M.M. (2006). The epidemiology and impact of traumatic brain injury: a brief overview. J. Head Trauma Rehabil. 21, 375–378 [DOI] [PubMed] [Google Scholar]
- 4. Prins M.L., and Giza C.C. (2012). Repeat traumatic brain injury in the developing brain. Int. J. Dev. Neurosci. 30, 185–190 [DOI] [PubMed] [Google Scholar]
- 5. Tagliaferri F., Compagnone C., Korsic M., Servadei F., and Kraus J. (2006). A systematic review of brain injury epidemiology in Europe. Acta Neurochir. (Wien) 148, 255–268 [DOI] [PubMed] [Google Scholar]
- 6. Shi H.Y., Hwang S.L., Lee K.T., and Lin C.L. (2013). In-hospital mortality after traumatic brain injury surgery: a nationwide population-based comparison of mortality predictors used in artificial neural network and logistic regression models. J. Neurosurg. 118, 746–752 [DOI] [PubMed] [Google Scholar]
- 7. Gardner R.C., Burke J.F., Nettiksimmons J., Kaup A., Barnes D.E., and Yaffe K. (2014). Dementia risk after traumatic brain injury vs nonbrain trauma: the role of age and severity. JAMA Neurol. 71, 1490–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barnes D.E., Kaup A., Kirby K.A., Byers A.L., Diaz-Arrastia R., and Yaffe K. (2014). Traumatic brain injury and risk of dementia in older veterans. Neurology 83, 312–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen A.J., Novakovic-Agopian T., Nycum T.J., Song S., Turner G.R., Hills N.K., Rome S., Abrams G.M., and D'Esposito M. (2011). Training of goal-directed attention regulation enhances control over neural processing for individuals with brain injury. Brain 134, 1541–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Greig N.H., Tweedie D., Rachmany L., Li Y., Rubovitch V., Schreiber S., Chiang Y.H., Hoffer B.J., Miller J., Lahiri D.K., Sambamurti K., Becker R.E., and Pick C.G. (2014). Incretin mimetics as pharmacologic tools to elucidate and as a new drug strategy to treat traumatic brain injury. Alzheimers Dement. 10, S62–S75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tweedie D., Rachmany L., Rubovitch V., Zhang Y., Becker K.G., Perez E., Hoffer B.J., Pick C.G., and Greig N.H. (2013). Changes in mouse cognition and hippocampal gene expression observed in a mild physical- and blast-traumatic brain injury. Neurobiol. Dis. 54, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goldstein L.E., Fisher A.M., Tagge C.A., Zhang X.L., Velisek L., Sullivan J.A., Upreti C., Kracht J.M., Ericsson M., Wojnarowicz M.W., Goletiani C.J., Maglakelidze G.M., Casey N., Moncaster J.A., Minaeva O., Moir R.D., Nowinski C.J., Stern R.A., Cantu R.C., Geiling J., Blusztajn J.K., Wolozin B.L., Ikezu T., Stein T.D., Budson A.E., Kowall N.W., Chargin D., Sharon A., Saman S., Hall G.F., Moss W.C., Cleveland R.O., Tanzi R.E., Stanton P.K., and McKee A.C. (2012). Chronic traumatic encephalopathy in blast-exposed military veterans and a blast neurotrauma mouse model. Sci. Transl. Med. 4, 134ra60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moppett I.K. (2007). Traumatic brain injury: assessment, resuscitation and early management. Br. J. Anaesth. 99, 18–31 [DOI] [PubMed] [Google Scholar]
- 14. LaPlaca M.C., Simon C.M., Prado G.R., and Cullen D.K. (2007). CNS injury biomechanics and experimental models. Prog. Brain Res. 161, 13–26 [DOI] [PubMed] [Google Scholar]
- 15. Barkhoudarian G., Hovda D.A, and Giza C.C. (2011). The molecular pathophysiology of concussive brain injury. Clin. Sports Med. 30, 33–48 [DOI] [PubMed] [Google Scholar]
- 16. Greve M.W., and Zink B.J. (2009). Pathophysiology of traumatic brain injury. Mt. Sinai J. Med. 76, 97–104 [DOI] [PubMed] [Google Scholar]
- 17. Morganti-Kossmann M.C., Rancan M., Stahel P.F., and Kossmann T. (2002). Inflammatory response in acute traumatic brain injury: a double-edged sword. Curr. Opin. Crit. Care 8, 101–105 [DOI] [PubMed] [Google Scholar]
- 18. Schmidt O.I., Heyde C.E., Ertel W., and Stahel P.F. (2005). Closed head injury–an inflammatory disease? Brain Res. Brain Res. Rev. 48, 388–399 [DOI] [PubMed] [Google Scholar]
- 19. Biomarkers Definitions Working Group. (2001). Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin. Pharmacol. Ther. 69, 89–95 [DOI] [PubMed] [Google Scholar]
- 20. Bettermann K., Slocomb J.E., Shivkumar V., and Lott M.E. (2012). Retinal vasoreactivity as a marker for chronic ischemic white matter disease? J. Neurol. Sci. 322, 206–210 [DOI] [PubMed] [Google Scholar]
- 21. Sharma R., and Laskowitz D.T. Biomarkers in traumatic brain injury. (2012). Curr. Neurol. Neurosci. Rep. 12, 560–569 [DOI] [PubMed] [Google Scholar]
- 22. Papa L., Stiell I.G., Clement C.M., Pawlowicz A., Wolfram A., Braga C., Draviam S., and Wells G.A. (2012). Performance of the Canadian CT Head Rule and the New Orleans Criteria for predicting any traumatic intracranial injury on computed tomography in a United States Level I trauma center. Acad. Emerg. Med. 19, 2–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Topolovec-Vranic J., Pollmann-Mudryj M.A., Ouchterlony D., Klein D., Spence J., Romaschin A., Rhind S., Tien H.C., and Baker A.J. (2011). The value of serum biomarkers in prediction models of outcome after mild traumatic brain injury. J. Trauma 71, S478–S486 [DOI] [PubMed] [Google Scholar]
- 24. De Kruijk J.R., Twijnstra A., and Leffers P. (2001). Diagnostic criteria and differential diagnosis of mild traumatic brain injury. Brain. Inj. 15, 99–106 [DOI] [PubMed] [Google Scholar]
- 25. Berger R.P., Pierce M.C., Wisniewski S.R., Adelson P.D., and Kochanek P.M. (2002). Serum S100B concentrations are increased after closed head injury in children: a preliminary study. J. Neurotrauma 19, 1405–1409 [DOI] [PubMed] [Google Scholar]
- 26. Giacoppo S. B.ramanti P., Barresi M., Celi D., Foti Cuzzola V., Palella E., and Marino S. (2012). Predictive biomarkers of recovery in traumatic brain injury. Neurocrit Care 16, 470–477 [DOI] [PubMed] [Google Scholar]
- 27. Persson L., Hårdemark H.G., Gustafsson J., Rundström G., Mendel-Hartvig I., Esscher T., and Påhlman S. (1987). S-100 protein and neuron-specific enolase in cerebrospinal fluid and serum: markers of cell damage in human central nervous system. Stroke 18, 911–918 [DOI] [PubMed] [Google Scholar]
- 28. Mussack T., Kirchhoff C., Buhmann S., Biberthaler P., Ladurner R., Gippner-Steppert C., Mutschler W., and Jochum M. (2006). Significance of Elecsys S100 immunoassay for real-time assessment of traumatic brain damage in multiple trauma patients. Clin. Chem. Lab. Med. 44, 1140–1145 [DOI] [PubMed] [Google Scholar]
- 29. Rothoerl RD., and Woertgen C. (2001). High serum S100B levels for trauma patients without head injuries. Neurosurgery 49, 1490–1493 [DOI] [PubMed] [Google Scholar]
- 30. Anderson RE., Hansson LO., Nilsson O., Dijlai-Merzoug R., and Settergren G. (2001). High serum S100B levels for trauma patients without head injuries. Neurosurgery 48,1255–1260 [DOI] [PubMed] [Google Scholar]
- 31. Romner B., and Ingebrigtsen T. (2001). High serum S100B levels for trauma patients without head injuries. Neurosurgery 49, 1490–1493 [DOI] [PubMed] [Google Scholar]
- 32. Stalnacke B.M., Ohlsson A., Tegner Y., and Sojka P. (2006). Serum concentrations of two biochemical markers of brain tissue damage S-100B and neurone specific enolase are increased in elite female soccer players after a competitive game. Br. J. Sports Med. 40, 313–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mondello S., Muller U., Jeromin A., Streeter J., Hayes R.L., and Wang K.K. (2011). Blood-based diagnostics of traumatic brain injuries. Expert Rev. Mol. Diagn 11, 65–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kovesdi E., Lückl J., Bukovics P., Farkas O., Pál J., Czeiter E., Szellár D., Dóczi T., Komoly S., and Büki A. (2010). Update on protein biomarkers in traumatic brain injury with emphasis on clinical use in adults and pediatrics. Acta Neurochir. 152, 1–17 [DOI] [PubMed] [Google Scholar]
- 35. Metting Z., Wilczak N., Rodiger L.A., Schaaf J.M., and van der Naalt J. (2012). GFAP and S100B in the acute phase of mild traumatic brain injury. Neurology 78, 1428–1433 [DOI] [PubMed] [Google Scholar]
- 36. Berger R.P., Adelson P.D., Pierce M.C., Dulani T., Cassidy L.D., and Kochanek P.M. (2005). Serum neuron-specific enolase, S100B, and myelin basic protein concentrations after inflicted and noninflicted traumatic brain injury in children. J. Neurosurg 103, 161–168 [DOI] [PubMed] [Google Scholar]
- 37. Zurek J., Bartlova L., and Fedora M. (2011). Hyperphosphorylated neurofilament NF-H as a predictor of mortality after brain injury in children. Brain Inj. 25, 221–226 [DOI] [PubMed] [Google Scholar]
- 38. Blennow K., Hampel H., Weiner M., and Zetterberg H. (2010). Cerebrospinal fluid and plasma biomarkers in Alzheimer disease. Nat. Rev. Neurol. 6, 131–144 [DOI] [PubMed] [Google Scholar]
- 39. Pollock N.J., Mirra S.S., Binder L.I., Hansen L.A., and Wood J.G. (1986). Filamentous aggregates in Pick's disease, progressive supranuclear palsy, and Alzheimer's disease share antigenic determinants with microtubule-associated protein tau. Lancet 2, 1211. [DOI] [PubMed] [Google Scholar]
- 40. Hampel H., Buerger K., Zinkowski R., Teipel S.J., Goernitz A., Andreasen N., Sjoegren M., DeBernardis J., Kerkman D., Ishiguro K., Ohno H., Vanmechelen E., Vanderstichele H., McCulloch C., Moller H.J., Davies P., and Blennow K. (2004). Measurement of phosphorylated tau epitopes in the differential diagnosis of Alzheimer disease: a comparative cerebrospinal fluid study. Arch. Gen. Psychiatry 61, 95–102 [DOI] [PubMed] [Google Scholar]
- 41. Hall S., Öhrfelt A., Constantinescu R., Andreasson U., Surova Y., Bostrom F., Nilsson C., Håkan W., Decraemer H., Någga K., Minthon L., Londos E., Vanmechelen E., Holmberg B., Zetterberg H., Blennow K., and Hansson O. (2012). Accuracy of a panel of 5 cerebrospinal fluid biomarkers in the differential diagnosis of patients with dementia and/or parkinsonian disorders. Arch. Neurol. 27, 1–8 [DOI] [PubMed] [Google Scholar]
- 42. Randall J., Mörtberg E., Provuncher G.K., Fournier D.R., Duffy D.C., Rubertsson S., Blennow K., Zetterberg H., and Wilson D.H. (2013). Tau proteins in serum predict neurological outcome after hypoxic brain injury from cardiac arrest: results of a pilot study. Resuscitation 84, 351–356 [DOI] [PubMed] [Google Scholar]
- 43. Kondo A., Shahpasand K., Mannix R., Qiu J., Moncaster J., Chen C.H., Yao Y., Lin Y.M., Driver J.A., Sun Y., Wei S., Luo M.L., Albayram O., Huang P., Rotenberg A., Ryo A., Goldstein L.E., Pascual-Leone A., McKee A.C., Meehan W., Zhou X.Z., and Lu K.P. (2015). Antibody against early driver of neurodegeneration cis P-tau blocks brain injury and tauopathy. Nature 523, 431–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Albayram O., Kondo A., Mannix R., Smith C., Tsai C.Y., Li C, Herbert M.K., Qiu J., Monuteaux M., Driver J., Yan S., Gormley W., Puccio A.M., Okonkwo D.O., Lucke-Wold B., Bailes J., Meehan W., Zeidel M., Lu K.P., and Zhou X.Z. (2017). Cis P-tau is induced in clinical and preclinical brain injury and contributes to post-injury sequelae. Nat. Commun. 8, 1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Riederer B.M., Zagon I.S., and Goodman S.R. (1986). Brain spectrin (240/235) and brain spectrin (240/235E): two distinct spectrin subtypes with different locations within mammalian neural cells. J. Cell Biol. 102, 2088–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pike B.R, Flint J., Dutta S., Johnson E., Wang K.K., and Hayes R.L. (2001). Accumulation of non-erythroid α II-spectrin and calpain-cleaved α II-spectrin breakdown products in cerebrospinal fluid after traumatic brain injury in rats. J. Neurochem. 78, 1297–1306 [DOI] [PubMed] [Google Scholar]
- 47. Pineda J.A, Lewis S.B., Valadka A.B., Papa L., Hannay H.J., Heaton S.C., Demery J.A., Liu M.C., Aikman J.M., Akle V., Brophy G.M., Tepas J.J., Wang K.K., Robertson C.S., and Hayes R.L. (2007). Clinical significance of α II-spectrin breakdown products in cerebrospinal fluid after severe traumatic brain injury. J. Neurotrauma 24, 354–366 [DOI] [PubMed] [Google Scholar]
- 48. Czeiter E., Mondello S., Kovacs N., Sandor J., Gabrielli A., Schmid K., Tortella F., Wang K.K., Hayes R.L., Barzo P., Ezer E., Doczi T., and Buki A. (2012). Brain injury biomarkers may improve the predictive power of the IMPACT outcome calculator. J. Neurotrauma 29, 1770–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Farkas O., Polgár B., Szekeres-Barthó J., Dóczi T., Povlishock J.T., and Büki A. (2005). Spectrin breakdown products in the cerebrospinal fluid in severe head injury—preliminary observations. Acta Neurochir. (Wien) 147, 855–861 [DOI] [PubMed] [Google Scholar]
- 50. Mondello S., Robicsek S.A., Gabrielli A., Brophy G.M., Papa L., Tepas J., Robertson C., Buki A., Scharf D., Jixiang M., Akinyi L., Muller U., Wang K.K., and Hayes R.L. (2010). α II-spectrin breakdown products (SBDPs): diagnosis and outcome in severe traumatic brain injury patients. J. Neurotrauma 27, 1203–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wilkinson K.D., Lee K.M., Deshpande S., Duerksen-Hughes P., Boss J.M., and Pohl J. (1989). The neuron-specific protein PGP 9.5 is a ubiquitin carboxyl-terminal hydrolase. Science 246, 670–673 [DOI] [PubMed] [Google Scholar]
- 52. Papa L., Lewis L.M., Silvestri S., Falk J.L., Giordano P., Brophy G.M., Demery J.A., Liu M.C., Mo J., Akinyi L., Mondello S., Schmid K., Robertson C.S., Tortella F.C., Hayes R.L., and Wang K.K. (2012). Serum levels of ubiquitin C-terminal hydrolase distinguish mild traumatic brain injury from trauma controls and are elevated in mild and moderate traumatic brain injury patients with intracranial lesions and neurosurgical intervention. J. Trauma Acute Care Surg. 72, 1335–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Théry C., Zitvogel L., and Amigorena S. (2002). Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2, 569–579 [DOI] [PubMed] [Google Scholar]
- 54. Vlassov A.V., Magdaleno S., Setterquist R., and Conrad R. (2012). Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochimica et Biophysica Acta 1820, 940–948 [DOI] [PubMed] [Google Scholar]
- 55. Subra C., Laulagnier K., Perret B., and Record M. (2007). Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie 89, 205–212 [DOI] [PubMed] [Google Scholar]
- 56. Subra C., Grand D., Laulagnier K., Stella A., Lambeau G., Paillasse M., De Medina P., Monsarrat B., Perret B., Silvente-Poirot S., Poirot M., and Record M. (2010). Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J. Lipid Res. 51, 2105–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Piccin A., Murphy W.G., and Smith O.P. (2007). Circulating microparticles: pathophysiology and clinical implications. Blood Rev. 21, 157–1571 [DOI] [PubMed] [Google Scholar]
- 58. Parolini I., Federici C., Raggi C., Lugini L., Palleschi S., De Milito A., Coscia C., Iessi E., Logozzi M., Molinari A., Colone M., Tatti M., Sargiacomo M., and Fais S. (2009). Microenvironmental pH is a key factor for exosome traffic in tumor cells. J. Biol. Chem. 284, 34211–34222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nolte-'t Hoen E.N., Buschow S.I., Anderton S.M., Stoorvogel W., and Wauben M.H. (2009). Activated T cells recruit exosomes secreted by dendritic cells via LFA-1. Blood 113, 1977–1981 [DOI] [PubMed] [Google Scholar]
- 60. Muntasell A., Berger A.C., and Roche P.A.T. (2007). Cell-induced secretion of MHC class II-peptide complexes on B cell exosomes. EMBO J. 26, 4263–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Raposo G., Nijman H.W., Stoorvogel W., Liejendekker R., Harding C.V., Melief C.J., and Geuze H.J. (1996). B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 183, 1161–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Admyre C., Johansson S.M., Qazi K.R., Filén J.J., Lahesmaa R., Norman M., Neve E.P., Scheynius A., and Gabrielsson S. (2007). Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 179, 1969–1978 [DOI] [PubMed] [Google Scholar]
- 63. Skog J., Würdinger T., van Rijn S., Meijer D.H., Gainche L., Sena-Esteves M., Curry W.T., Jr, Carter B.S., Krichevsky A.M., and Breakefield X.O. (2008). Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Johnstone R.M., Bianchini A., and Teng K. (1989) Reticulocyte maturation and exosome release: transferrin receptor containing exosomes shows multiple plasma membrane functions. Blood 74, 1844–1851 [PubMed] [Google Scholar]
- 65. Gupta A., and Pulliam L. (2014). Exosomes as mediators of neuroinflammation. J. Neuroinflammation 11, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kapustin A.N., Schoppet M., Schurgers L.J., Reynolds J.L., McNair R., Heiss A., Jahnen-Dechent W., Hackeng T.M., Schlieper G., Harrison P., and Shanahan C.M. (2017). Prothrombin loading of vascular smooth muscle cell-derived exosomes regulates coagulation and calcification. Arterioscler. Thromb. Vasc. Biol. 37, e22–e32 [DOI] [PubMed] [Google Scholar]
- 67. Salem K.Z., Moschetta M., Sacco A., Imberti L., Rossi G., Ghobrial I.M., Manier S., and Roccaro A.M. (2016). Exosomes in tumor angiogenesis. Methods Mol. Biol. 1464, 25–34 [DOI] [PubMed] [Google Scholar]
- 68. Luketic L., Delanghe J., Sobol P.T., Yang P., Frotten E., Mossman K.L., Gauldie J., Bramson J., and Wan Y. (2007). Antigen presentation by exosomes released from peptide-pulsed dendritic cells is not suppressed by the presence of active CTL. J. Immunol. 179, 5024–5032 [DOI] [PubMed] [Google Scholar]
- 69. Melo S.A., Sugimoto H., O'Connell J.T., Kato N., Villanueva A., Vidal A., Qiu L., Vitkin E., Perelman L.T., Melo C.A., Lucci A., Ivan C., Calin G.A., and Kalluri R. (2014). Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 26, 707–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chistiakov D.A., and Chistiakov A.A. (2017). α-Synuclein-carrying extracellular vesicles in Parkinson's disease: deadly transmitters. Acta Neurol. Belg. 117, 43–51 [DOI] [PubMed] [Google Scholar]
- 71. Bobrie A., Colombo M., Raposo G., and Thery C. (2011). Exosome secretion: molecular mechanisms and roles in immune responses. Traffic 12, 1659–1668 [DOI] [PubMed] [Google Scholar]
- 72. Simons M., and Raposo G. (2009). Exosomes–vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 21, 575–581 [DOI] [PubMed] [Google Scholar]
- 73. Potolicchio I., Carven G.J., Xu X., Stipp C., Riese R.J., Stern L.J., and Santambrogio L. (2005). Proteomic analysis of microglia-derived exosomes: metabolic role of the aminopeptidase CD13 in neuropeptide catabolism. J. Immunol. 175, 2237–2243 [DOI] [PubMed] [Google Scholar]
- 74. Faure J., Lachenal G., Court M., Hirrlinger J., Chatellard-Causse C., Blot B., Grange J., Schoehn G., Goldberg Y., Boyer V., Kirchhoff F., Raposo G., Garin J., and Sadoul R. (2006). Exosomes are released by cultured cortical neurones. Mol. Cell Neurosci. 31, 642–648 [DOI] [PubMed] [Google Scholar]
- 75. Taylor A.R., Robinson M.B., Gifondorwa D.J., Tytell M., and Milligan C.E. (2007). Regulation of heat shock protein 70 release in astrocytes: role of signaling kinases. Dev. Neurobiol. 67, 1815–1829 [DOI] [PubMed] [Google Scholar]
- 76. Schneider A., and Simons M. (2012). Exosomes: vesicular carriers for intercellular communication in neurodegenerative disorders. Cell Tissue Res. 352, 33–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bobrie A., Colombo M., Raposo G., and Thery C. (2011). Exosome secretion: molecular mechanisms and roles in immune responses. Traffic 12, 1659–1668 [DOI] [PubMed] [Google Scholar]
- 78. Keller S., Sanderson M.P., Stoeck A., and Altevogt P. (2006). Exosomes: from biogenesis and secretion to biological function. Immunol. Lett. 107, 102–108 [DOI] [PubMed] [Google Scholar]
- 79. van Niel G., Porto-Carreiro I., Simoes S., and Raposo G. (2006). Exosomes: a common pathway for a specialized function. J. Biochem. 140, 13–21 [DOI] [PubMed] [Google Scholar]
- 80. Pegtel D. M., Peferoen L., and Amor S. (2014). Extracellular vesicles as modulators of cell-to-cell communication in the healthy and diseased brain. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 369, pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Livshits M.A., Khomyakova E., Evtushenko E.G., Lazarev V.N., Kulemin N,A., Semina S.E., Generozov E.V., and Govorun V.M. (2015). Isolation of exosomes by differential centrifugation: theoretical analysis of a commonly used protocol. Sci. Rep. 5, 17319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Witwer K., Buzas E., Bemis L., Bora A., Lässer C., Lotvall J., Nolte-'t Hoen E.N., Piper M.G., Sivaraman S., Skog J., Théry C., Wauben M.H., and Hochberg F. (2013). Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2, doi: 10.3402/jev.v2i0.20360, eCollection 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Miranda K.C., Bond D.T., Levin J.Z., Adiconis X., Sivachenko A., Russ C., Brown D., Nusbaum C., and Russo L.M. (2014). Massively parallel sequencing of human urinary exosome/microvesicle RNA reveals a predominance of non-coding RNA. PLoS One 9, e96094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Li P., Kaslan M., Lee S.H., Yao J., and Gao Z. (2017). Progress in exosome isolation techniques. Theranostics 7, 789–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Taylor D., Lyons K., and Gercel-Taylor C. (2002). Shed membrane fragment-associated markers for endometrial and ovarian cancers. Gynecol. Oncol. 84, 443–448 [DOI] [PubMed] [Google Scholar]
- 86. Lozano-Ramos I., Bancu I., Oliveira-Tercero A., Armengol M., Menezes-Neto A., del Portillo H., Lauzurica-Valdemoros R., and Borràs F.E. (2015). Size-exclusion chromatography-based enrichment of extracellular vesicles from urine samples. J. Extracell. Vesicles 4, 27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zeringer E., Barta T., Li M., and Vlassov A.V. (2015). Strategies for isolation of exosomes. Cold Spring Harb. Protocol. 4, 319–323 [DOI] [PubMed] [Google Scholar]
- 88. Batrakova E.V., and Kim M.S. (2015). Using exosomes, naturally-equipped nanocarriers, for drug delivery. J. Control Release 219, 396–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Heinemann M.L., Ilmer M., Silva L.P., Hawke D.H., Recio A., Vorontsova M.A., and Vykoukal J. (2014). Benchtop isolation and characterization of functional exosomes by sequential filtration. J. Chromatogr. A. 1371, 125–135 [DOI] [PubMed] [Google Scholar]
- 90. Nordin J., Lee Y., Vader P., Mäger I., Johansson H., Heusermann W., Wiklander O.P., Hällbrink M., Seow Y., Bultema J.J., Gilthorpe J., Davies T., Fairchild P.J., Gabrielsson S., Meisner-Kober N.C., Lehtiö J., Smith C.I., Wood M.J., and El Andaloussi S. (2015). Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties. Nanomedicine 11, 879–883 [DOI] [PubMed] [Google Scholar]
- 91. Muller L., Hong C-S., Stolz D.B., Watkins S.C., and Whiteside T.L. (2014). Isolation of biologically-active exosomes from human plasma. J. Immunol. Methods 411, 55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Niu Z., Pang RTK., Liu W., Li Q., Cheng R., and Yeung W.S.B. (2017). Polymer-based precipitation preserves biological activities of extracellular vesicles from an endometrial cell line. PLoS One 12, e0186534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Rider M.A., Hurwitz S.N., and Meckes D.G., Jr. (2016). ExtraPEG: a polyethylene glycol-based method for enrichment of extracellular vesicles. Sci. Rep. 6, 23978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Helwa I., Cai J., Drewry M.D., Zimmerman A., Dinkins M.B., Khaled M.L., Seremwe M., Dismuke W.M., Bieberich E., Stamer W.D., Hamrick M.W., and Liu Y. A. (2017). Comparative study of serum exosome isolation using differential ultracentrifugation and three commercial reagents. PLoS One 12, e0170628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Taylor D.D., and Shah S. (2015). Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods 87, 3–10 [DOI] [PubMed] [Google Scholar]
- 96. Lener T., Gimona M., Aigner L., Börger V., Buzas E., Camussi G., Chaput N., Chatterjee D., Court F.A., Del Portillo H.A., O'Driscoll L., Fais S., Falcon-Perez J.M., Felderhoff-Mueser U., Fraile L., Gho Y.S., Görgens A., Gupta R.C., Hendrix A., Hermann D.M., Hill A.F., Hochberg F., Horn P.A., de Kleijn D., Kordelas L., Kramer B.W., Krämer-Albers E.M., Laner-Plamberger S., Laitinen S., Leonardi T, Lorenowicz M.J., Lim S.K., Lötvall J., Maguire C.A., Marcilla A., Nazarenko I., Ochiya T., Patel T., Pedersen S., Pocsfalvi G., Pluchino S., Quesenberry P., Reischl I.G., Rivera F.J., Sanzenbacher R., Schallmoser K., Slaper-Cortenbach I., Strunk D., Tonn T., Vader P., van Balkom B.W., Wauben M., Andaloussi S.E., Théry C., Rohde E., and Giebel B. (2015). Applying extracellular vesicles based therapeutics in clinical trials–an ISEV position paper. J. Extracell. Vesicles 4, 10..3402/jev.v4.30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. György B., Hung M.E., Breakefield X.O., and Leonard J.N. (2015). therapeutic applications of extracellular vesicles: clinical promise and open questions. Ann. Rev. Pharmacol. Toxicol. 55, 439–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Mustapic M., Eitan E., Werner J.K., Berkowitz S.T., Lazaropoulos M.P., Tran J., Goetzl E.J., and Kapogiannis D. (2017). Plasma extracellular vesicles enriched for neuronal origin: a potential window into brain pathologic processes. Front. Neurosci. 11, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Yamada T., Inoshima Y., Matsuda T., and Ishiguro N. (2012). Comparison of methods for isolating exosomes from bovine milk. J. Vet. Med. Sci. 74, 1523–1525 [DOI] [PubMed] [Google Scholar]
- 100. Fauré J., Lachenal G., Court M., Hirrlinger J., Chatellard-Causse C., Blot B., Grange J., Schoehn G., Goldberg Y., Boyer V., Kirchhoff F., Raposo G., Garin J., and Sadoul R. (2006). Exosomes are released by cultured cortical neurones. Mol. Cell. Neurosci. 31, 642–648 [DOI] [PubMed] [Google Scholar]
- 101. Crescitelli R., Lässer C., Szabó T.G., Kittel A., Eldh M., Dianzani I., Buzás E.I., and Lötvall J. (2013). Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J. Extracell. Vesicles 2, 20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Abels E.R., and Breakefield X.O. (2016). Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cell. Mol. Neurobiol. 36, 301–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Raposo G., and Stoorvogel W. (2013). Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200, 373–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Yáñez-Mó M., Siljander P.R., Andreu Z., Zavec A.B., Borràs F.E., Buzas E.I., Casal E., Cappello F., Carvalho J., Colás E., Cordeiro-da Silva A., Fais S., Falcon-Perez J.M., Ghobrial I.M., Giebel B., Gimona M., Graner M., Gursel I., Gursel M., Heegaard N.H., Hendrix A., Kierulf P., Kokubun K., Kosanovic M., Kralj-Iglic V., Krämer-Albers E.M., Laitinen S., Lässer C., Lener T., Ligeti E., Linē A., Lipps G., Llorente A., Lötvall J., Manček-Keber M., Marcilla A., Mittelbrunn M., Nazarenko I., Nolte-'t Hoen E.N., Nyman T.A., O'Driscoll L., Olivan M., Oliveira C., Pállinger É., Del Portillo H.A., Reventós J., Rigau M., Rohde E., Sammar M., Sánchez-Madrid F., Santarém N., Schallmoser K., Ostenfeld M.S., Stoorvogel W., Stukelj R., Van der Grein S.G., Vasconcelos M.H., Wauben M.H., and De Wever O. (2015). Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 4, 27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ban L.A., Shackel N.A., and McLennan S. V. (2016). Extracellular vesicles: a new frontier in biomarker discovery for non-alcoholic fatty liver disease. Int. J. Mol. Sci. 17, 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kalamvoki M., and Deschamps T. (2016). Extracellular vesicles during Herpes Simplex Virus type 1 infection: an inquire. Virol. J. 13, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Redman C., Tannetta D.S., Dragovic R.A., Gardiner C., Southcombe J.H., Collett G.P., and Sargent I.L. (2012). Review: does size matter? Placental debris and the pathophysiology of pre-eclampsia. Placenta 33, S48–S54 [DOI] [PubMed] [Google Scholar]
- 108. Mørk M., Pedersen S., Botha J., Lund S.M., and Kristensen S.R. (2016). Preanalytical, analytical, and biological variation of blood plasma submicron particle levels measured with nanoparticle tracking analysis and tunable resistive pulse sensing. Scand. J. Clin. Lab. Investig. 76, 1–12 [DOI] [PubMed] [Google Scholar]
- 109. Budnik V., Ruiz-Cañada C., and Wendler F. (2016). Extracellular vesicles round off communication in the nervous system. Nat. Rev. Neurosci. 17, 160–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Andreu Z., and Yáñez-Mó M. (2014). Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 5, 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Guitart K., Loers G., Buck F., Bork U., Schachner M., and Kleene R. (2016). Improvement of neuronal cell survival by astrocyte-derived exosomes under hypoxic and ischemic conditions depends on prion protein. Glia 64, 896–910 [DOI] [PubMed] [Google Scholar]
- 112. Frohlich D., Kuo W.P., Fruhbeis C., Sun J.J., Zehendner C.M., Luhmann H.J., Pinto S., Toedling J., Trotter J., and Krämer-Albers E.M. (2014). Multifaceted effects of oligodendroglial exosomes on neurons: impact on neuronal firing rate, signal transduction and gene regulation. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 369, pii: , doi: 10.1098/rstb.2013.0510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Asai H., Ikezu S., Tsunoda S., Medalla M., Luebke J., Haydar T., Wolozin B., Butovsky O., Kügler S., and Ikezu T. (2015). Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat. Neurosci. 18, 1584–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kosaka N., Iguchi H., Yoshioka Y., Takeshita F., Matsuki Y., and Ochiya T. (2010). Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 285, 17442–17452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Sharples R.A., Vella L.J., Nisbet R.M., Naylor R., Perez K., Barnham K.J., Masters C.L., and Hill A.F. (2008). Inhibition of gamma-secretase causes increased secretion of amyloid precursor protein C-terminal fragments in association with exosomes. FASEB J. 22, 1469–1478 [DOI] [PubMed] [Google Scholar]
- 116. Chivet M., Javalet C., Laulagnier K., Blot B., Hemming F.J., and Sadoul R. (2014). Exosomes secreted by cortical neurons upon glutamatergic synapse activation specifically interact with neurons. J. Extracell. Vesicles 3, 24722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. An K., Klyubin I., Kim Y., Jung J.H., Mably A.J., O'Dowd S.T., Lynch T., Kanmert D., Lemere C.A., Finan G.M., Park J.W., Kim T.W., Walsh D.M., Rowan M.J., and Kim J.H. (2013). Exosomes neutralize synaptic-plasticity-disrupting activity of Aβ assemblies in vivo. Mol. Brain 6, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Schneider A., and Simons M. (2013). Exosomes: vesicular carriers for intercellular communication in neurodegenerative disorders. Cell Tissue Res. 352, 33–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Colombo E., Borgiani B., Verderio C., and Furlan R. (2012). Microvesicles: novel biomarkers for neurological disorders. Front. Physiol. 3, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Abner E.L., Jicha G.A., Shaw L.M., Trojanowski J.Q., and Goetzl E.J. (2016). Plasma neuronal exosomal levels of Alzheimer's disease biomarkers in normal aging. Ann. Clin. Transl. Neurol. 3, 399–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Fiandaca M.S., Kapogiannis D., Mapstone M., Boxer A., Eitan E., Schwartz J.B., Abner E.L, Petersen R.C., Federoff H.J., Miller B.L., and Goetzl E.J. (2015). Identification of preclinical Alzheimer's disease by a profile of pathogenic proteins in neurally derived blood exosomes: A case-control study. Alzheimers Dement. 11, 600–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Kapogiannis D., Boxer A., Schwartz J.B., Abner E.L., Biragyn A., Masharani U., Frassetto L., Petersen R.C., Miller B.L., and Goetzl E.J. (2015). Dysfunctionally phosphorylated type 1 insulin receptor substrate in neural-derived blood exosomes of preclinical Alzheimer's disease. FASEB J. 29, 589–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Goetzl E.J., Boxer A., Schwartz J.B., Abner E.L., Petersen R.C., Miller B.L., Carlson O.D., Mustapic M., and Kapogiannis D. (2015). Low neural exosomal levels of cellular survival factors in Alzheimer's disease. Ann. Clin. Transl. Neurol. 2:769–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Goetzl E.J., Kapogiannis D., Schwartz J.B., Lobach I.V., Goetzl L., Abner E.L., Jicha G.A., Karydas A.M., Boxer A., and Miller B.L. (2016). Decreased synaptic proteins in neuronal exosomes of frontotemporal dementia and Alzheimer's disease. FASEB J. 30, 4141–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Stern R.A., Tripodis Y., Baugh C.M., Fritts N.G., Martin B.M., Chaisson C., Cantu R.C., Joyce J.A., Shah S., Ikezu T., Zhang J., Gercel-Taylor, and Taylor D.D. (2016). Preliminary study of plasma exosomal tau as a potential biomarker for chronic traumatic encephalopathy. J. Alzheimers Dis. 51, 1099–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Gill J., Mustapic M., Diaz-Arrastia R., Lange R., Gulyani S., Diehl T., Motamedi V., Osier N., Stern R.A., Kapogiannis D. (2018). Higher exosomal tau, amyloid-beta 42 and IL-10 are associated with mild TBIs and chronic symptoms in military personnel. Brain Inj. 32, 1277–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Goetzl E.J., Mustapic M., Kapogiannis D., Eitan E., Lobach I.V., Goetzl L., Schwartz J.B., and Miller B.L. (2016). Cargo proteins of plasma astrocyte-derived exosomes in Alzheimer's disease. FASEB J. 30, 3853–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Nigro A., Colombo F., Casella G., Finardi A., Verderio C., and Furlan R. (2016). Myeloid extracellular vesicles: messengers from the demented brain. Front. Immunol. 7, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J.J., and Lotvall J.O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 [DOI] [PubMed] [Google Scholar]
- 130. Alvarez-Erviti L., Seow Y., Schapira A.H., Gardiner C., Sargent I.L., Wood M.J., and Cooper J.M. (2011). Lysosomal dysfunction increases exosome-mediated alpha-synuclein release and transmission. Neurobiol. Dis. 42, 360–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Kim S.H., Lechman E.R., Bianco N., Menon R., Keravala A., Nash J., Mi Z., Watkins S.C., Gambotto A., and Robbins P.D. (2005). Exosomes derived from IL-10-treated dendritic cells can suppress inflammation and collagen-induced arthritis. J. Immunol. 174, 6440–6448 [DOI] [PubMed] [Google Scholar]
- 132. Zhang Y., Chopp M., Meng Y., Katakowski M., Xin H., Mahmood A., and Xiong Y. (2015). Effect of exosomes derived from multipluripotent mesenchymal stromal cells on functional recovery and neurovascular plasticity in rats after traumatic brain injury. J. Neurosurg. 122, 856–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Kim D.K., Nishida H., An S.Y., Shetty A.K., Bartosh T.J., and Prockop D.J. (2016). Chromatographically isolated CD63+CD81+ extracellular vesicles from mesenchymal stromal cells rescue cognitive impairments after TBI. PNAS 113, 170–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Zhang Y., Chopp M., Zhang Z.G., Katakowski M., Xin H., Qu C., Ali M., Mahmood A., and Xiong Y. (2016). Systemic administration of cell-free exosomes generated by human bone marrow derived mesenchymal stem cells cultured under 2D and 3D conditions improves functional recovery in rats after traumatic brain injury. Neurochem. Int.111, 69–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Laker S.R. (2011). Epidemiology of concussion and mild traumatic brain injury. PM R. 3, S354–S358 [DOI] [PubMed] [Google Scholar]
- 136. Voss J.D., Connolly J., Schwab K.A., and Scher A.I. (2015). Update on the epidemiology of concussion/mild traumatic brain injury. Curr. Pain Headache Rep. 19, 32. [DOI] [PubMed] [Google Scholar]
- 137. Echlin P.S., Tator C.H., Cusimano M.D., Cantu R.C., Taunton J.E., Upshur R.E., Czarnota M., Hall C.R., Johnson A.M., Forwell L.A., Driediger M., and Skopelja E.N. (2010). Return to play after an initial or recurrent concussion in a prospective study of physician-observed junior ice hockey concussions: implications for return to play after a concussion. Neurosurg. Focus. 29, E5. [DOI] [PubMed] [Google Scholar]
- 138. Moyron R.B., Gonda A., Selleck M.J., Luo-Owen X., Catalano R.D., O'Callahan T., Garberoglio C., Turay D., and Wall N.R. (2017). Differential protein expression in exosomal samples taken from trauma patients. Proteomics Clin. Appl. 11, 9–10 [DOI] [PubMed] [Google Scholar]
- 139. Doolan A.W., Day D.D., Maerlender A.C., Goforth M., and Gunnar Brolinson P. (2012). A review of return to play issues and sports-related concussion. Ann. Biomed. Eng. 40, 106–113 [DOI] [PubMed] [Google Scholar]
- 140. Agoston D.V., Shutes-David A., and Peskind E.R. (2017) Biofluid biomarkers of traumatic brain injury. Brain Inj. 31, 1195–1203 [DOI] [PubMed] [Google Scholar]
- 141. Papa L., Lewis L.M., Falk J.L., Zhang Z., Silvestri S., Giordano P., Brophy G.M., Demery J.A., Dixit N.K., Ferguson I., Liu M.C., Mo J., Akinyi L., Schmid K., Mondello S., Robertson CS., Tortella FC., Hayes R.L., and Wang K.K. (2012). Elevated levels of serum glial fibrillary acidic protein breakdown products in mild and moderate traumatic brain injury are associated with intracranial lesions and neurosurgical intervention. Ann. Emerg. Med. 59, 471–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Žurek J., and Fedora M. (2012). The usefulness of S100B, NSE, GFAP, NF-H, secretagogin and Hsp70 as a predictive biomarker of outcome in children with traumatic brain injury. Acta Neurochir. (Wien) 154, 93–103 [DOI] [PubMed] [Google Scholar]
- 143. Pelinka L.E., Toegel E., Mauritz W., and Red H. (2003). Serum S 100 B: a marker of brain damage in traumatic brain injury with and without multiple trauma. Shock 19, 195–200 [DOI] [PubMed] [Google Scholar]
- 144. Thelin E.P., Jeppsson E., Frostell A., Svensson M., Mondello S., Bellander B.M., and Nelson D.W. (2016). Utility of neuron-specific enolase in traumatic brain injury; relations to S100B levels, outcome, and extracranial injury severity. Crit. Care 8, 20, 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Mondello S., Sorinola A., Czeiter E., Vámos Z., Amrein K., Synnot A., Donoghue E., Sándor J., Wang K.K.W., Diaz-Arrastia R., Steyerberg E.W., Menon D.K., Mass A.I.R., and Buki A. (2018). Blood-based protein biomarkers for the management of traumatic brain injuries in adults presenting to emergency departments with mild brain injury: a living systematic review and meta-analysis. J Neurotrauma. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Liu M.C., Akle V., Zheng W., Kitlen J., O'Steen B., Larner S.F., Dave J.R., Tortella F.C., Hayes R.L., and Wang KK. (2006). Extensive degradation of myelin basic protein isoforms by calpain following traumatic brain injury. J. Neurochem. 98, 700–712 [DOI] [PubMed] [Google Scholar]
- 147. Vajtr D., Benada O., Linzer P., Sámal F., Springer D., Strejc P., Beran M., Průša R., and Zima T. (2012). Immunohistochemistry and serum values of S-100B, glial fibrillary acidic protein, and hyperphosphorylated neurofilaments in brain injuries. Soud Lek. 57, 7–12 [PubMed] [Google Scholar]
- 148. Randall J., Mörtberg E., Provuncher G.K., Fournier D.R., Duffy D.C., Rubertsson S., Blennow K., Zetterberg H., and Wilson D.H. (2013). Tau proteins in serum predict neurological outcome after hypoxic brain injury from cardiac arrest: results of a pilot study. Resuscitation 84, 351–356 [DOI] [PubMed] [Google Scholar]
- 149. Ross S.A., Halliday M.I., Campbell G.C., Byrnes D.P., and Rowlands B.J. (1994). The presence of tumour necrosis factor in CSF and plasma after severe head injury. Br. J. Neurosurg. 8, 419–425 [DOI] [PubMed] [Google Scholar]
- 150. Antunes A.A., Sotomaior V.S., Sakamoto K.S., de Camargo Neto C.P., Martins C., and Aguiar L.R. (2010). Interleukin-6 plasmatic levels in patients with head trauma and intracerebral hemorrhage. Asian J. Neurosurg. 5, 68–77 [PMC free article] [PubMed] [Google Scholar]
- 151. Mondello S., Papa L., Buki A., Bullock M.R., Czeiter E., Tortella F.C., Wang K.K., and Hayes R.L. (2011). Neuronal and glial markers are differently associated with computed tomography findings and outcome in patients with severe traumatic brain injury: a case control study. Crit. Care. 15, R156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Dash P.K., Zhao J., Hergenroeder G., and Moore A.N. (2010). Biomarkers for the diagnosis, prognosis, and evaluation of treatment efficacy for traumatic brain injury. Neurotherapeutics 7, 100–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Papa L., Akinyi L., Liu M.C., Pineda J.A., Tepas J.J, Oli M.W., Zheng W., Robinson G., Robicsek S.A., Gabrielli A., Heaton S.C., Hannay H.J., Demery J.A., Brophy G.M., Layon J., Robertson C.S., Hayes R.L., and Wang K.K. (2010). Ubiquitin C-terminal hydrolase is a novel biomarker in humans for severe traumatic brain injury. Crit. Care Med. 38, 138–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Pineda J.A., Lewis S.B., Valadka A.B., Papa L., Hannay H.J., Heaton S.C., Demery J.A., Liu MC, Aikman J.M., Akle V., Brophy G.M., Tepas J.J., Wang K.K., Robertson C.S., and Hayes R.L. (2007). Clinical significance of α II-spectrin breakdown products in cerebrospinal fluid after severe traumatic brain injury. J. Neurotrauma 24, 354–366 [DOI] [PubMed] [Google Scholar]
- 155. Bayir H., Marion D.W., Puccio A.M., Wisniewski S.R., Janesko K.L., Clark R.S., and Kochanek P.M. (2004): Marked gender effect on lipid peroxidation after severe traumatic brain injury in adult patients. J. Neurotrauma 21, 1–8 [DOI] [PubMed] [Google Scholar]
- 156. Wagner A.K., McCullough E.H., Niyonkuru C., Ozawa H., Loucks T.L., Dobos J.A., Brett C.A., Santarsieri M., Dixon C.E., Berga S.L., and Fabio A. (2011). Acute serum hormone levels: characterization and prognosis after severe traumatic brain injury. J. Neurotrauma 28, 871–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Colombo M., Moita C., van Niel G., Kowal J., Vigneron J., Benaroch P., Manel N., Moita L.F., Théry C., and Raposo G. (2013). Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 126(Pt. 24), 5553–5565 [DOI] [PubMed] [Google Scholar]
- 158. Ostrowski M., Carmo N.B., Krumeich S., Fanget I., Raposo G., Savina A., Moita C.F., Schauer K., Hume A.N., Freitas R.P., Goud B., Benaroch P., Hacohen N., Fukuda M., Desnos C., Seabra M.C., Darchen F., Amigorena S., Moita L.F., and Thery C. (2010). Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 12, 19–30 [DOI] [PubMed] [Google Scholar]
- 159. Binotti B., Jahn R., and Chua J.J. (2016). Functions of rab proteins at presynaptic sites. Cells 5, E7. [DOI] [PMC free article] [PubMed] [Google Scholar]