Abstract
Background
Disc degeneration (DD) is one of the common diseases worldwide, which deeply influences normal life and leads to excruciating pain. However, an effective treatment for DD is still not identified.
Method
The present study systemically examined the effect of melatonin on annulus fibrosus (AF) cells of patients with DD.
Results
Melatonin had the effect of promoting proliferation, inducing autophagy, and suppressing apoptosis on AF cells of patients with DD. Moreover, melatonin contributed to the translation and transcription of autophagy-related protein ATG7 and inhibited the function of miR-106a-5p in AF cells. In addition, the results suggested that miR-106a-5p mediated the expression of ATG7 by directly binding to its 3′UTR in AF cells.
Conclusion
This research not only gained a deep insight of melatonin mode of action, but also indicated its potential target signaling pathway in AF cells.
Keywords: disc degeneration, melatonin, ATG7, miR-106a-5p
Introduction
Disc degeneration (DD) is a complicated chronic process, which leads to the dysfunction of intervertebral discs (IVD).1 Nowadays, degenerative disc disease is emerging as a common disorder worldwide. However, the major treatment for this disease is still relying on surgical intervention, which increases the risk for adjacent DD.2 Therefore, an effective agent in the treatment for DD is urgently needed.
IVD are not only essential for the stability and flexibility of the spine, but also critical for resisting tension and bearing weight.3 Both the nucleus pulposus (NP) and annulus fibrosus (AF) regions are essential for maintaining the function of IVD.4
A previous report has indicated that the degeneration of the AF often leads to the dysfunction of IVD, which subsequently develops into disc herniation.5 Therefore, the stability of AF is critical for maintaining the health of IVD.
It has been confirmed that autophagy regulates the cell death pathway, which plays a key role in the degradation of cellular constituents.6 Moreover, a previous report has demonstrated that autophagy inhibits the apoptosis of AF cells and prevents the degeneration of IVD.7 Therefore, the autophagy process is essential for maintaining the function of AF cells. However, the precise mechanism still needs to be further explored.
Autophagy is highly mediated by autophagy-related genes (ATGs), such as ATG5, ATG7, and ATG12. Growing evidence has indicated that MicroRNAs (miRs) play a key role in the autophagy pathway. miRs are a group of small noncoding RNAs, which have served as a valuable post-transcriptional mediator by binding to the 3′ untranslated regions (UTRs) of the targeting mRNAs.8,9 miR-375 and miR-20a-5p have been identified to inhibit autophagy and reduce cell viability via regulating ATG7.10,11 Moreover, miR-200b has suppressed autophagy through inhibiting ATG12.12 However, the correlation between ATGs and miRs in AF cells is still not identified fully.
Melatonin has been reported as an endogenous neuro-hormone, which promotes proliferation and differentiation of neural stem cells.13 Moreover, it has been confirmed that melatonin inhibits the expression of miR-23a in hepatic metabolic diseases.14 In addition, melatonin suppressed the function of miR-34a in the regulation of neonatal brain inflammation.15 A previous report has elaborated that melatonin promotes type I collagen synthesis in human bone cells, which is the major fibrillary collagen of AF.16 Moreover, melatonin inhibits the apoptosis of neuronal cells.17,18 Additionally, melatonin regulates traumatic optic neuropathy via upregulating autophagy.19 Therefore, targeting autophagy is a promising approach for mitigating DD.20 However, the biological effect of melatonin on DD16,21,22 is still not clear.
In this research, we systematically analyzed the effect of melatonin on AF cells of patients with DD. Our results not only gained a deep insight into the biological function of melatonin but also provided evidences to indicate its possible signaling pathway in AF cells of patients with DD.
Materials and methods
Cell culture
Annular tissues of patients with DD and healthy volunteers were washed three times with sterile saline solution under aseptic conditions. Then, all samples were transferred to a sterile centrifuge tube and cut into pieces smaller than 1 mm3. Next, type II collagenase (0.2%) and trypsin solution (0.25%) were used to digest samples followed by adding DMEM (Trueline, Nashville, TN, USA) that contained 10% fetal bovine serum (FBS; Thermo Fisher Scientific). Cells were dispersed by mechanical shaking. Then, cells were cultured in T25 cell culture media (replaced every 3 days) at 37°C, in a 5% CO2 incubator after counting. Subcultures were performed when the primary culture of cells were 90% confluent. After that, cells were washed twice using sterile PBS solution and digested by trypsin (0.25%) at 37°C for 2 minutes. The passage 2 cells were treated with melatonin (Aladdin, Shanghai, People’s Republic of China) with different concentrations, including 10, 25, 50, 100, and 200 µmol/L, and used for subsequent analysis.
All patients were informed and gave written consent. This study was in accordance with the Declaration of Helsinki. Moreover, our research was approved by the independent ethics committee of Peking University Third Hospital.
Immunofluorescence detection
Immunofluorescent detection of light chain 3 (LC3) associations with autophagosomes was carried out as follows. In brief, cells were fixed using paraformaldehyde (4%) for 30 minutes after incubation with nigericin. Then, samples were washed using PBS (0.02 mol) for 3 minutes three times at 25°C. After that, cells were blocked using BSA (1%; Solarbio, Beijing, People’s Republic of China) for 1 hour at room temperature. Subsequently, cells were incubated with the rabbit anti-LC3 antibody (abcam, UK) in PBS overnight at 4°C followed by goat anti-rabbit IgG (H+L) (Beyotime, Haimen, People’s Republic of China) for 1 hour at room temperature. Images were acquired by an ECLIPSE Ni microscope and a digital image analyzer (NIKON, Tokyo, Japan). Three replicates were needed for each analysis.
RNA extraction and real-time PCR
Total RNA from different samples were extracted by using TRIzol Reagent (Thermo Fisher Scientific, Waltham, MA, USA). Then, the cDNA synthesis kit (Thermo Fisher Scientific) was used to reverse transcribe RNA into complementary DNA (cDNA) according to the instructions of the manufacturer. GAPDH was used to normalize the gene expression and measured using the 2−ΔΔCt method. Three replicates were needed for each analysis. Details of the primers used in this study are provided in the Supplementary materials, primer sequence information.
Cell transfection
The miR-106a-5p mimics, inhibitor, and negative control (NC) were obtained from GenePharma (Shanghai, People’s Republic of China). Cells were grown to 80% confluence prior for transfection. Lipofectamine 2000 (Thermo Fisher Scientific) was used to transfect the miR-106a-5p mimics or inhibitor into AF cells according to the protocol of the manufacturer. Then 48 hours post-transfection, the efficacy of transfection was measured by qRT-PCR. The sequence information is provided in Table S1. Three replicates were needed for each analysis.
Western blot
Total protein lysates were extracted by using RIPA lysis buffer (JRDUN, Shanghai, People’s Republic of China) with EDTA-free Protease inhibitor Cocktail (Roche, Germany). The protein concentration was quantified by an Enhanced BCA Protein Assay kit (Thermo Fisher Scientific). Equal amounts of protein (25 µg) were fractionated on 10% SDS-PAGE and transferred to a nitrocellulose membrane (Millipore, Billerica, MA, USA) overnight. After being blocked with 5% non-fat dry milk for 1 hour at 25°C, the membranes were probed at 4°C overnight with the primary antibodies, followed by secondary antibody anti-mouse IgG (1:1,000; Beyotime) for 1 hour at 37 °C. An enhanced chemiluminescence system (Tanon, Shanghai, People’s Republic of China) was used to detect protein content. The detailed information of primary antibodies is provided in Table S2. Three replicates were needed for each analysis.
Cell proliferation
CCK-8 assay kits (SAB, College Park, MD, USA) were uti-lized to determine cell proliferation according to the protocol of the manufacturer. Briefly, cells transfected as indicated were seeded in 96-well plates and cultured for 0, 24, 48, and 72 hours, CCK-8 solution (1:10) was mixed in each well and incubated for 1 hour. A microplate reader (Pulangxin, Beijing, People’s Republic of China) was used to measure the OD value at 450 nm. Triplicates were performed at each time point.
Cell apoptosis
In brief, an Annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit (Beyotime) was used to stain AF cells according to the instructions of the manufacturer after 48 hours of viral infection. Then, a flow cytometer (BD, San Diego, CA, USA) was used to detect the cells. Three replicates were needed for each analysis.
Dual-luciferase reporter assays
The ATG7 3′-UTR, containing the wild type or mutated target sequences of miR-106a-5p was cloned into Dual-Luciferase Expression Vector pGL3-Promoter (Promega, Madison, WI, USA). AF cells were transfected with NC or miR-106a-5p mimic in addition to the wild-type or mutant luciferase reporter, and incubated for 48 hours. Then, cells were harvested after 48 hours of transfection and then detected by using the Dual-GLO Luciferase Assay Kit (Promega). Three replicates were needed for each analysis.
Statistical analysis
GraphPad Prism software Version 7.0 (CA, USA) was uti-lized for the statistical analyses. Data were presented as the mean±SD of at least three samples. Statistical significance was determined by ANOVA for multiple comparisons and was accepted by a P-value<0.05.
Results
Melatonin stimulated proliferation, induced autophagy, and inhibited apoptosis in AF cells of patients with DD
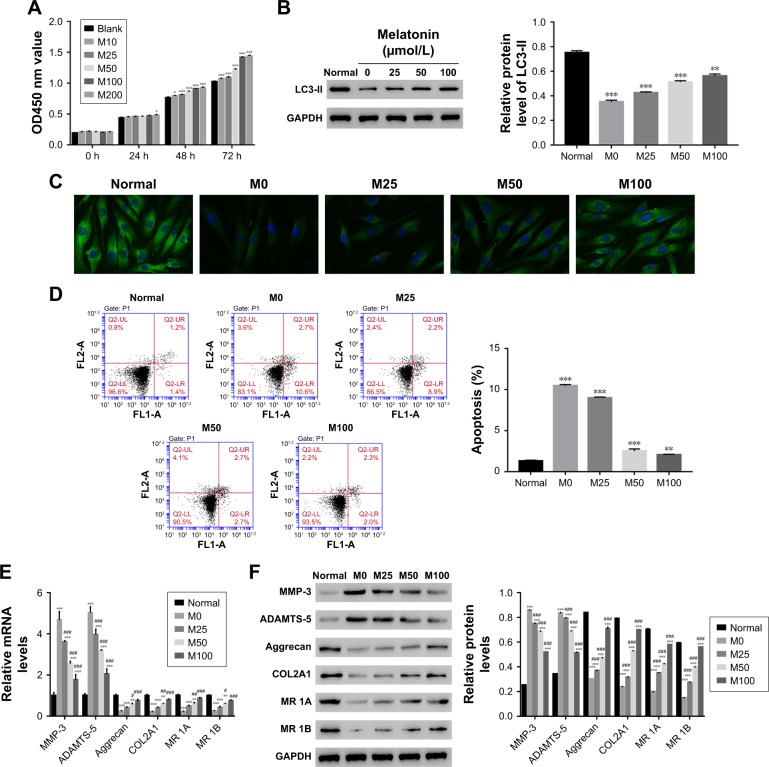
To further examine the correlation between melatonin and DD, AF cells of patients with DD were cultured in the presence of melatonin with different concentrations, including 10, 25, 50, 100, and 200 µmol/L. The untreated cells acted as the blank control. CCK-8 assay was utilized to examine the effect of melatonin in the proliferation on AF cells. As the concentration of melatonin increased, the cell proliferation rate was gradually increased (Figure 1A). These results suggest that melatonin contributed to the proliferation of AF cells of patients with DD.
Figure 1.
Overload of melatonin stimulated proliferation, induced autophagy, and apoptosis in AF cells of patients with DD. (A) Cell proliferation was detected 0, 24, 48, and 72 hours after treatment in AF cells of patients with DD treated with different melatonin concentrations. (B) Relative protein level of LC3-II in AF cells of patients with DD treated with 0, 25, 50, and 100 µmol/L of melatonin. (C) Representative images of immunofluorescent detection of LC3-II (green) in normal AF cells and AF cells of patients with DD. (D) The cell apoptosis profile of AF cells. (E and F) stand for the mRNA and protein level of MMP-3, ADAMTS-5, Aggrecan, COL2A1, and melatonin receptor 1A/1B in melatonin cultured cells.
Notes: *P<0.05 vs control, **P<0.01 vs control, ***P<0.001 vs control; #P<0.05 vs M0, ##P<0.01 vs M0, ###P<0.001 vs M0. Magnification of the image in (C) is 400×.
Abbreviations: AF, annulus fibrosus; DD, disc degeneration; LC3, light chain 3; M0, M25, M50, and M100, Melatonin of 0, 25, 50, and 100 µmol/L; MR 1A/1B, melatonin receptor 1A/1B.
LC3-II has been reported as an autophagy-associated protein.21 In this study, the protein level of LC3-II was significantly decreased in AF cells of patients with DD compared with that of normal AF cells. Moreover, it was easily identified that melatonin promoted the expression of LC3-II in AF cells of patient with DD, which was consistent with the result of LC3-II immunofluorescent detection (Figure 1B and C). In addition, the cell apoptosis rate of AF cells as indicated was much higher than that of normal AF cells. Interestingly, the cell apoptosis rate was deeply suppressed by melatonin (Figure 1D). All these results indicated that melatonin induced autophagy and inhibited apoptosis in AF cells of patients with DD.
In addition, the matrix metabolism is critical for the development of nucleus pulposus. The dysfunction of matrix metabolism leads to intervertebral disc degeneration.22,23 A previous report has indicated that melatonin inhibits the remodeling of the extracellular matrix (ECM) in nucleus pulposus (NP).24
In this study, we also examined the effect of melatonin on matrix biomarkers in AF cells of patients with DD, including catabolic factors (MMP-3, ADAMTS-5) and anabolic genes (Aggrecan and COL2A1). As shown in Figure 1E and F, both the mRNA and protein of MMP-3 and ADAMTS-5 were deeply suppressed by melatonin. Meanwhile, the level of anabolic genes, Aggrecan, and COL2A1, were promoted by melatonin in AF cells as indicated. Taken together, all these results demonstrated that melatonin inhibited the degradation of ECM in AF cells as indicated. In addition, the level of melatonin receptors 1A and 1B was also significantly upregulated in AF cells of patients with DD.
Melatonin promoted the translation of autophagy-related proteins in AF cells of patients with DD
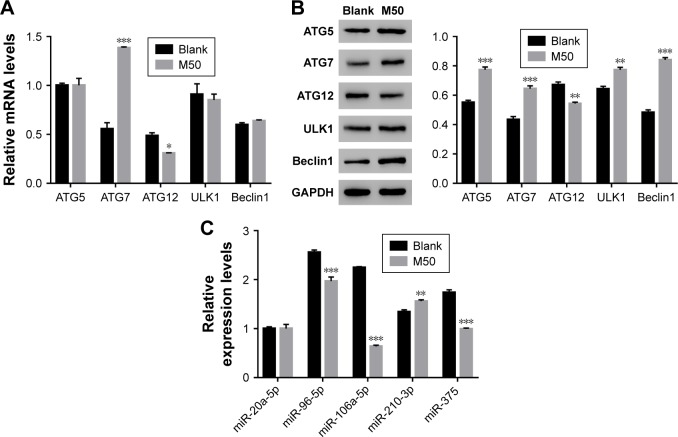
Next, quantitative real-time polymerase chain reaction (qRT-PCR) and Western blot were performed to quantify the mRNA and protein content of autophagy-related proteins, including ATG5, ATG7, ATG12, ULK1, and Beclin. The mRNA levels of ATG5, ULK1, and Beclin showed no significant difference between control and melatonin-treated AF cells (Figure 2A). However, the expression of ATG7 was significantly promoted by melatonin in AF cells of patients with DD. Interestingly, the expression of ATG12 was slightly inhibited by melatonin in AF cells. Moreover, nearly all indicated proteins (except for ATG12) were significantly upregulated in melatonin cultured cells (Figure 2B). Taken together, all these results indicated that melatonin affected the especially ATG7.
Figure 2.
High level of melatonin affected the level of autophagy-related proteins and microRNAs in AF cells of patients with DD. (A and B) Show the mRNA and protein levels of ATG5, ATG7, ATG12, ULK1, and Beclin 1 in AF cells of patients with DD and treated with melatonin of 50 µmol/L. (C) The mRNA levels of miR-20a-5p, miR-96-5p, miR-106a-5p, miR-210-3p, and miR-375 in AF cells of patients with DD and treated with melatonin of 50 µmol/L.
Notes: *P<0.05 vs control; **P<0.01 vs control, ***P<0.001 vs control.
Abbreviations: AF, annulus fibrosus; DD, disc degeneration; M50, melatonin of 50 µmol/L.
miR-106a-5p regulated ATG7 expression by directly binding to its 3′UTRin AF cells of patients with DD
Some highly conserved regions in 3′UTR of ATG7 were investigated by using the Targetscan database (http://www.targetscan.org), which might have served as the binding sites for miRs. Then, we examined the level of five miRs that might target ATG7 in AF cells, including miR-20a-5p, miR-96-5p, miR-106a-5p, miR-210-3p, and miR-375. As shown in Figure 2C, most of them were downregulated in melatonin-treated AF cells, especially miR-106a-5p.
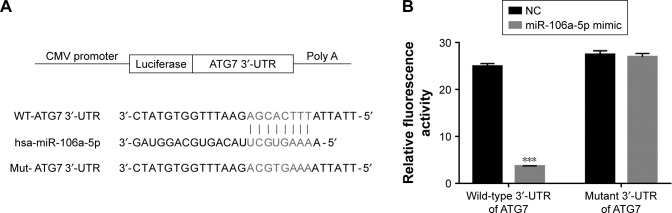
To further investigate the connection between miR-106a-5p and ATG7, luciferase reporters that contained the wild type (WT) or mutant 3′UTR of ATG7 were constructed (Figure 3A). As shown in Figure 3B, the luciferase activity of the wild type reporter was deeply abolished by the miR-106a-5p mimic. However, the luciferase activity of the mutant reporter showed no significant difference between miRs NC or miR-106a-5p mimic-transfected cells. Therefore, miR-106a-5p regulated the expression of ATG7 by directly binding to its 3′UTR in AF cells of patients with DD.
Figure 3.
miR-106a-5p directly binds to the 3′-UTR region of ATG7. (A) Schematic of the luciferase construct with the ATG7 3′-UTR containing an miR-106a-5p binding sequence. (B) Dual-luciferase assays of miR-106a-5p and ATG7 3′-UTR. Wild-type or mutant 3′-UTR of ATG7 was cloned into luciferase reporter vectors. AF cells were transfected with NC or miR-106a-5p mimic in addition to the wild-type or mutant luciferase reporter vector and incubated for 48 hours.
Note: ***P<0.001 vs NC.
Abbreviations: AF, annulus fibrosus; NC, negative control; UTR, untranslated region.
Melatonin inhibited the function of miR- 106a-5p in AF cells of patients with DD
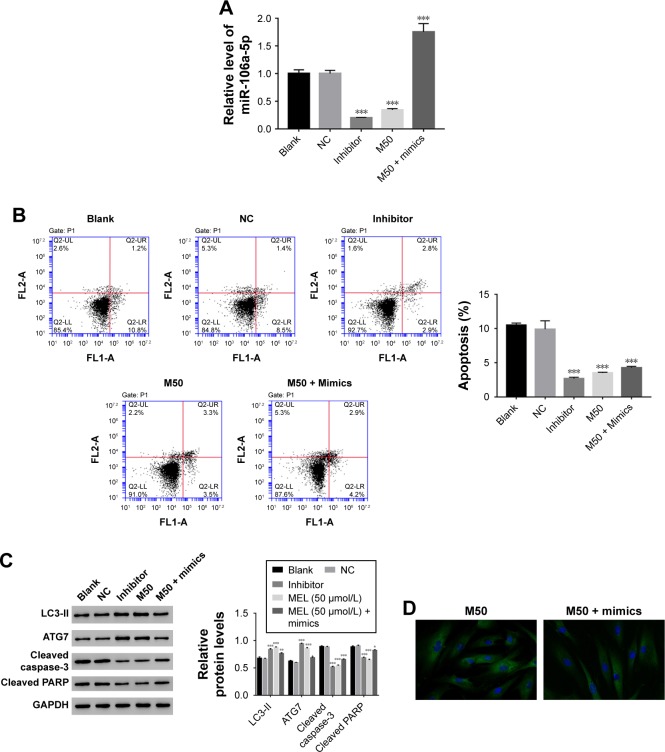
Then, we utilized gain or loss of function assay by transiently transfecting the miR-106a-5p mimic, inhibitor, and NC into AF cells. As shown in Figure 4A, all the transfected sequences functioned well in AF cells as expected. Moreover, our result suggested that miR-106a-5p was a pro-apoptotic element. However, melatonin deeply suppressed the function of miR-106a-5p in the cell apoptosis process (Figure 4B). Moreover, Western blot was performed to determine the protein levels of LC3-II, ATG7, cleaved caspase-3, and cleaved poly-ADP-ribose polymerases-1 (PARP). As shown in Figure 4C, the protein content of LC3-II was significantly upregulated in miR-106a-5p inhibitor transfected cells. Moreover, the protein content of LC3-II showed no significant difference between melatonin and miR-106a-5p mimics transfected cells. These results indicated that melatonin relieved the suppression of miR-106a-5p on ATG7. Meanwhile, cleaved caspase-3 and cleaved PARP-1 were positively correlated with miR-106a-5p. Moreover, the analysis of LC3-II immunofluorescent detection also obtained similar results (Figure 4D). Taken together, all these results demonstrated that melatonin suppressed the function of miR-106a-5p in AF cells of patients with DD.
Figure 4.
Melatonin inhibited the effect of miR-106a-5p in AF cells of patients with DD. (A) The expression level of miR-106a-5p was examined in AF cells that transfected with negative control, inhibitor, and treated with M50 or M50 + mimics, respectively. (B) The cell apoptosis profile of AF cells of patients with DD. (C) The relative protein level of LC3-II, ATG7, cleaved caspase-3, and cleaved PARP in AF cells of patients with DD. (D) Representative images of immunofluorescent detection of LC3-II (green) in AF cells of patient with DD.
Notes: *P<0.05 vs control, **P<0.01 vs control, and ***P<0.001 vs control. Magnification of the images is 400×.
Abbreviations: AF, annulus fibrosus; DD, disc degeneration; LC3, light chain 3; M50, melatonin of 50 µmol/L; PARP, poly-ADP-ribose polymerases-1.
Discussion
DD is a major risk for back pain in advanced age, which has caused huge socioeconomic implications.25 With the rising of the aging population, it is imperative to develop novel therapeutic approaches for the treatment of DD. The present study systematically analyzed the effect of melatonin on AF cells of patients with DD. Although the phenotype of the AF cells would change under monolayer culture conditions, and was different from the in-vivo condition of the IVD, our results gained a deep understanding into the effect of melatonin on AF cells.
Autophagy is an essential process for lysosomal degradation, which plays a key role in cell development.26 Moreover, autophagy has been reported as a protective mechanism against apoptosis in AF cells and IVD degeneration.7 In the present analysis, our result indicated that melatonin promoted the autophagy and inhibited the apoptosis of AF cells of patients with DD. Although the precise mechanism of disc degeneration is still not fully understood, our analysis indicated that melatonin might be a promising agent in the prevention of IVD.
It has been reported that ATG7 is essential for maintaining the homeostasis of axons and preventing their degeneration.27 Previous report has indicated that ATG7 is negatively correlated with the activity of caspase-3.28 In this study, we also obtained similar results. Therefore, melatonin might suppress the apoptosis of AF cells through inhibiting the activity of caspase-3 and PARP.
Moreover, PARP-1 was cleaved by activated caspases. Cleaved PARP-1 is reported as an apoptotic marker, which is responsible for DNA repair and cell viability.29 Besides that, PARP is an important regulator in the beginning of autophagy.30 Our results also indicated a negative correlation between ATG7 and cleaved PARP. Therefore, ATG7 was also involved in the regulation of the cell apoptosis process. Further analyses are needed to investigate the detailed correlation among ATG7, caspase-3, and PARP in AF cells.
It has been identified that miR-106a-5p suppresses cell metastases and promotes apoptosis.31–33 In this study, melatonin deeply inhibited the function of miR-106a-5p in AF cells of patients with DD. Moreover, our results demonstrated that miR-106a-5p suppressed the expression of ATG7 by directly binding to its 3′UTR in AF cells of patients with DD. However, this effect was deeply released by melatonin. Therefore, our results indicated miR-106a-5p might be an upstream component in the ATG7 signaling pathway. Melatonin might benefit DD through suppressing the miR-106a-5p/ATG7 signaling pathway.
Conclusion
The present study investigated the effect of melatonin on AF cells of patients with DD. Our results not only demonstrated that melatonin was a promising agent in the treatment for DD, but also indicated the possible signaling pathway it targets in AF cells.
Supplementary materials
Primer sequence information
- Homo sapiens autophagy related 5 (ATG5), transcript variant 2, mRNA
- Primer F 5′ GGCTGAGTGAACATCTGAG 3′
- Primer R 5′ CCCAGTTGCCTTATCTGAC 3′
- Pos: 976–1208
- Amplified product: Size: 233 bps
- Homo sapiens autophagy related 7 (ATG7), transcript variant 2, mRNA
- Primer F 5′ ACCCAGTGACGCCAGATTTC 3′
- Primer R 5′ AGGCAGGCACAGATGCTATG 3′
- Pos: 2704–2808
- Amplified product: Size: 105 bps
- Homo sapiens autophagy related 12 (ATG12), transcript variant 5, mRNA
- Primer F 5′ AGCGTTTCGGTCTTGTTG 3′
- Primer R 5′ TGAGGTCAGTCAGGAGTTTG 3′
- Pos: 2277–2495
- Amplified product: Size: 219 bps
- Homo sapiens unc-51 like autophagy activating kinase 1 (ULK1), mRNA
- Primer F 5′ GTCACACGCCACATAACAG 3′
- Primer R 5′ TTCCCAGGACTCAGGATTC 3′
- Pos: 4782–4981
- Amplified product: Size: 200 bps
- Homo sapiens beclin 1 (beclin 1), transcript variant 2, mRNA
- Primer F 5′ AGGGATGGAAGGGTCTAAG 3′
- Primer R 5′ GGGCTGTGGTAAGTAATGG 3′
- Pos: 161–315
- Amplified product: Size: 155 bps
- Homo sapiens glyceraldehyde-3-phosphatedehydrogenase (GAPDH), transcript variant 2, mRNA
- Primer F 5′AATCCCATCACCATCTTC 3′
- Primer R 5′AGGCTGTTGTCATACTTC 3′
- Pos: 436–653
- Amplified product: Size: 218 bps
- hsa-miR-106a-5p MIMAT0000103
- RT-Primer 5′ GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTACCT 3′
- PCR Primer:
- Primer F 5′ CGCGAAAAGTGCTTACAGTGC 3′
- Primer R 5′ AGTGCAGGGTCCGAGGTATT 3′
- hsa-miR-20a-5p MIMAT0000075
- RT-Primer
- 5′ GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTACCT 3′
- PCR Primer:
- Primer F 5′ GCGCGTAAAGTGCTTATAGTGC 3′
- Primer R 5′ AGTGCAGGGTCCGAGGTATT 3′
- hsa-miR-210-3p MIMAT0000267
- RT-Primer
- 5′ GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCAGCC 3′
- PCR Primer:
- Primer F 5′ CGCTGTGCGTGTGACAGC 3′
- Primer R 5′ AGTGCAGGGTCCGAGGTATT 3′
- hsa-miR-375 MI0000783
- RT-Primer
- 5′ GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCCTCA 3′
- PCR Primer: Primer F 5′ GTTCGTTCGGCTCGCG 3′
- Primer R 5′ AGTGCAGGGTCCGAGGTATT 3′
- hsa-miR-4802 MI0017450
- RT-Primer
- 5′ GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTGCTT 3′
- PCR Primer:
- Primer F 5′ GGAAACCTTCAAGCAGGCC 3′
- Primer R 5′ AGTGCAGGGTCCGAGGTATT 3′
- hsa-miR-96-5p MIMAT0000095
- RT-Primer
- 5′ GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGCAAA 3′
- PCR Primer:
- Primer F 5′ GCGTTTGGCACTAGCACATT 3′
- Primer R 5′ AGTGCAGGGTCCGAGGTATT 3′
- Homo sapiens RNA, U6 small nuclear 1 (RNU6-1), small nuclear RNA
- Primer F 5′ CTCGCTTCGGCAGCACA 3′
- Primer R 5′ AACGCTTCACGAATTTGCGT 3′
- Pos: 4–97
- Amplified product: Size: 94 bps
Table S1.
The sequence information for analysis hsa-miR-106a-5p
| Sequence name | Sequence |
|---|---|
| microRNA inhibitor NC | 5′-CAGUACUUUUGUGUAGUACAA-3′ |
| hsa-miR-106a-5p MIMAT0000103 | 5′-AAAAGUGCUUACAGUGCAGGUAG-3′ |
| hsa-miR-106a-5p inhibitor | 5′-CUACCUGCACUGUAAGCACUUUU-3′ |
| hsa-miR-106a-5p mimics | 5′-AAAAGUGCUUACAGUGCAGGUAG-3′ |
Abbreviation: NC, negative control.
Table S2.
The primary antibodies information
| Antibody name | Source | Dilution factor |
|---|---|---|
| LC3-II | Abcam, UK | 1:1,000 |
| ATG5 | Abcam, UK | 1:1,000 |
| ATG7 | Abcam, UK | 1:10,000 |
| ATG12 | Abcam, UK | 1:1,000 |
| ULK1 | Abcam, UK | 1:10,000 |
| Beclin 1 | Abcam, UK | 1:2,000 |
| Caspase-3 | Abcam, UK | 1:1,000 |
| PARP | Abcam, UK | 1:1,000 |
| GAPDH | CST, USA | 1:1,000 |
Acknowledgments
This study was supported by the fund for Forstering Young Scholar of Peking University Health Science Center (NO BMU2017PY017).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Michael AA, Roughley PJ. What is intervertebral disc degeneration, and what causes it? Spine. 2006;31(18):2151–2161. doi: 10.1097/01.brs.0000231761.73859.2c. [DOI] [PubMed] [Google Scholar]
- 2.Eck JC, Humphreys SC, Hodges SD. Adjacent-segment degeneration after lumbar fusion: a review of clinical, biomechanical, and radiologic studies. Am J Orthop. 1999;28:336–340. [PubMed] [Google Scholar]
- 3.Shu CC, Smith MM, Smith SM, Dart AJ, Little CB, Melrose J. A histopathological scheme for the quantitative scoring of intervertebral disc degeneration and the therapeutic utility of adult mesenchymal stem cells for intervertebral disc regeneration. Int J Mol Sci. 2017;18(5) doi: 10.3390/ijms18051049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Little JP, Pearcy MJ, Tevelen G, Evans JH, Pettet G, Adam CJ. The mechanical response of the ovine lumbar anulus fibrosus to uniaxial, biaxial and shear loads. J Mech Behav Biomed Mater. 2010;3(2):146–157. doi: 10.1016/j.jmbbm.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Daly C, Ghosh P, Jenkin G, Oehme D, Goldschlager T. A review of animal models of intervertebral disc degeneration: pathophysiology, regeneration, and translation to the clinic. Biomed Res Int. 2016;2016:5952165. doi: 10.1155/2016/5952165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290(5497):1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen C, Yan J, Jiang LS, Dai LY. Autophagy in rat annulus fibrosus cells: evidence and possible implications. Arthritis Res Ther. 2011;13(4):R132. doi: 10.1186/ar3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang Y, Yan W, He X, et al. miR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology. 2012;143(1):177–187.e8. doi: 10.1053/j.gastro.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Yu Y, Zhang J, Jin Y, et al. MiR-20a-5p suppresses tumor proliferation by targeting autophagy-related gene 7 in neuroblastoma. Cancer Cell Int. 2018;18:5. doi: 10.1186/s12935-017-0499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan B, Chen L, Feng B, Chen Y, Song H. MiR-200b regulates autophagy associated with chemoresistance in human lung adenocarcinoma. Oncotarget. 2015;6(32):32805–32820. doi: 10.18632/oncotarget.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu J, Zhao S-D, Liu H-J, Hao A-J. Melatonin promotes proliferation and differentiation of neural stem cells subjected to hypoxia in vitro. J Pineal Res. 2011;51:104–112. doi: 10.1111/j.1600-079X.2011.00867.x. [DOI] [PubMed] [Google Scholar]
- 14.Kim S-J, Kang HS, Lee J-H, et al. Melatonin ameliorates ER stress-mediated hepatic steatosis through miR-23a in the liver. Bio-chem Biophys Res Commun. 2015;458(3):462–469. doi: 10.1016/j.bbrc.2015.01.117. [DOI] [PubMed] [Google Scholar]
- 15.Carloni S, Favrais G, Saliba E, et al. Melatonin modulates neonatal brain inflammation through endoplasmic reticulum stress, autophagy, and miR-34a/silent information regulator 1 pathway. J Pineal Res. 2016;61(3):370–380. doi: 10.1111/jpi.12354. [DOI] [PubMed] [Google Scholar]
- 16.Nakade O, Koyama H, Ariji H, Yajima A, Kaku T. Melatonin stimulates proliferation and type I collagen synthesis in human bone cells in vitro. J Pineal Res. 1999;27:106–111. doi: 10.1111/j.1600-079x.1999.tb00603.x. [DOI] [PubMed] [Google Scholar]
- 17.Mayo JC, Sainz RM, Uria H, Isaac Antolin MME, Rodriguez C. Melatonin prevents apoptosis induced by 6-hydroxydopamine in neuronal cells_ Implications for Parkinson’s disease. J Pineal Res. 1998;24:179–192. doi: 10.1111/j.1600-079x.1998.tb00531.x. [DOI] [PubMed] [Google Scholar]
- 18.Cagnoli CM, Atabay C, Kharlamova E, Manev H. Melatonin protects neurons from singlet oxygen-induced apoptosis. J Pineal Reurvch. 1995;18:222–226. doi: 10.1111/j.1600-079X.1995.tb00163.x. [DOI] [PubMed] [Google Scholar]
- 19.Wei J, Ma LS, Liu DJ, Guo J, Jiang WK, Yu HJ. Melatonin regulates traumatic optic neuropathy via targeting autophagy. Eur Rev Med Pharmacol Sci. 2017;21(21):4946–4951. [PubMed] [Google Scholar]
- 20.Zhang SJ, Yang W, Wang C, et al. Autophagy: A double-edged sword in intervertebral disc degeneration. Clin Chim Acta. 2016;457:27–35. doi: 10.1016/j.cca.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 21.Holt SV, Wyspianska B, Randall KJ, James D, Foster JR, Wilkinson RW. The development of an immunohistochemical method to detect the autophagy-associated protein LC3-II in human tumor xenografts. Toxicol Pathol. 2011;39(3):516–523. doi: 10.1177/0192623310396903. [DOI] [PubMed] [Google Scholar]
- 22.Liu W, Xia P, Feng J, et al. MicroRNA-132 upregulation promotes matrix degradation in intervertebral disc degeneration. Exp Cell Res. 2017;359(1):39–49. doi: 10.1016/j.yexcr.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 23.Walker MH, Anderson DG. Molecular basis of intervertebral disc degeneration. Spine Jl. 2012;15(4):318–330. doi: 10.1016/j.spinee.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 24.Li Z, Li X, Chen C, Chan MTV, Wu WKK, Shen J. Melatonin inhibits nucleus pulposus (NP) cell proliferation and extracellular matrix (ECM) remodeling via the melatonin membrane receptors mediated PI3K-Akt pathway. J Pineal Res. 2017;63(3) doi: 10.1111/jpi.12435. [DOI] [PubMed] [Google Scholar]
- 25.Vasiliadis ES, Pneumaticos SG, Evangelopoulos DS, Papavassiliou AG. Biologic treatment of mild and moderate intervertebral disc degeneration. Mol Med. 2014;20:400–409. doi: 10.2119/molmed.2014.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komatsu M, Wang QJ, Holstein GR, et al. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci U S A. 2007;104(36):14489–14494. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baydas G, Reiter RJ, Akbulut M, Tuzcu M, Tamer S. Melatonin inhibits neural apoptosis induced by homocysteine in hippocampus of rats via inhibition of cytochrome c translocation and caspase-3 activation and by regulating pro- and anti-apoptotic protein levels. Neuroscience. 2005;135(3):879–886. doi: 10.1016/j.neuroscience.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 29.Casao A, Mata-Campuzano M, Ordas L, Cebrian-Perez JA, Muino-Blanco T, Martinez-Pastor F. Cleaved PARP-1, an apoptotic marker, can be detected in ram spermatozoa. Reprod Domest Anim. 2015;50(4):688–691. doi: 10.1111/rda.12549. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez-Vargas JM, Ruiz-Magana MJ, Ruiz-Ruiz C, et al. ROS-induced DNA damage and PARP-1 are required for optimal induction of starvation-induced autophagy. Cell Res. 2012;22(7):1181–1198. doi: 10.1038/cr.2012.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan YJ, Wei LL, Wu XJ, Huo FC, Mou J, Pei DS. MiR-106a-5p inhibits the cell migration and invasion of renal cell carcinoma through targeting PAK5. Cell Death Dis. 2017;8(10):e3155. doi: 10.1038/cddis.2017.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhi F, Zhou G, Shao N, et al. miR-106a-5p inhibits the proliferation and migration of astrocytoma cells and promotes apoptosis by targeting FASTK. PLoS One. 2013;8(8):e72390. doi: 10.1371/journal.pone.0072390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He QY, Wang GC, Zhang H, et al. miR-106a-5p suppresses the proliferation, migration, and invasion of osteosarcoma cells by targeting HMGA2. DNA Cell Biol. 2016;35(9):506–520. doi: 10.1089/dna.2015.3121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
The sequence information for analysis hsa-miR-106a-5p
| Sequence name | Sequence |
|---|---|
| microRNA inhibitor NC | 5′-CAGUACUUUUGUGUAGUACAA-3′ |
| hsa-miR-106a-5p MIMAT0000103 | 5′-AAAAGUGCUUACAGUGCAGGUAG-3′ |
| hsa-miR-106a-5p inhibitor | 5′-CUACCUGCACUGUAAGCACUUUU-3′ |
| hsa-miR-106a-5p mimics | 5′-AAAAGUGCUUACAGUGCAGGUAG-3′ |
Abbreviation: NC, negative control.
Table S2.
The primary antibodies information
| Antibody name | Source | Dilution factor |
|---|---|---|
| LC3-II | Abcam, UK | 1:1,000 |
| ATG5 | Abcam, UK | 1:1,000 |
| ATG7 | Abcam, UK | 1:10,000 |
| ATG12 | Abcam, UK | 1:1,000 |
| ULK1 | Abcam, UK | 1:10,000 |
| Beclin 1 | Abcam, UK | 1:2,000 |
| Caspase-3 | Abcam, UK | 1:1,000 |
| PARP | Abcam, UK | 1:1,000 |
| GAPDH | CST, USA | 1:1,000 |