Abstract
Background
Previous studies have suggested that breast parenchymal texture features may reflect the biologic risk factors associated with breast cancer development. Therefore, combining the characteristics of normal parenchyma from the contralateral breast with radiomic features of breast tumors may improve the accuracy of digital mammography in the diagnosis of breast cancer.
Purpose
To determine whether the addition of radiomic analysis of contralateral breast parenchyma to the characterization of breast lesions with digital mammography improves lesion classification over that with radiomic tumor features alone.
Materials and Methods
This HIPAA-compliant, retrospective study included 182 patients (age range, 25–90 years; mean age, 55.9 years ± 14.9) who underwent mammography between June 2002 and July 2009. There were 106 malignant and 76 benign lesions. Automatic lesion segmentation and radiomic analysis were performed for each breast lesion. Radiomic texture analysis was applied in the normal regions of interest in the contralateral breast parenchyma to assess the mammographic parenchymal patterns. The classification performance of both individual features and the output from a Bayesian artificial neural network classifier was evaluated with the leave-one-patient-out method by using the area under the receiver operating characteristic curve (AUC) as the figure of merit in the task of differentiating between malignant and benign lesions.
Results
The performance of the combined lesion and parenchyma classifier in the differentiation between malignant and benign mammographic lesions was better than that with the lesion features alone (AUC = 0.84 ± 0.03 vs 0.79 ± 0.03, respectively; P = .047). Overall, six radiomic features—spiculation, margin sharpness, size, circularity from the tumor feature set, and skewness and power law beta from the parenchymal feature set—were selected more than 50% of the time during the feature selection process on the combined feature set.
Conclusion
Combining quantitative radiomic data from tumors with contralateral parenchyma characterizations may improve diagnostic accuracy for breast cancer.
© RSNA, 2019
Online supplemental material is available for this article.
See also the editorial by Shaffer in this issue.
Summary
Combining radiomic analysis of breast tumors with that of parenchyma may improve diagnostic accuracy for breast cancer.
Key Points
■ The addition of contralateral breast parenchyma analysis in the characterization of breast tumors at full-field digital mammography through quantitative radiomics improves lesion classification compared with using radiomic tumor features alone (area under the receiver operating characteristic curve, 0.84 vs 0.79, respectively; P = .047).
■ Because breast parenchyma may reflect the biologic risk factors associated with breast cancer development, yielding the stromal parenchyma as an indicator of precancer, the combination of parenchyma and tumor characteristics may provide a stronger predictive model of malignancy.
Introduction
Computer-aided diagnosis of breast cancer has been investigated since the early 1990s (1–4). Floyd et al (5) used an artificial neutral network to predict breast cancer from mammographic findings with an improved sensitivity and specificity compared with radiologists’ performance. Huo et al (1) developed a computer-aided diagnostic method to quantify the spiculation measure of breast lesions, achieving high diagnostic accuracy. Sahiner et al (6) applied a rubber band–straightening transform method in the breast mass lesion classification task. The results from these studies showed that computer-aided diagnostic systems can help radiologists improve their diagnostic decision making (4,7).
Breast imaging has also been used to evaluate a woman’s risk for breast cancer. The relationship between mammographic parenchymal patterns and the risk of developing breast cancer has been studied extensively. In 1976, Wolfe (8) proposed a relationship between the radiographic appearance of the breast parenchyma and breast cancer risk based on the visual assessment of the breast. Other researchers (9,10) have also investigated mammographic parenchymal patterns by assessing the extent of fibroglandular tissue on mammograms. In addition, computerized radiographic texture analysis (11–13) has been performed to characterize the breast parenchymal patterns in the assessment of breast cancer risk. These studies demonstrated an association between breast cancer risk and mammographic patterns characterized by quantitative texture features. Thus, parenchymal texture features have been used as imaging markers for the assessment of breast cancer risk and their association with breast cancer development (11,12,14,15).
In addition to their utility in risk assessment, parenchymal texture features can help evaluate or predict the stage and subtype of breast cancer (16). This suggests that breast parenchymal texture features may reflect the biologic risk factors associated with breast cancer development because parenchymal stromal cells play an important role in breast tumor formation and development (17,18). Because the stromal parenchyma may be indicative of a precancerous state, a combination of parenchymal texture and tumor characteristics may yield stronger predictive models. Previous studies (14,19) have also found a high correlation between parenchymal texture features extracted from the left and right breast due to left-right breast symmetry.
We hypothesized that by combining characteristics of normal parenchyma from the contralateral breast with mammographic radiomic features of breast tumors, the accuracy of digital mammography in the diagnosis of breast cancer may be improved (Fig 1). We used normal breast parenchyma from the contralateral breast instead of the ipsilateral breast because parenchyma in the ipsilateral breast may be affected by the presence of breast lesions. The purpose of our study was to assess whether the addition of radiomic analysis of contralateral breast parenchyma to the characterization of breast lesions with digital mammography improves lesion classification over that with radiomic tumor features alone.
Figure 1:
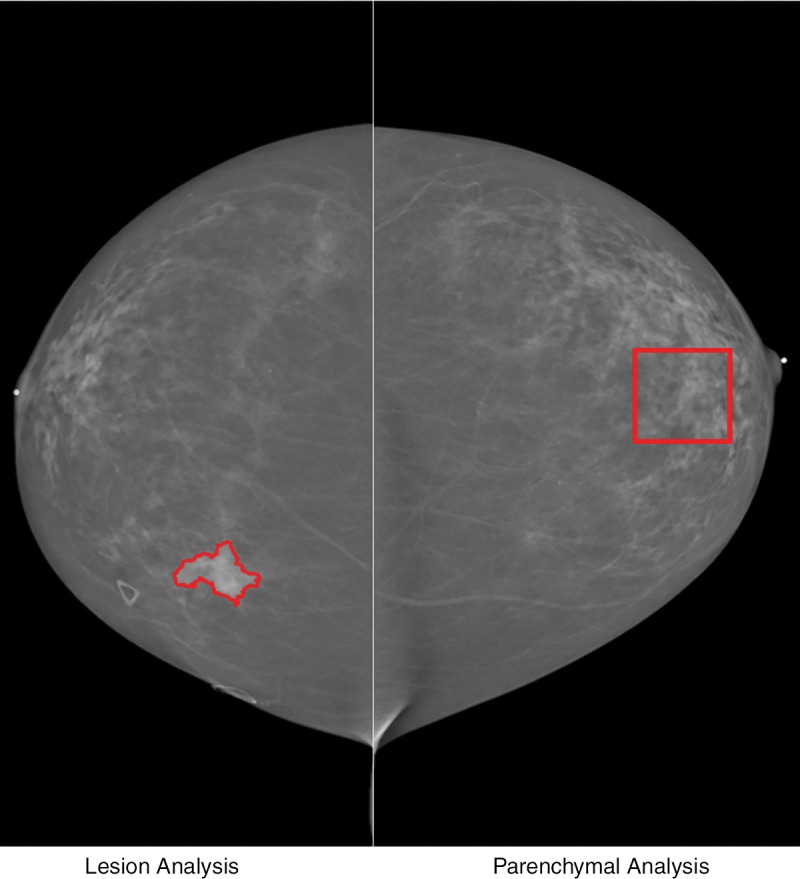
Illustration of computerized analysis method used on digital mammograms. Quantitative imaging analysis was performed in both lesion (left) and region of interest from normal contralateral breast (right). Segmentation outline was obtained from a dual-stage computerized segmentation method.
Materials and Methods
Data Set
This retrospective study was approved by the institutional review board and compliant with the Health Insurance Portability and Accountability Act. The requirement to obtain informed consent was waived. Images were obtained at the University of Chicago Medical Center (Chicago, Ill) from June 2002 to July 2009. The full-field digital mammograms were acquired by using a Senographe 2000D system (GE Medical Systems, Waukesha, Wis) at 12-bit quantization level and 100-μm pixel size. Images were obtained in 182 patients (age range, 25–90 years; mean age ± standard deviation, 55.9 years ± 14.9). We used the “For Presentation” format (ie, processed data) for our study.
The data set included 182 biopsy-proven breast lesions, 106 of which were malignant and 76 of which were benign. The malignant lesions ranged in diameter from 0.5 to 3.3 cm (mean, 1.9 cm ± 0.7), and the benign lesions ranged in diameter from 0.5 to 2.8 cm (mean, 1.5 cm ± 0.6). The patients had no previous cancer. Their contralateral mammograms were reviewed by a breast radiologist (D.S.) with 9 years of experience and were used only if no detectable abnormalities were observed. If there were multiple lesions, only the index lesion was used in our study. All patients were required to have standard mammographic views of the contralateral breast for inclusion in the computerized parenchymal analysis.
Each breast lesion had been assigned a Breast Imaging Reporting and Data System (BI-RADS) score by a dedicated breast radiologist at the time of the initial clinical image interpretation. The standardized BI-RADS lexicon enables malignancy risk stratification for each descriptor and helps guide management decisions. BI-RADS category 4 and 5 lesions are recommended for biopsy and carry a malignancy risk of 2% to more than 95%. The computer code referred to in the following section is available from the authors upon request.
Radiomic Analysis of Breast Lesions
The methods for computerized analysis of breast lesions at mammography have been described in detail elsewhere (2,20). Briefly, several steps are involved in the analysis. First, each breast lesion center was indicated by a radiologist, who in this study was a breast radiologist (D.S.). Second, breast lesions were automatically segmented from the parenchymal background by using a dual-stage segmentation method (20), which included an initial segmentation with a radial gradient index–based method followed by an active contour model. Third, radiomic lesion features (Appendix E1, Table E1 [online]), in terms of mathematical descriptors (21,22), were automatically extracted from the image data of lesions and surrounding tissues. Finally, subsequent classification was performed to differentiate between malignant and benign lesions by using a Bayesian artificial neural network (BANN) classifier (23).
A total of 32 features (Appendix E1, Table E1 [online]) were extracted from each tumor; these were used to characterize the tumor size, shape, spiculation, margin sharpness, radial gradient index, and texture (1,21,22). The size feature “effective diameter” is defined as the diameter of a circle yielding the same area as the segmented lesion. Shape features include circularity, irregularity, and compactness of the breast tumor. To quantify the degree of spiculation of a mass lesion, the full width at half maximum of the normalized edge gradient distribution relative to the radial direction is calculated. Margin sharpness is defined as the average gradient along the margin of the lesion. Radial gradient index corresponds to the average proportion of the gradient direction and thus contains shape information. Density features are radiomic features related to radiographic density information of the tumor and parenchyma, such as average gray value of the lesion and contrast. Texture features are calculated on the basis of the gray-level co-occurrence matrix. These features are used to quantify the different characteristics of a lesion, such as gray-level dependence, homogeneity, randomness, brightness, and variation.
Radiomic Analysis of Breast Parenchymal Patterns
The method used for computerized parenchymal texture analysis has been detailed in previous publications (24,25). Briefly, parenchymal analysis was performed on the normal contralateral breast, where each contralateral mammogram had no breast abnormalities present when reviewed by the breast radiologist (D.S.). Regions of interest (ROIs) of 256 × 256 pixels (25.6 × 25.6 mm) were manually selected by a computer scientist (L.L., with 18 years of experience in breast cancer image analysis) from the central breast region immediately behind the nipple in the craniocaudal view of the full-field digital mammographic images (Fig 2) because they usually included the dense parts of the breast (24). ROIs were selected such that regions along the skin line that contain subcutaneous fat were not included.
Figure 2:
Digital mammogram of normal contralateral breast shows selection of region of interest from central breast region behind nipple.
Quantitative radiomic texture features were extracted from each ROI to characterize the mammographic parenchymal patterns. The intraclass correlation coefficients from our previous study (26) on reliability of the reader’s ROI placement indicated high reliability of ROI selection and parenchymal analysis. A total of 45 texture features (Appendix E1, Table E2 [online]) were calculated from each individual ROI to characterize the local composition, contrast, homogeneity, and coarseness of breast parenchyma (22,25). These features are based on (a) gray-level histogram analysis to characterize the local tissue composition; (b) fractal analysis, including box-counting and Minkowski methods; (c) edge-frequency analysis; (d) Fourier analysis, including root mean square variation, first moment of power spectrum, and power law beta from power spectral analysis; and (e) textural analysis using neighborhood gray-tone difference matrix and gray-level co-occurrence matrix to characterize the special relationship among gray levels.
Classifier Development
The radiomic features were standardized with zero mean and unit variance across all cases before input to the classification tasks. Stepwise feature selection (27) was used with multiple linear regression analysis to select the subset of features for the classification task by using the Wilks Λ criterion. To reduce database bias, stepwise feature selection and classification were conducted concurrently within a leave-one-case-out cross-validation manner. Features were selected from each cross-validation iteration and used in the same iteration for classification with a BANN classifier. Forward feature addition and backward feature removal were both involved in the feature selection process. The stepwise feature selection process was automatically stopped when no statistically significant improvement was obtained by adding or removing features in each leave-one-case-out cross-validation iteration on the training data set. Note that in each iteration, the data set was split into two data sets: N − 1 cases for training and one case for testing (N is the total number of cases in our study). The BANN classifier was trained by using the N − 1 training cases with the selected features and then tested on one test case. This process was repeated N times for each classification task: that is, in each leave-one-case-out cross-validation iteration, feature selection was conducted only on the corresponding training cases; thus, a different feature set could result at each iteration.
However, to appreciate the important characteristics of the tumor and parenchyma, we also examined features that were selected more than 50% of the time during the iterative process. This same feature selection method was performed throughout our study.
To evaluate the additive role of contralateral parenchyma texture analysis to lesion characterization, classifications were performed by using (a) breast tumor features alone and (b) radiomic features extracted from both breast tumor and contralateral parenchymal stroma.
Statistical Analysis
Receiver operating characteristic analysis (28) was conducted to assess the performance of each classifier, with the area under the receiver operating characteristic curve (AUC) serving as the figure of merit in the task of differentiating between malignant and benign lesions by using output from the relevant BANN classifier as a decision variable. The statistical significance for the difference between classifiers’ performance in AUC was assessed by using software (ROCKIT; http://metz-roc.uchicago.edu/MetzROC/software). The Holm-Bonferroni method (29) was applied to adjust for multiple comparisons (αT = .05).
Results
The data set included 182 biopsy-proven breast lesions, 106 of which were malignant and 76 of which were benign, as described in Table 1. Of the 76 benign lesions, 69 were classified as BI-RADS category 4 and seven as BI-RADS category 5. Of the 106 malignant lesions, 30 were classified as BI-RADS category 4 and 76 as BI-RADS category 5.
Table 1:
Baseline Characteristics of Study Population
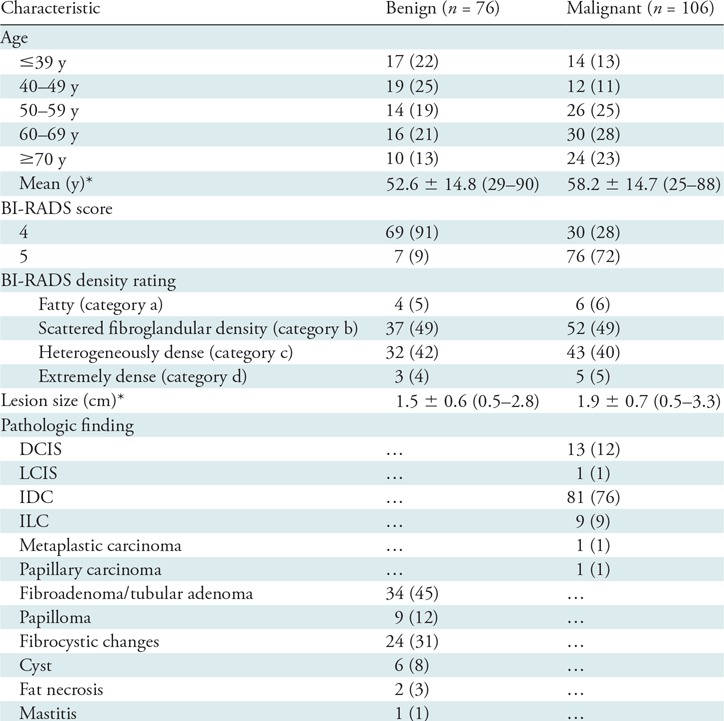
Note.—Except where indicated, data are numbers of patients, with percentages in parentheses. BI-RADS = Breast Imaging Reporting and Data System, DCIS = ductal carcinoma in situ, IDC = invasive ductal carcinoma, ILC = invasive lobular carcinoma, LCIS = lobular carcinoma in situ.
*Data are means ± standard deviations, with ranges in parentheses.
Radiomic Analysis of Breast Lesions
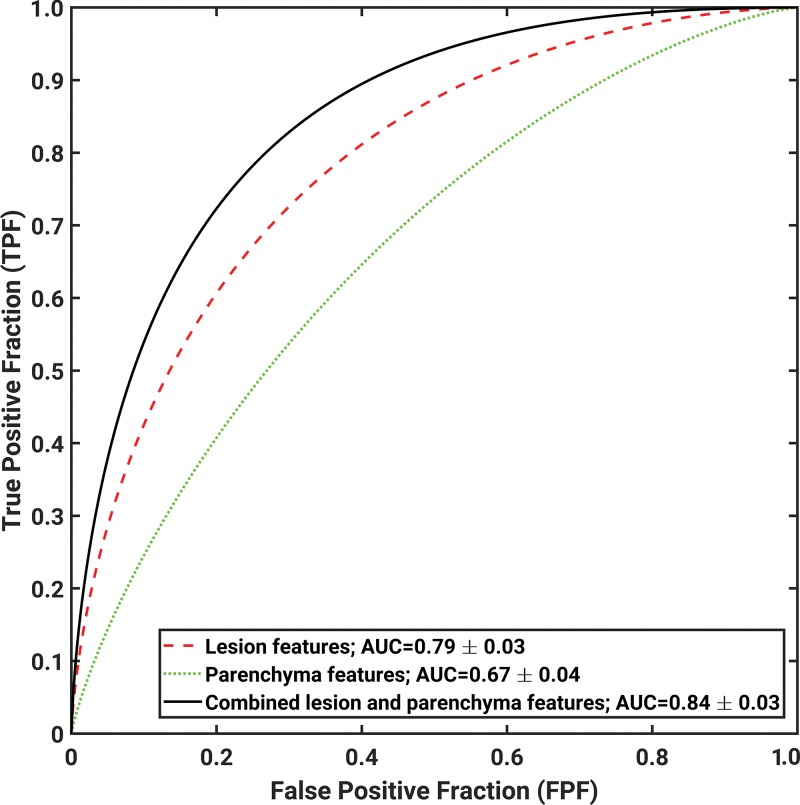
When we evaluated the tumor characteristics alone, an AUC of 0.79 ± 0.03 was obtained by using the BANN classifier outputs in the differentiation between malignant and benign lesions (Table 2, Fig 3). Individual radiomic lesion features had poor to moderate classification performance in lesion characterization, with AUCs ranging from 0.53 to 0.75. Four features were selected more than 50% of the time during the feature selection process: spiculation, margin sharpness, size, and circularity.
Table 2:
Classification Performance with Breast Tumor Features, Parenchymal Features, and Combined Features in the Differentiation of Malignant from Benign Breast Lesions
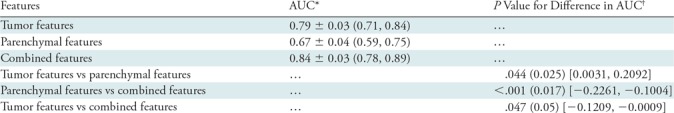
Note.—The Holm-Bonferroni method was implemented to correct for multiple comparisons. AUC = area under the receiver operating characteristic curve.
*Data are means ± standard deviations. Numbers in parentheses are 95% confidence intervals.
†Numbers in parentheses are the significance level, and numbers in brackets are the 95% confidence intervals for the difference in AUC.
Figure 3:
Receiver operating characteristic curves indicate performance of computerized analysis in differentiation between malignant and benign lesions by using lesion features alone, parenchymal features alone, and combined lesion and parenchymal features as decision variables. AUC = area under the receiver operating characteristic curve.
Radiomic Analysis of Breast Parenchymal Patterns
When we evaluated the parenchymal features alone, an AUC of 0.67 ± 0.04 was obtained by using the BANN classifier outputs in the differentiation between malignant and benign lesions (Table 2, Fig 3). Edge gradient, contrast, power law beta, and two texture features based on gray-level co-occurrence matrix were the most frequently selected features.
Radiomic Analysis of Combined Features
When we assessed the additive role of parenchymal stroma in breast cancer diagnosis, the classification performance with combined lesion and parenchyma classifier was better than that with lesion features alone (AUC = 0.84 ± 0.03 vs 0.79 ± 0.03, respectively; P = .047) (Table 2, Fig 3). Overall, six radiomics features—spiculation, margin sharpness, size, circularity from the tumor feature set, skewness, and power law beta from the parenchymal feature set—were selected more than 50% of the time during the feature selection process on the combined feature set.
We also investigated the additive role of the parenchyma to the cancer diagnosis relative to patient age, BI-RADS rating, and breast density. The results followed the same trend as the analysis on the entire data set (Appendix E1, Table E3 [online]). Given the reduction in the data sets as one subcategorizes the cases on the basis of patient age, BI-RADS rating, and breast density, one must be careful in interpreting the statistical evaluations; thus, we report just the classification performances in terms of AUCs to demonstrate the similar trends.
Discussion
In our study, we compared breast lesion classification with use of radiomic tumor features alone and with a combination of tumor and parenchymal texture features. We assessed the additive role of the breast parenchyma features in cancer diagnosis. By combining tumor features with normal parenchymal texture features, we observed improved (P = .047) lesion classification performance (AUC, 0.84) compared with that seen with use of tumor features alone (AUC, 0.79).
We are aware of various publications on the use of lesion radiomic data in the classification of malignant and benign masses, and we are aware of the various publications on the use of parenchymal features to predict cancer risk. Given the different databases and image analysis algorithms, some studies have higher AUCs and some have lower AUCs. However, to the best of our knowledge, ours is the first study to combine both breast tumor and contralateral parenchyma characteristics in the prediction of breast lesion malignancy.
The results from our study suggest that parenchymal stroma plays an important role in breast tumor formation. Several studies (8,30,31) have linked mammographic parenchymal patterns to the risk of developing breast cancer. Breast cancer is a complex disease involving interactions between tumor cells and host stromal cells. In addition, studies (32,33) also found that parenchyma stromal cells play an important role in breast tumor formation and development, acting as the stroma fostering tumor growth (34,35). These studies provide a biologic basis that supports our finding that combining both tumor and stromal characteristics may improve the accuracy of digital mammography in the diagnosis of breast cancer.
Our study has some limitations. The small data set limited us to performing leave-one-case-out cross-validation as opposed to using large independent training and testing sets. In addition, radiomic analysis of the parenchyma was performed only on standard-view images of the contralateral breast, thus limiting the cases collected. In addition, the parenchymal texture features were extracted only from the central breast region of the contralateral breast. Furthermore, mammographic images used in our study were in the “For Presentation” format. The “For Processing” format (ie, raw data) may have been better suited for our study because the raw data could have been preprocessed to minimize the effect due to different preprocessing methods. However, we did not have access to the “For Processing” data; thus, we chose to use the “For Presentation” format (ie, processed data). All mammograms used in our study were acquired with mammographic machines from the same manufacturer. We expect that this would minimize the effect due to different preprocessing methods from different manufacturers. In addition, many clinical practices use only the “For Presentation” format for interpretation; thus, those may be the only ones available for analysis.
In future studies, we plan to evaluate the breast parenchyma symmetric to the tumor location on the contralateral breast, the entire contralateral breast parenchyma, and parenchyma around the breast lesion to further understand the role of parenchyma stroma in the assessment of lesion malignancy. In addition, we will perform quantitative imaging analysis on both raw and processed images from various manufacturers of digital mammography units to assess the robustness of the methods for parenchyma analysis and breast tumor characterization and to allow for more generalization and flexibility in different clinical settings. Further study with a larger data set is needed to validate the findings from our study.
In summary, quantitative imaging analysis of both breast lesions and normal parenchyma can be conducted to characterize the tumor and stroma, yielding a diagnostic signature. Combining radiomic analysis of breast tumors with that of parenchyma may improve the accuracy of digital mammography in the diagnosis of breast cancer.
APPENDIX
Disclosures of Conflicts of Interest: H.L. disclosed no relevant relationships. K.R.M. disclosed no relevant relationships. L.L. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: receive royalties from Hologic. Other relationships: disclosed no relevant relationships. D.S. disclosed no relevant relationships. M.L.G. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: has patents planned, pending, or issued; institution has patents planned, pending, or issued; institution receives royalties; receives royalties from Hologic, GE Medical Systems, MEDIAN Technologies, Riverain Medical, Mitsubishi, and Toshiba; is a stockholder in R2 Technology/Hologic; is cofounder, equity holder, and scientific advisor in Quantitative Insights; institution has stock/stock options. Other relationships: disclosed no relevant relationships.
Abbreviations:
- AUC
- area under the receiver operating characteristic curve
- BANN
- Bayesian artificial neural network
- BI-RADS
- Breast Imaging Reporting and Data System
- ROI
- region of interest
References
- 1.Huo Z, Giger ML, Vyborny CJ, et al. Analysis of spiculation in the computerized classification of mammographic masses. Med Phys 1995;22(10):1569–1579. [DOI] [PubMed] [Google Scholar]
- 2.Li H, Giger ML, Yuan Y, et al. Evaluation of computer-aided diagnosis on a large clinical full-field digital mammographic dataset. Acad Radiol 2008;15(11):1437–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varela C, Timp S, Karssemeijer N. Use of border information in the classification of mammographic masses. Phys Med Biol 2006;51(2):425–441. [DOI] [PubMed] [Google Scholar]
- 4.Lo JY, Baker JA, Kornguth PJ, Floyd CE, Jr. Computer-aided diagnosis of breast cancer: artificial neural network approach for optimized merging of mammographic features. Acad Radiol 1995;2(10):841–850. [DOI] [PubMed] [Google Scholar]
- 5.Floyd CE, Jr, Lo JY, Yun AJ, Sullivan DC, Kornguth PJ. Prediction of breast cancer malignancy using an artificial neural network. Cancer 1994;74(11):2944–2948. [DOI] [PubMed] [Google Scholar]
- 6.Sahiner B, Chan H-P, Petrick N, Helvie MA, Goodsitt MM. Computerized characterization of masses on mammograms: the rubber band straightening transform and texture analysis. Med Phys 1998;25(4):516–526. [DOI] [PubMed] [Google Scholar]
- 7.Chan HP, Sahiner B, Helvie MA, et al. Improvement of radiologists’ characterization of mammographic masses by using computer-aided diagnosis: an ROC study. Radiology 1999;212(3):817–827. [DOI] [PubMed] [Google Scholar]
- 8.Wolfe JN. Breast patterns as an index of risk for developing breast cancer. AJR Am J Roentgenol 1976;126(6):1130–1137. [DOI] [PubMed] [Google Scholar]
- 9.Brisson J, Diorio C, Mâsse B. Wolfe’s parenchymal pattern and percentage of the breast with mammographic densities: redundant or complementary classifications? Cancer Epidemiol Biomarkers Prev 2003;12(8):728–732. [PubMed] [Google Scholar]
- 10.Boyd NF, Martin LJ, Stone J, Greenberg C, Minkin S, Yaffe MJ. Mammographic densities as a marker of human breast cancer risk and their use in chemoprevention. Curr Oncol Rep 2001;3(4):314–321. [DOI] [PubMed] [Google Scholar]
- 11.Manduca A, Carston MJ, Heine JJ, et al. Texture features from mammographic images and risk of breast cancer. Cancer Epidemiol Biomarkers Prev 2009;18(3):837–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei J, Chan HP, Wu YT, et al. Association of computerized mammographic parenchymal pattern measure with breast cancer risk: a pilot case-control study. Radiology 2011;260(1):42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gastounioti A, Conant EF, Kontos D. Beyond breast density: a review on the advancing role of parenchymal texture analysis in breast cancer risk assessment. Breast Cancer Res 2016;18(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huo Z, Giger ML, Wolverton DE, Zhong W, Cumming S, Olopade OI. Computerized analysis of mammographic parenchymal patterns for breast cancer risk assessment: feature selection. Med Phys 2000;27(1):4–12. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Giger ML, Lan L, et al. Computerized analysis of mammographic parenchymal patterns on a large clinical dataset of full-field digital mammograms: robustness study with two high-risk datasets. J Digit Imaging 2012;25(5):591–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sala E, Solomon L, Warren R, et al. Size, node status and grade of breast tumours: association with mammographic parenchymal patterns. Eur Radiol 2000;10(1):157–161. [DOI] [PubMed] [Google Scholar]
- 17.Korkaya H, Liu S, Wicha MS. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J Clin Invest 2011;121(10):3804–3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weigelt B, Bissell MJ. Unraveling the microenvironmental influences on the normal mammary gland and breast cancer. Semin Cancer Biol 2008;18(5):311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Byng JW, Boyd NF, Little L, et al. Symmetry of projection in the quantitative analysis of mammographic images. Eur J Cancer Prev 1996;5(5):319–327. [DOI] [PubMed] [Google Scholar]
- 20.Yuan Y, Giger ML, Li H, Suzuki K, Sennett C. A dual-stage method for lesion segmentation on digital mammograms. Med Phys 2007;34(11):4180–4193. [DOI] [PubMed] [Google Scholar]
- 21.Huo Z, Giger ML, Vyborny CJ, Wolverton DE, Schmidt RA, Doi K. Automated computerized classification of malignant and benign masses on digitized mammograms. Acad Radiol 1998;5(3):155–168. [DOI] [PubMed] [Google Scholar]
- 22.Chen W, Giger ML, Li H, Bick U, Newstead GM. Volumetric texture analysis of breast lesions on contrast-enhanced magnetic resonance images. Magn Reson Med 2007;58(3):562–571. [DOI] [PubMed] [Google Scholar]
- 23.Bishop CM. Neural networks for pattern recognition. Oxford, England: Clarendon Press/Oxford University Press, 1995. [Google Scholar]
- 24.Li H, Giger ML, Huo Z, et al. Computerized analysis of mammographic parenchymal patterns for assessing breast cancer risk: effect of ROI size and location. Med Phys 2004;31(3):549–555. [DOI] [PubMed] [Google Scholar]
- 25.Li H, Giger ML, Lan L, Janardanan J, Sennett CA. Comparative analysis of image-based phenotypes of mammographic density and parenchymal patterns in distinguishing between BRCA1/2 cases, unilateral cancer cases, and controls. J Med Imaging (Bellingham) 2014;1(3):031009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gierach GL, Li H, Loud JT, et al. Relationships between computer-extracted mammographic texture pattern features and BRCA1/2 mutation status: a cross-sectional study. Breast Cancer Res 2014;16(4):424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Golugula A, Lee G, Madabhushi A. Evaluating feature selection strategies for high dimensional, small sample size datasets. In: 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE, 2011; 949–952. [DOI] [PubMed] [Google Scholar]
- 28.Pesce LL, Metz CE. Reliable and computationally efficient maximum-likelihood estimation of “proper” binormal ROC curves. Acad Radiol 2007;14(7):814–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat 1979;6(2):65–70. [Google Scholar]
- 30.McCormack VA, dos Santos Silva I. Breast density and parenchymal patterns as markers of breast cancer risk: a meta-analysis. Cancer Epidemiol Biomarkers Prev 2006;15(6):1159–1169. [DOI] [PubMed] [Google Scholar]
- 31.Saftlas AF, Szklo M. Mammographic parenchymal patterns and breast cancer risk. Epidemiol Rev 1987;9(1):146–174. [DOI] [PubMed] [Google Scholar]
- 32.Shekhar MP, Pauley R, Heppner G. Host microenvironment in breast cancer development: extracellular matrix-stromal cell contribution to neoplastic phenotype of epithelial cells in the breast. Breast Cancer Res 2003;5(3):130–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiseman BS, Werb Z. Stromal effects on mammary gland development and breast cancer. Science 2002;296(5570):1046–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fidler IJ, Poste G. The “seed and soil” hypothesis revisited. Lancet Oncol 2008;9(8):808. [DOI] [PubMed] [Google Scholar]
- 35.Mao Y, Keller ET, Garfield DH, Shen K, Wang J. Stromal cells in tumor microenvironment and breast cancer. Cancer Metastasis Rev 2013;32(1-2):303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.