Abstract
Male ornaments and other sex-specific traits present some of the most dramatic examples of evolutionary innovations. Comparative studies of similar but independently evolved traits are particularly important for identifying repeated patterns in the evolution of these traits. Male-specific modifications of the front legs have evolved repeatedly in Drosophilidae and other Diptera. The best understood of these novel structures is the sex comb of Drosophila melanogaster and its close relatives. Here, we examine the evolution of another male foreleg modification, the sex brush, found in the distantly related Drosophila immigrans species group. Similar to the sex comb, we find that the origin of the sex brush correlates with novel, spatially restricted expression of the doublesex (dsx) transcription factor, the primary effector of the Drosophila sex determination pathway. The diversity of Dsx expression patterns in the immigrans species group closely reflects the differences in the presence, position, and size of the sex brush. Together with previous work on sex comb evolution, these observations suggest that tissue-specific activation of dsx expression may be a common mechanism responsible for the evolution of sexual dimorphism and particularly for the origin of novel male-specific ornaments.
INTRODUCTION
Some of the most striking examples of morphological variation can be seen in the differences between males and females of the same species. Even among closely related animals, sex-specific characters can differ dramatically. This simple observation implies that new sexual characters are gained, and old ones are lost, during the evolution of any animal lineage. Elucidating the genetic basis of these changes is essential to understand the origin of sexual dimorphism and biodiversity (Arnoult et al., 2013; Gompel, Prud’homme, Wittkopp, Kassner, & Carroll, 2005; Gotoh et al., 2014; Kijimoto, Moczek, & Andrews, 2012; Kopp, Duncan, & Carroll, 2000). Going beyond individual examples, we need to identify general patterns in the evolution of sexual dimorphism. How predictable are the genetic changes that give rise to new sex-specific traits? This question places a premium on research metamodels where similar traits have evolved independently in multiple lineages, each of which is amenable to experimental study (Kopp 2009).
The forelegs of true flies (Diptera) often show dramatic male-specific modifications (Hardy 1965; Sivinski 1979; Eberhard 2001; Ingram et al 2007; Stark & O’Grady, 2010; Daugeron, Plant, Winkler, Stark, & Baylac, 2011). Structures composed of modified bristles are especially common and have evolved independently in many Dipteran lineages. In the family Drosophilidae alone, examples include a variety of bristle brushes and bristle-filled spoon structures in Hawaiian Drosophila (Stark & O’Grady, 2010), pincushion-like outgrowths covered with fine bristles in Zaprionus (Tsakas and Chassagnard 1990; Chassagnard and Tsacas 1993), the thick, often massive “sex combs” in the Drosophila melanogaster and obscura species groups (Kopp, 2011; Hanna-Alava, 1958), and the densely packed bristle brush in Drosophila immigrans (Sturtevant, 1942) and its relatives (Figure 1). Repeated origin of male-specific bristle modifications in the same body region (most commonly, the tarsi of the front legs) suggests that across Diptera, the forelegs have been a target of sexual selection. It is theorized that ancestral Neoptera mated with females perched on top of males (Huber et al., 2007; McAlpine, 1981). This position places the male foreleg on the substrate, allowing it less opportunity to interact with the female. In many Diptera, however, there has been a shift to a mating posture with the male on top the female, allowing the male forelegs to interact with and potentially grasp the female. This shift in mating position may have contributed to the repeated evolution of sexually dimorphic structures in the Dipteran forelegs.
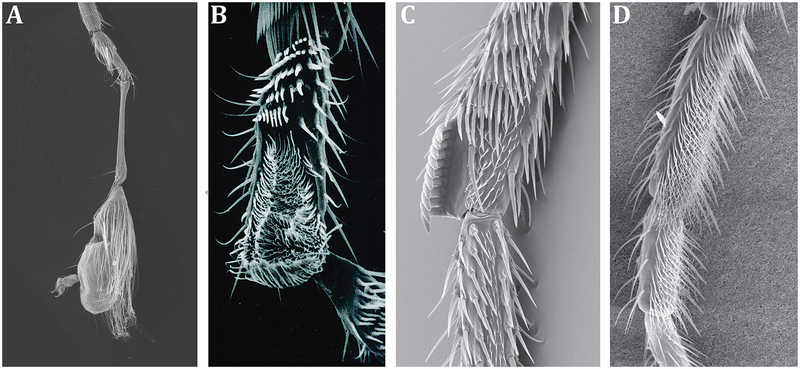
Figure 1. Many groups of Diptera have independently evolved diverse male-specific ornaments on front legs.
Scanning electron micrographs of adult male forelegs of A) Empis (C.) jaschhoforum (Daugeron et al., 2011), B) Zaprionus inermis (Museum of Paris), C) Drosophila melanogaster (Kopp & True, 2002), D) Drosophila immigrans. Reproduced with permission from Daugeron et al., 2011; Stark & O’Grady, 2010; and the Museum of Paris.
Independent evolution of morphologically distinct male-specific structures from homologous tissues presents an excellent opportunity to investigate the general rules that may govern the origin of novel sex-specific traits. The best studied male foreleg modification is the sex comb of the Drosophila melanogaster and obscura species groups: a regular array of enlarged bristles on the first and second tarsal segments (Figure 1). Sex combs play an important role in male courtship and mating success (Hurtado-Gonzales, Gallaher, Warner, & Polak, 2015; Ng & Kopp, 2008; Spieth, 1952). They develop as a male-specific modification of deeply conserved, sexually monomorphic transverse bristle rows (TBRs) that cover the ventral-anterior side of the foreleg (Tokunaga, 1962; Kopp, 2011). Like most other sexually dimorphic external structures in Drosophila, sex comb development requires the function of the gene doublesex (dsx), a transcription factor that acts downstream of the sex determination pathway to control sex-specific cell differentiation (Baker & Ridge, 1980; Hildreth, 1965). dsx expression in the foreleg is activated at the late larval and early pupal stage by Sex combs reduced (Scr), the HOX gene that controls the identity of the first thoracic segment (Tanaka, Barmina, Sanders, Arbeitman, & Kopp, 2011). In males, the male-specific isoform of Dsx upregulates Scr; the resulting feedback loop stabilizes the expression of both genes, which are jointly required to specify sex comb position and morphology. Females express a different Dsx isoform, which does not activate Scr and is not capable of inducing sex comb development (Tanaka et al., 2011).
In Drosophila, dsx is only expressed in cells that undergo sex-specific development (Camara, Whitworth, & Doren, 2008; Robinett, Vaughan, Knapp, & Baker, 2010). Expression in the male foreleg is restricted to the presumptive sex comb and sex-specific chemosensory organs (Tanaka et al., 2011; Mellert, Robinett, & Baker, 2012). In species that primitively lack sex combs, Dsx is not expressed in the homologous leg region, indicating that the origin of the sex comb coincides with the evolution of a novel dsx expression domain (Tanaka et al., 2011). Given the central role of dsx in sexual development, this suggests a general model where changes in the spatial regulation of dsx are often a necessary first step in the evolution of sexual dimorphism (Kopp, 2012). For a new sex-specific structure to evolve in ancestrally monomorphic tissue, dsx expression must first be turned on in that tissue. Subsequent diversification in the size, position, and morphology of the new structure is likely to require further evolutionary changes in dsx regulation.
To test the generality of this mechanism, we decided to apply the metamodel approach (Kopp, 2009) by focusing on another male-specific foreleg modification that evolved independently in a distantly related group of Drosophila. The immigrans and melanogaster species groups diverged >20 million years ago (Izumitani, Kusaka, Koshikawa, Toda, & Katoh, 2016; Russo, Mello, Frazão, & Voloch, 2013). The “sex brush” of D. immigrans develops in the same leg region as the sex comb (ventral-anterior surface of the first and second tarsal segment of the male foreleg), but has a very different morphology (Markow & O’Grady Patrick M, 2006; Spieth, 1952; Wilson, Wheeler, Harget, & Kambysellis, 1969) (Figure 2). As expected of a sexually dimorphic structure, the cells that produce the sex brush also express Dsx (Tanaka et al., 2011). Here, we use phylogenetic analysis to reconstruct sex brush origin and diversification, and test for correlation between morphological evolution and the evolution of Dsx expression. We find that, similar to the sex comb, the origin of the sex brush coincides with the appearance of a new domain of Dsx expression, and that Dsx patterns in the developing forelegs correlate closely with the diversity of sex brush morphologies across species.
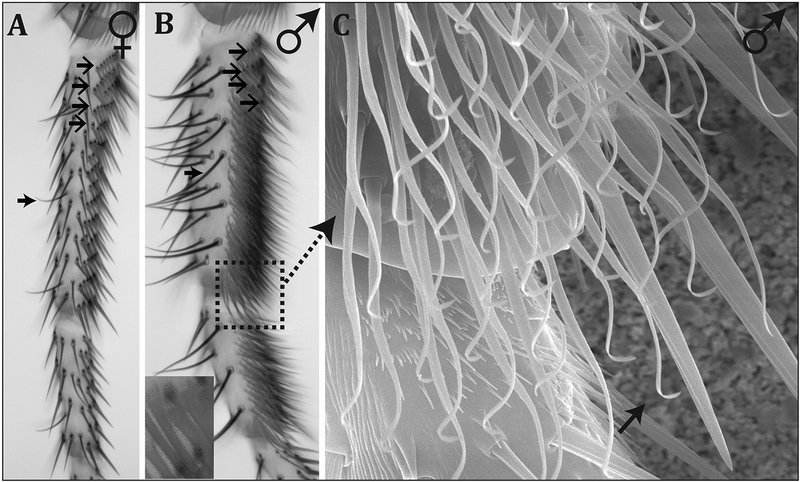
Figure 2. The male-specific sex brush of D. immigrans is composed of densely packed, hook-shaped bristles.
First and second tarsal segments of D. immigrans female (A) and male (B, C) forelegs. A) The first tarsal segment of D. immigrans females is covered by transverse bristle rows (TBRs, open face arrows). Anterior/dorsal (left) of the TBRs are chemosensory bristles (closed face arrow). Note that all TBR bristles have bracts. B) In males, the ventral/anterior surfaces of the first and second tarsal segments are mostly covered by the sex brush, which is composed of densely packed, bractless bristles with hooked tips. The most proximal (top) rows of bristles are composed of a mix of TBR (open faced arrows) and sex brush bristles (inset). Chemosensory bristles (closed face arrow) are larger and more numerous in males. The dashed box indicates the region shown in panel C. C) SEM image of the distal first tarsal segment showing the sex brush. The arrow highlights a single sex brush bristle, hooked at the distal end.
MATERIALS AND METHODS
Drosophila Strains:
To study sex brush evolution in the immigrans species group, we used the following subset of described species, representing all major subgroups: D. immigrans (Sturtevant, 1921), D. formosana (Duda, 1926; Sturtevant, 1927), D. ruberrima (De Meijere, 1911), D. hypocausta (Osten Sacken, 1882), D. siamana (Hihara & Lin, 1984; Ikeda, Hihara, Asada, Fujiwara, & Lin, 1983), D. rubida (Mather, 1960), D. neohypocausta (Lin & Tseng, 1973), D. nasuta (Lamb, 1914), D. albomicans (Duda, 1926), D. sulfurigaster (Duda, 1926), and D. kepulauana (Wilson et al., 1969). D. quadrilineata (De Meijere, 1911) was used as an outgroup. Stocks were obtained from both the Ehime University Drosophila Stock Center (D. albomicans KM55–5, D. sulfrigaster 5112-M, D. formosana 292.8 (E-13401), D. ruberrima OKNH8 (E-14601), D. rubida 23.01 CAR-316, D. siamana 1741.00-TC (E-22701), D. neohypocausta 1881.02-TC (E-22501), D. quadrilineata TMU (E14402) and the UC San Diego Drosophila Stock Center (D. immigrans 15111–1731.08, D. hypocausta 15115–1871.05, D. kepulauana 15112–1761.03, D. nasuta 15112–1781.00).
Phylogenetic Reconstruction:
We used partial genomic sequences of nine nuclear loci: Alcohol dehydrogenase (Adh), Dopa decarboxylase (Ddc), extra sexcombs (esc), kinase suppressor of ras (ksr), Phospho-glucose isomerase (Pgi), Triose phosphate isomerase (Tpi), Xanthine dehydrogenase (Xdh), Amylase-related (Amyrel), and Glycerol 3 phosphate dehydrogenase (Gpdh) (Supplementary Table 1). Some of the sequences used in this study were generated by previous phylogenetic and population-genetic studies (Da Lage et al., 2007; Huang et al., 2002; Katoh, Nakaya, Tamura, & Aotsuka, 2007) and were obtained from GenBank. Sequences of D. albomicans, D. nasuta, D. sulfurigaster, and D. immigrans were extracted from the whole genome sequences of these species, kindly provided by Doris Bachtrog and Michael Eisen. The remaining sequences were amplified using the primers shown in Supplementary Table 2; these primers were modified from a previous study of the melanogaster species group phylogeny (Kopp, 2006).
DNA was extracted from 20 flies of each species using live strains maintained in our laboratory. Voucher specimens have been preserved in ethanol. Amplified DNA was either gel-purified and sequenced directly using the amplification primers (Supplementary Table 2) or ligated into pCRII (Invitrogen) and sequenced using the vector forward and reverse primers M13F and M13R. ABI chromatograms were examined by eye. Heterozygous nucleotide positions, if present, were represented by IUPAC ambiguity codes. Sequences were aligned and tiled using MUSCLE (Edgar 2004) through the program Geneious (Kearse et al., 2012). The lengths of available sequences varied from species to species, and the missing nucleotide positions at the ends of fragments were coded as gaps. To ensure correct alignments, all coding sequences were translated into proteins. Exons were determined by alignment to D. melanogaster cDNA. Due to difficulties in establishing homology, intronic sequences were removed from the dataset. The edited alignments were then partitioned by codon for analyses.
A published Adh sequence from D. siamana (GenBank accession AY044125) was found to closely resemble D. albomicans, which conflicted with the rest of our loci. We re-sequenced this locus from our stock of D. siamana. The resulting sequence is similar to the one found in Izumitani et al., 2016, GenBank accession AB261135. Our sequence was used as the representative sequence of D. siamana in further analysis.
In order to determine the best nucleotide substitution models, we used the program Partitionfinder (Lanfear, Calcott, Ho, & Guindon, 2012). Most of the codon partitions fit a GTR+G substitution model; while Adh codon position 2, Amyrel codon position 2, Ddc codon position 3, Pgi codon position 2, Tpi codon position 2, Xdh codon position 2 fit a TVM+I+G substitution model; GpDH codon position 2, esc codon position 2 fit a F81 substitution model; and GpDH codon position 3, ksr codon position 3 fit a HKY+G substitution model. D. quadrilineata has been repeatedly found to be basal, or closely related, to the immigrans lineage, making it a suitable outgroup (Da Lage et al., 2007; Huang et al., 2002; Izumitani et al., 2016; Katoh et al., 2007; Yan, Zeng, Yang, & Qian, 2006). Partitioned alignments were analyzed using BEASTv1.8.0 (Drummond, Suchard, Xie, & Rambaut, 2012) or MrBayes v3.2.1 (Ronquist et al., 2012), BEAST ran for 100,000,000 generations with the first 10,000,000 generations discarded. Trees were sampled every 1,000 generations (.xml file in Supplementary Text 1). MrBayes ran for 10,000,000 generations with 4 chains. Trees were sampled every 200 generations. The logs were analyzed in Tracerv1.6; for all parameters, the Effective Sample Sizes (ESS) were over 4000. Visual inspection of the traces indicated good mixing, and analysis of several independent chains showed convergence in tree topology and ancestral state reconstruction.
We used BayesTraits Version 3 (Pagel, Meade, & Barker, 2004), with the tree generated above, to test for correlation between the sex brush and Dsx expression. We performed three runs of both the independent and dependent models with a stepping stone model (250 stones each sampled 10,000 iterations). We averaged the log marginal likelihood from ten runs and calculated the Log Bayes Factors for these two models.
Immunohistochemistry and Microscopy:
Samples for immunohistochemistry were synchronized at pupariation, aged for 24 hours or longer depending on the species-specific rate of development, and processed and imaged as described in Mellert et al., 2012 and Tanaka et al., 2011. The primary antibodies used were rat anti-DsxCommon, 1:50 (Sanders and Arbeitman 2008), monoclonal mouse anti-Dsx[DBD], 1:10 (a gift from C. Robinett and B. Baker, Janelia Farm Research Campus, HHMI and available from the Developmental Studies Hybridoma Bank at the University of Iowa, DsxDBD), and mouse anti-Scr 6H4.1, 1:10 (Developmental Studies Hybridoma Bank, University of Iowa). The secondary antibodies were AlexaFluor 488 and 594, used at 1:200 (Invitrogen, Carlsbad, CA). Confocal images were collected on an Olympus FV1000 laser scanning confocal microscope with a 40X lens, with the gain adjusted for the dynamic range of each sample. In species where no Dsx expression was found, the gain was increased to detect even very weak signal, leading to higher background intensity in these samples.
For light microscopy, adult legs were dissected in water and mounted in PVA Mounting Medium (BioQuip) until fully cleared. Images were taken under bright field illumination with a 20X lens on a Leica DM500B microscope with a Leica DC500 camera. For scanning electron microscopy, adult flies were dehydrated in 100% ethanol, processed by critical point drying, coated with gold, and imaged on a Philips XL30 SEM microscope.
RESULTS
The Drosophila immigrans male-specific sex brush is composed of densely packed, hook-shaped bristles.
Although several studies have mentioned modified bristles on the foreleg of D. immigrans males (Spieth, 1952; Sturtevant, 1942), neither the morphology nor the interspecific diversity of the sex brush have been examined previously. In D. immigrans females, the chaetotaxy of the first tarsal segment of the foreleg is very similar to other Drosophila species. The ventral-anterior side of this segment is covered by regularly spaced transverse bristle rows (TBRs), which consist of bracted mechanosensory bristles (Figure 2A). In D. immigrans males, the sex brush replaces most of the TBR field, leaving only a few normal bristles in the most proximal TBRs. In addition, the sex brush occupies the ventral-anterior side of the second tarsal segment (Figure 2B). In contrast to females, the bristles comprising the male sex brush are not organized into linear rows, lack bracts, and are packed extremely densely: on the first tarsal segment, there are ~265 sex brush bristles in males, compared to ~55 TBR bristles in females. While the TBR bristles are straight with pointed tips, the much thinner sex brush bristles become curly and hooked near the tip (Figure 2C).
The sex brush of D. immigrans develops from an area of Dsx and Scr co-expression.
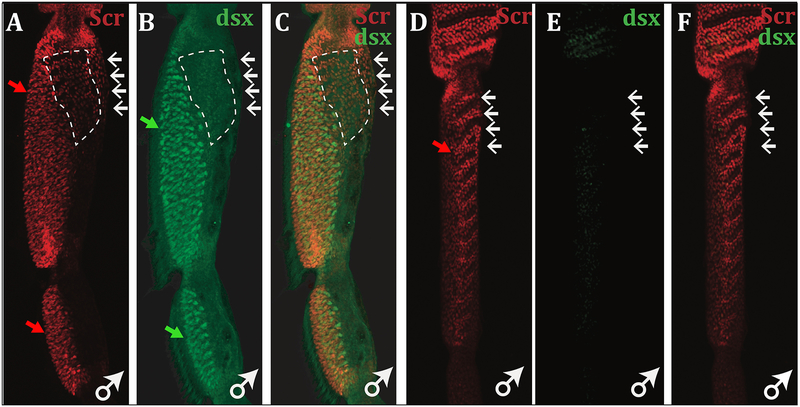
The sex comb of D. melanogaster and its relatives is established by coexpression of the HOX gene Sex combs reduced (Scr) and the sex determination gene doublesex (dsx) (Tanaka et al., 2011). Similarly, in the forelegs of D. immigrans male pupae, we find that Scr and Dsx expression patterns overlap on the ventral-anterior side of the first and second tarsal segments, where the sex brush will develop (Figure 3A–C). At the proximal end of the first tarsal segment, Scr expression is seen without Dsx, presumably corresponding to the remaining sexually monomorphic TBR bristles. Although Scr is expressed in a wider region than Dsx, Scr levels are highest in the Dsx expression domain (Figure 3A–C). In D. albomicans, an immigrans group species that lacks the sex brush and has unmodified TBRs in both sexes, we do not observe strong Dsx expression and, in contrast to D. immigrans, Scr expression is much weaker in the tarsus than in the distal tibia (Figure 3D–F). This observation suggests that, similar to the sex comb (Tanaka et al., 2011), Scr and Dsx co-expression in D. immigrans males may be maintained by a positive feedback loop and is required for the development of the male sex brush.
Figure 3. Overlapping Scr and Dsx expression in the sex brush primordium.
The first and second tarsal segments of the pupal male forelegs are shown. Scr (red) is expressed in both TBR and sex brush regions, while Dsx expression (green), assayed with the anti-DsxCommon antibody (Sanders & Arbeitman, 2008), is limited to the sex brush. Expression of Dsx and Scr are highlighted by closed face green and red arrows respectively. As in the sex combs of D. melanogaster and its relatives (Tanaka et al., 2011), Dsx is expressed predominantly in bristle precursor cells, whereas Scr is present mainly in the surrounding epithelial cells. A-C) D. immigrans male pupal forelegs at 48 hrs after pupariation. Scr expression that does not overlap Dsx likely represents the proximal TBR bristles (open faced arrow). Scr expression is lower in regions where Dsx is absent (dashed white box). D-F) D. albomicans male pupal forelegs at 30 hrs after pupariation. Scr expression can still be seen in the TBRs, but Dsx is not expressed in the corresponding region.
Revised phylogeny of the immigrans species group
The independent origin of the sex comb and sex brush in distantly related Drosophila lineages allows us to test whether novel Dsx expression is generally necessary for the gain and diversification of a new sex-specific trait. Our survey revealed extensive variation in the presence and morphology of the sex brush in the immigrans species group. Although previous studies have investigated phylogenetic relationships in this group, most of these studies used small non-overlapping datasets and produced conflicting results (Da Lage et al., 2007; Huang et al., 2002; Izumitani et al., 2016; Katoh et al., 2007; Yan et al., 2006). The branching order of the immigrans, nasuta and hypocausta subgroups, and the placement of D. ruberrima and D. neohypocausta, have remained controversial. Since resolving these relationships is essential for reconstructing sex brush evolution, we undertook a more robust phylogenetic analysis.
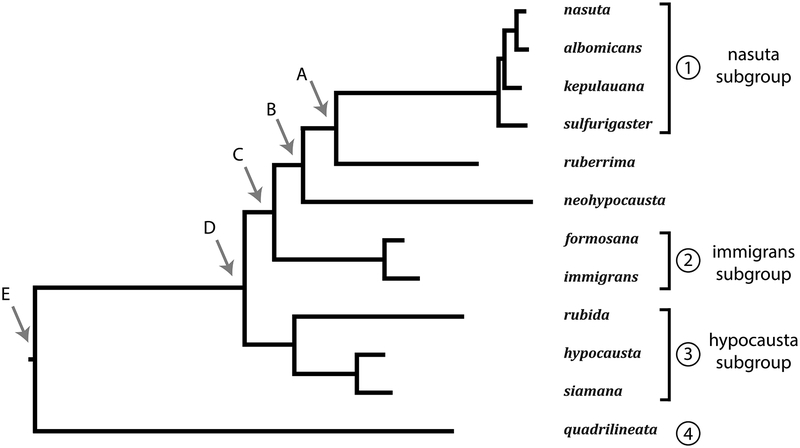
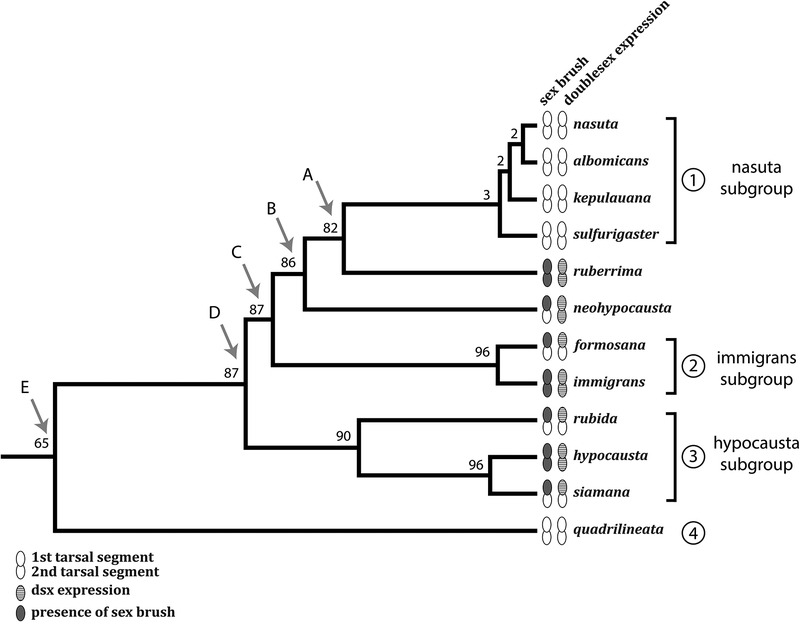
We generated an updated phylogeny using nine loci from twelve immigrans group species using Bayesian inference in BEAST (Drummond et al., 2012) and MrBayes (Ronquist et al., 2012) (Figure 4). Our analysis resulted in a fully resolved and well supported tree with posterior probabilities over 99% at all nodes. This tree identifies three lineages in the immigrans species group: the nasuta subgroup, including D. nasuta, D. albomicans, D. kepulauana, and D. sulfurigaster; the immigrans subgroup, including D. immigrans and D. formosana; and the hypocausta subgroup proper, including D. hypocausta, D. siamana, and D. rubida. Although D. ruberrima is traditionally assigned to the immigrans subgroup (Wilson et al., 1969), our analysis places it as sister to the nasuta subgroup (Node A, Figure 4). D. neohypocausta, described originally as part of the hypocausta subgroup (Osten Sacken, 1882), is more closely related to the nasuta subgroup and D. ruberrima than to the hypocausta subgroup (Node B, Figure 4). The nasuta and immigrans subgroups cluster together (Node C, Figure 4), followed by the branching of the hypocausta subgroup (Node D, Figure 4), and finally D. quadrilineata in the most basal position.
Figure 4. Phylogeny of the Drosophila immigrans species group.
Phylogenetic tree produced by Bayesian analysis in MrBayes. Posterior probabilities at all nodes are >99%. This tree identifies three monophyletic clades: nasuta subgroup (1), immigrans subgroup (2), and hypocausta subgroup proper (3). Although D. ruberrima is traditionally assigned to the immigrans subgroup (De Meijere, 1911), our analysis places it as sister group to the nasuta subgroup (Node A). D. neohypocausta was originally described as part of the hypocausta subgroup (Osten Sacken, 1882), but here it is also placed closer to the nasuta subgroup (node B). The immigrans subgroup forms a monophyletic clade with the nasuta subgroup + D. ruberrima + D. neohypocausta (node C), and the hypocausta subgroup occupies the most basal position in the immigrans species group (node D). D. quadrilineata was used as an outgroup to root the tree.
Sex brush diversity in the immigrans species group
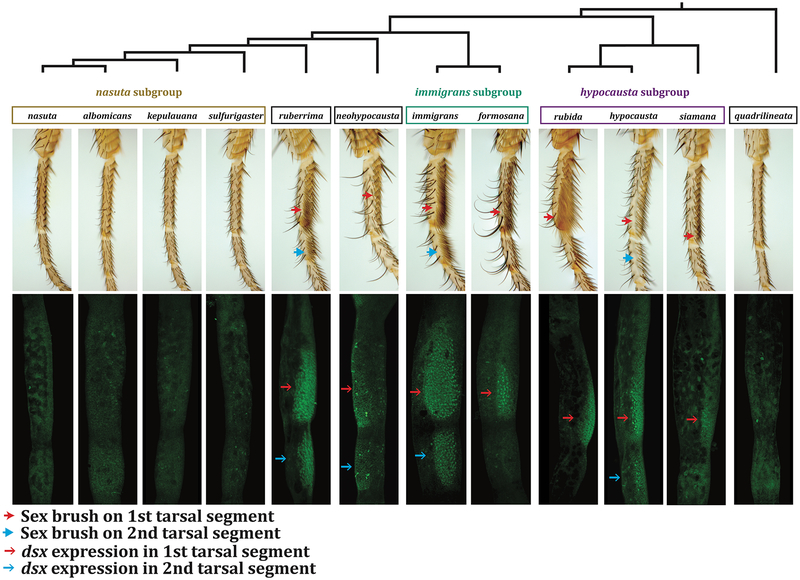
We defined a species as having a sex brush if the foreleg TBRs contained at least a few male-specific bractless bristles. By this lenient definition, only the nasuta species subgroup and D. quadrilineata lack a sex brush (Figure 5); these species do not show overt sexual dimorphism in the morphology of TBRs (Supplementary Figure 1). However, all examined species show different degrees of sex-specificity in the number and size of chemosensory bristles, (Supplementary Figure 1), indicating that the front leg has the developmental capacity for sexual dimorphism. In the remaining immigrans group species, sex brushes vary widely in size and morphology (Figure 5). The largest and most densely packed sex brushes are found in the immigrans subgroup, D. rubida (hypocausta subgroup), and in D. ruberrima, which is sister to the brush-less nasuta subgroup. In these species, the sex brush replaces most of the TBRs in males. In D. hypocausta, D. siamana, and D. neohypocausta, sex brushes are smaller and look more similar to normal TBRs, but contain some supernumerary, irregularly arranged, bractless bristles. D. neohypocausta and D. siamana have male-specific bristles in only a subset of 3–5 TBRs (Supplementary Figure 1). In closely related species, the sex brush differs in both position and size. For example, the sex brush occupies first and second tarsal segments in D. immigrans, but is only found on the first segment of its sister species, D. formosana. In the hypocausta subgroup, D. hypocausta has small sex brushes on the first and second tarsal segments, while D. rubida has a very well developed sex brush but only on the first segment (Figure 5). Neither the species with the largest sex brushes, nor the ones with the smallest, cluster together. Rather, each clade within the immigrans species group shows a diversity of sex brush types.
Figure 5. Dsx expression in the D. immigrans species group correlates with sex brush morphology.
Adult male phenotypes and phylogenetic relationships are shown in the top row. The bottom row shows Dsx expression, assayed by the Dsx[DBD] antibody (Mellert et al., 2012), in male pupal legs at 24 hours after pupariation. Dsx immunostaining (green) is seen in sex brush bristles (arrows). In most species the position and size of Dsx expression corresponds with the sex brush. In D. neohypocausta, Dsx expression is seen throughout the first and second tarsal segments, yet only two bractless bristles, suggestive of a rudimentary sex brush, are found on the first tarsal segment (red arrow in the top row). More intense Dsx staining correlates with the positions of chemosensory bristles (see also Supplementary Figure 1).
Dsx expression correlates with sex brush origin and diversification
To test whether Dsx expression correlates with sex brush morphology, we stained the male pupal forelegs of immigrans group species with an antibody against the highly conserved DM domain of Dsx (Mellert et al., 2012). To confirm cross-reactivity, we stained the larval testes of several immigrans group species, and found Dsx expression similar to that in D. melanogaster (data not shown). Dsx expression in the leg corresponds well to the size and position of the sex brush (Figure 5). In species that lack a sex brush (the nasuta subgroup and D. quadrilineata), no Dsx expression is seen. In species with sex brushes on both first and second tarsal segments, Dsx is expressed in both segments (Figure 5). In species with a sex brush on only the first tarsal segment, Dsx expression is also confined to that segment (Figure 5). The size of the Dsx expression domain within the first tarsal segment also correlates with the size of the sex brush. For example, Dsx expression is most extensive in D. immigrans, D. rubida, and D. ruberrima, and much more restricted in D. hypocausta and D. siamana (Figure 5). The one exception is D. neohypocausta, where Dsx expression is present in both first and second tarsal segments, but only a few weakly differentiated male-specific bristles are seen in the first segment, and none in the second. This pattern may be related to an early specification of male-specific chemosensory bristles, which are especially large in this species (Supplementary Figure 1).
We used BEAST (Drummond et al., 2012) to estimate ancestral character states across the immigrans species group phylogeny, while accommodating for uncertainty in tree topology (Figure 6). Our analysis indicates strong but not overwhelming probability (87%) that the last common ancestor of the immigrans species group had a male sex brush and associated Dsx expression (Node D, Figure 6). Although the phylogenetic distribution of the sex brush could in principle be explained either by multiple gains or by a single gain followed by reduction and loss, the latter model appears significantly more likely. In particular, the nasuta subgroup probably lost the sex brush secondarily.
Figure 6. Phylogenetic reconstruction of sex brush evolution.
BEAST was used to estimate the ancestral state of the sex brush and Dsx expression at each node of the phylogeny. Grey ovals show the presence of a sex brush, and striped ovals show the presence of Dsx expression, in extant species. Open ovals represent the lack of sex brush or Dsx expression. The numbers at each internal node indicate posterior probabilities that the last common ancestor of the corresponding clade had a sex brush, which are identical to the probabilities of expressing Dsx. The posterior probabilities were estimated in BEAST using a Speciation: Birth-death model with incomplete sampling.
We then used BayesTraits (Pagel et al., 2004) to test for correlation between the presence of the sex brush and Dsx expression. We found strong evidence that the sex brush and Dsx expression were correlated (Log Bayes Factor of 7.74). This suggests that Dsx expression is indeed associated with sex brush evolution – and, more broadly, supports our model that changes in Dsx expression are vital to the origin and loss of sexually dimorphic traits.
DISCUSSION:
To search for general patterns in the evolution of sexually dimorphic traits, we focused on the evolution of doublesex expression. dsx is the primary transcription factor responsible for inducing sex-specific differentiation of somatic cells in insects (Kopp, 2012). For example, dsx has been implicated in the development and evolution of sexually dimorphic traits such as male horns in the Onthophagus dung beetles (Kijimoto et al., 2012; Ledón-Rettig, E, Zattara, & Moczek, 2017) (Kijimoto et al., 2012; Ledón-Rettig et al., 2017) and Trypoxylus dichotomus (Ito et al., 2013), exaggerated mandibles in the stag beetle Cyclommatus metallifer (Gotoh et al., 2014), reduced male wings in the jewel wasp Nasonia vitripennis (Loehlin et al., 2010), the genital morphology of Bombyx mori (Xu et al., 2017), and sex-specific mimetic color patterns in Papilio swallowtail and Agraulis vanilla butterflies (Komata, Lin, Iijima, Fujiwara, & Sota, 2016; Kunte et al., 2014; Martin et al., 2014; Nishikawa et al., 2015).
Earlier work on the sex combs of D. melanogaster and its relatives suggested a model where the origin of a new sex-specific structure in a previously monomorphic tissue depends on evolutionary changes in the spatial regulation of dsx (Tanaka et al., 2011). Given the central role of dsx in sexually dimorphic cell differentiation, for any sex-specific structure to evolve, Dsx must be expressed in the cells that give rise to this structure. In tissues that already express Dsx, sexual dimorphism can evolve with relative ease if dsx acquires new downstream targets. In contrast, if a tissue is ancestrally monomorphic and does not express Dsx, changes in dsx regulation that result in its de novo expression in that tissue must be a necessary first step.
Here, we tested the generality of this model by focusing on a different sex-specific innovation – the sex brush that evolved in the Drosophila immigrans species group, which is distantly related to D. melanogaster. Similar to sex combs, we found abundant phenotypic diversity and a strong correlation between Dsx expression and sex brush morphology (Figure 4). In D. quadrilineata, the most basal species that lacks a sex brush, Dsx is not expressed in the foretarsal TBRs. In another similarity with sex combs, the foreleg-specific HOX gene Scr is coexpressed with Dsx and upregulated in the developing sex brush. Thus, it appears that independent evolution of male-specific foreleg modifications may have a similar genetic basis in distantly related lineages. We predict that a similar pattern will be found in other Dipteran families.
One of the striking features of the D. immigrans sex brush is the hooked shape of the bristles. Although the selective forces that led to this bristle morphology are unknown, D. immigrans does not go through some of the stereotypical courtship steps found in most other Drosophila, such as licking. Instead, after circling and tapping the female’s abdomen, the male lunges at the female, grabbing her abdomen with his forelegs, and attempts to inseminate the female (Spieth, 1952). Spieth conjectured that modified foreleg bristles in D. immigrans, as well as other foreleg modifications found throughout Drosophila, enable males to grasp the female more effectively during courtship.
This function may explain the frequent origin of new male-specific structures on the forelegs of Drosophila and other Diptera. For example, male flies in the genus Zaprionus have pincushion-like outgrowths on their front tarsi, covered with thin, densely packed bristles reminiscent of the sex brush bristles of D. immigrans and its relatives (Figure 1). These structures also express Dsx (Tanaka et al., 2011). Like D. immigrans, Zaprionus species have shortened courtship compared to other Drosophilidae (Spieth, 1952). The phylogenic position of Zaprionus within Drosophilidae is contentious, with some studies suggesting that Zaprionus and the immigrans species group are closely related (Yassin et al., 2010). If so, it is possible that the modified foreleg bristles of Zaprionus and the immigrans group, and the associated domains of Dsx expression, share a common origin.
The sex comb and sex brush share similar positions on the foreleg; this allows us to investigate the genetic constraints on establishing new sex-specific structures. However, the morphology of male-specific bristles differs greatly between the comb and the brush. Identifying the downstream targets of dsx in these two structures may illuminate the genetic and cellular pathways that transform a deeply conserved bristle type (TBRs) into two radically different shapes.
Supplementary Material
Supplementary Figure 1 The sex brush is male-specific in the D. immigrans species group. The first and second tarsal segments of the adult male (top two rows) and female forelegs (bottom two rows) are shown. Phylogenetic relationships are indicated above. The second and fourth rows from the top show the distal first tarsal segment at higher magnification. Red and blue arrows indicate the presence of a sex brush on the first and second tarsal segments, respectively. In addition to the sex brush, males of different species vary in the number and size of chemosensory bristles (long upwardly curved bristles). The female TBRs do not show much variation among species. The sex brushes of D. siamana and D. neohypocausta are rudimentary, with only a few supernumerary, bractless bristles found adjacent to the otherwise monomorphic TBRs in males.
Supplementary Table 1. Loci for immigrans group phylogeny References or Genbank codes for each locus used in the phylogeny.
Supplementary Table 2. Primers for immigrans group phylogeny A list of the forward and reverse primers used to generate the dataset for the immigrans species group phylogeny. Alignment length represents the size of the aligned coding sequence used in phylogenetic reconstruction.
Supplemental Text 1 The nexus file run in MrBayes to generate the phylogeny seen in Figure 5.
Supplemental Text 2 The .xml file generated in BEAUti and run in BEAST to infer the ancestral state of the sex brush and Dsx expression seen in Figure 6.
ACKNOWLEDGEMENTS:
We thank the UC San Diego and Ehime University Drosophila species stock centers for Drosophila strains, B. Baker, B. Oliver, C. Robinett, and the Developmental Studies Hybridoma Bank for antibodies, D. Bachtrog for sharing the genomic sequences of D. albomicans, D. nasuta, and D. sulfurigaster, M. Eisen for the D. immigrans genome sequence, and Michael May, Jiansi Gao, and Brian Moore for their advice on phylogenetic analysis. This work was funded by NIH grant R01GM105726 to AK and NSF DDIG Fellowship 1501148 to GR.
REFERENCES:
- Arnoult L, Su KFY, Manoel D, Minervino C, Magriña J, Gompel N, & Prud’homme B (2013). Emergence and Diversification of Fly Pigmentation Through Evolution of a Gene Regulatory Module. Science, 339, 1423–1426. 10.1126/science.1233749 [DOI] [PubMed] [Google Scholar]
- Baker BS, & Ridge KA (1980). Sex and the single cell. I. On the action of major loci affecting sex determination in Drosophila melanogaster. Genetics, 94(2), 383–423. Retrieved from http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1214149&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camara N, Whitworth C, & Van Doren M. (2008). The Creation of Sexual Dimorphism in the Drosophila Soma CURRENT TOPICS IN DEVELOPMENTAL BIOLOGY (Vol. 83). Elsevier Inc. 10.1016/S0070-2153(08)00403-1 [DOI] [PubMed] [Google Scholar]
- Da Lage J-L, Kergoat GJ, Maczkowiak F, Silvain J-F, Cariou M-L, & Lachaise D (2007). A phylogeny of Drosophilidae using the Amyrel gene: questioning the Drosophila melanogaster species group boundaries. Journal of Zoological Systematics and Evolutionary Research, 45(1), 47–63. 10.1111/j.1439-0469.2006.00389.x [DOI] [Google Scholar]
- De Meijere JCH (1911). Studien uber sudostasiatische Dipteren. VI. Tijdschrift Voor Entomologie, 54, 258–432. [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, & Rambaut A (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution, 29(8), 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda O (1926). Die orientalischen und australischen Drosophiliden-Arten (Dipteren) des Ungarischen National-Museums zu Budapest. I. Nachtrag. Annales Historico-Naturales Musei Nationalis Hungarici [Termeszettudomanyi Muzeum Evkonyve], 23, 241–250. [Google Scholar]
- Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32(5), 1792–7. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompel N, Prud’homme B, Wittkopp PJ, Kassner VA, & Carroll SB (2005). Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature, 433(7025), 481–487. 10.1038/nature03235 [DOI] [PubMed] [Google Scholar]
- Gotoh H, Miyakawa H, Ishikawa A, Ishikawa Y, Sugime Y, Emlen DJ, … Miura T (2014). Developmental Link between Sex and Nutrition; doublesex Regulates Sex-Specific Mandible Growth via Juvenile Hormone Signaling in Stag Beetles. PLoS Genetics, 10(1), 1–9. 10.1371/journal.pgen.1004098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna-Alava A (1958). Morphology and chaetotaxy of the legs of Drosophila melanogaster. Journal of Morphology, 103(2), 281–310. [Google Scholar]
- Hihara F, & Lin F-J (1984). A new species of Drosophila hypocausta subgroup of species from Malaysia and Thailand (Diptera-Drosophilidae-Drosophila)..pdf. Bulletin of the Institute of Zoology, Academia Sinica [Tung Wu Yen Chiu so Chi K’an], 23(2), 205–2. [Google Scholar]
- Hildreth PE (1965). Doublesex, Recessive Gene That Transforms Both Males and Females of Drosophila Into Intersexes. Genetics, 51(April), 659–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Hao L, Liu S, Li L, Zhang W-X, & Dai Z-H (2002). Phylogenetic position of Chinese endemic Drosophila curviceps species subgroup in the Drosophila immigrans group. Yi Chuan Xue Bao = Acta Genetica Sinica, 29(5), 417–23. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/12043569 [PubMed] [Google Scholar]
- Huber BA, Sinclair BJ, Schmitt M, Huber BA, Sinclair BJ, & Schmitt M (2007). The evolution of asymmetric genitalia in spiders and insects, 82, 647–698. 10.1111/j.1469-185X.2007.00029.x [DOI] [PubMed] [Google Scholar]
- Hurtado-Gonzales J, Gallaher W, Warner A, & Polak M (2015). Microscale laser surgery demonstrates the grasping function of the male sex combs in drosophila melanogaster and drosophila bipectinata. Ethology, 121(1), 45–56. 10.1111/eth.12316 [DOI] [Google Scholar]
- Ikeda H, Hihara F, Asada N, Fujiwara K, & Lin FJ (1983). Reproductive isolation among three species belonging to the Drosophila hypocausta subgroup of the immigrans species group, with a description of a new species In: The overseas scientific expedition for the collection of drosophilid flies, 1971–1982. Tokyo, 18–28. [Google Scholar]
- Ito Y, Harigai A, Nakata M, Hosoya T, Araya K, Oba Y, … Niimi T (2013). The role of doublesex in the evolution of exaggerated horns in the Japanese rhinoceros beetle. EMBO Reports, 14(6), 561–567. 10.1038/embor.2013.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumitani HF, Kusaka Y, Koshikawa S, Toda MJ, & Katoh T (2016). Phylogeography of the Subgenus Drosophila (Diptera: Drosophilidae): Evolutionary History of Faunal Divergence between the Old and the New Worlds. PloS One, 11(7), e0160051 10.1371/journal.pone.0160051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, Nakaya D, Tamura K, & Aotsuka T (2007). Phylogeny of the Drosophila immigrans species group (Diptera: Drosophilidae) based on Adh and Gpdh sequences. Zoological Science, 24(9), 913–21. 10.2108/zsj.24.913 [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, … Drummond A (2012). Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28(12), 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijimoto T, Moczek AP, & Andrews J (2012). Diversification of doublesex function underlies of beetle horns. Proceedings of the National Academy of Sciences of the United States of America, 109(50), 20526–20531. 10.1073/pnas.1118589109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komata S, Lin C, Iijima T, Fujiwara H, & Sota T (2016). Identification of doublesex alleles associated with the female-limited Batesian mimicry polymorphism in Papilio memnon. Nature Publishing Group, 6(October), 1–7. 10.1038/srep34782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp A (2006). Basal relationships in the Drosophila melanogaster species group. Molecular Phylogenetics and Evolution, 39(3), 787–798. 10.1016/j.ympev.2006.01.029 [DOI] [PubMed] [Google Scholar]
- Kopp A (2009). Metamodels and phylogenetic replication: A systematic approach to the evolution of developmental pathways. Evolution, 63(11), 2771–2789. 10.1111/j.1558-5646.2009.00761.x [DOI] [PubMed] [Google Scholar]
- Kopp A (2012). Dmrt genes in the development and evolution of sexual dimorphism. Trends in Genetics : TIG, 28(4), 175–84. 10.1016/j.tig.2012.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp A, Duncan I, & Carroll SB (2000). Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature, 408, 553–559. [DOI] [PubMed] [Google Scholar]
- Kopp A, & True JR (2002). Evolution of male sexual characters in the oriental Drosophila melanogaster species group. Evolution & Development, 4(4), 278–91. 10.1046/j.1525-142X.2002.02017.x [DOI] [PubMed] [Google Scholar]
- Kunte K, Zhang W, Tenger-Trolander A, Palmer DH, Martin A, Reed RD, … Kronforst MR (2014). doublesex is a mimicry supergene. Nature, 507(7491), 229–232. 10.1038/nature13112 [DOI] [PubMed] [Google Scholar]
- Lamb CG (1914). The Percy Sladen Trust expedition to the Indian Ocean in 1905. XV. Diptera: Heteroneuridae, Ortalidae, Trypetidae, Sepsidae, Micropezidae, Drosophilidae, Geomyzidae, Milichidae. Transactions of the Linnean Society of London, 16, 307–372. [Google Scholar]
- Lanfear R, Calcott B, Ho SYW, & Guindon S (2012). PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution, 29(6), 1695–1701. [DOI] [PubMed] [Google Scholar]
- Ledón-Rettig C, Zattara EE, & Moczek AP (2017). Asymmetric interactions between doublesex and tissue- and sex-specific target genes mediate sexual dimorphism in beetles. Nature Communications, 8, 1–10. 10.1038/ncomms14593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F-J, & Tseng H-C (1973). The Drosophila immigrans species group in Taiwan with descriptions of five new species. Bulletin of the Institute of Zoology, Academia Sinica, 12, 13–26. [Google Scholar]
- Loehlin DW, Oliveira DCSG, Edwards R, Giebel JD, Michael E, Cattani MV, … Beukeboom LW (2010). Non-Coding Changes Cause Sex-Specific Wing Size Differences between Closely Related Species of Nasonia. PLoS Genetics, 6(1), 1–9. 10.1371/journal.pgen.1000821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow TA, & O’Grady Patrick M (2006). Drosophila: A guide to species identification and use. Statewide Agricultural Land Use Baseline 2015, 1 10.1017/CBO9781107415324.004 [DOI] [Google Scholar]
- Martin A, Mcculloch KJ, Patel NH, Briscoe AD, Gilbert LE, & Reed RD (2014). Multiple recent co-options of Optix associated with novel traits in adaptive butterfly wing radiations Multiple recent co-options of Optix associated with novel traits in adaptive butterfly wing radiations. EvoDevo, 5(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather WB (1960). Additions to the Drosophila Fauna of Australia. University of Queensland Papers. Department of Zoology, 1, 229–239. [Google Scholar]
- McAlpine JF (1981). Morphology and terminology – Adults In McAlpine JF, Peterson BV, Shewell GE, Teskey HJ, R. VJ, & Wood DM (Eds.), Manual of Nearctic Diptera (27th ed., pp. 9–63). Agriculture Canada Monograph. [Google Scholar]
- Mellert DJ, Robinett CC, & Baker BS (2012). Doublesex Functions Early and Late in Gustatory Sense Organ Development. PLoS ONE, 7(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CS, & Kopp A (2008). Sex combs are important for male mating success in Drosophila melanogaster. Behavior Genetics, 38(2), 195–201. 10.1007/s10519-008-9190-7 [DOI] [PubMed] [Google Scholar]
- Nishikawa H, Iijima T, Kajitani R, Yamaguchi J, Ando T, Suzuki Y, … Ozaki K (2015). A genetic mechanism for female-limited Batesian mimicry in Papilio butterfly. Nature Publishing Group, 47(4), 405–409. 10.1038/ng.3241 [DOI] [PubMed] [Google Scholar]
- Osten Sacken CR (1882). Diptera from the Philippine Islands brought home by Dr. Carl Semper. Deutsche Entomologische Zeitschrift, 24, 83–120, 187–252. [Google Scholar]
- Pagel M, Meade A, & Barker D (2004). Bayesian Estimation of Ancestral Character States on Phylogenies. Systematic Biology, 53(5), 673–684. 10.1080/10635150490522232 [DOI] [PubMed] [Google Scholar]
- Robinett CC, Vaughan AG, Knapp J-M, & Baker BS (2010). Sex and the single cell. II. There is a time and place for sex. PLoS Biology, 8(5), e1000365 10.1371/journal.pbio.1000365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo CAM, Mello B, Frazão A, & Voloch CM (2013). Phylogenetic analysis and a time tree for a large drosophilid data set (Diptera: Drosophilidae). Zoological Journal of the Linnean Society, 169(4), 765–775. 10.1111/zoj.12062 [DOI] [Google Scholar]
- Sanders LE, & Arbeitman MN (2008). Doublesex establishes sexual dimorphism in the Drosophila central nervous system in an isoform-dependent manner by directing cell number. Developmental Biology, 320, 378–390. 10.1016/j.ydbio.2008.05.543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieth HT (1952). Mating behavior within the genus Drosophila (Diptera). Bulletin of the AMNH, 99, 395–474. [Google Scholar]
- Stark JB, & O’Grady PM (2010). Morphological Variation in the Forelegs of the Hawaiian Drosophilidae. I. The AMC Clade. Journal of Morphology, 271(1), 86–103. 10.1002/jmor.10783 [DOI] [PubMed] [Google Scholar]
- Sturtevant AH (1921). The North American Species of Drosophila..pdf. Carnegie Institute of Washington Publication, 301, 1–150. [Google Scholar]
- Sturtevant AH (1927). Philippine and other Oriental Drosophilidae..pdf. Philippine Journal of Science, 32, 361–374. [Google Scholar]
- Sturtevant AH (1942). The classification of the genus Drosophila, with descriptions of nine new species. University of Texas Publication, 4213(4213), 5–51. [Google Scholar]
- Tanaka K, Barmina O, Sanders LE, Arbeitman MN, & Kopp A (2011). Evolution of sex-specific traits through changes in HOX-dependent doublesex expression. PLoS Biology, 9(8), e1001131 10.1371/journal.pbio.1001131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga C (1962). Cell lineage and differentiation on the male foreleg of Drosophila melanogaster. Developmental Biology, 4(3), 489–516. [DOI] [PubMed] [Google Scholar]
- Wilson FD, Wheeler MR, Harget M, & Kambysellis M (1969). Cytogenetic Relations in the Drosophila nasuta Subgroup of the immigrans Group of Species. The University of Texas Publication, 6918, 207–253. [Google Scholar]
- Xu J, Zhan S, Chen S, Zeng B, Li Z, James AA, … Huang Y (2017). Sexually dimorphic traits in the silkworm, Bombyx mori, are regulated by doublesex. Insect Biochemistry and Molecular Biology, 80, 42–51. 10.1016/j.ibmb.2016.11.005 [DOI] [PubMed] [Google Scholar]
- Yan XY, Zeng QT, Yang Y, & Qian YH (2006). Molecular phylogenetic relationships of Drosophila immigrans species group based on H2a-H2b spacer and Cyt b sequences. Fen Zi Xi Bao Sheng Wu Xue Bao, 39(3), 277–80. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/16944604 [PubMed] [Google Scholar]
- Yassin A, Da Lage JL, David JR, Kondo M, Madi-Ravazzi L, Prigent SR, & Toda MJ (2010). Polyphyly of the Zaprionus genus group (Diptera: Drosophilidae). Molecular Phylogenetics and Evolution, 55(1), 335–339. 10.1016/j.ympev.2009.09.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 The sex brush is male-specific in the D. immigrans species group. The first and second tarsal segments of the adult male (top two rows) and female forelegs (bottom two rows) are shown. Phylogenetic relationships are indicated above. The second and fourth rows from the top show the distal first tarsal segment at higher magnification. Red and blue arrows indicate the presence of a sex brush on the first and second tarsal segments, respectively. In addition to the sex brush, males of different species vary in the number and size of chemosensory bristles (long upwardly curved bristles). The female TBRs do not show much variation among species. The sex brushes of D. siamana and D. neohypocausta are rudimentary, with only a few supernumerary, bractless bristles found adjacent to the otherwise monomorphic TBRs in males.
Supplementary Table 1. Loci for immigrans group phylogeny References or Genbank codes for each locus used in the phylogeny.
Supplementary Table 2. Primers for immigrans group phylogeny A list of the forward and reverse primers used to generate the dataset for the immigrans species group phylogeny. Alignment length represents the size of the aligned coding sequence used in phylogenetic reconstruction.
Supplemental Text 1 The nexus file run in MrBayes to generate the phylogeny seen in Figure 5.
Supplemental Text 2 The .xml file generated in BEAUti and run in BEAST to infer the ancestral state of the sex brush and Dsx expression seen in Figure 6.