Abstract
Involvement of biofilm formation process during phosphate (P) solubilization by rhizobacterial strains is not clearly understood. Scanning electron microscopic observations revealed prominent biofilm development on tricalcium phosphate as well as on four different rock phosphate granules by two P solubilizing rhizobacteria viz. Burkholderia tropica P4 and B. unamae P9. Variation in the biofilm developments were also observed depending on the total P content of insoluble P used. Biofilm quantification suggested a strong correlation between the amounts of available P and degrees of biofilm formation. Lower concentrations of soluble P directed both the organisms towards compact biofilm development with maximum substratum coverage. Variation in the production of extracellular polymeric substances (EPS) in the similar pattern also suggested its close relationship with biofilm formation by the isolates. Presence of BraI/R quorum sensing (QS) system in both the organisms were detected by PCR amplification and sequencing of two QS associated genes viz. braR and rsaL, which are probably responsible for biofilm formation during P solubilization process. Overall observations help to hypothesize for the first time that, biofilm on insoluble P granules creates a close environment for better functioning of organic acids secreted by Burkholderia strains for maximum P solubilization during P deficient conditions.
Introduction
Biofilm formation can be defined as a process whereby microbial cells irreversibly attach to and grow on a surface with the production of extracellular organic polymeric matrix resulting in an alteration in the phenotype of the microorganisms with respect to their growth rate and gene transcription. For bacterial organisms, the process of biofilm formation is advantageous as it offers protection to the producing organisms from antibiotics1, disinfectants or dynamic environmental conditions. It also helps them to survive in nutrient deficient or oligotrophic conditions2. Microbial surface attachment is considered as an important factor during competition for carbon sources and trace elements in the growth limiting conditions especially in the marine environment3. About 99% of the world population of bacteria produce diverse biofilm structures during various stages of their growth4. Bacterial biofilms may form on a wide variety of surfaces, including living tissues, indwelling medical devices, industrial or potable water system piping, or natural aquatic systems5. Different types of noncellular materials like mineral crystals, corrosion particles, clay or silt particles, or blood components, may also be found in the biofilm matrix depending on the environment of biofilm development5. Biofilms in natural environments are commonly found on coating around various particles that occur in soils, lake, river and others. In soil, microbial biofilms play a crucial role in the formation and stability of soil aggregates, weathering of different minerals as well as degradation and sequestration of organic carbon6. Biofilm formation in drinking water pipe network is a major cause for deterioration of water quality and also for operational problems. It is responsible for the increase of pathogenic bacterial load, decrease of dissolved oxygen, change of taste and odor, and red or black coloration of supplied drinking water7. On the other hand, biological wastewater treatment is getting much attention in the recent years and bacterial biofilms impart significant roles for such treatments. Based on bacterial biofilm different types of biofilters have been designed which are useful for successful removal of organic and inorganic pollutants from the wastewater. Another important aspect of microbial biofilms in natural environment is their ability to stabilize sediment which is known as biostabilization. It gives protection to the sediments from erosion. Even after erosion microbial biofilm promote sediment aggregation by altering the floc properties of sediments8.
Production of extracellular polymeric substances (EPS) is one of the important features of most of the biofilm forming bacteria and it is considered as the major factor that influences biofilm formation9. More than 90% of the dry mass in a mature biofilm is represented by EPS components which include different polysaccharides, proteins, nucleic acids, lipids and other biomolecules10. EPS is responsible for bacterial cells adhesion to surfaces and is also responsible for maintaining the three-dimensional architecture and morphology of the biofilm matrix11,12. The extensive production of EPS occurs during the specific adhesion stage of biofilm development for successful attachment. EPS also protect the bacterial cells from various stresses like antimicrobials, oxidation and metallic cations. In addition, EPS also helps to retain quorum sensing (QS) signaling molecules, different extracellular enzymes and other metabolic products which ultimately supports cell-cell communication and degradation of substances13.
Competition for nutrients and other growth requirements is definitely an important driving force for the biofilm development. Increased cell density favors chemical signals for communicating with the responding cells for social interactions in biofilms. Furthermore, the expression of different adhesins, their cognate receptors, and exopolymeric components by individual cell types within a biofilm community can contribute to overall biofilm development14,15. Bacteria communicate with neighbors and monitor their population density by producing and sensing signaling molecules in a process called quorum sensing (QS)16. Many bacteria are capable of using a QS mechanism to regulate biofilm formation and other social activities17. The concentration of the signaling molecule increases alongside the bacterial population density and, when it reaches a significant level, bacteria respond and modulate target gene expression. In Gram negative bacteria, QS system involves the production and response to a signaling molecule known as acylated homoserine lactone (AHL). On the other hand peptide-based signal molecules like linear and crystallized oligopeptides have been reported in case of Gram-positive bacteria18. In addition, some other signaling molecules like gamma-butyrolactones (GBLs) of Streptomyces spp. and the A-signaling amino acids of Myxococcus xanthus had also been reported and were well studied19. The signal molecules are either recognized by specific extracytoplasmic sensor proteins or they can transit across the cell membrane and interact and directly modulates the function of a cytoplasmic target protein. In addition to biofilm formation, QS regulation also plays vital roles in many other processes which include interaction of bacteria with higher organisms, regulation of virulence, conjugation, expression and regulation of genes related to the production of toxins, enzymes, antibiotics and other secondary metabolites18. In Gram negative bacteria although QS circuit has been identified for several species but in most of the cases it resembles the canonical QS circuits (lux operon) of Vibrio fischeri. QS circuit in Gram negative bacteria contains genes homologous to V. fischeri luxI and luxR genes. The product of luxI is responsible for biosynthesis of a specific AHL molecule which binds with the receptor protein LuxR. LuxR-AHL complex then activates the target gene transcription18.
In the genus Burkholderia, QS system has been extensively studied. The species of BCC complex share a conserved QS system known as CepI/R20 that produces and responds to N-octanoyl homoserine lactone (C8-AHL). CepI/R QS system regulates virulence and several important phenotypes like biofilm formation and siderophore production21–23. On the other hand Suárez-Moreno et al.24 reported BraI/R quorum sensing system which produces and responds mainly to C12-3-oxo-AHL and stringently regulated by a repressor RsaL. Later on, they have also reported that BraI/R QS system is related to EPS production and biofilm formation by some plant associated Burkholderia spp.16.
Although a large number of soil bacteria were found to solubilize insoluble P but the role of plant associated Burkholderia spp. as potent P solubilizers have been reported in the recent years. In the present study two potent P solubilizing bacterial (PSB) strains viz. Burkholderia tropica P4 and Burkholderia unamae P9 were selected which were isolated from the rhizospheric soil of Lycopodiella cernua (L.) Pic. Serm. grown in the P deficient lateritic soil of West Bengal, India25. They were found effective to release appreciable amounts of soluble P from insoluble tricalcium phosphate (TCP) or different rock phosphates25. Earlier study also suggested a decrease of medium pH by both the isolates during P solubilization process. Although secretion of organic acids by Gram negative bacteria were considered as one of the key mechanisms for inorganic P solubilization, involvement of biofilm formation, EPS production and quorum sensing process during P solubilization were not studied earlier especially in these bacterial species. Thus for finding out the detail mechanisms of P solubilization by the isolates, the present study was focused on their biofilm forming properties during solubilization of insoluble P granules. In addition, EPS production by these organisms was also quantified for understanding their role in biofilm formation as well as in P solubilization processes. Lastly, the presence of QS systems in both the bacterial strains was detected which probably helps in biofilm formation during solubilization of insoluble P by them.
A large number of P solubilizing bacteria has been reported earlier which were able to produce good amount of soluble P in vitro but most of the cases their efficiencies in the actual field condition were not satisfactory. Investigation on their biofilm forming properties will be helpful for their proper application as well as for the preparation of suitable formulations to get satisfactory results. In addition, bioprocessing of rock phosphate ores using biofilm forming microorganisms is mentioned earlier26 but this field is not well studied till date due to lack of sufficient information. Application of biofilm forming Burkholderia spp. for such bioprocessing using appropriate bioreactors will also be a promising approach for the production of soluble P by avoiding conventional acid digestion methods.
Results
Biofilm formation on insoluble P granules
In order to observe the biofilm formation by the strains B. tropica P4 and B. unamae P9 on the granules of TCP as well as four different rock phosphates viz. Jordan Rock Phosphate (JRP), Purulia Rock Phosphate (PRP), Udaypur Rock Phosphate (URP) and Mussoorie Rock Phosphate (MRP), bacteria were allowed to grow for 48 hours at 28 °C and observed for their physical interactions. Scanning electron microscopic (SEM) observation indicated improved adhesions of bacterial cells on insoluble P granules. Both the strains were effectively produced compact biofilm structures on TCP (Fig. 1A,D) as well as on rock phosphate granules. In addition to firm attachments with insoluble P granules, prominent cell to cell adhesions were also observed for both the strains. Production of extracellular polymeric matrix surrounding those biofilm structures were also observed during SEM study. Although formation of biofilms was found for all the different insoluble P granules, but the extent of biofilm formation varied depending upon the nature of insoluble P granules used in the National Botanical Research Institute Phosphate Growth Medium devoid of Yeast Extract (NBRIY) broth. Among the four different rock phosphates, maximum bacterial attachments as well as cell to cell adhesions were observed on MRP granules for both the isolates (Fig. 1C,F). Very dense extracellular polymeric matrix surrounding the biofilm structures was also found under the higher magnification of SEM for MRP granules in comparison to other rock phosphate used (Fig. 1C,F). On the other hand, comparatively reduced bacterial adherences with thin EPS matrices were noticed for TCP (Fig. 1A,D) and Jordan Rock Phosphate (JRP) granules (Fig. 1B–E).
Figure 1.
Scanning electron micrographs of biofilm formation by bacterial strains on insoluble phosphate granules. (A–C) B. tropica P4; (D–E) B. unamae P9; (A,D) on TCP; (B,E) on JRP; (C,F) on MRP.
Variation in biofilm formation at different concentrations of available phosphate
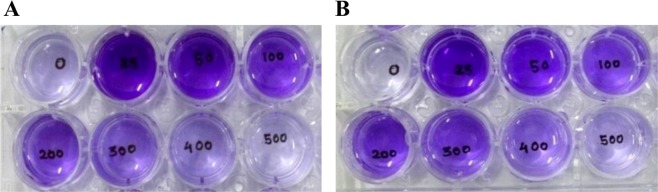
Degrees of biofilm formation by the two species of Burkholderia were quantified using different concentrations of soluble P (K2HPO4) in NBRIY medium for finding out the relationship of biofilm formation with available P. When the crystal violet stain was extracted using 33% acetic acid from different wells of 24 wells culture plates, visual differences in biofilm formation at different concentrations of K2HPO4 were noticed for both the strains (Fig. 2A,B). UV-VIS spectrophotometric measurements of optical densities (OD) of crystal violet solutions indicated maximum biofilm formation by both the strains in the presence of 25 μg/ml of K2HPO4 (Fig. 3). Maximum OD of 2.135 ± 0.15 and 1.86 ± 0.17 were recorded for B. tropica P4 and B. unamae P9 respectively when 25 μg/ml of K2HPO4 was used. Biofilm formation significantly (P < 0.001) decreased along with increased concentrations of K2HPO4 for the strains (Fig. 3) and OD of crystal violet solutions suggested almost no biofilm formation by them in the presence of K2HPO4 at a concentration of 500 μg/ml. In the complete absence of K2HPO4 in the growth medium, biofilm formations were also negligible for both the organisms. Similar pattern of biofilm formation was also observed for both the strains when the K2HPO4 was replaced with different concentrations of Na2HPO4.
Figure 2.
Crystal violet staining of biofilm developed by bacterial strains in the presence of different concentration of K2HPO4 (0–500 µg/ml). (A) B. tropica P4; (B) B. unamae P9.
Figure 3.

Quantification of biofilm development and planktonic cell growth of bacterial strains at different concentrations of K2HPO4 (0–500 µg/ml).
Planktonic cell densities at different concentrations of available P
Planktonic cell densities were also determined at different concentrations of available P in NBRIY medium for both the strains. Counting of colony forming units (CFUs) after plating showed that unlike biofilm formation planktonic cell densities increased significantly (P < 0.001) along with increased concentration of soluble P (K2HPO4) for both B. tropica P4 and B. unamae P9 (Fig. 3). Maximum numbers of planktonic cells were observed when 500 µg/ml of K2HPO4 was used in NBRIY broth. Reduced planktonic cell densities were also observed in the complete absence of K2HPO4 in the medium.
Biofilm morphologies at different concentrations of available P
When the polyester films placed in the wells of 24 wells plates were subjected to light microscopic studies after crystal violet staining, prominent structural variations of biofilm morphologies were observed depending on the concentrations of K2HPO4 used in NBRIY broth. Under light microscope, compact biofilm structures were observed for both the strains at lower concentrations of K2HPO4 in comparison to higher concentrations. Thick biofilm structures with maximum substratum coverage were observed in the presence of 25 µg/ml of K2HPO4 (Fig. 4A,E). Both the thickness as well as substratum coverage markedly decreased along with increased concentrations of K2HPO4 for B. tropica P4 (Fig. 4B–D) as well as for B. unamae P9 (Fig. 4F–H). Almost no patchy microcolonies were observed for both the strains above 300 µg/ml of K2HPO4 (4D,H). On the other hand biofilm structures were also not noticed in the absence of K2HPO4 in the medium.
Figure 4.
Biofilm morphologies of B. tropica P4 (A–D) and B. unamae P9 (E–H) at different concentrations of K2HPO4 (µg/ml) as observed under light microscope: (A,E) 25 µg/ml; (B,F)- 50 µg/ml; (C,G) 100 µg/ml; (D,H) 500 µg/ml.
EPS production by bacterial isolates in the presence of different insoluble P
When the EPS production by the strains were quantified in terms of dry weights as well as total carbohydrate contents in the presence of different types of insoluble P in culture medium, maximum EPS production was observed in the presence of MRP followed by URP (Table 1). B. tropica P4 and B. unamae P9 produced 5.01 ± 0.29 g/l and 4.86 ± 0.32 g/l of EPS respectively in the presence of MRP as insoluble P source (Table 1). On the other hand EPS production significantly (P < 0.001) decreased in the presence of TCP for both the strain (Table 1). Reduced EPS production was also observed in the presence of JRP and PRP in comparison to MRP. In terms of total carbohydrate also EPS production was almost two times higher in MRP (0.68 ± 0.04 g/l for P4 and 0.59 ± 0.04 g/l for P9) than TCP (0.32 ± 0.04 g/l for P4 and 0.28 ± 0.03 g/l for P9) for both B. tropica P4 as well as B. unamae P9 (Table 1).
Table 1.
EPS production in terms of dry weight as well as total carbohydrate (TC) by isolated bacterial strains during solubilization of different insoluble P.
| Insoluble P used in NBRIY broth | EPS production by isolated bacterial strains after 48 hours of incubation | |||
|---|---|---|---|---|
| B. tropica P4 | B. unamae P9 | |||
| Dry wt. (g/l) | TC (g/l) | Dry wt (g/l) | TC (g/l) | |
| TCP | 2.75 ± 0.25 | 0.32 ± 0.04 | 2.68 ± 0.27 | 0.28 ± 0.03 |
| JRP | 2.82 ± 0.27 | 0.36 ± 0.03 | 2.92 ± 0.16 | 0.40 ± 0.07 |
| PRP | 3.22 ± 0.22 | 0.48 ± 0.06 | 3.45 ± 0.25 | 0.45 ± 0.05 |
| URP | 4.69 ± 0.17 | 0.54 ± 0.06 | 4.34 ± 0.29 | 0.51 ± 0.06 |
| MRP | 5.01 ± 0.29 | 0.68 ± 0.04 | 4.86 ± 0.32 | 0.59 ± 0.04 |
EPS production in the presence of different concentrations of K2HPO4
Like biofilm formation, EPS production by both the strains also showed similar pattern when culture medium was supplemented with different concentrations of K2HPO4. Maximum EPS production was observed in the presence of K2HPO4 at a concentrations of 25 µg/ml for B. tropica P4 (5.65 ± 0.28 g/l) as well as B. unamae P9 (5.02 ± 0.34 g/l) which decreased gradually (P < 0.001) along with increased concentrations of K2HPO4. In terms of total carbohydrate contents also EPS productions by the isolates showed similar patterns (Fig. 5).
Figure 5.
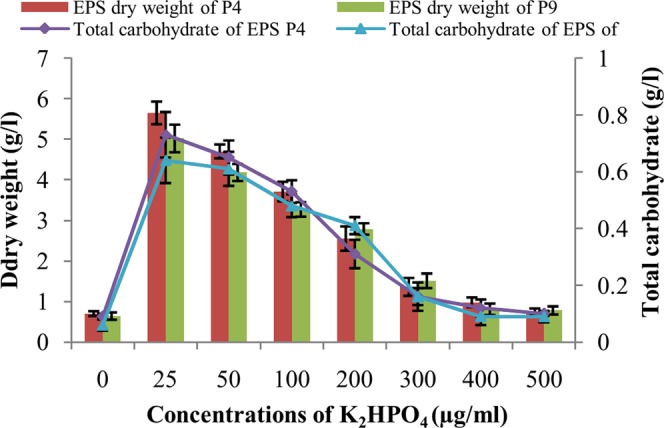
Production of EPS by B. tropica P4 and B. unamae P9 in terms of dry weight as well as total carbohydrate content in the presence of different concentrations of K2HPO4.
Presence of BraI/R QS system in B. tropica P4 and B. unamae P9
In both the organisms BraI/R QS system was detected by amplifying two genes i.e. braR and rsaL using the primers described earlier16. When the amplified DNA were resolved on 1.2% agarose gel single band of nearly 720 bp for braR gene was detected for both B. tropica P4 as well as B. unamae P9 (Fig. 6A). On the other hand nearly 225 bp bands were observed for rsaL gene of the strains (Fig. 6B). Nucleotide sequence of each gene was generated using forward and reverse sequence data and confirmed as braR and rsaL gene by BLAST analysis on NCBI GenBank database.
Figure 6.
Amplified DNA for braR and rsaL gene on 1.2% agarose gel: (A) lane 1–100 bp DNA ladder; lane 2- braR of B. tropica P4; lane 3- braR of B. unamae P9; (B) lane 1–100 bp DNA ladder; lane 2- rsaL of B. tropica P4; lane 3- rsaL of B. unamae P9.
Clustal Omega (1.2.1) multiple sequence alignment program revealed that braR of B. tropica P4 showed 79.50% identity with B. kururiensis M130 and 93.79% identity with B. unamae MTI-641T where as braR of B. unamae P9 showed 80.90% identity with B. kururiensis M130 and 96.12% identity with B. unamae MTI-641T. On the other hand rsaL nucleotide sequence of B. tropica P4 showed 74.32% identity with B. kururiensis M130 and 85.79% identity with B. unamae MTI-641T where as the rsaL nucleotide sequence of B. unamae P9 showed 73.63% identity with B. kururiensis M130 and 84.62% identity with B. unamae MTI-641T.
Discussion
In environment several microorganisms aggregate at solid-liquid interfaces to form adherent multicellular structures which is generally known as biofilm. During the development of biofilm, bacterial cells attach to each other and often these cells adhere to a solid surface. Sometimes these adherent cells remained embedded within an extracellular matrix generally made up of polysaccharides and other macromolecules. Microorganisms produce biofilm to conquer different environmental stresses such as nutrient limitation, desiccation and predatory grazing2. In pathogenic microorganisms, biofilm generally serve as one of the important protecting mechanisms against several antimicrobials27. In addition to various drawbacks of microbial biofilm development, there are also several positive aspects28. As for example, biofilm formation by several bacterial strains on crop plant rhizosphere confer protection against various fungal diseases29.
The present work demonstrated biofilm formation by two plant growth promoting bacterial strains during solubilization of insoluble P. Our initial observations suggested maximum biofilm formation by both the strains after 48 hours of incubation. After 48 hours of incubation, formation of prominent biofilm structures by both the strains of Burkholderia on all the insoluble P granules was observed using scanning electron microscopic technique. Both the strains were found to attach firmly to the surfaces of rock phosphate granules which can withstand even after repeated washing. Surface roughness of rock phosphate granules probably plays major role for their stable attachments. Cell surface hydrophobicity, presence of different appendages and degrees of EPS production have also been considered as important factors for bacterial attachments30. It has been reported that three different types of forces viz. hydrogen bonds, electrostatic interaction and london dispersion forces are involved in biofilm formation process31. In case of B. mallei and B. pseudomallei, role of trimeric autotransporter adhesins has also been suggested for their attachment and biofilm formation during environmental as well as virulence-associated interactions32,33.
Although maximum biofilms formation by the isolates was detected after 48 hours of incubation, maximum release of soluble P was detected after 72 hours of incubation for most of the insoluble P used25. When TCP was used as sole P source, after 48 hours of incubation at 28 °C, 370.69 ± 8.79 µg/ml and 463.03 ± 15.17 µg/ml of soluble P were released by B. tropica P4 and B. unamae P9 respectively. After 72 hours of incubation the amount of soluble P increased to 580.56 ± 13.38 µg/ml and 517.12 ± 17.15 µg/ml for B. tropica P4 and B. unamae P9 respectively25. The decrease of medium pH by both the strains (pH 3.35 ± 0.20 by P4 and pH 3.50 ± 0.17 by P9 after 72 hours of incubation) was also correlated with release of soluble P in the medium. Initial pH of 6.5–7.0 was found effective for maximum bacterial growth and they decrease medium pH to less than pH 3.5 within 72 hours of incubation. They were also able to survive in these lower pH with sufficient CFUs. The current observation suggested that biofilm formation by the strains within 48 hours of incubation probably creates a close environment to secrete organic acid by them which was responsible for maximum release of soluble P after 72 hours of incubation. Most of the PSB solubilize inorganic P by secreting different types of organic acids like gluconic acid, 2-keto gluconic acid, citric acid, malic acid, oxalic acid, lactic acid and others34,35. IR spectrophotometric analysis of partially purified solubilizing principles suggested the production of carboxylic acids by both the isolates. Appearances of peaks at 1620.09 cm−1 and 3382.91 cm−1 for P4 (see Supplementary Fig. S1) and at 1757.03 cm−1 and 3492.85 cm−1 for P9 (see Supplementary Fig. S2) suggested the presence of C=H stretch and O-H stretch respectively for carboxylic acids. The major soluble form of mineral P is the monovalent anion phosphate H2PO4− which generally produces at lower pH. Organic acid produced by PSB decreases the pH of surrounding environment which ultimately helps to release P ions by H+ substitution for the cation bound to P36. Gram negative bacteria generally produce gluconic acid and 2-keto gluconic acid during P solubilization process. Gluconic acid is produced mainly in the bacterial periplasmic space by direct oxidation of glucose with the help of pyrrolo quinoline quinone (PQQ) dependent membrane bound enzyme glucose dehydrogenase (m-GDH). PQQ act as a redox cofactor for the enzyme m-GDH37. Gluconic acid may further oxidized to 2-keto gluconic acid by the enzyme gluconate dehydrogenase38.
Comparison between the degrees of biofilm formation on different insoluble P granules revealed maximum biofilm formation by both the organisms on MRP granules. It has been reported earlier that P content in MRP was low (17% P2O5) in comparison to other rock phosphates39. Therefore, release of appreciable amount of soluble P from low P content MRP granules required maximum efforts, and that may be fulfilled by producing maximum biofilm coverage. On the other hand JRP contained 34% P2O5, and the strains B. tropica P4 and B. unamae P9 produced 514.80 ± 11.33 µg/ml and 464.11 ± 12.15 µg/ml of soluble P by using JRP25. Therefore, less biofilm development may also be correlated with its high P content and easy release of soluble P in appreciable amount.
By assuming the role of available P on the extent of biofilm formation by the isolates, quantification of biofilm formation was performed in the presence of different concentrations of soluble P (K2HPO4). Maximum biofilm formation by both the strains under P limitation conditions, suggested that the P solubilizing bacterial strains attach to the surfaces of insoluble P granules and release more soluble P for overcoming the stress generated by P limitation. Upregulation of alkaline phophatase activity in phosphorous limited biofilm has been reported by Huang et al.40. P limitation also enhanced the biofilm formation of plant pathogenic bacteria Agrobacterium tumefaciens2. Reduced planktonic cell densities at lower K2HPO4 concentrations also supported the positive correlation between P limitation and development of bacterial biofilm. On the other hand in case of Escherichia coli impairment in biofilm formation was observed in the presence of high concentration of (>25 mM) of P in the culture medium. Addition of higher concentration of P to the medium caused disassembly of 24 hours preformed biofilm41. It has been found that PolyP degradation is an important factor for biofilm formation at stationary phase. Excess P concentration in the culture media was responsible for high PolyP level up to stationary phase which ultimately impaired biofilm formation in E. coli in a PhoB dependent manner42. Effect of environmental P concentration has also been noticed in the biofilm formation pattern on uropathogenic E. coli isolates and implication of P as a possible physiological signal was highlighted to regulate biofilm phenotype in E. coli species43. It has also been reported that bacterial stringent response plays important roles in response to low level of specific nutrients including amino acids, glucose, phosphate, iron and fatty acids44. It also imparts significant function in antibiotic tolerance and biofilm formation. Some intercellular alarmons viz. guanosine tetraphosphate and guanosine pentaphosphate [collectively known as (p)ppGpp] are considered as universal signal molecules during that type of response45. Significant increase in (p)ppGpp levels was described in several stress conditions and depended on the presence of two ribosome associated protein RelA and SpoT that act as (p)ppGpp synthase and hydrolases. (p)ppGpp tunes the RNA polymerase for specific regulation of promoters of the gene associated with bacterial stress tolerance46. Use of other growth requirements in same concentrations during quantification experiments strongly suggested involvement of P source in biofilm formation processes for both the strains of Burkholderia. On the other hand, similar patterns of biofilm formation after alteration of phosphate source (K2HPO4) with Na2HPO4 clearly demonstrated that biofilm formation process was dependent only on the concentration of P and independent on the nature of P source. This type of independency during biofilm formation on the nature of P source was also observed for plant pathogenic bacteria Agrobacterium tumefaciens2.
Although maximum biofilm formation by the strains P4 and P9 was observed in the presence of 25 µg/ml of K2HPO4, both of them were unable to form biofilm in the absence of soluble P due to reduced bacterial growth in the absence of essential macroelement phosphorous. Morphological variations of biofilm structures under light microscope also supported their tendencies towards biofilm development at P limitation conditions. Little bacterial growth was detected in the absence of K2HPO4 due to trace amount of soluble P contamination in other nutrients supplied. There are four major steps in biofilm formation process which include attachment to the surface, growth of bacteria, biofilm maturation and dispersion of cells from matured biofilm. Among them, initial attachment and bacterial growth impart critical role for overall biofilm development and can be considered as rate limiting steps in biofilm formation process. To support the sufficient bacterial growth and biofilm formation, 25 µg/ml of K2HPO4 was supplied in the NBRIY medium. When the experiments were performed using rock phosphate granules, rock phosphates were used after repeated washing to remove trace amount of soluble P. The organisms were able to form biofilms after initial solubilization of rock phosphates to fulfill their P requirement. Maximum biofilms development after 48 hours also supported their initial requirement of P for maximum growth and biofilm development which ultimately help for maximum (>500 µg/ml) release of soluble P. For better yield, crop plants require more than 60 ppm of soluble P but in agricultural soil P concentration is very low (nearly 1 ppm) which cannot fulfill the need of crop plants47. The present observation suggested that the Burkholderia strains can be used in combination with rock phosphates to fulfill the P demand in agricultural soil for better crop production.
The biosynthesis of EPS serves many functions like promotion of bacterial attachments to solid surfaces and formation as well as maintenance of microcolony and mature biofilm structure6. In addition, EPS enhances the resistance of biofilm structures to environmental stresses and sometime also help the bacteria for accumulating the nutrients48. In the present endeavor, increase of EPS production along with biofilm development by both the strains, strongly indicated the direct involvement of EPS during biofilm formation at P deficient conditions. Involvement of EPS production in biofilm formation has been reported by several workers for various bacterial species9. Quantification in terms of dry weight and total carbohydrate content also suggested variation in EPS production in the presence of different insoluble P as observed during SEM study. Maximum amount of EPS production in the presence of MRP, directs the bacterial organisms to develop more condensed biofilm structures to release sufficient soluble P from MRP granules. Due to sufficient P content in TCP and JRP, bacterial organisms faced less difficulties to produce significant amount of soluble P, which ultimately suppressed the necessities of EPS production for the development of condensed biofilm structures. Highest degrees of EPS production in the presence of lowest concentration of K2HPO4 also suggested its strong correlation with available P. According to Chunha et al.49, EPS production was not essential in Burkholderia cepacia complex for biofilm initiation but it plays major role for the establishment of thick biofilm structures. Significant amount of EPS production was also reported for four bacterial strains under the genus Enterobacter, Arthrobacter and Azotobacter having TCP solubilization potential50. A branched acetylated heptasaccharide known as cepacian was detected in different members of Burkholderia as the major exopolysaccharide which is important for their survival at different environmental conditions like desiccation, metal ion stress etc.51.
In most of the Gram negative bacteria AHL dependent quorum sensing system plays significant role in biofilm formation. In the present study presence of BraI/R QS system was detected in B. tropica P4 and B. unamae P9. As the primer sets were effective to amplify the braR and rsaL gene from 20 species of Burkholderia24, therefore they were used to detect the presence of BraI/R QS system in both the bacterial strains. Suárez-Moreno et al.16 also reported that biofilm formation and EPS productions were regulated by BraI/R QS system in some plant associated Burkholderia spp. Transcriptome analysis also suggested that BraI/R regulon is species specific and associated with EPS production in Burkholderia xenovorans LB400 and B. phymatum STM8152. Involvement of similar type of QS system was also reported in regulation of siderophore production in B. cepacia21,22. Although no such direct correlation between siderophore production and P solubilization have been reported, better iron phosphate solubilizing potential was observed in siderophore producing B. cepacia25. In the present observation it was found that biofilm formation and EPS production by B. tropica P4 as well as B. unamae P9 were highly correlated with available P in the medium. Although further studies are required, the present observation suggested that there may be some positive relation of this QS system which regulates the biofilm formation or EPS production during P solubiliziation by the isolates.
In developing countries like India, China, Bangladesh, Vietnam, Cambodia, Laos and others, agriculture sector is considered as one of the important pillars of economy for their development. In terms of agricultural aspects phosphorus is the second major macronutrient that required for growth and development of crop plants. The high cost of chemical fertilizers is a great problem for developing countries53. Environmental pollution is another major concern regarding the excessive application of chemical fertilizers. Application of P solubilizing microbes is considered as a suitable alternative in this regard. The observed correlation between biofilm formation and P solubilizing process will be helpful to apply the species of Burkholderia along with rock phosphate granules in the appropriate field condition as biofertilizer to fulfill the P demands of crop plants for their better production. Suitable formulation can be prepared based on present study for their proper application. When the P solubilizing potential of the strains were studied at different temperatures, both the strains were able to release sufficient amount of soluble P from TCP at low temperature like 20 °C (>330 µg/ml) as well as high temperature like 40 °C (>300 µg/ml)25 which also confirmed their applicability at temperature fluctuating countries. Both the organisms were also found to grow in comparatively higher pH (pH 9) and were able to produce prominent zones of P solubilization which also indicated their applicability in highly basic soils. On the other hand, bioprocessing of rock phosphate for the production of soluble P may provide an energy efficient and environmentally favorable alternative in comparison to current acid digestion technology for industrial production of P fertilizers26. Selective release of soluble P by bacterial biofilms from rock phosphates also reduces the extraction of undesirable ore contaminants like radioactive toxic materials. Biofilm forming properties of the isolates will be applicable to design proper bioreactors for the processing of rock phosphates following bioprocessing technology. Fixed bed biofilm reactors and contact based biofilm reactors can be used with suitable modification for their application in this purpose. Based on our study it can be suggested that rock phosphates will be used as solid phase and lower concentration of soluble P will be maintained within the bioreactors for the development of maximum biofilm and release of soluble P. In addition to bioprocessing of rock phosphates, the isolates can also be used for P recycling from the sludge produced during wastewater treatment. During chemical processing of wastewater a huge amount of sludge with precipitated P is produced. This sludge with high P contents is considered as a cheap source of P especially in developing countries with limited rock phosphate sources. Effective P solubilizing microorganisms can be used to solubilize and recycle the P from the wastewater sludge which will fulfill the P demands in the agricultural fields of developing countries. Therefore the isolates B. tropica P4 and B. unamae P9 may also be helpful for bioprocessing of sludge for releasing soluble P which can be used as biofertilizer for better crop production.
Methods
Microbial strains and culture condition
Two potent P solubilizing bacterial strains viz. Burkholderia tropica P4 and B. unamae P9, isolated from the rhizospheric soil of Lycopodiella cernua (L.) Pic. Serm25 were taken in the present study. The bacterial strains P4 and P9 were previously identified on the basis of 16 S rDNA sequence homologies25. Both the strains were maintained in NBRIY medium and preserved at 4 °C. Glycerol stocks were also prepared for long time preservation of the strains. Both the strains were grown at 28 °C in BOD incubator (Instrumentation India, Kolkata, India).
Study of biofilm formation on insoluble P granules by P4 and P9
Biofilm formation during P solubilization by both the strains viz. B. tropica P4 and B. unamae P9 were studied using TCP as well as four different rock phosphates viz. JRP, PRP, URP and MRP.
Culture conditions and development of bacterial biofilm
Both the bacterial strains were grown in nutrient broth at 28 °C with mild shaking (120 rpm). 100 µl of starter cultures (OD620nm = 0.5) were added to 100 ml of NBRIY broth in 250 ml Erlenmeyer flasks supplemented with different insoluble P at a concentration of 5 g/l. Sterilized polyester chips (2 cm × 2 cm) were placed within each flask and incubated at 28 °C without shaking. At every 24 hours of intervals polyester chips were removed carefully from all the flasks without any agitation and then slowly washed with sterilized distilled water for removing the planktonic cells and finally air dried aseptically in front of laminar air flow (Klenzaids, Mumbai, India).
Study of bacterial biofilms under scanning electron microscope
All the polyester chips containing layers of insoluble P granules and bacterial cells were prepared for SEM study54. They were prefixed with 2% glutaraldehyde in 20 mM Na-P buffer (pH 6.5) plus 5% dimethyl sulphoxide (DMSO) for 30 minutes. After pre-fixation the samples were gently washed with sterilized distilled water and post fixed with osmium tetraoxide dissolved in 50 mM Na-P buffer (pH 6.5). All the samples were then dehydrated using a series of alcohol grades (30–100%) retaining them for 10 minutes in every dilution. The polyester chips were cut into small pieces (4 mm × 4 mm) and placed on metal stubs. They were coated with gold using an ion sputter (Coater IB-2, Gike Engineering, Japan) and finally observed using SEM (HITACHI S-530, Japan). Biofilm formation patterns by the strains on TCP as well as on four different rock phosphates were compared.
Study of biofilm formation by isolated bacterial strains in relation to available P
In order to ascertain the relations of biofilm formation by P4 and P9 with phosphate availability in their surrounding environment, the insoluble P source of the NBRIY medium was replaced with different concentrations of soluble P i.e. dipotassium phosphate (K2HPO4). In the presence of different amounts of soluble P, biofilm formation were quantified, biofilm morphologies were studied and the planktonic cell densities were also determined to correlate the biofilm forming tendencies of the isolates in relation to available P.
Quantification of biofilm formation
Two ml of NBRIY broth (pH 7.0) supplemented with different concentrations of K2HPO4 were taken in separate wells of 24 wells polystyrene cell culture plates and were inoculated with 20 μl of bacterial culture (OD620nm = 0.5). Uninoculated wells for each concentration were treated as control. All the plates were then kept undisturbed in a BOD incubator (Instrumentation India, Kolkata, India) at 28 °C for 48 hours. After that broth were decanted from each well without disturbing the biofilms and the wells were washed twice with sterilized distilled water carefully to remove the planktonic cells. After air drying in front of laminar airflow, Two ml of 0.1% crystal violet solution was added to each well55. After 10 minutes of incubation at room temperature, the stain was removed from the wells and the plates were finally washed with sterilized distilled water for 3–4 times and again air dried. One ml of 33% acetic acid was applied to each well and then incubated for 30 minutes for extracting the crystal violet from adhered cells56. Optical densities were then measured at 595 nm using UV-VIS spectrophotometer (SPECTRASCAN UV-2700, Thermo Scientific, India). For confirming the role of P concentration on biofilm formation process, similar quantification experiments were also performed by replacing the P source (K2HPO4) with Na2HPO4.
Estimation of planktonic cell densities
For determining the planktonic cell densities, 100 μl of bacterial culture was taken from each well before decanting the broth and sprayed on nutrient agar (NA) plates after serial dilution using sterilized distilled water. Plates were incubated at 28 °C for 24 hours and numbers of CFUs were counted.
Study of biofilm morphologies
In order to observe the morphological variation in biofilm structures in relation with available P, sterilized polyester chips (2 cm × 1 cm) were placed in slanting position within the wells of cell culture plates. Two ml of NBRIY broth (pH 7.0) containing different concentrations of K2HPO4 was added to each well and inoculated with bacterial strains. Plastic chips were removed carefully after 48 hours of incubation at 28 °C, washed slowly with sterilized distilled water and air dried. All the polyester chips with bacterial biofilm were stained with 0.1% crystal violet, air dried and finally observed under light microscope (LEICA DM2500, Germany).
Study of EPS production by bacterial strains
EPS production by both the strains were studied in 100 ml of NBRIY broth either supplemented with different insoluble P or soluble P i.e. K2HPO4. EPS production was quantified in terms of dry weight as well as total carbohydrate content. Bacterial cultures were centrifuged at 1000 rpm for 20 minutes to get the cell free supernatants (CFS). EPS from CFS was precipitated by adding same volume of ice cold ethanol followed by incubation at 4 °C for overnight57. Precipitated EPSs were separated by centrifugation at 7000 rpm for 20 minutes. To measure the dry weight, EPSs were taken in petriplates and dried at 60 °C. On the other hand for measuring the total carbohydrate, precipitated EPSs were resuspended in 5 ml of sterilized distilled water. One ml of sample was taken to measure the total carbohydrate by phenol sulfuric acid method58 using sucrose as standard.
EPS production in the presence of different insoluble phosphates
EPS production by both the bacterial strains were studied in the presence of TCP as well as four different rock phosphates viz. JRP, PRP, MRP and URP. Each bacterial strain was inoculated separately to 100 ml NBRIY broth (pH 7.0) containing different insoluble P (5 g/l) in 250 ml Erlenmeyer flasks. The EPS produced by the strains were quantified after 48 hours of incubation at 28 °C. EPS production by all the bacterial isolates were quantified in terms of dry weight as well as total carbohydrate contents as mentioned.
EPS production in the presence of different concentrations of soluble P
In order to determine the relation of EPS production by bacterial strains with available P in the media EPS production were also quantified in the presence of different concentrations of K2HPO4. Similar to biofilm formation experiments, insoluble P source of NBRIY broth (pH 7.0) was replaced by K2HPO4. Each bacterial strain was inoculated separately to 100 ml NBRIY broth (pH 7.0) containing different concentrations of K2HPO4 (0, 25, 50, 100, 200, 300, 400, 500 μg/ml) in 250 ml Erlenmeyer flasks and incubated at 28 °C with mild shaking (120 rpm). Dry weight and total carbohydrate contents of EPS were determined after 48 hours of incubation as mentioned.
Detection of bacterial quorum sensing system
The presence of BraI/R quorum sensing system, found in many plant associated Burkholderia sp.24 was detected for the strain P4 and P9 by PCR amplification followed by sequencing of genes (braR and rsaL) related with QS systems. braR gene of both of the bacterial strains (B. tropica P4 and B. unamae P9) was amplified at an annealing temperature of 62 °C. Following primer pairs24 were used for the amplification of braR gene for both the bacterial strains- pQEbraRfw: 5′-GGGGATCCTCGCCGATACTGGCCGCATC-3′ pQEbraRrv: 5′-GGGAAGCTTTCAGCCCGGATCTATAAGGCC -3′. On the other hand rsaL gene of both the organisms was also amplified at an annealing temperature of 48 °C. Following primer pairs17 were used for the amplification of this gene for the strains- braLFw: 5′-TTGTTGAAATAAAGTCCCAG-3′ braLRv: 5′-CTGGAAAATCACTGGCA-3′. Amplified DNA was resolved on 1.2% agarose gel and purified using Xcelgen DNA Gel/PCR Purification Miniprep kit following manufacturer instructions. The purified PCR products were sequenced from Xcelric Genomics, India. Sequencing were done in forward and reverse direction in separate reactions. Each reaction mixtures were prepared with template DNA, specific primer, water and BigDye sequencing buffer and BigDye Terminator v3.1 Ready Reaction Mix (Applied Biosystems) of required volumes. Each reaction was heated to 96 °C for one minute, followed by 25 cycle at 96 °C for 10 sec, 50 °C for 5 sec and 60 °C for 4 min. The product was purified following ethanol precipitation method and pelleted by centrifugation. The pellet was washed with 70% ethanol and then air dried. The air dried samples were rehydrated using 15 µl of formamide and then used for sequencing using ABI 3730xl genetic analyzer (Applied Biosystems, USA). Sequence alignment program BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) was used for assembling the nucleotide sequences. Nucleotide sequences were used for carrying out BLAST with the NCBI GeneBank database (http://www.ncbi.nlm.nih.gov/) and aligned by Clustal Omega (1.2.1) multiple sequence alignment program59.
Statistical analysis
All the experiments were repeated and the data represented were means of at least three replicates. Means and standard deviations were calculated by using Microsoft Excel program version 2007. Student t-test was performed to test the significance of difference between the mean values of EPS production (both in terms of dry weight and total carbohydrate) in the presence of different insoluble P sources. Single factor ANOVA was performed at 0.05 significant levels to assess the variance of biofilm formation, planktonic cell densities and EPS production in the presence of different concentrations of soluble P.
Supplementary information
Acknowledgements
R.G. is thankful to Department of Science and Technology, Govt. of India for providing INSPIRE Fellowship.
Author Contributions
R.G. & N.C.M. planned all the experiments. R.G. performed the experiments of Figs 1–7. S.B. performed some experiments for Fig. 1 and Fig. 4. R.G. wrote the manuscript. N.C.M. reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41726-9.
References
- 1.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Sci. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 2.Danhorn T, Hentzer M, Givskov M, Parsek MR, Fuqua C. Phosphorus limitation enhances biofilm formation of the plant pathogen Agrobacterium tumefaciens through the PhoR-PhoB regulatory system. J. Bacteriol. 2004;186:4492–4501. doi: 10.1128/JB.186.14.4492-4501.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dang H, Lovellc CR. Microbial Surface Colonization and Biofilm Development in Marine Environments. Microbiol. Mol. Biol. Rev. 2016;80(1):92–138. doi: 10.1128/MMBR.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalton HM, March PE. Molecular genetics of bacterial attachment and biofouling. Curr. Opin. Biotechnol. 1998;9:252–255. doi: 10.1016/S0958-1669(98)80055-4. [DOI] [PubMed] [Google Scholar]
- 5.Donlan RM. Biofilms: microbial life on surfaces. Emerg. Infect. Diseases. 2002;8(9):881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma W, et al. Bacillus subtilis biofilm development in the presence of soil clay minerals and iron oxides. npj Biofilms and Microbiomes. 2017;4:1–9. doi: 10.1038/s41522-017-0013-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulamattathil SG, Bezuidenhout C, Mbewe M. Biofilm formation in surface and drinking water distribution systems in Mafikeng, South Africa. S. Afr. J. Sci. 2014;110(11/12):Art. #2013–0306. doi: 10.1590/sajs.2014/20130306. [DOI] [Google Scholar]
- 8.Chen XD, et al. Stabilizing effects of bacterial biofilms: EPS penetration and redistribution of bed stability down the sediment profile. J. Geophys. Res.: Biogeosci. 2017;122(12):3113–3125. doi: 10.1002/2017JG004050. [DOI] [Google Scholar]
- 9.Czaczyk K, Myszka K. Biosynthesis of extracellular polymeric substances (EPS) and its role in microbial biofilm formation. Pol. J. Environ. Stud. 2007;16(6):799–806. [Google Scholar]
- 10.Vu B, Chen M, Crawford RJ, Ivanova EP. Bacterial extracellular polysaccharides involved in biofilm formation. Molecules. 2009;14:2535–2554. doi: 10.3390/molecules14072535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mattos-Guaral DIA, Formiga LCD, Periera GA. Cell surface components and adhesion in. Corynebacterium diphtheriae. Microb. Infect. 2000;2:1507. doi: 10.1016/S1286-4579(00)01305-8. [DOI] [PubMed] [Google Scholar]
- 12.Langille ESE, Geesey GG, Weiner RM. Polysaccharide– specific probes inhibit adhesion of Hyphomonas rosenbergii strain VP-6 to hydrophilic surfaces. J. Ind. Microbiol. Biotechnol. 2000;25:81. doi: 10.1038/sj.jim.7000033. [DOI] [Google Scholar]
- 13.Toyofuku M, et al. Enivironmental factors that shape biofilm formation. Biosci. Biotechnol Biochem. 2016;80(1):7–12. doi: 10.1080/09168451.2015.1058701. [DOI] [PubMed] [Google Scholar]
- 14.Watnick P, Kolter R. Biofilm, city of microbes. J. Bacteriol. 2000;182:2675–2679. doi: 10.1128/JB.182.10.2675-2679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davey ME, O’Toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 2000;64:847–867. doi: 10.1128/MMBR.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suárez-Moreno ZR, et al. Commonalities and differences in regulation of N-Acyl homoserine lactone quorum sensing in the beneficial plant-associated Burkholderia species cluster. Appl. Environ. Microbiol. 2010;76(13):4302–4317. doi: 10.1128/AEM.03086-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parsek MR, Greenberg EP. Sociomicrobiology: The connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001;55:165–99. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 19.Visick KL, Fuqua C. Decoding Microbial Chatter: Cell-Cell Communication in Bacteria. J. Bact. 2005;187(16):5507–5519. doi: 10.1128/JB.187.16.5507-5519.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gotschlich A, et al. Synthesis of multiple N-acylhomoserine lactones is wide-spread among the members of the Burkholderia cepacia complex. Syst. Appl. Microbiol. 2001;24(1):1–14. doi: 10.1078/0723-2020-00013. [DOI] [PubMed] [Google Scholar]
- 21.Eberl L. Quorum sensing in the genus Burkholderia. Int. J. Med. Microbiol. 2006;296:103–110. doi: 10.1016/j.ijmm.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 22.Huber B, et al. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology. 2001;147:2517–2528. doi: 10.1099/00221287-147-9-2517. [DOI] [PubMed] [Google Scholar]
- 23.Venturi V, Friscina A, Bertani I, Devescovi G, Aguilar C. Quorum sensing in the Burkholderia cepacia complex. Res. Microbiol. 2004;155:238–244. doi: 10.1016/j.resmic.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 24.Suárez-Moreno ZR, Caballero-Mellado J, Venturi V. The new group of non-pathogenic plant-associated nitrogen-fixing Burkholderia spp. shares a conserved quorum-sensing system, which is tightly regulated by the RsaL repressor. Microbiology. 2008;154(7):2048–59. doi: 10.1099/mic.0.2008/017780-0. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh R, Barman S, Mukherjee R, Mandal NC. Role of phosphate solubilizing Burkholderia spp. for successful colonization and growth promotion of Lycopodium cernuum L. (Lycopodiaceae) in lateritic belt of Birbhum district of West. Bengal, India. Microbiol. Res. 2016;183:80–91. doi: 10.1016/j.micres.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 26.Goldstein, A. H. Bioprocessing of rock phosphate ore: essential technical considerations for the development of a successful commercial technology. Proceedings of the 4th International Fertilizer Association Technical Conference (Paris: IFA;) 220 (2000).
- 27.Hall CW, Mah TF. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017;41(3):276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- 28.Robertson SR, McLean RJC. Beneficial biofilms. AIMS Bioengineering. 2015;2(4):437–448. doi: 10.1078/1438-4221-00196. [DOI] [Google Scholar]
- 29.Maddula VS, Pierson EA, Pierson LS. Altering the ratio of phenazines in Pseudomonas chlororaphis (aureofaciens) strain 30-84: effects on biofilm formation and pathogen inhibition. J. Bacteriol. 2008;190:2759–2766. doi: 10.1128/JB.01587-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donlan RM. Biofilms: Microbial life on surfaces. Emerging infect. Dis. 2002;8(9):881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flemming HC, Wingender J. Relevance of microbial extracellular polymeric substances (EPSs)-Part I: Structural and ecological aspects. Water Sci. tehnol. 2001;43(6):1–8. doi: 10.2166/wst.2001.0326. [DOI] [PubMed] [Google Scholar]
- 32.Lafontaine ER, Balder R, Michel F, Hogan RJ. Characterization of an autotransporter adhesion protein shared by Burkholderia mallei and Burkholderia pseudomallei. BMC Microbiol. 2014;14:92. doi: 10.1186/1471-2180-14-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lazar Adler NR, et al. Identification of a predicted trimeric autotransporter adhesin required for biofilm formation of Burkholderia pseudomallei. Plos One. 2013;8(11):e79461. doi: 10.1371/journal.pone.0079461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muhammad A, et al. Phosphate-solubilizing bacteria nullify the antagonistic effect of soil calcification on bioavailability of phosphorus in alkaline soils. Sci. Rep. 2017;7:16131. doi: 10.1038/s41598-017-16537-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bolan NS, Naidu R, Mahimairaja S, Baskaran S. Influence of low-molecular weight organic acids on the solubilization of phosphates. Biol. Fertil. Soils. 1994;18:311–319. doi: 10.1007/BF00570634. [DOI] [Google Scholar]
- 36.Goldstein, A. H. Involvement of the quinoprotein glucose dehydrogenase in the solubilization of exogenous phosphates by Gram-negative bacteria. In: Torriani-Gorini, A., Yagiland, E., Silver, S. (eds) Phosphate in microorganisms: Cellular and molecular biology. ASM Press, Washington (DC), pp 197–203 (1994).
- 37.Duine J, Frank J, van Zeeland J. Glucose dehydrogenase from Acinetobacter calcoaceticus: A ‘quinoprotein’. FEBS Lett. 1979;15:443–446. doi: 10.1016/0014-5793(79)80584-0. [DOI] [PubMed] [Google Scholar]
- 38.Kumar C, Yadav K, Archana G, Naresh Kumar G. 2013. 2-ketogluconic acid secretion by incorporation of Pseudomonas putida KT 2440 gluconate dehydrogenase (gad) operon in Enterobacter asburiae PSI3 improves mineral phosphate solubilization. Curr. Microbiol. 2013;67(3):388–394. doi: 10.1007/s00284-013-0372-z. [DOI] [PubMed] [Google Scholar]
- 39.Pal, R. Studies on Phosphate Solubilization and Crop Productivity by Some Common Diazotrophs Isolated from Rhizosphere. (Ph.D. thesis). Visva-Bharati (A central University), India (2006).
- 40.Huang CT, Xu KD, McFeters GA, Stewart PS. Spatial patterns of alkaline phosphatase expression within bacterial colonies and biofilms in response to phosphate starvation. Appl. Environ. Microbiol. 1998;6:1526–1531. doi: 10.1128/aem.64.4.1526-1531.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grillo-Puertas M, Villegas JM, Rintoul MR, Rapisarda A. Polyphosphate degradation in stationary phase triggers biofilm formation via LuxS quorum sensing system in Escherichia coli. Plos One. 2012;7(11):e50368. doi: 10.1371/journal.pone.0050368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grillo-Puertas M, Rintoul MR, Rapisarda VA. PhoB activation in non-limiting phosphate condition by the maintenance of high polyphosphate levels in the stationary phase inhibits biofilm formation in Escherichia coli. Microbiol. 2016;162:1000–1008. doi: 10.1099/mic.0.000281. [DOI] [PubMed] [Google Scholar]
- 43.Grillo-Puertas M, Martínez-Zamora MG, Rintoul MR, Soto SM, Rapisarda VA. Environmental phosphate differentially affects virulence phenotypes of uropathogenic Escherichia coli isolates causative of prostatitis. Virulence. 2015;6(6):608–617. doi: 10.1080/21505594.2015.1059561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cornforth DM, Foster KR. Competition sensing: the social side of bacterial stress responses. Nat. Rev. Microbiol. 2013;11(4):285–93. doi: 10.1038/nrmicro2977. [DOI] [PubMed] [Google Scholar]
- 45.Atkinson GC, Tenson T, Hauryliuk V. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One. 2011;6:e23479. doi: 10.1371/journal.pone.0023479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Zborníková E, Rejman D, Gerdesa K. Novel (p)ppGpp Binding and Metabolizing Proteins of Escherichia coli. MBio. 2018;9(1):e02188–17. doi: 10.1128/mBio.02188-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krishnaraj PU, Dahale S. Mineral Phosphate Solubilization: Concepts and Prospects in Sustainable Agriculture. Proc. Indian Natn. Sci. Acad. 2014;80(2):389–405. doi: 10.16943/ptinsa/2014/v80i2/55116. [DOI] [Google Scholar]
- 48.Pontefract RD. Bacterial adherence: its consequences in food processing. Can. Inst. Food Sci. Technol. J. 1991;24:113. doi: 10.1016/S0315-5463(91)70033-3. [DOI] [Google Scholar]
- 49.Cunha MV, et al. Studies on the Involvement of the Exopolysaccharide Produced by Cystic Fibrosis-Associated Isolates of the Burkholderia cepacia Complex in Biofilm Formation and in Persistence of Respiratory Infections. J. Clin. Microbiol. 2004;42(7):3052–3058. doi: 10.1128/JCM.42.7.3052-3058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yi Y, Huang W, Ge Y. Exopolysaccharide: a novel important factor in the microbial dissolution of tricalcium phosphate. World J. Microbiol. Biotechnol. 2008;24:1059–1065. doi: 10.1007/s11274-007-9575-4. [DOI] [Google Scholar]
- 51.Ferreira AS, et al. Distribution of Cepacian Biosynthesis Genes among Environmental and Clinical Burkholderia Strains and Role of Cepacian Exopolysaccharide in Resistance to Stress Conditions. Appl. Environ. Microbiol. 2010;76(2):441–450. doi: 10.1128/AEM.01828-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coutinho BG, et al. Regulon Studis and In Planta Role of the BraI/R Quorum Sensin System in the Plant Beneficial Burkholderia Cluster. Appl. Environ. Microbiol. 2013;79(14):4421–4431. doi: 10.1128/AEM.00635-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.López-Bellido L, Muñoz-Romero V, López-Bellido RJ. Nitrate accumulation in the soil profile: long-term effects of tillage, rotation and N rate in a mediterranean vertisol. Soil Till. Res. 2013;130:18–23. doi: 10.1016/j.still.2013.02.002. [DOI] [Google Scholar]
- 54.Mandal V, Sen SK, Mandal NC. Production and partial characterization of an inducer dependent novel antifungal compound(s) by Pediococcus acidilactici LAB5. J. Sci. Food Agr. 2013;93:2445–2453. doi: 10.1002/jsfa.6055. [DOI] [PubMed] [Google Scholar]
- 55.Djordjevic D, Weidmann M, McLandsbrough LA. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 2002;68:2950–2958. doi: 10.1128/AEM.68.6.2950-2958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adetunji VO, Isola TO. Crystal violet binding assay for assessment of biofilm formation by Listeria monocytogenes and Listeria spp on wood, steel and glass surfaces. Global. Veterinaria. 2011;6(1):06–10. [Google Scholar]
- 57.Ohno N, Miura N, Nakajima M, Yadomae T. Antitumor 1-3-glucan from cultured fruit body of Sparasis crispa. Biol. Pharm. Bull. 2000;23:866–872. doi: 10.1248/bpb.23.866. [DOI] [PubMed] [Google Scholar]
- 58.Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956;28:350–356. doi: 10.1021/ac60111a017. [DOI] [Google Scholar]
- 59.Sievers F, et al. Fast scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.