Abstract
Schnyder corneal dystrophy (SCD) is an autosomal dominant disease characterized by germline variants in UBIAD1 introducing missense alterations leading to deposition of cholesterol in the cornea, progressive opacification, and loss of visual acuity. UBIAD1 was recently shown to synthesize menaquinone-4 (MK-4, vitamin K2, but causal mechanisms of SCD are unknown. We report a novel c.864G>A UBIAD1 mutation altering glycine 177 to glutamic acid (p.G177E) in six SCD families, including four families from Finland who share a likely founder mutation. We observed reduced MK-4 synthesis by UBIAD1 altered by SCD mutations p.N102S, p.G177R/E, and p.D112N, and molecular models showed p.G177-mutant UBIAD1 disrupted transmembrane helices and active site residues. We show UBIAD1 interacts with HMGCR and SOAT1, enzymes catalyzing cholesterol synthesis and storage, respectively, using yeast two-hybrid screening and immunoprecipitation. Docking simulations indicate cholesterol binds to UBIAD1 in the substrate binding cleft and binding overlaps with GGPP binding, a MK-4 substrate, suggesting potential competition between these metabolites. Impaired MK-4 synthesis is a biochemical defect identified in SCD suggesting UBIAD1 links vitamin K and cholesterol metabolism through physical contact between enzymes and metabolites. Our data suggests a role for endogenous MK-4 in maintaining cornea health and visual acuity.
Keywords: Schnyder crystalline corneal dystrophy, vision, eye, TERE1, HMGCR, SOAT1, ACAT, cholesterol, menadione, phospholipid, genetic disease, protein model
Introduction
Schnyder corneal dystrophy, formerly known as Schnyder crystalline corneal dystrophy (MIM# 121800) [Van Went and Wibaut, 1924; Schnyder, 1929, 1939; Weiss et al., 2008] is an autosomal dominant eye disease characterized by abnormal deposition of cholesterol and phospholipids in the cornea [Weiss et al., 1992; Weiss, 1992; Rodrigues et al., 1987]. The resultant corneal opacification is progressive and bilateral resulting in visual loss. Patients demonstrate a characteristic pattern of corneal opacification dependent on age and 50% have crystalline corneal cholesterol deposits [Weiss, 1992; Weiss, 2007]. Non-crystalline SCD has a more subtle presentation with only corneal haze and consequently may be more difficult to diagnose. Light scattering due to the corneal deposits has been postulated to cause glare and visual loss, and the significant visual morbidity of the disease is underscored by the number of patients requiring ocular surgery: 20 of 37 (54%) individuals 50 years of age and younger, and 10 of 13 (77%) individuals 70 and younger reported having had corneal transplantation surgery [Weiss, 2007].
In 2007, two groups independently identified mutations in SCD patients in the UbiA prenyltransferase domain containing protein 1 (UBIAD1, MIM# 611632) gene on chromosome 1p36, which was named after bacterial (E. coli) UbiA protein and was predicted to encode a prenyltranferase that had no prior connection to corneal dystrophy [Orr et al., 2007; Weiss et al., 2007]. To date, 23 nucleotide changes resulting in 22 missense mutations have been characterized in 45 SCD families with the alteration of asparagine (N) to serine (S) at residue 102 (p.N102S) being most frequent in 17/45 (38%) of families. Comparison with E. coli proteins, UbiA and menA, revealed a highly conserved putative UBIAD1 active site containing the p.N102 residue, suggesting an alteration in enzymatic function as likely causal to disease [Weiss et al., 2008; Nickerson et al., 2010]. UBIAD1 was recently shown to catalyze the prenylation of plant phylloquinone (PK) to produce MK-4, the predominant form of hormonally active vitamin K in humans and an important cofactor in blood clotting and bone metabolism [Nakagawa et al., 2010]. Vitamin K2 was recently shown to serve as a mitochondrial electron carrier during ATP generation by the electron transport chain [Vos et al., 2012] but the effects of SCD mutations on vitamin K2 synthesis have not been examined.
This study sought to further elucidate the genetic, enzymatic, and metabolic pathways contributing to SCD. We identify a novel UBIAD1 founder mutation in four large SCD families in which the disease was documented in photographs from the late 1800s, prior to the first published article on SCD. We show significantly reduced MK-4 synthesis due to SCD alteration of UBIAD1, and physical association of UBIAD1 with enzymes involved in cholesterol synthesis and storage, providing direct links between UBIAD1 and cholesterol metabolism that are likely relevant to SCD disease pathology.
Methods and Materials
Human Subjects and Samples
Institutional Review Board approval was obtained from the University of Massachusetts Medical Center, Wayne State University School of Medicine [Weiss et al., 2007] and the study was exempt as determined by the National Institutes of Health Office of Human Subjects Research. Affected probands were recruited by J.S.W., self-referred, or were referred by other ophthalmologists. Family members provided informed consent under the Declaration of Helsinki, family history, and blood samples, and were subjected to an ophthalmologic examination of visual acuity and slit lamp characterization of both corneas. DNAs from 300 healthy individuals were obtained as controls from the Dean lab, the Coriell Institute for Medical Research (Camden, NJ), and as previously described [Weiss et al., 2007; Weiss et al., 2008; Nickerson et al., 2010].
DNA Isolation, PCR, Sequencing, and Variant Annotation
Genomic DNA was isolated using Puregene (Gentra/Qiagen, Valencia CA). PCR utilized Fast Start PCR Master Mix (Roche, Mannheim, Germany), and sequencing was done with Big Dye v.3.1 (Applied Biosystems, ABI, Foster City, CA) on a 3730 Genetic Analyzer (ABI) as previously described (Supp. Fig. 1) [Weiss et al., 2007; Weiss et al., 2008; Nickerson et al., 2010]. Chromatograms were analyzed using Sequencher v4.8 (GeneCodes, Ann Arbor, MI). Variants were annotated to Genbank reference cDNA NM_013319.2 and reference protein sequence NP_037451. Nucleotide numbering reflects cDNA numbering with +1 corresponding to the first base of the 5’ untranslated region. The novel variant in the new SCD families described in this study have been submitted to the Leiden Open Variation Database under the UBIAD1 gene, <www.lovd.nl/UBIAD1>.
UBIAD1 Biosynthesis of MK-4
UBIAD1 synthesis of MK-4 was assayed similarly to Nakagawa et al. (2010) except microsomes from Epstein-Barr virus-immortalized patient B-cell lysates rather than insect cells were examined. Washed cell pellets were resuspended in homogenization buffer containing 100 mM Tris-HCl and protease inhibitor cocktail (Complete Mini, EDTA free; Roche), disrupted by freeze–thaw and sonication on ice, centrifuged at 3,000g for 10 minutes to remove unbroken cells and nuclei, and cell lysates were centrifuged at 105,000g (Beckman Coulter, Inc., Brea, CA) for 60 minutes. The resulting pellet was resuspended in homogenization buffer and used as microsomes. Western blotting was used to normalize protein amounts with an affinity-purified rabbit polyclonal antibody raised against a UBIAD1 peptide starting at amino acid 31, N-CPEQDRLPQRSWRQK-COOH (MRL Co., Ltd, Woburn, MA) [Nakagawa et al., 2010]. The peroxidase-conjugated secondary antibody was rabbit immunoglobulin raised in donkey (Santa Cruz Biotechnology, Santa Cruz, CA) incubated for 1.5 hour and detected with an electrochemiluminescent detection system (Nakalai Tesque, Inc., Kyoto, Japan). Reaction mixtures contained K3-d8, 1 mM geranylgeranyl diphosphate (GGPP), 1 mM dithiothreitol in 100 mM Tris-HCl (pH 8.5), and 90 μg of microsomal protein. Reactions were stopped after 1 hour by the addition of ethanol and were extracted and subjected to liquid-chromatography/atmospheric-pressure chemical ionization tandem mass spectrometry (LC-APCI-MS/MS) to quantitate MK-4-d7 converted from K3-d8 in multiple reaction monitoring (MRM) chromatograms as previously described [Nakagawa et al., 2010].
Protein Homology and Modeling
Full length UBIAD1 protein sequences from19 species were aligned using Clustal 2.0.11 then were subjected to local optimization of the alignment in sequential, overlapping regions [Nickerson et al., 2010]. A 3D model of UBIAD1 derived from an experimentally validated structural model of E. coli UbiA enzyme [Bräuer et al., 2008] was previously used to examine docking of farnesyl-diphosphate and 1,4-dihydroxy aryl compounds and release of the resulting reaction products from UBIAD1 [Nickerson et al., 2010].
The quality of the three dimensional (3D)-model of UBIAD1 was checked with PROCHECK [Laskowski et al., 1993], PROSA II [Sippl, 1990], and Errat2 (http://nihserver.mbi.ucla.edu/ERRATv2/) [Colovos and Yeates, 1993]. Analyses with PROCHECK evaluated the model for stereo-chemical parameters, e.g. the Ramachandran plot quality examined the backbone dihedral angles (Supp. Fig. S2). Despite two or three outlier residues in wt (Supp. Fig. S2A) and G177E (Supp. Fig. S2B) mutant protein, respectively, all stereo-chemical criteria of the model analyzed by PROCHECK meet or exceed comparable criteria derived from an X-ray structure with a resolution of 2Å. The graphical analysis with PROSA II examined energy z-scores of the Cα-Cβ pairs and a –4.45 indicates a native-like folded structure. A combined energy z-score including surface potentials was not considered due to membrane binding/localization of the enzyme. The ligands (menadione, GGPP, and cholesterol) were docked using the GOLD-suite 5.0.1 (Verdonk, et al., 2003, Hartshorn et al., 2007). For all ligands, 30 docking runs with standard settings of GOLD were performed.
No structural changes in the wild type (wt) enzyme were observed upon introduction of new ligands to the active site, validating the robustness of docking arrangements already published. A UBIAD1 p.G177E mutation was introduced in silico and the structure was refined by 20 simulated annealing molecular dynamics cycles using YASARA (www.yasara.org) [Krieger et al. 2009]. Finally, MK-4 was docked to the wild-type and p.G177E mutant enzyme using molecular dynamics simulations for more than 6 nanoseconds which included membrane lipids (phosphatidylethanolamine) surrounding the protein and a water box above and below the membrane region with periodic boundary conditions.
Yeast Two-Hybrid Screening
Y2H reagents and techniques are highly standardized and have been described [Garrus et al., 2001]. Briefly, bait constructs expressing either full-length (1–338 aa) or partial (102–314 aa) UBIAD1 were fused to the C-terminus of the Gal4 DNA binding domain and used to transform PNY200 yeast. Human prostate cancer, lung, and kidney cDNA libraries fused to the C-terminus of the Gal4 activation domain were used to transform BK100 yeast containing a multiple reporter system. Mated diploid yeast colonies containing interacting bait/prey cDNAs were selected by amino acid complementation and sequenced.
Immunoprecipitation
HEK293T cells were cultured at 37°C in DMEM with 10% fetal bovine serum, 100 μg/ml penicillin, and 100 μg/ml streptomycin; grown to ~60% confluency, then transfected with a 5:1 (μl:μg) ratio of polyethylenimine (1 mg/ml, Polysciences Inc., Warrington, PA) to DNA (3xFlag-UBIAD1, Myc-HMGCR, SOAT1-Myc, or the indicated combinations). Forty-eight hours post-transfection, cells were washed with cold PBS and harvested on ice in the presence of 1 ml lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.5 mM EDTA, 0.1% NP-40, and 1x protease inhibitor cocktail). Cells were scraped, lysed through one freeze-thaw cycle, and centrifuged 10 minutes at 17,500g. The supernatant was incubated for 2 hours at 4°C with either 25 μl Protein G Sepharose 4 Fast Flow (GE Heathcare, Waukesha, WI) and 2 μg of anti-Myc-tag antibody (#2276, Cell Signaling Technology, Danvers, MA), or with 25 μl anti-Flag M2 agarose (Sigma, St. Louis, MO). The resin was washed 3x with wash buffer (50 mM Tris, pH 7.4, 150 mM NaCl), centrifuged 30 seconds at 1500g, and incubated at 95°C for 5 minutes in 2x LDS (Invitrogen, Carlsbad, CA) containing reducing agent. Protein samples were divided, resolved on 4–12% Bis-Tris SDS-PAGE gels (Invitrogen, Carlsbad, CA), transferred to nitrocellulose membranes (Protran, GE Heathcare), and probed with 1:1000 dilutions of either anti-Flag or anti-Myc. Secondary antibodies were conjugated to horseradish peroxidase (Thermo Fisher Scientific, Waltham, MA; Jackson ImmunoResearch Laboratories, West Grove, PA) and were visualized using an ECL substrate (Pierce/Thermo Fisher Scientific), and X-OMAT Blue film (Eastman Kodak, Rochester, NY).
Co-localization of UBIAD1 and HMGCR
HEK293T cells were grown on cover slips to ~50% confluency in two separate 10 cm2 dishes and transfected with 3xFLAG-UBIAD1 and MYC-HMGCR or empty vector. After 48 hours, cells were fixed with 3.7% formaldehyde for 15 minutes, permeabilized with 0.1% Triton X-100 for 10 minutes and blocked with 10% goat serum for 30 minutes, all prepared in PBS. Primary antibodies were applied at a dilution of 1:100 in 10% goat serum for 1 hour. Secondary antibodies conjugated to either 568 or 488 fluorophore (Invitrogen, Carlsbad, CA) were applied at a dilution of 1:1000 for 1 hour and Hoechst 33342 was applied at a concentration of 2 ng/μl for 10 minutes. Three PBS washes were performed between each new application. Cells were imaged with a Laser Scanning Microscope 710 and the Zen software package (Carl Zeiss Microscopy, Oberkochen, Germany).
Results
Mutation of UBIAD1 in SCD Families from Finland, Japan, and Turkey
The UBIAD1 locus encodes four transcripts (Supp. Fig. S1A) [Weiss et al., 2007] and all SCD-associated alterations have been observed in reference transcript, NM_013319.2, encoding a two-exon, 338 amino acid membrane protein. Protein coding sequences and splice junctions were examined using PCR and sequencing as previously described [Orr et al., 2007; Weiss et al., 2007] in probands from four, six generation SCD families originating in Finland, from a self-described Swede-Finn cultural/ethnic group [Weiss, 1992]. A novel G>A alteration at nucleotide 864 (n.864) in transcript NM_013319.2 was observed in the first coding base of exon 2 that introduced a nonconservative substitution of glutamic acid (E) for glycine (G) at amino acid 177 (p.G177E, Fig. 1). A n.864 G/A was also detected in probands from two additional SCD families from Japan and Turkey [Köksal et al., 2004] (Table 1). The alteration was not observed in DNAs from 300 control individuals (600 chromosomes).
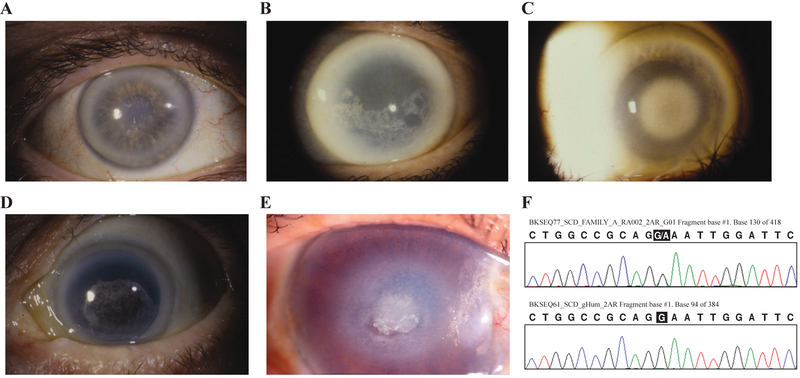
Figure 1.
Proband corneas and sequencing. A: Family A, a 51 year old female (IV:29, Fig. 2A) with central corneal opacity with crystals, mid peripheral haze, and arcus lipoides. B: Family B, a 64 year old female (IV:19, Fig. 2B) with paracentral crystalline deposits, diffuse corneal haze, and arcus lipoides. C: Family C, a 72 year old female (III:6, Fig. 2C) with central corneal opacification, mid peripheral haze, and arcus lipoides. D: Family D, an 81 year old female (III:11, Fig. 2D) with central corneal opacity and crystals, mid peripheral haze, and arcus lipoides. E: Family JJ, a 73 year old female (II:2, Fig. 2E) with diffuse corneal haze and central disciform opacity with subepithelial crystalline deposits, mid peripheral haze, and peripheral arcus lipoides. A cornea photo of the Family W proband has been published [Köksal et al., 2004]. F: Representative sequences of the n.864 G/A mutation from the Family A proband (top) and an unaffected donor (bottom). Sequences from other probands were similar.
Table 1.
UBIAD1 Mutations in SCD Familiesa
| Family | Ethnicityb | Gene mutationc | Proteind | Exon | Loope | Reference |
|---|---|---|---|---|---|---|
| GG | Irish-French Canadian | 623 G>A | A97T | 1 | 1 | Nickerson et al., 2010 |
| Case C | Chinese | 624 G>A | G98S | 1 | 1 | Jing et al., 2009 |
| F123 | Italian | 639 A>G | N102S | 1 | 1 | Orr et al., 2007 |
| BB1 | British | 639 A>G | N102S | 1 | 1 | Weiss et al., 2007 |
| BB2 | British | 639 A>G | N102S | 1 | 1 | Weiss et al., 2007 |
| Q | American | 639 A>G | N102S | 1 | 1 | Weiss et al., 2007 |
| U | American | 639 A>G | N102S | 1 | 1 | Weiss et al., 2007 |
| Y | German | 639 A>G | N102S | 1 | 1 | Weiss et al., 2007 |
| 1 | Irish | 639 A>G | N102S | 1 | 1 | Yellore et al., 2007 |
| BB | Czechoslavakian | 639 A>G | N102S | 1 | 1 | Weiss et al., 2008 |
| DD | Chinese-Taiwan | 639 A>G | N102S | 1 | 1 | Weiss et al., 2008 |
| K | German | 639 A>G | N102S | 1 | 1 | Weiss et al., 2008 |
| L | American | 639 A>G | N102S | 1 | 1 | Weiss et al., 2008 |
| R | American | 639 A>G | N102S | 1 | 1 | Weiss et al., 2008 |
| CC | Japan | 639 A>G | N102S | 1 | 1 | Nickerson et al., 2010 |
| II | American | 639 A>G | N102S | 1 | 1 | Nickerson et al., 2010 |
| KK | American | 639 A>G | N102S | 1 | 1 | Nickerson et al., 2010 |
| MM | American | 639 A>G | N102S | 1 | 1 | Nickerson et al., 2010 |
| N | American | 639 A>G | N102S | 1 | 1 | Nickerson et al., 2010 |
| LL | French-British-CAf | 668 G>A | D112N | 1 | 1 | Nickerson et al., 2010 |
| F122 | Indian | 669 A>G | D112G | 1 | 1 | Orr et al., 2007 |
| H | American | 687 A>G | D118G | 1 | 1 | Weiss et al., 2008 |
| F105 | Spanish?-Canadian | 689 A>G | R119G | 1 | 1 | Orr et al., 2007 |
| 3 | African-American | 689 A>G | R119G | 1 | 1 | Yellore et al., 2007 |
| 2 | Egyptian | 695 C>G | L121V | 1 | 1 | Yellore et al., 2007 |
| BB3 | British | 695 C>G | L121F | 1 | 1 | Weiss et al., 2008 |
| O | American | 695 C>G | L121F | 1 | 1 | Weiss et al., 2008 |
| Case A | Saudi Arabian | 695 C>G | L121F | 1 | 1 | Al-Ghadeer et al, 2011 |
| AA | Native American | 699 T>A | V122E | 1 | 1 | Nickerson et al., 2010 |
| F1 | Finnish | 699 T>G | V122G | 1 | 1 | Nickerson et al., 2010 |
| K1 | German | 845 T>C | S171P | 1 | 2 | Weiss et al., 2008 |
| Case 1 | Chinese | 845 T>C | S171P | 1 | 2 | Mehta et al., 2009 |
| Case 1 | Japanese | 855 A>G | Y174C | 1 | 2 | Kobayashi et al, 2009 |
| F115 | Scottish | 858 C>T | T175I | 1 | 2 | Orr et al., 2007 |
| J | Hungarian-American | 858 C>T | T175I | 1 | 2 | Weiss et al., 2008 |
| T | American | 863 G>C | G177R | 1 | 2 | Weiss et al., 2007 |
| X | Chinese-Taiwan | 863 G>A | G177R | 1 | 2 | Weiss et al., 2008 |
| Z | Kosovar | 863 G>A | G177R | 1 | 2 | Weiss et al., 2008 |
| A | Swede-Finn | 864 G>A | G177E | 2 | 2 | This publication |
| B | Swede-Finn | 864 G>A | G177E | 2 | 2 | This publication |
| C | Swede-Finn | 864 G>A | G177E | 2 | 2 | This publication |
| D | Swede-Finn | 864 G>A | G177E | 2 | 2 | This publication |
| JJ | Japanese | 864 G>A | G177E | 2 | 2 | This publication |
| W | Turkish | 864 G>A | G177E | 2 | 2 | This publication |
| Case 4 | Japanese | 876 A>G | K181R | 2 | 2 | Kobayashi et al, 2009 |
| G | German-American | 890 G>A | G186R | 2 | 2 | Weiss et al., 2008 |
| EE | Chinese-Taiwan | 897 T>A | L188H | 2 | 2 | Nickerson et al., 2010 |
| F118 | Canadian | 1029 A>G | N232S | 2 | 3 | Orr et al., 2007 |
| Case 6 | Japanese | 1031 A>C | N233H | 2 | 3 | Kobayashi et al, 2009 |
| FF | African-American | 1042 C>G | D226E | 2 | 3 | Weiss et al., 2008 |
| SS | American | 1052 G>A | D240N | 2 | 3 | Weiss et al., 2010 |
Descending order 5’ to 3’ nucleotide number in the Reference Sequence.
Ethnicity is given if known, otherwise location of proband is listed.
Location of mutation in RefSeq NM_013319.2.
Predicted effect of genetic mutation on protein NP_037451.
Loop, see Figure 3B.
CA, Canadian Native American.
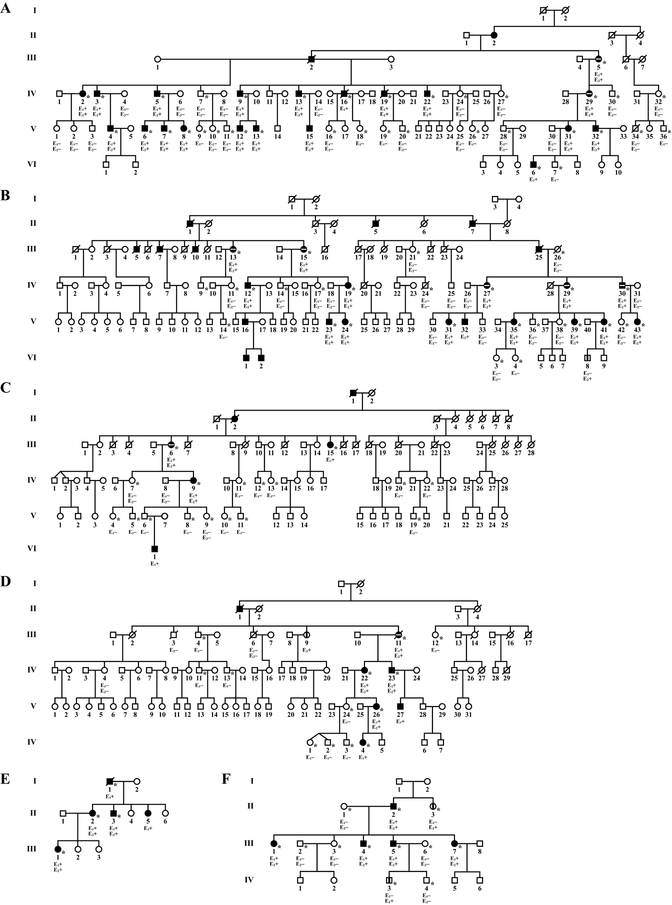
Participating members of the six SCD families (Fig. 2) were examined for association of the variant with disease (Supp. Table S1). Seventy family members were clinically evaluated as affected, 62 were clinically unaffected, and 98 individuals were sequenced including 47 affected and 51 unaffected individuals. Fifty-one individuals were mutation positive including all 47 affected and four unaffected subjects: VI-8, Family B; III-9, Family D; and II-3 and IV-3, Family W (Fig.2).
Figure 2.
SCD families with UBIAD1 p.G177E. A-F: SCD Swede-Finn families A-D, Japanese Family JJ, and Turkish Family W, respectively. Circles indicate females; squares, males; filled symbols, affected individuals; horizontal white line in a filled symbol, cornea transplant surgery (penetrating keratoplasty); vertical line, clinically or self-reported negative for SCD but positive for a p.G177E mutation; E1, clinical examination: (+) affected, (−) unaffected; E2, genetic analysis: (+) G>A alteration, (−) wild type; asterisks, examined by J.S.W.
A Founder Mutation in Swede-Finn SCD Families
The four Swede-Finn families had a common surname of Johnson, had ancestry from towns located within 60 kilometers of each other on the southwest coast of Finland [Weiss, 1992], and shared a common haplotype of chromosome 1p36 microsatellite markers and single nucleotide polymorphisms extending across the UBIAD1 locus [Theendakara et al., 2004], indicating a high probability that disease in these families occurred due to a founder mutation. Characteristics of SCD in affected individuals revealed that virtually all exhibited similar, previously described corneal changes dependent on age [Weiss, 1992; Weiss, 2007]. Within each family, however, there were individuals with significant differences in clinical manifestation of the disease. Corneal crystals were found in some but not all affected members and crystals were observed in only one eye of some patients. Thus, individuals with acrystalline (diffuse deposition) and crystalline forms of disease co-occurred within a family and differences in the disease phenotype may be due to additional, unknown genetic or environmental factors.
Protein Structure and Conservation of p.G177
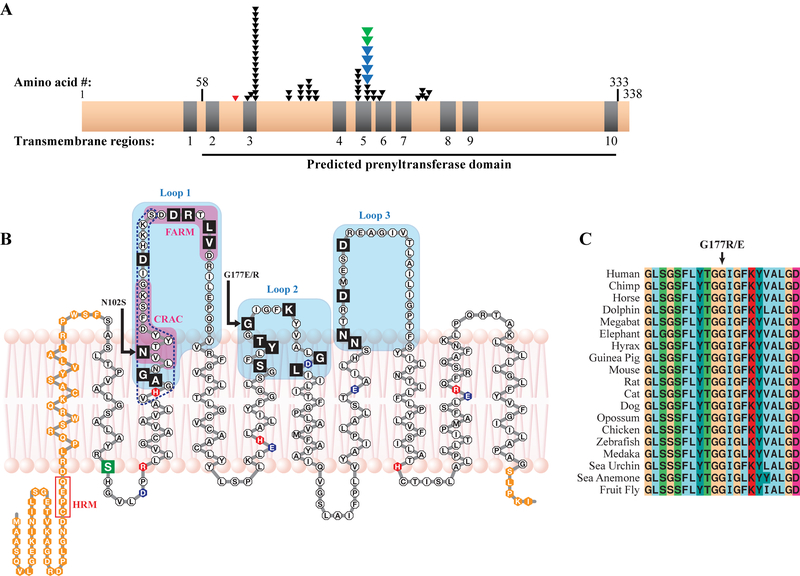
UBIAD1 has 10 transmembrane (TM) helices including two recently identified helices (TM 1 and 2) in the N-terminus of the protein [Fredericks et al., 2011] and all SCD mutations occur in the prenyltransferase domain that spans TM helices 2–10 (Fig. 3A). The new mutation in Swede-Finn, Japanese, and Turkish SCD families are indicated. UBIAD1 p.N102 and p.G177 are the two most frequently altered codons, in 17/51 (33%) and 9/51 (18%) of SCD families, respectively (Table 1). Both residues occupy similar positions in a lipid bilayer-embedded model of UBIAD1, near the membrane-aqueous interface where TM helices 3 and 5 emerge from the membrane into Loops 1 (p.N102) and 2 (p.G177), respectively (Fig. 3B). The frequency of familial mutations declines with distance from p.N102 towards the C-terminus; 30 families had alterations in Loop 1 (63%), 14 in Loop 2 (29%), and 4 in Loop 3 (8%). Mutation of p.G177 to both positive (arginine [R], p.G177R, SCD families T, X, and Z, Table 1) and negative residues (glutamic acid, SCD families A-D, JJ, and W) suggests loss of uncharged glycine may be critical for disease. This appears to be similar to other codons altered by multiple mutations where loss of the native residue appears to be critical for disease, i.e. p.D112N/G, p.L121F/V, and p.V122E/G [Nickerson et al., 2010].
Figure 3.
SCD mutations and the UBIAD1 protein. A: Distribution of SCD family mutations on a linear protein model. Nine of ten predicted TM helices (grey) are in a predicted prenyltransferase domain (horizontal line, bottom) containing 19/19 residues altered in SCD families (one arrow per family), including p.G177E mutations in Swede-Finn (blue arrows) and Japanese and Turkish (green arrows) SCD families. A germline variant, p.S75F, is shown (red arrow). B: UBIAD1 in a lipid bilayer. SCD mutations (black boxes) occur in Loops 1–3 (shaded), including mutation hotspots asparagine 102 and glycine 177 (arrows). A predicted active site based on E.coli menA (dotted line, Weiss et al., 2008) and CRAC and FARM domains (Fredericks et al., 2011) are indicated. Acidic (blue), basic (red), a p.S75F SNP (green), and residues outside the predicted prenyltransferase domain (orange) are indicated. C: Complete conservation of codon p.G177 in 19 species.
Protein homology indicated p.G177 was completely conserved in 19 species from sea urchin to human (Fig. 3C) and was located within Loop 2 on the same side of the membrane as all SCD mutations so far characterized [Weiss et al., 2008]. The presence of a Cholesterol Recognition Amino Acid Consensus (CRAC) sequence and a First Aspartate-Rich Motif (FARM) indicate likely cholesterol and farnesyl binding sites, respectively [Fredericks et al., 2011; Epand, 2006]. A previously identified putative ligand/polyprenyldiphosphate active site based upon E.coli menA is also indicated (dotted line, Fig. 3C) [Weiss et al., 2008].
SCD Mutation of UBIAD1 Impairs MK-4 Synthesis
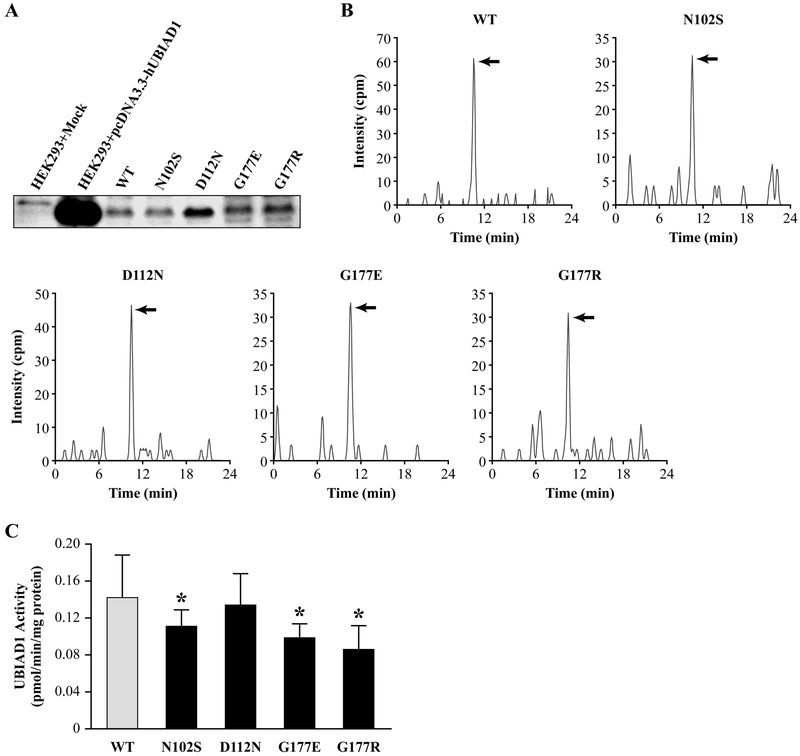
We examined the effect of SCD mutations on UBIAD1 catalyzed MK-4 synthesis to determine whether there was a detectable biochemical impact of these alterations. During protein expression studies, we observed proteolytic cleavage of the p.G177E mutant protein (but not wt protein) in the microsomal fraction from baculovirus-infected insect Spodoptera frugiperda (Sf-9) cells (unpublished data) and sought to develop an alternative to the published method [Nakagawa et al., 2010]. Intact protein was observed in a wide variety of mammalian cell lines, mouse tissues, and microsomes isolated from SCD patient B cell lysates, confirming specificity of the anti-SCD antibody [Nakagawa et al., 2010] and absence of proteolytic cleavage of p.G177E mutant UBIAD1 outside of insect cells (Fig. 4A). Therefore, we analyzed normalized human microsomal fractions from a donor not affected by SCD and SCD patient B cell lysates as described in the Methods and Materials. Measurements of deuterium-labeled MK-4 (MK-4-d7) revealed that p.N102S, p.D112N, p.G177E, and p.G177R mutation of UBIAD1 lowered MK-4 biosynthetic activity converting K3-d8 to MK-4-d7 by 22%, 6%, 30%, and 39%, respectively, compared to wt UBIAD1, and p.N102S and both alterations of p.G177 were significant (p<0.05, Fig. 4).
Figure 4.
MK-4 biosynthesis by wt and SCD mutant UBIAD1. A: Endogenous and transiently expressed UBIAD1 in HEK293 cells, and WT and SCD mutant UBIAD1 in cell lysates from unaffected (normal) and SCD mutant subjects. Protein amounts were normalized using an UBIAD1-specific antibody. B: MRM chromatograms from LC-APCI-MS/MS analysis of MK-4-d7 (arrows) converted from PK-d7 or K3-d8 in the microsomal fractions of cell lysates from WT and SCD mutant subjects. C: Quantitation of MK-4-d7 synthesis. Values are means ± SD (n=3) and those significantly different from wt UBIAD1 activity are indicated (*p<0.05, Student’s t-test).
Molecular Modeling Indicates Conformational Changes Due to Mutation of p.G177
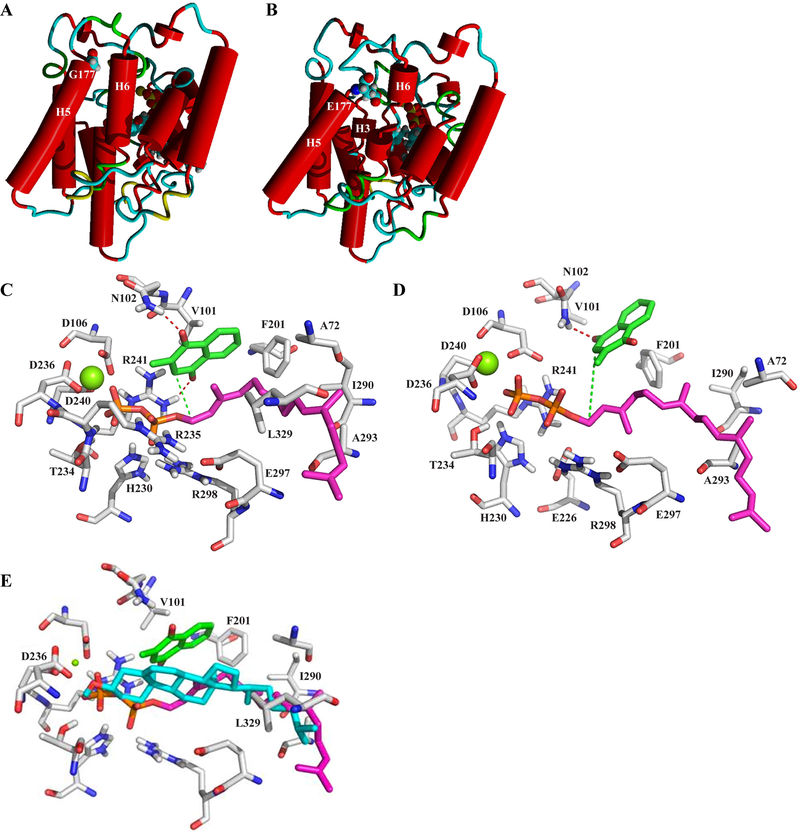
Molecular modeling of UBIAD1 to assess structure-function relationships of wt and SCD mutant UBIAD1 has been used to evaluate protein structure and substrate binding [Nickerson et al., 2010]. For example, docking simulations led to the prediction that the function of UBIAD1 might be related to vitamin K synthesis, which was recently experimentally confirmed [Nakagawa et al., 2010] and supported the robustness of the model. Moreover, in silico alterations to active site residues provided effective predictions of biochemical effects caused by SCD mutations such as p.N102S [Weiss et al., 2007] and p.D240N [Weiss et al., 2010], i.e. native residues recognized menadione (p.N102) or a divalent magnesium cation (p.D240) in the active site and allowed entry of the diphosphate moiety of geranylgeranyldiphosphate (GGPP) [Nickerson et al., 2010].
The structure of wt UBIAD1 shows p.G177 near the end of TM helix 5 when substrates menadione (green) and GGPP (magenta) are docked to simulate formation of MK-4 (Fig. 5A). The p.G177E mutant protein (Fig. 5B) shows striking structural perturbations in the arrangement of TM helices. Molecular dynamics simulations after replacement of glycine 177 with glutamic acid introduces steric hindrances causing a partial destruction of helix 6 that altered the orientation of helix 3, which includes the active site residue, p.N102. This is indicated by both the simulated annealing dynamics refinement as well as in a 6 ns molecular dynamics simulation of p.G177E mutant and wt protein in a membrane environment (Supp. Fig. S3). After 3 ns simulation time considerably more alterations of the tertiary structure of the mutant protein than the wt are observed (increased separation of dotted and solid lines). Inspection of structural alterations to the active site show for those residues which are closer to the p.G177E mutation, such as p.N102, the average root-mean-square deviation (RMSD) is 1.92 Å in the wt protein and 3.70 Å in the mutant; and for serine 61 the wt RMSD is 1.22 and the p.G177E mutant protein RMSD is 4.62. Residues of other helices (further away from p.G177E) such as arginine 298 and arginine 235 are almost unchanged in their RMSD.
Figure 5.
Molecular modeling and substrate docking simulations. A: 3D model of the native protein showing p.G177 at the top of helix 5 in loop 2 and docked substrates, GGPP and menadione, partly visible behind and to the right of helix 6. Mg2+ required to activate the diphosphate for prenyl transfer is shown (green ball). B: Structural changes induced by a p.G177E in silico substitution. C, D: Details of altered interactions (dotted lines) between UBIAD1 active site residues and substrates menadione (green) and GGPP (magenta) in native (panel C) and p.G177E (panel D) proteins. Mg2+ (green ball), diphosphate moiety of GGPP (red and orange). E: Overlap of GGPP and cholesterol in the UBIAD1 binding cleft. GGPP (magenta) and menaquinone (green) or cholesterol (cyan) was independently docked to UBIAD1 and substrate positions were compared. Binding sites overlapped as shown, indicating potential competitive occupancy of the substrate binding cleft. UBIAD1 carbon residues (grey), oxygen (red), and the GGPP diphosphate (red/orange) are highlighted. Orientation of substrates is similar to Fig. 5C and D.
The p.G177E mutation causes structural changes to the active site clearly visible at higher magnification (Fig. 5C, D). We mapped out charged and uncharged UBIAD1 residues interacting with diphosphate and geranylgeranyl moieties, respectively, of GGPP and a complexed Mg2+ cation. Hydrogen bonds (red dotted lines) are predicted between menadione and wt UBIAD1 residues, p.N102 and p.R241, and p.D240 (an SCD mutated residue) interacted with GGPP (Fig. 5C). The catalytic reaction is favored by proximity (3.8 Å) between the C1-atom of GGPP and the prenylation position of menadione (green dotted line). Upon p.G177E substitution (Fig. 5D), there is a loss of the hydrogen bond between menadione and p.R241, and the distance between GGPP and menadione increased to 5.2 Å, preventing close contact of substrates required for the reaction to proceed. The model predicted deleterious effects due to a p.G177E change that are sufficient to explain the experimentally-observed loss of vitamin K biosynthesis.
UBIAD1 Binds HMGCR and SOAT1
Diagnosis of SCD is based on characteristic findings of abnormal deposition of cholesterol and lipids in the cornea. It is unclear how decreased synthesis of MK-4 due to mutation of UBIAD1 connects to cholesterol deposition in SCD. We sought to identify UBIAD1 binding partners to provide additional details about protein function that could be connected to a signaling pathway or some aspect of cellular physiology. We performed yeast-two hybrid (Y2H) screens of human prostate cancer, and lung and kidney tissue cDNA prey libraries since UBIAD1 is widely expressed [McGarvey et al., 2001], including in these tissues according to expressed sequence tags in the Unigene database (www.ncbi.nlm.nih.gov/unigene). We used full length (amino acid (aa) 1–338) and the C-terminal region (aa 102–314) of UBIAD1 as bait fragments.
Multiple, independent cDNA clones were isolated as prey fragments from 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), sterol O-acyltransferase (SOAT1), and vascular endothelial growth factor A (VEGF-A) (Table 2). HMGCR is an enzyme catalyzing a rate-limiting step in cholesterol biosynthesis and is the target of statins (24–26), and SOAT1 (a.k.a. acyl-coenzyme A:cholesterol acyltransferase, ACAT) esterifies cholesterol for intracellular storage [Lu et al., 2011; Nissen et al., 2006; Peters et al., 2011; Chang et al., 1997]. VEGF-A is a secreted endothelial cell mitogen that promotes cell migration and angiogenesis [Murphy and Fitzgerald, 2001].
Table 2.
Identification of Proteins that Interact with UBIAD1 by Y2H
| Bait | Prey | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Round 1: | |||||||||||||
| Entrez Gene | GenBank | GenProt | 5’ aa | 3’ aa | Prey name | Entrez Gene | GenBank | GenProt | 5’ aa | 3’ aa | Prey library | ||
| UBIAD1 | 29914 | NM_013319 | NP_037451 | 1 | 338 | HMGCR (888 aa) | 3-Hydroxy-3-methyl-glutaryl-Coenzyme A reductase | 3156 | NM_000859 | NP_000850 | −18 | 285 | Prostate Cancer |
| 1 | 338 | HMGCR | −25 | 359 | Prostate Cancer | ||||||||
| 1 | 338 | HMGCR | −11 | 361 | Prostate Cancer | ||||||||
| 1 | 338 | HMGCR | −22 | 390 | Prostate Cancer | ||||||||
| 1 | 338 | HMGCR | −30 | 391 | Prostate Cancer | ||||||||
| 1 | 338 | HMGCR | −22 | 411 | Prostate Cancer | ||||||||
| 1 | 338 | HMGCR | −22 | 440 | Prostate Cancer | ||||||||
| 1 | 338 | HMGCR | −5 | 443 | Prostate Cancer | ||||||||
| 1 | 338 | HMGCR | −20 | 495 | Prostate Cancer | ||||||||
| 1 | 133 | SOAT1 (550 aa) | Sterol O-acyltransferase 1 | 6646 | NM_003101 | NP_003092 | −1 | 305 | Lung | ||||
| 1 | 133 | SOAT1 | −36 | 330 | Prostate Cancer | ||||||||
| 102 | 314 | VEGFA (412 aa) | Vascular endothelial growth factor A, isoform a | 7422 | NM_001025366 | NP_001020537 | 77 | 315 | Lung | ||||
| 102 | 338 | VEGFA | 73 | 314 | Kidney | ||||||||
| 102 | 338 | VEGFA (327 aa) | Vascular endothelial growth factor A, isoform f; VEGF121 | 7422 | NM_001025370 | NP_001020541 | 66 | 325 | Prostate Cancer | ||||
| Round 2: | |||||||||||||
| UBIAD1 | 29914 | NM_013319 | NP_037451 | 1 | 338 | HMGCR | 3156 | NM_000859 | NP_000850 | −14 | 514 | Lung | |
| 1 | 338 | HMGCR | −23 | 344 | Prostate Cancer | ||||||||
| 1 | 338 | HMGCR | −25 | 361 | Prostate Cancer | ||||||||
| 1 | 338 | HMGCR | −20 | 391 | Prostate Cancer | ||||||||
| 1 | 338 | HMGCR | −15 | 485 | Prostate Cancer | ||||||||
| 102 | 314 | HMGCR | −20 | 361 | Prostate Cancer | ||||||||
| 1 | 133 | SOAT1 | 6646 | NM_003101 | NP_003092 | 62 | 309 | Prostate Cancer | |||||
Interactors were identified in 2 independent rounds of screening or were independently identified from multiple libraries. All entries represent independently-isolated interacting clones. aa, amino acids in the isoform examined.
Overlapping HMGCR clones were isolated with a minimal prey fragment containing HMGCR residues 1–285, indicating UBIAD1 may be specifically interacting with a Sterol Sensing 5 Transmembrane BOX motif (IPR000731) encompassing residues 60–218. The motif has been observed in proteins with roles in cholesterol biosynthesis and cholesterol-linked signaling [Epand, 2006; Ikonen, 2008]. The minimum region of UBIAD1 observed to interact with HMGCR was a single clone with residues 102–314 encompassing 17/19 UBIAD1 residues mutated in SCD and most of the prenyltransferase domain. Interaction between UBIAD1 and SOAT1 reinforced the observation of the UBIAD1-HMGCR interaction, and increased the significance of a previously described interaction between UBIAD1 and apolipoprotein E (apoE), a cholesterol transport protein [McGarvey et al., 2005; Fredericks et al., 2011]. To insure against false positive interactions, bait and prey clones were reversed and plated under stringent conditions. The interactions were confirmed by directed Y2H liquid culture assays resulting in high β-galactosidase activities (>300 fold over background, data not shown). Prey did not interact with a panel of non-specific baits (False Positive Specificity Assay, ProNet [Garrus et al., 2001]) and UBIAD1 did not interact with other cholesterol-binding, membrane proteins that were present during screening (data not shown). Y2H cDNA prey libraries were not available from cornea tissue and additional studies of UBIAD1 protein binding partners in cornea are planned.
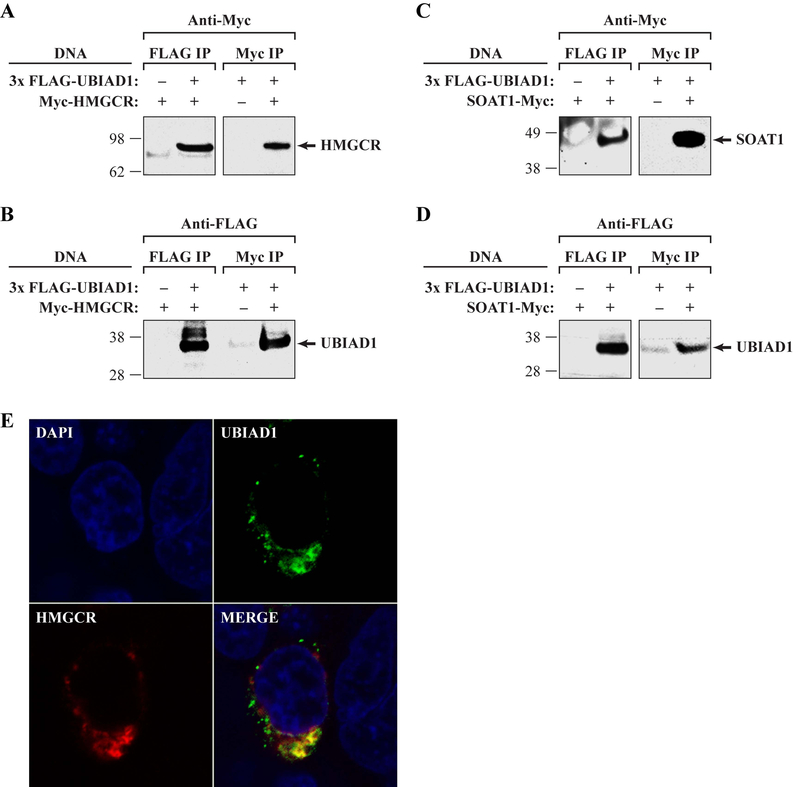
We confirmed the interaction of UBIAD1 with HMGCR and SOAT1 by immunoprecipitation (IP) of expressed, tagged proteins in HEK293T cells (Fig. 6), similarly to validation of the UBIAD1-apoE interaction reported by McGarvey et al. (2005). In two independent experiments, an N-terminal 3xFlag tagged UBIAD1 co-precipitated HMGCR and SOAT1. These interactions were not detected using an anti-Flag antibody alone. Furthermore, two independent experiments showed that reverse IP using N-terminal Myc tagged HMGCR and SOAT1 were able to co-precipitate 3xFlag tagged UBIAD1. Further support for the interaction was obtained by confocal microscopy co-localization of expressed tagged UBIAD1 and HMGCR proteins that revealed substantial overlap of signal in HEK293T cells (Fig. 6E).
Figure 6.
Confirmation of HMGCR and SOAT1 interactions with UBIAD1. A,C: Anti-Myc to examine Flag and Myc IPs after transient transfection with indicated DNAs (left). The expected HMGCR molecular weight (MW, 97 kDa) and markers are indicated (left). B,D: Antibodies against a Flag tag were used to examine Flag and Myc IPs after transient transfection with the indicated DNAs (left). The expected UBIAD1 MW was 37 kDa. E: HEK293T cells expressing both 3xFlag-UBIAD1 and Myc-HMGCR were stained with mouse anti-Flag and rabbit Anti-Myc antibodies and secondary antibodies conjugated to either AlexaFluor488 (anti-mouse) or AlexaFluor568 (anti-rabbit). Representative images are shown; untransfected cells showed no signal (unpublished data).
Molecular Modeling Indicates Cholesterol Binds to UBIAD1
Substrate docking simulations show cholesterol binding to UBIAD1 with high affinity as indicated by a high GOLD docking score of 49.8, and binding occurred in the same binding cleft as GGPP, a UBIAD1 substrate for MK-4 synthesis (Fig. 5E). As shown in Fig. 5E, the hydrophobic cholesterol moiety nicely overlaps with the same hydrophobic binding site of geranylgeranyl in GGPP, whereas the oxygen atom of the hydroxyl group on cholesterol forms attractive electrostatic interactions with the divalent magnesium ion in the same region where the GGPP diphosphate binds.
Discussion
This study identified a novel DNA mutation, n.864 G>A in transcript NM_013319.2, that introduced a missense amino acid substitution, p.G177E, in the UBIAD1 prenyltransferase in 51 members of six families with SCD, including four, six-generation Swede-Finn families originating from a common region in southwest Finland bordering the Bay of Bothnia (Table 1, Supp. Table S1) [Weiss et al., 1992]. The mutation is likely causal to disease as it segregated with clinically diagnosed SCD in 47/47 individuals, was not observed in 300 control individuals, and altered a codon previously linked to disease by a p.G177R mutation observed in SCD families, T, X, and Z, from Taiwan, Kosovo, and the United States, respectively (Table 1).
The Swede-Finn SCD families are historically significant as family members first identified in Massachusetts (by J.S.W.) were traced through genealogic records to a 60 kilometer region in Finland. A review of family photographs from the late 1800s revealed affected individuals and documented occurrence of the disease three decades before the seminal description of SCD in 1924 [Van Went and Wibaut, 1924]. The n.864 G>A alteration likely arose once as a founder event in an ancestor of the current Swede-Finn families as the families shared a common geographic region of origin, a common surname (Johnson) among descendants, and a common haplotype across the UBIAD1 locus [Shearman et al., 1996; Theendakara et al., 2004]. The mutation likely occurred independently in SCD families JJ and W, from Japan and Turkey [Köksal et al., 2004], respectively, due to distinct ethnicity and geography.
Four individuals that carried a p.G177E alteration were clinically unaffected. Subject VI-8 in Family B was young at the time of the clinical exam, which was likely prior to the onset of corneal cholesterol deposition. Subject III-9 in Family D was not examined by an ophthalmologist and self-reported as unaffected but this diagnosis may have been due to a lack of awareness of more subtle indications of SCD. Two unaffected patients (II-3 and IV-3) in Family W may have been misdiagnosed due to previously described difficulties of detecting SCD in patients with non-crystalline forms of the disease [Weiss et al., 2007]. Alternately, the disease may have low penetrance in some individuals, which might be expected since highly variable corneal deposition was observed between members of the same family and also between corneas of the same patient.
UBIAD1 p.N102 and p.G177 codons are the most frequently altered codons and are altered in 17/51 (33%) and 9/51 (18%) of SCD families. Mutant p.N102S, p.G177E, and p.G177R UBIAD1 significantly decreased MK-4 biosynthetic activity by 22%, 30%, and 39%, respectively, compared to wt enzyme (p<0.05, Fig. 4). Though not significant, the D112N SCD mutation also impaired MK-4 synthesis (a 6% decrease). The decreased catalytic activity of the newly described G177E mutation is effectively explained by molecular modeling and substrate docking simulations that showed substitution of the native glycine residue (48 van der Waals volume) with glutamic acid (109 van der Waals volume) at position 177 destabilized transmembrane helices and increased hydrogen bonding from 3.8 to 5.2Å between docked substrates GGPP and menadione (Fig. 5). This data indicates that decreased MK-4 synthesis by SCD-mutant UBIAD1 is a consistent biochemical defect associated with the accumulation of cholesterol observed in SCD patient corneas.
Identification of HMGCR and SOAT1, key enzymes for cholesterol biosynthesis [Jo and DeBose-Boyd, 2010; Ikonen, 2008; Chang et al., 2006] and storage [Lu et al., 2011; Nissen et al., 2006; Peters et al., 2011; Chang et al., 1997], respectively, as UBIAD1 binding partners (Fig. 6), in addition to a previously described interaction between UBIAD1 and the cholesterol transport protein, apoE [Fredericks et al., 2011; McGarvey et al., 2005], indicates complex molecular links between enzymes catalyzing vitamin K synthesis and cholesterol metabolism. A multi-protein complex containing UBIAD1 and HMGCR, SOAT1, or apoE may explain the dominant inheritance pattern observed in SCD if a mutant UBIAD1 subunit impairs the function of the complex even in the presence of native UBIAD1. Physical interaction between these proteins may modify enzyme function, promote post-translational modifications of complex subunits, improve catalytic efficiency through proximity of enzymes and concentration of substrates, or allow regulatory control by substrates or reaction products of one enzyme on other enzymes of the complex [Conrado et al., 2008; Jørgensen et al., 2005].
Substrate docking simulations showed that cholesterol binds to UBIAD1 with high affinity in the same binding cleft as GGPP, indicating potential competition for UBIAD1 by these molecules and a potential regulatory role for HMGCR and cholesterol on UBIAD1 biosynthesis of MK-4 (Fig. 5E). UBIAD1 or its product, MK-4, may also influence cholesterol synthesis, storage, or transport. These predictions remain to be tested experimentally. MK-4 has been shown to significantly decrease serum cholesterol and triglycerides in rats [Sogabe et al., 2011] and vitamin K-dependent clotting factors have been correlated with serum cholesterol and triglyceride levels [Constantino et al., 1977; Hoffman et al., 1994] but a detailed mechanism for these effects is currently not known.
The data supports several possible mechanisms whereby SCD mutation of UBIAD1 could cause cholesterol accumulation in the cornea. MK-4 could alter gene expression through physical interactions with the orphan nuclear receptor (SXR, a.k.a. nuclear receptor subfamily NR1l2) and its ortholog, PXR, which in turn, regulate the nuclear receptor, liver X receptor (LXR) by binding response elements in promoters [Tabb et al., 2003; Fredericks et al., 2011; reviewed in Calkin and Tontonoz, 2012]. Treatment of osteosarcoma and osteoblastic cells with vitamin K2 resulted in altered expression of 21 genes [Tabb et al., 2003; Ichikawa et al., 2006; 2007] including increased expression of five genes encoding proteins with known associations to cholesterol homeostasis, ABCB1/MDR1, AKR1C1, AGTR1, CD24, and CYP3A4 (Ingenuity Pathway Analysis, Ingenuity, Redwood City, CA). Decreased MK-4 due to SCD mutation of UBIAD1 may down-regulate transcription of these genes to cause cholesterol accumulation. Cholesterol esterification may be affected as the angiotensin II receptor, type 1 (AGTR1) transcriptionally regulates SOAT1 and overexpression of ABCB1 increases cholesterol esterification by trafficking plasma membrane cholesterol to endoplasmic reticulum [Luker et al., 1999; Kanome et al., 2008]. Of additional interest in cornea and bone tissues, MK-4 up-regulated transcription of seven genes involved in collagen and extracellular matrix deposition (TSK, ELN, MATN2, COL6A1, TNFRSF11B, SPP1, and MGP; Ingenuity Pathway Analysis) though the significance of this is currently unclear.
Secondly, SCD mutations could affect interactions between UBIAD1 and apoE affecting cholesterol entry or efflux from cornea cells [Weiss et al., 2007; Fredericks et al. 2011]. Cornea have been shown to actively take up and store cholesterol [Burns et al., 1978] and apoE has been implicated in both delivery of cholesterol to cells and cholesterol efflux through reverse cholesterol transport [Kruth et al., 1994; Zhang et al., 1996; Fredericks et al. 2011]. Intriguingly, apoE also participates in delivery of phylloquinone (vitamin K1) to cells, which is a substrate for UBIAD1 synthesis of MK-4 [Lamon-Fava et al., 1998; Newman et al., 2002; reviewed in Shearer and Newman, 2008]. Lamon-Fava et al. (1998) suggest that phylloquinone in high density lipoproteins might be exchanged for cholesteryl ester thus providing an extracellular parallel to the intracellular interactions between UBIAD1 and HMGCR and SOAT1.
Finally, SCD mutant UBIAD1 or decreased MK-4 could alter highly regulated endoplasmic reticulum-associated degradation (ERAD) of HMGCR, which is cell specific and complex [reviewed in Jo and DeBose-Boyd, 2010]. A perturbation in ERAD of HMGCR due to altered SCD mutant UBIAD1-HMGCR interactions could reduce HMGCR degradation thereby promoting continued cholesterol synthesis and accumulation. A role for UBIAD1 in HMGCR degradation may be relevant to disparate results of UBIAD1 localization to both mitochondria and endoplasmic reticulum [Nickerson et al., 2010; Nakagawa et al., 2010]. Additional experiments to test these predictions are in progress.
Vitamin K was originally identified by experiments depleting dietary cholesterol in chickens resulting in hemorrhages and uncontrolled bleeding. This was due to co-depletion of dietary vitamin K [Dam, 1935] which was thought to be obtained solely from plants and gut microbiota. The current study indicates endogenous, intracellular MK-4 produced by UBIAD1 has a physiologic role in maintaining corneal health and visual acuity that is distinct from the role of dietary vitamin K in blood clotting. Further delineation of the physiologic role of endogenous MK-4, such as its participation in the electron transport chain in mitochondria [Vos et al., 2012], remain to be described. Lastly, the observations of decreased MK-4 synthesis by SCD mutant UBIAD1 suggests MK-4 supplements, potentially delivered by topical administration to the eyes, might be useful as a therapy to treat opacified cornea, or at least the continued accumulation of cholesterol and lipids, that are observed in SCD.
Supplementary Material
Supp. Table S1. Summary of analysis of SCD families with a p.G177E alteration
Supp. Fig. S1. UBIAD1 transcripts and PCR assay amplicons
Supp. Fig. S2. Ramachandran plots of wt and G177E mutant UBIAD1
Supp. Fig. S3. Root-mean-square deviation (RMSD) in Å of Cα-atoms from a 6 ns molecular dynamics simulation.
Supp. Fig. S4. Structures of molecules described in the article
Acknowledgements
We thank participating physicians and SCD patients and their families, and appreciate critical review and discussion by Drs. Berton Zbar and Timothy Veenstra (NCI), Richard Snyder (University of Florida), Tara Moore (University of Ulster, UK), and William Fredericks (University of Pennsylvania), and by members of the Dean laboratory. We benefited from technical assistance from Drs. Lawrence R. Sternberg and Luis Rodriguez, and Mary Thompson, David W. Wells, Andrew C. Warner, Lisa Garland, Christina Ruiz, and Jiro Wada. This work was supported in part by grants from the Midwest Eye Banks and Research to Prevent Blindness [to J.S.W.], by the National Cancer Institute, National Institutes of Health [Contract No. HHSN261200800001E], and the National Cancer Institute Intramural Research Program [to M.L.N., B.N.K., and M.D.]. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare there are no conflicts of interest to report.
Grant Sponsor: Funding was obtained in part from the Midwest Eye Banks and Research to Prevent Blindness [to J.S.W.], and from the National Cancer Institute, National Institutes of Health [Contract No. HHSN261200800001E] and the National Cancer Institute Intramural Research Program [to M.L.N., B.N.K., and M.D.]
References
- Al-Ghadeer H, Mohamed JY, Khan AO. 2011. Schnyder corneal dystrophy in a Saudi Arabian family with heterozygous UBIAD1 mutation (p.L121F). Middle East Afr J Ophthalmol 18:61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräuer L, Brandt W, Schulze D, Zakharova S, Wessjohann L. 2008. A structural model of the membrane-bound aromatic prenyltransferase UbiA from E. coli. Chembiochem 9:982–992. [DOI] [PubMed] [Google Scholar]
- Calkin AC and Tontonaz P. 2012. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat Rev Molec Cell Biol 13:213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TY, Chang CC, Cheng D. 1997. Acyl-coenzyme A:cholesterol acyltransferase. Annu Rev Biochem 66:613–638. [DOI] [PubMed] [Google Scholar]
- Chang TY, Chang CC, Ohgami N, Yamauchi Y. 2006. Cholesterol sensing, trafficking, and esterification. Annu Rev Cell Dev Biol 22:129–157. [DOI] [PubMed] [Google Scholar]
- Colovos C, Yeates TO. 1993. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 2:1511–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrado RJ, Varner JD, DeLisa MP. 2008. Engineering the spatial organization of metabolic enzymes: mimicking nature’s synergy. Curr Opin Biotechnol 19:492–499. [DOI] [PubMed] [Google Scholar]
- Constantino M, Merskey C, Kudzma DJ, Zucker MB. 1977. Increased activity of vitamin K-dependent clotting factors in human hyperlipoproteinaemia-association with cholesterol and triglyceride levels. Thromb Haemost 38:465–474. [PubMed] [Google Scholar]
- Dam H 1935. The antihaemorrhagic vitamin of the chick: Occurrence and chemical nature. Nature 135:652–653. [Google Scholar]
- Epand RM. 2006. Cholesterol and the interaction of proteins with membrane domains. Prog Lipid Res 45:279–294. [DOI] [PubMed] [Google Scholar]
- Fredericks WJ, McGarvey T, Wang H, Lal P, Puthiyaveettil R, Tomaszewski J, Sepulveda J, Labelle E, Weiss JS, Nickerson ML, Kruth HS, Brandt W, Wessjohann LA, Malkowicz SB. 2011. The bladder tumor suppressor protein TERE1 (UBIAD1) modulates cell cholesterol: Implications for tumor progression. DNA Cell Biol 11:851–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Côté M, Rich RL, Myszka DG, Sundquist WI. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55–65. [DOI] [PubMed] [Google Scholar]
- Gaynor MG, Zhang W, Weiss JS, Skarlatos SI, Rodrigues MM, Kruth HS. 1996. Accumulation of HDL apolipoproteins accompanies abnormal cholesterol accumulation in Schnyder’s corneal dystrophy. Arterioscler Thromb Vasc Biol 16: 992–999. [DOI] [PubMed] [Google Scholar]
- Hartshorn MJ, Verdonk ML, Chessari G, Brewerton SC, Mooij WTM, Mortenson PN, Murray CW. 2007. Diverse, high-quality test set for the validation of protein-ligand docking performance. J Med Chem 50:726–741. [DOI] [PubMed] [Google Scholar]
- Hoffman CJ, Lawson WE, Miller RH, Hultin MB. 1994. Correlation of vitamin K-dependent clotting factors with cholesterol and triglycerides in healthy young adults. Arterioscler Thromb 14:1737–1740. [DOI] [PubMed] [Google Scholar]
- Ichikawa T, Horie-Inoue K, Ikeda K, Blumberg B, Inoue S. 2006. Steroid and xenobiotic receptor SXR mediates vitamin K2-activated transcription of extracellular matrix-related genes and collagen accumulation in osteoblastic cells. J Biol Chem 281:16927–16934. [DOI] [PubMed] [Google Scholar]
- Ichikawa T, Horie-Inoue K, Ikeda K, Blumberg B, Inoue S. 2007. Vitamin K2 induces phosphorylation of protein kinase A and expression of novel target genes in osteoblastic cells. J Molec Endocrin 39:239–247. [DOI] [PubMed] [Google Scholar]
- Ikonen E 2008. Cellular cholesterol trafficking and compartmentalization. Nat Rev Mol Cell Biol 9:125–138. [DOI] [PubMed] [Google Scholar]
- Jing Y, Liu C, Xu J, Wang L. 2009. A novel UBIAD1 mutation identified in a Chinese family with Schnyder crystalline corneal dystrophy. Mol Vis 15:1463–1469. [PMC free article] [PubMed] [Google Scholar]
- Jo Y, DeBose-Boyd RA. 2010. Control of cholesterol synthesis through regulated ER-associated degradation of HMG CoA Reductase. Crit Rev Biochem Mol Biol 45:185–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen K, Rasmussen AV, Morant M, Nielsen AH, Bjarnholt N, Zagrobelny M, Bak S, Møller BL. 2005. Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr Opin Plant Biol 8:280–291. [DOI] [PubMed] [Google Scholar]
- Kanome T, Watanabe T, Nishio K, Takahashi K, Hongo S, Miyazaki A. 2008. Angiotensin II upregulates acyl-CoA:cholesterol acyltransferase-1 via the angiotensin II Type 1 receptor in human monocyte-macrophages. Hypertens Res 31:1801–1810. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Fujiki K, Murakami A, Sugiyama K. 2009. In vivo laser confocal microscopy findings and mutational analysis for Schnyder’s crystalline corneal dystrophy. Ophthalmology 116:1029–1037. [DOI] [PubMed] [Google Scholar]
- Köksal M, Kargi S, Gürelik G, Akata F. 2004. Phototherapeutic keratectomy in Schnyder crystalline corneal dystrophy. Cornea 23:311–313. [DOI] [PubMed] [Google Scholar]
- Krieger E, Joo K, Lee J, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K. 2009. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins 77 suppl 9:114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruth HS, Skarlatos SI, Gaynor PM, Gamble W. 1994. Production of cholesterol-enriched nascent high density lipoproteins by human monocytederived macrophages is a mechanism that contributes to macrophage cholesterol efflux. J Biol Chem 269:24511–24518. [PubMed] [Google Scholar]
- Lamon-Fava S, Sadowski JA, Davidson KW, O’Brien ME, McNamara JR, Schaefer EJ. 1998. Plasma lipoproteins as carriers of phylloquinone (vitamin K1) in humans. Am J Clin Nutr 67:1226–1231. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW Moss DS, Thornton JM. 1993. Procheck - a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. [Google Scholar]
- Lu Z, Yuan Z, Miyoshi T, Wang Q, Su Z, Chang CC, Shi W. 2011. Identification of SOAT1 as a quantitative trait locus gene on mouse chromosome 1 contributing to hyperlipidemia. PLoS ONE 6:e25344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker GD, Nilsson KR, Covey DF, Piwnica-Worms D. 1999. Multidrug resistance (MDR1) P-glycoprotein enhances esterification of plasma membrane cholesterol. J Biol Chem 12:6979–6991. [DOI] [PubMed] [Google Scholar]
- McGarvey TW, Nguyen T, Tomaszewski JE, Monson FC, Malkowicz SB. 2001. Isolation and characterization of the TERE1 gene, a gene down-regulated in transitional cell carcinoma of the bladder. Oncogene 20:1042–1051. [DOI] [PubMed] [Google Scholar]
- McGarvey TW, Nguyen TB, Malkowicz SB. 2005. An interaction between apolipoprotein E and TERE1 with a possible association with bladder tumor formation. J Cell Biochem 95:419–428. [DOI] [PubMed] [Google Scholar]
- Mehta JS, Vithana EN, Venkataraman D, Venkatraman A, Yong VH, Aung T, Tan DT. 2009. Surgical management and genetic analysis of a Chinese family with the S171P mutation in the UBIAD1 gene, the gene for Schnyder corneal dystrophy. Br J Ophthalmol 93:926–931. [DOI] [PubMed] [Google Scholar]
- Murphy JF, Fitzgerald DJ. 2001. Vascular endothelial growth factor induces cyclooxygenase-dependent proliferation of endothelial cells via the VEGF-2 receptor. FASEB J 15:1667–1669. [DOI] [PubMed] [Google Scholar]
- Nakagawa K, Hirota Y, Sawada N, Yuge N, Watanabe M, Uchino Y, Okuda N, Shimomura Y, Suhara Y, Okano T. 2010. Identification of UBIAD1 as a novel human menaquinone-4 biosynthetic enzyme. Nature 468:117–121. [DOI] [PubMed] [Google Scholar]
- Newman P, Bonello F, Wierzbicki AS, Lumb P, Savidge GF, Shearer MJ. 2002. The uptake of lipoprotein-borne phylloquinone (vitamin K1) by osteoblasts and osteoblast-like cells: Role of heparin sulfate proteoglycans and apolipoprotein E. J Bone Miner Res 17:426–433. [DOI] [PubMed] [Google Scholar]
- Nickerson ML, Kostiha BN, Brandt W, Fredericks W, Xu KP, Yu FS, Gold B, Chodosh J, Goldberg M, Lu da W, Yamada M, Tervo TM, et al. 2010. UBIAD1 mutation alters a mitochondrial prenyl-transferase to cause Schnyder corneal dystrophy. PLoS ONE 5:e10760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen SE, Tuzcu EM, Brewer HB, Sipahi I, Nicholls SJ, Ganz P, Schoenhagen P, Waters DD, Pepine CJ, Crowe TD, Davidson MH, Deanfield JE, et al. 2006. Effect of ACAT inhibition on the progression of coronary atherosclerosis. N Engl J Med 354:1253–1263. [DOI] [PubMed] [Google Scholar]
- Orr A, Dubé M-P, Marcadier P, Jiang H, Federico A, George S, Seamone C, Andrews D, Dubord P, Holland S, Provost S, Mongrain V, et al. 2007. Mutations in the UBIAD1 gene, encoding a potential prenyl-transferase, are causal for Schnyder crystalline corneal dystrophy. PLoS ONE 2:e685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BJ, Pett H, Klungel OH, Stricker BH, Psaty BM, Glazer NL, Wiggins KL, Bis JC, de Boer A, Maitland-van der Zee AH. 2011. Genetic variability within the cholesterol lowering pathway and the effectiveness of statins in reducing the risk of MI. Atherosclerosis 217:458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues MM, Kruth HS, Krachmer JH, Willis R. 1987. Unesterified cholesterol in Schnyder’s corneal crystalline dystrophy. Am J Ophthalmol 104:157–163. [DOI] [PubMed] [Google Scholar]
- Schnyder WF. 1929. Mitteilung uber einen neuen typus von familiarer hornhauterkrankung. Schweiz Med Wschr 10:559–571. [Google Scholar]
- Schnyder WF. 1939. Scheibenformige kristalllenlagerungen in der hornhautmitte als erbleiben. Klin Monatsbl Augenheilkd 103:494–502. [Google Scholar]
- Shearer MJ, Newman P. 2008. Metabolism and cell biology of vitamin K. Thromb Haemost 100:530–547. [PubMed] [Google Scholar]
- Shearman AM, Hudson TJ, Andresen JM, Wu X, Sohn RL, Haluska F, Housman DE, Weiss JS. 1996. The gene for Schnyder’s crystalline corneal dystrophy maps to human chromosome 1p34-p36. Hum Mol Genet 5:1667–1672. [DOI] [PubMed] [Google Scholar]
- Sippl MJ. 1990. Calculation of conformational ensembles from potentials of mean force - an approach to the knowledge-based prediction of local structures in globular-proteins. J Mol Biol. 213:859–883. [DOI] [PubMed] [Google Scholar]
- Sogabe N, Maruyama R, Baba O, Hosoi T, Goseki-Sone M. 2011. Effects of long-term vitamin K(1) (phylloquinone) or vitamin K(2) (menaquinone-4) supplementation on body composition and serum parameters in rats. Bone 48:1036–1042. [DOI] [PubMed] [Google Scholar]
- Tabb MM, Sun A, Zhou C, Grun F, Errandi J, Romero K, Pham H, Inoue S, Mallick S, Lin M, Forman BM, Blumberg B. 2003. Vitamin K2 regulation of bone homeostasis is mediated by the steroid and xenobiotic receptor SXR. J Biol Chem 278:43919–43927. [DOI] [PubMed] [Google Scholar]
- Theendakara V, Tromp G, Kuivaniemi H, White PS, Panchal S, Cox J, Winters RS, Riebeling P, Tost F, Hoeltzenbein M, Tervo TM, Henn W, et al. 2004. Fine mapping of the Schnyder’s crystalline corneal dystrophy locus. Hum Genet 114:594–600. [DOI] [PubMed] [Google Scholar]
- Van Went JM, Wibaut F. 1924. En Zeldzame erfelijke hoornvliessandoening. Niederl Tijdschr Geneesks 68:2996–2997. [Google Scholar]
- Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. 2003. Improved protein-ligand docking using GOLD. Proteins 52:609–623. [DOI] [PubMed] [Google Scholar]
- Vos M, Esposito G, Edirisinghe JN, Vilain S, Haddad DM, Slabbaert JR, Van Meensel S, Schaap O, De Strooper B, Meganathan R, Morais VA, Verstreken P. 2012. Vitamin K2 is a mitochondrial electron carrier that rescues Pink1 deficiency. Science 336:1306–1310. [DOI] [PubMed] [Google Scholar]
- Weiss JS, Rodrigues MM, Kruth HS, Rajagopalan S, Radar DJ, Kachadoorian H. 1992. Panstromal Schnyder’s corneal dystrophy: Ultrastructural and histochemical studies. Ophthalmology 99:1072–1081. [DOI] [PubMed] [Google Scholar]
- Weiss JS. 1992. Schnyder’s dystrophy of the cornea: A Swede-Finn connection. Cornea 11:93–101. [DOI] [PubMed] [Google Scholar]
- Weiss JS. 2007. Visual morbidity in thirty-four families with Schnyder crystalline corneal dystrophy. Trans Am Ophthalmol Soc 105:616–648. [PMC free article] [PubMed] [Google Scholar]
- Weiss JS, Kruth HS, Kuivaniemi H, Tromp G, White PS, Winters RS, Lisch W, Henn W, Denninger E, Krause M, Wasson P, Ebenezer N, et al. 2007. Mutations in the UBIAD1 gene on chromosome short arm 1, region 36, cause Schnyder crystalline corneal dystrophy. Invest Ophthalmol Vis Sci 48:5007–5012. [DOI] [PubMed] [Google Scholar]
- Weiss JS, Kruth HS, Kuivaniemi H, Tromp G, Karkera J, Mahurkar S, Lisch W, Dupps WJ Jr, White PS, Winters RS, Kim C, Rapuano CJ, et al. 2008. Genetic analysis of 14 families with Schnyder crystalline corneal dystrophy reveals clues to UBIAD1 protein function. Am J Med Genet 146A:271–283. [DOI] [PubMed] [Google Scholar]
- Weiss JS, Møller HU, Lisch W, Kinoshita S, Aldave AJ, Belin MW, Kivelä T, Busin M, Munier FL, Seitz B, Sutphin J, Bredrup C, et al. 2008. The IC3D classification of the corneal dystrophies Cornea Suppl 2:S1–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss JS, Wiaux C, Yellore V, Raber I, Eagle R, Mequio M, Aldave A. 2010. Newly reported p.Asp240Asn mutation in UBIAD1 suggests central discoid corneal dystrophy is a variant of Schnyder corneal dystrophy. Cornea 29:777–780. [DOI] [PubMed] [Google Scholar]
- Yellore VS, Khan MA, Bourla N, Rayner SA, Chen MC, Sonmez B, Momi RS, Sampat KM, Gorin MB, Aldave AJ. 2007. Identification of mutations in UBIAD1 following exclusion of coding mutations in the chromosome 1p36 locus for Schnyder crystalline corneal dystrophy. Mol Vis 13:1777–1782. [PubMed] [Google Scholar]
- Zhang WY, Gaynor PM, Kruth HS. 1996. Apolipoprotein E produced by human monocyte-derived macrophages mediates cholesterol efflux that occurs in the absence of added cholesterol acceptors. J Biol Chem 271:28641–28646. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp. Table S1. Summary of analysis of SCD families with a p.G177E alteration
Supp. Fig. S1. UBIAD1 transcripts and PCR assay amplicons
Supp. Fig. S2. Ramachandran plots of wt and G177E mutant UBIAD1
Supp. Fig. S3. Root-mean-square deviation (RMSD) in Å of Cα-atoms from a 6 ns molecular dynamics simulation.
Supp. Fig. S4. Structures of molecules described in the article